Abstract
Epigenetic aging biomarkers are associated with increased morbidity and mortality. We evaluated if occupational exposure to three established chemical carcinogens is associated with acceleration of epigenetic aging. We studied workers in China occupationally exposed to benzene, trichloroethylene (TCE) or formaldehyde by measuring personal air exposures prior to blood collection. Unexposed controls matched by age and sex were selected from nearby factories. We measured leukocyte DNA methylation (DNAm) in peripheral white blood cells using the Infinium HumanMethylation450 BeadChip to calculate five epigenetic aging clocks and DNAmTL, a biomarker associated with leukocyte telomere length and cell replication. We tested associations between exposure intensity and epigenetic age acceleration (EAA), defined as the residuals of regressing the DNAm aging biomarker on chronological age, matching factors, and potential confounders. Median differences in EAA between exposure groups were tested using a permutation test with exact p-values. Epigenetic clocks were strongly correlated with age (Spearman r>0.8) in all three occupational studies. There was a positive exposure-response relationship between benzene and the Skin-Blood Clock EAA biomarker: median EAA was −0.91 years in controls (n=44), 0.78 years in workers exposed to <10 ppm (n=41; mean benzene=1.35 ppm; p=0.034 vs. controls), and 2.10 years in workers exposed to ≥10 ppm (n=9; mean ppm=27.3; p=0.019 vs. controls; ptrend =0.0021). In the TCE study, control workers had a median Skin-Blood Clock EAA of −0.54 years (n=71) compared to 1.63 years among workers exposed to <10 ppm of TCE (n=27; mean TCE = 4.22 ppm; p=0.035). We observed no evidence of associations with formaldehyde exposure (39 controls, 31 exposed). Occupational benzene exposure was associated with increased epigenetic age acceleration measured by the Skin-Blood Clock. For TCE, there was some evidence of epigenetic age acceleration for lower exposures compared to controls. Our results suggest that some chemical carcinogens may accelerate epigenetic aging.
Keywords: Epigenetic age, DNA methylation, occupational health, benzene, formaldehyde, trichloroethylene
1. INTRODUCTION
It has been hypothesized that behavioral and environmental factors, such as exercise and diet as well as chemical exposures, affect the rate of aging. As a result, it is of high interest to develop accurate biomarkers that quantitatively capture biological aging rates, as opposed to chronological age. For example, ample research has documented that chromosomal telomere length reflects aging, is associated with age-related diseases and predicts mortality (Ehrlenbach et al., 2009). Recent progress in high throughput technologies has led to the development of several DNA methylation-based aging biomarkers, or epigenetic clocks, with different utilities and properties. Epigenetic clocks have been constructed using high dimensional DNA methylation measurements of CpG sites across the human genome to build prediction models of chronological age with high accuracy (r≥0.8) (Horvath and Raj, 2018; Jylhävä et al., 2017).
For example, Hannum and colleagues built a blood-based epigenetic clock for adults that provided an accurate aging biomarker associated with mortality but dependent on age-related leukocyte composition (Hannum et al., 2013; Marioni et al., 2015). A multi-tissue clock, or the Horvath pan tissue clock, was designed across multiple tissues and cells and performs well across the entire human lifespan (Horvath, 2013). From these clocks, one can derive both Extrinsic and Intrinsic epigenetic age acceleration (EEAA and IEAA), which reflect biological aging that is dependent and independent of known age-related immune cell changes, respectively (Lu et al., 2019a). Slower EEAA has previously been associated with certain social and behavioral factors, such as greater fish intake, moderate alcohol consumption, higher education and income and greater fruit and vegetable intake (Quach et al., 2017). For environmental exposures, ambient air pollution, organochlorine pesticides and tobacco smoke exposure have been associated with greater epigenetic age acceleration (Dhingra et al., 2018; Lind et al., 2018; de Prado-Bert et al., 2021). Second generation clocks were subsequently developed to better predict aging, morbidity and mortality (Levine, 2020). For example, the PhenoAge Clock outperformed the previous epigenetic aging biomarkers at predicting all-cause mortality, cancer, health span, physical functioning and Alzheimer’s disease (Levine et al., 2018), and the GrimAge Clock outperformed first-generation clocks at predicting mortality and age-related morbidity (Lu et al., 2019b). Another epigenetic clock, the Skin-Blood Clock, has proven valuable in ex vivo studies, tracking closely with replicative senescence, and is more accurate when applied to blood-derived samples (Horvath et al., 2018; Sturm et al., 2019). Finally, a DNA methylation-based biomarker of leukocyte telomere length (DNAmTL) was developed, which is more strongly associated with mortality, coronary heart disease, heart failure and smoking compared to measured telomere length (Lu et al., 2019a). DNAmTL reflects in part cell replication rather than the actual length of the telomeres, which might point to a greater sensitivity from environmental chemical exposures.
Novel epigenetic biomarkers of biological aging have been associated with increased morbidity and mortality risk and certain lifestyle factors (Amenyah et al., 2020; Phang et al., 2020; Wagner et al., 2020). However, beyond pesticides and air pollution, there is very limited evidence of the impact of occupational exposures on these epigenetic clocks, particularly among second-generation ones that more accurately reflect morbidity and mortality risk. Here, we explored the relationship between well characterized occupational exposure to benzene, trichloroethylene (TCE) and formaldehyde and Epigenetic Age Acceleration (EAA). These three compounds were chosen due to their pervasive use in industrial settings around the world and known carcinogenic and non-carcinogenic health effects (Baan et al., 2009; Guha et al., 2012; IARC Working Group on the Evaluation of Carcinogenic Risks to Humans, 2018; Loomis et al., 2017). For example, benzene exposure increases risk for lymphoid neoplasms and may play a role in pathologic aging through decreasing cellular expression of P53, a tumor suppressor gene possibly involved in premature aging (Bloemen et al., 2004; Vigneron and Vousden, 2010; Wilbur et al., 2008). We hypothesized that workers exposed to these three different carcinogens would exhibit EAA due to DNA methylation-induced alterations. Epigenetic dysregulation is a key characteristic of human carcinogens (Smith et al., 2020), and we hypothesized that EAA reflects this property across chemical carcinogens. We have previously reported that all three chemicals were associated with a reduction in peripheral WBC count and specific WBC subtypes (Bassig et al., 2016) and that both benzene and formaldehyde were associated with increased cytogenetic alterations in progenitor cells cultured from peripheral blood (Bassig et al., 2016; Zhang et al., 2009, 2010).
2. MATERIALS AND METHODS
2.1. Study Participants
2.1.1. Benzene study:
This study was conducted in manufacturing factories within a region of Tianjin, China, as described elsewhere (Lan et al., 2004). Briefly, benzene-exposed shoe workers were selected along with unexposed control workers matched by age and sex from three clothes-manufacturing factories. Individual benzene and toluene exposure were monitored in workers wearing 3M™ organic vapor monitors. Individual full-shift air monitoring took place every 1–2 months over a 16-month period in the larger factory with lower benzene levels and 5 times in the factory with higher exposures, resulting in the successful collection of 2,783 workplace samples. Analyses of the ambient monitors were performed by Gas Chromatography with a Flame Ionization Detector (GC-FID). Other hydrocarbons were also monitored; they were generally below 5 ppm (Lan et al., 2004). We analyzed benzene exposed workers (mean: 6.02 ppm, SD=12.90) vs. controls (<0.035 ppm, or below the limit of detection). In sensitivity analyses, we further categorized workers into three groups by benzene levels measured during the month before phlebotomy: controls (<0.035 ppm; n=48), lower exposure group (mean 1.35 ppm, SD= 1.93; n=41) and a higher exposure group (mean 27.3 ppm SD=18.5; n=9). The analysis was performed for both classifications. Participation was voluntary, and written informed consent was obtained from all volunteers. Blood samples were collected from June 2000 during the first year of the study to June 2001. The study was approved by the Institutional Review Boards at the United States National Cancer Institute and the Chinese Academy of Prevention Medicine, China (Lan et al., 2004).
2.1.2. TCE Study:
This study was conducted in the Guangdong Province of China between June and July 2006, as described elsewhere (Lan et al., 2010). Exposed workers were sampled from six factories that reported the use of TCE in manufacturing processes, had no detectable benzene, styrene, ethylene oxide, formaldehyde, or epichlorohydrin levels and had low to negligible levels of other chlorinated solvents. Unexposed control participants were sampled from four factories that were in the same geographical region as the factories that used TCE and were frequency-matched by sex and age (± 5 years) to exposed workers. Participation was voluntary, and all subjects provided written informed consent. The study excluded any participant with a history of cancer, radiotherapy, chemotherapy, or a previous occupation with notable exposure to benzene, styrene, butadiene, and/or ionizing radiation. Levels of TCE were measured during the month before phlebotomy with full-shift personal air exposure measurements (two to three per subject) taken in a three-week time-period using Dräger tubes and 3M™ organic vapor monitoring badges. Study participants were categorized into three groups of TCE exposure levels based on an 8-hour time-weighted average: unexposed controls (<0.005 ppm, n=68), a lower exposure group (mean 4.22 ppm, SD=2.56, n=25) and a higher exposure group (mean 38.7 ppm, SD=47.07, n=35) (Bassig et al., 2016). Informed consent was obtained from all subjects and the study was approved by the Institutional Review Boards at the United States National Cancer Institute and the Guangdong National Poison Control Center in China. Telomere length qPCR measurements were obtained using methods described in the previous study (Bassig et al., 2014).
2.1.3. Formaldehyde study:
In this study, we identified one factory that produced formaldehyde-melamine resins and one factory that used formaldehyde-melamine resins to manufacture plastic utensils. We monitored formaldehyde levels in these factories during an initial screening and found no other exposures to known or suspected leukemogens or hematotoxicants (e.g., benzene, phenol and chlorinated solvents). We selected control workers from three workplaces in the same geographic region as factories with formaldehyde exposure and enrolled workers who had comparable demographic and socioeconomic characteristics and who were engaged primarily in manufacturing. We monitored formaldehyde levels during full shifts (>240 min) on three working days over a three-week period using UMEx 100 diffusion samplers (SKC Inc., Eighty Four, PA). Exposure to other organic compounds was monitored with 3M organic vapor monitors, and no hydrocarbons were detected in any samples, as previously described (Zhang et al., 2010). Mean (standard deviation=SD) among formaldehyde levels in control workers was 0.02 ppm (0.007) and 1.25 ppm (0.62) among the exposed group. Participants volunteered for this study, informed consent was obtained from all subjects, and the study was approved by the Institutional Review Boards at the United States National Cancer Institute and the Guangdong National Poison Control Center in China (Bassig et al., 2016; Zhang et al., 2010).
2.2. Epigenetic Aging Biomarkers
We collected blood samples via venipuncture from all study participants and isolated DNA. We measured DNA methylation in all three studies utilizing the HumanMethylation450 BeadChips (Illumina HM450K arrays) and applied standard processing pipelines and quality controls as described previously (Phillips et al., 2019). We calculated all epigenetic aging biomarkers utilizing the Horvath’s new online calculator (http://dnamage.genetics.ucla.edu/). The outcomes of interest were EAA and age-adjusted DNAmTL (DNAmTLAdj), defined as the residuals of epigenetic aging and DNAmTL biomarkers linearly regressed on chronological age and pre-selected confounding covariates.
We calculated five different epigenetic clocks: Horvath’s Pan-Tissue Clock, Skin-Blood Clock, Hannum’s Clock, PhenoAge (Levine’s Clock) and GrimAge. Two additional measures of EAA were calculated that reflect different aspects of immunological aging: Extrinsic Epigenetic Age Acceleration (EEAA) is derived from Hannum’s Clock, which upweights the contributions of blood cell composition known to change with age (naïve cytotoxic T cells, cytotoxic T cells, and plasmablasts), and Intrinsic Epigenetic Age Acceleration (IEAA) is derived from the Pan-Tissue Clock, which is independent of age-related changes in leukocyte composition (Chen et al., 2016). Although Hannum’s EAA measure is known to be correlated with blood cell counts, its extrinsic EAA derived measure has been shown to predict all-cause mortality more strongly. Similarly, while the pan-tissue EAA is generalizable across tissues, correlations with leukocyte cell counts remain; however, its derived measure of intrinsic EAA has been shown to better reflect systemic cell-type aging, as it is preserved across tissues and cells collected from the same individual (Chen et al., 2016; Marioni et al., 2015). Therefore, these two measures provide a nuanced understanding of biological aging related to age related immune changes (EEAA) and biological aging across cells and tissues independent of immune changes (IEAA).
In addition, we tested associations with estimates of DNAmTL, adjusted for age and confounders. In this paper, we will generally refer to the adjusted DNAmTL biomarker as a measure of epigenetic age acceleration. However, this biomarker is a predictor for telomere length in units kb, and therefore, it is not necessarily a measure of telomere length itself but rather correlated with it.
On average, we expect the age acceleration (adjusted residuals) for the cohort of interest to be zero for all epigenetic clocks. However, a particular subgroup may on average have negative or positive values for the EAA measures in which case the subgroup may be viewed as being, on average, biologically younger or older than those not in the subgroup, even after adjusted for chronological age and confounders. When comparing the EAA of multiple subgroups, one should interpret the EAA values relative to the other subgroups (i.e., compare the differences in the EAA values between subgroups).
We modeled the residuals as a measure of biological age acceleration, as opposed to the difference in predicted and chronological age for each individual, as these measures become uncorrelated with predicted age and chronological age. Additionally, to calculate epigenetic residuals measures, we adjusted residuals models for BMI, smoking, recent alcohol consumption and self-reported recent infection. We did not adjust the EEAA and IEAA measures for confounders other than regressing chronological age. In sensitivity analyses, we tested age acceleration residual associations uncorrected for confounders. The results were similar but correcting for potential confounders generally improved the precision of estimates.
2.3. Statistical Analyses
We described each study with means and proportions for the variables analyzed and evaluated accuracy of epigenetic aging biomarkers via their empirical correlation with chronological age as well as scatterplots of all six epigenetic aging biomarkers across all three studies. Additionally, for the TCE study, raw measurements of the DNA telomere length were compared to the DNAmTL predictions via stratified scatter plots and correlations. To test for differences in epigenetic age acceleration (EAA) measures between exposure groups, we employed non-parametric statistical tests for differences in the median and distribution between the exposure groups. We defined the main test statistic of interest as the difference in the median value of age acceleration between exposure groups. We employed permutation-based tests to obtain exact p-values for the difference in the median age acceleration measure for each pair of exposure groups across all epigenetic aging biomarkers. The median test allows for exact inference and is preferable for its robustness against outliers, which were prevalent in the predictions of the epigenetic clocks. It should be noted that EAA residuals are obtained by performing a linear regression on all the data. To specifically test the null hypothesis of conditional independence, we derived and applied a conditional permutation test, whereby the exposure group labels are randomly permuted and the null permutation distribution of the median difference in residualized epigenetic age is computed and obtained quasi-exact p-values for the null hypothesis. The residualized epigenetic age is defined as the residuals of the linear regression of the unadjusted epigenetic age outcome on chronological age and the selected confounders (BMI, smoking, recent alcohol consumption and self-reported recent infection), omitting the exposure group assignment. By quasi-exact, we mean that the test has exact finite-sample type-1 error control against the null hypothesis of marginal independence and asymptotically exact type-1 error control against the null hypothesis of conditional independence. This and the permutation nature of the test suggest the method has better finite-sample control than asymptotic model-based methods. For robustness, we used conditional quantile regression for the EAA measures, adjusting for the pre-selected confounders of age, alcohol intake, sex, BMI, and smoking history in the models as sensitivity analyses for epigenetic clocks. To assess the continuous association between exposure and age acceleration, we performed Spearman’s test of correlation, which is a nonparametric test of monotonicity, and report p-values. We considered a p-value<0.05 as statistically significant monotonic association between exposure and age acceleration. We perform the trend test (ptrend) across all participants, pooling across the control and exposed participants.
We expected that both the median permutation test and quantile regression estimates would give similar results and control type I errors. We report 95% confidence intervals (95% CIs) for differences in medians using exposure-group stratified bootstraps along with exact p-values. We did not adjust for multiple testing but provide estimates and measures of uncertainty for interpretability. All analyses were performed with the R statistical software (https://www.R-project.org/).
3. RESULTS
3.1. Participant Characteristics:
A total of 98 workers were analyzed in the original benzene study: 48 were controls and 50 participants were exposed to benzene with a mean exposure of 6.02 ppm (SD=12.91). In this study, the mean chronological age was 31.2 (SD=8.3) years. In the TCE study, a total of 128 workers were analyzed. Of these, 68 were controls exposed to <0.0047 ppm TCE, 25 were exposed to levels between >0.005 to <10 ppm with an average of 4.22 ppm (SD=2.56) and 35 were exposed to levels ≥10 ppm, with an average of 38.7 ppm (SD=47.07). The mean age of study participants was 26.9 (SD=6.9 years). Participant characteristics for the benzene and TCE studies are described in Table 1. In the formaldehyde study, 39 control workers were exposed on average to 0.02 ppm (SD=0.007), while 31 exposed workers had a mean exposure of 1.25 ppm (SD=0.62). In this current study, the mean age was 30.5 (6.8) years. Participant characteristics for the formaldehyde study are shown in Supplementary Table S1.
Table 1.
Participant characteristics for the benzene study and the trichloroethylene (TCE) study.
| Benzene Study | TCE Study | ||||
|---|---|---|---|---|---|
| Controls | Exposed | Controls | Exposed | ||
| Subject Demographics | (n=48) | (n=50) | (n=68) | >0.005 to <10 ppm (n=25) | ≥10 ppm (n =35) |
| Age, mean (SD) | 31.2 (8.3) | 29.1 (7.34) | 27.9 (7.1) | 24.1 (5.1) | 27.3 (7.7) |
| Body mass index, mean (SD) | 22.8 (4.6) | 22.1 (3.29) | 21.3 (2.7) | 21.1 (2.1) | 21.8 (3.1) |
| Sex, n (%) | |||||
| Female | 32 (67%) | 28 (56%) | 54 (79%) | 15 (60%) | 27 (77%) |
| Male | 16 (33%) | 22 (44%) | 14 (21%) | 10 (40%) | 8 (23%) |
| Current smoker, n (%) | |||||
| Yes | 11 (23%) | 9 (18%) | 27 (40%) | 10 (40%) | 13 (38%) |
| No | 37 (77%) | 41 (82%) | 41 (60%) | 15 (60%) | 22 (62%) |
| Recently infected, n(%) | |||||
| Yes | 5 (8%) | 4 (8%) | 17(25%) | 4(16%) | 6 (17%) |
| No | 43(92%) | 46 (92%) | 51 (75%) | 21 (84%) | 29 (83%) |
| Drinks alcohol, n (%) | |||||
| Yes | 16 (33%) | 19 (38%) | 26 (41%) | 7 (28%) | 14 (40%) |
| No | 32 (67%) | 31 (62%) | 42 (59%) | 18 (72%) | 21 (60%) |
| Exposure (ppm) | Benzene Exposure | TCE Exposure | |||
| mean (SD) | <0.035 (LOD) | 6.02 (12.91) | <0.0047 (0) | 4.22 (2.56) | 38.7 (47.07) |
| range | --- | (0.20, 72.87) | (0.0045, 0.0046) | (0.4451, 9.300) | (10.35, 228.92) |
3.2. Performance of Epigenetic Biomarkers:
In the benzene study, correlations between epigenetic aging biomarkers and chronological age were high, ranging from r=0.93 for the Skin-Blood and Grim Age Clocks to r=−0.64 for DNAmTL estimates. For the TCE study, epigenetic clock predictions for chronological age were also high, ranging from r=0.92 for the Skin-Blood Clock to r=−0.73 for DNAmTL. A slightly lower performance was observed in the formaldehyde study, with the highest correlation of r=0.88 observed in the Skin-Blood Clock to r=−0.68 for DNAmTL (Figure 1). Across all studies, the Skin-Blood Clock had the highest correlation, while the DNAmTL showed the lowest absolute correlations. For the TCE study, qPCR measurements of the DNA telomere length were available and compared to the DNAmTL estimated using Pearson’s test of correlation. Across all TCE study participants, the correlation was moderate between DNAmTL and qPCR measured leukocyte telomere length (r=0.31; p=0.0019) (Supplementary Figure S1-A), but similar in magnitude to the original DNAmTL publication. We found that the control and lower exposure group’s DNA telomere length measurements respectively had a moderate correlation of r=0.47 (p=0.0026) and r=0.51 (p=0.0087) with DNAmTL, while the higher exposure group had a nonsignificant correlation of r=−0.0051 (p=0.98) (Supplementary Figure S1-B). We additionally compared the difference in correlation between the exposure groups using the Fischer-transform test of difference in correlation. We found that the correlation of the control and lower exposure groups were not significantly different from each other (p=0.59), but both the correlations comparing the higher exposure group to the control (p=0.044) and lower (p=0.02) exposure groups were significantly different (Supplementary Table S2).
Figure 1.
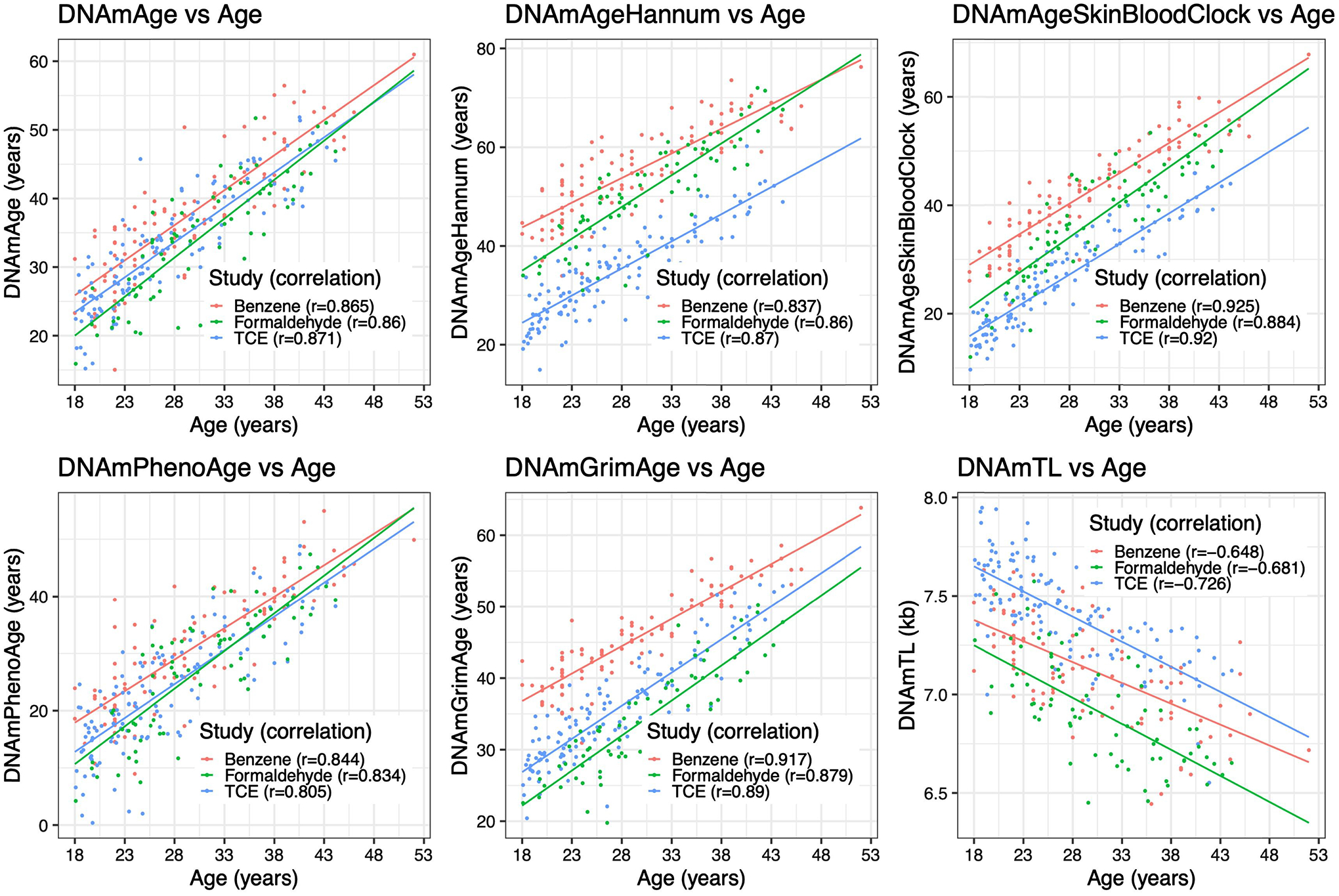
Scatterplots and regression lines of chronological age (years) versus epigenetic aging biomarkers, color-coded by study type.
3.3. Correlation between age acceleration measures:
In Supplementary Table S3, we report the Spearman correlation matrix between the age acceleration measures of all clocks (residuals of predicted age based on the adjusted linear model). All age acceleration measures were uncorrelated with age. We found that IEAA and the Horvath Pan-Tissue Clock are highly correlated (r=0.94), and the EEAA and the Hannum EAA were also highly correlated (r=0.95). This is expected since the IEAA is derived from the Horvath Pan-Tissue Clock, and similarly, the EEAA measure is derived from the Hannum-Blood Clock. As expected, the residuals of DNAmTL were negatively correlated with all EAA measures.
3.4. Epigenetic Aging Biomarkers and Benzene Exposure:
When comparing unexposed controls (<0.035 ppm, n=48) to benzene exposed workers (n=50), we observed that the benzene exposed group had a median of 1.75 years greater EAA for the Skin-Blood Clock (95% CI: 0.27, 3.28; p=0.012) and 0.09 kb shorter DNAmTLAdj (95% CI: −0.18, −0.001; p=0.023) relative to controls (Table 2). No significant results were found for the median EAA of the other clocks. Results for all epigenetic aging biomarkers and their association with benzene exposure are summarized in Table 2. The results for difference in mean EAA between the control and exposed groups using adjusted quantile regression can be found in Supplementary Table S4A, for controls vs. exposed, and Supplementary Table S4B, for controls vs. lower vs. higher exposure groups. Similar to the median permutation test results, unexposed controls were observed to have a median EAA adjusted difference for the Skin-Blood Clock that is 1.74 years greater than the exposed group (p=0.008), and median DNAm-age adjusted telomere length 0.10 kb shorter than the exposed group (p=0.037). No significant differences were observed for the remaining mean EAA measures.
Table 2.
Median epigenetic aging biomarkers and differences among workers exposed to benzene and controls. Bootstrapped 95% confidence intervals and p-values based on the median difference permutation test are also provided. Bold values represent p<0.05.
| Median Epigenetic Age Acceleration* | Difference Exposed vs. Controls | |||
|---|---|---|---|---|
| Epigenetic Aging Biomarker | Exposed | Controls | Difference (95% CI) | Exact p-value |
| Horvath Pan tissue Clock | 0.46 | −0.74 | 1.20 (−1.15, 3.62) | 0.34 |
| Hannum Clock | 0.52 | −0.35 | 0.87 (−2.98, 2.51) | 0.68 |
| Skin-Blood Clock | 0.84 | −0.91 | 1.75 (0.27, 3.28) | 0.012 |
| Grim Age Clock | −0.06 | 0.78 | −0.84 (−1.59, 0.40) | 0.09 |
| PhenoAge Clock | −0.06 | −0.21 | 0.15 (−3.12, 2.24) | 0.16 |
| DNAm Telomere Length Adjusted (DNAmTLAdj) | −0.03 | 0.06 | -0.09 (−0.18, −0.001) | 0.023 |
| Intrinsic Epigenetic Age Acceleration (IEAA) | −0.30 | 0.80 | −1.10 (−2.40, 1.92) | 0.47 |
| Extrinsic Epigenetic Age Acceleration (EEAA) | −0.58 | −0.27 | −0.31 (−4.10, 3.93) | 0.93 |
Age acceleration is defined as the residuals of the regression of the marker on age and possible confounders, and therefore, it may be negative. The age acceleration measure value for a specific exposure group should be interpreted only relative to the age acceleration measure values of the other exposure groups.
To assess the association of higher benzene exposure and age acceleration, we further classified the exposure groups into controls (<0.035 ppm, n=48), a lower exposure group (<10 ppm, mean 1.35 ppm, n=41) and higher exposure group (≥10 ppm, mean 27.3 ppm, n=9). When comparing the lower exposure to the control group, EAA for the Skin-Blood Clock remained increased by 1.69 years (95% CI: −0.09, 3.17; p=0.034). The higher exposure group had a median increase of 3.01 years in EAA for the Skin-Blood Clock (95% CI: 0.01, 4.98; p=0.019) relative to controls. Median adjusted DNAmTL residual was lower in the higher exposure group by −0.25 kb (95% CI: −0.47, 0.06; p=0.0038) and by −0.09 kb (95% CI: −0.15, 0.02; p=0.06) for the lower exposure group relative to controls. Benzene results across the three exposure groups are summarized in Table 3. When testing for trends across all participants, we found a significant increasing trend in EAA for the Skin-Blood Clock (p=0.0021) and a significant decreasing trend in DNAmTLAdj (p=0.0213). Associations for EAA for each clock across exposure groups are shown in Figure 2. No further significant trends were reported.
Table 3.
Medians and differences in medians for Epigenetic Age Acceleration biomarkers among controls, workers exposed to <10 ppm, and workers exposed to ≥10 ppm of benzene. Bootstrapped 95% confidence intervals and p-values for the median difference permutation test are also provided. Trend test between exposure and age acceleration across all data. Bold values represent p<0.05.
| Median Epigenetic Age Acceleration* | Differences (95% CI) P-value | |||||
|---|---|---|---|---|---|---|
| Epigenetic Aging Biomarker | Controls (n=48; <0.035 ppm) | <10 ppm (n=41; 1.35 ppm) | ≥10 ppm (n=9; 27.3 ppm) | <10 ppm vs. Control | ≥10 ppm vs. Control | Trend test p-value |
| Horvath Pan tissue Clock | −0.74 | 0.02 | 1.67 | 0.76 (−1.89, 3.61) p=0.61 | 2.41 (−0.67, 4.60) p=0.16 | p=0.35 |
| Hannum Clock | −0.35 | −1.69 | 3.27 | −1.34 (−3.67, 1.97) p=0.37 | 3.62 (−2.52, 6.41) p=0.13 | p=0.62 |
| Skin-Blood Clock | −0.91 | 0.78 | 2.10 | 1.69 (−0.09, 3.17) p=0.034 | 3.01 (0.01, 4.98) p=0.019 | p=0.0021 |
| Grim Age Clock | 0.78 | −0.14 | 0.53 | −0.92 (−1.96, 0.31) p=0.07 | −0.25 (−1.84, 1.74) p=0.82 | p=0.55 |
| PhenoAge Clock | −0.21 | −0.50 | 2.86 | −0.29 (−3.60, 2.05) p=0.85 | 3.07 (−5.47, 6.26) p=0.13 | p=0.90 |
| DNAm Telomere Length Adjusted (DNAmTLAdj) | 0.06 | −0.02 | −0.18 | −0.09 (−0.15, 0.02) p=0.06 | -0.25 (−0.47, 0.06) p=0.004 | p=0.021 |
| Intrinsic Epigenetic Age Acceleration (IEAA) | 0.80 | −0.54 | 0.84 | −1.34 (−2.72, 1.60) p=0.39 | 0.04 (−2.30, 3.09) p=0.96 | p=0.46 |
| Extrinsic Epigenetic Age Acceleration (EEAA) | −0.27 | −1.69 | 4.07 | −1.42 (−4.77, 3.46) p=0.59 | 4.34 (−3.05, 9.08) p=0.19 | p=0.70 |
Age acceleration is defined as the residuals of the regression of the marker on age and possible confounders, and therefore, it may be negative. The age acceleration measure value for a specific exposure group should be interpreted only relative to the age acceleration measure values of the other exposure groups.
Figure 2.
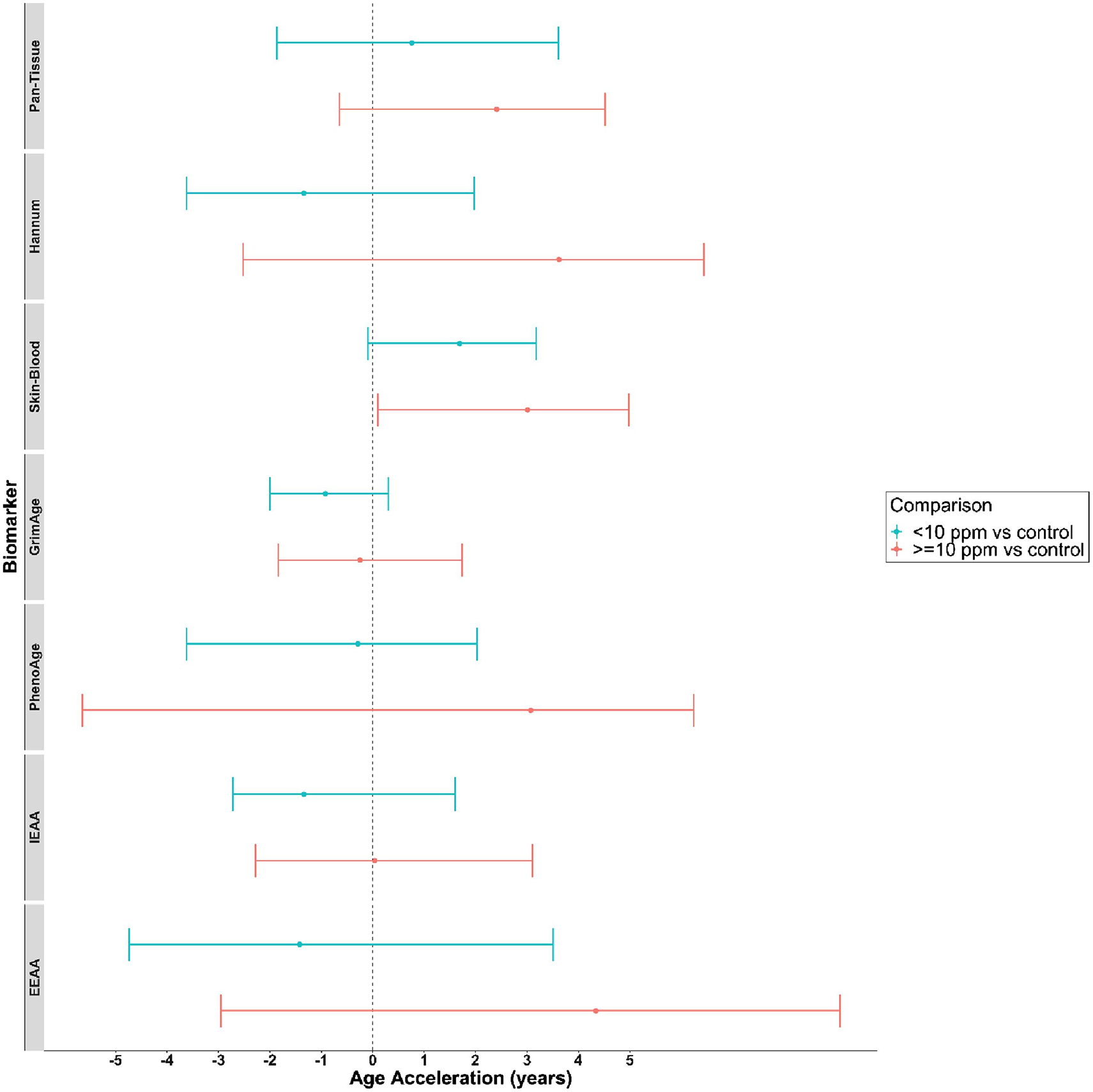
Median difference in Epigenetic Age Acceleration (adjusted residuals) between Benzene exposure groups; <10 ppm and ≥10 ppm compared to controls and 95% bootstrap confidence intervals for all epigenetic clocks. Point estimates that are to the right of the dashed line correspond with increased age acceleration of the exposure group relative to control.
3.5. Epigenetic Aging Biomarkers and TCE:
In the TCE study, no significant differences were observed when comparing controls to all TCE exposed workers (Supplementary Table S5) and no consistent exposure-response patterns were present. When comparing controls, lower and higher exposure groups certain exposed groups were significantly different from controls. Namely, control workers had a median Skin-Blood Clock EAA of −0.54 years (n=71) compared to 1.63 years among workers exposed to <10 ppm of TCE (n=27; mean TCE = 4.22 ppm; p=0.035) (Table 4). The results for the adjusted median difference in epigenetic aging between exposure groups estimated with quantile regression can be found in Supplementary Table S6, with consistent estimate of age acceleration observed between workers exposed to <10 ppm and controls of 2.13 years (95% CI: 0.37, 4.84).
Table 4.
Medians and differences in medians for Epigenetic Age Acceleration biomarkers among workers with TCE exposure ≥10ppm (mean 37.4 ppm), <10 ppm (mean 4.38 ppm) and controls. Bootstrapped 95% confidence intervals and p-values for the median difference permutation test are also provided. Final column shows the results of a trend test (Spearman test of correlation) between exposure and age acceleration across all data. Bold values represent p<0.05.
| Median Epigenetic Age Acceleration* | Median Differences (95 %CI) P-value | |||||
|---|---|---|---|---|---|---|
| Epigenetic Aging Biomarker | Controls (n=71) | <10 ppm (n=27) | ≥10 ppm (n=37) | < 10 ppm vs. Controls | ≥10 ppm vs. Controls | Trend Test p-value |
| Horvath Pan tissue Clock | −0.35 | 0.59 | −0.16 | 0.94 (−1.47, 3.35) p=0.51 | 0.19 (−2.96, 1.54) p=0.91 | p=0.70 |
| Hannum Clock | −0.07 | 2.13 | −1.11 | 2.20 (−2.47, 3.75) p=0.07 | −1.04 (−2.75, 0.35) p=0.19 | p=0.25 |
| Skin-Blood Clock | −0.54 | 1.63 | −0.39 | 2.17 (−0.68, 5.01) p=0.035 | 0.15 (−1.27, 1.68) p=0.88 | p=0.45 |
| Grim Age Clock | −0.02 | 0.09 | 0.67 | 0.11 (−1.78, 2.00) p=0.93 | 0.69 (−0.67, 1.78) p=0.33 | p=0.51 |
| PhenoAge Clock | 0.21 | −0.57 | 1.22 | −0.78 (−4.70, 6.06) p=0.64 | 1.01 (−1.07, 3.49) p=0.33 | p=0.72 |
| DNAm Telomere Length Adjusted (DNAmTLAdj) | 0.02 | 0.04 | −0.06 | 0.02 (−0.08, 0.08) p=0.89 | −0.08 (−0.15, 0.04) p p=0.08 | p=0.41 |
| Intrinsic Epigenetic Age Acceleration (IEAA) | −0.60 | 0.39 | 0.56 | 0.99 (−1.00, 3.10) p=0.39 | 1.16 (−1.34, 2.69) p=0.33 | p=0.13 |
| Extrinsic Epigenetic Age Acceleration (EEAA) | 0.11 | 0.62 | −0.83 | 0.51 (−2.95, 3.91) p=0.53 | −0.94 (−2.84, 0.42) p=0.24 | p=0.34 |
Age acceleration is defined as the residuals of the regression of the marker on age and possible confounders, and therefore, it may be negative. The age acceleration measure value for a specific exposure group should be interpreted only relative to the age acceleration measure values of the other exposure groups.
3.6. Epigenetic Aging Biomarkers for Formaldehyde:
No statistically significant differences were observed between formaldehyde exposed workers relative to controls by permutation testing or estimates of quantile regression. The results are shown in Supplemental Table S7.
4. DISCUSSION
In this study, we investigated associations between occupational exposure to air pollutants benzene, trichloroethylene (TCE) and formaldehyde for six epigenetic aging biomarkers and their measures of biological age acceleration. We observed that exposure to benzene accelerated the Hannum and Skin-Blood epigenetic clocks and was also associated with shorter DNAm estimates of telomere length, which more accurately reflects cell replication rather than actual telomere length. For the TCE study, epigenetic age acceleration was observed among workers exposed to <10 ppm of TCE relative to controls for the Skin-Blood Clock.
The Skin-Blood Clock was developed using DNA methylation measurements in white blood cells, leukocytes. It is therefore not surprising that benzene affects this clock, as benzene exposure damages the hematopoietic stem cells of the bone marrow and inhibits the development of leukocytes (Smith, 2010). The shorter DNAm estimates of telomere length we observed after occupational benzene exposure are consistent with this conclusion, as they reflect lowered proliferation of white blood cell precursors. Given the universal nature of benzene exposure as a result of it being present in gasoline, fire smoke and smoke from cigarettes, pipes and other tobacco products (IARC Working Group on the Evaluation of Carcinogenic Risks to Humans, 2018), the fact that it may accelerate biological aging is another reason for concern and appropriate environmental and occupational regulations. A previous report on telomere length in WBCs measured by qPCR in workers exposed to relatively high levels of benzene found that benzene was associated with longer telomere length (Bassig et al., 2014). As such, the observation in our study on DNAm estimates of telomere length might more likely reflect cellular proliferation, as previously noted, rather than telomere length itself.
We did not observe associations with formaldehyde exposure in these relatively highly exposed workers, even though we have previously reported that it has been associated with lower white blood cell counts, though to a lesser extent than benzene. We also previously demonstrated that formaldehyde exposure was associated with an increase in cytogenetic alterations important for myeloid leukemia in progenitor cells cultured from peripheral blood (Lan et al., 2010; Zhang et al., 2010) in the same study population evaluated in the current report. It is possible that our report represents a true null finding. Alternatively, epigenetic markers evaluated here might be less sensitive to relatively high formaldehyde exposure, and as such, a larger sample size in a future study would be needed to identify these more subtle effects.
TCE exposure has been associated with a decrease in total peripheral lymphocyte counts and all major lymphocyte subsets. It does not appear to directly affect peripheral blood cells derived from myeloid progenitors, and it alters several other biomarkers, consistent with an immunotoxic effect (Lan et al., 2010). Overall, TCE was not consistently associated with epigenetic clocks, but a significant association was observed at lower levels of exposure for the Skin-Blood Clock compared to controls workers. Further studies will be needed to examine the relationship between TCE exposure and accelerated aging.
We observed excellent performance for the Skin-Blood Clock at predicting chronological age, and this clock appeared to be the most sensitive to occupational benzene and TCE exposure in terms of EAA. Given the higher accuracy, it is possible that this clock could be a more suitable epigenetic biomarker to study the effect of environmental exposures on biological aging in epidemiological studies that collect and analyze DNA methylation from leukocytes. Our results are consistent with those from a study in children that found that exposure to indoor particulate matter and parental smoking were associated with increased EAA for the Skin-Blood Clock (estimates of 0.07 (0.02–0.12) and 0.15 (0.01–0.29), respectively), and this biomarker outperformed other epigenetic age measures (de Prado-Bert et al., 2021). We additionally report that at very high TCE exposure (≥10 ppm), the DNAmTL biomarker becomes uncorrelated with qPCR measures of leukocyte telomere length, which might be explained by cell replication disruptions at high exposure levels. Future studies should interpret findings on this biomarker in light of this limitation.
This study contributes to the literature about the application of aging biomarkers within the field of environmental health sciences by researching the effects of occupational chemical exposures. Existing studies have shown that exposure to certain pesticides and air pollutants, such as particulate matter and black carbon, were associated with increased EAA with the Horvath clock as well as EEAA (Dhingra et al., 2018). However, in some studies, metal exposure to cadmium and cobalt and pesticide exposure to organophosphates have not been associated with aging biomarkers (Dhingra et al., 2018). Occupational chemical exposures generally occur at an intensity or duration higher than that of the average exposure in public, and since EAA has been shown to correlate closely with cancer and mortality risk, it is important to understand how these factors can increase risk for diseases on an epigenetic level.
While second-generation clocks such as GrimAge and PhenoAge are calibrated to predict mortality and morbidity we did not see evidence of age acceleration with benzene, TCE or formaldehyde. There might be several reasons for this including, low statistical power, different toxicity pathways and the relevance of biomarkers used to construct these clocks. For example, the GrimAge clock uses multiple plasma proteins and self-reported smoking as predictor while the PhenoAge clock leverages clinical measures associated with mortality. These clocks are predictive of all cause mortality and morbidity, not specific to a given pathway therefore the lack of association could be due to non-specific training of the clocks. The Skin-Blood Clock retains similar properties to the original pan-tissue and second-generation clocks in that it predicts mortality and is associated with several nutritional exposures with the added benefit of improved accuracy. The increased precision might be a contributing factor to its shown sensitivity across chemical exposures.
This study has some limitations. Although our previous reports from these studies of workers exposed to benzene, TCE and formaldehyde have found associations with multiple endpoints (e.g., a decrease in peripheral lymphocyte counts) (Bassig et al., 2016), the sample sizes are still relatively small. If epigenetic aging markers are associated with all three chemicals but are less sensitive than these previously studied biomarkers, then larger sample sizes might be needed to identify these effects. Of note, we computed multiple complementary epigenetic aging biomarkers across three different studies and observed consistency of performance for the observed associations with age, lending confidence to the quality of the epigenetic aging biomarker data. However, the age acceleration measure might carry greater error, requiring larger sample size to detect small associations. Although we controlled for some potential confounders in the statistical models, residual confounding is still possible, which could lead to overestimates or underestimates of associations. Several efforts were made to ensure no other solvents or potential hydrocarbons were present, and this was supported by the monitoring data. However, exposure to other carcinogens or factors that altered the clocks is possible. Lastly, we conducted multiple statistical tests and did not adjust for multiple testing, so findings by chance are possible.
In conclusion, our results provide evidence that benzene and TCE exposures influence biomarkers of epigenetic aging. Few studies have evaluated the role of occupational and environmental exposures on biomarkers of biological aging beyond classical measures of telomere length. Future research should continue to evaluate potential mediation of epigenetic aging biomarker as environmental drivers of disease susceptibility.
Supplementary Material
Highlights:
Occupational exposures were associated with DNA methylation aging biomarkers.
Benzene and trichloroethylene exposures increased epigenetic age acceleration.
Epigenetic aging biomarkers capture impacts of environmental chemicals on health.
Funding details:
This project was supported by the Superfund Research Center at UC Berkeley National Institute of Environmental Health Sciences (Research Triangle Park, NC) Grant P42ES004705 and National Institute of Environmental Health Sciences (Research Triangle Park, NC) Grant R01ES031259, National Institute on Aging (Bethesda, MD) Grant R03AG067064 and Intramural funds from the National Cancer Institute (Bethesda, MD). Funding sources had no roles in the study design, statistical analysis, or decision to publish.
Abbreviations:
- DNAm
DNA methylation
- DNAmTL
DNA methylation-based biomarker of leukocyte telomere length
- EAA
epigenetic age acceleration
- EEAA
extrinsic epigenetic age acceleration
- IEAA
intrinsic epigenetic age acceleration
- TCE
trichloroethylene
Footnotes
Declaration of Competing Financial Interest:
Martyn T. Smith is retained as a consultant and expert witness in U.S. litigation involving benzene, trichloroethylene, formaldehyde and their associations with kidney injury and cancer. All other authors have no disclosures.
Data Statement:
Reasonable requests for access to data can be sent to the corresponding author, who will evaluate all inquiries.
REFERENCES
- 1.Amenyah SD, Ward M, Strain JJ, McNulty H, Hughes CF, Dollin C, Walsh CP, and Lees-Murdock DJ (2020). Nutritional Epigenomics and Age-Related Disease. Curr. Dev. Nutr 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baan R, Grosse Y, Straif K, Secretan B, El Ghissassi F, Bouvard V, Benbrahim-Tallaa L, Guha N, Freeman C, Galichet L, et al. (2009). A review of human carcinogens--Part F: chemical agents and related occupations. Lancet Oncol. 10, 1143–1144. [DOI] [PubMed] [Google Scholar]
- 3.Bassig BA, Zhang L, Cawthon RM, Smith MT, Yin S, Li G, Hu W, Shen M, Rappaport S, Barone-Adesi F, et al. (2014). Alterations in Leukocyte Telomere Length in Workers Occupationally Exposed to Benzene. Environ. Mol. Mutagen 55, 673–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bassig BA, Zhang L, Vermeulen R, Tang X, Li G, Hu W, Guo W, Purdue MP, Yin S, Rappaport SM, et al. (2016). Comparison of hematological alterations and markers of B-cell activation in workers exposed to benzene, formaldehyde and trichloroethylene. Carcinogenesis 37, 692–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bloemen LJ, Youk A, Bradley TD, Bodner KM, and Marsh G (2004). Lymphohaematopoietic cancer risk among chemical workers exposed to benzene. Occup. Environ. Med 61, 270–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen BH, Marioni RE, Colicino E, Peters MJ, Ward-Caviness CK, Tsai P-C, Roetker NS, Just AC, Demerath EW, Guan W, et al. (2016). DNA methylation-based measures of biological age: meta-analysis predicting time to death. Aging 8, 1844–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dhingra R, Nwanaji-Enwerem JC, Samet M, and Ward-Caviness CK (2018). DNA methylation age – environmental influences, health impacts, and its role in environmental epidemiology. Curr. Environ. Health Rep 5, 317–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ehrlenbach S, Willeit P, Kiechl S, Willeit J, Reindl M, Schanda K, Kronenberg F, and Brandstätter A (2009). Influences on the reduction of relative telomere length over 10 years in the population-based Bruneck Study: introduction of a well-controlled high-throughput assay. Int. J. Epidemiol 38, 1725–1734. [DOI] [PubMed] [Google Scholar]
- 9.Guha N, Loomis D, Grosse Y, Lauby-Secretan B, El Ghissassi F, Bouvard V, Benbrahim-Tallaa L, Baan R, Mattock H, Straif K, et al. (2012). Carcinogenicity of trichloroethylene, tetrachloroethylene, some other chlorinated solvents, and their metabolites. Lancet Oncol. 13, 1192–1193. [DOI] [PubMed] [Google Scholar]
- 10.Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, Klotzle B, Bibikova M, Fan J-B, Gao Y, et al. (2013). Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol. Cell 49, 359–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horvath S (2013). DNA methylation age of human tissues and cell types. Genome Biol. 14, 3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horvath S, and Raj K (2018). DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat. Rev. Genet 19, 371–384. [DOI] [PubMed] [Google Scholar]
- 13.Horvath S, Oshima J, Martin GM, Lu AT, Quach A, Cohen H, Felton S, Matsuyama M, Lowe D, Kabacik S, et al. (2018). Epigenetic clock for skin and blood cells applied to Hutchinson Gilford Progeria Syndrome and ex vivo studies. Aging 10, 1758–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.IARC Working Group on the Evaluation of Carcinogenic Risks to Humans (2018). Benzene (Lyon (FR): International Agency for Research on Cancer).
- 15.Jylhävä J, Pedersen NL, and Hägg S (2017). Biological Age Predictors. EBioMedicine 21, 29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lan Q, Zhang L, Li G, Vermeulen R, Weinberg RS, Dosemeci M, Rappaport SM, Shen M, Alter BP, Wu Y, et al. (2004). Hematotoxicity in workers exposed to low levels of benzene. Science 306, 1774–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lan Q, Zhang L, Tang X, Shen M, Smith MT, Qiu C, Ge Y, Ji Z, Xiong J, He J, et al. (2010). Occupational exposure to trichloroethylene is associated with a decline in lymphocyte subsets and soluble CD27 and CD30 markers. Carcinogenesis 31, 1592–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levine ME (2020). Assessment of Epigenetic Clocks as Biomarkers of Aging in Basic and Population Research. J. Gerontol. A. Biol. Sci. Med. Sci 75, 463–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levine ME, Lu AT, Quach A, Chen BH, Assimes TL, Bandinelli S, Hou L, Baccarelli AA, Stewart JD, Li Y, et al. (2018). An epigenetic biomarker of aging for lifespan and healthspan. Aging 10, 573–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lind PM, Salihovic S, and Lind L (2018). High plasma organochlorine pesticide levels are related to increased biological age as calculated by DNA methylation analysis. Environ. Int 113, 109–113. [DOI] [PubMed] [Google Scholar]
- 21.Loomis D, Guyton KZ, Grosse Y, El Ghissassi F, Bouvard V, Benbrahim-Tallaa L, Guha N, Vilahur N, Mattock H, Straif K, et al. (2017). Carcinogenicity of benzene. Lancet Oncol. 18, 1574–1575. [DOI] [PubMed] [Google Scholar]
- 22.Lu AT, Seeboth A, Tsai P-C, Sun D, Quach A, Reiner AP, Kooperberg C, Ferrucci L, Hou L, Baccarelli AA, et al. (2019a). DNA methylation-based estimator of telomere length. Aging 11, 5895–5923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu AT, Quach A, Wilson JG, Reiner AP, Aviv A, Raj K, Hou L, Baccarelli AA, Li Y, Stewart JD, et al. (2019b). DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging 11, 303–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marioni RE, Shah S, McRae AF, Chen BH, Colicino E, Harris SE, Gibson J, Henders AK, Redmond P, Cox SR, et al. (2015). DNA methylation age of blood predicts all-cause mortality in later life. Genome Biol. 16, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Phang M, Ross J, Raythatha JH, Dissanayake HU, McMullan RL, Kong Y, Hyett J, Gordon A, Molloy P, and Skilton MR (2020). Epigenetic aging in newborns: role of maternal diet. Am. J. Clin. Nutr 111, 555–561. [DOI] [PubMed] [Google Scholar]
- 26.Phillips RV, Rieswijk L, Hubbard AE, Vermeulen R, Zhang J, Hu W, Li L, Bassig BA, Wong JYY, Reiss B, et al. (2019). Human exposure to trichloroethylene is associated with increased variability of blood DNA methylation that is enriched in genes and pathways related to autoimmune disease and cancer. Epigenetics 14, 1112–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Prado-Bert P, Ruiz-Arenas C, Vives-Usano M, Andrusaityte S, Cadiou S, Carracedo Á, Casas M, Chatzi L, Dadvand P, González JR, et al. (2021). The early-life exposome and epigenetic age acceleration in children. Environ. Int 155, 106683. [DOI] [PubMed] [Google Scholar]
- 28.Quach A, Levine ME, Tanaka T, Lu AT, Chen BH, Ferrucci L, Ritz B, Bandinelli S, Neuhouser ML, Beasley JM, et al. (2017). Epigenetic clock analysis of diet, exercise, education, and lifestyle factors. Aging 9, 419–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith MT (2010). Advances in understanding benzene health effects and susceptibility. Annu. Rev. Public Health 31, 133–148 2 p following 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith MT, Guyton KZ, Kleinstreuer N, Borrel A, Cardenas A, Chiu WA, Felsher DW, Gibbons CF, Goodson WH, Houck KA, et al. (2020). The Key Characteristics of Carcinogens: Relationship to the Hallmarks of Cancer, Relevant Biomarkers, and Assays to Measure Them. Cancer Epidemiol. Biomark. Prev. Publ. Am. Assoc. Cancer Res. Cosponsored Am. Soc. Prev. Oncol 29, 1887–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sturm G, Cardenas A, Bind M-A, Horvath S, Wang S, Wang Y, Hägg S, Hirano M, and Picard M (2019). Human aging DNA methylation signatures are conserved but accelerated in cultured fibroblasts. Epigenetics 14, 961–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vigneron A, and Vousden KH (2010). p53, ROS and senescence in the control of aging. Aging 2, 471–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wagner MA, Erickson KI, Bender CM, and Conley YP (2020). The Influence of Physical Activity and Epigenomics On Cognitive Function and Brain Health in Breast Cancer. Front. Aging Neurosci 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilbur S, Wohlers D, Paikoff S, Keith L, and Faroon O (2008). ATSDR evaluation of health effects of benzene and relevance to public health. Toxicol. Ind. Health 24, 263–398. [DOI] [PubMed] [Google Scholar]
- 35.Zhang L, Steinmaus C, Eastmond DA, Xin XK, and Smith MT (2009). Formaldehyde exposure and leukemia: a new meta-analysis and potential mechanisms. Mutat. Res. Mutat. Res 681, 150–168. [DOI] [PubMed] [Google Scholar]
- 36.Zhang L, Tang X, Rothman N, Vermeulen R, Ji Z, Shen M, Qiu C, Guo W, Liu S, Reiss B, et al. (2010). Occupational exposure to formaldehyde, hematotoxicity, and leukemia-specific chromosome changes in cultured myeloid progenitor cells. Cancer Epidemiol. Biomark. Prev. Publ. Am. Assoc. Cancer Res. Cosponsored Am. Soc. Prev. Oncol 19, 80–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


