Abstract
Multidrug-resistant strain Acinetobacter baumannii BM4454 was isolated from a patient with a urinary tract infection. The adeB gene, which encodes a resistance-nodulation-cell division (RND) protein, was detected in this strain by PCR with two degenerate oligodeoxynucleotides. Insertional inactivation of adeB in BM4454, which generated BM4454-1, showed that the corresponding protein was responsible for aminoglycoside resistance and was involved in the level of susceptibility to other drugs including fluoroquinolones, tetracyclines, chloramphenicol, erythromycin, trimethoprim, and ethidium bromide. Study of ethidium bromide accumulation in BM4454 and BM4454-1, in the presence or in the absence of carbonyl cyanide m-chlorophenylhydrazone, demonstrated that AdeB was responsible for the decrease in intracellular ethidium bromide levels in a proton motive force-dependent manner. The adeB gene was part of a cluster that included adeA and adeC which encodes proteins homologous to membrane fusion and outer membrane proteins of RND-type three-component efflux systems, respectively. The products of two upstream open reading frames encoding a putative two-component regulatory system might be involved in the regulation of expression of the adeABC gene cluster.
During the last 20 years hospital-acquired infections caused by multidrug-resistant, gram-negative bacilli have increased considerably to become a significant health problem. Acinetobacter spp. are ubiquitous, nonfermentative, gram-negative bacilli which play a significant role in the colonization and infection of patients in intensive care units. Acinetobacter baumannii is the predominant species associated with outbreaks of nosocomial infections (3). This opportunistic microorganism may cause epidemic pneumonia, urinary tract infections, septicemia, and meningitis (24). Few antibiotics are effective for the treatment of Acinetobacter infections because of the numerous mechanisms of resistance accumulated by isolates of this bacterial genus and the frequency of multidrug-resistant strains. Acinetobacter infections are thus often very difficult to treat, and combination therapy is usually required for effective treatment (3). Aminoglycosides can be used successfully in combination with an effective β-lactam, and combinations of a β-lactam with either a fluoroquinolone or rifampin have also been proposed. However, treatment failure and death caused by Acinetobacter infections or underlying diseases are common (3).
Resistance of Acinetobacter to β-lactams is partially intrinsic due to the synthesis of a species-specific cephalosporinase (11, 33). However, additional plasmid- or transposon-borne β-lactamase genes can be acquired (5, 9). Mutations in the gyrA gene have been associated with high-level resistance to fluoroquinolones in this organism (3, 34). Aminoglycoside resistance is also common in Acinetobacter and results primarily from inactivation of the antibiotic by specific modifying enzymes. Three classes of aminoglycoside-inactivating enzymes (acetyltransferases, phosphotransferases, and adenylyltransferases) have been identified in Acinetobacter (15). Acinetobacter haemolyticus and related species are intrinsically resistant to aminoglycosides by synthesis of a chromosomally encoded specific N-acetyltransferase [AAC(6′)] (14, 30). In contrast, aminoglycoside-resistant A. baumannii isolates usually result from the acquisition of genes encoding modifying enzymes (15, 33). Aminoglycoside resistance mediated by an efflux system has not yet been reported in Acinetobacter; the only evidence for an efflux mechanism is the export of phosphonium ions in Acinetobacter calcoaceticus (18).
Efflux systems are widely found in microorganisms and confer resistance to various compounds, including antibiotics, by extrusion of the drug. The ATP-dependent multidrug transporters use ATP as a source of energy, whereas the secondary multidrug transporters are sensitive to agents that dissipate the proton motive force, suggesting that they mediate the efflux of the toxic compounds from the cell in a coupled exchange with protons. These secondary multidrug transporters can be subdivided into distinct families: the major facilitator (MF) superfamily, the small multidrug resistance (SMR) superfamily, the multidrug and toxic compound extrusion (MATE) superfamily, and the resistance-nodulation-cell division (RND) family (27). Most of the multidrug transporters belonging to the RND family interact with a membrane fusion protein (MFP) and an outer membrane protein (OMP) to allow drug transport across both the inner and the outer membranes of gram-negative bacteria (32, 37). The secondary structure of RND-type efflux proteins was proposed to consist of 12 transmembrane segments (TMSs), with two long loops between TMSs 1 and 2 and TMSs 7 and 8 (27, 32). The trimeric form of the OMP generates a continuous, solvent-accessible “channel-tunnel” that spans both the outer membrane and the periplasmic space (36, 13). MFP could be involved in either the bringing of the inner and outer membranes closer or the stabilization of the OMP structure (37, 23). Recently, three RND-type efflux pumps have been demonstrated to be involved in aminoglycoside resistance: AmrAB-OprA, responsible for intrinsic aminoglycoside and macrolide resistance in Burkholderia pseudomallei (20); MexXY, which exports aminoglycosides, tetracycline, and erythromycin from Pseudomonas aeruginosa (19, 28, 35); and AcrD in Escherichia coli (29).
A. baumannii BM4454, isolated from a patient with a urinary tract infection, was resistant to multiple antibiotics with a phenotype of aminoglycoside resistance, suggesting that resistance to this class of drugs could be due to active efflux. The present study was undertaken to identify the molecular mechanism that confers the particularly broad-spectrum aminoglycoside resistance of A. baumannii BM4454. A three-component efflux system that included an RND multidrug transporter was shown to be involved in resistance by insertional inactivation.
MATERIALS AND METHODS
Plasmids, strains, and growth conditions.
A. baumannii BM4454 was isolated in 1999 from a patient with a urinary tract infection at the Hôpital Saint-Michel in Paris, France. The strains were grown in brain heart infusion broth and agar (Difco Laboratories, Detroit, Mich.) at 37°C. For BM4454-1, growth media were supplemented with ticarcillin (80 μg/ml) in order to maintain selection pressure.
Susceptibility testing.
Antibiotic susceptibility was tested by disk diffusion on Mueller-Hinton agar (Bio-Rad, Marnes-la-Coquette, France). The MICs of antibiotics were determined by the method of Steers et al. (31) with 104 CFU per spot on agar after 24 h of incubation. Susceptibility to ethidium bromide and to safranin O was tested on Mueller-Hinton agar drug gradients.
DNA manipulations.
Total and plasmid DNAs were prepared as described previously (4). Purification of plasmid DNA was performed by using the Wizard minipreps DNA kit (Promega, Madison, Wis.). Restriction with endonucleases was done according to the supplier's recommendations (Life Technologies Inc., Gaithersburg, Md.). Extraction of DNA fragments separated by agarose gel electrophoresis was carried out by using the Sephaglas BandPrep Kit (Pharmacia Biotech, Saint-Quentin en Yvelines, France).
PCR was performed in a GeneAmp PCR system 2400 (Perkin-Elmer Cetus, Norwalk, Conn.) with Pfu DNA polymerase (Stratagene, La Jolla, Calif.) according to the manufacturers' recommendations. Annealing steps were performed at 50 or 55°C with degenerate or specific primers (Unité de Chimie Organique, Institut Pasteur, Paris, France), respectively.
For Southern hybridization, DNA fragments were transferred from the agarose gel to a Hybond-N+ membrane (Amersham International, Little Chalfont, England) by vacuum with a Trans Vac TE80 apparatus (Hoefer Scientific Instruments, San Francisco, Calif.). The amplification products used to generate the probes were labeled with [α-32P]dCTP (3,000 Ci/mmol; Amersham Radiochemical Center, Amersham, England) with a nick translation kit (Amersham).
Detection and insertion-inactivation of adeB.
The adeB gene was detected by PCR amplification of A. baumannii BM4454 total DNA with degenerate oligodeoxynucleotides O1 [5′-GT(A/T)GA(T/C)GA(T/C)GC(A/T)AT(A/T)GT(A/T)GT-3′] and O2 [5′-A(A/G)(A/T)(C/G)(A/T)(A/T)GTCAT(A/T)A(A/G)(A/T)AT(A/T)GG-3′], which were complementary to the conserved motifs D and C of the RND protein family, respectively, and which were designed by taking into account the genetic codon preference of Acinetobacter.
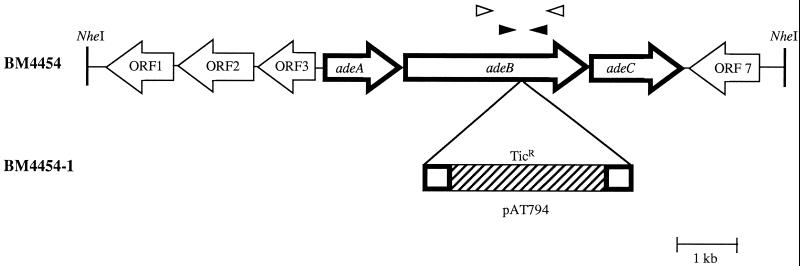
A fragment internal to the adeB gene of BM4454 was amplified with specific oligodeoxynucleotides O3 (5′-GTATGAATTGATGCTGC-3′) and O4 (5′-CACTCGTAGCCAATACC-3′) (O3 from positions 6199 to 6213 and O4 from positions 7177 to 7161, with the numbering given 5′ to 3′, according to the sequence with GenBank accession no. AF370885). The 979-bp amplification product was cloned into SmaI-linearized pUC18, a suicide vector in Acinetobacter (25), to generate pAT794. Plasmid pAT794 DNA was introduced into BM4454 by electrotransformation, and transformants were selected for ticarcillin resistance. Total DNA from the three clones stably resistant to ticarcillin was obtained and was analyzed by PCR with the M13 reverse primer and the M13 (−20) forward primers and with two specific primers complementary to the flanking regions of the pAT794 insert in the BM4454 chromosome. Total DNA from the three transformants was restricted with ClaI and NdeI and was analyzed by Southern hybridization with probes specific for the blaTEM-1 gene of pUC18 and for the 3′ end of the adeB gene downstream from the O3-O4 internal fragment. Transformant BM4454-1, the only one that contained a single copy of pAT794 in the adeB gene, was selected for further studies.
Determination of the sequence of the ade gene cluster.
To obtain plasmid pAT796, total DNA from BM4454-1 was digested with NheI (Fig. 1), self-ligated, and used to transform E. coli Top 10 competent cells with selection on ticarcillin. DNA sequencing was performed with specific primers with a CEQ 2000 DNA Analysis System automatic sequencer (Beckman Instruments, Inc., Palo Alto, Calif.).
FIG. 1.
Schematic representation of the ade gene clusters from A. baumannii BM4454 and BM4454-1 and their environments. Open arrows indicate the direction of transcription. The genes encoding the three-component Ade efflux system are represented by boldface arrows. In pAT794, which conferred ticarcillin resistance (TicR), the sequence of pUC18 is hatched. Open arrowheads represent degenerate oligodeoxynucleotides O1 and O2 used to detect the adeB gene, and closed arrowheads represent specific oligodeoxynucleotides O3 and O4 used to generate pAT794. NheI restriction sites are indicated by vertical lines.
Accumulation of ethidium bromide.
The kinetics of ethidium bromide accumulation in BM4454 and BM4454-1 cells were monitored by a fluorimetric assay slightly modified from that described previously (8). Cells were grown to an optical density at 600 nm of 0.5, pelleted carefully at room temperature, resuspended to an A600 of 0.2 in sodium phosphate buffer (pH 7.0), and returned to 37°C. Ethidium bromide was added at a final concentration of 2 μg/ml, and after 420 s of incubation, carbonyl cyanide m-chlorophenylhydrazone (CCCP) was added at a final concentration of 100 μM. The change in fluorescence intensity (λexcite, 530 nm; λemit, 600 nm), which is proportional to the quantity of intracellular dye, was recorded on an SFM 25 spectrofluorimeter (Bio-Tek Kontron Instruments, Saint-Quentin en Yvelines, France) for 700 s following the addition of ethidium bromide.
Computer analysis of sequence data.
Nucleotide sequence data were analyzed with the GCG sequence analysis software package (version 7; Genetics Computer Group, Madison, Wis.). Amino acid sequences were analyzed at the website of the National Center for Biotechnology Information (www.ncbi.nih.gov/gorf/gorf.html). The GenBank and protein databases were screened for sequence similarities.
Nucleotide sequence accession number.
The 10,629-bp sequence of BM4454-1 has been deposited in the GenBank data library (GenBank, Los Alamos, N.M.) under accession no. AF370885.
RESULTS
Resistance phenotype of A. baumannii BM4454.
A. baumannii BM4454 was resistant to aminoglycosides, tetracyclines, fluoroquinolones, and macrolides-lincosamides-streptogramins (MLS), in addition to β-lactams, to which this species has intrinsic resistance. The strain was resistant to low levels of all aminoglycosides tested, but curiously, it was more susceptible to kanamycin than to the others members of this drug class. Moreover, both 2′-N-ethylnetilmicin and 6′-N-ethylnetilmicin were similarly inactive against BM4454. This resistance phenotype did not correspond to any of the phenotypes conferred by a known aminoglycoside-modifying enzyme or any combination of known aminoglycoside-modifying enzymes. In our experience, clinical isolates of A. baumannii, such as BM4454, with a broad aminoglycoside resistance phenotype are relatively common, but aminoglycoside-modifying activity has never been detected in such strains by the phosphocellulose paper-binding assay (data not shown). In addition, these strains exhibit an uncommonly low level of susceptibility to norfloxacin compared to their level of susceptibility to pefloxacin. Taken together, these data suggested that aminoglycoside resistance in BM4454 could be due to active efflux of the drugs, which certainly affected other compounds. Additionally, high-level resistance to fluoroquinolones and MLS suggested that BM4454 had developed multiple mechanisms for antibiotic resistance.
Detection and inactivation of Ade efflux system.
In order to detect a putative efflux protein in A. baumannii BM4454, degenerate oligonucleotides O1 and O2, complementary to the conserved motifs D and C of the RND protein family, respectively (26), were used to amplify total DNA from BM4454, and the sequence of the PCR product of the expected size (ca. 1,700 bp) was determined. Comparison of the sequence with those in the GenBank database revealed a high degree of identity (approximately 50%) with internal regions of structural genes encoding RND proteins. Two specific primers, O3 and O4, deduced from this sequence, were used to amplify an internal 979-bp fragment that was ligated into pUC18 to generate recombinant suicide plasmid pAT794. Plasmid pAT794 DNA was introduced into BM4454 by electrotransformation, and transformants stably resistant to ticarcillin were found to be susceptible to aminoglycosides by disk diffusion. They were analyzed by PCR, sequence determination, and Southern hybridization. The resulting data (data not shown) allowed selection of a clone, BM4454-1, with a single integrated copy of pAT794 in the partly characterized gene, following a homologous recombination event. This indicated that the gene encoded an RND-type protein involved in aminoglycoside resistance in BM4454 and was named adeB, for Acinetobacter drug efflux. The adeB gene was detected by dot hybridization in the six A. baumannii strains tested but not in the four A. haemolyticus strains tested (data not shown).
Cloning and characterization of the genes encoding the Ade efflux system.
Plasmid pAT796, which contained approximately 12 kb of DNA flanking the pAT794 insertion site, was obtained by restriction of BM4454-1 genomic DNA, followed by self-ligation and transformation into E. coli with selection on ticarcillin. The sequence of 10.6 consecutive kb of DNA was determined, and the search for stop codons in the three reading frames in each DNA strand revealed the presence of seven complete open reading frames (ORF) (Fig. 1). A search of the nr database indicated that the deduced products of the three adjacent genes, adeA (ORF4; nucleotides 3439 to 4629), adeB (ORF5; nucleotides 4629 to 7736), and adeC (ORF6; nucleotides 7813 to 9210) were highly similar to proteins constituting three-component multidrug efflux systems of the RND type. The AdeB protein consisted of 1,035 amino acids and exhibited a high degree of identity (approximately 50%) with RND proteins (Table 1). The hydropathy profile of the deduced AdeB sequence revealed a pattern that was duplicated in the N- and C-terminal halves of the protein and 12 hydrophobic domains which may correspond to the transmembrane segments of the typical RND proteins (data not shown). AdeA was homologous to MFPs with 35 to 40% identity (Table 1), whereas AdeC was most similar (40% identity) to the OprM OMP from P. aeruginosa.
TABLE 1.
Levels of identity between the putative proteins deduced from the adeA and adeB genes and those from RND-type efflux systems
| Sequence compared | % Identity
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AcrA | AcrB | MtrC | MtrD | MexA | MexB | MexC | MexD | MexX | MexY | AmrA | AmrB | AcrD | |
| AdeA | 38 | 39 | 37 | 37 | 34 | 37 | |||||||
| AdeB | 49 | 47 | 47 | 53 | 46 | 46 | 45 | ||||||
Three other adjacent ORFs (ORF1, ORF2, and ORF3) located upstream from adeA and in the opposite orientation were identified (Fig. 1). Proteins encoded by ORF2 and ORF3 were highly similar to various two-component regulatory systems. The deduced proteins from ORF2 and ORF3 exhibited, on average, 30 and 35% amino acid identities with sensors and response regulators, respectively. The putative protein encoded by ORF3 consisted of 228 amino acids, which is the typical length for transcriptional regulators. In contrast, the product of ORF2 was approximately 100 amino acids shorter at its N terminus than typical bacterial histidine kinases. No significant homology was found in the database with the full-length sequence deduced from ORF1, located at the 3′ end of ORF2 and similarly oriented. However, the C-terminal region of the deduced sequence of ORF1 exhibited 25% identity with a putative transcriptional termination-antitermination factor from Mycobacterium tuberculosis. Finally, the deduced amino acid sequence of ORF7, identified downstream from adeC but in the opposite orientation (Fig. 1), shared a modest degree of identity with numerous hypothetical, but functionally unidentified, proteins from various microorganisms.
Substrate specificity of Ade pump.
The functional role of AdeB was studied by comparing the inhibitory activities of various antibiotics and toxic compounds against BM4454 and its derivative, BM4454-1, in which the adeB gene had been disrupted. Determination of the MICs of various structural classes of antibiotics (Table 2) indicated that all antibiotics tested, including aminoglycosides, fluoroquinolones, cefotaxime, erythromycin, tetracycline, chloramphenicol, and trimethoprim, were substrates for the AdeB pump since MICs were 4 to more than 32 times higher for BM4454 than for BM4454-1. Disk diffusion indicated that BM4454 was also significantly more resistant than BM4454-1 to minocycline. In contrast, the activities of rifampin, sulfonamides, amoxicillin, and ceftazidime remained unchanged (data not shown). Moreover, BM4454 was able to grow on much higher concentrations of ethidium bromide than BM4454-1 was, but both strains were inhibited at the same concentration of the dye safranin O.
TABLE 2.
Antibiotic susceptibilities of A. baumannii strains
| Strain | MIC (μg/ml)a
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| KAN | GEN | TOB | NET | AMK | CTX | TET | ERY | CHL | TMP | SPX | OFX | NOR | PEF | |
| BM4454 | 4 | 8 | 2 | 16 | 8 | 16 | 64 | 64 | 512 | 64 | 64 | 64 | 512 | 256 |
| BM4454-1 | 1 | ≤0.25 | ≤0.25 | 0.5 | 1 | 4 | 8 | 8 | 128 | 4 | 2 | 4 | 64 | 16 |
Abbreviations: AMK amikacin; CHL, chloramphenicol; CTX, cefotaxime; ERY, erythromycin; GEN, gentamicin; KAN, kanamycin; NOR, norfloxacin; NET, netilmicin; OFX, ofloxacin; PEF, pefloxacin; SPX, sparfloxacin; TET, tetracycline; TOB, tobramycin; TMP, trimethoprim.
Accumulation of ethidium bromide.
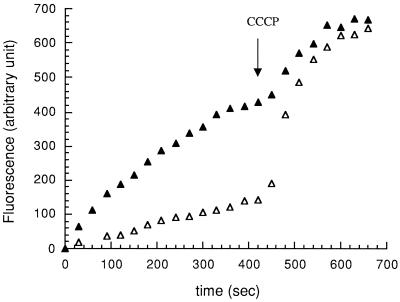
To confirm that the difference in drug susceptibility between BM4454 and BM4454-1 was due to an efflux mechanism, the level of accumulation of ethidium bromide was measured and was found to be approximately three times lower in BM4454 than in BM4454-1 after 400 s (Fig. 2). Dissipation of the membrane proton motive force by addition of the protonophore CCCP at 420 s increased the level of accumulation of ethidium bromide in both strains, resulting in similar concentrations 300 s after CCCP addition.
FIG. 2.
Ethidium bromide accumulation in A. baumannii BM4454 (▵) and BM4454-1 (▴). At 420 s CCCP was added to the bacterial suspensions at a final concentration of 100 μM.
DISCUSSION
Until recently, antibiotic resistance by active efflux of the drug was limited to hydrophobic and amphiphilic compounds, including quinolones, tetracyclines, macrolides, β-lactams, chloramphenicol, and rifampin (22). In 1999, involvement of the AmrB RND-type efflux pump in the intrinsic aminoglycoside resistance of B. pseudomallei was reported (20). Subsequently, disruption of mexY in P. aeruginosa (28, 19, 35) and acrD in E. coli (29) was shown to result in aminoglycoside hypersusceptibility. Moreover, multidrug transporters belonging to other families, including the ABC (17), SMR (12), MF (6, 2), and MATE (21) superfamilies, have been shown to be able to export aminoglycosides when their structural genes are cloned into a multicopy plasmid. We have characterized the adeABC gene cluster (Fig. 1) from multidrug-resistant clinical isolate A. baumannii BM4454, which exhibited an aminoglycoside resistance pattern that could not be explained by production of one or a combination of known aminoglycoside-modifying enzymes. The proteins deduced from the adeA, adeB, and adeC genes were highly similar to MFPs, the RND proteins, and OMPs, respectively, suggesting that these genes encode a three-component efflux system. Disruption of the adeB gene in BM4454 demonstrated that the corresponding protein was responsible for aminoglycoside resistance and also contributed to the multiple-antibiotic-resistance phenotype of this strain. The proteins most similar to AdeA and AdeB were MtrC from Neisseria gonorrhoeae and MexD from P. aeruginosa, respectively (Table 1), which have not been reported to have an ability to transport aminoglycosides.
Aminoglycosides, as well as cefotaxime, tetracyclines, erythromycin, chloramphenicol, trimethoprim, fluoroquinolones, and ethidium bromide, were found to be substrates for AdeB (Table 2). Thus, this efflux pump can apparently recognize a wide spectrum of substrates including hydrophobic, amphiphilic, and hydrophilic molecules which can be either positively charged or neutral. Among the aminoglycosides, kanamycin and amikacin appeared to be less effectively transported than other compounds by AdeABC. These two aminoglycosides contain the largest number of hydroxyl substituents and, consequently, are the most hydrophilic. In the same way, the hydrophobic fluoroquinolones ofloxacin and sparfloxacin appeared to be slightly better substrates than the more hydrophilic fluoroquinolones norfloxacin and pefloxacin. However, an additional mechanism must be involved in fluoroquinolone resistance in BM4454, and this is probably modification of the intracellular target, since BM4454-1 still exhibited high-level resistance to these molecules. This combination of mechanisms could explain the very high level of fluoroquinolone resistance of the clinical isolate.
Efflux mediated by AdeABC was confirmed by comparing the levels of intracellular accumulation of ethidium bromide by BM4454 and BM4454-1 (Fig. 2). Accumulation experiments were not carried out with radiolabeled aminoglycosides in order to test the effect of the protonophore CCCP, since this molecule dissipates the proton motive force essential for both efflux by RND pumps and uptake of aminoglycosides. The results indicate that disruption of adeB increases ethidium bromide accumulation in a CCCP-dependent fashion, supporting a proton motive force-dependent efflux mechanism for AdeB-mediated aminoglycoside resistance. Moreover, in BM4454-1 the rate of accumulation of ethidium bromide was increased in the presence of CCCP, suggesting that other efflux pumps dependent on the proton motive force may contribute to ethidium bromide efflux in A. baumannii. This is not surprising given that ethidium bromide is an efficient and common substrate for RND multidrug transporters, many of which may coexist in a single bacterium, as is illustrated by the multiple Acr and Mex efflux systems of E. coli and P. aeruginosa, respectively.
The expression of multidrug transporters is commonly controlled by specific regulatory proteins, whose structural genes are most often adjacent to those encoding the efflux system (10, 1, 16). For example, the mexZ (28) and amrR (20) genes encoding putative transcriptional repressors are located upstream from the mexXY and amrAB clusters, respectively. No such sequences were found in the environment of the adeABC genes, but three divergently transcribed ORFs were identified in the upstream region. Comparison of the sequences of the deduced proteins with those in a protein database suggested that they could code for a two-component regulatory system and a protein partly homologous to a putative transcriptional terminator-antiterminator from Mycobacterium. The genetic organization and sequence homology of ORF1, ORF2, and ORF3 suggested that the corresponding products may be involved in the regulation of adeABC expression, similar to the regulation of the norA gene in Staphylococcus aureus (7). The presence of multiple 11-bp imperfect inverted repeat sequences in the 5′ end of the adeA gene is compatible with this hypothesis. The absence of the parental strain of A. baumannii BM4454 makes it difficult to characterize the event responsible for the emergence of antibiotic resistance in this clinical isolate. In order to study the possible involvement of the proteins encoded by ORF1, ORF 2, and ORF3 in adeABC transcriptional regulation, transcriptional fusions and gene inactivation experiments are in progress.
ACKNOWLEDGMENTS
We thank M. Chignard and S. Dulong for help with ethidium bromide accumulation experiments.
This work was supported in part by a Bristol-Myers Squibb Unrestricted Biomedical Research Grant in Infectious Diseases. S.M. was a recipient of a doctoral fellowship from the Ministère de l'Education Nationale de la Recherche et de la Technologie and from the Fondation pour la Recherche Médicale.
REFERENCES
- 1.Ahmed M, Borsch C M, Taylor S S, Vazquez-Laslop N, Neyfakh A A. A protein that activates expression of a multidrug efflux transporter upon binding the transporter substrates. J Biol Chem. 1994;269:28506–28513. [PubMed] [Google Scholar]
- 2.Ainsa J A, Blokpoel M C, Otal I, Young D B, De Smet K A, Martin C. Molecular cloning and characterization of Tap, a putative multidrug efflux pump present in Mycobacterium fortuitum and Mycobacterium tuberculosis. J Bacteriol. 1998;180:5836–5843. doi: 10.1128/jb.180.22.5836-5843.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergogne-Bérézin E, Towner K J. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin Microbiol Rev. 1996;9:148–165. doi: 10.1128/cmr.9.2.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Devaud M, Kayser F H, Bachi B. Transposon-mediated multiple antibiotic resistance in Acinetobacter strains. Antimicrob Agents Chemother. 1982;22:323–329. doi: 10.1128/aac.22.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edgar R, Bibi E. MdfA, an Escherichia coli multidrug resistance protein with an extraordinarily broad spectrum of drug recognition. J Bacteriol. 1997;179:2274–2280. doi: 10.1128/jb.179.7.2274-2280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fournier B, Aras R, Hooper D C. Expression of the multidrug resistance transporter NorA from Staphylococcus aureus is modified by a two-component regulatory system. J Bacteriol. 2000;182:664–671. doi: 10.1128/jb.182.3.664-671.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giraud E, Cloeckaert A, Kerboeuf D, Chaslus-Dancla E. Evidence for active efflux as the primary mechanism of resistance to ciprofloxacin in Salmonella enterica serovar Typhimurium. Antimicrob Agents Chemother. 2000;44:1223–1228. doi: 10.1128/aac.44.5.1223-1228.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldstein F W, Labigne-Roussel A, Gerbaud G, Carlier C, Collatz E, Courvalin P. Transferable plasmid-mediated antibiotic resistance in Acinetobacter. Plasmid. 1983;10:138–147. doi: 10.1016/0147-619x(83)90066-5. [DOI] [PubMed] [Google Scholar]
- 10.Hagman K E, Pan W, Spratt B G, Balthazar J T, Judd R C, Shafer W M. Resistance of Neisseria gonorrhoeae to antimicrobial hydrophobic agents is modulated by the mtrRCDE efflux system. Microbiology. 1995;141:611–622. doi: 10.1099/13500872-141-3-611. [DOI] [PubMed] [Google Scholar]
- 11.Hood J, Amyes S G B. The chromosomal β-lactamases of the genus Acinetobacter: enzymes which challenge our imagination. In: Towner K J, Fewson C A, editors. The biology of Acinetobacter. New York, N.Y: Plenum Publishing Corp.; 1991. pp. 117–132. [Google Scholar]
- 12.Jack D L, Storms M L, Tchieu J H, Paulsen I T, Saier M H. A broad-specificity multidrug efflux pump requiring a pair of homologous SMR-type proteins. J Bacteriol. 2000;182:2311–2313. doi: 10.1128/jb.182.8.2311-2313.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koronakis V, Sharff A, Koronakis E, Luisi B, Hughes C. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature. 2000;405:914–919. doi: 10.1038/35016007. [DOI] [PubMed] [Google Scholar]
- 14.Lambert T, Gerbaud G, Galimand M, Courvalin P. Characterization of an Acinetobacter haemolyticus aac(6′)-Ig gene encoding an aminoglycoside 6′-N-acetyltransferase which modifies amikacin. Antimicrob Agents Chemother. 1993;37:2093–2100. doi: 10.1128/aac.37.10.2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lambert T, Rudant E, Bouvet P, Courvalin P. Molecular basis of aminoglycoside resistance in Acinetobacter spp. J Med Microbiol. 1997;46:731–735. [Google Scholar]
- 16.Ma D, Alberti M, Lynch C, Nikaido H, Hearst J E. The local repressor AcrR plays a modulating role in the regulation of acrAB genes of Escherichia coli by global stress signals. Mol Microbiol. 1996;19:101–112. doi: 10.1046/j.1365-2958.1996.357881.x. [DOI] [PubMed] [Google Scholar]
- 17.Masuda N, Sakagawa E, Ohya S, Gotoh N, Tsujimoto H, Nishino T. Substrate specificities of MexAB-OprM, MexCD-OprJ, and MexXY-OprM efflux pumps in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2000;44:3322–3327. doi: 10.1128/aac.44.12.3322-3327.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Midgley M, Iscandar N S, Daves E A. The interaction of phosphonium ions with Acinetobacter calcoaceticus: evidence for the operation of an efflux system. Biochim Biophys Acta. 1986;856:45–49. [Google Scholar]
- 19.Mine T, Morita Y, Kataoka A, Mizushima T, Tsuchiya T. Expression in Escherichia coli of a new multidrug efflux pump, MexXY, from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1999;43:415–417. doi: 10.1128/aac.43.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moore R A, Deshazer D, Reckseidler S, Weissman A, Woods D E. Efflux-mediated aminoglycoside and macrolide resistance in Burkholderia pseudomallei. Antimicrob Agents Chemother. 1999;43:465–470. doi: 10.1128/aac.43.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morita Y, Kodama K, Shiota S, Mine T, Kataoka A, Mizushima T, Tsuchiya T. NorM, a putative multidrug efflux protein of Vibrio parahaemolyticus and its homolog in Escherichia coli. Antimicrob Agents Chemother. 1998;42:1778–1782. doi: 10.1128/aac.42.7.1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nikaido H. Antibiotic resistance caused by gram-negative multidrug efflux pumps. Clin Infect Dis. 1998;27(Suppl. 1):32–41. doi: 10.1086/514920. [DOI] [PubMed] [Google Scholar]
- 23.Nikaido H. How do exported proteins and antibiotics bypass the periplasm in gram-negative bacterial cells? Trends Microbiol. 2000;8:481–483. doi: 10.1016/s0966-842x(00)01864-3. [DOI] [PubMed] [Google Scholar]
- 24.Noble W C. Hospital epidemiology of Acinetobacter infection. In: Towner K J, Fewson C A, editors. The biology of Acinetobacter. New York, N.Y: Plenum Publishing Corp.; 1991. pp. 53–62. [Google Scholar]
- 25.Palmen R, Vosman B, Buijsman P, Breek C K, Hellingwerf K J. Physiological characterization of natural transformation in Acinetobacter calcoaceticus. J Gen Microbiol. 1993;139:295–305. doi: 10.1099/00221287-139-2-295. [DOI] [PubMed] [Google Scholar]
- 26.Paulsen I T, Brown M H, Skurray R A. Proton-dependent multidrug efflux systems. Microbiol Rev. 1996;60:575–608. doi: 10.1128/mr.60.4.575-608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Putman M, van Veen H W, Konings W N. Molecular properties of bacterial multidrug transporters. Microbiol Mol Biol Rev. 2000;64:672–693. doi: 10.1128/mmbr.64.4.672-693.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramos Aires J, Köhler T, Nikaido H, Plésiat P. Involvement of an active efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob Agents Chemother. 1999;43:2624–2628. doi: 10.1128/aac.43.11.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosenberg E Y, Ma D, Nikaido H. AcrD of Escherichia coli is an aminoglycoside efflux pump. J Bacteriol. 2000;182:1754–1756. doi: 10.1128/jb.182.6.1754-1756.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rudant E, Bouvet P, Courvalin P, Lambert T. Phylogenetic analysis of proteolytic Acinetobacter strains based on the sequence of genes encoding aminoglycoside 6′-N-acetyltransferases. Syst Appl Microbiol. 1999;22:59–67. doi: 10.1016/S0723-2020(99)80028-9. [DOI] [PubMed] [Google Scholar]
- 31.Steers E, Foltz E L, Graves B S, Riden J. An inocula replicating apparatus for routine testing of bacterial susceptibility to antibiotics. Antibiot Chemother (Basel) 1959;9:307–311. [PubMed] [Google Scholar]
- 32.Tseng T T, Gratwick K S, Kollman J, Park D, Nies D H, Goffeau A, Saier M H. The RND permease superfamily: an ancient, ubiquitous and diverse family that includes human disease and development proteins. J Mol Microbiol Biotechnol. 1999;1:107–125. [PubMed] [Google Scholar]
- 33.Vila J, Marcos A, Marco F, Abdalla S, Vergara Y, Reig R, Gomez-Lus R, Jimenez de Anta T. In vitro antimicrobial production of beta-lactamases, aminoglycoside-modifying enzymes, and chloramphenicol acetyltransferase by and susceptibility of clinical isolates of Acinetobacter baumannii. Antimicrob Agents Chemother. 1993;37:138–141. doi: 10.1128/aac.37.1.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vila J, Ruiz J, Goni P, Marcos A, Jimenez de Anta T. Mutation in the gyrA gene of quinolone-resistant clinical isolates of Acinetobacter baumannii. Antimicrob Agents Chemother. 1995;39:1201–1203. doi: 10.1128/aac.39.5.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Westbrock-Wadman S, Sherman D R, Hickey M J, Coulter S N, Zhu Y Q, Warrener P, Nguyen L Y, Shawar R M, Folger K R, Stover C K. Characterization of a Pseudomonas aeruginosa efflux pump contributing to aminoglycoside impermeability. Antimicrob Agents Chemother. 1999;43:2975–2983. doi: 10.1128/aac.43.12.2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wong K K, Brinkman F S, Benz R S, Hancock R E. Evaluation of a structural model of Pseudomonas aeruginosa outer membrane protein OprM, an efflux component involved in intrinsic antibiotic resistance. J Bacteriol. 2001;183:367–374. doi: 10.1128/JB.183.1.367-374.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zgurskaya H I, Nikaido H. Multidrug resistance mechanisms: drug efflux across two membranes. Mol Microbiol. 2000;37:219–225. doi: 10.1046/j.1365-2958.2000.01926.x. [DOI] [PubMed] [Google Scholar]