Abstract
The chromosome segregation 1-like (CSE1L) protein, which regulates cellular mitosis and apoptosis, was previously found to be overexpressed in colorectal cancer (CRC) cells harboring mutations. Therefore, regulating CSE1L expression may confer chemotherapeutic effects against CRC. The gut microflora can regulate gene expression in colonic cells. In particular, metabolites produced by the gut microflora, including the short-chain fatty acid butyrate, have been shown to reduce CRC risk. Butyrates may exert antioncogenic potential in CRC cells by modulating p53 expression. The present study evaluated the association between CSE1L expression and butyrate treatment from two non-transformed colon cell lines (CCD-18Co and FHC) and six CRC cell lines (LS 174T, HCT116 p53+/+, HCT116 p53−/−, Caco-2, SW480 and SW620). Lentiviral knockdown of CSE1L and p53, reverse transcription-quantitative PCR (CSE1L, c-Myc and p53), western blotting [CSE1L, p53, cyclin (CCN) A2, CCNB2 and CCND1], wound healing assay (cell migration), flow cytometry (cell cycle analysis) and immunofluorescence staining (CSE1L and tubulin) were adopted to verify the effects of butyrate on CSE1L-expressing CRC cells. The butyrate-producing gut bacteria Butyricicoccus pullicaecorum was administered to mice with 1,2-dimethylhydrazine-induced colon tumors before the measurement of CSE1L expression. The effects of B. pullicaecorum on CSE1L expression were then assessed by immunohistochemical staining for CSE1L and p53 in tissues from CRC-bearing mice. Non-cancerous colon cells with the R273H p53 mutation or CRC cells haboring p53 mutations were found to exhibit significantly higher CSE1L expression levels. CSE1L knockdown in HCT116 p53−/− cells resulted in G1-and G2/M-phase cell cycle arrest. Furthermore, in HCT116 p53−/− cells, CSE1L expression was already high at interphase, increased at prophase, peaked during metaphase before declining at cytokinesis but remained relatively high compared with that in HCT116 expressing wild-type p53. Significantly decreased expression levels of CSE1L were also observed in HCT116 p53−/− cells that were treated with butyrate for 24 h. In addition, the migration of HCT116 p53−/− cells was significantly decreased after CSE1L knockdown or butyrate treatment. Tumors with more intense nuclear p53 staining and weaker CSE1L staining were found in mice bearing DMH/DSS-induced CRC that were administered with B. pullicaecorum. Taken together, the results indicated that butyrate can impair CSE1L-induced tumorigenic potential. In conclusion, butyrate-producing microbes, such as B. pullicaecorum, may reverse the genetic distortion caused by p53 mutations in CRC by regulating CSE1L expression levels.
Keywords: colorectal cancer, chromosome segregation 1-like, butyricicoccus pullicaecorum, short-chain fatty acid, p53 tumor suppressor
Introduction
The overexpression of chromosome segregation 1-like (CSE1L), also known as cellular apoptosis susceptibility protein, has been previously reported to correlate positively with the progression of various malignacies, such as gastric cancer and colorectal cancer (CRC) (1–5). However, this CSE1L-induced risk of CRC tumorigenisis can be suppressed by the activation of wild-type p53 expression (6). It has been also reported that different chemotherapeutic agents, including 5-fluorouracil, cisplatin, and paclitaxel, can mediate differential apoptotic effects in CSE1L-overexpressing CRC cancer cells (7,8). The therapeutic efficacy of drugs against CRC can be reduced through the suppression of paclitaxel-induced apoptosis in CSE1L-expressing CRC cells (8,9). Therefore, CSE1L knockdown may improve CRC treatment (8).
Gut microbes can regulate the gene expression profile in colonic cells, which may in turn alter the course of CRC (10,11). Short-chain fatty acids (SCFAs) derived from microbial metabolism in the gut, including acetate, propionate and butyrate, are key for the maintenance of intestinal homeostasis (12,13). SCFAs can induce cell apoptosis and cell cycle arrest to reduce cancer risk (14), rendering them to be potential chemotherapeutic agents (15). Butyrate-producing microorganisms in the gut have been reported to prevent necrotic enteritis and reduce pathogen abundance (16–18). Therefore, dysbiosis caused by the overpopulation of detrimental microbes and underpopulation of beneficial butyrate-producing microbes in the gut may confer clinical significance in CRC. However, the effects of butyrate in CRC and the molecular mechanism underlying such effects remain unclear (19). Butyrate has been previously documented to downregulate the expression of a number of genes, including placenta specific 8 protein, toll-like receptor 4 and glucose 6-phosphatase (10,20–22). It can attenuate the lipopolysaccharide-induced inflammation of intestinal epithelial cells whilst exerting antioncogenic potential in LS1034 or WiDr human CRC cells by promoting p53 gene expression (23–25). In particular, patients with inflammatory bowel disease or CRC were found to have lower concentrations of the butyrate-producing microbe butyricicoccus pullicaecorum in their stools (10,26). In addition, the culture supernatant of B. pullicaecorum is rich in butyrates and can strengthen intestinal barrier function (17,26), which supports the pharmabiotic potential B. pullicaecorum for clinical application (10,20,27,28). However, the possible association between CSE1L and/or the butyrate-producing B. pullicaecorum in the development of CRC remain poorly understood.
A previous study has indicated that suppression of CSE1L expression in CRC cells may improve CRC treatment (8). Butyrate-producing microorganisms in the gut have been reported to confer potentially anti-CRC effects (17). Accordingly, a possible strategy to alter CRC physiology would be to decrease CSE1L expression in CRC cells by either B. pullicaecorum administration or butyrate supplementation. However, information concerning the potential effects remain unavailable. Therefore, the aim of the present study was to explore the potential role of butyrate in the molecular events mediated by CSE1L in CRC cell lines with different p53 genotypes. Knockdown of CSE1L in HCT116 p53−/− cells, knockdown of p53 in HCT116 p53+/+ and CRC cell lines with very distinct differences in the p53 status were applied to evaluate the effects of CSE1L on the expression levels of wild-type p53. To examine the molecular significance of butyrate supplementation on CSE1L expression and CRC alleviation, cellular physiology of buyrate-treated HCT116 p53−/− cells in vitro and the colon tumors from mice after B. pullicaecorum administration in vivo were used. In this manner, the potential role of B. pullicaecorum and CSE1L in CRC were investigated and clarified.
Materials and methods
Induction of CRC in mice
A total of 17 male BALB/cByJNarl mice aged 4–6 weeks, weighing 22.7±0.6 g, were provided by the National Laboratory Animal Center (National Applied Research Laboratories, Taipei, Taiwan). All animal experiments were conducted in compliance with ARRIVE (Animal Research: Reporting of In Vivo Experiments) guidelines (29). The protocols followed the principles of Reduction, Refinement and Replacement and were approved by the Institutional Animal Care and Use Committees of Cathay General Hospital (approval no. 107-008; Taipei, Taiwan). Mice (at n=3–5 per plastic cage) were housed in an individually ventilated cage rack system (Tecniplast Group) and had free access to food and drinking water under the following conditions: 50±10% humidity, 12-h light/dark cycle and 23±2°C temperature. All efforts were made to minimize the number of mice and their suffering. The mice were classified into the following groups as previously described (17): i) Control group (n=5), consisting of mice that did not receive any chemical or B. pullicaecorum administration; ii) 1,2-dimethylhydrazine (DMH; cat. no. D0741; Tokyo Chemical Industry Co., Ltd.)/dextran sulphate sodium (DSS; cat. no. D5144; Tokyo Chemical Industry Co., Ltd.) group (n=6), consisting of mice that received DMH (20 mg/kg) once at the beginning of the experiment through intraperitoneal injection, followed by 1 week of normal water and 1 week of DSS (30 g/l) in the drinking water, with two cycles of additional DSS treatment (2 weeks of normal water + 1 week of DSS (30 g/l) in drinking water); and iii) DMH/DSS/B. pullicaecorum group (n=6), consisting of mice that received DMH/DSS in the same manner as DMH/DSS group, but were treated with B. pullicaecorum every 7 days during the experiment. The body weight of each mouse was monitored once a week. All mice were euthanized with CO2 in a cage when they showed weakness and rapid weight loss of 15–20% or at the end of the experiment. The duration of this animal experiment was 2–3 months. The CO2 flow rate was set to displace 30% of the cage volume per minute. Immobility for >2 min and lack of spontaneous breathing were used to confirm animal death before the colon samples were collected.
B. pullicaecorum administration by oral gavage
The molecular effects of B. pullicaecorum on colon tumor formation was evaluated. B. pullicaecorum (3.125×107 colony-forming units in 100 µl of modified peptone yeast extract broth) was administered by oral gavage. B. pullicaecorum (cat. no. BCRC-81109; https://catalog.bcrc.firdi.org.tw/BcrcContent?bid=81109) and the growth medium (modified peptone yeast extract broth; cat. no. 967; Bioresource Collection and Research Center) were purchased from the Bioresource Collection and Research Center (Hsinchu, Taiwan) and cultured for 3 days under anaerobic conditions (10% CO2 and 90% N2) at 37°C as described previously (17).
Cell lines and reagents
In total, two human colon cell lines, CCD-18Co [cat. no. CRL-1459; American Type Culture Collection (ATCC)], which harbors wild-type p53 and FHC (cat. no. CRL-1831; ATCC), which expresses the R273H p53 mutant (30), were acquired as non-transformed colon cells (30,31). In addition, three human CRC cell lines (LS 174T, cat. no. CL-188; T84, cat. no. CCL-248; HCT116 p53+/+, cat. no. CCL-247; all from ATCC) expressing wild-type p53, two human CRC cell lines (SW480, cat. no. CCL-228; SW620, cat. no. CCL-227; both from ATCC) carrying the R273H and P309S double p53 mutation (32), in addition to two p53-null cell lines [Caco-2, cat. no. HTB-37, ATCC; HCT116 p53−/−, a gift from Professor Bert Vogelstein (School of Medicine, Johns Hopkins University, Baltimore, USA)] (33,34), were used as tumorigenic cancer cells (35–38). These cell lines were expanded in complete media [medium suggested for each cell line by the ATCC, 10% FBS (Asia Bioscience Co., Ltd.) and 1X antibiotic/antimycotic solution (cat. no. 15240-062; Thermo Fisher Scientific, Inc.)] in a humidified chamber with 95% air and 5% CO2 at 37°C with some exceptions. Briefly, four cell lines (FHC, T84, HCT116 p53+/+ and HCT116 p53−/−) were cultured with DMEM (cat. no. 12800-017; Thermo Fisher Scientific, Inc.) whereas three cell lines (CCD-18Co, LS 174T and Caco-2) were cultured with MEM (cat. no. 41500-034; Thermo Fisher Scientific). In addition, SW480 and SW620 cell lines were cultured with Leibovitz's L-15 medium (cat. no. 11415-064; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS and 1X antibiotic/antimycotic solution, but maintained under 100% atmospheric air (without CO2) in a humidified incubator at 37°C.
To measure the expression of CSE1L in cells after treatment with 5-fluouracil (5-FU; cat. no. F6627; Merck KGaA) or sodium buyrate (NaB; cat. no. B5887; Merck KGaA), a total of 5×105 cells were cultured for 24 h at 37°C and before chemicals were added as follows: HCT116 p53+/+ cells with 5-FU (40 µM) for 24 h at 37°C, whereas HCT116 p53−/−, SW480 and SW620 cells with NaB (5 mM) for 24 or 48 h at 37°C.
To differentiate Caco-2 cells into a polarized enterocyte-like monolayer, cells were seeded at 8×105 cells per well and cultured to confluence for 21 days at 37°C, with changes of fresh MEM supplemented with 20% FBS and 1% antibiotic/antimycotic solution every 1–2 days (39,40).
Lentiviral knockdown of p53 and CSE1L
All lentiviral particles were obtained from the RNA Technology Platform and Gene Manipulation Core (https://rnai.genmed.sinica.edu.tw/index.html). Briefly, lentiviral particles were packaged in 293T cells (cat. no. CRL-3216; ATCC) using the 2nd generation system, with the combined ratio of lentiviral construct, packaging plasmid and envelope plasmid at 1 µg: 900 ng:100 ng. The 293T cells were then cultured in DMEM supplemented with 10% FBS and 0.1X antibiotic/antimycotic solution in a humidified chamber with 95% air and 5% CO2 at 37°C for 40 h. The cultured media were spun (300 x g for 5 min) to remove any packaging cells and supernatant containing viral paricles were collected. In total, two lentiviral constructs, namely pLKO.1_ p53 (clone ID: TRCN0000003753) encoding a short hairpin RNA (shRNA) targeting p53 (shp53) and pLKO.1_CSE1L (clone ID: TRCN0000061789) targeting CSE1L (shCSE1L), were used for p53 knockdown in HCT116 p53+/+ cells and CSE1L knockdown in HCT116 p53−/− cells. The control pLKO.1-luciferase (Luc; clone ID: TRCN0000072249) vector targeting Luc was used as the negative control (shLuc-HCT116 p53+/+ for shp53 or shLuc-HCT116 p53−/− for shCSE1L). A total of 1.25×105 cells/well were cultured in six-well plates for 24 h at 37°C, before subsequent lentiviral infections (multiplicity of infection, 3) were performed to knock down the expression of target genes in the cells. Subsequently, medium containing 2 mg/ml puromycin (Thermo Fisher Scientific, Inc.) was used to select and maintain the stable clones. After a 48-h incubation at 37°C, transfection efficiency was determined using reverse transcription-quantitative PCR (RT-qPCR).
RNA isolation, cDNA synthesis and gene quantitation
Total RNA was extracted from the parental CRC cell lines (CCD-18Co, FHC, LS 174T, T84, Caco-2, HCT116 p53+/+, HCT116 p53−/−, SW480 and SW620) and their derived cells, using the Easy Pure Total RNA Spin kit (cat. no. RT050; Bioman Scientific, Co., Ltd.) according to the manufacturer's protocol. Single-stranded cDNA was generated from 1 µg total RNA in the presence of an oligo (dT)12 primer using the High-Capacity cDNA Reverse Transcription kit (cat. no. 4368813; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. mRNA expression levels were quantified through qPCR using the LightCycler® TaqMan Master mix (cat. no. 04535286001; Roche Diagnostics GmbH) with a specific thermocycling profile (95°C for 10 min, followed by 50 cycles at 95°C for 10 sec and 60°C for 20 sec) as described in a previous study (10,17). Primer sequences and probe numbers were CSE1L (Universal Probe: #27): forward, 5′-GTTGTCTACCGCCTGTCCA-3′ and reverse, 5′-AAATGCAGTTTAAAGCAGTGTCA-3′; c-Myc (Universal Probe: #34): forward, 5′-CACCAGCAGCGACTCTGA-3′ and reverse, 5′-ACTCTGACCTTTTGCCAGGA-3′; p53 (Universal Probe: #12): forward, 5′-AGGCCTTGGAACTCAAGGAT-3′ and reverse, 5′-CCCTTTTTGGACTTCAGGTG-3′ and GAPDH (Universal Probe: #60): forward, 5′-CTCTGCTCCTCCTGTTCGAC-3′ and reverse 5′-ACGACCAAATCCGTTGACTC-3′. Expression levels were quantified using the 2−ΔΔCq method and normalized to the expression level of GAPDH (41). The human reference cDNA (HRC; cat. no. 636692; Takara Bio, Inc.) was used as an expression control. Gene expression data were obtained after performing ≥ three independent experiments with similar results.
Preparation of whole cell extracts and nuclear/cytosol fractions for western blotting
Whole-cell extracts from shLuc-HCT116 p53−/− and shCSE1L-HCT116 p53−/− cells were prepared using PRO-PREP Protein Extraction Solution (Intron Biotechnology, Inc.) in the presence of a protease inhibitor (cat. no. P8340; Merck KGaA) according to the manufacturer's protocols. Cell fractionation was performed using NE-PER Nuclear and Cytoplasmic Extraction Reagents kit (cat. no. 78833; Thermo Fisher Scientific, Inc.) to isolate the different protein fractions from the cytoplasm and nuclei, according to the manufacturer's protocols. The purity of non-nuclear and nuclear fractions was determined using specific protein markers, namely Tubulin and TATA-box binding protein (TBP), respectively. Each protein concentration was then determined using a Bio-Rad Protein Assay reagent (cat. no. 500-0006; Bio-Rad Laboratories, Inc.). Next, 30 µg of protein per lane was denatured at 95°C for 10 min, separated using 12% SDS-PAGE in 1X NuPAGE LDS Sample Buffer (Thermo Fisher Scientific, Inc.) and then transferred onto 0.2-µm PolyScreen 2 PVDF Transfer membranes (PerkinElmer, Inc.). The membranes were blocked with 3% bovine serum albumin (cat. no. ALB001.100; BioShop Canada, Inc.) for 1 h at room temperature and incubated with the following primary antibodies for 1 h at room temperature: Anti-CSE1L (1:1,000; cat. no. 22219-1-AP; Proteintech Group, Inc.), anti-p53 (1:500; cat. no. NCL-L-p53-DO7; Leica Biosystems, Inc.), anti-cyclin A2 (CCNA2; 1:2,000; cat. no. 4656P; Cell Signaling Technology, Inc.), anti-cyclin B2 (CCNB2; 1:2,000; cat. no. ab185622; Abcam), anti-cyclin D1 (CCND1; 1:1,000; cat. no. 2978; Cell Signaling Technology, Inc.), anti-tubulin (1:1,000; cat. no. sc-5286, Santa Cruz Biotechnology, Inc.), anti-TBP (1:1,000; cat. no. 22006-1-AP, Proteintech Group, Inc.) and anti-GAPDH (1:5,000; cat. no. 60004-1-Ig; Proteintech Group, Inc.). Expression of GAPDH was used as the endogenous control gene. After incubation of the primary antibodies, the membranes were incubated with a HRP-conjugated anti-mouse IgG (H&L) secondary antibody (1:5,000; cat. no. ab6808; Abcam) HRP-conjugated antirabbit IgG secondary antibody (1:5,000; cat. no. L3012; Signalway Antibody LLC) for 60 min at room temperature. Protein bands were visualized using Western Lightning Chemiluminescence Reagent Plus (PerkinElmer, Inc.) and an AlphaView software (version 3.2.2) of the FluorChem FC2 Imaging System (Cell Biosciences, Inc.) according to the manufacturers' protocols (Alpha Innotech FluorChem FC2 Imaging System; Cell Biosciences, Inc.).
Cell cycle analysis and immunofluorescent staining
shLuc-HCT116 p53−/− cells and shCSE1L HCT116 p53−/− cells were starved under low-serum conditions (0.5%) for 24 h at 37°C and then cultured in complete medium for 24 h at 37°C. They were then fixed in 70% prechilled ethanol for >1 h at −20°C, washed twice with PBS, incubated with 1 µg/ml RNase A for 1 h at 37°C and stained with 5 µg/ml propidium iodide for 1 h at room temperature. Light emission at 585 nm from propidium iodide-stained nuclei was detected using a BD FACScan flow cytometer (BD Bioscienes). The percentages of cells (from 1×104 cells) at different phases of cell cycle were determined using FlowJo software (v. 8.7; FlowJo LLC).
A total of 1.5×104 HCT116 p53−/− cells and HCT116 p53+/+ cells for immunofluorescence staining were cultured on 12-mm cover slips (SPL Life Sciences). The cells were probed with a diluted anti-CSE1L antibody (1:50; cat. no. 22219-1-AP; Proteintech Group, Inc.) or anti-Tubulin antibody (1:50; cat. no. sc-5286; Santa Cruz Biotechnology, Inc.) for 16 h at 4°C after fixation with 4% paraformaldehyde (Merck KGaA) in PBS for 10 min at room temperature, permeabilization with 0.1% Triton X-100 (Merck KGaA) in PBS for 35 min at room temperature, and blocking with 1.5% normal horse serum blocking solution (cat. no. S-2000-20; Vector Laboratories, Inc.; Maravai LifeSciences) in 10 ml PBS for 30 min at room temperature. Next, the FITC-conjugated secondary antibody (1:200; cat. no. AP132F; Merck KGaA) for CSE1L and the Cy3-conjugated secondary antibody (1:200; cat. no. AP124C; Merck KGaA) for α-tubulin were incubated for 1 h at room temperature. Nuclear DNA was stained with 1 µg/ml DAPI (cat. no. 71-03-01; Kirkegaard & Perry Laboratories Inc.) for 15 min at room temperature. The stained samples were dehydrated through an ascending ethanol series and air-dried for 10 min at room temperature before being mounted in VECTASHIELD® HardSet™ Antifade Mounting Medium (cat. no. H-1400; Vector Laboratories, Inc.; Maravai LifeSciences). They were then observed using a Nikon Eclipse 80i fluorescence microscope at ×200 magnification (Nikon Corporation) before fluorescence was quantified from > five fields of views (10 views for HCT116 p53+/+ cells and five views for HCT116 p53−/− cells) using Adobe Photoshop (version CS6; Adobe Systems, Inc.).
Immunohistochemical staining for mouse tissues
Mouse colorectal samples were fixed with 4% paraformaldehyde in PBS for 10 min at room temperture and embedded in paraffin. Paraffin sections (3–5 µm thickness) were obtained and then processed using the avidin-biotin-immunoperox-idase method to measure the expression of CSE1L and p53. Immunohistochemical staining was performed on an auto-mated BenchMark GX slide stainer (Roche Diagnostics) in a closed and fixed program, which included deparaffinization with EZ Prep solution (cat. no. 950-102; Ventana Medical Systems) at 75°C for 8 min, antigen retrieval with Cell Conditioning 1 solution (cat. no. 950-124; Ventana Medical Systems) at 95°C for 64 min, incubation with primary anti-body (at 37°C for 32 min) followed by HQ Universal Linker (cat. no. 253-4580; Ventana Medical Systems) at 37°C for 8 min and HRP Multimer (cat. no. 253-4581; Ventana Medical Systems) at 37°C for 8 min and visualization by DAB. The Optiview DAB IHC detection kit (cat. no. 760-700; Roche Diagnostics) was used as a detection system. All sections were counterstained with Hematoxylin II at 25°C for 8 min (cat. no. 790-2208; Ventana Medical Systems) and Bluing Reagent at 25°C for 4 min (cat. no. 760-2037; Ventana Medical Systems). Anti-CSE1L (1:50; cat. no. 22219-1-AP; Proteintech Group, Inc.) or anti-p53 (1:50; cat. no. BS-8687R, Thermo Fisher Scientific, Inc.) were hybridized to detect target protein. A pathologist (CYL) observed and categorized the immunohistochemically stained sections using a Nikon Eclipse 80i fluorescence microscope at ×200 magnification by light microscopy (Nikon Corporation).
Cell migration assay
The shLuc-HCT116 p53−/− and shCSE1L-HCT116 p53−/− cells were grown to confluence on six-well plates before a wound was made by scraping across the cell monolayer with a 30 gauge needle (outer diameter, 300 µm). The motility of the cells at the edge of a scratch wound in the presence or absence of NaB (5 mM) was then analyzed. Cells at the wound edge were imaged using a bright-field/phase-contrast microscope at ×200 magnification. Repeat images were taken after wounding in medium with low serum (DMEM, 1% FBS and 1% antibiotic/antimyotic solution) at 37°C for 16 h and followed with complete media for indicated time (0, 4 and 8 h). Serum-free media was first tested for this assay but HCT p53−/− cells could not survive in this condition, which necesitated the use of 1% for maintenance followed by complete medium (10% FBS) for the assay. It is predicted that the extent of interference due to cell proliferation would be minimal, as the doubling time of HCT116 cells is ~18 h and the maximum duration of the wound healing assay in the present study was 8 h. ImageJ (version 1.45s; National Institutes of Health) was used to measure the migration distance at each time point (42). Next, the cell migration efficiency after 8 h of cultivation was assessed using the recovery ratio according to the reduction of wound area (the percentage of cell area difference, relative to the inititial time point of 0 h) (43). In total, three or four independent sets of experiments were performed for each assay.
Statistical analysis
The difference in gene expression, cell phase and cell migration between the groups was assessed. A unpaired student's t test was used to compare two groups whereas one-way analysis of variance was performed to compare among ≥ three different groups. All ANOVA analyses were followed with Bonferroni post hoc testing. The statistical analyses were performed using SPSS (v. 22.0; IBM Corp). Data are presented as the mean ± standard error of the mean. P<0.05 was considered to indicate a statistically significant difference.
Results
CSE1L mRNA expression levels in the different colonic and CRC cell lines
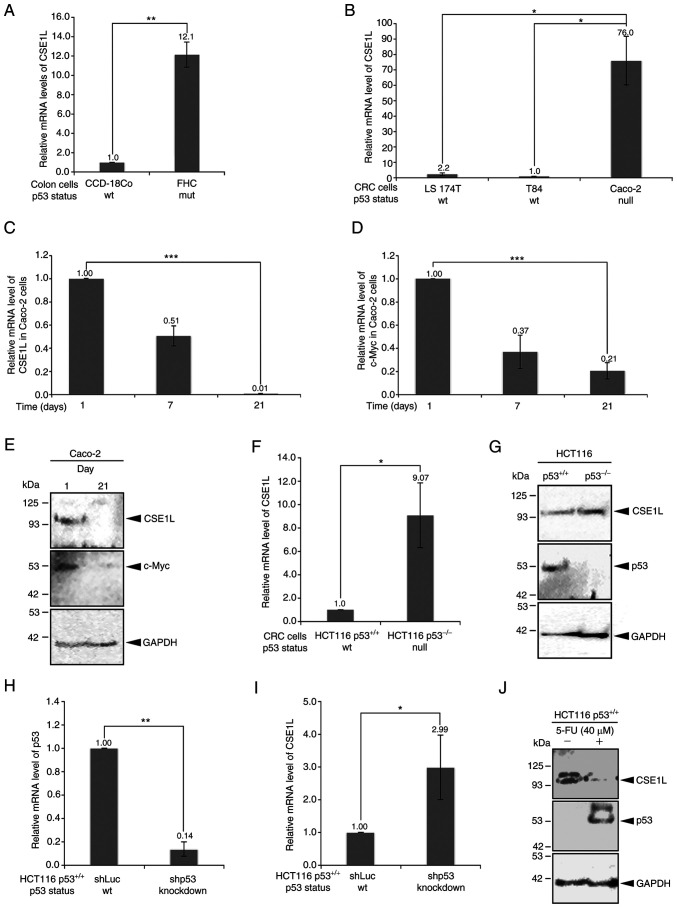
The expression levels of CSE1L in different colonic and CRC cell lines were quantified (Fig. 1). CSE1L expression levels in the two non-transformed cell lines, CCD-18Co and FHC, varied significantly according to their different p53 mutation statuses (Fig. 1A). Briefly, the CCD-18Co cells with the wild-type p53 expressed the lower levels of CSE1L compared with those in FHC cells with the R273H p53 mutant. Among the CRC cell lines, Caco-2 cells haboring p53 mutations exhibited significantly higher CSE1L expression levels compared with those in LS 174T and T84 cells, both of which express wild-type p53 (Fig. 1B). However, CSE1L and c-Myc mRNA expression levels both simulataneously and progressively reduced in Caco-2 cells as their confluency increased (Fig. 1C and D). In addition, the protein expression levels of CSE1L and c-Myc were markedly decreased in Caco-2 cells following proliferation to confluence on day 21 compared with those in cells on day 1 (Fig. 1E).
Figure 1.
Different CSE1L expression levels in the colon cell lines. (A) Relative mRNA expression levels of CSE1L in CCD-18Co and FHC cells. (B) Relative mRNA expression levels of CSE1L in the CRC cell lines. (C) Relative mRNA expression levels of CSE1L in overconfluent Caco-2 cells. (D) Relative mRNA expression levels of c-Myc in overconfluent Caco-2 cells. (E) Protein expression level of CSE1L and c-Myc in overconfluent Caco-2 cells. (F) Relative mRNA expression levels of CSE1L in HCT116 cells with or without p53 expression. (G) Protein expression level of CSE1L in HCT116 cells with or without p53 expression. (H) Knockdown efficacy of p53 by mRNA level in HCT116 p53+/+ cells. (I) Relative mRNA expression levels of CSE1L in HCT116 cells without or with p53 knockdown. (J) Protein expression levels of CSE1L in 5-FU-treated wild-type HCT116 cells. *P<0.05, **P<0.01 and ***P<0.001. CRC, colorectal cancer; 5-FU, 5-fluouracil; shLuc, lentiviral construct targeting luciferase; shp53, lentiviral construct targeting p53; wt, wild-type; sh, short hairpin; CSE1L, chromo-some segregation 1-like protein.
Higher CSE1L protein expression levels were also observed in HCT116 cells not expressing p53 (Fig. 1F and G) or in HCT116 cells following p53 knockdown (Fig. 1H and I). Compared with those in HCT116 p53+/+ cells, either mRNA (Fig. 1F) or protein (Fig. 1G) expression levels of CSE1L were markedly higher in HCT116 p53−/− cells. This differential expression was also observed in HCT116 p53+/+ cells with p53 expression knocked down. After p53 was significantly knocked down in HCT116 p53+/+ cells compared with that in shLuc-transfected cells (Fig. 1H), the expression level of CSE1L mRNA was also significantly increased (Fig. 1I). A similar finding could also be made on p53 protein expression in HCT116 p53+/+ cells after 5-FU (40 µM) treatment for 24 h, which was increased (Fig. 1J). In addition, the expression of CSE1L was markedly reduced in the 5-FU-treated HCT116 p53+/+ cells compared with that in their untreated counterparts (Fig. 1J).
Cell cycle regulation of p53-null CRC cells by CSE1L expression
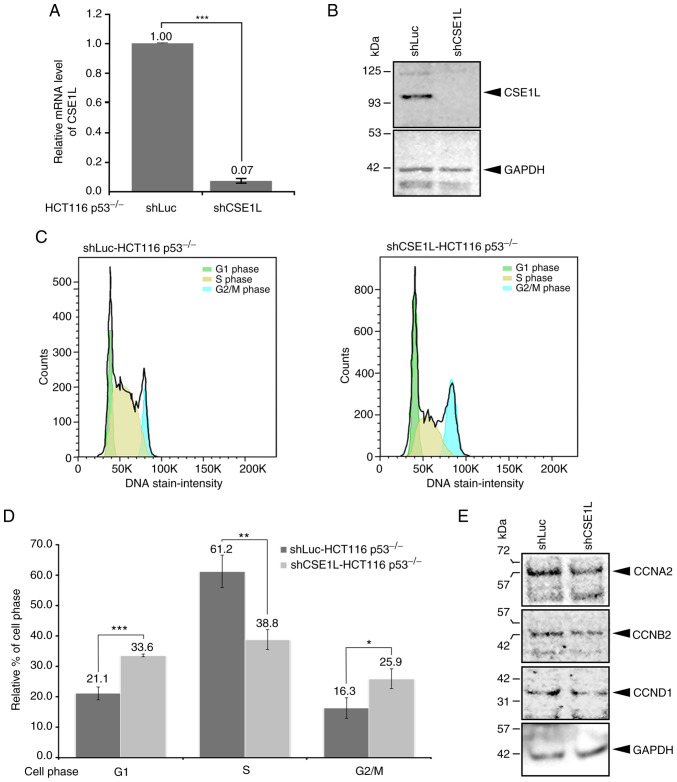
To understand the molecular significance of CSE1L expression in CRC cells, CSE1L expression was knocked down in HCT116 p53−/− cells, which was achieved to significant levels compared with that in the shLuc-HCT116 p53−/− cells (Fig. 2A). In addition, shCSE1L-HCT116 p53−/− cells expressed markedly lower expression levels of CSE1L protein (Fig. 2B). Compared with those in shLuc-HCT116 p53−/− cells, the cell populations in various phases of cell the cycle were altered in the shCSE1L-HCT116 p53−/− cells (Fig. 2C). Specifically, the percentage of shCSE1L-HCT116 p53−/− cells in S phase was significantly decreased, whereas that in the G1 and G2/M phases was significantly increased, compared with those of shLuc-HCT116 p53−/− cells (Fig. 2C and D). Supporting this, shCSE1L-HCT116 p53−/− cells also expressed markedly lower protein levels of cell cycle regulators CCNA2, CCNB2 and CCND1 compared with those in shLuc-HCT116 p53−/− cells (Fig. 2E).
Figure 2.
Cellular changes in HCT116 p53−/− cells after knocking down CSE1L expression. (A) Knockdown efficacy of CSE1L in HCT116 p53−/− cells. (B) Protein expression levels of CSE1L in HCT116 p53−/− cells without or with CSE1L knockdown. (C) Population of HCT116 p53−/− cells in the various phases of cell cycle without or with CSE1L knockdown. (D) Percentages of shCSE1L-HCT116 p53−/− cells in the various phases of cell cycle without or with knockdown of CSE1L expression were quantified. (E) Protein expression levels of CCNA2, CCNB2 and CCND1 in HCT116 p53−/− cells without or with CSE1L knockdown. *P<0.05, **P<0.01 and ***P<0.001. HCT116 p53−/−, p53-null HCT116 cells. shLuc, lentiviral construct targeting luciferase; shCSE1L, lentiviral construct targeting CSE1L; sh, short hairpin; CSE1L, chromosome segregation 1-like protein; CCNA2, cyclin A2; CCNB2, cyclin B2; CCND1, cyclin D1.
Dynamic expression of CSE1L in HCT116 CRC cells during mitosis
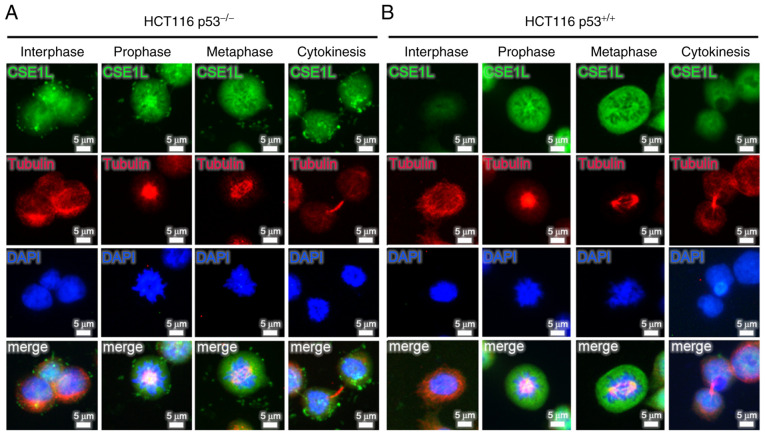
Analysis of CSE1L expression during different phases of mitosis in HCT116 cells revealed that expression profile of CSE1L changed dynamically throughout mitosis (Fig. 3). Both HCT116 p53−/− cells and HCT116 p53+/+ cells expressed high levels of CSE1L at prophase and metaphase. However, the signals for CSE1L in HCT116 p53−/− cells were stronger compared with those in HCT116 p53+/+ cells during interphase and cytokinesis. As shown in Fig. 3A for HCT116 p53−/− cells, CSE1L emerged at interphase, increased at prophase, peaked during metaphase before declining at the cytokinesis stage. The dynamic expression profiles of CSE1L in HCT116 p53−/− cells during mitosis was subsequently analyzed, with the highest levels of CSE1L expression found at prophase and metaphase (Fig. S1). By contrast, as shown in Fig. 3B, low levels of CSE1L expression were detected during interphase and cytokinesis in HCT116 p53+/+ cells, which increased markedly at prophase before peaking at metaphase.
Figure 3.
Dynamic expression profiles of CSE1L throughout the mitotic phase of the cell cycle in HCT116 cells of different p53 statuses. Immunofluorescence microscopy was used to evaluate the localization of endogenously expressed CSE1L in separate (A) HCT116 p53+/+ cells and (B) HCT116 p53−/− cells at distinct phases of mitosis. CSE1L are presented in green. DAPI (nuclear DNA) is presented in blue. Tubulin is presented in red. Merge represents CSE1L + Tubulin + DAPI. Scale bar, 5 µm. HCT116 p53−/−, p53-null HCT116 cells; CSE1L, chromosome segregation 1-like protein.
Reduced CSE1L expression in butyrate-treated HCT116 p53−/− cells and colon tumors in mice treated with B. pullicaecorum administration
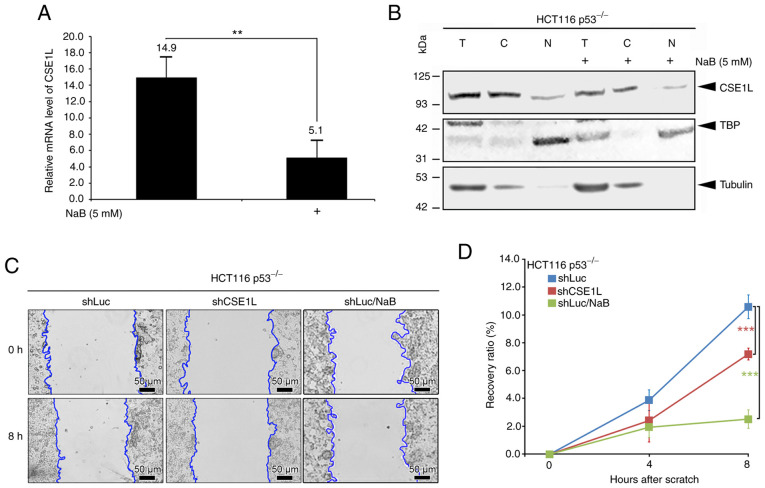
HCT116 p53−/− cells treated with 5 mM NaB for 24 h exhibited lower expression levels of of both CSE1L mRNA (Fig. 4A) and protein in the total cell lysate (Fig. 4B) compared with those in cells not treated with NaB. In addition, NaB treatment reduced the mRNA expression of CSE1L in both the SW480 and SW620 cell lines (with the p53 mutant) after 24 and 48 h (Fig. S2). In the cytosolic and nuclear compartments of HCT116 p53−/− cells, the expression levels of CSE1L also decreased as a result of 5 mM NaB treatment (Fig. 4B). Furthermore, the recovery ratio in the migration of shCSE1L-HCT116 p53−/− or NaB-treated-shLuc-HCT116 p53−/− cells was significantly decreased compared with that in the control shLuc-HCT116 p53−/− cells (Fig. 4C and D).
Figure 4.
Effects of butyrate on CSE1L expression in HCT116 p53−/− cells. (A) Relative mRNA expression levels of CSE1L in cells without or with butyrate treatment. (B) Protein expression levels of CSE1L in the nucleus or cytosol of cells following butyrate treatment. (C) Migration changes and (D) Recovery ratios of cells without or with CSE1L knockdown or butyrate treatment. Blue line presents the edge of cell migration. Scale bars, 50 µm. **P<0.01 and ***P<0.001. CSE1L, chromosome segregation 1-like protein; TBP, TATA-binding protein; NaB, sodium butyrate; T, total cell lysate; C, cytosolic part; N, nuclear part; shLuc, lentiviral construct targeting luciferase; shCSE1L, lentiviral construct targeting CSE1L; sh, short hairpin.
CSE1L expression in colon tumors of mice with B. pullicaecorum administration
H&E staining and immunohistochemical analysis of p53 and CSE1L expression was performed in the mouse intestinal tissues. Reactivity was almost absent in control healthy intestinal tissues (Fig. 5). Compared with those in the control mice, colon tumors were induced in mice by DMH/DSS treatment (Fig. 5). In the colons of mice following DMH/DSS treatment without B. pullicaecorum administration, histological sections showed exophytic tumors exhibiting irregular and complex dysplastic glands, indicating intramucosal adenocarcinoma (red arrows in the middle panel of Fig. 5). Weak nuclear staining of p53 and increased expression of CSE1L were observed in large tumors with high-grade dysplasia (Fig. 5). By contrast, in mouse tissues treated with B. pullicaecorum, the histological sections revealed polypoid lesions consisting mostly of low-grade adenomas, representing the early stage of neoplasia (red arrows in the right panel of Fig. 5). Immunohistochemical analysis showed positive nuclear staining for p53 and a low intensity CSE1L signal were detected in precancerous tumors with low-grade dysplasia from DMH/DSS-treated mice that were administered with butyrate-producing B. pullicaecorum through oral gavage.
Figure 5.
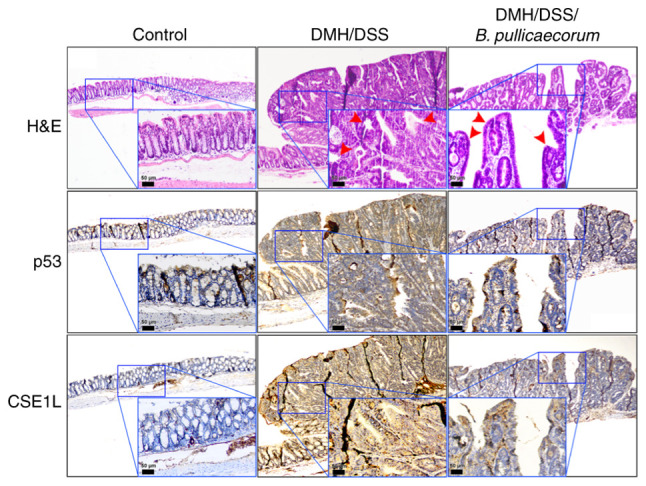
Representative immunohistochemical staining images for the expression of p53 and CSE1L proteins in the colon tissues. Colon tissues were sectioned from the following different groups: Control group, consisting of mice that did not receive any chemical treatment or B. pullicaecorum admin-istration; DMH/DSS group, consisting of mice that received DMH through intraperitoneal injection and DSS in their drinking water but did not receive B. pullicaecorum; and DMH/DSS/B. pullicaecorum group, consisting of mice that received DMH/DSS and B. pullicaecorum. Colon tumors exhibited weak nuclear staining of p53 and markedly increased expression of CSE1L. Insets show the magnified views of the boxed areas. Red arrows indicate the intramucosal adenocarcinoma in DMH/DSS group (middle panel) and the low-grade adenoma in DMH/DSS/B. pullicaecorum group (right panel). Scale bar, 50 µm for inset. CSE1L, chromosome segregation 1-like protein; DMH, 1,2-dimethylhydrazine; DSS, dextran sulphate sodium; B. pullicaecorum, Butyricicoccus pullicaecorum.
Discussion
CSE1L overexpression was previously found to associate with the progression of a number of gastrointestinal cancers, including esophageal cancer, gastric cancer, hepatocellular carcinoma and CRC (1,2,44,45). Furthermore, CSE1L can promotes the nuclear distribution of the transcriptional coactivator with PDZ-binding motif to enhance the malignancy of human cancer tissues from osteosarcoma, glioma and lung cancer (46). Therefore, understanding the molecular mechanism underlying the effects of CSE1L may facilitate the optimization of cancer therapy (47).
CSE1L and p53 serve antagonistic effects on cell cycle regulation (6,48). In CRC, whilst ~50% all samples harbor p53 mutations that have been shown to be associated with poor prognosis and chemoresistance (49,50), others have reported that the majority of CRC tissues are positive for CSE1L expression (4,8,9). In the present study, in the colon cell lines tested, which were either cells from the normal colon or from cancer tissues, they were found to express varing levels of CSE1L. The colon cell lines haboring mutant p53 proteins (FHC and Caco-2) exhibited relatively high CSE1L expression levels. In addition, p53-null HCT116 cells or HCT116 cells with p53 expression knocked down were found to express higher levels of CSE1L. Conversely, an overexpression of wild-type p53 in CRC cells by 5-FU treatment reduced CSE1L expression. These results provide evidence that changing the functionality of p53 in CRC cells can alter the expression of CSE1L.
Overexpression of CSE1L in CRC has been associated with tumor development and malignancy (8,51,52), such that CSE1L knockdown can inhibit the growth and metastasis of CRC tumors (2,53). Pimiento et al (8) previously reported that CSE1L knockdown may represent a potential target for CRC treatment. This finding is consistent with that in the present study. Differentiation of Caco-2 cells into a polarized enterocyte-like monolayer was shown reduce the extent of malignancy (39). Decreasing CSE1L expression levels were accompanied by reduced c-Myc expression as the confluency of Caco-2 cells increased. In addition, CSE1L knockdown in HCT116 p53−/− cells, specifically shCSE1L-HCT116 p53−/− cells in the present study, was found to arrest cell cycle progression at the G1 phase whilst inhibiting DNA replication at S phase. These results would agree with immunofluorescent images of cells under different mitotic phases, which indicated that the CSE1L-expressing HCT116 p53−/− cells would potentiate the expression of CSE1L at prophase and metaphase. A lack of CSE1L upregulation in the HCT116 p53−/− cells, such as shCSE1L-HCT116 p53−/− cells or butyrate-treated HCT116 p53−/− cells, thereby impeded CRC cell cycle progression or migration. Depletion of cyclins caused by CSE1L knockdown also suggested that the cell cycle was arrested at the G1 phase. As previously reported by Ye et al (49) in breast cancer cells, this form of cell cycle arrest may not be only caused by reduced cyclin expression but also by the upregulated expression of the cytochrome P450 family of proteins (54). It will be necessary to examine the expression of the cytochrome P450 superfamily of proteins in the different CRC cell lines following the manipulation of CSE1L expression to clarify the significance of this relationship. Taken together, results from the present study imply that CSE1L knockdown can impede CRC progression. Since CSE1L has been reported to be a potential target for CRC treatment (8,55), methods that can decrease the expression of CSE1L in CRC may serve clinical potential.
Butyrates can regulate intestinal barrier function and has potential clinical application for human gastrointestinal diseases (10,56,57). In addition, it has been applied in combination with other chemotherapeutic agents, such as irinotecan, for CRC treatment (23). In the present study, the expression of CSE1L was reduced after the treatment of CRC cells with butyrate in vitro) or after the administration of the butyrate-producing B. pullicaecorum to CRC-bearing mice in vivo). Butyrate also reduced the CSE1L expression levels in CRC cells carrying p53 mutations, such as SW480 cells and SW620 cells. Therefore, butyrate may also display anticancer properties by downregulating the expression of CSE1L, in addition to butyrate also exhibiting synergistic anticancer effects with p53 (58,59). In combination with the present results, CSE1L knockdown may mitigate CRC malignancy and that butyrate may reduce the expression of CSE1L further.
In the present study, the results demonstrated that butyrate could reduce the expression of CSE1L in CRC cells in not only the in vitro cell modesl, but also an in vivo animal model. Administration of B. pullicaecorum was previously shown to improve the clinical outcome of CRC and colitis (17,26). Tumors with more intense nuclear staining of p53 and weaker CSE1L staining were especially found in mice bearing DMH/DSS-induced CRC that were administered with B. pullicaecorum. Pathologically, these tumors were diagnosed to be precancerous with low-grade dysplasia. However, the present study may not have completely elucidated the precise mechanism by which B. pullicaecorum regulates CSE1L expression or how the differential CSE1L expression can arrest cell cycle progression in CRC. In the future, a further in vivo study is required to evaluate the prognosis of mice with CSE1L overexpression after B. pullicaecorum administration.
In conclusion, high CSE1L expression levels is associated with the malignancy of CRC, where reduced CSE1L expression in CRC cells may hinder proliferation or improve cancer outcomes. As depicated in Fig. 6, CSE1L represents a potential target for CRC treatment, such that the reduction of CSE1L expression or activity can be achieved by butyrate treatment or B. pullicaecorum administration. This is because butyrate can repress CSE1L-induced tumorigenic potential, whereby butyrate-producing microbes, such as B. pullicaecorum, may reverse the genetic distortion caused by p53 mutations in CRC by regulating CSE1L expression. Therefore, CSE1L-induced CRC growth may be impaired by butyrate supplementation or B. pullicaecorum administration.
Figure 6.
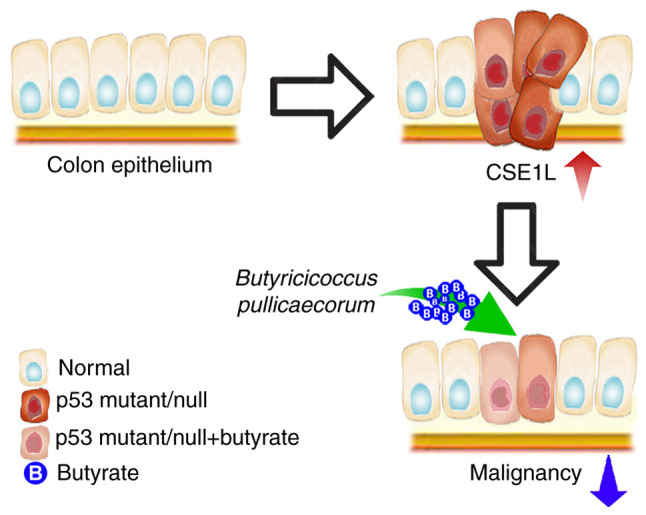
Effect of butyrate supplementation and CSE1L overexpression on the genetic distortion caused by p53 mutations in colorectal cancer. Butyrate supplementation downregulates expression of CSE1L in p53-mutated CRC cells to mitigate the maligancy of CRC. CSE1L, chromosome segregation 1-like protein.
Supplementary Data
Acknowledgements
Not applicable.
Funding Statement
The present study was supported by grants from the Cathay General Hospital and Taipei Medical University (grant nos. 106CGH-TMU-03 and 107CGH-TMU-02). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
CCC, RNY and CJH were involved in the study conception and design. HHS, YAK, PYL, CYL, KWC and CJH performed experiments and analysis of data. CCC, CJH, WYK, CYL and WCK were involved in the study conception and design, performing the experiments and analysis of data. RNY and CJH can confirm the authenticity of all the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The animal experiment in this study was performed in accordance with the principles of replacement, refinement and reduction and were approved (approval no. 107-008) by the Institutional Animal Care and Use Committees of Cathay General Hospital (Taipei, Taiwan).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Jiang K, Neill K, Cowden D, Klapman J, Eschrich S, Pimiento J, Malafa MP, Coppola D. Expression of CAS/CSE1L, the cellular apoptosis susceptibility protein, correlates with neoplastic progression in barrett's esophagus. Appl Immunohistochem Mol Morphol. 2018;26:552–556. doi: 10.1097/PAI.0000000000000464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Y, Yuan S, Liu J, Wang Y, Zhang Y, Chen X, Si W. CSE1L silence inhibits the growth and metastasis in gastric cancer by repressing GPNMB via positively regulating transcription factor MITF. J Cell Physiol. 2020;235:2071–2079. doi: 10.1002/jcp.29107. [DOI] [PubMed] [Google Scholar]
- 3.Liao CF, Lin SH, Chen HC, Tai CJ, Chang CC, Li LT, Yeh CM, Yeh KT, Chen YC, Hsu TH, et al. CSE1L, a novel microvesicle membrane protein, mediates Rastriggered microvesicle generation and metastasis of tumor cells. Mol Med. 2012;18:1269–1280. doi: 10.2119/molmed.2012.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tai CJ, Su TC, Jiang MC, Chen HC, Shen SC, Lee WR, Liao CF, Chen YC, Lin SH, Li LT, et al. Correlations between cytoplasmic CSE1L in neoplastic colorectal glands and depth of tumor penetration and cancer stage. J Transl Med. 2013;11:29. doi: 10.1186/1479-5876-11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tunccan T, Duzer S, Dilek G, Yuksel UM, Cetiner H, Kılıc C, Ant A, Duran AB. The role of CSE1L expression in cervical lymph node metastasis of larynx tumors. Braz J Otorhinolaryngol. 2021;87:42–46. doi: 10.1016/j.bjorl.2019.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanaka T, Ohkubo S, Tatsuno I, Prives C. hCAS/CSE1L associates with chromatin and regulates expression of select p53 target genes. Cell. 2007;130:638–650. doi: 10.1016/j.cell.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 7.Liao CF, Luo SF, Shen TY, Lin CH, Chien JT, Du SY, Jiang MC. CSE1L/CAS, a microtubule-associated protein, inhibits taxol (paclitaxel)-induced apoptosis but enhances cancer cell apoptosis induced by various chemotherapeutic drugs. BMB Rep. 2008;41:210–216. doi: 10.5483/BMBRep.2008.41.3.210. [DOI] [PubMed] [Google Scholar]
- 8.Pimiento JM, Neill KG, Henderson-Jackson E, Eschrich SA, Chen DT, Husain K, Shibata D, Coppola D, Malafa M. Knockdown of CSE1L gene in colorectal cancer reduces tumorigenesis in vitro. Am J Pathol. 2016;186:2761–2768. doi: 10.1016/j.ajpath.2016.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tai CJ, Hsu CH, Shen SC, Lee WR, Jiang MC. Cellular apoptosis susceptibility (CSE1L/CAS) protein in cancer metastasis and chemotherapeutic drug-induced apoptosis. J Exp Clin Cancer Res. 2010;29:110. doi: 10.1186/1756-9966-29-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang CC, Shen MH, Chen SK, Yang SH, Liu CY, Guo JW, Chang KW, Huang CH. Gut butyrate-producing organisms correlate to placenta specific 8 protein: Importance to colorectal cancer progression. J Adv Res. 2020;22:7–20. doi: 10.1016/j.jare.2019.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, Huang D, Chen KY, Cui M, Wang W, Huang X, Awadellah A, Li Q, Friedman A, Xin WW, et al. Fucosylation deficiency in mice leads to colitis and adenocarcinoma. Gastroenterology. 2017;152:193–205. doi: 10.1053/j.gastro.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Venegas DP, De la Fuente MK, Landskron G, González MJ, Quera R, Dijkstra G, Harmsen HJM, Faber KN, Hermoso MA. Short chain fatty acids (scfas)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front Immunol. 2019;10:277. doi: 10.3389/fimmu.2019.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Puertollano E, Kolida S, Yaqoob P. Biological significance of short-chain fatty acid metabolism by the intestinal microbiome. Curr Opin Clin Nutr Metab Care. 2014;17:139–144. doi: 10.1097/MCO.0000000000000025. [DOI] [PubMed] [Google Scholar]
- 14.Gill PA, van Zelm MC, Muir JG, Gibson PR. Review article: Short chain fatty acids as potential therapeutic agents in human gastrointestinal and inflammatory disorders. Aliment Pharmacol Ther. 2018;48:15–34. doi: 10.1111/apt.14689. [DOI] [PubMed] [Google Scholar]
- 15.Kannen V, Parry L, Martin FL. Phages enter the fight against colorectal cancer. Trends Cancer. 2019;5:577–579. doi: 10.1016/j.trecan.2019.08.002. [DOI] [PubMed] [Google Scholar]
- 16.Boesmans L, Valles-Colomer M, Wang J, Eeckhaut V, Falony G, Ducatelle R, Van Immerseel F, Raes J, Verbeke K. Butyrate producers as potential next-generation probiotics: Safety assessment of the administration of butyricicoccus pullicaecorum to healthy volunteers. mSystems. 2018;3:e00094–e00018. doi: 10.1128/mSystems.00094-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang SC, Shen MH, Liu CY, Pu CM, Hu JM, Huang CJ. A gut butyrate-producing bacterium Butyricicoccus pullicaecorum regulates short-chain fatty acid transporter and receptor to reduce the progression of 1,2-dimethylhydrazine-associated colorectal cancer. Oncol Lett. 2020;20:327. doi: 10.3892/ol.2020.12190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eeckhaut V, Wang J, Van Parys A, Haesebrouck F, Joossens M, Falony G, Raes J, Ducatelle R, Van Immerseel F. The probiotic butyricicoccus pullicaecorum reduces feed conversion and protects from potentially harmful intestinal microorganisms and necrotic enteritis in broilers. Front Microbiol. 2016;7:1416. doi: 10.3389/fmicb.2016.01416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu X, Wu Y, He L, Wu L, Wang X, Liu Z. Effects of the intestinal microbial metabolite butyrate on the development of colorectal cancer. J Cancer. 2018;9:2510–2517. doi: 10.7150/jca.25324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang YC, Ku WC, Liu CY, Cheng YC, Chien CC, Chang KW, Huang CJ. Supplementation of probiotic butyricicoccus pullicaecorum mediates anticancer effect on bladder urothelial cells by regulating butyrate-responsive molecular signatures. Diagnostics (Basel) 2021;11:2270. doi: 10.3390/diagnostics11122270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kazemi Sefat NA, Mohammadi MM, Hadjati J, Talebi S, Ajami M, Daneshvar H. Sodium butyrate as a histone deacetylase inhibitor affects toll-like receptor 4 expression in colorectal cancer cell lines. Immunol Invest. 2019;48:759–769. doi: 10.1080/08820139.2019.1595643. [DOI] [PubMed] [Google Scholar]
- 22.Ji X, Zhou F, Zhang Y, Deng R, Xu W, Bai M, Liu Y, Shao L, Wang X, Zhou L. Butyrate stimulates hepatic gluconeogenesis in mouse primary hepatocytes. Exp Ther Med. 2019;17:1677–1687. doi: 10.3892/etm.2018.7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Encarnacao JC, Pires AS, Amaral RA, Gonçalves TJ, Laranjo M, Casalta-Lopes JE, Gonçalves AC, Sarmento-Ribeiro AB, Abrantes AM, Botelho MF. Butyrate, a dietary fiber derivative that improves irinotecan effect in colon cancer cells. J Nutr Biochem. 2018;56:183–192. doi: 10.1016/j.jnutbio.2018.02.018. [DOI] [PubMed] [Google Scholar]
- 24.Nakano K, Mizuno T, Sowa Y, Orita T, Yoshino T, Okuyama Y, Fujita T, Fujita NO, Matsukawa Y, Tokino T, et al. Butyrate activates the WAF1/Cip1 gene promoter through Sp1 sites in a p53-negative human colon cancer cell line. J Biol Chem. 1997;272:22199–22206. doi: 10.1074/jbc.272.35.22199. [DOI] [PubMed] [Google Scholar]
- 25.Russo I, Luciani A, De Cicco P, Troncone E, Ciacci C. Butyrate attenuates lipopolysaccharide-induced inflammation in intestinal cells and crohn's mucosa through modulation of antioxidant defense machinery. PLoS One. 2012;7:e32841. doi: 10.1371/journal.pone.0032841. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Eeckhaut V, Machiels K, Perrier C, Romero C, Maes S, Flahou B, Steppe M, Haesebrouck F, Sas B, Ducatelle R, et al. Butyricicoccus pullicaecorum in inflammatory bowel disease. Gut. 2013;62:1745–1752. doi: 10.1136/gutjnl-2012-303611. [DOI] [PubMed] [Google Scholar]
- 27.Marteau P. Butyrate-producing bacteria as pharmabiotics for inflammatory bowel disease. Gut. 2013;62:1673. doi: 10.1136/gutjnl-2012-304240. [DOI] [PubMed] [Google Scholar]
- 28.Eeckhaut V, Ducatelle R, Sas B, Vermeire S, Van Immerseel F. Progress towards butyrate-producing pharmabiotics: Butyricicoccus pullicaecorum capsule and efficacy in TNBS models in comparison with therapeutics. Gut. 2014;63:367. doi: 10.1136/gutjnl-2013-305293. [DOI] [PubMed] [Google Scholar]
- 29.Kilkenny C, Browne W, Cuthill IC, Emerson M, Altman DG, NC3Rs Reporting Guidelines Working Group Animal research: Reporting in vivo experiments: The ARRIVE guidelines. J Gene Med. 2010;12:561–563. doi: 10.1002/jgm.1473. [DOI] [PubMed] [Google Scholar]
- 30.Soucek K, Gajduskova P, Brazdova M, Hýzd'alová M, Kocí L, Vydra D, Trojanec R, Pernicová Z, Lentvorská L, Hajdúch M, et al. Fetal colon cell line FHC exhibits tumorigenic phenotype, complex karyotype, and TP53 gene mutation. Cancer Genet Cytogenet. 2010;197:107–116. doi: 10.1016/j.cancergencyto.2009.11.009. [DOI] [PubMed] [Google Scholar]
- 31.Hayashi Y, Tsujii M, Kodama T, Akasaka T, Kondo J, Hikita H, Inoue T, Tsujii Y, Maekawa A, Yoshii S, et al. p53 functional deficiency in human colon cancer cells promotes fibroblast-mediated angiogenesis and tumor growth. Carcinogenesis. 2016;37:972–984. doi: 10.1093/carcin/bgw085. [DOI] [PubMed] [Google Scholar]
- 32.Rochette PJ, Bastien N, Lavoie J, Guerin SL, Drouin R. SW480, a p53 double-mutant cell line retains proficiency for some p53 functions. J Mol Biol. 2005;352:44–57. doi: 10.1016/j.jmb.2005.06.033. [DOI] [PubMed] [Google Scholar]
- 33.Huang CJ, Yang SH, Huang SM, Lin CM, Chien CC, Chen YC, Lee CL, Wu HH, Chang CC. A predicted protein, KIAA0247, is a cell cycle modulator in colorectal cancer cells under 5-FU treatment. J Transl Med. 2011;9:82. doi: 10.1186/1479-5876-9-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ishimine M, Lee HC, Nakaoka H, Orita H, Kobayashi T, Mizuguchi K, Endo M, Inoue I, Sato K, Yokomizo T. The Relationship between TP53 Gene status and carboxylesterase 2 expression in human colorectal cancer. Dis Markers. 2018;2018:5280736. doi: 10.1155/2018/5280736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abu El Maaty MA, Strassburger W, Qaiser T, Dabiri Y, Wölfl S. Differences in p53 status significantly influence the cellular response and cell survival to 1,25-dihydroxyvitamin D3-metformin cotreatment in colorectal cancer cells. Mol Carcinog. 2017;56:2486–2498. doi: 10.1002/mc.22696. [DOI] [PubMed] [Google Scholar]
- 36.Li DD, Sun T, Wu XQ, Chen SP, Deng R, Jiang S, Feng GK, Pan JX, Zhang XC, Zeng YX, Zhu XF. The inhibition of autophagy sensitises colon cancer cells with wild-type p53 but not mutant p53 to topotecan treatment. PLoS One. 2012;7:e45058. doi: 10.1371/journal.pone.0045058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhat UG, Gartel AL. Differential sensitivity of human colon cancer cell lines to the nucleoside analogs ARC and DRB. Int J Cancer. 2008;122:1426–1429. doi: 10.1002/ijc.23239. [DOI] [PubMed] [Google Scholar]
- 38.Kralj M, Husnjak K, Körbler T, Pavelić J. Endogenous p21WAF1/CIP1 status predicts the response of human tumor cells to wild-type p53 and p21WAF1/CIP1 overexpression. Cancer Gene Ther. 2003;10:457–467. doi: 10.1038/sj.cgt.7700588. [DOI] [PubMed] [Google Scholar]
- 39.Huang CJ, Lee CL, Yang SH, Chien CC, Huang CC, Yang RN, Chang CC. Upregulation of the growth arrest-specific-2 in recurrent colorectal cancers, and its susceptibility to chemo-therapy in a model cell system. Biochim Biophys Acta. 2016;1862:1345–1353. doi: 10.1016/j.bbadis.2016.04.010. [DOI] [PubMed] [Google Scholar]
- 40.Leoni BD, Natoli M, Nardella M, Bucci B, Zucco F, D'Agnano L, Felsani A. Differentiation of Caco-2 cells requires both transcriptional and post-translational down-regulation of Myc. Differentiation. 2012;83:116–127. doi: 10.1016/j.diff.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 41.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 42.Jonkman JE, Cathcart JA, Xu F, Bartolini ME, Amon JE, Stevens KM, Colarusso P. An introduction to the wound healing assay using live-cell microscopy. Cell Adh Migr. 2014;8:440–451. doi: 10.4161/cam.36224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ye S, Zhou HB, Chen Y, Li KQ, Jiang SS, Hao K. Crizotinib changes the metabolic pattern and inhibits ATP production in A549 non-small cell lung cancer cells. Oncol Lett. 2021;21:61. doi: 10.3892/ol.2020.12323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Winkler J, Roessler S, Sticht C, DiGuilio AL, Drucker E, Holzer K, Eiteneuer E, Herpel E, Breuhahn K, Gretz N, et al. Cellular apoptosis susceptibility (CAS) is linked to integrin beta1 and required for tumor cell migration and invasion in hepatocellular carcinoma (HCC) Oncotarget. 2016;7:22883–22892. doi: 10.18632/oncotarget.8256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu JH, Hong DF, Song YM, Sun LF, Wang ZF, Wang JW. Suppression of cellular apoptosis susceptibility (CSE1L) inhibits proliferation and induces apoptosis in colorectal cancer cells. Asian Pac J Cancer Prev. 2013;14:1017–1021. doi: 10.7314/APJCP.2013.14.2.1017. [DOI] [PubMed] [Google Scholar]
- 46.Nagashima S, Maruyama J, Honda K, Kondoh Y, Osada H, Nawa M, Nakahama KI, Yuasa MI, Kagechika H, Sugimura H, et al. CSE1L promotes nuclear accumulation of transcriptional coactivator TAZ and enhances invasiveness of human cancer cells. J Biol Chem. 2021;297:100803. doi: 10.1016/j.jbc.2021.100803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Snijders AM, Mao JH. Multi-omics approach to infer cancer therapeutic targets on chromosome 20q across tumor types. Adv Mod Oncol Res. 2016;2:215–223. doi: 10.18282/amor.v2.i4.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Behrens P, Brinkmann U, Wellmann A. CSE1L/CAS: Its role in proliferation and apoptosis. Apoptosis. 2003;8:39–44. doi: 10.1023/A:1021644918117. [DOI] [PubMed] [Google Scholar]
- 49.Li H, Zhang J, Tong JHM, Chan AWH, Yu J, Kang W, To KF. Targeting the oncogenic p53 mutants in colorectal cancer and other solid tumors. Int J Mol Sci. 2019;20:5999. doi: 10.3390/ijms20235999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iacopetta B. TP53 mutation in colorectal cancer. Hum Mutat. 2003;21:271–276. doi: 10.1002/humu.10175. [DOI] [PubMed] [Google Scholar]
- 51.Wang X, Ren Y, Ma S, Wang S. Circular RNA 0060745, a novel circRNA, promotes colorectal cancer cell proliferation and metastasis through miR-4736 sponging. Onco Targets Ther. 2020;13:1941–1951. doi: 10.2147/OTT.S240642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ma S, Yang D, Liu Y, Wang Y, Lin T, Li Y, Yang S, Zhang W, Zhang R. LncRNA BANCR promotes tumorigenesis and enhances adriamycin resistance in colorectal cancer. Aging (Albany NY) 2018;10:2062–2078. doi: 10.18632/aging.101530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cheng DD, Lin HC, Li SJ, Yao M, Yang QC, Fan CY. CSE1L interaction with MSH6 promotes osteosarcoma progression and predicts poor patient survival. Sci Rep. 2017;7:46238. doi: 10.1038/srep46238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ye M, Han R, Shi J, Wang X, Zhao AZ, Li F, Chen H. Cellular apoptosis susceptibility protein (CAS) suppresses the proliferation of breast cancer cells by upregulated cyp24a1. Med Oncol. 2020;37:43. doi: 10.1007/s12032-020-01366-w. [DOI] [PubMed] [Google Scholar]
- 55.Li KK, Yang L, Pang JC, Chan AKY, Zhou L, Mao Y, Wang Y, Lau KM, Poon WS, Shi Z, Ng HK. MIR-137 suppresses growth and invasion, is downregulated in oligodendroglial tumors and targets CSE1L. Brain Pathol. 2013;23:426–439. doi: 10.1111/bpa.12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beaumont M, Paes C, Mussard E, Knudsen C, Cauquil L, Aymard P, Barilly C, Gabinaud B, Zemb O, Fourre S, et al. Gut microbiota derived metabolites contribute to intestinal barrier maturation at the suckling-to-weaning transition. Gut Microbes. 2020;11:1–19. doi: 10.1080/19490976.2020.1747335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Silva JPB, Navegantes-Lima KC, Oliveira ALB, Rodrigues DVS, Gaspar SLF, Monteiro VVS, Moura DP, Monteiro MC. Protective mechanisms of butyrate on inflammatory bowel disease. Curr Pharm Des. 2018;24:4154–4166. doi: 10.2174/1381612824666181001153605. [DOI] [PubMed] [Google Scholar]
- 58.Zhao Y, Shi L, Hu C, Sang S. Wheat bran for colon cancer prevention: The synergy between phytochemical alkylresorcinol C21 and intestinal microbial metabolite butyrate. J Agric Food Chem. 2019;67:12761–12769. doi: 10.1021/acs.jafc.9b05666. [DOI] [PubMed] [Google Scholar]
- 59.Pant K, Mishra AK, Pradhan SM, Nayak B, Das P, Shalimar D, Saraya A, Venugopal SK. Butyrate inhibits HBV replication and HBV-induced hepatoma cell proliferation via modulating SIRT-1/Ac-p53 regulatory axis. Mol Carcinog. 2019;58:524–532. doi: 10.1002/mc.22946. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.