Abstract.
Significance
Fluorescence resonance energy transfer (FRET) sensors offer enormous benefits when studying neurophysiology through confocal microscopy. Yet, their use for fiber-based in vivo recordings is hampered by massive confounding effects and has therefore been scarcely reported.
Aim
We aim to investigate whether in vivo fiber-based lactate recordings in the rodent brain are feasible with FRET sensors and implement a correction algorithm for the predominant hemodynamic artifact.
Approach
We performed fiber-based FRET recordings of lactate (Laconic) and calcium (Twitch-2B) simultaneously with functional MRI and pharmacological MRI. MR-derived parameters were applied to correct hemodynamic artifacts. Results of FRET measurements were validated by local field potential, magnetic resonance spectroscopy, and blood analysis.
Results
Hemodynamic artifacts dominated fiber-based in vivo FRET measurements with both Laconic and Twitch-2B. Our MR-based correction algorithm enabled to remove the artifacts and detect lactate and calcium changes during sensory stimulation or intravenous lactate injections.
Conclusions
In vivo fiber-based lactate recordings are feasible using FRET-based sensors. However, signal corrections are required. MR-derived hemodynamic parameters can successfully be applied for artifact correction.
Keywords: multimodal fMRI, fluorescence, fiber-based, fluorescence resonance energy transfer, blood oxygenation level-dependent, CBV
1. Introduction
Lactate has long been merely considered a waste product of anaerobic metabolism that, however, can be utilized to indicate pathologic conditions, such as cancer1,2 or inflammatory and autoimmune diseases.3,4 Evidence is growing that brain lactate plays important role in energy metabolism, maintaining homeostatic functions, and acting as a signaling molecule. Investigating the underlying mechanisms is a matter of current research.5,6 In vivo, cerebral lactate measurements remain challenging since physiologic lactate levels are low compared with other metabolites.7 Cerebral lactate can be detected by using magnetic resonance spectroscopy (MRS),8–10 microdialysis,11–13 or cyclic voltammetry.14,15 However, only a bulk signal or extracellular lactate can be measured using these methods. In addition, for MRS and microdialysis, only a low temporal resolution is achievable. These shortcomings particularly impede the investigation of cerebral energy metabolism. Resolving the distinct roles of neurons and astrocytes requires cell-specific detection of lactate levels with high temporal resolution.16,17 A genetically encoded lactate sensor, Laconic,18 has recently been developed and was successfully used for cell-specific in vivo lactate recordings in mice using two-photon microscopy.19,20
Combining functional magnetic resonance imaging (fMRI) and fluorescence recordings of genetically encoded lactate sensors has the potential to provide insight into cerebral lactate metabolism by complementing local lactate levels with a whole-brain view of hemodynamic activity. Although magnetic resonance (MR) compatible microscopes have been developed21,22 and cortex-wide calcium imaging has been successfully performed during fMRI,23 fiber-based fluorescence recordings24 have found widespread application for simultaneous fluorescence-fMRI recordings.25–34
Laconic is a fluorescence resonance energy transfer (FRET) sensor. Generally, FRET metabolic sensors consist of a sensing domain and two fluorophores, referred to as donor and acceptor, which are capable of FRET from the blue-shifted donor to the red-shifted acceptor.35 The sensor signal is obtained as the ratio of light emitted by each of the fluorophores. In contrast to single-wavelength fluorescence sensors, FRET requires the detection of light from both fluorophores at different wavelengths. Natsubori et al.36 successfully performed in vivo fiber-based fluorescence measurements in animals expressing a FRET-based calcium sensor. Yet, potential confounders for fiber-based FRET recordings such as altered cerebral blood flow or oxygenation changes have not been investigated in detail, as compared with reports for both fiber-based fluorescence measurements33 and microscopy23,37,38 using single-wavelength detection. For fiber-based lactate detection, it appears to be of particular importance to correct for such potential artifacts due to the low cerebral lactate level.
In this study, we investigated whether in vivo fiber-based FRET recordings enable the detection of changes in lactate level. We first instituted a FRET-based calcium sensor, Twitch-2B,39 with a known impulse response function (IRF) for investigating the hemodynamic artifact. As MR contrast is intrinsically sensitive to hemodynamic changes,40 we then used MR-derived parameters to correct hemodynamic artifacts in fiber-based fluorescence recordings.
2. Materials and Methods
Animal experiments were carried out according to the German Animal Welfare Act and were approved by the State Agency for Nature, Environment and Consumer Protection of North Rhine-Westphalia, Germany (LANUV approval IDs 84-02.04.2015.A427, 84-02.04.2016.A135, and 81-02.04.2018.A426). Experiments were performed on 43 adult female Fischer rats [ months, ]. Rats were housed in groups of two to five animals under a regular light/dark schedule (12/12 h) with food and water ad libitum.
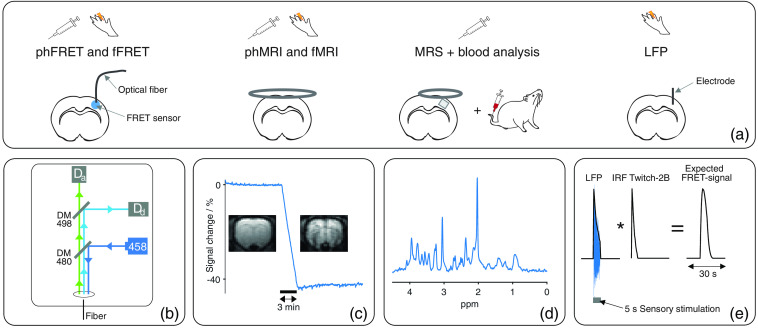
This study consisted of fiber-based optical recordings in animals expressing the genetically encoded FRET-based calcium (Twitch-2B) or lactate (Laconic) sensor in the sensory cortex, fMRI, and pharmacological MRI (phMRI) to understand and correct hemodynamic artifacts and local field potential (LFP) recordings, MRS measurements as well as blood analysis for interpretation of FRET signals [Fig. 1(a)].
Fig. 1.
(a) Study design: functional () and pharmacological (ph) experiments were performed using fiber-based FRET recordings, MRI, MRS, blood analysis, or LFP recordings. For FRET recordings, a fiber was implanted in the S1Fl of Twitch-2B- or Laconic-expressing animals. For MRI and MRS, a 2- and 1-cm surface coil were used, respectively. A cortical voxel was investigated using MRS. For blood analysis, blood was sampled from the tail vein. LFP signals were recorded as difference between the electric currents of S1Fl and cerebellum using implanted electrodes. (b) Schematic of fiber-based FRET setup: a laser (458 nm) emitted excitation light (dark blue). Excitation light was reflected by a dichroic mirror (CWL: 480 nm) and coupled into a fiber. Fluorescent light emitted by the FRET sensors was separated using a second dichroic mirror (CWL: 498 nm): Fluorescent light emitted by donor fluorophores (light blue) was reflected and detected by an APD (). Light emitted from acceptor fluorophores (green) transmitted to the mirror and was detected by a second APD (). (c) Exemplary MRI measurement during contrast agent injection: signal decreases roughly 40% altering contrast notably. (d) Exemplary in vivo NMR spectrum. (e) Calculation of expected FRET ratios of Twitch-2B-expressing animals: Envelope of LFP response (blue) to sensory stimulation was convolved with the IRF of Twitch-2B.39
We performed fiber-based in vivo functional FRET (fFRET) fluorescence measurements simultaneously with fMRI (fMRI-FRET) upon short sensory stimulation. Additionally, pharmacological FRET (phFRET) was conducted in two separate experiments: first, contrast-agent-free EPI measurements (phEPI) were performed simultaneously with phFRET (phEPI-FRET). Second, pharmacological cerebral blood volume-weighted (CBV) MR measurements were conducted (phCBV). We estimated the expected fFRET signal for Twitch-2B expressing animals using LFP measurements. For interpretation of phFRET measurements, we examined whether the injected metabolite penetrates the blood-brain barrier using blood analysis and MRS measurements. An overview of all individual experiments is shown in Table S1 in the Supplemental Materials.
2.1. Animal Handling
Animal preparation and surgical procedures were performed under isoflurane anesthesia (5% induction, 2% to 3% maintenance, in ). Blood sampling was performed under 1.8% isoflurane anesthesia. All other experiments were performed under medetomidine sedation, which was induced by a subcutaneous bolus injection of and followed by a continuous infusion of . After medetomidine bolus, isoflurane was discontinued within 10 to 15 min and experiments were executed at least 40 min after bolus injection. Medetomidine sedation was supplemented by 0.4% isoflurane in eight pharmacological experiments. This attenuated the increase in respiration rate due to lactate infusion (Table S1 in the Supplemental Materials).
About 30 min prior to stereotaxic surgery for virus injection, fiber or electrode implantation, the animals received analgesia [Metacam ( s.c.) or Metamizol ( s.c.)].
16 animals were ventilated (MRI-1 Ventilator, CWE, Inc., Ardmore, Pennsylvania, United States). Three of them received the muscle relaxant Pancuronium ( bolus followed by continuous injection of ). The end-expiratory was continuously monitored in ventilated animals using a analyzer (Micro CapStar End-Tidal Analyzer, CWE, Inc., Ardmore, Pennsylvania, United States). Respiration rate of ventilated animals was set to 53 breaths per minute (bpm). The respiratory minute volume was between 95 and . Respiration rate of spontaneously breathing animals was . Animals were placed on a temperature-controlled bed and fixated with bite and ear bars. Rectal temperature was held at .
2.2. Functional and Pharmacological Measurements
For functional measurements, electrical paw stimulation was used. For this purpose, electrodes were inserted into the forepaw contralateral to the implanted fiber/electrode. 1 ms pulses were applied at a frequency of 9 Hz and an amplitude of 1.5 mA according to a block paradigm (5-s stimulation, 25-s rest, and six repetitions). For LFP, recordings of one stimulation period were sufficient.
For pharmacological measurements, a 500 mM solution of l-lactate was injected via a lateral tail vein catheter. For phFRET, phMRI and MRS 2.5 or lactate were injected over 180 s (Table S1 in the Supplemental Materials). For blood analysis, a lower lactate concentration (250 mM) was used and lactate was injected in 150 s. This ensured that the maximum detectable lactate level of the detection device (Sec. 2.6) was not exceeded. As control, phFRET and phMRI measurements were performed by injecting saline over 180 s.
2.3. Fiber-Based FRET Fluorescence Recordings
2.3.1. Animal preparation for fiber-based fluorescence recordings
Preparation procedures for fiber-based fluorescence recordings have been described in detail previously.41 AAVs encoding Twitch-2B or Laconic were injected into forelimb region of primary somatosensory cortex (S1Fl) at least 4 weeks prior to fluorescence recordings. Histology confirmed that FRET sensors were mainly expressed in neurons. Detailed information about the viral vectors is given in the supplement (Fig. S1 and Table S2 in the Supplemental Materials). Immediately before the fluorescence experiment, an optical fiber ( diameter) was implanted above the sensor expressing region and glued to the skull.
Laconic/pcDNA3.1(-) was a gift from Luis Felipe Barros (Addgene plasmid # 44238;42 RRID:Addgene_44238). pAAV.hSyn1.Twitch-2B.WPRE.SV40 was a gift from Oliver Griesbeck (Addgene plasmid # 100040;43 RRID:Addgene_100040).
2.3.2. Procedure of fiber-based fluorescence recordings
A custom-built setup was used for fiber-based fluorescence recordings [Fig. 1(b)]. This setup was similar to setups commonly used for fiber-based fluorescence recordings, which mainly consist of a light source, a dichroic mirror, and a light detector. To enable FRET-based fluorescence recordings a second pair of dichroic mirror and light detector was added to detect the light emitted from donor and acceptor fluorophores separately.
The excitation and emission spectra of the fluorophores of Twitch-2B and Laconic (Figs. S2(a) and S2(b) in the Supplemental Materials) resulted in two specific requirements for the optical setup. First, the excitation light wavelength should be near the maximum of the excitation spectrum of donor fluorophores (around 450 nm44,45). Second, a dichroic mirror was required to separate blue light (around 480 nm44,45) emitted by the donor fluorophores from green light emitted by acceptor fluorophores (around 530 nm46).
In detail, the setup was composed as follows [Fig. S2(c) and Table S3 in the Supplemental Materials]. Excitation light was supplied by a blue laser (458 nm), reduced by neutral density filters, and reflected by a dichroic mirror [central wavelength (CWL): 480 nm]. A collimator focused the excitation light into the implanted optical fiber and the sensor expressing region was constantly illuminated via the fiber (25 to at the fiber tip). Fluorescence light emitted by the donor and acceptor fluorophores was collected by the fiber, passed the dichroic mirror, a notch filter (CWL: 457 nm), and was separated by a second dichroic mirror (CWL: 498 nm). The light emitted by donor fluorophores was reflected by the mirror and went through a bandpass filter (CWL: 479 nm), whereas the light emitted by acceptor fluorophores was transmitted to the mirror and passed a longpass filter (cut-on wavelength: 480 nm). The separated fluorescence light was focused by two lenses and detected by two avalanche photodiodes (APDs). Fluorescence signals were recorded at 2 kHz using a multifunction data acquisition device (PCIe-6363, National Instruments, Austin, Texas, United States) and a custom-written LabView script (National Instruments).
It should be noted that a portion of the excitation light was reflected at the fiber instead of being collimated. Although the used notch filter removed most of this reflected light, some light fell onto the detectors. The resulting technical background noise was calibrated for each laser light intensity and was subtracted from the detected fluorescence signals.
2.4. MR Measurements
All MR measurements were acquired on a 9.4 T Bruker Biospec 94/20 small animal scanner equipped with a gradient system (Bruker BioSpin GmbH, Ettlingen, Germany), a linearly polarized resonator, and a receive-only surface coil with 2 cm (MRI) or 1 cm (MRS) diameter. First, an anatomical image was acquired to identify the slice position for echo-planar imaging (EPI) or the voxel position for spectroscopy: 2D Turbo spin echo sequence (RARE), , RARE factor 8, Matrix , field of view (FOV) , slice thickness 1.2 mm, nine contiguous slices. Subsequently, homogenization in the area of interest was achieved using the MAPSHIM Bruker routine.
2.4.1. GE-EPI and CBV measurements
Gradient-echo-EPI (GE-EPI) and CBV measurements were performed using a single-shot gradient-echo EPI sequence (TR 1 s, same FOV as anatomy; GE-EPI: TE 18.0 ms, Matrix , flip angle 60 deg, nine slices; CBV: TE 8.2 ms, Matrix , flip angle 52 deg, three slices). Measurements lasted 3 min for functional measurements and 5 min for pharmacological measurements (2-min baseline, 3-min injection).
For CBV measurements the contrast agent Molday Ion (30-nm particle size, BioPAL, Worcester, Massachusetts, United States), containing superparamagnetic iron oxide nanoparticles, was injected into the catheterized tail veins of nine animals (15 mg Fe/kg). Injections were performed during 25 min lasting CBV-EPI measurement [10-min baseline, 3-min injection, 12-min baseline; Fig. 1(c)]. Subsequently, a pharmacological measurement (phCBV) was performed. Four animals received the lactate solution, and five animals were given saline injections.
2.4.2. MRS
Localized spectra were acquired from a cortical volume-of-interest () using a LASER sequence47 [, 2048 points, VAPOR water suppression,48 90 or 150 averages; Fig. 1(d)]. A chemical shift displacement artifact of 15% was expected between the water and lipid signals. Water linewidths were after homogenization. Series of spectra were performed before (up to five), during (one or two spectra), and after the intravenous infusion of the lactate solution.
2.5. LFP Measurements
For LFP recordings, two custom-made silver electrodes ( diameter) were implanted in the brain of 10 animals. One electrode was placed in the S1Fl and the reference electrode was positioned in the cerebellum. Electrodes were glued to the skull using dental cement. LFP signals were recorded as the difference between the electric currents detected by the electrodes using a differential amplifier (DPA-2FX, NPI Electronics, Tamm, Germany) with an implemented amplifier and bandpass filter (HP: 3 Hz and LP: 30 Hz). Signals were acquired at 2 kHz using the same data acquisition device and LabView script as for the fluorescence recordings.
2.6. Blood Analysis
Blood lactate levels were obtained in three animals before and after infusion of the l-lactate solution under isoflurane anesthesia. blood was sampled from the lateral tail vein, using heparinized glass capillaries and analyzed using a blood analyzer (Vet Scan i-STAT1, Abaxis, Union City, California, United States). Under medetomidine sedation, blood sampling was omitted due to peripheral vasoconstriction.
2.7. Data Preprocessing
Analysis was done using MATLAB (Release 2018b, The MathWorks, Inc., Natick, Massachusetts, United States) if not explicitly specified otherwise. EPI datasets of GE EPI and CBV measurements were realigned using SPM 12 (Statistical Parametric Mapping, Functional Imaging Laboratory, Wellcome Trust Centre for Neuroimaging, London, United Kingdom). The first five scans of each dataset were discarded to avoid presteady-state artifacts. In each dataset, a region of interest (ROI) was positioned in S1FL with the implanted fiber within the ROI.
For fMRI blood oxygenation level-dependent (BOLD) data a voxelwise Mann–Whitney -test49 was performed within the chosen ROI to find significantly activated voxels. The -test examined whether the signals of rest and stimulation periods differed significantly after Bonferroni correction (). To take the delayed onset of the hemodynamic response into account, the stimulation paradigm was shifted by 2 s. Signal changes were classified as positive if the stimulation signal was larger than the signal of the rest periods. The sum of voxels with positive activation was averaged across stimulation trials. The resulting time courses were normalized to the baseline and linear signal drifts were removed using the function “detrend.”
Time courses of phMRI measurements were determined by summing up the signals of all voxels within the ROI. The resulting time courses were normalized to the baseline and downsampled to a temporal resolution of 4 s. Subsequently, phCBV time courses were exposed to a further correction: Empirically, the MR signal decreased during contrast injection. However, the signal drop () from each animal deviated slightly from this value. This deviation was corrected by multiplying each CBV signal by the factor .
Fluorescence recordings of donor and acceptor signals were normalized to their baseline and downsampled to the temporal resolution of the MR measurements (1 s for fMRI and 4 s for phMRI). FRET time courses with a negative signal drift and functional data with an oscillating baseline were excluded from analysis.
NMR spectra were analyzed using LCModel50 and a dedicated simulated basis-set of metabolite spectra based on the density matrix formalism using a spin system simulation software implemented in MATLAB. Relative metabolite concentrations were derived using total Creatine () as an internal reference of concentration. The macromolecule and lipid background signals were parameterized as proposed by Lopez-Kolkovsky et al.51 An additional constraint was set to ensure that the background remained stable throughout experiments: per animal only those spectra were examined, for which the total background (sum of all macromolecules and lipids) did not deviate by from the background of the first spectrum in the time series. Spectra exhibiting Cramer–Rao lower bounds for lactate were excluded from analysis.
Spectra were used to calculate the relative increase of cortical lactate level caused by i.v. lactate injection for each animal. Lactate concentrations measured before injection were averaged. A rise in lactate level can be expected during the lactate injection and in the first minutes after the injection.20 It was assumed that the maximum lactate concentration of the three spectra recorded during and after injection corresponds to the rise of the lactate level caused by the injection. This maximum lactate concentration was determined for each animal and normalized to the preinjection concentration.
2.8. Calculation of FRET-Signals Expected in Twitch-2B-Expressing Animals
Our method for estimating the expected FRET signal of Twitch-2B-expressing animals during sensory stimulation is based on two assumptions: first, the expected FRET signal corresponds to the convolution of the IRF of Twitch-2B and the neuronal response to the applied sensory stimulation. Second, the measured LFP response reflects the neuronal response. Accordingly, the convolution of the measured LFP response and the IRF of Twitch-2B was calculated to determine the expected fFRET signal of Twitch-2B-expressing animals during one cycle of the sensory stimulation [Fig. 1(e)].
For this purpose, the IRF of Twitch-2B was taken from the study of Thestrup et al.39 with a temporal resolution of 0.5 s. This time resolution was chosen because it was close to the resolution of the fMRI data (1 s) and the time to peak (ttp) of the IRF of Twitch-2B could still be resolved. The same temporal resolution was used to extract the measured LFP response to sensory stimulation: first, LFP responses to sensory stimulation were averaged across 10 animals. Subsequently, the envelope of the averaged LFP response was obtained with the desired time resolution of 0.5 s. Finally, the convolution of the envelope of the LFP response and the IRF of Twitch-2B was calculated.
2.9. Correction of the Hemodynamic Artifact
Ma et al.38 have shown that hemodynamic artifacts in the measured fluorescence signal can be modeled by an absorption term as shown in Eq. (1). It consists of the true fluorescence signal (), the change in the absorption coefficients of the excitation () and the emitted light (), as well as the light traveling path length . For fiber-based recordings, all possible light paths can be combined since the light coming from all emitting cells is measured
| (1) |
We extended the absorption model by introducing a proportionality constant between changes in an absorption coefficient and changes in the longitudinal relaxations rate
| (2) |
This correlation is justified, considering that the relaxation rate is also dependent on hemodynamics. For small changes in relaxivity, the relative MR signal depends on the echo time TE and the change in relaxation rate 52,53
| (3) |
Summarizing Eqs. (2) and (3) show that changes in an absorption coefficient are proportional to the relative MR signal . The absorption coefficient can be described by a proportionality constant , which depends on the echo time TE
| (4) |
Combining all fixed constants (, ) of Eq. (1) into one absorption parameter , yields the following equation:
| (5) |
In FRET measurements, this equation applies to the signals of donor and acceptor, each with its own proportionality constant (, ).
2.9.1. Fitting procedure
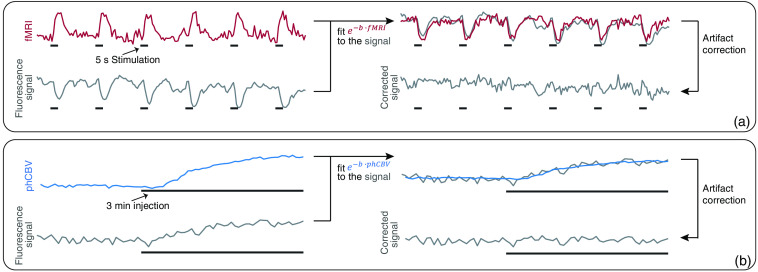
To determine the proportionality constants, the absorption term was fitted to the measured fluorescence signals by adjusting the absorption parameter . For fitting of fFRET time courses, the simultaneously recorded fMRI signal was inserted as MR signal [Fig. 2(a)]. For fitting of phFRET-signals, time courses of the averaged CBV signals were used [Fig. 2(b)].
Fig. 2.
Principle of MRI-based correction method: to each measured fluorescence time course an absorption term was fitted via optimizing the absorption parameter . For fitting fFRET data, the simultaneously measured fMRI signal was used. For phFRET recordings, the averaged CBV signal was used. Correction of the artifact was performed by dividing the fluorescence signal by the resulting absorption term. Correction procedure for (a) an exemplary fFRET fluorescence and (b) an exemplary phFRET time course.
The fit was optimized by minimizing the sum of squared residuals using the function “fmincon.” The optimization procedure was repeated for 100 starting points to find the global minimum. Starting points were generated with the function “multistart.” The fitting procedure showed the best results when omitting time points with the baseline signal. Therefore, for fFRET, only active time points were considered in the calculation of the least-squares deviation (stimulation start until 2 s after the end of stimulation). For phFRET, only time points during injections were used.
In the first step, the fluorescence signal of the acceptor fluorophore was fitted. Limits for the parameter were set to . Afterward donor signals were fitted. Limits of the parameter were set according to the parameter (fFRET: ; phFRET: ). Limiting the parameter prevented parts of the actual fluorescence signal were removed by artifact correction.
2.9.2. Artifact correction and FRET-signal calculation
For artifact correction, fluorescence signals were divided by the absorption terms resulting from the fitting procedure. Finally, FRET signals (ratios of uncorrected or corrected fluorescence signals) were calculated according to the specifications of the sensor proteins (Twitch-2B: acceptor signals divided by donor signals,39 Laconic: donor signals divided by acceptor signals18). FRET signals were reported as percent change from baseline.
For phFRET measurements, the change in FRET signal during injection was determined for each FRET-signal time course (i.e., the difference in FRET signal from injection end and injection start). The first three and the last three data points recorded during each injection were averaged and the difference was calculated. In addition, the signal-to-noise ratio (SNR) was determined for each FRET-signal time course by calculating the ratio of the detected signal change and the standard deviation of the period before injection. Only when the SNR was equal or larger than an arbitrarily chosen threshold (), the detected change was considered to be an actual alteration of the FRET signal.
2.10. Data Representation
Throughout the text, results are stated as mean ± standard deviation, or minima and maxima are specified. Figures with time courses show averages and standard deviation. In the case of wide variability of time courses, individual signal courses are shown. If not specified otherwise, individual data clusters are displayed as scatterplots and boxplots. In boxplots the central mark represents the median, boxes include the 25th and 75th percentiles, whiskers extend to the last data points within the interquartile, and outliers are indicated as crosses.
Before testing for statistical differences, data were subjected to a Kolmogorov–Smirnov test for normal distribution. Since none of our data were normally distributed, a nonparametric test (-test) was used for further statistical testing.
3. Results
3.1. fMRI-FRET
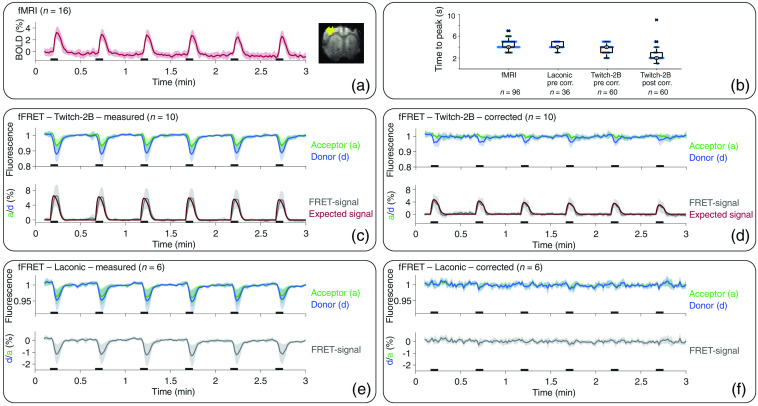
To assess whether fiber-based in vivo detection of calcium and lactate levels is feasible, we performed simultaneous fMRI-FRET measurements with rats expressing either Twitch-2B or Laconic as sensors, respectively. We performed functional measurements with electrical paw stimulation. fMRI activation maps and time courses () showed a distinct response to stimulation with a ttp of and an onset around 1 s after the start of each stimulation train [Figs. 3(a) and 3(b)]. Fluorescence time courses () of Twitch-2B-expressing animals showed signal changes starting directly with stimulation onset [Fig. 3(c)]. Acceptor signals increased while donor signals decreased. After , a signal decrease of both fluorophores was observed. FRET signals of uncorrected fluorescence data started to rise directly with stimulation onset and peaked at ttp of [Fig. 3(b)]. For artifact correction, the absorption term was fitted to the fluorescence signals. For each measurement, the simultaneously recorded fMRI signal was inserted for . Application of this correction removed major components of the signal decrease [Fig. 3(d)] and the remaining FRET signal had a ttp of [Fig. 3(b)]. To compare these data with the theoretically expected signal in artifact-free fluorescence recordings of Twitch-2B-expressing animals, we calculated a reference by convolving the neuronal response with the known IRF of the Twitch-2B sensor. LFP measurements were used to identify the neuronal response to stimulation. The resulting time course showed a ttp of 2.0 s and started to rise directly with stimulation onset. Comparing this time course to our measured data, it became evident that prior to correction, the measured FRET signal deviated from the expected signal curve [Fig. 3(c)]. After applying the correction, FRET-signal time courses agreed well with the expected time course [Fig. 3(d)].
Fig. 3.
Averaged time courses of fMRI-FRET with standard deviation, exemplary activation map, and ttp: (a) averaged fMRI signal and exemplary activation map. Time courses showed a clear response to each stimulation period (black bars). (b) Overlay of scatter plots and boxplots of ttp’s of fMRI and FRET signals. Note the temporal resolution of 1 s. (c) Time courses of the donor (blue), acceptor (green), and the FRET signal (gray) of Twitch-2B-expressing animals before and (d) after correction. The expected time course of the FRET signal (red) was normalized to the maxima of the actual FRET signal. (e) Fluorescence and FRET-signal time courses of Laconic-expressing animals before and (f) after correction of hemodynamic artifacts.
Fluorescence measurements () of donor and acceptor fluorophores in Laconic-expressing animals showed a signal decrease starting around 1 s after the start of each stimulation train, resembling the signal decrease observed for Twitch-2B-expressing animals [Fig. 3(e)]. The amplitude of signals from donor fluorophores was larger compared with signals from acceptor fluorophores. Accordingly, a decrease was evident in FRET signals of fluorescence data. Ttp of FRET signals was [Fig. 3(b)], similar to ttp observed in fMRI measurements. Application of the MR-based correction algorithm removed the signal drops in fluorescence time courses of Laconic expressing animals [Fig. 3(f)]. The averaged time course of corrected FRET signals showed no pronounced signal change triggered by stimulation. However, a -test revealed significant differences between stimulation and the first 5 s of rest periods, for the averaged corrected FRET signals (, Bonferroni corrected). This difference was not found for the uncorrected data (, Bonferroni corrected).
3.2. phMRI + phFRET
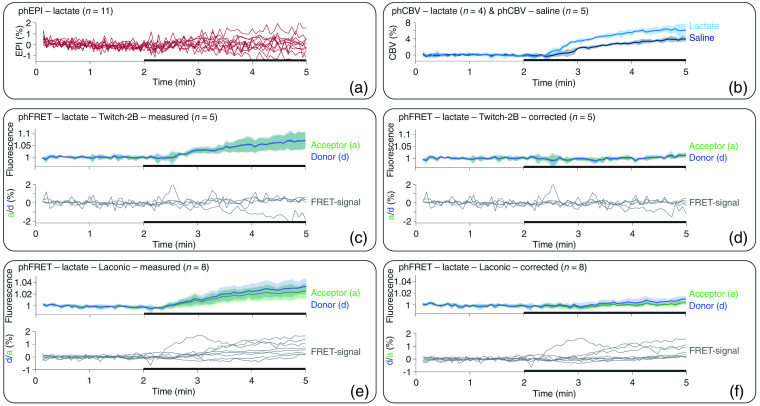
To better assess the sensitivity of FRET-based lactate detection, we performed phMRI and phFRET measurements during lactate injection in rats expressing Laconic or Twitch-2B. Signal changes in phEPI measurements upon lactate injection were low () and showed large variability, and no clear signal trend was evident [Fig. 4(a)]. Therefore, we deemed the phEPI data unsuitable for MR-based correction of hemodynamic artifacts in fluorescence measurements. In contrast, phCBV measurements showed a substantial signal increase of around 6% and 4% for both lactate () and saline () injections, respectively [Fig. 4(b)]. Therefore, phCBV measurements were used for MR-based correction of the fluorescence recordings, and the average of the phCBV signals was inserted into the absorption term for the correction procedure.
Fig. 4.
Averaged time courses of phMRI and phFRET measurements for lacate and saline injections. Individual signal time courses are shown for data sets with strongly varying time courses. (a) phEPI signals (red) showed large variations and no clear trend for injection of lactate (black bars). (b) phCBV signal increased during the injection of both lactate (light blue) and saline (dark blue). (c) Time courses of the donor (blue), acceptor (green), and the FRET signal (gray) in Twitch-2B-expressing animals during lactate injection prior to and (d) after artifact correction (right). (e) Fluorescence and FRET-signal time courses of Laconic-expressing animals during lactate injection before and (f) after correction.
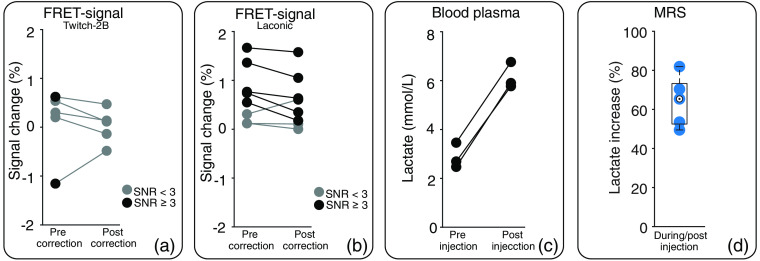
Fluorescence time courses of Twitch-2B-expressing animals showed a signal increase during lactate injection [Fig. 4(c)], with roughly similar magnitudes for donor and acceptor. Only for two animals, the corresponding FRET signal showed an SNR higher than three [Fig. 5(a)] and the FRET-signal change was considered. After application of the correction, fluorescence signals and FRET signal remained constant during lactate injection [Figs. 4(d) and 5(a)]. In Twitch-2B-expressing animals, fluorescence signals also increased during saline injection (), which was completely removed by the correction (Fig. S3 in the Supplemental Materials).
Fig. 5.
FRET-signal change and increase in lactate level caused by lactate injection. Data points of individual animals are connected by lines. Individual change of the FRET signal in (a) Twitch-2B-expressing and (b) Laconic-expressing animals upon lactate injection, before (left) and after (right) application of the correction algorithm. Only values with (black) were considered as FRET-signal change. Values with (gray) were classified as no signal change. (c) Lactate level in blood plasma before (left) and after injection (right) of lactate () in animals. (d) Relative increase in cortical lactate level caused by lactate injection () as determined by MRS in animals.
In Laconic-expressing animals (), prior to correction, both fluorescence signals and FRET-signal time courses showed an increase during lactate injection [Fig. 4(e)]. After correction, the acceptor signal remained stable, whereas the donor signal increased, resulting in a net increase of the FRET signal [Fig. 4(f)]. However, changes in FRET signals showed a large variability between individual Laconic-expressing animals [Fig. 5(b)]. Five-time courses showed an increase in the FRET signal () before applying the MR-based correction. The signal change was between 0.5% and 1.7%. This increase persisted after correction, albeit with slightly reduced values between 0.2% and 1.6%. One of three-time courses that showed no signal change before correction showed a signal increase of 0.6% after correction, whereas the FRET signals of the other two measurements remained unchanged.
3.3. Blood Analysis and MRS
To verify that lactate injection leads to a sufficient increase in lactate levels in blood and brain, blood analysis and MRS experiments were performed, respectively. Blood analysis showed a doubling of the lactate blood plasma level from to , caused by lactate injection [Fig. 5(c)]. One outlier was found in preinjection values of the MRS data using the interquartile range method. Spectra of this animal were excluded from further analysis. Cortical lactate level of remaining animals () increased by during i.v. lactate injection [Fig. 5(d)].
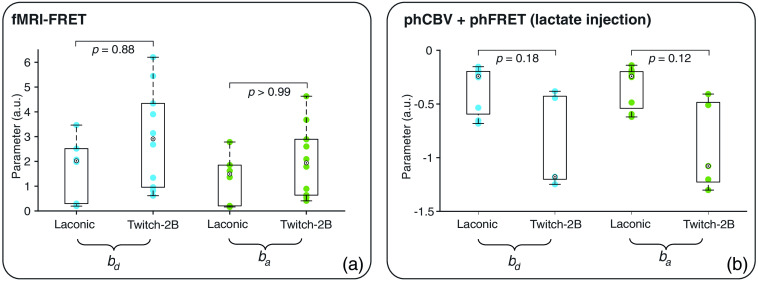
3.4. Absorption Parameters
Absorption parameters and were calculated by fitting the absorption term to the measured fluorescence separately for donor and acceptor signals, respectively. The calculated parameters ranged from 0.2 to 6.2 for fMRI-FRET measurements [Fig. 6(a)] and from to [Fig. 6(b)] for phCBV and phFRET measurements with lactate injection. Overall, the absorption parameters showed a large variability with standard deviations ranging from 0.5 to 0.8. Although not significant (-test, , Bonferroni corrected), absolute parameters for measurements with Twitch-2B-expressing animals tended to be larger than those from Laconic-expressing animals. We, therefore, conclude that the absorption strength also depends on the used FRET sensor.
Fig. 6.
Absorption parameters and for signals of donor (blue) and acceptor (green) fluorophores for (a) fMRI-FRET and (b) phCBV and phFRET measurements during lactate injection. A -test was used to test for significant differences.
4. Discussion
In this study, we performed fiber-based lactate and calcium recordings in combination with fMRI and CBV measurements. Fluorescence signals were measured in the cortex of rats expressing the genetically encoded FRET sensors Laconic and Twitch-2B. When comparing time courses of fluorescence, fMRI, and CBV signals, recorded upon forepaw stimulation and i.v. injections of lactate or saline, we identified prominent effects of hemodynamic artifacts. Artifacts were also present in the ratios of fluorescence signals that corresponded to the calcium and the lactate signals for Twitch-2B and Laconic, respectively. The application of an MR parameter-based correction algorithm yielded results that were consistent with expected signal changes based on LFP recordings, MRS, and blood analysis.
4.1. Performance of the MR-Based Correction Algorithm
In vivo calcium recordings using Twitch-2B verified that fiber-based FRET measurements are possible and showed that hemodynamic artifact correction is necessary. FRET-based calcium measurements are well suited to examine hemodynamic artifacts. Cortical calcium dynamics are well-understood and closely linked to neuronal activity.25,26,54,55 Since the IRF of Twitch-2B is known,39 we were able to calculate the expected calcium response to electrical paw stimulation using LFP recordings. Regarding phFRET recordings, we did not expect any calcium release-eliciting effect for saline or lactate injections. Calcium signals of uncorrected FRET data showed a significant deviation from the expected signals for both functional and pharmacological measurements. The high agreement of the corrected FRET signals with expected signals demonstrated the suitability of the presented MR-based correction method.
4.2. Fiber-Based Lactate Recordings are Feasible In Vivo
We detected changes in lactate level with fiber-based FRET measurements using Laconic, for both sensory stimulation and lactate injections, when the correction algorithm was applied. Uncorrected fluorescence signals clearly matched the BOLD fMRI time courses, which was particularly evident from similar values of onset and ttp. We, therefore, conclude that mainly a hemodynamic artifact was detected. Using the correction algorithm these artifacts were successfully corrected, revealing a small but significant change in lactate signal upon electrical paw stimulation. Previously, lactate changes during electrical brain stimulation or brain needle prick were investigated using microdialysis. Caesar et al.12 reported that a change in lactate level was detectable only after 30-s stimulation. In contrast, other studies showed that a shorter duration of stimulation was sufficient to cause a slow lactate rise lasting several minutes, and starting 5 s to 1 min after the stimulation onset.11,13 We did not detect a further lactate increase during resting periods. Hu and Wilson11 additionally reported an initial dip in the lactate level that started immediately with stimulation. This initial dip coincides with the lactate change detected in our study.
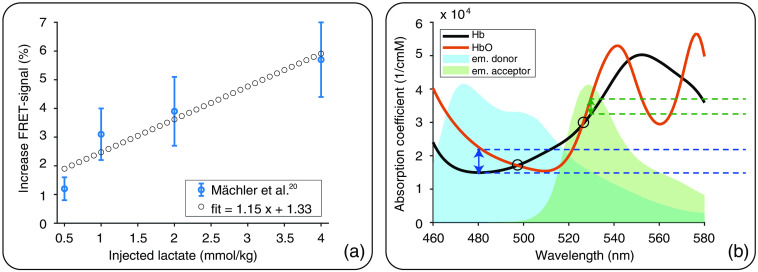
Furthermore, our fiber-based measurements demonstrated a lactate increase of up to 1.6% after i.v. injection. The rise in lactate level was verified by MRS and blood analysis. MRS measurements showed that the cortical lactate level increased around 60% to 70% during injection of lactate. Based on blood analysis, it was estimated that the blood lactate level was increased fivefold after injecting the lactate volume used for MRS (). However, blood vessels represent only around 2% to 3% of the cortical tissue.56,57 Accordingly, only 15% to 20% of the lactate increase detected by MRS can originate from the increase in blood lactate level in vessels. We, therefore, attribute part of the detected signal change during lactate injection to an increase in intra- and extracellular lactate levels. Histological validation found sensor expression exclusively in neurons, suggesting that signal changes represented an increase in neuronal lactate level. Mächler et al.20 found an increase in neuronal lactate level upon i.v. lactate injection in mice using two-photon microscopy. According to that study, an increase in neuronal lactate level of around 4% can be expected for i.v. injection of lactate [Fig. 7(a)]. However, a substantially lower signal is expected for measurements using fiber-based detection since it is not possible to examine the signal from individual cells. Instead, a bulk signal is detected from all FRET sensor-expressing cells as well as background signal from the neuropil and auto-fluorescence from tissue.
Fig. 7.
(a) Mächler et al.20 showed the increase in FRET signal for Laconic-expressing mice (blue points, average ± standard deviation). According to a linear fit (dotted line), an increase in FRET signal of around 4% can be expected for injection of 2.5 mmol/kg lactate. (b) Absorption coefficients of Hb (black) and HbO (red) are wavelength-dependent.58 For the blue and green light emitted by the donor (blue double-headed arrow and dashed lines) and acceptor (green double-headed arrow and dashed lines) fluorophores of Twitch-2B and Laconic, the absorption coefficient of HbO is greater than that of Hb. At the isosbestic points 479 and 526 nm (circles) absorption coefficients of Hb and HbO are identical. Each point lies within the emission spectra of one of the fluorophores44,46 (blue and green background; arbitrary units). If narrow bandpass filters were used, centered at the isosbestic wavelengths, oxygenation change-related artifacts could be minimized. Full absorption and emission spectra of Twitch-2B and Laconic are given in Figs. S2(a) and S2(b) in the Supplemental Materials.
4.3. Hemodynamic Artifacts in FRET Recordings
Hemodynamic changes lead to artifacts in fluorescence recordings since hemoglobin in blood absorbs light. Furthermore, hemodynamic changes modulate MR contrast. However, the generic term “hemodynamics changes” comprises various effects such as changes in blood oxygenation, changes in blood volume as well as blood dilution. Here, we observed that in functional recordings, the effect of oxygenation dominated hemodynamic artifacts (Sec. 4.3.1), whereas in pharmacological measurements oxygenation remained constant and the combination of dilution and volume change was prominent (Sec. 4.3.2).
4.3.1. Oxygenation affects fFRET
We observed hemodynamic artifacts in fFRET measurements and identified changes in oxygenation caused by neuronal activity as their major source. This notion is supported by the following observations: A hemodynamic contribution to the response to sensory stimulation was indicated by the observation of similar values of ttp of fMRI- and FRET signals when uncorrected fluorescence signals were analyzed. Oxygenation changes as the origin of the artifact were concluded from both the positive sign of the absorption parameter as well as from the larger artifact amplitudes in donor signals as compared with acceptor signals, as explained in the following.
During fMRI measurements, the change in blood oxygenation has a strong effect on contrast, which is known as the BOLD effect.59,60 Our fMRI measurements showed typical BOLD time courses. Onset and ttp corresponded to values expected according to a cortical hemodynamic response function optimized for rats.49 Changes in blood oxygenation also affect the detected fluorescence signal, due to different absorption coefficients of oxygenated (HbO) and deoxygenated hemoglobin (Hb). For the FRET sensors used in this study, the absorption coefficients are larger for HbO than for Hb.58 Thus, an increase in blood oxygenation will elicit a decrease in fluorescence signals and an increase in the MR signal. Consequently, the absorbance parameter of the presented absorbance term has a positive sign when deoxygenation of the blood occurs.
To examine this artifact in more detail, we compared the fluorescence spectra of the FRET sensors [Figs. S2(a) and S2(b) in the Supplemental Materials] with the absorption coefficients of HbO and Hb [Fig 7(b)]. This comparison demonstrated that the difference in absorption coefficients between HbO and Hb is twice as large for the emission of the donor fluorophore (around 480 nm) compared with the acceptor fluorophore (around 530 nm). Thus, a larger artifact in the donor indicates a change of oxygenation. Due to the different amplitudes in signals of donor and acceptor fluorophores, the artifact cannot be eliminated by calculating the FRET ratio of the uncorrected fluorescence data. However, the artifact may be minimized by measuring fluorescence at two isosbestic points of Hb and HbO. The two isosbestic points at 479 and 526 nm lie within the emission spectra of donor and acceptor fluorophores, respectively [Fig. 7(b)]. Using narrow bandpass filters centered at these wavelengths might render signal correction unnecessary. However, the remaining fluorescence signal may be too low for fiber-based FRET recordings.
4.3.2. Dilution and blood volume changes affect phFRET
The observed hemodynamic artifacts in fiber-based phFRET recordings were attributed to dilution effects and cerebral blood volume changes, and not to oxygenation changes. Both a decrease in blood volume and blood dilution lead to a decrease in hemoglobin in the ROI and are, therefore, expected to cause a decrease in absorption of fluorescence signals. It is important to note that this absorption change is independent of the wavelength since the ratio of Hb and HbO remains constant, and the resulting hemodynamic artifact is equal in donor and acceptor signals. CBV measurements are similarly affected by a decrease in blood volume and blood dilution, as both are associated with a local decrease of iron nanoparticles. Accordingly, blood dilution and decreasing blood volume lead to an increase in both CBV and fluorescence signals. Consequently, the absorbance parameter has a negative sign for effects resulting from blood volume changes or dilution.
In our phFRET measurements using Twitch-2B, amplitudes of donor and acceptor signals were equal during the injections, and absorption parameters had negative signs. We assume that these phFRET measurements represented the pure hemodynamic artifact since we expected no change in calcium level during either lactate or saline injections. We, therefore, conclude that hemodynamic artifacts in phFRET recordings were induced by dilution effects and blood volume changes and oxygenation changes could be excluded as an artifact source. This result was of practical relevance for the analysis of MRS data. Due to the constant oxygenation, no correction of BOLD-induced line-narrowing10,61,62 was necessary.
While it can be reasonably assumed that i.v. injection of any liquid into the blood causes similar dilution effects, our phCBV measurements showed a higher signal increase during lactate injection than during saline injection. This observation suggests that injection of lactate leads to an additional decrease in blood volume. This notion is supported by evidence that a rise in lactate during physical exercise causes a decrease in blood volume63 and that replacing some blood with Ringer’s lactate solution causes a significant decrease in blood volume.64
4.4. Artifact Correction Is Necessary
We found large variations in absorption parameters and signal amplitude of lactate between animals. These variations may originate from the fiber location relative to pial and penetrating vessels within the cortex65,66: if the implanted fiber is close to the surface of the cortex or one of the diving vessels, the hemodynamic artifact will be larger than for a fiber remote from the closest blood vessel. Furthermore, our results show that the strength of the absorption also depends on the FRET sensors used. Therefore, artifact correction is required separately for each animal and no general correction function can be calculated.
Confocal fluorescence imaging often uses reflectance measurements to correct the fluorescence signal.23,37,38 However, reflectance and fluorescence light need to have the same wavelength for effective correction. Therefore, this method is not applicable to fiber-based recordings since a bulk signal is measured, making it impossible to distinguish between reflectance and fluorescence light. A recent preprinted study presented a correction method for fiber-based recordings, where fluorescent signals from several fluorescence dyes were recorded simultaneously using fiber and a spectrometer.33 However, fiber-based detection usually does not employ spectrometers, requiring substantial modifications to perform corrections based on reflectance measurements. The MR-based method presented in this study provides a feasible alternative, for combined fluorescence-MRI measurements.25–32,34 It is also readily applicable with separate MRI measurements, as shown here for phCBV. Furthermore, our correction method can be applied to recordings of any fluorescence sensor. A potential limitation of our proposed method is the required downsampling of fluorescence data to the temporal resolution of MR scans, which may cause a loss of kinetic information. However, this was not the case for our study since the fluorescence signals had slow dynamics in all experiments. The half decay time of Twitch-2B is 2.8 s.67 Further, the response of cerebral lactate to electrical stimulation is on the timescale of seconds,14,15 and finally, a very slow but constant signal change is expected for phFRET upon a 3-min injection. For other fluorescence recordings with fast kinetics (e.g., calcium imaging with GCaMP6f68), line scanning MRI could be used for correction, enabling temporal resolutions of 50 ms.69,70
Interestingly, hemodynamic artifacts are often not reported in fiber-based studies, although their impact on fluorescence measurements has already been demonstrated in the 1990s.71 Their magnitude and importance may vary with the choice of detection wavelength and amplitude of fluorescence signal relative to the artifact. Our measurements clearly show that the absorption properties of blood lead to prominent hemodynamic artifacts in fiber-based FRET measurements and require proper correction.
5. Conclusions
Fiber-based fluorescence recordings with genetically encoded FRET-based lactate and calcium sensors are possible in the rodent brain. Hemodynamic artifacts are present in both functional and pharmacological FRET measurements and must be corrected for each animal individually. We introduced an MR-based correction algorithm that effectively removes these artifacts for both simultaneous and separate fluorescence-MRI measurements.
Supplementary Material
Acknowledgments
We thank Leo Hebbelmann for assisting surgery. This work was funded by the Deutsche Forschungsgemeinschaft (DFG), Project number 406818964, the Agence National de Recherche (ANR) project number ANR-18-CE92-0054–01 and the Interdisziplinäres Zentrum für klinische Forschung Münster (IZKF PIX).
Biographies
Henriette Lambers is a PhD candidate at the University of Münster. She is working in the Translational Research Imaging Center (TRIC) at the University Hospital Münster. She received her BSc and MSc degrees in physics from the University of Münster in 2015 and 2017, respectively. Her research focuses on multimodal neuroimaging by combining functional MRI with optical readouts.
Lydia Wachsmuth received her PhD from the University of Frankfurt in 1999. Following a postdoc at the Institute of Medical Physics in Erlangen, she worked for several years in industry and performed small animal imaging in Translational Biomedical Research. Currently, she is a research assistant at the Translational Imaging Center (TRIC) at the University Hospital Münster. Her research interests include neuroimaging, in particular, functional MRI in the healthy rodent brain and animal models of neurological disorders.
Dominik Thomas received his BSc degree in computer science and his BSc and MSc degrees in physics from the University of Münster in 2018, 2016, and 2020, respectively. Currently, he is a PhD candidate with the Steel and Hydraulic Steel Structures research group at the Helmut Schmidt University/University of the Federal Armed Forces Hamburg and is researching methods for automatic infrastructure inspection via UAS within the MISDRO project.
Fawzi Boumezbeur received his bachelor’s degree in chemistry from Ecole Supérieure de Physique et Chimie de la Ville de Paris in 2000 and his master’s and PhD degrees in physics from the University Paris-Sud in 2001 and 2005, respectively. He is currently a research director at NeuroSpin (CEA, University Paris-Saclay). He is an expert in multimodal NMR imaging and spectroscopy at high and ultra-high magnetic fields on rodents, primates, and humans. His research team is dedicated to develop new translational bioimaging technologies and apply them to further our understanding of brain metabolism in normal and pathological conditions with a focus on neurodegenerative and psychiatric diseases.
Vanessa Hoesker is a biology research technician with a strong interest in biomedical research. She was part of the Translational Research Imaging Center (TRIC) at the University Hospital Münster between April 2018 and February 2022.
Bruno Pradier is a neurobiologist and obtained his PhD from the University of Bonn, Germany. As a postdoctoral fellow, he studied synaptic physiology in the brainstem and the spinal cord at Brown University, United States. At the University of Münster, Germany, he joined an interdisciplinary team of researchers combining behavioral science and neuroimaging. His current research focuses on multimodal imaging in mice to investigate the central processing of touch and pain.
Cornelius Faber is a professor for experimental magnetic resonance at the University of Münster since 2008. He graduated in physics (Diplom), received his PhD both from the University of Bayreuth, and completed his habilitation for experimental physics from the University of Würzburg, in 1996, 1999, and 2007, respectively. His group at the Translational Research Imaging Center (TRIC) at the University Hospital Münster is devoted to developing novel MR methods to push the limits of sensitivity and resolution. His major interests are in neuroimaging using multimodal MRI, and in molecular and cellular MRI including infection imaging.
Disclosures
No conflicts of interest, financial or otherwise, are declared by the authors.
Contributor Information
Henriette Lambers, Email: h.lambers@uni-muenster.de.
Lydia Wachsmuth, Email: lydia.wachsmuth@uni-muenster.de.
Dominik Thomas, Email: dominik.thomas@hsu-hh.de.
Fawzi Boumezbeur, Email: fawzi.boumezbeur@cea.fr.
Vanessa Hoesker, Email: Vanessa.Hoesker@ukmuenster.de.
Bruno Pradier, Email: bruno.pradier@uni-muenster.de.
Cornelius Faber, Email: faberc@uni-muenster.de.
Code, Data, and Material Availability
All data and codes used in this study are available upon direct request.
References
- 1.Warburg O., “On the origin of cancer cells,” Science 123(3191), 309–314 (1956). 10.1126/science.123.3191.309 [DOI] [PubMed] [Google Scholar]
- 2.Hirschhaeuser F., Sattler U. G. A., Mueller-Klieser W., “Lactate: a metabolic key player in cancer,” Cancer Res. 71(22), 6921–6925 (2011). 10.1158/0008-5472.CAN-11-1457 [DOI] [PubMed] [Google Scholar]
- 3.Haas R., et al. , “Lactate regulates metabolic and proinflammatory circuits in control of T cell migration and effector functions,” PLoS Biol. 13(7), e1002202 (2015). 10.1371/journal.pbio.1002202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pucino V., et al. , “Lactate at the crossroads of metabolism, inflammation, and autoimmunity,” Eur. J. Immunol. 47(1), 14–21 (2017). 10.1002/eji.201646477 [DOI] [PubMed] [Google Scholar]
- 5.Gladden L. B., “Lactate metabolism: a new paradigm for the third millennium,” J. Physiol. 558(1), 5–30 (2004). 10.1113/jphysiol.2003.058701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Magistretti P. J., Allaman I., “Lactate in the brain: from metabolic end-product to signalling molecule,” Nat. Rev. Neurosci. 19(4), 235–249 (2018). 10.1038/nrn.2018.19 [DOI] [PubMed] [Google Scholar]
- 7.Tkáč I., et al. , “Highly resolved in vivo NMR spectroscopy of the mouse brain at 9.4 T,” Magn. Reson. Med. 52(3), 478–484 (2004). 10.1002/mrm.20184 [DOI] [PubMed] [Google Scholar]
- 8.Boretius S., et al. , “Halogenated volatile anesthetics alter brain metabolism as revealed by proton magnetic resonance spectroscopy of mice in vivo,” Neuroimage 69, 244–255 (2013). 10.1016/j.neuroimage.2012.12.020 [DOI] [PubMed] [Google Scholar]
- 9.Just N., et al. , “Characterization of sustained BOLD activation in the rat barrel cortex and neurochemical consequences,” Neuroimage 74, 343–351 (2013). 10.1016/j.neuroimage.2013.02.042 [DOI] [PubMed] [Google Scholar]
- 10.Just N., Faber C., “Probing activation-induced neurochemical changes using optogenetics combined with functional magnetic resonance spectroscopy: a feasibility study in the rat primary somatosensory cortex,” J. Neurochem. 150(4), 402–419 (2019). 10.1111/jnc.14799 [DOI] [PubMed] [Google Scholar]
- 11.Hu Y., Wilson G. S., “A temporary local energy pool coupled to neuronal activity: fluctuations of extracellular lactate levels in rat brain monitored with rapid-response enzyme-based sensor,” J. Neurochem. 69(4), 1484–1490 (1997). 10.1046/j.1471-4159.1997.69041484.x [DOI] [PubMed] [Google Scholar]
- 12.Caesar K., et al. , “Glutamate receptor-dependent increments in lactate, glucose and oxygen metabolism evoked in rat cerebellum in vivo,” J. Physiol. 586(5), 1337–1349 (2008). 10.1113/jphysiol.2007.144154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rogers M. L., et al. , “Continuous online microdialysis using microfluidic sensors: dynamic neurometabolic changes during spreading depolarization,” ACS Chem. Neurosci. 4(5), 799–807 (2013). 10.1021/cn400047x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith S. K., et al. , “Carbon-fiber microbiosensor for monitoring rapid lactate fluctuations in brain tissue using fast-scan cyclic voltammetry,” Anal. Chem. 90(21), 12994–12999 (2018). 10.1021/acs.analchem.8b03694 [DOI] [PubMed] [Google Scholar]
- 15.Forderhase A. G., et al. , “Simultaneous voltammetric detection of glucose and lactate fluctuations in rat striatum evoked by electrical stimulation of the midbrain,” Anal. Bioanal. Chem. 412, 6611–6624 (2020). 10.1007/s00216-020-02797-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barros L. F., “Metabolic signaling by lactate in the brain,” Trends Neurosci. 36(7), 396–404 (2013). 10.1016/j.tins.2013.04.002 [DOI] [PubMed] [Google Scholar]
- 17.Mason S., “Lactate shuttles in neuroenergetics-homeostasis, allostasis and beyond,” Front. Neurosci. 11, 43 (2017). 10.3389/fnins.2017.00043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.San Martín A., et al. , “A genetically encoded FRET lactate sensor and its use to detect the warburg effect in single cancer cells,” PLoS One 8(2), e57712 (2013). 10.1371/journal.pone.0057712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sotelo-Hitschfeld T., et al. , “Channel-mediated lactate release by K+-stimulated astrocytes,” J. Neurosci. 35(10), 4168–4178 (2015). 10.1523/JNEUROSCI.5036-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mächler P., et al. , “In vivo evidence for a lactate gradient from astrocytes to neurons,” Cell Metab. 23(1), 94–102 (2016). 10.1016/j.cmet.2015.10.010 [DOI] [PubMed] [Google Scholar]
- 21.Cui M., et al. , “A proof-of-concept study for developing integrated two-photon microscopic and magnetic resonance imaging modality at ultrahigh field of 16.4 tesla,” Sci. Rep. 7(1), 1–7 (2017). 10.1038/s41598-017-02864-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wapler M. C., et al. , “MR-compatible optical microscope for in-situ dual-mode MR-optical microscopy,” PLoS One 16(5), e0250903 (2021). 10.1371/journal.pone.0250903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lake E. M. R., et al. , “Simultaneous cortex-wide fluorescence Ca2+ imaging and whole-brain fMRI,” Nat. Methods 17(12), 1262–1271 (2020). 10.1038/s41592-020-00984-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adelsberger H., Garaschuk O., Konnerth A., “Cortical calcium waves in resting newborn mice,” Nat. Neurosci. 8(8), 988–990 (2005). 10.1038/nn1502 [DOI] [PubMed] [Google Scholar]
- 25.Schulz K., et al. , “Simultaneous BOLD fMRI and fiber-optic calcium recording in rat neocortex,” Nat. Methods 9(6), 597–602 (2012). 10.1038/nmeth.2013 [DOI] [PubMed] [Google Scholar]
- 26.Schwalm M., et al. , “Cortex-wide BOLD fMRI activity reflects locally-recorded slow oscillation-associated calcium waves,” Elife 6, e27602 (2017). 10.7554/eLife.27602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He Y., et al. , “Ultra-slow single-vessel BOLD and CBV-based fMRI Spatiotemporal dynamics and their correlation with neuronal intracellular calcium signals,” Neuron 97(4), 925–939.e5 (2018). 10.1016/j.neuron.2018.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schlegel F., et al. , “Fiber-optic implant for simultaneous fluorescence-based calcium recordings and BOLD fMRI in mice,” Nat. Protoc. 13(5), 840–855 (2018). 10.1038/nprot.2018.003 [DOI] [PubMed] [Google Scholar]
- 29.Wang M., et al. , “Brain-state dependent astrocytic Ca2+ signals are coupled to both positive and negative BOLD-fMRI signals,” Proc. Natl. Acad. Sci. U. S. A. 115(7), E1647–E1656 (2018). 10.1073/pnas.1711692115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Y., et al. , “MRI-guided robotic arm drives optogenetic fMRI with concurrent Ca2+ recording,” Nat. Commun. 10(1), 2536 (2019). 10.1038/s41467-019-10450-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Alst T. M., et al. , “Anesthesia differentially modulates neuronal and vascular contributions to the BOLD signal,” Neuroimage 195, 89–103 (2019). 10.1016/j.neuroimage.2019.03.057 [DOI] [PubMed] [Google Scholar]
- 32.Pais-Roldán P., et al. , “Indexing brain state-dependent pupil dynamics with simultaneous fMRI and optical fiber calcium recording,” Proc. Natl. Acad. Sci. U. S. A. 117(12), 6875–6882 (2020). 10.1073/pnas.1909937117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang W.-T., et al. , “Spectral fiber-photometry derives hemoglobin-absorption changes for accurate measurement of fluorescent sensor activity,” bioRxiv, 2021.08.23.457372 (2021). [DOI] [PMC free article] [PubMed]
- 34.Chao T. H., Zhang W., Hsu L., “Computing hemodynamic response functions from concurrent spectral fiber-photometry and fMRI data,” Neurophotonics 9, 032205 (2022). 10.1117/1.NPh.9.3.032205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bajar B. T., et al. , “A guide to fluorescent protein FRET pairs,” Sensors (Switzerland) 16(9), 1488 (2016). 10.3390/s16091488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Natsubori A., et al. , “Ventrolateral striatal medium spiny neurons positively regulate food-incentive, goal-directed behavior independently of D1 and D2 selectivity,” J. Neurosci. 37(10), 2723–2733 (2017). 10.1523/JNEUROSCI.3377-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kozberg M. G., et al. , “Rapid postnatal expansion of neural networks occurs in an environment of altered neurovascular and neurometabolic coupling,” J. Neurosci. 36(25), 6704–6717 (2016). 10.1523/JNEUROSCI.2363-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma Y., et al. , “Wide-field optical mapping of neural activity and brain haemodynamics: considerations and novel approaches,” Philos. Trans. R. Soc. B Biol. Sci. 371(1705) (2016). 10.1098/rstb.2015.0360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thestrup T., et al. , “Optimized ratiometric calcium sensors for functional in vivo imaging of neurons and T lymphocytes,” Nat. Methods 11(2), 175–182 (2014). 10.1038/nmeth.2773 [DOI] [PubMed] [Google Scholar]
- 40.Buxton R. B., “The physics of functional magnetic resonance imaging (fMRI),” Rep. Prog. Phys. 76(9), 096601 (2013). 10.1088/0034-4885/76/9/096601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmid F., et al. , “Assessing sensory versus optogenetic network activation by combining (o)fMRI with optical recordings,” J. Cereb. Blood Flow Metab. 36(11), 1885–1900 (2016). 10.1177/0271678X15619428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.San Martín A., et al. , “Addgene plasmid # 44238,” http://n2t.net/addgene:44238 (2013).
- 43.Thestrup T., et al. , “Addgene plasmid # 100040,” http://n2t.net/addgene:100040 (2014).
- 44.Markwardt M. L., et al. , “An improved cerulean fluorescent protein with enhanced brightness and reduced reversible photoswitching,” PLoS One 6(3), e17896 (2011). 10.1371/journal.pone.0017896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ai H. W., et al. , “Directed evolution of a monomeric, bright and photostable version of Clavularia cyan fluorescent protein: structural characterization and applications in fluorescence imaging,” Biochem. J. 400(3), 531–540 (2006). 10.1042/BJ20060874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nagai T., et al. , “A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications,” Nat. Biotechnol. 20(1), 87–90 (2002). 10.1038/nbt0102-87 [DOI] [PubMed] [Google Scholar]
- 47.Garwood M., DelaBarre L., “The return of the frequency sweep: designing adiabatic pulses for contemporary NMR,” J. Magn. Reson. 153(2), 155–177 (2001). 10.1006/jmre.2001.2340 [DOI] [PubMed] [Google Scholar]
- 48.Tkáč I., et al. , “In vivo NMR spectroscopy of rat brain at 1 ms echo time,” Magn. Reson. Med. 41(4), 649–656 (1999). [DOI] [PubMed] [Google Scholar]
- 49.Lambers H., et al. , “A cortical rat hemodynamic response function for improved detection of BOLD activation under common experimental conditions,” Neuroimage 208, 116446 (2020). 10.1016/j.neuroimage.2019.116446 [DOI] [PubMed] [Google Scholar]
- 50.Provencher S. W., “Estimation of metabolite concentrations from localized in vivo proton NMR spectra,” Magn. Reson. Med. 30(6), 672–679 (1993). 10.1002/mrm.1910300604 [DOI] [PubMed] [Google Scholar]
- 51.Lopez-Kolkovsky A. L., Mériaux S., Boumezbeur F., “Metabolite and macromolecule T1 and T2 relaxation times in the rat brain in vivo at 17.2T,” Magn. Reson. Med. 75(2), 503–514 (2016). 10.1002/mrm.25602 [DOI] [PubMed] [Google Scholar]
- 52.Davis T. L., et al. , “Calibrated functional MRI: mapping the dynamics of oxidative metabolism,” Proc. Natl. Acad. Sci. U. S. A. 95(4), 1834–1839 (1998). 10.1073/pnas.95.4.1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Buxton R. B., et al. , “Modeling the hemodynamic response to brain activation,” Neuroimage 23(suppl. 1), 220–233 (2004). 10.1016/j.neuroimage.2004.07.013 [DOI] [PubMed] [Google Scholar]
- 54.Berridge M. J., Lipp P., Bootman M. D., “The versatility and universality of calcium signalling,” Nat. Rev. Mol. Cell Biol. 1(1), 11–21 (2000). 10.1038/35036035 [DOI] [PubMed] [Google Scholar]
- 55.Grienberger C., Konnerth A., “Imaging calcium in neurons,” Neuron 73(5), 862–885 (2012). 10.1016/j.neuron.2012.02.011 [DOI] [PubMed] [Google Scholar]
- 56.Stewart P. A., et al. , “Ultrastructural concomitants of hypoxia-induced angiogenesis,” Acta Neuropathol. 93(6), 579–584 (1997). 10.1007/s004010050654 [DOI] [PubMed] [Google Scholar]
- 57.Schlageter K. E., et al. , “Microvessel organization and structure in experimental brain tumors: microvessel populations with distinctive structural and functional properties,” Microvasc. Res. 58(3), 312–328 (1999). 10.1006/mvre.1999.2188 [DOI] [PubMed] [Google Scholar]
- 58.Dervieux E., et al. , “Measuring hemoglobin spectra: searching for carbamino-hemoglobin,” J. Biomed. Opt. 25(10), 105001 (2020). 10.1117/1.JBO.25.10.105001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ogawa S., et al. , “Oxygenation‐sensitive contrast in magnetic resonance image of rodent brain at high magnetic fields,” Magn. Reson. Med. 14(1), 68–78 (1990). 10.1002/mrm.1910140108 [DOI] [PubMed] [Google Scholar]
- 60.Ogawa S., et al. , “Intrinsic signal changes accompanying sensory stimulation: functional brain mapping with magnetic resonance imaging,” Proc. Natl. Acad. Sci. U. S. A. 89(13), 5951–5955 (1992). 10.1073/pnas.89.13.5951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mangia S., et al. , “Sensitivity of single-voxel 1H-MRS in investigating the metabolism of the activated human visual cortex at 7 T,” Magn. Reson. Imaging 24(4), 343–348 (2006). 10.1016/j.mri.2005.12.023 [DOI] [PubMed] [Google Scholar]
- 62.Xu S., et al. , “Metabolic alterations in focally activated primary somatosensory cortex of -chloralose-anesthetized rats measured by MRS at 11.7 T,” Neuroimage 28(2), 401–409 (2005). 10.1016/j.neuroimage.2005.06.016 [DOI] [PubMed] [Google Scholar]
- 63.Davis J. A., et al. , “Effect of plasma volume loss during graded exercise testing on blood lactate concentration,” J. Physiol. Sci. 57(2), 95–99 (2007). 10.2170/physiolsci.RP014406 [DOI] [PubMed] [Google Scholar]
- 64.Jacob M., et al. , “The intravascular volume effect of Ringer’s lactate is below 20%: a prospective study in humans,” Crit. Care 16(3), R86 (2012). 10.1186/cc11344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xiong B., et al. , “Precise cerebral vascular atlas in stereotaxic coordinates of whole mouse brain,” Front. Neuroanat. 11(128), 1–17 (2017). 10.3389/fnana.2017.00128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schmid F., et al. , “Vascular density and distribution in neocortex,” Neuroimage 197, 792–805 (2019). 10.1016/j.neuroimage.2017.06.046 [DOI] [PubMed] [Google Scholar]
- 67.Li E. S., Saha M. S., “Optimizing calcium detection methods in animal systems: a sandbox for synthetic biology,” Biomolecules 11(3), 343 (2021). 10.3390/biom11030343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen T. W., et al. , “Ultrasensitive fluorescent proteins for imaging neuronal activity,” Nature 499(7458), 295–300 (2013). 10.1038/nature12354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu X., et al. , “Deciphering laminar-specific neural inputs with line-scanning fMRI,” Nat. Methods 11(1), 55–58 (2014). 10.1038/nmeth.2730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Albers F., et al. , “Line scanning fMRI reveals earlier onset of optogenetically evoked BOLD response in rat somatosensory cortex as compared to sensory stimulation,” Neuroimage 164(Dec. 2016), 144–154 (2018). 10.1016/j.neuroimage.2016.12.059 [DOI] [PubMed] [Google Scholar]
- 71.Malonek D., Grinvald A., “Interactions between electrical activity and cortical microcirculation revealed by imaging spectroscopy: implications for functional brain mapping,” Science 272(5261), 551–554 (1996). 10.1126/science.272.5261.551 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.