Abstract
Photoaffinity labeling (PAL) in combination with a chemical probe to covalently bind its target upon UV irradiation has demonstrated considerable promise in drug discovery for identifying new drug targets and binding sites. In particular, carbene-mediated photoaffinity labeling (cmPAL) has been widely used in drug target identification owing to its excellent photolabeling efficiency, minimal steric interference and longer excitation wavelength. Specifically, diazirines, which are among the precursors of carbenes and have higher carbene yields and greater chemical stability than diazo compounds, have proved to be valuable photolabile reagents in a diverse range of biological systems. This review highlights current advances of cmPAL in medicinal chemistry, with a focus on structures and applications for identifying small molecule–protein and macromolecule–protein interactions and ligand-gated ion channels, coupled with advances in the discovery of targets and inhibitors using carbene precursor-based biological probes developed in recent decades.
Photoaffinity labeling (PAL) in combination with a chemical probe to covalently bind its target upon UV irradiation has demonstrated considerable promise in drug discovery for identifying new drug targets and binding sites.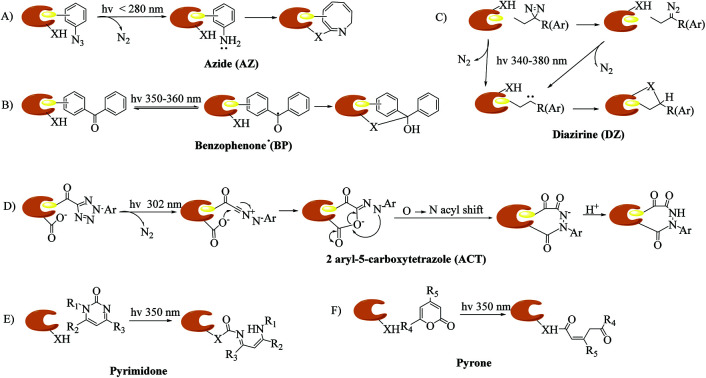
1. Introduction
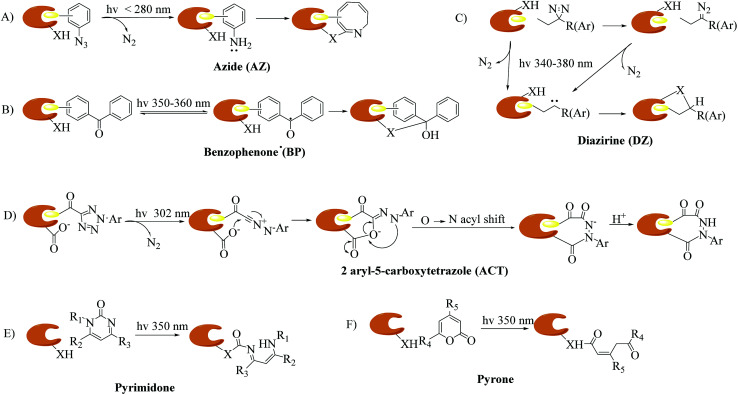
The identification of targets of active molecules and natural products plays an important role in biomedical research and drug discovery. As a result of current advances in mass spectrometry and bioinformatics used in chemical probes, increasing numbers of biological targets have been found and become clinical drug candidates. However, most of them still remain unknown because there is little understanding of the mode of action between probes and relevant targets. The concept of photoaffinity labeling (PAL) was originally introduced by Frank Westheimer in the early 1960s,1 and since then it has gradually emerged as a powerful tool for the identification and localization of targets and corresponding interaction sites in complicated biological systems,2 especially in some cases such as low-abundance proteins and low-affinity interactions that fail to survive disruptive washing steps.3,4 Four common types of photoreactive group are used in PAL, namely, aryl azides (AZs), benzophenones (BPs), diazirines (DAs)5,6 (New) and 2-aryl-5-carboxytetrazoles (ACTs),7,8 which are classified in accordance with their photochemically generated reactive species, namely, nitrenes, diradicals, carbenes and carboxy-nitrile imines. In addition, some photoreactive components of natural molecules that can undergo photolysis to form highly reactive intermediates, such as steroid enones (e.g., pyrones and pyrimidones9), various aryl chlorides, and several thioethers, have been utilized as probes to avoid complications associated with fully synthetic probes.10 The mechanisms of different types of photoaffinity labeling are simply illustrated in Fig. 1. In particular, carbenes are highly reactive species with extremely short lifetimes11 that can not only react with various nucleophilic residues, but also be inserted into C–H or O–H bonds. Carbene-mediated photoaffinity labeling (cmPAL) has been increasingly used in target identification owing to the small size of carbenes, high crosslinking efficiency, long excitation wavelength, excellent chemical stability, less disruption of the interaction interface and minimal damage to biological samples. There are two major types of carbene precursor used in cmPAL, namely, diazirine and diazo compounds, which have occupied a uniquely important place in PAL. In particular, in comparison with the latter, diazirines generate relatively more reactive carbenes12,13 and have either relatively high thermal and chemical stability2 or a much shorter lifetime when activated by photoirradiation. The potential of carbene precursors as photoreactive groups, especially 3H-aryldiazirines with an absorption wavelength ranging from 340 to 380 nm, was first revealed in 1973 by Jeremy Knowles.14 Since then, carbene precursor analogues of several biologically important ligands such as amino acids, nucleic acids, lipids, carbohydrates and steroids have been used as photoprobes15 for ligand-binding sites to facilitate the formation of a covalent bond in biological systems upon photoirradiation at a certain wavelength. The boom in cmPAL, as well as the recent development of various diazirine–compatible reactions12 and the increasing commercial availability of diazirine derivatives, has inspired equally innovative advances in research into small molecule–protein and macromolecule–protein interactions and ligand-gated ion channels. In the last few decades, a great deal of effort has been dedicated to characterizing many classes of enzymes such as kinases, γ-secretases, methyltransferases, metalloproteinases and histone deacetylases (HDACs) by using cmPAL in combination with chemical probes, namely, affinity-based probes (AfBPs), in which cmPAL is used to create covalent interactions between the probe and an enzyme that does not employ a catalytic nucleophilic amino acid within the ligand-binding site. Interestingly, a large number of these enzymes are relevant to human diseases and thus provide opportunities for the potential diagnosis and therapy of cancer.
Fig. 1. Proposed mechanisms of different types of photoaffinity labeling: (A) azide (AZ), (B) benzophenone (BP), (C) diazirine (DZ), (D) 2-aryl-5-carboxytetrazole (ACT), and two “more natural” types, namely, (E) pyrimidone, and (F) pyrone. X = C, N, O, or S.
Several excellent reviews have extensively summarized the field of cmPAL and its application in the identification of targets of natural products and bioactive compounds within the past decade.15–20 In this highlight, we will briefly introduce current developments in cmPAL in medicinal chemistry, with a focus on synthetic methods and applications for understanding biological molecule–protein interactions and ligand-gated ion channels, as well as the discovery of targets and inhibitors using different diazirinyl biological probes that were developed recently. By presenting specific examples, we show here that cmPAL provides researchers with a distinctive set of chemical tools to embark on the study of the identification of targets of drug candidates with an unknown mode of action and the functions of many of the uncharacterized classes of enzymes that populate eukaryotic and prokaryotic proteomes.
2. Carbene-mediated PAL (cmPAL)
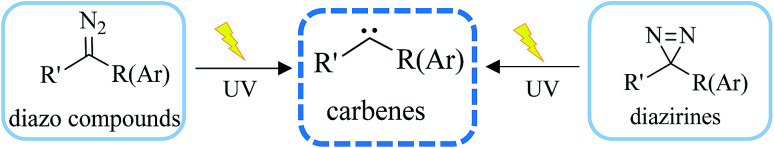
Carbenes are more reactive species than nitrenes with extremely short lifetimes that can not only be inserted into C–H or O–H bonds in proteins, but also react with various nucleophilic residues. As major precursors of carbenes, diazo compounds and diazirines (Fig. 2) have occupied a uniquely important place in photoaffinity labeling. Upon exposure to UV light at a certain wavelength (350–380 nm), carbenes generated by diazo compounds or diazirines are capable of spontaneously forming a covalent bond with a neighboring molecule via insertion into bonds such as C–H, O–H, N–H and C–C to stabilize the interaction between a biological target and a probe. The use of cmPAL in enzyme modification was first described in 1962,1 when Westheimer et al. reported the use of a diazoacetyl group to inactivate chymotrypsin. Since then, there has been a steady increase in the use of cmPAL for understanding enzymes, in particular those with previously uncharacterized functions, because of its distinct advantages. The employment of both types of carbene precursor in AfBPs is illustrated below.
Fig. 2. Structure of carbenes derived from diazo compounds or diazirines upon UV irradiation.
2.1. Diazo compounds
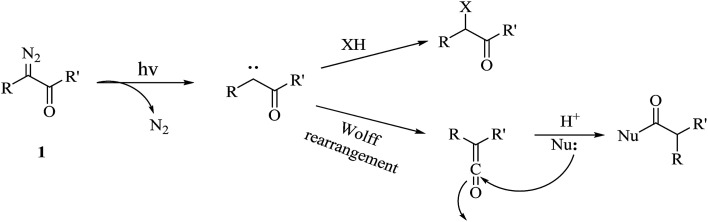
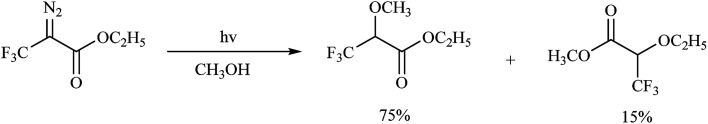
The diazo group has been widely used in the field of organic chemistry for over a century owing to its diverse reactivity, including alkylation, carbene generation, nucleophilic addition, homologation, and ring expansion.21,22 Similarly, the diazo group is particularly attractive as a photoprobe and has enabled novel modifications of proteins and nucleic acids23 (New) because of its small size and ease of introduction within a short time.24,25 The first example of PAL was achieved with a diazoacetate.1 Upon photoirradiation, the diazoketone 1 fragments into molecular nitrogen and a highly reactive carbene, which can undergo either a photolabeling process via bond insertion26 or a Wolff rearrangement,27 followed by nucleophilic attack on the resulting ketene (Fig. 3). Both of these products can react with neighboring functional groups and thus have a negative impact on accurate photolabeling.13 To reduce the probability of a Wolff rearrangement and stabilize the carbene, electron-withdrawing groups can be introduced. This conclusion has been confirmed by Chowdhry et al., who demonstrated that the photolysis of ethyl diazotrifluoropropanoate in methanol causes much less rearrangement and results in an efficient insertion reaction with the O–H bond (Scheme 1).28 With high stability and a maximum UV absorption at around 360 nm, diazo compounds hold promising potential for use in photolabeling studies.29,30 This strategy has been a powerful biochemical technique for mapping the architecture of chymotrypsin,1 revealing antibody-binding sites,31 examining the structure of lipid membranes,32 and identifying isoprenoid-binding sites in proteins.33
Fig. 3. Photochemical reaction of diazoketone 1 for photoaffinity labeling, including an insertion reaction and a Wolff rearrangement.
Scheme 1. Photochemical reaction of ethyl diazotrifluoropropanoate.
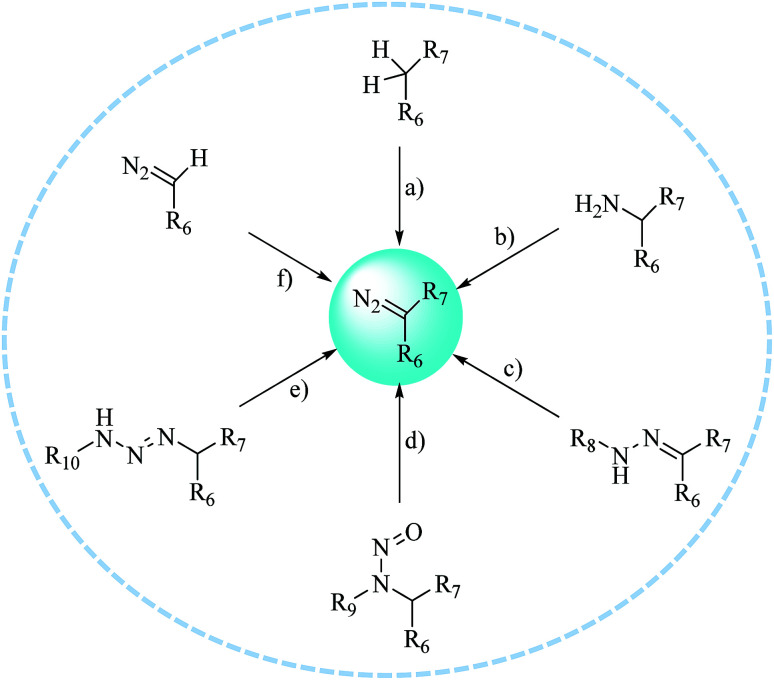
The first diazo compound was synthesized in the 19th century.34,35 As a result of their broad reactivity and coexisting functional groups, diazo compounds can present a challenge with respect to their preparation and purification. Here, we have summarized several typical synthetic methods for preparing diazo compounds (Fig. 4).36 Among these, the preparation of diazo compounds via the fragmentation of triazenes was originally described by Baumgarten37 and has been increasingly used in recent years because of its mild synthetic conditions, and thus facilitated more biological applications such as the labeling of proteins and use as tunable reactants in 1,3-dipolar cycloaddition reactions with cycloalkynes. A further effort was made by Myers et al., who used azides as substrates and employed mild tetrahydrofuran–water conditions to synthesize a variety of alkyl acyl triazenes. The ability of Staudinger-type phosphines to produce diazo compounds also revealed that these acyl triazenes undergo thermal or base-catalyzed fragmentation to form α-diazo compounds in high yields.36 Similarly, Chou et al. described another transformation that utilized an activated phosphinoester to convert an azide into a diazo compound in a phosphate buffer at neutral pH and room temperature. As azido groups can be easily introduced into and retained in their ligands, the conversion between azides and diazo compounds tolerates the presence of functional groups relevant to chemical biology.38
Fig. 4. Typical methods for preparing diazo compounds: (a) diazo transfer; (b) diazotization; (c) decomposition or oxidation of hydrazones; (d) rearrangement of N-alkyl-N-nitroso compounds; (e) fragmentation of 1,3-disubstituted alkyl aryl triazenes; and (f) elaboration of more readily available diazo compounds.
2.2. Diazirines
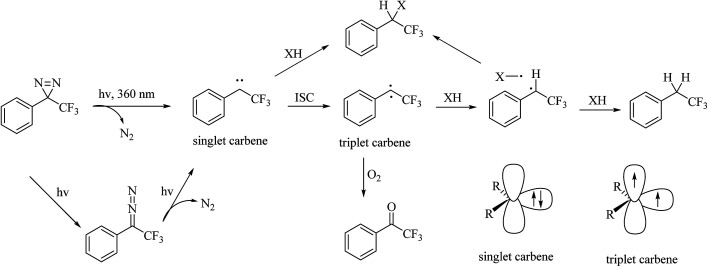
Diazirines were first chemically synthesized in 1960,39,40 and their structures were confirmed in 1962.41,42 A diazirine molecule is a three-membered ring system containing two nitrogen atoms and one carbon atom, which can be further categorized into two types, namely, aromatic and aliphatic compounds, depending upon whether the diazirine ring is attached directly to an aromatic ring or to an aliphatic carbon atom, respectively. Diazirines can be decomposed into molecular nitrogen and a very short-lived singlet carbene under the influence of light. In addition to generating carbenes, they can undergo isomerization to be transformed into linear diazo compounds (>30%), which also generate carbenes or carbocations for labeling the nucleophilic residues of proteins. A proportion of the highly reactive singlet carbenes immediately form a covalent bond with X–H (X = C, N, O, or S) or C–C bonds derived from a neighboring molecule via insertion under stable physiological conditions. The other singlet carbenes can be transformed into triplet carbenes, in which two electrons with parallel spins occupy two different orbitals via intersystem crossing (ISC). Singlet and triplet carbenes exhibit similar reactivity in comparison with the corresponding nitrenes. The intrinsic efficient reactivity of the singlet carbene with O–H bonds leads to scavenging of the reactive species by water. The triplet carbene is initially converted into a radical intermediate, which either undergoes an insertion reaction like the singlet carbene when it reacts with an X–H bond or abstracts a second hydrogen atom from a different C–H bond, which results in reduction. The triplet carbene can be oxidized to the corresponding ketone by molecular oxygen.13 In summary, the reactive species in diazirine photolabeling are primarily the singlet carbene and the diazo compound, of which the proportion depends upon the chemical nature of the diazirine.15 Another main drawback of diazirines is their complicated synthetic methods. Despite all these factors, diazirines have increasingly been employed in cmPAL as photoreactive reagents owing to their unique advantages, which include smaller sizes, longer excitation wavelengths (350–380 nm) and higher photocrosslinking efficiency than other photoreactive groups. On the other hand, in comparison with diazo compounds, diazirines generate relatively more highly reactive carbenes12,13 and have either relatively high thermal and chemical stability2 or a much shorter lifetime when activated by photoirradiation. Hence, diazirines have largely superseded diazo compounds as photoreactive reagents on account of their improved chemical stability in the dark and higher photochemical responsiveness. Furthermore, the recent development of various diazirine-compatible reactions,12 coupled with the increasing commercial availability of diazirine derivatives, now allows their more widespread use in cmPAL. Diazirines have been increasingly regarded as among the most promising photolysis reagents. Attention has been given to different types of diazirinyl analogues with different biologically important ligands such as amino acids, nucleic acids, lipids, and so on, which are discussed below.
2.2.1. Aromatic diazirines
The potential of diazirines as photoreactive groups, in particular that of 3H-aryldiazirines (Fig. 5), was first revealed in 1973 by Jeremy Knowles.14 Aromatic diazirine derivatives, especially trifluoromethylphenyl diazirine (TPD) derivatives,43 in which the diazirine group is directly attached to an aromatic ring, have been developed to address some of the limitations associated with alkyldiazirines, one of which is low crosslinking efficiency because of the rapid isomerization of the photogenerated carbene intermediates into diazo isomers.43 Furthermore, several studies have indicated that aromatic diazirines tend to photogenerate more carbenes than alkyldiazirines.44–46 In comparison with other diazirines, TPDs display much higher chemical stability under a wide variety of conditions, including strongly acidic, strongly basic, oxidizing and reducing agents.12 This is because the electron-withdrawing nature of the trifluoromethyl group and phenyl group located at the α-position of the diazirine moiety can not only prevent the carbene from undergoing rearrangements but also strongly stabilize the photogenerated diazo isomer under normal photolysis conditions. With a maximum absorption at around 360 nm, aromatic diazirines minimize damage to the targeted biological system. Aromatic diazirines are quite bulky in comparison with small diazirines but can be incorporated into molecules with a structure similar to that of naturally occurring compounds to address this issue; thus, the biological activity of the modified ligand and the crosslinking efficiency of cmPAL-based probes are not impaired too greatly. In general, aromatic diazirines preferentially react with O–H bonds rather than C–H bonds.47,48 One shortcoming of TPDs is that aromatic diazirines can react with N–H bonds and generate enamines as a side product, which are subject to further hydrolysis with concomitant loss of the labeling targets.49 Collectively, TPDs are currently preferred as the most promising photolabeling reagents that have been developed, although their synthetic routes have been intricate thus far.
Fig. 5. Possible pathways and intermediates formed after photolysis of 3-aryl-3H-diazirines and representation of the electronic states of the carbene.
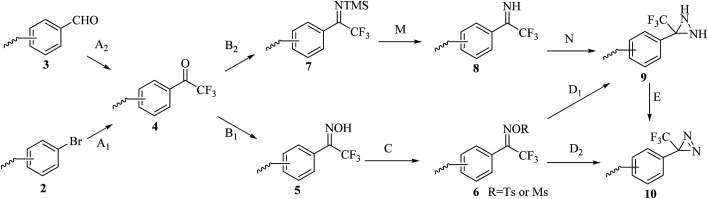
Although TPDs appear to be the most promising photoreactive reagents for cmPAL, their introduction into different chemical probes in a simple and convenient manner remains challenging and constitutes the major hindrance to their practical use.2,12 A great deal of work has been invested in improving their synthesis (Scheme 2). The general strategy for preparing an aromatic diazirine analogue 10 comprises the conversion of a ketone derivative 4, which can be obtained from an aromatic bromide 2 or an aromatic formaldehyde 3, into the corresponding ethanone oxime 5. The hydroxy group in 5 can be activated to form a good leaving group by using either p-tosyl chloride (TsCl) or mesyl chloride (MsCl) to give a tosyl oxime 6. After treatment with NH3 (l), the tosyl oxime 6 is converted into a diaziridine 9 and then oxidized to 10. Another synthetic strategy includes the direct conversion of the tosyl oxime 6 into an aromatic diazirine under the following conditions: either using liquid NH3 at 80 °C or with LiNH2 (ref. 50) (Table 1). The diaziridine can also be made using an ammonia-free synthetic method, which significantly requires less time to prepare TPDs.51 The phenyl ketone 4 reacts with lithium bis(trimethylsilyl)amide to give an N-TMS-ketimine 7, which is then subjected to solvolysis with methanol to provide an imine 8 in good yields, followed by treatment with o-mesitylenesulfonyl hydroxylamine, which was synthesized according to the methodology reported by Tamura et al.,52 to give the corresponding diaziridine. Details of the reagents and conditions in the syntheses of the aromatic diazirine 10 are summarized in Table 1.
Scheme 2. Synthetic routes to the aromatic diazirine 10 from the aromatic bromide 2 or the aromatic formaldehyde 3.
Summary of steps in the synthesis of aromatic diazirines. MsCl: mesyl chloride; Py: pyridine; pTsCl: p-tosyl chloride; DMAP: N,N-dimethyl-4-aminopyridine.
| Step | Synthetic conditions | Ref. |
|---|---|---|
| A1 | n-BuLi, ether, CF3COOEt, −78 °C, 2 h | 49,53 |
| n-BuLi, THF, CF3COOMe, −78 °C, 2 h | 54 | |
| Mg, CF3COR, R = piperidinyl | 55 | |
| Mg/CF3COOH or n-BuLi/CF3COR, R = OMe, piperidinyl | 20 | |
| A2 | (1) TMS-CF3, catalytic TBAF, THF; then aqueous HCl | 56 |
| (2) Dess-Martin periodinane, TFA, CH2Cl2 | ||
| (1) TMS-CF3, K2CO3, DMA, rt, 2 h | 57 | |
| (2) 1 M HCl, rt, 1 h | ||
| (3) Dess-Martin, CH2Cl2, rt, 8 h | ||
| B1 | NH2OH, NaOH, EtOH, reflux, 21 h | 49 |
| NH2OH·HCl, pyridine, EtOH, 60 °C, 4 h | 53–58 | |
| NH2OH·HCl, NaOH, EtOH, reflux, 16 h or pyridine, 70 °C, 3 h | 59 | |
| B2 | 1 M LiN(TMS)2 in THF, toluene, 0 °C | 51 |
| C | MsCl, TEA | 49 |
| TsCl, pyridine, reflux | 2,53,54,56,59 | |
| TsCl, TEA, N,N-dimethylaminopyridine, CH2Cl2, 0 °C, 45 min | 55 | |
| TsCl, DMAP, TEA, CH2Cl2, rt, 45 min | 57 | |
| D1 | NH3 (l), CH2Cl2, −78 °C | 49,54,55 |
| NH3 (l), ether, −78 °C→rt or rt | 53,56,57,59 | |
| NH3 (l), 80 °C, 20 h | 58 | |
| D2 | NH3 (l), LiNH2, rt, 11 h | 50,57,59 |
| M | MeOH, 18 h | 51 |
| N | (1) 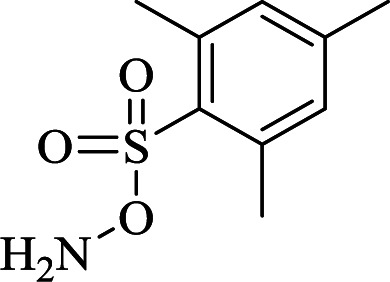 in THF, 0 °C in THF, 0 °C |
51 |
| (2) piperidine | ||
| E | Ag2O, ether, 23 °C, 3.5 h | 49,54 |
| tert-BuOCl, TEA | 2,55 | |
| I2, TEA, CH2Cl2 or MeOH, 0 °C | 56,58,59 | |
| MnO2, ether or CH2Cl2, rt | 53,57 | |
| (COCl)2, DMSO, DCM, −78 °C | 60 |
2.2.2. Alkyldiazirines
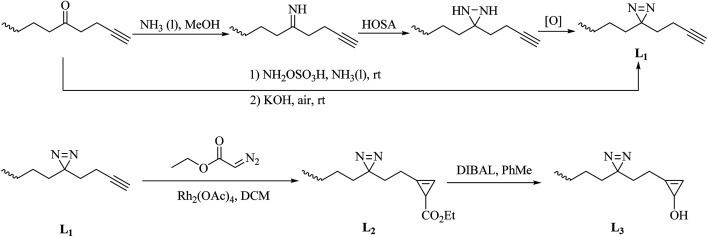
Increasing numbers of researchers are using alkyldiazirines for photolabeling studies, despite initial apprehensions due to their extremely small size, although TPDs appear to come closest to satisfying the chemical and biological criteria required for ideal photoprobes.43 Typically, 3H-diazirine, which is the simplest form of diazirine, when the ring system has been directly attached to alkyl carbon atoms has been used to probe different states of proteins.61–63 The use of alkyldiazirines has been limited in the past, mainly for the reasons that they are prone to generate smaller amounts of carbenes upon UV irradiation and have moderate crosslinking efficiency owing to the side products from rearrangement reaction. Furthermore, the diazo compounds derived from alkyldiazirines are liable to generate carbocations that can react with nucleophilic side chains in amino acids, which leads to non-specific photolabeling. Nevertheless, all these limitations of alkyldiazirines can be compensated for by their extremely compact sizes, which make them highly attractive in cases in which the drug–target interactions are sensitive to the size of the photoprobe. Furthermore, one of the most essential challenges in cmPAL is that the probe, which is a derivative of the drug or natural product, must retain most of the original biological activities of the original compound and exhibit little alteration in binding to the native target after modification. A smaller probe size is desirable to minimize the chance that the crosslinking group will disrupt the interaction interface and cause a significant decrease in affinity with the target molecule. This therefore calls for the development of “minimalist” linkers in which both the photoactivatable group and the detection moiety are made as small as possible so as to provide a chemically suitable modality for detecting covalent protein–probe interactions in vivo. Since the concept of the “minimalist linker” was first introduced by Yao et al. in 2013,64 both a diazirine and a clickable moiety have been concurrently incorporated into a single alkyl chain (L1–3, Scheme 3).64,65 This strategy has been demonstrated to be a remarkably efficient method for cmPAL and cell imaging. Very recently, a suite of “minimalist linkers” containing alkyldiazirine units and different bioorthogonal tags have been developed to find targets of BRD4 inhibitors and obtain better insights into how a tag might affect the overall performance of a probe66 (New). In another case, a derivative of a non-steroidal anti-inflammatory drug (NSAID), namely, a photo-NSAID, containing a “minimalist linker” was used to study NSAIDs. Such a strategy not only enables the characterization of binding site hotspots for NSAIDs but can also map global binding sites for virtually any molecule of interest67 (New).
Scheme 3. Schematic of the syntheses of the minimalist alkyne linkers L1–3: HOSA: hydroxylamine-O-sulfonic acid; [O]: Ag2O, I2/TEA or CrO3; DIBAL: diisobutylaluminium hydride.
In a similar way to aromatic diazirines, the main strategy for the synthesis of alkyldiazirines comprises starting from an aliphatic ketone, modifying it to form an alkyldiazirine and preparing diazirine precursors without oximation and tosylation. The syntheses of three commonly used alkyldiazirine-based linkers are described here. An alkyl ketone directly reacts with ammonia and an aminating reagent such as a chloramine or hydroxylamine-O-sulfonic acid to give the corresponding diaziridine. After oxidation of the corresponding diaziridine with a variety of oxidants, such as Ag2O, I2/TEA or CrO3, the desired diazirine L1 is prepared (Scheme 3). In addition, a simple base-mediated one-pot reaction can also be used to prepare L1. It was demonstrated that the intermediate can be directly converted into the diazirine after the addition of KOH under air68,69 (New). The synthesis of L2 from L1 was accomplished by treatment with commercially available ethyl diazoacetate and a catalytic amount of Rh2(OAc)4, and further reduction of the C-3 ester in L2 by DIBAL afforded L3 in an excellent yield65 (Scheme 3).
3. Applications of cmPAL for biological molecule–protein interactions and ligand-gated ion channels
The chemical and thermal stability of carbene precursors allows sophisticated derivatization of the corresponding biological probes, which can be further categorized according to the type of parent ligand molecule. The development of different cmPAL labels enabled a simple strategy for linking photoreactive groups, especially diazirines, to important classes of biological ligands such as amino acids, nucleic acids, lipids, carbohydrates, steroids and other small molecules to produce the corresponding diazirinyl photoprobes. Specific examples of the use of cmPAL for studying small molecule–protein and macromolecule–protein interactions and ligand-gated ion channels are discussed.
3.1. Protein–protein interactions
Because PAL can detect participants in non-covalent interactions spatioselectively, photoreactive amino acids have emerged as a critical tool for studying protein–protein interactions (PPIs) directly in a complex proteome70 (New). The similarity between photoreactive amino acids and the corresponding natural amino acids makes them elude the stringent identity control mechanisms during protein synthesis and be directly incorporated into proteins during cell culture by the native mammalian translation machinery for protein biosynthesis.71 Several diazirine analogues of amino acids have been widely incorporated into proteins to probe PPIs, which thereby allows the global identification of interacting proteins in living cells.
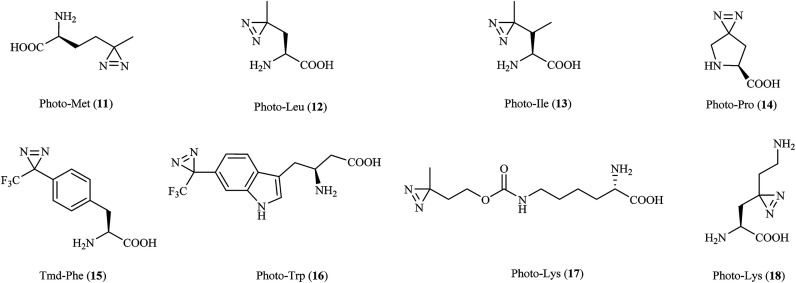
Three diazirine-containing amino acids, namely, photo-Met, photo-Leu and photo-Ile (11–13, Fig. 6), which resemble natural methionine, leucine and isoleucine except for the presence of a diazirine ring in the side chain, were efficiently incorporated into proteins by mammalian cells to study membrane protein complexes in living cells,71,72 covalently detect MH2–MH2 domain interactions in proteins,73 inhibit the expression of a vascular cell adhesion molecule (VCAM) in a fungal cyclodepsipeptide,74 and map the protein interaction network of human protein kinase D2 (PKD2), which is an enzyme that belongs to the family of serine/threonine kinases and is involved in multiple biological processes.75 It was demonstrated that insights from structural proteomics can be used to generate CaM-insensitive mutants of targets of CaM for functional studies in vitro or ideally in vivo via the incorporation of synthetic photo-Met.76
Fig. 6. Diazirine analogues of amino acids.
In another study, a derivative of L27-11 containing photo-Pro (14, Fig. 6) was reported to label LptD, which is an outer-membrane protein widely distributed in Gram-negative bacteria, against Pseudomonas aeruginosa77 and enable the study of its essential function in the bacterium.78 Similarly, two proline-based diazirinyl amino acids, namely, JB-95 (ref. 79) and DYN5,80 were developed to study several β-barrel OM proteins, including BamA and LptD,79 and find the potential target, which was found to be the sensor ParS.80
Another synthetic amino acid with a trifluoromethyldiazirine moiety attached to phenylalanine, namely, Tmd-Phe (15, Fig. 6), was first prepared by Nassal in 1984.81 After the successful identification of calmodulin (CaM) using Tmd-Phe in a calcium-dependent manner,82 Tippmann et al. extended the use of Tmd-Phe to label the amber codon, which gave a highly efficient and precise response in E. coli.83 In a subsequent study, in order to find novel complexes of proteins with Grb2 and map PPIs in living cells, Tmd-Phe was incorporated into multiple sites (N103-L112) in the SH2 domains of Grb2 (ref. 84) and was specifically crosslinked with EGFR after stimulation by EGF.85 In addition, some Tmd-Phe derivatives also have sufficient affinity for the sweet taste receptor to identify the binding sites of ligands in the sweet taste receptor.86
Photo-Trp (16, Fig. 6) is a new photoactivatable amino acid designed to be incorporated into tryptophan-containing peptides for cmPAL. Two types of photo-Trp, namely, 5- and 6-trifluoromethyldiazirinyltryptophan, acted as skeletons for the comprehensive synthesis of various bioactive indole metabolites and were used in biological functional analysis via cmPAL.87 Furthermore, a Boc-protected version of photo-Trp, which carried a diazirinyl substituent at the 6-position of the indole ring, was used to target the cytotoxic marine natural product hemiasterlin.88
Two lysine-based diazirinyl analogues (17 and 18, Fig. 6) were developed to efficiently photocrosslink a test model protein in vitro and in vivo89 and identify proteins that recognize PTMs of lysine, including ‘readers’ and ‘erasers’ of histone modifications,90 respectively. In another case, genetically encoded photo-lysine was used as an unmodified “control probe” in conjunction with the photoaffinity label crotonyllysine to study the specificity of an effector of interest toward the corresponding PTM of lysine, which enabled the investigation of PPIs mediated by the lysine PTM between the full-length H3 protein and its epigenetic regulatory effectors.91
From these studies, we can see that the development of new photoreactive amino acids of which the structures and properties closely resemble those of natural amino acids may help to address difficulties in the identification of PPIs in living cells.
3.2. Nucleic acid–protein interactions
Since nucleic acid analogues that carried a diazirine unit were successfully used for cmPAL, increasing numbers of photolabile nucleoside, nucleotide and oligonucleotide analogues containing diazirines have been developed for studying nucleic acid–protein interactions.
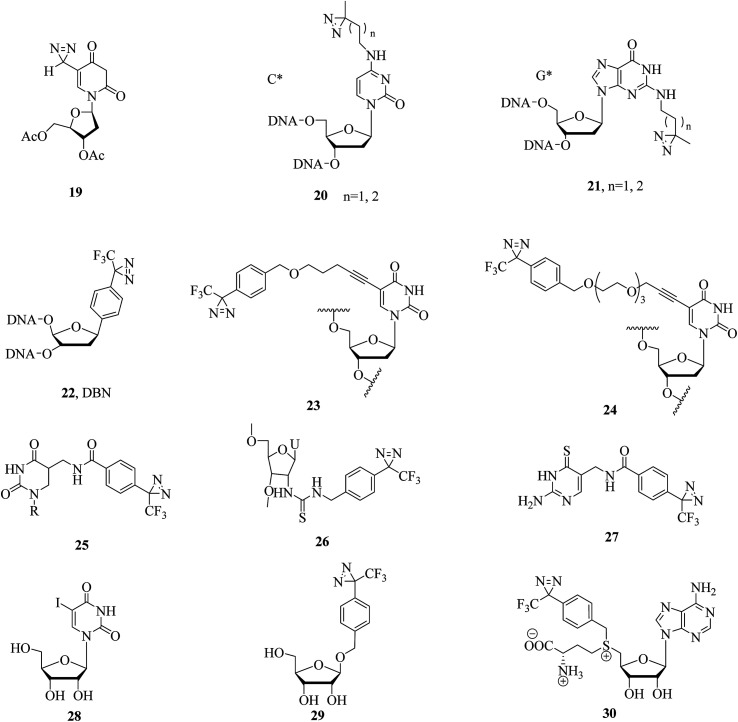
The simplest nucleoside derivative 19 (Fig. 7), which consisted of deoxyuridine substituted at the 5-position with a 3H-diazirine group, was synthesized by Taranenko et al., who used this so-called “zero-length” diazirinyl nucleoside to investigate nucleic acid–protein interactions.92 In addition, two nucleotide compounds comprising cytosine (C*, 20, Fig. 7) and guanine (G*, 21, Fig. 7) with diazirine units were developed by He et al., who described a simple and rapid solid-phase synthetic method for the incorporation of diazirine into the major and minor grooves of DNA for efficient photocrosslinking to proteins.93 Similarly, a diazirine-based nucleoside analogue 22 (DBN, Fig. 7) was successfully incorporated into DNA by solid-phase oligonucleotide synthesis, efficiently formed inter-strand crosslinks in dsDNA upon photoirradiation and thus enabled a wide range of applications in biotechnology, such as probing nucleic acid–nucleic acid and protein–nucleic acid interactions and in phototherapy.94 A different application of photolabile nucleic acids was described by Carell et al., who synthesized two thymidine analogues 23 and 24 (Fig. 7) by attaching TPD groups to the 5-position of the nucleobase uridine to avoid disturbing the duplex structure of DNA. After cmPAL, two DNA repair enzymes (Rad14, which is the yeast homologue of human XPA protein, and Fpg/MuM-DNA glycosylase from Lactococcus lactis) were identified,95 and an optimal photocrosslinker that exerted a significant effect on the yield and efficiency of photocrosslinking in the human nuclear proteome was found to be involved in processing the lesion,96 respectively.
Fig. 7. Structures of diazirinyl nucleic acid analogues and the 5-iodouridine RNA derivative 28.
Several derivatives that bear a diazirine ring at different positions in RNA have been synthesized, and their photoaffinity efficiencies have been tested. Protocols for the synthesis and photocrosslinking of diazirine-containing RNAs have been described by Sergiev et al., who chose Escherichia coli ribosomes as a model for testing photoactivated RNA analogues 25–27 (Fig. 7), which can be incorporated into RNA by T7 transcription.97–99 The TPD moiety in the nucleotide analogue 25 is located in the major groove of the mRNA helix and is in contact with ribosomal 16S RNA in helix region 28.97 Upon UV irradiation, the 4-thiouridine (s4U) analogue 26 was non-specifically crosslinked to either single- or double-stranded RNA regions, which enabled an understanding of the structure and function of 5S rRNA and its environment in the ribosome.98 The 6-thioguanosine diazirinyl analogue 27 was crosslinked at the 3′-end of 16S rRNA (nucleotides 1496–1542), which performed photochemical crosslinking within the E. coli ribosome complex.99 In another study, some photolabeled siRNAs (small interfering RNAs) with TPD moieties in the 3′-overhang region were reported and compared to a 5-iodouridine RNA derivative 28 (Fig. 7) by Ueno et al., which enabled much more sensitive detection and would serve as a powerful tool for understanding the mechanism of the assembly of the RISC (RNA-induced silencing complex) by Argonaute proteins.100 Similarly, a photoreactive miRNA-145 probe 29 containing photolabeled microRNA (Fig. 7), which has sufficient gene-silencing activity even if the guide strands of the miRNAs are modified with the analogue 29 in some cases,101 was found to specifically label the target mRNAs, namely, FSCN1 and KLF4, upon UV-A irradiation in DLD-1 human colon cancer cells.102 An S-adenosyl-l-methionine (AdoMet) analogue 30 (Fig. 7), which can be enzymatically transferred to the target of methyltransferase, namely, the mRNA cap, with high efficiency, was demonstrated to be the optimal photochemical reagent for enzymatic transfer and photocrosslinking to a directly interacting protein.103
These photolabeled nucleic acid analogues, in combination with cmPAL and mass spectrometry methods, should enable more scientists to characterize proteins that are specifically involved in DNA or RNA processes in detail.
3.3. Lipid–protein interactions
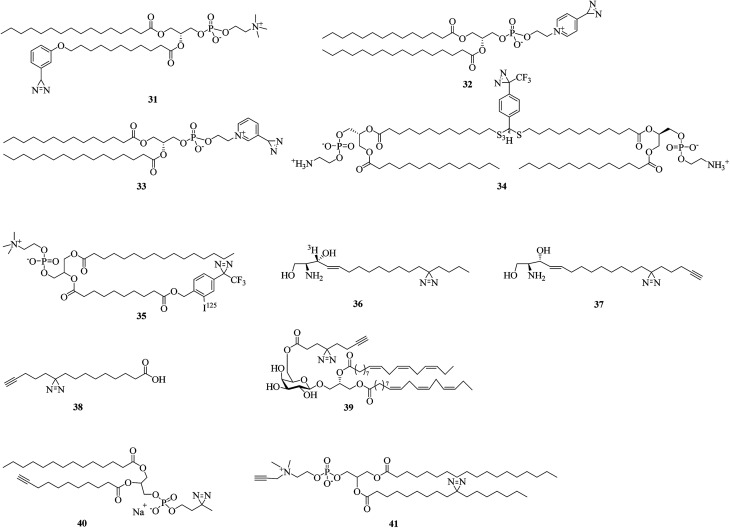
Diazirine analogues of lipids that bear photoreactive diazirine groups in either the polar head or the hydrophobic part can be incorporated into a lipid bilayer to study lipid–protein interactions. When exposed to light, photogenerated highly reactive carbenes can covalently crosslink with membrane proteins or surrounding lipids. Since the first aliphatic diazirine-based phospholipid was reported by Khorana et al. in 1980,104 various hydrophobic diazirinyl analogues of lipids have been developed for labeling hydrophobic biomembrane regions owing to their higher labeling efficiency in hydrophobic environments in comparison with an aqueous medium.12,105
A pioneering study by Ross using diazirinyl lipid analogues 31–33 (Fig. 8) bearing diazirine groups either in the fatty acyl chain or in the polar head provided a direct method for investigating the biomembrane of glycophorin A. Further results of labeling using the phospholipid 31 showed that the Glu-70 residue is embedded in the membrane.106 In addition, in combination with radiolabeling, two lipid analogues 34 and 35 containing TPD units (Fig. 8) were designed to study the interactions between the influenza virus and target membrane lipids,107 mitochondrial proteins and phosphatidylcholine.108 The successful confirmation that the HA2 subunit of influenza hemagglutinin was inserted into the target membrane prior to fusion107 and the highly specific labeling of a 70 kDa protein108 indicated the potential of these two photolipid analogues as tools for proteome analysis.
Fig. 8. Diazirine analogues of lipids.
As structural components of membranes, sphingolipids can act as intracellular second messengers. In order to understand how sphingolipids affect the function of proteins within the membrane bilayer of living cells, Haberkant et al. developed a radioactively labeled photosphingosine 36 (Fig. 8)109 and two clickable photosphingosines 37 (Fig. 8)110 and 38 (Fig. 8)111 bearing an alkyldiazirine moiety in the fatty acyl chain. A bifunctional sphingosine such as 38 can not only be metabolically incorporated into all major classes of glycerolipids, as well as into proteins via fatty acylation in living cells, but can also be conjugated to a coumarin caging group via a carbamate linkage at the amino group to give a trifunctional sphingosine for studying different aspects of lipid biology in the context of living cells.112 Another clickable fatty acid-based probe 39 (Fig. 8) was reported by Liu et al., who designed a fatty acid-based probe bearing diazirine and terminal alkyne groups, which enables the identification or visualization of the respective crosslinked protein–glycolipid complex.113
Similarly, in order to discover more new protein–lipid interactions in which proteins bind to phosphatidylcholine (PC), which is the most abundant type of phospholipid in most lipoproteins and membranes, via the fatty acid tails, two photoactivatable and clickable phosphatidylcholine analogues 40 and 41 (Fig. 8) were prepared.114,115 Compound 40, which can be incorporated into recombinant high-density lipoprotein particles or membrane bilayers, facilitated the identification of residues critical for the docking of proteins with the lipid surface of lipoproteins or cell membranes.114 The other PC analogue 41 was used in a double incorporation strategy for the metabolic synthesis of bifunctional PCs to provide an obvious improvement in global mapping of genuine protein–lipid interactions in living cells. It is evident that this strategy not only facilitates the successful biosynthesis of bifunctional 41 but also identifies many high-confidence PC-binding proteins.115
It is evident that alkyldiazirine analogues of lipids bearing diazirines in the fatty acyl chain have been increasingly used to study protein–lipid interactions in biological membranes in recent years owing to their smaller size and close structural resemblance to lipid derivatives.
3.4. Carbohydrate–protein interactions
Carbohydrates constitute integral components of the cell machinery in the forms of polysaccharides, proteoglycans, glycoproteins, glycolipids, and glycosylated secondary metabolites and play crucial roles in a wide range of biological and pathological processes.116 However, there are still many carbohydrate–protein interactions that remain unknown owing to transient and low-affinity interactions.117 The use of carbene-mediated photocrosslinking of sugars represents a powerful strategy for investigating carbohydrate–protein interactions in their native context. Several efforts have been made to develop diazirinyl carbohydrate analogues to provide the first step in elucidating the biological roles of carbohydrates.
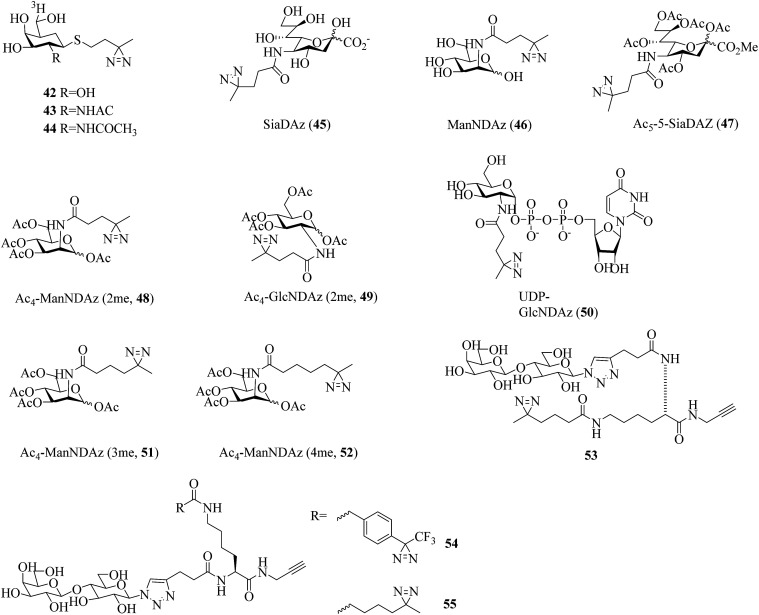
In a related study, Kuhn et al. prepared two thioglycoside carbohydrate analogues 42 (Fig. 9)118 and 43 (Fig. 9)119 bearing a diazirine photoreactive group for photocrosslinking β-galactosidase and β-hexosaminidase, respectively. Under UV light, the group can generate a reactive carbene that can undergo an insertion reaction with a nearby amino acid residue within the enzyme. A tritium label [3H] was used to radiolabel the captured target. Similarly, a diazirinyl carbohydrate analogue [3H]-1-ATB-GalNAc 44 (Fig. 9), in which the R group was replaced by an acetamide group, has been employed as a useful photocrosslinking reagent to label Glu-355, which is highly conserved and located within a region of Hex B.120
Fig. 9. Diazirine analogues of carbohydrates.
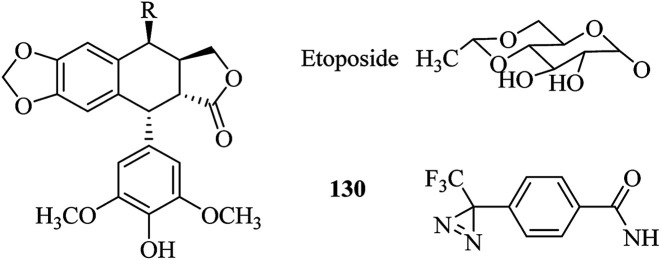
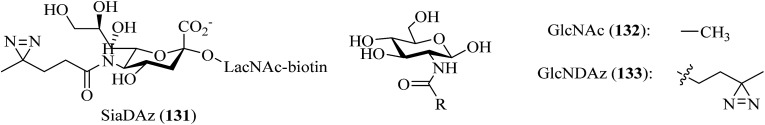
A great deal of effort has been made by Kohler et al., who designed and synthesized a series of diazirine-containing photosugars, typically, a sialic acid analogue SiaDAz (45, Fig. 9) and an N-acetylmannosamine analogue ManNDAz (46, Fig. 9), which were metabolically incorporated into cell surfaces for the covalent detection of transient interactions between sialic acids and sialic acid-recognizing proteins via cmPAL. With the help of a commercially available sialidase that removed the natural sialic acid Neu5Ac from cell surfaces while leaving SiaDAz-modified glycoconjugates intact, the utility of SiaDAz was greatly improved, and this process can be applied generally in the case of sialic acid-mediated interactions and will facilitate the identification of binding partners of sialic acid.121 In addition, a fully acetylated sialic acid photosugar Ac5-SiaDAz (47, Fig. 9) and a mannosamine precursor Ac4-ManNDAz (48, Fig. 9) bearing a diazirine photocrosslinker on the N-acyl (C-5) side chain122 were metabolically incorporated into cell surface glycoconjugates to identify and characterize the binding partners of the glycoconjugates.123 These two photosugars 47 and 48, as well as the control molecule, namely, the N-acetylglucosamine analogue Ac4-GlcNDAz (49, Fig. 9), displayed improved membrane permeability and produced photoreactive sialyl glycans in various types of cell. To demonstrate the photocrosslinking utility of these metabolically incorporated photosugars, the sialic acid-dependent oligomerization of CD22 was examined. After cells were cultured with either ManNDAz or 5-SiaDAz, photocrosslinking of CD22 was observed, whereas the control was not crosslinked, which confirmed that the crosslinking depended on the incorporation of 5-SiaDAz.122 Using a similar metabolic labeling approach, cultured cells were induced to produce a modified nucleotide sugar donor UDP-GlcNDAz 50 (Fig. 9) and to transfer Ac4-GlcNDAz 49 to proteins that are normally modified with an O-GlcNAc group. Subsequent photocrosslinking resulted in the selective covalent capture of O-GlcNDAz-modified nucleoporins and nuclear transport factors, which indicated that modification with an O-GlcNAc group is intimately associated with the recognition events that occur during nuclear transport.124 Another two diazirine-modified ManNAc analogues 51 and 52 (Fig. 9) were synthesized, in which the number of methylene groups separating the carbonyl and diazirine groups in the N-acyl side chain was varied in order to guarantee an optimal distance for the efficient capture of binding partners of sialoside. The smallest diazirine-modified sialic acid precursor 48 was shown to be very efficiently metabolized and competed well with endogenous sialic acid.125
As a model carbohydrate ligand, lactose is known to bind to peanut agglutinin (PNA, Kd = 770 μM), which belongs to the lectin family of carbohydrate-binding proteins. A set of lactose-based analogues bearing different photoreactive groups, such as aryl azide, benzophenone, alkyldiazirine and trifluoromethylphenyldiazirine (TPD), were synthesized to directly compare their efficiency and selectivity in photocrosslinking a carbohydrate-binding protein. Despite the low yield of crosslinking, the diazirinyl lactose analogue (53, Fig. 9) displayed high ligand-dependent reactivity that was consistent with the ideal mechanism of PAL.126 Although the crosslinking efficiency of the TPD derivative (54, Fig. 9) was higher than that of the alkyldiazirinyl derivative (55, Fig. 9) when it reacted with a single binding protein, the latter exhibited significantly more selective PAL of a binding protein in a cell lysate than the corresponding TPD derivative.127
In summary, diazirine analogues of carbohydrates can be metabolically incorporated into a cell surface and further used to covalently detect carbohydrate–protein interactions upon UV irradiation or as model ligands to compare different photocrosslinkers.
3.5. Steroid–protein interactions
Several photolabile steroid analogues that incorporate diazirines into the tetracyclic steroid nucleus have been developed over the past decade.
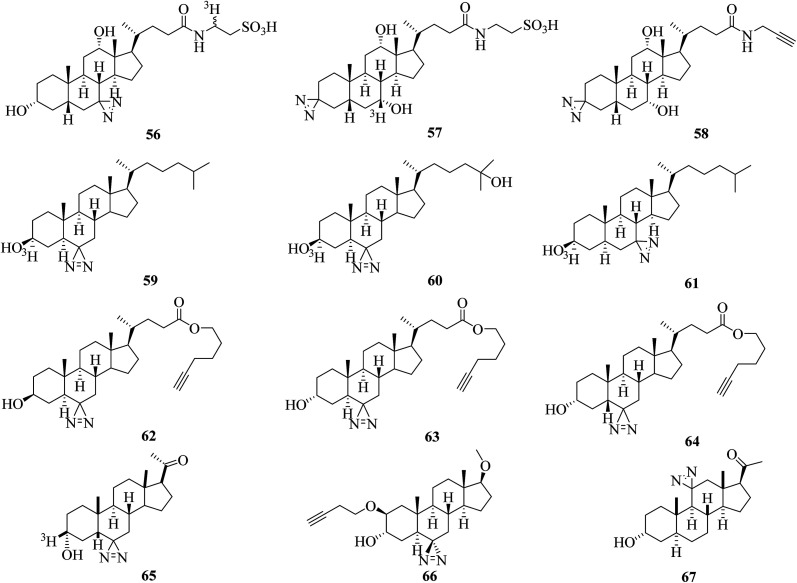
Typically, a radiolabeled diazirinyl analogue of a bile salt 56 (Fig. 10), which belongs to a class of important endogenous metabolites that consist of a steroid core and a side chain with a carboxyl group,128 was reported to detect virtually all bile salt-binding receptors129 and was used to investigate the taurocholate transport system in hepatocytes.130 Similarly, another photolabile bile acid derivative 57 (Fig. 10)131 and its analogue 58 (Fig. 10)132 with a diazirine group attached at the 3-position of the steroid nucleus were synthesized to minimize the possibility that any receptor sites might elude detection131 and to map proteins that interact with bile acids and are involved in many important physiological processes.132
Fig. 10. Diazirine analogues of steroids.
More efforts have been made to study cholesterol, which is a precursor of steroid hormones and bile acids and is necessary for the functional activity of several membrane proteins. Two photoactivatable cholesterol analogues 59 and 60 (Fig. 10), in which the hydrogen atom at C-6 was replaced by a diazirine moiety, were developed to investigate cholesterol-binding proteins in neuronal cells133 and study oxysterol-binding protein-related proteins (ORPs).134 The results obtained by photocrosslinking showed that synaptophysin was a major specific cholesterol-binding protein in PC12 cells and brain synaptic vesicles,133 and the strengths of the signals obtained for different ORPs do not necessarily reflect the order of their relative affinities for native 25OH.134 In another study, a cholesterol analogue 61 (Fig. 10) was demonstrated to be an effective reagent for studying cholesterol–protein interactions involved in intracellular cholesterol trafficking135 and can be used to label Niemann-Pick type C (NPC) 1 protein, which plays important roles in transporting cholesterol and other lipids out of late endosomes by means of vesicular trafficking.136 Similarly, more than 250 cholesterol-binding proteins in HeLa cells were identified using three clickable diazirinyl sterol analogues 62–64 (Fig. 10), which differ in the diastereomeric relationship between the C-3 alcohol group and C-5 hydrogen atom attached to the cholesterol core.137
In other cases, in order to understand the mode of action of a neurosteroid produced from cholesterol, three photolabile neurosteroid analogues 65–67 (Fig. 10) were described.138–140 By using the radiolabeled analogue 65 in which a diazirine moiety is attached to the C-6 position, the protein VDAC-1 was successfully identified as a neuroactive steroid-binding protein in the rat brain that may modulate the function of the GABAA receptor.138 With a similar structure to 65, the clickable analogue 66 exhibited selective compartmentalization in the Golgi apparatus with a preferential effect on proximal inhibition.139 The C-11-modified photoprobe 67, which exhibited potency equal to or higher than that of alphaxalone as a general anesthetic and potentiator of GABAAR responses, was used to discover neuroactive steroid–GABAAR binding sites.140
All in all, these diazirinyl steroid analogues can be used to globally map steroid–protein interactions directly in living cells. The possibility that any receptor sites will elude detection can be minimized by attaching the photolabile diazirine group at different positions in the molecule.
3.6. Ligand-gated ion channels
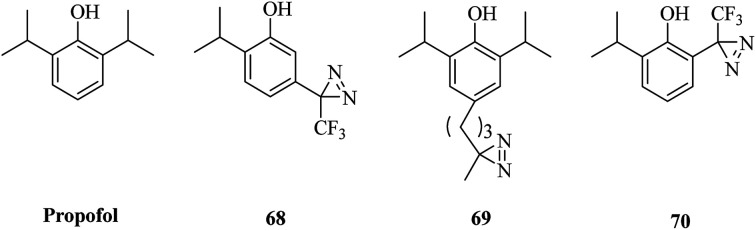
Propofol (Fig. 11) is the most commonly used sedative-hypnotic drug for difficult procedures and acts by potentiating GABAA (γ-aminobutyric acid type A) receptors, but where it binds to these receptors is not known. A set of propofol-based photolabeling reagents bearing diazirine moieties have been developed to identify its potential binding sites over the past decade.
Fig. 11. Structures of propofol and its diazirinyl analogues.
In a related study, a propofol analogue m-azipropofol (AziPm, 68, Fig. 11), which contained an alkyldiazirine group in a meta-position relative to the hydroxyl group, was synthesized to identify targets and thereby mechanisms of propofol. This design not only precluded any possible intramolecular reaction of the reactive site with the phenolic hydroxyl group but also revealed a previously identified propofol-binding cavity.141 A further application was described that [3H]AziPm was used to identify propofol-binding sites in Torpedo nAChR (nicotinic acetylcholine receptors), which are members of the Cys-loop superfamily of pentameric ligand-gated ion channels and can be inhibited by propofol.142,143 After photocrosslinking, three binding sites in the transmembrane domain of nAChR, namely, the subunit helix bundle, ion channel and γ–α interface, were revealed.144 In comparison with AziPm 68, another photoactivatable propofol derivative p-4-AziC5-Pro (69, Fig. 11), which keeps the core structure of propofol intact by adding a reactive diazirine group at the para-position of the phenol ring, has different photoselectivity for amino acid residues and enables the possibility of studying the binding sites of propofol in more detail.145 In addition, an ortho-propofol diazirine analogue (o-PD, 70, Fig. 11) bearing a photoreactive group at the ortho-position of the phenol ring was synthesized to reveal unknown propofol-binding sites in membrane receptors and provides the ability to use a combination of deuterated and protiated variants of ortho-propofol diazirine. In combination with cmPAL, as well as high-resolution MS, a new binding site of propofol in GABAA receptors that consisted of both β3 homopentamers and α1β3 heteropentamers was identified.146
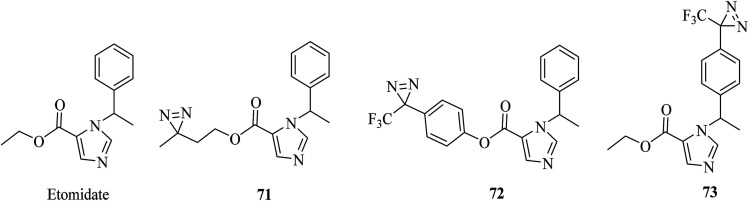
In a similar way to propofol, which acts by interacting with GABA type A receptors (GABAARs), several etomidate (Fig. 12)-based diazirine analogues, including azi–etomidate 71 (Fig. 12),147,148 TDBzl–etomidate 72 (Fig. 12)149 and pTFD–etomidate 73 (Fig. 12)150 were synthesized by Cohen et al. to identify binding domains in a ligand-gated ion channel147,149,150 and directly photolabel amino acids that contribute to an anesthetic-binding site in GABAAR.148 Photoaffinity labeling established that the analogue 71 identified two labeled amino acids, of which one was located in the membrane-spanning helix of α-subunit M1 (αMet236) and the other in the membrane-spanning helix of β-subunit M3 (βMet286).148 It was demonstrated that the agent 72, which contains a TPD moiety, not only exerts pharmacological actions on members of the Cys-loop ion channel superfamily of receptors149 but also acts as a positive allosteric modulator of nAChR rather than as an inhibitor,151 and site-directed mutagenesis suggests that S-TFD-etomidate acts at different sites in comparison with R-etomidate.150
Fig. 12. Etomidate and its diazirinyl analogues.
Photoactivatable propofol analogues that have an alkyldiazirine moiety at different positions of the phenol ring would present a more comprehensive view of the mechanism of propofol with membrane receptors. Furthermore, these different diazirinyl anesthetic analogues would provide a powerful tool for understanding the modes of action of the anesthetic on ligand-gated ion channels.
Most of the carbene precursor analogues described here retained similar or even better biological activity in comparison with the parent compounds, largely owing to the extremely small size of the diazirine photoreactive groups and the rational selection of the positions where these photolabile groups were incorporated within the probe. Despite the sophisticated derivatization of these biological probes, the minimal interference with the original biological activity, as well as the native interactions, makes them a robust tool for studying protein–protein, protein–lipid, protein–macromolecule, protein–steroid and other ligand–receptor interactions.
4. Applications of cmPAL for discovery of targets and inhibitors
Within the past two decades, activity-based protein profiling (ABPP) via the employment of irreversible chemical probes has been pursued as a useful strategy for studying the subcellular location and activities of many classes of enzymes.152–155 However, all of these must be active enzymes that contain a catalytic nucleophilic amino acid residue. In the case of low-affinity interactions between probes and inactive enzymes, the labeling usually fails to survive disruptive washing steps. The emerging technique of PAL, especially cmPAL in combination with chemical proteomics, does not necessarily label active enzymes and thus overcomes the limitations of non-covalent affinity reagents. With excellent photolabeling efficiency, minimal steric interference and longer excitation wavelengths, cmPAL when coupled with chemical probes has been increasingly developed to study various classes of enzymes and their inhibitors. Here, we introduce different applications of cmPAL for classes of enzymes and their inhibitors, including kinases, γ-secretase, methyltransferases, metalloproteinases, histone deacetylase, and so on.
4.1. Kinases
Kinases, especially protein kinases (PKs), are one of the most important classes of enzymes in human cells, play a key role in signal transduction and regulate a variety of cell processes. Of the more than 500 known PKs, many are potential therapeutic targets.156 To study potential cellular targets of kinases and their inhibitors, recent efforts have focused on cmPAL using a large panel of AfBPs, as well as mass spectrometry-based chemical profiling methods.
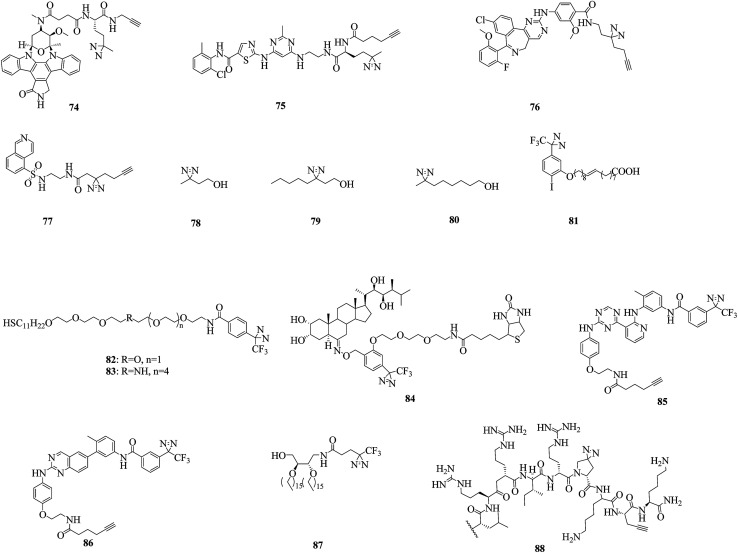
As a prototypical natural-product kinase inhibitor that competes with ATP, staurosporine can target at least 253 kinases.157 Significant efforts have been made by Yao et al., who developed a staurosporine-based clickable probe STS-1 (74, Fig. 13) bearing a diazirine group that was capable of proteome-wide profiling of potential cellular targets in live HepG2 cells for the first time.158 The same strategy was used to study dasatinib, which is a dual Src/Abl inhibitor and a promising therapeutic agent with oral bioavailability. By using the cell-permeable probe DA-2 (75, Fig. 13), as well as chemical proteomics methods, a number of previously unknown targets of dasatinib, including several serine/threonine kinases, were identified.159 In addition, since improved minimalist photocrosslinkers that possess two carbon atoms on each side of the diazirine ring flanked by a terminal alkyne and a functional group (CO2H/NH2/I) were first introduced into 12 well-known kinase inhibitors for cell-based proteomic profiling of potential cellular targets under native conditions,64 more potential targets of kinases have been found by using these minimalist alkyldiazirine alkyne linkers. Typically, photoaffinity probes 76 and 77 (Fig. 13) that contain a ‘‘minimalist’’ linker were capable of both live-cell imaging of the activities of aurora kinase A (AKA) and in situ proteomic profiling of potential off-targets of MLN8237, which is a highly potent and presumably selective inhibitor of AKA160 and can be used to study the mechanism of action of H8, which is an inhibitor of protein kinase A (PKA).161
Fig. 13. Representative applications of cmPAL for studying kinases.
An important signal transduction protein, namely, protein kinase C (PKC), has been proposed as the target of anesthetics such as alcohols and halothane. In a similar way to these, azialcohols such as 3-azioctanol, 7-azioctanol and 3-azibutanol (78–80, Fig. 13) can be used for photolabeling with PKCδC1B (the high-affinity phorbol-binding subdomain)162,163 and locate binding sites in adenylate kinase, which is an enzyme that catalyzes the transfer of a phosphoryl group.164 A TPD-based oleic acid probe 81 (Fig. 13) is also a useful tool for elucidating the functions of the specific activation of PKC by oleic acid.165
Much progress has been made in understanding the mode of action of serine/threonine kinases, which are involved in multiple cellular functions and pathological processes. In a sophisticated study, two thiol photoaffinity linkers 82 and 83 (Fig. 13) were synthesized to find the site where p62, which plays important roles in proteasomal or autophagosomal protein degradation, binds to p38, which is a serine/threonine kinase. Surface plasmon resonance (SPR) signals produced by the interaction between p38 and peptides derived from p62 were clearly detected, which enabled us to identify the binding site.166 Another important serine/threonine kinase, namely, protein kinase D2 (PKD2), was studied using two diazirine-modified photo-amino acids 11 and 12 (illustrated in Fig. 6), as well as a series of proteomics approaches such as cmPAL, trypsin digestion and label-free LC/MS analysis, which revealed several interaction partners, to map the protein interaction network of PKD2.75 In addition, a leucine-rich repeat (LRR) receptor serine/threonine kinase, namely, brassinosteroid insensitive 1 (BRI1), which is localized in the plasma membrane,167 was studied using biotin-tagged photoaffinity castasterone (BPCS, 84, Fig. 13), which revealed that brassinosteroids bind directly to the 94 amino acids that constitute ID-LRR22 in the extracellular domain of BRI1 and defined a new binding domain for steroid hormones.168
Other kinases, including type II kinases,169 diacylglycerol kinases (DGKs)170 and a bacterial sensor kinase, namely, ParS,80 have also been used as models to investigate the binding sites of their inhibitors or the structure of their catalytic domains. The use of two probes 85 and 86 directed to the active site (Fig. 13), which were derived from type II ATP-competitive inhibitors, in combination with stable-isotope labeling with amino acids in cell culture (SILAC) and mass spectrometry resulted in the identification of a diverse set of targets of protein kinases that were selectively enriched.169 Photoaffinity labeling of a probe 87 (Fig. 13) demonstrated that the crosslinking efficiency depended on the length of the linker between the photocrosslinker and the diacylglycerol-like moiety in the probe: probes that contained longer linkers could access the active site and be used as substrates.170 Similarly, a probe DYN5 (88, Fig. 13) that contained a diazirine-modified photo-proline in place of proline triggered a clear virulence response in Pseudomonas aeruginosa, which was consistent with the probes DYN4, DYN3, and DYN1, which contained an aryl azide moiety or a benzophenone moiety incorporated at the C-terminus of the sequence or in place of phenylalanine, respectively.80
4.2. γ-Secretase
A particularly challenging target for drug discovery is γ-secretase, which is an intramembrane aspartyl protease complex that cleaves amyloid precursor proteins (APPs) to release Aβ peptides, which probably play a causative role in the pathogenesis of Alzheimer's disease (AD). Hence, the inhibition of the processing of APPs using γ-secretase inhibitors is one of the most important strategies for the prevention and treatment of AD. Over the past several years, a number of photoprobes have been reported to provide a greater understanding of ligand-binding sites within γ-secretase.
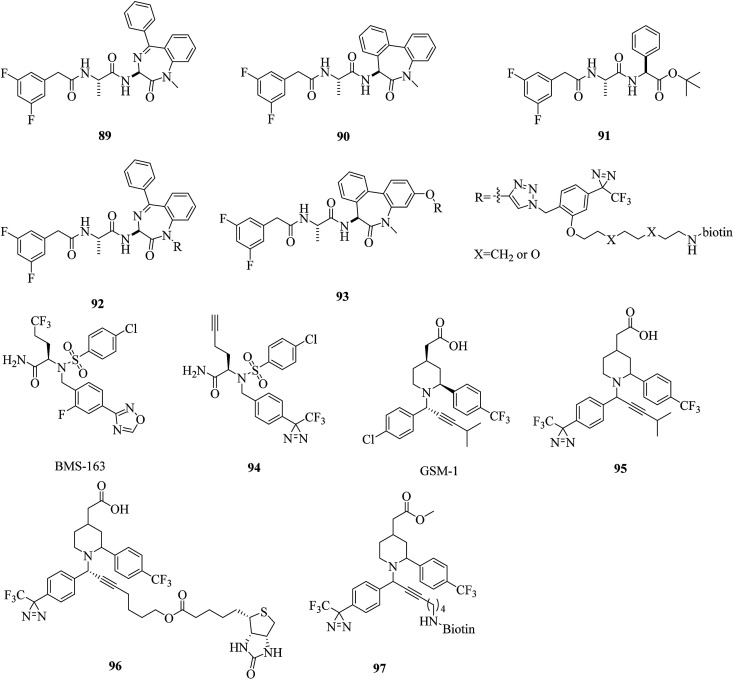
The caprolactam-type dipeptide γ-secretase inhibitors CE (89, Fig. 14) and DBZ (90, Fig. 14) are analogues of DAPT (91, Fig. 14), which is a compound that exhibits excellent γ-secretase-inhibitory activity in APP-transgenic mice.171 In order to elucidate the enzyme specificity of dipeptide γ-secretase inhibitors, Fuwa et al. developed a set of multifunctional molecular probes based on CE and DBZ by means of a copper(i)-mediated azide/alkyne fusion process. Despite their robust inhibitory activities against the production of Aβ, which were comparable to those of the parent compounds, two benzophenone probes with an ethylene glycol linker and phenyldiazirine groups (92 and 93, Fig. 14) were ineffective in photoaffinity labeling experiments, which reflected the difficulties in the “rational” design of an effective multifunctional photoprobe, even if sufficient information about the SARs of an affinity ligand moiety is provided.172 A similar case was described by Crump et al., who modified BMS-163 (Fig. 14), which is a non-selective γ-secretase inhibitor,173 with a clickable moiety and various photoreactive groups to target γ-secretase in vitro and in cells. Benzophenone probes based on BMS-163 covalently labeled PS1-NTF, whereas the diazirine-containing probe 163-DZ (94, Fig. 14) did not, which further demonstrates that benzophenone is preferred over diazirine as a photoreactive group for labeling γ-secretase.174
Fig. 14. Representative applications of cmPAL for studying γ-secretase.
In contrast, several highly potent acidic γ-secretase modulators (GSMs) that lowered Aβ42 levels in cellular assays with nanomolar modulatory activity were modified by Rennhack et al., who introduced photoreactive diazirine moieties into the acidic GSM scaffolds according to an analysis of their SARs. Most of these retained a strong ability to lower Aβ42 levels that was comparable to the potency of the parent compound and may be potential photoreactive reagents for crosslinking the N-terminal fragment of presenilin (PSEN, the catalytic subunit of the γ-secretase complex).175 In addition, Jumpertz et al. used GSM-based photoprobes such as AR80, AR243, and AR366 (95–97, Fig. 14) to investigate the molecular mechanism of GSMs. By using established cell-free conditions and living cells, PSEN was identified as the molecular target of potent acidic GSMs. Further studies indicated that potent acidic GSMs target PSEN to modulate the enzymatic activity of the γ-secretase complex. The inverse modulator AR366, which exhibits micromolar potency, represented an interesting tool for further investigations.176 Furthermore, these diazirinyl probes were successfully crosslinked to PSEN, which suggested that diazirine is a viable option for targeting γ-secretase.
4.3. Methyltransferases
Methyltransferases (MTs), which constitute one of the largest classes of enzymes in nature, can catalyze the methylation processes of many biomolecules such as DNA, RNA, and metabolites. With the help of various S-adenosylmethionine (SAM)-dependent MTs, methyl groups are extensively introduced into these biomolecules, so that the biomolecules are structurally and functionally diversified. The metabolic product of catalysis, namely, S-adenosyl-l-homocysteine (SAH), can then be further catabolized to homocysteine, converted into methionine, and reincorporated into SAM.177 Because methylation can control DNA transcription both directly via DNA methylation and indirectly via histone methylation, MTs can be mainly classified into two types, namely, DNA methyltransferases (DNMTs) and histone methyltransferases (HMTs), on the basis of the substrate of methylation in vertebrates. Given their important role in epigenetic control, lipid biosynthesis, protein repair, hormone inactivation, and tissue differentiation178 and the fact that a large fraction of the human MT family remains poorly understood, significant efforts have been made to identify and characterize MTs and their inhibitors in native biological systems over the past decade.
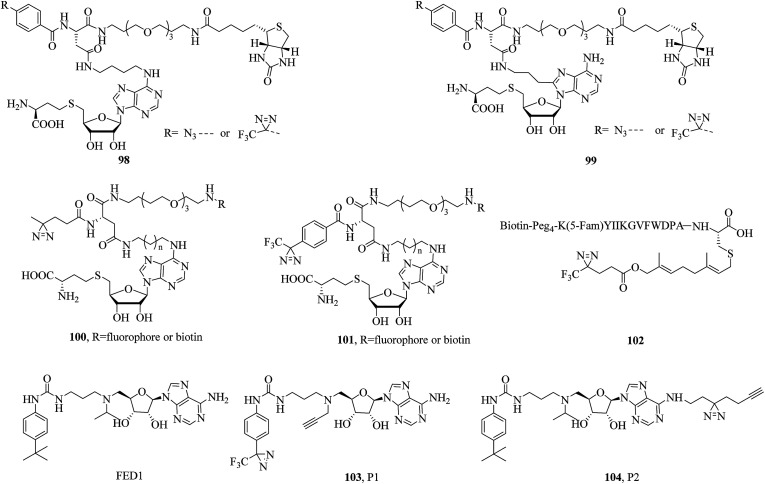
In a related study, He et al. expressed and purified E. coli DNA adenine methyltransferase (EcoDam), which is an enzyme that methylates the N-6 position of adenine in GATC sequences, to investigate its photocrosslinking to various diazirinyl DNA probes such as the cytosine- and guanine-based probes 20 and 21 (illustrated in Fig. 7). After crosslinking with EcoDam, good to excellent yields of the resulting DNA probes bearing a diazirine unit were observed, which would provide valuable information about how EcoDam binds to the corresponding DNA.93 Similarly, in order to carry out the selective photoinduced isolation of MTs, Dalhoff et al. synthesized a series of SAH-based capture compounds that carried a TPD moiety or an azide group (98 and 99, Fig. 15). When tested using purified MTs, these SAH-based capture compounds exhibited a certain applicability in the determination of the dissociation constant KD of MT-cofactor complexes and the isolation of SAH-binding enzymes from an E. coli cell lysate.179 In another case, Horning et al. used a suite of SAH photoaffinity probes bearing different photoexcitable groups, as well as chemical proteomics experiments, to specifically profile and enrich MTs from human cancer cell lysates. In addition to enriching MT-associated proteins, these SAH probes were used to successfully discover a covalent inhibitor of nicotinamide N-methyltransferase (NNMT), which is an enzyme implicated in cancer and metabolic disorders. Furthermore, study results revealed that SAH-based diazirinyl probes (100 and 101, Fig. 15) were capable of enriching a substantial fraction of human MTs, and the aliphatic diazirine group exhibited the broadest and most specific coverage of MTs from cancer cell proteomes among different photoreactive groups.180
Fig. 15. Representative applications of cmPAL for studying methyltransferases.
Isoprenylcysteine carboxyl methyltransferases (Icmts) are not only a class of SAM-dependent enzymes that catalyze the carboxymethylation process but are also localized in the membrane of the endoplasmic reticulum. Methylation catalyzed by Icmts can lead to the generation of a-factor, which is a naturally occurring farnesylated peptide found in yeast. In order to investigate methyltransferases, Vervacke et al. incorporated a diazirine-containing isoprenoid unit into a biotinylated and fluorescently labeled a-factor peptide analogue (102, Fig. 15). Then the probe was evaluated using a recombinant Icmt (Ste14p) that incorporated both a His10 tag for purification and a triply repeated myc tag to facilitate detection by immunoblotting analysis (His-Ste14p). The greater yield of photocrosslinked His-Ste14p, coupled with the ease of analysis achieved with the probe, suggested that this new class of isoprenoid analogues should be more widely used in studies of enzymes that act on terpene-derived molecules.181
DOT1L is not only the sole protein methyltransferase that methylates histone H3 on lysine 79 (H3K79) but is also a promising target for anticancer drugs. Yao et al. designed and synthesized two clickable AfBPs, namely, P1 and P2 (103 and 104, Fig. 15), which were based on a scaffold of FED1 (Fig. 15), which is a potent DOT1L inhibitor and structural mimic of SAM, to study potential cellular off-targets of FED1 in native cellular environments. In combination with quantitative LC-MS/MS, as well as chemical proteomics approaches, NOP2 (a putative ribosomal RNA methyltransferase) was confirmed to be probably a genuine off-target of FED1. Live-cell imaging experiments showed that most probes were not able to reach the cell nucleus, where functional DOT1L resides in mammalian cells, which provided a plausible explanation of the poor cellular activity of FED1.182
4.4. Metalloproteinases
Metalloproteinases (MPs), of which the catalytic mechanism involves a metal, are the most diverse of the four main types of protease, and more than 50 families have been classified to date. There are two subgroups of MPs, namely, metalloexopeptidases and metalloendopeptidases. Well-known metalloendopeptidases include matrix MPs (MMPs) and a disintegrin and MPs (ADAMs), both of which are catalytically active Zn2+-dependent proteins and have important physiological functions. MMPs play a central role in the degradation of extracellular matrix proteins (gelatin, elastin, and collagen).183–185 ADAMs are membrane-anchored MPs that contain both a cellular adhesion domain and an MMP domain and are involved in cell migration, muscle development, and fertilization, and so on.186,187 The biological importance of these two classes of enzymes has made the development of chemical tools for their study an important and active field of research in recent years.
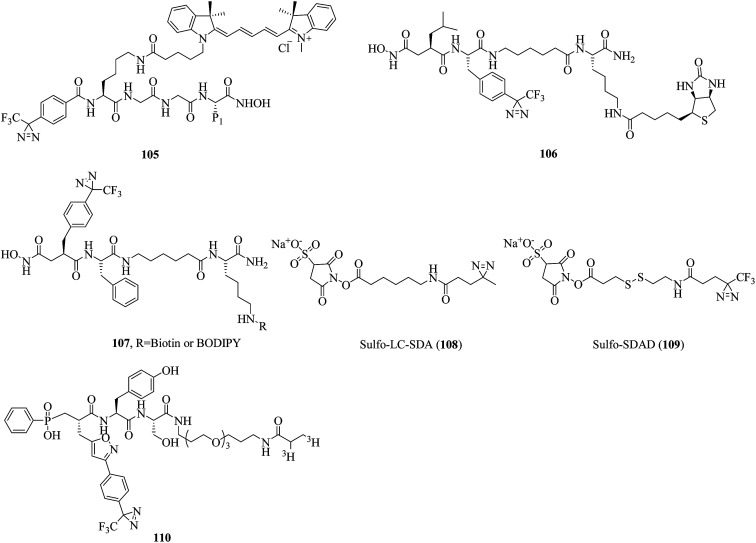
In a related study, Chan et al. reported a diazirinyl probe (105, Fig. 16) that carried a similar succinyl hydroxamate moiety as the zinc-binding group (ZBG) and a Cy3 dye as the substrate recognition unit for identifying catalytic sites of MPs. After a PAL experiment, the probe successfully targeted MMP-9 enzyme from a crude yeast extract with high sensitivity and selectivity. Furthermore, the extent of labeling was found to be dependent on the catalytic activity of the enzyme. Given the significant roles that many MPs play in a variety of diseases, this strategy may serve as a useful tool for potential therapeutics.188 In another case, Leeuwenburgh et al. synthesized a peptide hydroxamate analogue (106, Fig. 16) that featured both a biotin moiety and a TPD moiety via a solid-phase synthesis strategy. Upon incubation of purified recombinant ADAM-10 and subsequent photoaffinity labeling, the MP was covalently and irreversibly modified, which makes probe 106 a promising candidate for profiling ADAMs or MMPs.189 In addition, Geurink et al. modified a succinyl hydroxamate moiety by placing a TPD group at the P1′-position to make a more effective probe with which active MPs can be profiled and visualised. Evaluation using purified recombinant ADAM-17 revealed that positioning the photoactivatable group at the P1′-position, as in 107 (Fig. 16), makes a photoactivatable activity-based probe for MMP/ADAM comparatively more potent than previously. It was also demonstrated that the photoactivatable group should be directed towards the S1′ pocket, rather than the S2′ pocket.190
Fig. 16. Representative applications of cmPAL for studying metalloproteases.
Lozito et al. reported two heterobifunctional crosslinkers, namely, sulfo-LC-SDA (108, Fig. 16) and sulfo-SDAD (109, Fig. 16), which contained negatively charged sulfate groups to improve their solubility and diazirine units to identify specific interactions between TIMPs (tissue inhibitors of MPs) and MMPs contributed by MSCs (mesenchymal stem cells) and ECs (endothelial cells) in biological environments. It was demonstrated that MSCs inhibit both endogenous and exogenous MMPs via secreted TIMPs. Under control conditions, TIMP-2 and TIMP-1 protect the niche from MMP-2 and MMP-9, respectively. An investigation of the interactions between MMP-2 and TIMPs showed that TIMP-2 was the primary inhibitor of endogenous MMP-2. Nevertheless, under pathological stress MSCs act as a critical perivascular source of vessel-stabilizing TIMPs that are capable of protecting the vascular environment from disruption by proteases.191 In another example, Nury et al. reported a diazirinyl photoprobe (110, Fig. 16), which was based on a scaffold of a potent broad-spectrum phosphinic peptide inhibitor of MMPs, for detecting active forms of MMPs from biological fluids or tissue extracts. In the characterization of seven catalytic domains of human MMPs, the probe displayed high sensitivity in the detection of these MMPs. Furthermore, the probe permitted simultaneous detection with low background labeling in a complex proteome supplemented with four recombinant MMPs (MMP-2, -9, -12 and -13).192
4.5. Histone deacetylase
Histone deacetylases (HDACs) are hydrolases that catalyze the removal of acetyl groups from the ε-amino groups of lysine residues in histones or other cellular proteins, which allows the histones to wrap DNA more tightly. They thus play important roles in regulating various physiological and pathological processes.193–196 There are three classical HDACs, namely, Classes I, II, and IV, of which the activity is zinc-dependent. In addition, Class III consists of NAD+-dependent protein deacetylases known as sirtuins 1–7 (Sirt1–7).197 Many sophisticated studies of HDACs or HDAC inhibitors have been reported in the past three years.
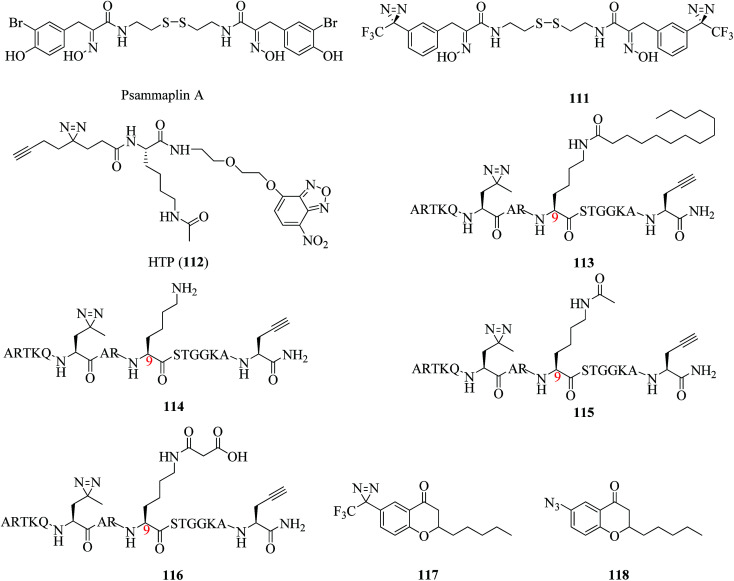
A related study was described by Hentschel et al., who developed the first photoreactive psammaplin A (111, Fig. 17), which was as potent an HDAC inhibitor (IC50 = 35 nM) as the parent compound psammaplin A (Fig. 17). Its very similar cytotoxicity profile to that of psammaplin A makes compound 111 a candidate for cmPAL, which may assist in the identification of new targets of psammaplin A and a better understanding of the mode of action with zinc-dependent HDACs.198 For the single-step detection and proteomic profiling of HDACs, Xie et al. developed a novel fluorescent probe (112, Fig. 17) by combining a Kac (N-acetyllysine) recognition unit, an O-NBD (nitrobenzoxadiazole) fluorophore and a minimalist photocrosslinker. It was a breakthrough finding that epigenetic readers and erasers can be readily identified and differentiated by using a single probe in combination with an in-gel fluorescence scanning technique and pull-down assays in comparison with traditional AfBPs. Furthermore, it was demonstrated that the probe can not only detect enzymatic activity but also directly identify protein targets in the native cellular environment. Such a strategy provides a robust tool for functional analysis of HDACs and facilitates future drug discovery in epigenetics.199
Fig. 17. Representative applications of cmPAL for studying histone deacetylases.
In another study, Liu et al. designed and synthesized a diazirinyl photoprobe (113, Fig. 17), which was based on a scaffold of a Lys9-myristoylated histone H3 peptide (H3K9Myr), and a K9-unmodified control probe (114, Fig. 17), which were coupled with a series of chemical biology approaches such as cmPAL, SDS-PAGE, and in-gel fluorescence scanning to examine human sirtuins as fatty acid deacylases in vitro and in cells. Sirt6, which is an enzyme that catalyzes the removal of the myristoyl group from the H3K9Myr peptide in vitro, was selectively captured by probe 113 but not by the unmodified control probe 114, which suggested that probe 113 can be used to examine the interactions between N-myristoylated lysine and its erasers. In the same way, another two classes of sirtuins, namely, Sirt2 and Sirt3, were also successfully labeled, which revealed their potential to recognize the myristoylation of lysine. As it is preferentially labeled by probe 113 in complex protein mixtures, Sirt2 exhibits stronger interactions with H3K9Myr than other tested sirtuins.200 In addition, Yang et al. developed two AfBPs, namely, 115 and 116 (Fig. 17), which were based on a histone H3 peptide in which the residue Lys9 was acetylated and malonylated, respectively, as well as the same control probe 114 to identify ‘erasers’ of histone lysine acylation. When reacted with recombinant human Sirt3 and Sirt5, probes 115 and 116 selectively labeled Sirt3 and Sirt5, respectively, whereas the control probe 114 failed to capture either of these enzymes, which suggested that probes 115 and 116 are ideal photoreactive reagents for the identification of their corresponding ‘erasers’. Furthermore, these two probes can specifically identify and enrich endogenous Sirt3 and Sirt5, respectively, from complex HeLa S3 lysates, which demonstrated that diazirine-based AfBPs are also useful tools for trapping erasers of histone lysine acylation from a complex biological environment.201 In brief, this strategy enables applications for both examining interactions between other PTMs and their erasers and revealing unknown cellular mechanisms controlled by sirtuins.
Similarly, according to an analysis of the SAR, a potent Sirt2 inhibitor was described by Seifert et al., who used two chroman-4-one-based AfBPs, namely, 117 and 118 (Fig. 17), which carried a diazirine moiety and an azide group, respectively, to identify the binding site of isoform-selective Sirt2 inhibitors. Of these probes, the diazirinyl probe 117, which displayed higher activity in the inhibition of Sirt2 (IC50 = 8.2 μM) than probe 118, which carried an azide group, was examined using recombinant Sirt2. The cmPAL experiments with the enzyme followed by tryptic digestion and LC-MS/MS analysis located the binding site of probe 117 in the tryptic peptide I175-K210, which includes the binding site of chroman-4-one-based Sirt2 inhibitors. Furthermore, the irradiation times (3 × 10 s) in the cmPAL experiments were reduced using a high-power LED to avoid undesirable labeling of Sirt2.202
4.6. Other enzymes
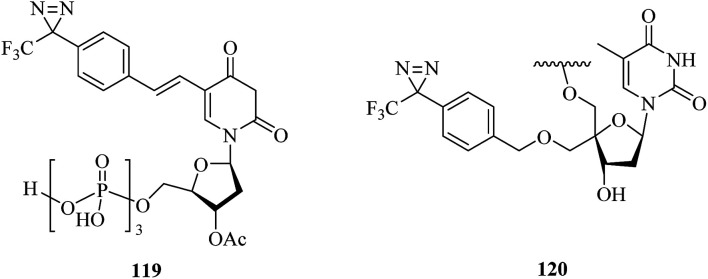
DNA polymerase β, which is an enzyme that interacts significantly with the sugar–phosphate backbone of DNA, is predominantly involved in DNA replication and carries out processive replication of chromosomal DNA during cell division in eukaryotes.203 A related study was reported by Yamaguchi et al., who incorporated some phosphoramidite derivatives of 119 (Fig. 18) into oligonucleotides via solid-phase synthesis to label mammalian DNA polymerase. Upon irradiation with near-UV light, the photolabile oligonucleotides were successfully crosslinked to recombinant rat DNA polymerase β, which suggested that they may be useful for extending the study of DNA–protein interactions.204 Similarly, Liebmann et al. reported the labeling of DNA polymerase β via the attachment of a light-sensitive TPD moiety to the C-4′ position of 2′-deoxyribose. Modification of the pentose moiety in the incorporated thymidine 120 (Fig. 18) enabled an investigation of the interactions in the minor groove of the helix. The new photoactive building block based on TPD selectively labeled DNA polymerase β even in the presence of BSA, which revealed that it would enable several applications in investigations of DNA backbone interactions in the minor groove.205
Fig. 18. Representative applications of cmPAL for studying DNA polymerase.
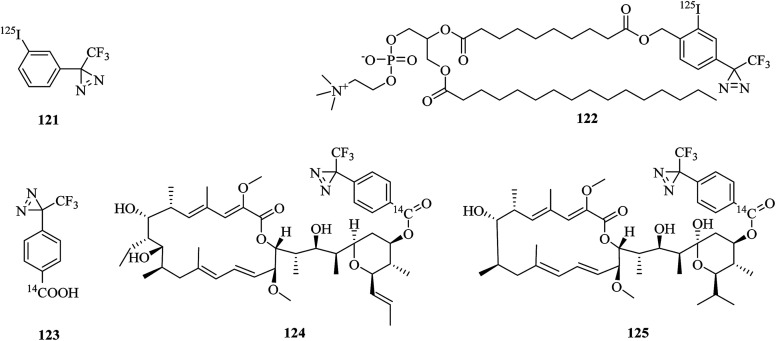
ATPases are enzymes that catalyze the decomposition of ATP into ADP and a free phosphate ion. There are different types of ATPase, which differ in function, structure (F-, V- and A-ATPases contain rotary motors) and the types of ion that they transport (Na/K-ATPases transport sodium and potassium ions across the plasma membrane).206–208 In a related study, Blanton et al. synthesized a radiolabeled diazirine probe [125I]TID (121, Fig. 19) and a phosphatidylcholine analogue [125I]TIDPC/16 (122, Fig. 19) to identify a lipid–protein interface, namely, the lipid-exposed transmembrane segments and transmembrane structural transitions of Torpedo Na/K-ATPase. A photolabeling experiment using probe 122 and Na/K-ATPase in combination with an analysis of the amino-terminal sequence of proteolytic fragments showed that the primary binding sites were localized in the hydrophobic segments M1, M3, M9, and M10, which constitute the major part of the lipid–protein interface in the α-subunit of Na/K-ATPase. Studies of the conformational sensitivity of probe 121 suggest that this radiolabeled diazirine probe would be a useful tool for structural characterization of the cation translocation pathway and conformationally dependent changes in the pathway.209 In addition, in order to study the binding sites of 21-deoxyconcanolide A and bafilomycin A1, which are two specific inhibitors of V-type ATPase (a proton translocation machine and pH regulator in almost all eukaryotic cells), Bender et al. incorporated a radiolabeled diazirine probe (123, Fig. 19) into two AfBPs (124 and 125, Fig. 19), which were based on a scaffold of 21-deoxyconcanolide A and bafilomycin A1, respectively. Both these AfBPs exhibited high inhibitory activity, which thus makes them ideal candidates for the further identification of the precise binding site of these potent V-ATPase inhibitors.210
Fig. 19. Representative applications of cmPAL for studying ATPases.
Another well-known enzyme is protein tyrosine phosphatase 1B (PTP1B), which belongs to the protein tyrosine phosphatase (PTP) family and is involved in downregulating the insulin receptor and can serve as a target of drugs for the treatment of type II diabetes.211 To better understand the mode of action of PTP1B inhibitors, a cell-based assay for directly measuring the enzyme occupancy of PTP1B by inhibitors using cmPAL was developed by Skorey et al., who synthesized two diazirine-based AfBPs, namely, 126 and 127 (Fig. 20), of which the most potent photoprobe exhibited an IC50 value of 0.2 nM for PTP1B. The photoaffinity assay of enzyme occupancy was successfully performed with both purified recombinant FLAG-tagged PTP1B and endogenous PTP1B in intact and lysed HepG2 cells, which indicated that these two compounds are powerful tools for research into PTP1B inhibitors.212
Fig. 20. Representative applications of cmPAL for studying protein tyrosine phosphatase 1B.
The removal of an N-terminal methionine residue from nascent polypeptides is dependent upon a unique class of proteases typified by the dinuclear metalloenzyme methionine aminopeptidase (MetAP) from Escherichia coli. Although the identification of human MetAP as the target of putative anticancer drugs reiterates the importance of this family of enzymes,213 the physiological function of MetAP1 (type 1 MetAP), however, has remained elusive. In a related study, Qiu et al. modified the potent MetAP1 inhibitor L134 (IC50 = 0.049 nM) with a TPD moiety and a photostable azido-acetyl group to prepare two trimodular photoaffinity probes (128 and 129, Fig. 21), which retained their inhibitory activity toward EcMetAP1, with nanomolar IC50 values. Then these probes were evaluated using purified recombinant EcMetAP1 and a crude cell lysate containing overexpressed MetAP1. Both of them were capable of labeling a 30 kDa polypeptide, which corresponded to the predicted molecular weight of EcMetAP1. In the presence of the competitive inhibitor L288, the signal intensities for the 30 kDa species were obviously reduced, whereas the signals for the 60 and 28 kDa species displayed no sensitivity to L288, which suggested that the 30 kDa polypeptide labeled by probe 128 was the native MetAP1. In addition, probe 129 was more effective than probe 128 despite the similarity in their IC50 values, which demonstrated that the length of the linker was a contributing factor. The trimodular probe 129 provided much more desirable flexibility for a prototype scaffold, which also allows improvements for different applications.214
Fig. 21. Representative applications of cmPAL for studying methionine aminopeptidase.
Etoposide (Fig. 22) is a widely used anticancer drug that targets topoisomerase II, which is an essential nuclear enzyme, with an unknown mode of binding. In order to identify the sites of binding between etoposide and topoisomerase II, Chee et al. developed a diazirinyl photoprobe (130, Fig. 22), which was based on a scaffold of etoposide. A set of characterizations of its photoreactivity and biological activities indicated that this photoaffinity label binds topoisomerase II with a similar affinity. It is worth mentioning that this diazirinyl photoprobe should be a promising tool for identifying the binding sites of etoposide in topoisomerase II.215
Fig. 22. Structures of 4′-demethyl-4b-podophyllotoxin derivatives.
Sialidases, which are also known as neuraminidases, are enzymes that catalyze the hydrolysis of terminal sialic acid residues in oligosaccharides, glycoproteins, and glycolipids.216 In order to improve the utility of the diazirine-based sialic acid analogue SiaDAz (131, Fig. 23), McCombs et al. screened a panel of sialidases to find one that would cleave natural sialic acids but remain inactive toward SiaDAz. The results of the study demonstrated that the crosslinking of SiaDAz-modified glycoconjugates can be enhanced by pre-treating cells with STNA (Salmonella typhimurium neuraminidase), which is a commercially available sialidase that removes the natural sialic acid Neu5Ac, but not SiaDAz. This strategy can be applied generally to sialic acid-mediated interactions and facilitate the identification of binding partners of sialic acid.121 In another study, Rodriguez et al. modified the N-acyl side chain of GlcNAc (132, Fig. 23) with an alkyldiazirine to prepare a photoprobe GlcNDAz (133, Fig. 23) and used the probe to examine the activity of recombinant human O-GlcNAc hydrolase (OGA), which is an enzyme that removes O-GlcNAc groups from serine and threonine residues in intracellular glycoproteins. Recombinant human OGA is unable to hydrolyze an O-GlcNDAz mimic or to remove an O-GlcNDAz group from a peptide, which provides new insights into the recognition of substrates by OGA, which is an important drug target.217
Fig. 23. Representative applications of cmPAL for studying sialidases.
In order to acquire a better understanding of the pharmacological effects of LW6 (Fig. 24) and its relation to HIF-1α and malate dehydrogenase 2 (MDH2), Naik et al. synthesized a probe as an LW6 mimic (134, Fig. 24), which contained a photoactive diazirine group. Both in vitro binding between the probe and recombinant human MDH2 via cmPAL and competitive inhibition of MDH2 activity with NADH, as in the case of LW6, were successfully confirmed, which suggested that the probe should provide a more comprehensive understanding of LW6 and MDH2.218 Similarly, Bongo et al. described the general use of a diazirinyl probe (135, Fig. 24) as a photoreactive mimic of α-amino acids, which can be cleaved upon treatment with a mild base to facilitate the retrieval of biotin-tagged proteins, as well as peptides, from avidin matrices. Upon irradiation with UV light, this diazirinyl probe specifically crosslinked glutamyl endopeptidase, l-glutamate dehydrogenase, glutamate oxaloacetate transaminase and l-glutamine synthetase, which are all enzymes that exhibit high affinity toward acidic α-amino acids. Its simple synthesis, coupled with its multifunctional nature, suggested that the probe would be a versatile photoreactive building block.219
Fig. 24. Structures of photoreactive acidic α-amino acid analogues.
CmPAL using a diazirinyl photoprobe to covalently bind its target in response to activation by light has become a frequently used tool for studying potential cellular targets of enzymes or their inhibitors and identifying the location and structure of binding sites. Recent efforts have mainly focused on cmPAL using a large panel of photoaffinity probes, as well as mass spectrometry-based chemical profiling methods, to identify binding sites and reveal unknown cellular mechanisms controlled by enzymes or corresponding inhibitors. Here, we have discussed different applications of cmPAL for classes of enzymes, including kinases, γ-secretase, methyltransferases, metalloproteinases, histone deacetylases, and so on. Although in some cases a benzophenone was the logical choice of label given the structure of the pharmacophore, it might be expected that diazirines will become the dominant photoreactive group and even develop into a versatile platform for labeling enzymes in the future as a development of synthetic methods involving the direct introduction of a minimalist crosslinker bearing an alkyldiazirine moiety and a terminal alkyne or TPD functionality into probe scaffolds.
5. Conclusions and outlook
As discussed in this review, the applications of cmPAL have established its uniquely important place in the identification of targets or binding sites in recent years. In particular, as precursors of carbenes, diazirines, which have been shown to be the most promising photoreactive reagents used for cmPAL, have enabled different types of sophisticated derivatization of diazirinyl probes according to the type of the parent ligand molecule. Supported by the development of analytical techniques, much more effort has been devoted to revealing unknown cellular mechanisms and drug–target binding sites controlled by various enzymes, especially inactive enzymes and corresponding inhibitors, by using a large panel of diazirine-based biological probes. Furthermore, the use of several probes for different classes of enzymes with different photoaffinity groups located in different regions may be necessary to provide a more comprehensive tool. In spite of the successful examples reported in this review, a continuing challenge in cmPAL involves the rational design of photoaffinity probes, which should retain the potency and specificity of the parent compounds, with less non-specific photolabeling and better accuracy in the proteome-wide identification of targets via MS analysis. Given these limitations, this is understandable, and clearly an urgent need still exists for the chemical biology community to develop more rational cmPAL-based probes for uncharacterized drug candidates and enzyme families relevant to various human diseases in particular. In addition, drugs and inhibitors need not be the only components of the proteome examined by cmPAL. Future research may provide insights into the development of cmPAL for investigating receptors and structural proteins.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
We wish to thank the financial support from the Key Technologies R&D Program (2014BAD23B01), the National Natural Science Foundation of China (21372052), and the Research Project of the Chinese Ministry of Education (213033A, 20135201110005).
References
- Singh A. Thornton E. R. Westheimer F. H. J. Biol. Chem. 1962;237:3006–3008. [PubMed] [Google Scholar]
- Hashimoto M. Hatanaka Y. Eur. J. Org. Chem. 2008:2513–2523. doi: 10.1002/ejoc.200701069. [DOI] [Google Scholar]
- Tanaka Y. Bond M. R. Kohler J. J. Mol. BioSyst. 2008;4:473–480. doi: 10.1039/B803218A. [DOI] [PubMed] [Google Scholar]
- Smith E. Collins I. Future Med. Chem. 2015;7:159–183. doi: 10.4155/fmc.14.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai K. Ozawa S. Yamaguchi T. Bioorg. Med. Chem. 2015;23:5319–5325. doi: 10.1016/j.bmc.2015.07.065. [DOI] [PubMed] [Google Scholar]
- Park J. Koh M. Koo J. Y. Lee S. Park S. B. ACS Chem. Biol. 2016;11:44–52. doi: 10.1021/acschembio.5b00671. [DOI] [PubMed] [Google Scholar]
- Tian Y. Jacinto M. P. Zeng Y. Yu Z. Qu J. Liu W. R. Lin Q. J. Am. Chem. Soc. 2017;139:6078–6081. doi: 10.1021/jacs.7b02615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herner A. Marjanovic J. Lewandowski T. M. Marin V. Patterson M. Miesbauer L. Ready D. Williams J. Vasudevan A. Lin Q. J. Am. Chem. Soc. 2016;138:14609–14615. doi: 10.1021/jacs.6b06645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battenberg O. A. Nodwell M. B. Sieber S. A. J. Org. Chem. 2011;76:6075–6087. doi: 10.1021/jo201281c. [DOI] [PubMed] [Google Scholar]
- Sumranjit J. Chung S. J. Molecules. 2013;18:10425–10451. doi: 10.3390/molecules180910425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford F. Yuzawa T. Platz M. S. Matzinger S. Fülscher M. J. Am. Chem. Soc. 1998;120:4430–4438. doi: 10.1021/ja9724598. [DOI] [Google Scholar]
- Blencowe A. Hayes W. Soft Matter. 2005;1:178–205. doi: 10.1039/B501989C. [DOI] [PubMed] [Google Scholar]
- Fleming S. A. Tetrahedron. 1995;51:12479–12520. doi: 10.1016/0040-4020(95)00598-3. [DOI] [Google Scholar]
- Smith R. A. G. Knowles J. R. J. Am. Chem. Soc. 1973;95:5072–5073. doi: 10.1021/ja00796a062. [DOI] [PubMed] [Google Scholar]
- Das J. Chem. Rev. 2011;111:4405–4417. doi: 10.1021/cr1002722. [DOI] [PubMed] [Google Scholar]
- Smith E. Collins I. Future Med. Chem. 2015;72:159–183. doi: 10.4155/fmc.14.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y. Peng L. Chem. Rev. 2013;113:7880–7929. doi: 10.1021/cr300419p. [DOI] [PubMed] [Google Scholar]
- Hashimoto M. Hatanaka Y. Eur. J. Org. Chem. 2008:2513–2523. doi: 10.1002/ejoc.200701069. [DOI] [Google Scholar]
- Dubinsky L. Krom B. P. Meijler M. M. Bioorg. Med. Chem. 2012;20:554–570. doi: 10.1016/j.bmc.2011.06.066. [DOI] [PubMed] [Google Scholar]
- Pan S. Zhang H. Wang C. Yao S. C. L. Yao S. Q. Nat. Prod. Rep. 2016;33:612–620. doi: 10.1039/C5NP00101C. [DOI] [PubMed] [Google Scholar]
- Aronoff M. R. Gold B. Raines R. T. Org. Lett. 2016;18:1538–1541. doi: 10.1021/acs.orglett.6b00278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M. E. Ferreira E. M. Stoltz B. M. Chem. Commun. 2006:1316–1318. doi: 10.1039/B517719G. [DOI] [PubMed] [Google Scholar]
- Mix K. A. Aronoff M. R. Raines R. T. ACS Chem. Biol. 2016;11:3233–3244. doi: 10.1021/acschembio.6b00810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayley H. Knowles J. R. Methods Enzymol. 1977;46:69–114. doi: 10.1016/s0076-6879(77)46012-9. [DOI] [PubMed] [Google Scholar]
- Chowdhry V. Westheimer F. H. Annu. Rev. Biochem. 1979;48:293–325. doi: 10.1146/annurev.bi.48.070179.001453. [DOI] [PubMed] [Google Scholar]
- Wolff L. Justus Liebigs Ann. Chem. 1902;325:129–195. doi: 10.1002/jlac.19023250202. [DOI] [Google Scholar]
- Kirmse W. Eur. J. Org. Chem. 2002:2193–2256. doi: 10.1002/1099-0690(200207)2002:14<2193::AID-EJOC2193>3.0.CO;2-D. [DOI] [Google Scholar]
- Chowdhry V. Vaughan R. Westheimer F. H. Proc. Natl. Acad. Sci. U. S. A. 1976;73:1406–1408. doi: 10.1073/pnas.73.5.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L. Alcaraz M. L. Klotz P. Kotzyba-Hibert F. Goeldner M. FEBS Lett. 1994;346:127–131. doi: 10.1016/0014-5793(94)00392-0. [DOI] [PubMed] [Google Scholar]
- Alcaraz M. L. Peng L. Klotz P. Goeldner M. J. Org. Chem. 1996;61:192–201. doi: 10.1021/jo951350k. [DOI] [Google Scholar]
- Converse C. A. Richards F. F. Biochemistry. 1969;8:4431–4436. doi: 10.1021/bi00839a031. [DOI] [PubMed] [Google Scholar]
- Gupta C. M. Costello C. E. Khorana H. G. Proc. Natl. Acad. Sci. U. S. A. 1979;76:3139–3143. doi: 10.1073/pnas.76.7.3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kale T. A. Distefano M. D. Org. Lett. 2003;5:609–612. doi: 10.1021/ol026752a. [DOI] [PubMed] [Google Scholar]
- Curtius T. Ber. Dtsch. Chem. Ges. 1890;23:3023–3033. doi: 10.1002/cber.189002302232. [DOI] [Google Scholar]
- Curtius T. J. Prakt. Chem. 1894;50:275–294. doi: 10.1002/prac.18940500125. [DOI] [Google Scholar]
- Myers E. L. Raines R. T. Angew. Chem., Int. Ed. Engl. 2009;48:2359–2363. doi: 10.1002/anie.200804689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgarten R. J. J. Org. Chem. 1967;32:484–485. doi: 10.1021/jo01288a059. [DOI] [Google Scholar]
- Chou H. H. Raines R. T. J. Am. Chem. Soc. 2013;135:14936–14939. doi: 10.1021/ja407822b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen S. R. Angew. Chem., Int. Ed. Engl. 1960;72:781–782. doi: 10.1002/ange.19600722119. [DOI] [Google Scholar]
- Schmitz E. Ohme R. Angew. Chem., Int. Ed. Engl. 1961;73:220–221. doi: 10.1002/ange.19610730616. [DOI] [Google Scholar]
- Pierce L. Dobyns V. J. Am. Chem. Soc. 1962;84:2651–2652. doi: 10.1021/ja00872a043. [DOI] [Google Scholar]
- Schmitz E. Ohme R. Schmidt R. D. Chem. Ber. 1962;95:2714–2717. doi: 10.1002/cber.19620951119. [DOI] [Google Scholar]
- Brunner J. Senn H. Richards F. M. J. Biol. Chem. 1980;255:3313–3318. [PubMed] [Google Scholar]
- Bonneau R. Liu M. T. H. J. Am. Chem. Soc. 1996;118:7229–7230. doi: 10.1021/ja9613217. [DOI] [Google Scholar]
- Akasaka T. Liu M. T. H. Niino Y. Maeda Y. Wakahara T. Okamura M. Kobayashi K. Nagase S. J. Am. Chem. Soc. 2000;122:7134–7135. doi: 10.1021/ja994242e. [DOI] [PubMed] [Google Scholar]
- Wakahara T. Niino Y. Kato T. Maeda Y. Akasaka T. Liu M. T. H. Kobayashi K. Nagase S. J. Am. Chem. Soc. 2002;124:9465–9468. doi: 10.1021/ja0116876. [DOI] [PubMed] [Google Scholar]
- Geurink P. P. Prely L. M. van der Marel G. A. Bischoff R. Overkleeft H. S. Top. Curr. Chem. 2012;324:85–114. doi: 10.1007/128_2011_286. [DOI] [PubMed] [Google Scholar]
- Preston G. W. Wilson A. J. Chem. Soc. Rev. 2013;42:3289–3301. doi: 10.1039/C3CS35459H. [DOI] [PubMed] [Google Scholar]
- Platz M. Admasu A. S. Kwiatkowski S. Cracker P. J. Imai N. Watt D. S. Bioconjugate Chem. 1991;2:337–341. doi: 10.1021/bc00011a008. [DOI] [PubMed] [Google Scholar]
- Wang L. Murai Y. Yoshida T. Ishida A. Masuda K. Sakihama Y. Hashidoko Y. Hatanaka Y. Hashimoto M. Org. Lett. 2015;17:616–619. doi: 10.1021/ol503630z. [DOI] [PubMed] [Google Scholar]
- Kumar A. B. Manetsch R. Synth. Commun. 2018;48:626–631. doi: 10.1080/00397911.2017.1354026. [DOI] [Google Scholar]
- Tamura Y. Minamikawa J. Sumoto K. Fujii S. Ikeda M. J. Org. Chem. 1973;38:1239–1241. doi: 10.1021/jo00946a045. [DOI] [PubMed] [Google Scholar]
- Jack T. Ruepp M. D. Thompson A. J. Mühlemann O. Lochner M. Chimia. 2014;68:239–242. doi: 10.2533/chimia.2014.239. [DOI] [PubMed] [Google Scholar]
- Mayer T. Maier M. E. Eur. J. Org. Chem. 2007:4711–4720. doi: 10.1002/ejoc.200700188. [DOI] [Google Scholar]
- Hatanaka Y. Hashimoto M. Kurihara H. Nakayama H. Kanaoka Y. J. Org. Chem. 1994;59:383–387. doi: 10.1021/jo00081a017. [DOI] [Google Scholar]
- Fang K. Hashimoto M. Jockusch S. Turro N. J. Nakanishi K. J. Am. Chem. Soc. 1998;120:8543–8544. doi: 10.1021/ja9817186. [DOI] [Google Scholar]
- Ishida A. Wang L. Tachrim Z. P. Suzuki T. Sakihama Y. Hashidoko Y. Hashimoto M. ChemistrySelect. 2017;2:160–164. doi: 10.1002/slct.201601675. [DOI] [Google Scholar]
- Wixe T. Almqvist F. Tetrahedron Lett. 2017;58:3350–3352. doi: 10.1016/j.tetlet.2017.07.031. [DOI] [Google Scholar]
- Wang L. Yoshida T. Muto Y. Murai Y. Tachrim Z. P. Ishida A. Nakagawa S. Sakihama Y. Hashidoko Y. Masuda K. Hatanaka Y. Hashimoto M. Eur. J. Org. Chem. 2015:3129–3134. doi: 10.1002/ejoc.201500184. [DOI] [Google Scholar]
- Kumar N. S. Young R. N. Bioorg. Med. Chem. 2009;17:5388–5395. doi: 10.1016/j.bmc.2009.06.048. [DOI] [PubMed] [Google Scholar]
- Richards F. M. Lamed R. Wynn R. Patel D. Olack G. Protein Sci. 2000;9:2506–2517. doi: 10.1110/ps.9.12.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig P. O. Ureta D. B. Delfino J. M. Protein Sci. 2002;11:1353–1366. doi: 10.1110/ps.4710102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ureta D. B. Craig P. O. Gómez G. E. Delfino J. M. Biochemistry. 2007;46:14567–14577. doi: 10.1021/bi7012867. [DOI] [PubMed] [Google Scholar]
- Li Z. Hao P. Li L. Tan C. Y. J. Cheng X. Chen G. Y. J. Sze S. K. Shen H. M. Yao S. Q. Angew. Chem., Int. Ed. 2013;52:8551–8556. doi: 10.1002/anie.201300683. [DOI] [PubMed] [Google Scholar]
- Li Z. Wang D. Li L. Pan S. Na Z. Tan C. Y. J. Yao S. Q. J. Am. Chem. Soc. 2014;136:9990–9998. doi: 10.1021/ja502780z. [DOI] [PubMed] [Google Scholar]
- Pan S. Jang S. Y. Wang D. Liew S. S. Li Z. Lee J. S. Yao S. Q. Angew. Chem., Int. Ed. Engl. 2017;56:11816–11821. doi: 10.1002/anie.201706076. [DOI] [PubMed] [Google Scholar]
- Gao J. Mfuh A. Amako Y. Woo C. M. J. Am. Chem. Soc. 2018;140:4259–4268. doi: 10.1021/jacs.7b11639. [DOI] [PubMed] [Google Scholar]
- Wang L. Tachrim Z. P. Kurokawa N. Ohashi F. Sakihama Y. Hashidoko Y. Hashimoto M. Molecules. 2017;22:1389. doi: 10.3390/molecules22081389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill J. R. Robertson A. A. B. J. Med. Chem. 2018 doi: 10.1021/acs.jmedchem.7b01561. [DOI] [Google Scholar]
- Murale D. P. Hong S. C. Haque M. M. Lee J. S. Proteome Sci. 2016;15:14. doi: 10.1186/s12953-017-0123-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchanek M. Radzikowska A. Thiele C. Nat. Methods. 2005;2:261–268. doi: 10.1038/nmeth752. [DOI] [PubMed] [Google Scholar]
- Markham K. Bai Y. Schmitt-Ulms G. Anal. Bioanal. Chem. 2007;389:461–473. doi: 10.1007/s00216-007-1385-x. [DOI] [PubMed] [Google Scholar]
- Vila-Perelló M. Pratt M. R. Tulin F. Muir T. W. J. Am. Chem. Soc. 2007;129:8068–8069. doi: 10.1021/ja072013j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKinnon A. L. Garrison J. L. Hegde R. S. Taunton J. J. Am. Chem. Soc. 2007;129:14560–14561. doi: 10.1021/ja076250y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häupl B. Ihling C. H. Sinz A. Proteomics. 2017;17:1600459. doi: 10.1002/pmic.201600459. [DOI] [PubMed] [Google Scholar]
- Lipstein N. Göth M. Piotrowski C. Pagel K. Sinz A. Jahn O. Expert Rev. Proteomics. 2017;14:223–242. doi: 10.1080/14789450.2017.1275966. [DOI] [PubMed] [Google Scholar]
- Srinivas N. Jetter P. Ueberbacher B. J. Werneburg M. Zerbe K. Steinmann J. Van der Meijden B. Bernardini F. Lederer A. Dias R. L. Misson P. E. Henze H. Zumbrunn J. Gombert F. O. Obrecht D. Hunziker P. Schauer S. Ziegler U. Kach A. Eberl L. Riedel K. DeMarco S. J. Robinson J. A. Science. 2010;327:1010–1013. doi: 10.1126/science.1182749. [DOI] [PubMed] [Google Scholar]
- Zerbe K. Moehle K. Robinson J. A. Acc. Chem. Res. 2017;50:1323–1331. doi: 10.1021/acs.accounts.7b00129. [DOI] [PubMed] [Google Scholar]
- Urfer M. Bogdanovic J. Lo Monte F. Moehle K. Zerbe K. Omasits U. Ahrens C. H. Pessi G. Eberl L. Robinson J. A. J. Biol. Chem. 2016;291:1921–1932. doi: 10.1074/jbc.M115.691725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright M. H. Fetzer C. Sieber S. A. J. Am. Chem. Soc. 2017;139:6152–6159. doi: 10.1021/jacs.7b01072. [DOI] [PubMed] [Google Scholar]
- Nassal M. J. Am. Chem. Soc. 1984;106:7540–7545. doi: 10.1021/ja00336a038. [DOI] [Google Scholar]
- Nakashima H. Hashimoto M. Sadakane Y. Tomohiro T. Hatanaka Y. J. Am. Chem. Soc. 2006;128:15092–15093. doi: 10.1021/ja066479y. [DOI] [PubMed] [Google Scholar]
- Tippmann E. M. Liu W. Summerer D. Mack A. V. Schultz P. G. ChemBioChem. 2007;8:2210–2214. doi: 10.1002/cbic.200700460. [DOI] [PubMed] [Google Scholar]
- Hino N. Oyama M. Sato A. Mukai T. Iraha F. Hayashi A. Kozuka-Hata H. Yamamoto T. Yokoyama S. Sakamoto K. J. Mol. Biol. 2011;406:343–353. doi: 10.1016/j.jmb.2010.12.022. [DOI] [PubMed] [Google Scholar]
- Yang Y. Song H. Chen P. R. IUBMB Life. 2016;68:879–886. doi: 10.1002/iub.1560. [DOI] [PubMed] [Google Scholar]
- Masuda K. Koizumi A. Misaka T. Hatanaka Y. Abe K. Tanaka T. Ishiguro M. Hashimoto M. Bioorg. Med. Chem. Lett. 2010;20:1081–1083. doi: 10.1016/j.bmcl.2009.12.029. [DOI] [PubMed] [Google Scholar]
- Murai Y. Masuda K. Sakihama Y. Hashidoko Y. Hatanaka Y. Hashimoto M. J. Org. Chem. 2012;77:8581–8587. doi: 10.1021/jo301552m. [DOI] [PubMed] [Google Scholar]
- Wartmann T. Lindel T. Eur. J. Org. Chem. 2013:1649–1652. doi: 10.1002/ejoc.201201726. [DOI] [Google Scholar]
- Chou C. Uprety R. Davis L. Chin J. W. Deiters A. Chem. Sci. 2011;2:480–483. doi: 10.1039/C0SC00373E. [DOI] [Google Scholar]
- Yang T. Li X. M. Bao X. Fung Y. M. Li X. D. Nat. Chem. Biol. 2016;12:70–72. doi: 10.1038/nchembio.1990. [DOI] [PubMed] [Google Scholar]
- Xie X. Li X. M. Qin F. Lin J. Zhang G. Zhao J. Bao X. Zhu R. Song H. Li X. D. Chen P. R. J. Am. Chem. Soc. 2017;139:6522–6525. doi: 10.1021/jacs.7b01431. [DOI] [PubMed] [Google Scholar]
- Taranenko M. Mtchedlidze M. Sumbatyan N. Korshunova G. Nucleosides, Nucleotides Nucleic Acids. 2003;22:715–717. doi: 10.1081/NCN-120022617. [DOI] [PubMed] [Google Scholar]
- Shigdel U. K. Zhang J. He C. Angew. Chem., Int. Ed. Engl. 2008;47:90–93. doi: 10.1002/anie.200703625. [DOI] [PubMed] [Google Scholar]
- Qiu Z. Lu L. Jian X. He C. J. Am. Chem. Soc. 2008;130:14398–14399. doi: 10.1021/ja805445j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winnacker M. Breeger S. Strasser R. Carell T. ChemBioChem. 2009;10:109–118. doi: 10.1002/cbic.200800397. [DOI] [PubMed] [Google Scholar]
- Winnacker M. Welzmiller V. Strasser R. Carell T. ChemBioChem. 2010;11:1345–1349. doi: 10.1002/cbic.201000244. [DOI] [PubMed] [Google Scholar]
- Sergiev P. V. Lavrik I. N. Wlasoff V. A. Dokudovskaya S. S. Dontsova O. A. Bogdanov A. A. Brimacombe R. RNA. 1997;3:464–475. [PMC free article] [PubMed] [Google Scholar]
- Sergiev P. Dokudovskaya S. Romanova E. Topin A. Bogdanov A. Brimacombe R. Dontsova O. Nucleic Acids Res. 1998;26:2519–2525. doi: 10.1093/nar/26.11.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavrik I. N. Sergive P. V. Bogdanov A. A. Zimmermann R. A. Dontsova O. A. Molecular Biology. 2004;38:799–805. doi: 10.1023/B:MBIL.0000043949.56555.03. [DOI] [Google Scholar]
- Kuboe S. Yoda M. Ogata A. Kitade Y. Tomari Y. Ueno Y. Chem. Commun. 2010;46:7367–7369. doi: 10.1039/C0CC02450C. [DOI] [PubMed] [Google Scholar]
- Nakamoto K. Ueno Y. J. Org. Chem. 2014;79:2463–2472. doi: 10.1021/jo402738t. [DOI] [PubMed] [Google Scholar]
- Nakamoto K. Minami K. Akao Y. Ueno Y. Chem. Commun. 2016;52:6720–6722. doi: 10.1039/C6CC01360K. [DOI] [PubMed] [Google Scholar]
- Muttach F. Mäsing F. Studer A. Rentmeister A. Chem.–Eur. J. 2017;23:5988–5993. doi: 10.1002/chem.201605663. [DOI] [PubMed] [Google Scholar]
- Erni B. Khorana H. G. J. Am. Chem. Soc. 1980;102:3888–3896. doi: 10.1021/ja00531a034. [DOI] [Google Scholar]
- Brunner J. Annu. Rev. Biochem. 1993;62:483–514. doi: 10.1146/annurev.bi.62.070193.002411. [DOI] [PubMed] [Google Scholar]
- Ross A. H. Ramachandran R. Robson R. J. Khorana H. G. J. Biol. Chem. 1982;257:4152–4161. [PubMed] [Google Scholar]
- Stegmann T. Delfino J. M. Richards F. M. Helenius A. J. Biol. Chem. 1991;266:18404–18410. [PubMed] [Google Scholar]
- Janssen M. J. F. W. van Voorst F. Ploeger G. E. J. Larsen P. M. Larsen M. R. de Kroon A. I. P. M. de Kruijff B. Biochemistry. 2002;41:5702–5711. doi: 10.1021/bi025550j. [DOI] [PubMed] [Google Scholar]
- Haberkant P. Schmitt O. Contreras F. X. Thiele C. Hanada K. Sprong H. Reinhard C. Wieland F. T. Brügger B. J. Lipid Res. 2008;49:251–262. doi: 10.1194/jlr.D700023-JLR200. [DOI] [PubMed] [Google Scholar]
- Haberkant P. Stein F. Höglinger D. Gerl M. J. Brugger B. Van Veldhoven P. P. Krijgsveld J. Gavin A. C. Schultz C. ACS Chem. Biol. 2016;11:222–230. doi: 10.1021/acschembio.5b00810. [DOI] [PubMed] [Google Scholar]
- Haberkant P. Raijmakers R. Wildwater M. Sachsenheimer T. Brügger B. Maeda K. Houweling M. Gavin A. C. Schultz C. van Meer G. Heck A. J. Holthuis J. C. Angew. Chem., Int. Ed. Engl. 2013;52:4033–4038. doi: 10.1002/anie.201210178. [DOI] [PubMed] [Google Scholar]
- Höglinger D. Nadler A. Haberkant P. Kirkpatrick J. Schifferer M. Stein F. Hauke S. Porter F. D. Schultz C. Proc. Natl. Acad. Sci. U. S. A. 2017;114:1566–1571. doi: 10.1073/pnas.1611096114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. Dong T. Zhou Y. Huang N. Lei X. Angew. Chem., Int. Ed. 2016;55:14330–14334. doi: 10.1002/anie.201608827. [DOI] [PubMed] [Google Scholar]
- Gu X. Huang Y. Levison B. S. Gerstenecker G. DiDonato A. J. Hazen L. B. Lee J. Gogonea V. DiDonato J. A. Hazen S. L. J. Biol. Chem. 2016;291:1890–18904. doi: 10.1074/jbc.M115.678334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D. Du S. Cazenave-Gassiot A. Ge J. Lee J. S. Wenk M. R. Yao S. Q. Angew. Chem., Int. Ed. 2017;56:5829–5833. doi: 10.1002/anie.201702509. [DOI] [PubMed] [Google Scholar]
- Sakurai K. Asian J. Org. Chem. 2015;4:116–126. doi: 10.1002/ajoc.201402209. [DOI] [Google Scholar]
- Bertozzi C. R. Kiessling L. L. Science. 2001;291:2357–2364. doi: 10.1126/science.1059820. [DOI] [PubMed] [Google Scholar]
- Kuhn C. S. Lehmann J. Jung G. Stevanović S. Carbohydr. Res. 1992;232:227–233. doi: 10.1016/0008-6215(92)80056-7. [DOI] [PubMed] [Google Scholar]
- Kuhn C. S. Lehmann J. Sandhoff K. Bioconjugate Chem. 1992;3:230–233. doi: 10.1021/bc00015a005. [DOI] [PubMed] [Google Scholar]
- Liessem B. Glombitza G. J. Knoll F. Lehmann J. Kellermann J. Lottspeich F. Sandhoff K. J. Biol. Chem. 1995;270:23693–23699. doi: 10.1074/jbc.270.40.23693. [DOI] [PubMed] [Google Scholar]
- McCombs J. E. Zou C. Parker R. B. Cairo C. W. Kohler J. J. ACS Chem. Biol. 2016;11:185–192. doi: 10.1021/acschembio.5b00775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y. Kohler J. J. J. Am. Chem. Soc. 2008;130:3278–3279. doi: 10.1021/ja7109772. [DOI] [PubMed] [Google Scholar]
- Bond M. R. Zhang H. Vu P. D. Kohler J. J. Nat. Protoc. 2009;4:1044–1063. doi: 10.1038/nprot.2009.85. [DOI] [PubMed] [Google Scholar]
- Yu S. H. Boyce M. Wands A. M. Bond M. R. Bertozzi C. R. Kohler J. J. Proc. Natl. Acad. Sci. U. S. A. 2012;109:4834–4839. doi: 10.1073/pnas.1114356109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond M. R. Zhang H. Kim J. Yu S. H. Yang F. Patrie S. M. Kohler J. J. Bioconjugate Chem. 2011;22:1811–1823. doi: 10.1021/bc2002117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai K. Ozawa S. Yamada R. Yasui T. Mizuno S. ChemBioChem. 2014;15:1399–1403. doi: 10.1002/cbic.201402051. [DOI] [PubMed] [Google Scholar]
- Sakurai K. Yasui T. Mizuno S. Asian J. Org. Chem. 2015;4:724–728. doi: 10.1002/ajoc.201500116. [DOI] [Google Scholar]
- Monte M. J. Marin J. J. Antelo A. Vazquez-Tato J. World J. Gastroenterol. 2009;15:804–816. doi: 10.3748/wjg.15.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer W. Kurz G. J. Lipid Res. 1983;24:910–923. [PubMed] [Google Scholar]
- von Dippe P. Levy D. J. Biol. Chem. 1983;258:8896–8901. [PubMed] [Google Scholar]
- Kramer W. Schneider S. J. Lipid Res. 1989;30:1281–1288. [PubMed] [Google Scholar]
- Zhuang S. Li Q. Cai L. Wang C. Lei X. ACS Cent. Sci. 2017;3:501–509. doi: 10.1021/acscentsci.7b00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele C. Hannah M. J. Fahrenholz F. Huttner W. B. Nat. Cell Biol. 2000;2:42–49. doi: 10.1038/71366. [DOI] [PubMed] [Google Scholar]
- Suchanek M. Hynynen R. Wohlfahrt G. Lehto M. Johansson M. Saarinen H. Radzikowska A. Thiele C. Olkkonen V. M. Biochem. J. 2007;405:473–480. doi: 10.1042/BJ20070176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz J. C. Thomas M. Wong E. Ohgami N. Sugii S. Curphey T. Chang C. C. Y. Chang T. Y. J. Lipid Res. 2002;43:1341–1347. [PubMed] [Google Scholar]
- Ohgami N. Ko D. C. Thomas M. Scott M. P. Chang C. C. Y. Chang T. Y. Proc. Natl. Acad. Sci. U. S. A. 2004;101:12473–12478. doi: 10.1073/pnas.0405255101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulce J. J. Cognetta A. B. Niphakis M. J. Tully S. E. Cravatt B. F. Nat. Methods. 2013;10:259–264. doi: 10.1038/nmeth.2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darbandi-Tonkabon R. Hastings W. R. Zeng C. M. Akk G. Manion B. D. Bracamontes J. R. Steinbach J. H. Mennerick S. J. Covey D. F. Evers A. S. J. Biol. Chem. 2003;278:13196–13206. doi: 10.1074/jbc.M213168200. [DOI] [PubMed] [Google Scholar]
- Jiang X. Shu H. J. Krishnan K. Qian M. Taylor A. A. Covey D. F. Zorumski C. F. Mennerick S. Neuropharmacology. 2016;108:193–206. doi: 10.1016/j.neuropharm.2016.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savechenkov P. Y. Chiara D. C. Desai R. Stern A. T. Zhou X. Ziemba A. M. Szabo A. L. Zhang Y. Cohen J. B. Forman S. A. Miller K. W. Bruzik K. S. Eur. J. Med. Chem. 2017;136:334–347. doi: 10.1016/j.ejmech.2017.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall M. A. Xi J. Lor C. Dai S. Pearce R. Dailey W. P. Eckenhoff R. G. J. Med. Chem. 2010;53:5667–5675. doi: 10.1021/jm1004072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flood P. Ramirez-Latorre J. Role L. Anesthesiology. 1997;86:859–865. doi: 10.1097/00000542-199704000-00016. [DOI] [PubMed] [Google Scholar]
- Violet J. M. Downie D. L. Nakisa R. C. Lieb W. R. Franks N. P. Anesthesiology. 1997;86:866–874. doi: 10.1097/00000542-199704000-00017. [DOI] [PubMed] [Google Scholar]
- Jayakar S. S. Dailey W. P. Eckenhoff R. G. Cohen J. B. J. Biol. Chem. 2013;288:6178–6189. doi: 10.1074/jbc.M112.435909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart D. S. Savechenkov P. Y. Dostalova Z. Chiara D. C. Ge R. Raines D. E. Cohen J. B. Forman S. A. Bruzik K. S. Miller K. W. J. Med. Chem. 2011;54:8124–8135. doi: 10.1021/jm200943f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip G. M. S. Chen Z. W. Edge C. J. Smith E. H. Dickinson R. Hohenester E. Townsend R. R. Fuchs K. Sieghart W. Evers A. S. Franks N. P. Nat. Chem. Biol. 2013;9:715–720. doi: 10.1038/nchembio.1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebell M. R. Nirthanan S. Husain S. S. Miller K. W. Cohen J. B. J. Biol. Chem. 2004;279:17640–17649. doi: 10.1074/jbc.M313886200. [DOI] [PubMed] [Google Scholar]
- Li G. D. Chiara D. C. Sawyer G. W. Husain S. S. Olsen R. W. Cohen J. B. J. Neurosci. 2006;26:11599–11605. doi: 10.1523/JNEUROSCI.3467-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husain S. S. Nirthanan S. Ruesch D. Solt K. Cheng Q. Li G. D. Arevalo E. Olsen R. W. Raines D. E. Forman S. A. Cohen J. B. Miller K. W. J. Med. Chem. 2006;49:4818–4825. doi: 10.1021/jm051207b. [DOI] [PubMed] [Google Scholar]
- Husain S. S. Stewart D. Desai R. Hamouda A. K. Li S. G. Kelly E. Dostalova Z. Zhou X. Cotten J. F. Raines D. E. Olsen R. W. Cohen J. B. Forman S. A. Miller K. W. J. Med. Chem. 2010;53:6432–6444. doi: 10.1021/jm100498u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nirthanan S. Chiara III G. Chiara D. C. Husain S. S. Cohen J. B. J. Biol. Chem. 2008;283:22051–22062. doi: 10.1074/jbc.M801332200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans M. J. Cravatt B. F. Chem. Rev. 2006;106:3279–3301. doi: 10.1021/cr050288g. [DOI] [PubMed] [Google Scholar]
- Barglow K. T. Cravatt B. F. Nat. Methods. 2007;4:822–827. doi: 10.1038/nmeth1092. [DOI] [PubMed] [Google Scholar]
- Fonović M. Bogyo M. Expert Rev. Proteomics. 2008;5:721–730. doi: 10.1586/14789450.5.5.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uttamchandani M. Li J. Sun H. Yao S. Q. ChemBioChem. 2008;9:667–675. doi: 10.1002/cbic.200700755. [DOI] [PubMed] [Google Scholar]
- Dancey J. Sausville E. A. Nat. Rev. Drug Discovery. 2003;2:296–313. doi: 10.1038/nrd1066. [DOI] [PubMed] [Google Scholar]
- Karaman M. W. Herrgard S. Treiber D. K. Gallant P. Atteridge C. E. Campbell B. T. Chan K. W. Ciceri P. Davis M. I. Edeen P. T. Faraoni R. Floyd M. Hunt J. P. Lockhart D. J. Milanov Z. V. Morrison M. J. Pallares G. Patel H. K. Pritchard S. Wodicka L. M. Zarrinkar P. P. Nat. Biotechnol. 2008;26:127–132. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- Shi H. Cheng X. Sze S. K. Yao S. Q. Chem. Commun. 2011;47:11306–11308. doi: 10.1039/C1CC14824A. [DOI] [PubMed] [Google Scholar]
- Shi H. Zhang C. J. Chen G. Y. J. Yao S. Q. J. Am. Chem. Soc. 2012;134:3001–3014. doi: 10.1021/ja208518u. [DOI] [PubMed] [Google Scholar]
- Su Y. Pan S. Li Z. Li L. Wu X. Hao P. Sze S. K. Yao S. Q. Sci. Rep. 2015;5:7724. doi: 10.1038/srep07724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiner P. Heydenreuter W. Stahl M. Korotkov V. S. Sieber S. A. Angew. Chem., Int. Ed. 2016;56:1396–1401. doi: 10.1002/anie.201605993. [DOI] [PubMed] [Google Scholar]
- Das J. Addona G. H. Sandberg W. S. Husain S. S. Stehle T. Miller K. W. J. Biol. Chem. 2004;279:37964–37972. doi: 10.1074/jbc.M405137200. [DOI] [PubMed] [Google Scholar]
- Das J. Zhou X. Miller K. W. Protein Sci. 2006;15:2107–2119. doi: 10.1110/ps.062237606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addona G. H. Husain S. S. Stehle T. Miller K. W. J. Biol. Chem. 2002;277:25685–25691. doi: 10.1074/jbc.M201303200. [DOI] [PubMed] [Google Scholar]
- Hashimoto M. Nabeta K. Murakami K. Bioorg. Med. Chem. Lett. 2003;13:1531–1533. doi: 10.1016/S0960-894X(03)00200-2. [DOI] [PubMed] [Google Scholar]
- Saito A. Kawai K. Takayama H. Sudo T. Osada H. Chem.–Asian J. 2008;3:1607–1612. doi: 10.1002/asia.200800099. [DOI] [PubMed] [Google Scholar]
- Wang Z. Y. Seto H. Fujioka S. Yoshida S. Chory J. Nature. 2001;410:380–383. doi: 10.1038/35066597. [DOI] [PubMed] [Google Scholar]
- Kinoshita T. Caño-Delgado A. Seto H. Hiranuma S. Fujioka S. Yoshida S. Chory J. Nature. 2005;433:167–171. doi: 10.1038/nature03227. [DOI] [PubMed] [Google Scholar]
- Ranjitkar P. Perera B. G. Swaney D. L. Hari S. B. Larson E. T. Krishnamurty R. Merritt E. A. Villén J. Maly D. J. J. Am. Chem. Soc. 2012;134:19017–19025. doi: 10.1021/ja306035v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eni S. E. Rowland M. Best M. D. RSC Adv. 2015;5:25457–25461. doi: 10.1039/C4RA16730A. [DOI] [Google Scholar]
- Dovey H. F. John V. Anderson J. P. Chen L. Z. de Saint Andrieu P. Fang L. Y. Freedman S. B. Folmer B. Goldbach E. Holsztynska E. J. Hu K. L. Johnson-Wood K. L. Kennedy S. L. Kholodenko D. Knops J. E. Latimer L. H. Lee M. Liao Z. Lieberburg I. M. Motter R. N. Mutter L. C. Nietz J. Quinn K. P. Sacchi K. L. Seubert P. A. Shopp G. M. Thorsett E. D. Tung J. S. Wu J. Yang S. Yin C. T. Schenk D. B. May P. C. Altstiel L. D. Bender M. H. Boggs L. N. Britton T. C. Clemens J. C. Czilli D. L. Dieckman-McGinty D. K. Droste J. J. Fuson K. S. Gitter B. D. Hyslop P. A. Johnstone E. M. Li W. Y. Little S. P. Mabry T. E. Miller F. D. Audia J. E. J. Neurochem. 2001;76:173–181. doi: 10.1046/j.1471-4159.2001.00012.x. [DOI] [PubMed] [Google Scholar]
- Fuwa H. Takahashi Y. Konno Y. Watanabe N. Miyashita H. Sasaki M. Natsugari H. Kan T. Fukuyama T. Tomita T. Iwatsubo T. ACS Chem. Biol. 2007;2:408–418. doi: 10.1021/cb700073y. [DOI] [PubMed] [Google Scholar]
- Crump C. J. Castro S. V. Wang F. Pozdnyakov N. Ballard T. E. Sisodia S. S. Bales K. R. Johnson D. S. Li Y. M. Biochemistry. 2012;51:7209–7211. doi: 10.1021/bi301137h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump C. J. Murrey H. E. Ballard T. E. Am Ende C. W. Wu X. Gertsik N. Johnson D. S. Li Y. M. ACS Chem. Neurosci. 2016;7:1166–1173. doi: 10.1021/acschemneuro.6b00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennhack A. Jumpertz T. Ness J. Baches S. Pietrzik C. U. Weggen S. Bulic B. Bioorg. Med. Chem. 2012;20:6523–6532. doi: 10.1016/j.bmc.2012.08.034. [DOI] [PubMed] [Google Scholar]
- Jumpertz T. Rennhack A. Ness J. Baches S. Pietrzik C. U. Bulic B. Weggen S. PLoS One. 2012;7:e30484. doi: 10.1371/journal.pone.0030484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S. C. Int. J. Biochem. Cell Biol. 2000;32:391–395. doi: 10.1016/S1357-2725(99)00139-9. [DOI] [PubMed] [Google Scholar]
- Petrossian T. C. Clarke S. G. Mol. Cell. Proteomics. 2011;10:M110.000976. doi: 10.1074/mcp.M110.000976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalhoff C. Hüben M. Lenz T. Poot P. Nordhoff E. Koster H. Weinhold E. ChemBioChem. 2010;11:256–265. doi: 10.1002/cbic.200900349. [DOI] [PubMed] [Google Scholar]
- Horning B. D. Suciu R. M. Ghadiri D. A. Ulanovskaya O. A. Matthews M. L. Lum K. M. Backus K. M. Brown S. J. Rosen H. Cravatt B. F. J. Am. Chem. Soc. 2016;138:13335–13343. doi: 10.1021/jacs.6b07830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vervacke J. S. Funk A. L. Wang Y. C. Strom M. Hrycyna C. A. Distefano M. D. J. Org. Chem. 2014;79:1971–1978. doi: 10.1021/jo402600b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B. Zhang H. Pan S. Wang C. Ge J. Lee J. S. Yao S. Q. Chem. Eur. J. 2016;22:7824–7836. doi: 10.1002/chem.201600259. [DOI] [PubMed] [Google Scholar]
- Chang C. Werb Z. Trends Cell Biol. 2001;11:27–43. doi: 10.1016/S0962-8924(01)82222-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foda H. D. Zucker S. Drug Discovery Today. 2001;6:478–482. doi: 10.1016/S1359-6446(01)01752-4. [DOI] [PubMed] [Google Scholar]
- Sternlicht M. D. Werb Z. Annu. Rev. Cell Dev. Biol. 2001;17:463–516. doi: 10.1146/annurev.cellbio.17.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seals D. F. Courtneidge S. A. Genes Dev. 2003;17:7–30. doi: 10.1101/gad.1039703. [DOI] [PubMed] [Google Scholar]
- Black R. A. White J. M. Curr. Opin. Cell Biol. 1998;10:654–659. doi: 10.1016/S0955-0674(98)80042-2. [DOI] [PubMed] [Google Scholar]
- Chan E. W. S. Chattopadhaya S. Panicker R. C. Huang X. Yao S. Q. J. Am. Chem. Soc. 2004;126:14435–14446. doi: 10.1021/ja047044i. [DOI] [PubMed] [Google Scholar]
- Leeuwenburgh M. A. Geurink P. P. Klein T. Kauffman H. F. van der Marel G. A. Bischoff R. Overkleeft H. S. Org. Lett. 2006;8:1705–1708. doi: 10.1021/ol060409e. [DOI] [PubMed] [Google Scholar]
- Geurink P. P. Klein T. Prèly L. Paal K. Leeuwenburgh M. A. van der Marel G. A. Kauffman H. F. Overkleeft H. S. Bischoff R. Eur. J. Org. Chem. 2010:2100–2112. doi: 10.1002/ejoc.200901385. [DOI] [Google Scholar]
- Lozito T. P. Tuan R. S. J. Cell. Physiol. 2011;226:385–396. doi: 10.1002/jcp.22344. [DOI] [PubMed] [Google Scholar]
- Nury C. Czarny B. Cassar-Lajeunesse E. Georgiadis D. Bregant S. Dive V. ChemBioChem. 2013;14:107–114. doi: 10.1002/cbic.201200583. [DOI] [PubMed] [Google Scholar]
- Wolffe A. P. Science. 1996;272:371–372. doi: 10.1126/science.272.5260.371. [DOI] [PubMed] [Google Scholar]
- Gregoretti I. V. Lee Y. M. Goodson H. V. J. Mol. Biol. 2004;338:17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- Corminboeuf C. Hu P. Tuckerman M. E. Zhang Y. J. Am. Chem. Soc. 2006;128:4530–4531. doi: 10.1021/ja0600882. [DOI] [PubMed] [Google Scholar]
- Gallinari P. Di Marco S. Jones P. Pallaoro M. Steinkuhler C. Cell Res. 2007;17:195–211. doi: 10.1038/sj.cr.7310149. [DOI] [PubMed] [Google Scholar]
- Smith B. C. Hallows W. C. Denu J. M. Chem. Biol. 2008;15:1002–1013. doi: 10.1016/j.chembiol.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentschel F. Raimer B. Kelter G. Fiebig H. H. Sasse F. Lindel T. Eur. J. Org. Chem. 2014:2120–2127. doi: 10.1002/ejoc.201301717. [DOI] [Google Scholar]
- Xie Y. Ge J. Lei H. Peng B. Zhang H. Wang D. Pan S. Chen G. Chen L. Wang Y. Hao Q. Yao S. Q. Sun H. J. Am. Chem. Soc. 2016;138:15596–15604. doi: 10.1021/jacs.6b07334. [DOI] [PubMed] [Google Scholar]
- Liu Z. Yang T. Li X. Peng T. Hang H. C. Li X. D. Angew. Chem., Int. Ed. Engl. 2015;54:1149–1152. doi: 10.1002/anie.201408763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T. Liu Z. Li X. D. Chem. Sci. 2015;6:1011–1017. doi: 10.1039/C4SC02328E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert T. Malo M. Lengqvist J. Sihlbom C. Jarho E. M. Luthman K. J. Med. Chem. 2016;59:10794–10799. doi: 10.1021/acs.jmedchem.6b01117. [DOI] [PubMed] [Google Scholar]
- Rothwell P. J. Waksman G. Adv. Protein Chem. 2005;71:401–440. doi: 10.1016/S0065-3233(04)71011-6. [DOI] [PubMed] [Google Scholar]
- Yamaguchi T. Suyama K. Narita K. Kohgo S. Tomikawa A. Saneyoshi M. Nucleic Acids Res. 1997;25:2352–2358. doi: 10.1093/nar/25.12.2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebmann M. Di Pasquale F. Marx A. ChemBioChem. 2006;7:1965–1969. doi: 10.1002/cbic.200600333. [DOI] [PubMed] [Google Scholar]
- Murata T. Yamato I. Kakinuma Y. Leslie A. G. Walker J. E. Science. 2005;308:654–659. doi: 10.1126/science.1110064. [DOI] [PubMed] [Google Scholar]
- Lee S. H. Rho J. Jeong D. Sul J. Y. Kim T. Kim N. Kang J. S. Miyamoto T. Suda T. Lee S. K. Pignolo R. J. Koczon-Jaremko B. Lorenzo J. Choi Y. Nat. Med. 2006;12:1403–1409. doi: 10.1038/nm1514. [DOI] [PubMed] [Google Scholar]
- Kaplan J. H. Annu. Rev. Biochem. 2002;71:511–535. doi: 10.1146/annurev.biochem.71.102201.141218. [DOI] [PubMed] [Google Scholar]
- Blanton M. P. McCardy E. A. Biochemistry. 2000;39:13534–13544. doi: 10.1021/bi0015461. [DOI] [PubMed] [Google Scholar]
- Bender T. Huss M. Wieczorek H. Grond S. von Zezschwitz P. Eur. J. Org. Chem. 2007:3870–3878. doi: 10.1002/ejoc.200700194. [DOI] [Google Scholar]
- Lo L. C. Pang T. L. Kuo C. H. Chiang Y. L. Wang H. Y. Lin J. J. J. Proteome Res. 2002;1:35–40. doi: 10.1021/pr015506a. [DOI] [PubMed] [Google Scholar]
- Skorey K. Waddleton D. Therien M. Leriche T. Anal. Biochem. 2006;349:49–61. doi: 10.1016/j.ab.2005.11.018. [DOI] [PubMed] [Google Scholar]
- Lowther W. T. Matthews B. W. Biochim. Biophys. Acta. 2000;1477:157–167. doi: 10.1016/S0167-4838(99)00271-X. [DOI] [PubMed] [Google Scholar]
- Qiu W. W. Xu J. Li J. Y. Li J. Nan F. J. ChemBioChem. 2007;8:1351–1358. doi: 10.1002/cbic.200700148. [DOI] [PubMed] [Google Scholar]
- Chee G. L. Yalowich J. C. Bodner A. Wu X. Hasinoff B. B. Bioorg. Med. Chem. 2010;18:830–838. doi: 10.1016/j.bmc.2009.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannappan R. Ando M. Furuhata K. Uda Y. Biol. Pharm. Bull. 2008;31:352–356. doi: 10.1248/bpb.31.352. [DOI] [PubMed] [Google Scholar]
- Rodriguez A. C. Kohler J. J. Med. Chem. Commun. 2014;5:1227–1234. doi: 10.1039/C4MD00164H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik R. Won M. Ban H. S. Bhattarai D. Xu X. Eo Y. Hong Y. S. Singh S. Choi Y. Ahn H. C. Lee K. J. Med. Chem. 2014;57:9522–9538. doi: 10.1021/jm501241g. [DOI] [PubMed] [Google Scholar]
- Bongo N. B. Tomohiro T. Hatanaka Y. Bioorg. Med. Chem. Lett. 2009;19:80–82. doi: 10.1016/j.bmcl.2008.11.013. [DOI] [PubMed] [Google Scholar]