Abstract
Corynebacterium bovis is an opportunistic pathogen of the skin of immunodeficient mice and is sensitive to oral antibiotics that reach therapeutic blood concentrations. However, prophylactic antibiotics are considered to be ineffective at preventing C. bovis infection. In addition, the effect of C. bovis on the skin microbiome (SM) of common immunodeficient mouse strains has yet to be characterized. Consequently, we evaluated whether oral prophylactic antibiotics prevent C. bovis infection after inoculation. An infectious dose of C. bovis was applied to the skin of Hsd:Athymic Nude (nude) and NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice. Mice were then housed individually and assigned randomly to receive either untreated drinking water (Cb+Abx− group) or prophylactic amoxicillin-clavulanic acid in the drinking water (0.375 mg/mL) for 14 d (Cb+Abx+ group). A third treatment group of each mouse strain was uninoculated and untreated (Cb−Abx−group). Mice from all groups were serially sampled by using dermal swabs to monitor C. bovis infection via quantitative real-time PCR and the SM via 16S rRNA sequence analysis. Fourteen days of prophylactic antibiotics prevented the perpetuation of C. bovis skin infection in both strains. Only the combination of C. bovis inoculation and oral antibiotics (Cb+Abx+) significantly affected the SM of NSG mice at day 14; this effect resolved by the end of the study (day 70). In mice that did not receive antibiotics, C. bovis significantly altered the SM of nude mice but not NSG mice at days 14 and 70. These findings demonstrate the potential benefit of prophylactic antibiotics for prevention of C. bovis infection. However, indirect effect of antibiotics on commensal bacteria and potential effects on xenograft models must be considered.
Abbreviations: Abx, prophylactic antibiotics; Cb, Corynebacterium bovis; GM, gastrointestinal (gut) microbiome; PDX, patient-derived xenograft; qPCR, quantitative real-time PCR; SM, skin microbiome
Immunodeficient mice are used for a wide variety of biomedical research spanning the fields of immunology, transplantation, infectious disease, and cancer. The lack of specific anatomic or cellular components of the innate and adaptive immune systems makes these mice invaluable for research. However, these same inherent features also make these mouse strains susceptible to opportunistic infections. For example, Corynebacterium bovis infects the skin of a variety of immunodeficient strains8,54 and mice with uncharacterized immune systems.28,29,43 Although the clinical signs and progression of skin disease varies among strains, C. bovis infection of immunodeficient mice can alter experimental outcomes in cancer research.63
Prophylactic antibiotics reportedly will not protect immunodeficient mice from C. bovis infection.5 However, this conclusion conflicts with work showing that C. bovis is sensitive to antibiotics commonly used in mice.17,64 These data indicate that the administration of amoxicillin in drinking water leads to plasma concentrations in mice that exceed the organism’s minimum inhibitory concentration to inhibit growth of 90% of the isolates evaluated in vitro.17,40 Given the known difficulty in eliminating this pathogen from immunodeficient mouse colonies, leveraging the antimicrobial susceptibility of C. bovis with antibiotics that are commonly used in rodents may allow prophylactic protection of immunodeficient mice in the event of C. bovis outbreak.
Antibiotics have been used in veterinary and human medicine for well over half a century. However, the growing recognition of the beneficial effects of host-associated microbiota on numerous aspects of host physiology and immunology demands that antibiotics be used cautiously. Therefore, investigators and veterinarians should be aware of treatment efficacy and the potential effect of an infectious agent and antimicrobial treatment on the commensal flora of the mouse strain.30 To date, the effects of C. bovis and oral antibiotics on the skin microbiome (SM) have not been investigated in common immune-impaired mouse strains.28,29 In addition, interest is growing in the influences of opportunistic and infectious agents on the skin microbiome during both health and disease.18 Consequently, the goals of this project were to investigate the use of prophylactic antibiotic therapy to prevent C. bovis infection in Hsd:Athymic Nude (nude) and NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice after acute inoculation. In addition, the response of the SM to C. bovis infection and antibiotic administration were evaluated in parallel.
Materials and Methods
Mice, housing, and husbandry.
Female nude mice (n = 15; age, 7 wk; strain no. 069, Envigo, Indianapolis, IN) and female NSG mice (n = 14; age, 7 wk; strain no. 005557, Jackson Laboratories, Bar Harbor, ME) were obtained and documented by each vendor to be free of ectoparasites, endoparasites and excluded viral and bacterial pathogens, including C. bovis. Mice were group-housed (5 mice/cage) and allowed to acclimate for 7 d prior to being individually housed for experimental manipulations. Mice were maintained under barrier conditions in autoclaved IVC (Allentown Caging, Allentown, NJ) containing aspen chip bedding (Envigo), a cotton square (Ancare, Bellmore, NY), and a rodent enrichment device (Mouse Igloo, BioServ, Flemmington, NJ) with 40 air changes per hour. Mice received irradiated rodent diet (2920X, Teklad Diet, Envigo) ad libitum and autoclaved, reverse-osmosis-purified water in an intracage 375-mL water bottle. The macroenvironment is electronically controlled to provide 22.2 ± 1°C, a 14:10 light:dark cycle, and 30% to 40% humidity with at least 12 air changes per hour. All studies were approved by the University of Colorado Denver Anschutz Medical Campus IACUC.
C. bovis inoculum and inoculation.
C. bovis isolate CUAMC1 was cultured from athymic nude mice at the University of Colorado Anschutz Medical Campus in 2014.17 Culture conditions of isolates for mouse inoculation were recently described in detail.47 Briefly, CUMAC1 was cryopreserved in media made from 10% nonfat milk (w/v) in water and stored at −80°C. Chips of frozen media were cultured on trypticase soy agar containing 5% sheep blood (catalog no. B21261X, Becton Dickinson, Sparks, MD) for 48 to 72 h at 37°C and ambient atmosphere. Individual colonies were collected and resuspended in heart infusion broth (catalog no. 238400, Becton Dickinson) containing 1% Tween 80 in an incubator with an elliptical shaker for 24 h at 32°C with 250 rotations per minute. After 24 h, the OD600 was compared with a previously generated standard curve to estimate the concentration (in cfu/mL) of the inoculum. After inoculation, serial dilutions of the inoculum in sterile PBS and colony plate counts on trypticase soy agar with 5% sheep blood were performed to confirm the final inoculation dose that mice received. Based on pilot studies, NSG mice required a higher inoculation dose than did nude mice to ensure all mice became infected (data not shown). Nude and NSG mice were anesthetized with isoflurane and then inoculated with doses of 2 × 106 cfu and 2 × 107 cfu, respectively. The inoculum was delivered in 50 µL of sterile PBS and distributed by using a sterile cotton swab. For nude mice, the distribution of inoculum focused on the scalp, ear pinnae, and dorsal back. Except for the dorsal back, these sites are difficult for a singly housed mouse to groom effectively, thus increasing inoculum contact time. For NSG mice, inoculation was distributed over the scalp, ear pinnae, periocular conjunctiva, muzzle, and commissures of the mouth. These locations were selected because most of these sites are not densely haired, and they are difficult for a singly house mouse to groom effectively.
Experimental design, inoculation, and antibiotic prophylaxis.
Athymic nude and NSG mice were each randomly divided into 3 treatment groups: no C. bovis inoculation and no antibiotics (Cb−Abx−, negative control, n = 5 nude and 4 NSG); C. bovis inoculation but no antibiotics (Cb+Abx−, positive control, n = 5/strain); and C. bovis inoculation with prophylactic antibiotics (Cb+Abx+, treatment, n = 5/strain, Figure 1). After C. bovis inoculation, mice in the Cb+Abx+ groups were given 0.375 mg/mL of amoxicillin-clavulanic acid (amoxicillin trihydrate-clavulanate potassium, Zoetis, Florham Park, NJ) in a nonacidified, autoclaved water bottle.17,47 With this formulation of amoxicillin-clavulanic acid, the water bottle contained 0.3 mg/mL of amoxicillin, resulting in a daily dose of 48 to 60 mg/kg of amoxicillin for a 25-g mouse drinking 4 to 5 mL of medicated water daily. The dose of amoxicillin-clavulanic acid was based on the demonstrated blood plasma concentration of amoxicillin achieved by using an oral amoxicillin concentration of 0.25 mg/mL in water.40 According to the cited report,40 the blood concentration exceeds the ampicillin susceptibility of C. bovis isolates obtained from cows and immunodeficient rodents.17,64 Ampicillin susceptibility can be correlated directly to amoxicillin because both antibiotics work very similarly in vitro.22
Figure 1.
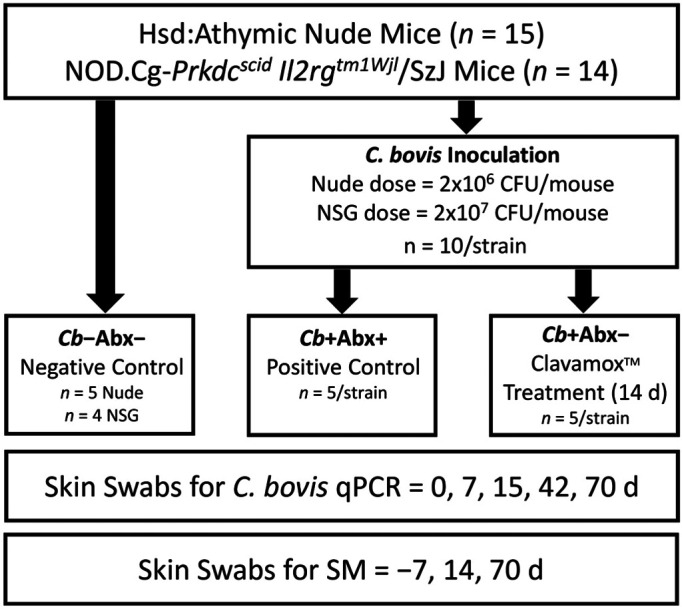
Illustration of the experimental design, allocation of each mouse strain into experimental groups, and the interval of sample collection for C. bovis–specific qPCR and skin microbiome (SM) analyses.
The prophylactic antibiotic group received antibiotic water for 14 d after C. bovis inoculation. This duration of treatment has been used previously to prevent C. bovis infection,5 and was demonstrated to be an adequate duration for the elimination of Rodentibacter pneumotropicus with antibiotic treatment.60 Antibiotic-containing water was not shaken after placement in the cage but was replaced after 7 d.40,41 The C. bovis status of each mouse was tested by using combined oral and skin swabs (BBL Culture Swab EZ, Becton Dickinson, Franklin Lakes, NJ) for C. bovis DNA and quantitative real-time PCR (qPCR) assay on days 0 (before inoculation), 7 (during treatment), 15 (the day after antibiotic discontinuation), 42, and 70 after inoculation. For SM analysis, skin swabs (PurFlock Ultra, Puritan, Guilford, ME) were collected from the lateral aspect of the mouse including the ear, neck, and axilla encompassing approximately 2 cm2 of surface area. In total, 40 strokes of the swab were made, including 10 from the ears, 10 from the neck, and 20 from the axilla, with the swab rotated constantly and under sufficient pressure to cause mild skin hyperemia but not abrasion. SM samples were collected on days −7, 14 (last day of antibiotics), and 70 after inoculation (Figure 1). All skin swabs were collected antemortem. When SM and C. bovis qPCR samples were collected on the same day, the SM sample was collected first to prevent contamination of the skin with oral bacterial flora.
Mouse cages were changed every 7 d for the first 14 d after C. bovis inoculation and then every 14 d until the end of the study. Weekly cage changes for the first 14 d minimized intracage C. bovis contamination after the initial skin inoculation.47 At the time of cage caging, mice were placed into new cages without transfer of any bedding, nesting material, or cage furniture. All cage manipulations were performed under a downdraft, HEPA-filtered animal transfer station (ATS2, Allentown Caging). All work surfaces were disinfected between cages and remained moist with Clidox-S (Pharmacal, Naugatuck, CT) at a dilution of 1:18:1. A glove-change technique for working with C. bovis–infected and -uninfected mice under experimental conditions has recently been described in detail.47 Briefly, gloves were changed between cages, prior to touching the mice or anything in the intracage environment.
Mice were euthanized by CO2 asphyxiation, followed by cervical dislocation at 70 d after inoculation with C. bovis, which is equivalent to 56 d after antibiotic discontinuation for that group. This time point was selected based on a previous study5 in which nude mice with previously established C. bovis infections and treatment with 4 wk of amoxicillin-medicated chow returned to C. bovis–positive culture status within 0 to 46 d (mean, 17 d). Another experiment in the same report5 placed naïve nude mice on amoxicillin-medicated chow for 2 wk, beginning at the same time as mice were housed in an enzootically C. bovis–infected room. After 2 wk of antibiotics, the time to infection after housing in the room was 20 to 55 d (mean, 34.5 d).5 Our endpoint was selected to exceed the previously demonstrated maximal duration for mice to become infected after completing therapeutic or prophylactic antibiotics.
At the time of euthanasia, skin from the dorsal head, including the ears and dorsal neck, was collected from a subset of mice of each strain in each treatment group and fixed in 10% buffered formalin. Hematoxylin and eosin staining was performed on transverse sections of the pinnae and dorsal neck skin. Sections were evaluated for histologic changes commonly associated with C. bovis infection, including hyperkeratosis and acanthosis.
SM and C. bovis qPCR analyses.
Skin-associated bacterial profiles were determined by broad-range amplification and sequence analysis of 16S rRNA genes using our previously described methods.20,46,65 In brief, DNA was extracted from swabs by using a QIAamp PowerFecal DNA extraction kit (Qiagen, Carlsbad, CA) according to the manufacturer’s protocol; cells were lysed by bead beating for 30 s at 6000 rpm by using a MagnaLyser (Roche Sequencing and Life Science, Indianapolis, IN). PCR amplicons were generated through 35 cycles using dual-indexed primers19 (27F-YM: 5′ AGA GTT TGA TYM TGG CTC AG 3′; 338R: 5′ TGC CTC CCG TAG GAG T 3′) that target approximately 300 base pairs of the 16S rRNA gene V1V2.21,34 PCR products were normalized by using a SequalPrep kit (Invitrogen, Carlsbad, CA), pooled, lyophilized, purified, and concentrated by using a DNA Clean and Concentrator Kit (Zymo, Irvine, CA). Pooled amplicons were quantified by using a Qubit Fluorometer 2.0 (Invitrogen). The pool was diluted to 4 nM and denatured with 0.2 N NaOH at room temperature. The denatured DNA was diluted to 15 pM and spiked with 25% of the Illumina PhiX control DNA prior to loading of the sequencer. Paired-end sequencing using a 600-cycle version 3 reagent kit was performed on the Illumina MiSeq platform via MiSeq Control Software and MiSeq Reporter (version 2.4 for both).
Paired-end sequences were sorted by sample via barcodes in the paired reads by using a Python script.20,46,65 Sorted paired-end sequence data and accompanying metadata were deposited in the NCBI Sequence Read Archive under BioProject ID PRJNA663095. The paired reads were assembled by using phrap,15,16 and pairs that did not assemble were discarded. Assembled sequence ends were trimmed over a moving window of 5 nucleotides until the average quality met or exceeded 20. Trimmed sequences with more than 1 ambiguity or shorter than 250 nucleotides were discarded. Potential chimeras identified via Uchime (usearch6.0.203_i86linux32)12 using the Schloss55 Silva reference sequences were removed from subsequent analyses. Assembled sequences were aligned and classified by using SINA (1.3.0-r23838)49 with the 418,497 bacterial sequences in Silva 115NR9950 as references configured to yield the Silva taxonomy; taxonomic assignments used the lowest-common-ancestor approach with default SINA settings. Closed-reference, operational taxonomic units were produced by binning sequences with identical Silva/SINA lowest-common-ancestor assignments. This process generated 6,727,469 quality-filtered 16S rRNA gene sequences for 88 samples (median, 63,154 sequences per sample; interquartile range, 20,029 to 119,267 sequences). All sequence libraries had a Good coverage score greater than 98.5%, indicating excellent depth of sequence coverage. For C. bovis–specific qPCR analysis, skin swabs were submitted to the University of Colorado Anschutz Medical Campus Quantitative PCR Core as previously described.38 DNA was prepared from these specimens by using the QIAamp DNA Mini extraction kit (Qiagen).
Statistics.
Mann–Whitney rank sum testing was performed on C. bovis DNA copies between C. bovis positive controls of each strain at time points on days 15, 42, and 70 after inoculation. A contingency table was created and the Fisher Exact test was performed on the number of C. bovis–positive mice after prophylactic antibiotic treatment as compared with C. bovis–positive controls for each strain.
R (version 3.6.3) and Explicet (version 2.10.552) software packages were used to visualize and analyze microbiome data. Differences in overall microbiota composition (that is, β-diversity) between groups were assessed by a permutation-based multiple analysis of the variance test, as implemented via the adonis2 function of the vegan R package.45 Dissimilarities were measured by using the Morisita–Horn index; P values were inferred through 106 label permutations. Shannon diversity indices (that is, assessment of α-diversity) were calculated for each sequence library through 1000 replicate samples at a rarefaction point of 5000 sequences. Between-group differences were assessed by ANOVA (across 3 or more groups) followed by Tukey Honest Significant Difference tests (for pairwise comparisons). Principal coordinates analysis used the weighted classic multidimensional scaling function of the vegan R package,45 with dissimilarities measured using the Morisita–Horn index. Ordination plots were drawn by using the ordiplot and ordiellipse commands of the vegan R package.45
Results
We assessed the use of prophylactic antibiotics for the prevention of C. bovis skin infections in immunodeficient nude and NSG mice. Both nude and NSG Cb−Abx− mice remained C. bovis–negative for the duration of the study despite being housed in the same room and manipulated in the same animal transfer station as C. bovis–inoculated mice. During the experiment, no clinical signs of C. bovis infection were seen in any mice of either strain or any experimental group.
Effect of prophylactic antibiotics on C. bovis infections.
Cb+Abx− nude mice were C. bovis–positive from day 7 after inoculation until the end of the study. As anticipated, no C. bovis was detectable on Cb−Abx− nude mice (Figure 2 A). With prophylactic antibiotics, all 5 Cb+Abx+ nude mice remained C. bovis–negative for the duration of the study (P ≤ 0.05; Figure 2 A).
Figure 2.
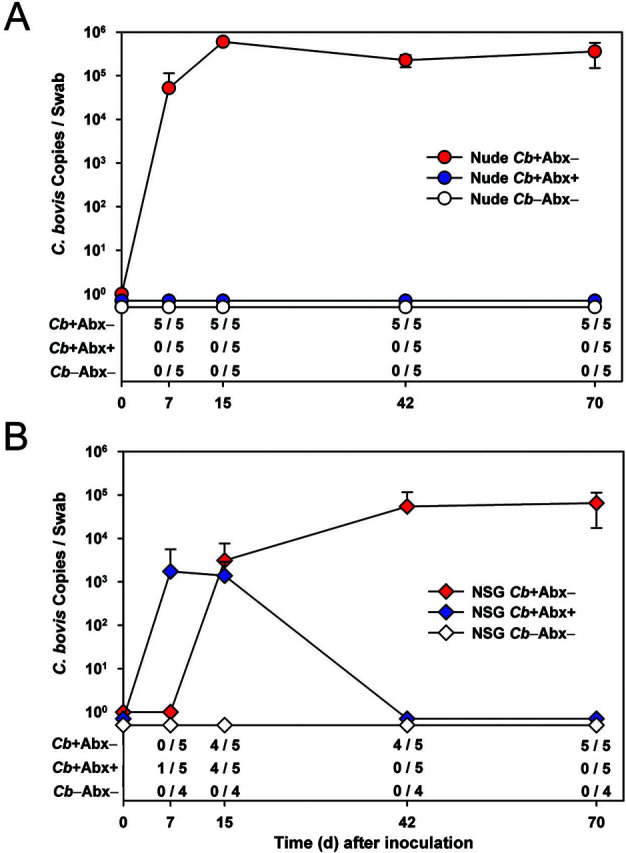
The upper portion of each graph depicts the absolute copy number of C. bovis DNA (1 log10 ± 1 SD) collected from the skin of (A) nude and (B) NSG mice. The limit of detection is 30 copies. The lower table of each figure shows the number of C. bovis qPCR-positive mice among the total number of mice per group (n = 4 or 5).
For Cb+Abx− NSG mice, 4 of 5 (80%) were infected by day 15 after inoculation. However, by the end of the study, all Cb+Abx− NSG mice were C. bovis–positive. During prophylactic antibiotic treatment, 1 of 5 (20%) Cb+Abx+ NSG mice was C. bovis–positive at 7 d after inoculation; the single positive mouse was then negative by 15 d after inoculation. At 15 d after inoculation, 4 of 5 (80%) Cb+Abx+ NSG mice were C. bovis–positive. However, at both 42 and 70 d after inoculation, all 5 Cb+Abx+ NSG mice were C. bovis–negative (P ≤ 0.05; Figure 2 B).
Higher numbers of C. bovis DNA were detected on the skin of nude mice as compared with NSG mice at 15, 42, and 70 d after inoculation (P ≤ 0.03; Figure 2 A compared with Figure 2 B). This outcome occurred despite the 10-fold higher inoculation dose used to infect NSG mice as compared with nude mice. For both strains, the copy number remained consistent between 42 and 70 d after inoculation. The C. bovis copy number (mean ± 1 SD) at 70 d after inoculation was 358,608 ± 209,540 copies for nude mice and 65,076 ± 47,693 copies for NSG mice (P = 0.03; Figure 2).
The SM of immunodeficient strains.
To investigate the extent to which C. bovis and oral amoxicillin alter the SM, we used 16S rRNA gene sequencing to assess skin microbiota at days −7, 14, and 70 (88 total samples). From these data, stacked bar charts were generated to visualize the data according to mouse strain, treatment group, and time point (Figure 3). SM profiling revealed Corynebacterium spp. on the skin of both mouse strains at the time of their arrival to our facility, but C. bovis in particular was not detected by using species-specific qPCR primers. Samples collected from the skin prominently included taxa that are most commonly associated with gut microbiota (GM; for example, Bifidobacterium, Bacteroides, Lactobacillus, Clostridia, Enterobacteriaceae); this was likely due to exposure to feces in the animal’s cage. The SM differed significantly (P < 1 × 10−6) in overall composition (β-diversity) between the 2 mouse strains (Figure 3).13 Nude mice exhibited a more even distribution of typical skin and gut taxa, whereas NSG mice were dominated by staphylococci. In addition, the SM of nude mice was significantly (P < 1 × 10−6) more α-diverse than that of NSG mice (Figure 4). The microbiota of Cb−Abx− nude mice shifted in overall composition from the time of arrival to the study’s end (P = 0.0078 and P = 0.0077 for day −7 compared with day 14 and day −7 compared with day 70, respectively; Figure 3). Likewise, Shannon diversity decreased in Cb−Abx− nude mice over the course of the study (day −7 compared with day 14, P = 0.032; day −7 compared with day 70, P < 1 × 10−6; Figure 4). In contrast, NSG mice did not exhibit such longitudinal shifts in overall composition or Shannon diversity from days −7 through 70 (P > 0.05 for all pairwise tests of α- and β-diversity). Finally, baseline overall composition and Shannon diversity were not different among mice that were subsequently randomized to the 3 treatment groups (P > 0.05 for all pairwise comparisons; Figures 3 and 4).
Figure 3.
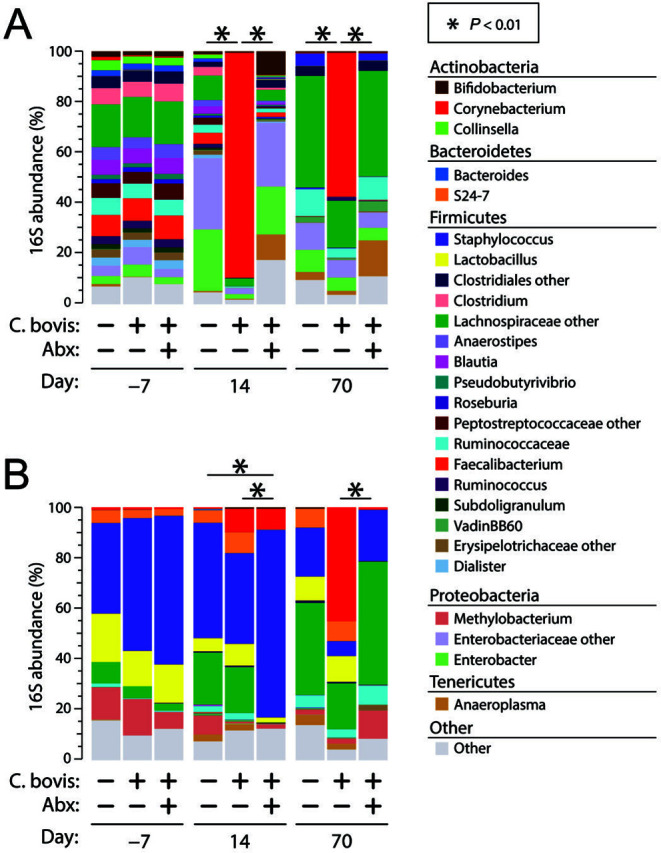
Bacterial profiles of skin microbiota. Bar charts summarize mean relative abundances (RA) of prominent genus-level bacterial taxa, stratified by mouse strain (panel A, nude; panel B, NSG), treatment group, and time point. Taxa with RA < 0.05% are collapsed into the Other category but were included in statistical tests. Significance was assessed through PERMANOVA tests at each time point. These tests were performed only among treatment arms within each time point (that is, comparisons were not made between time points). Each bar represents 5 mice for all groups, except NSG Cb−Abx− (n = 4).
Figure 4.
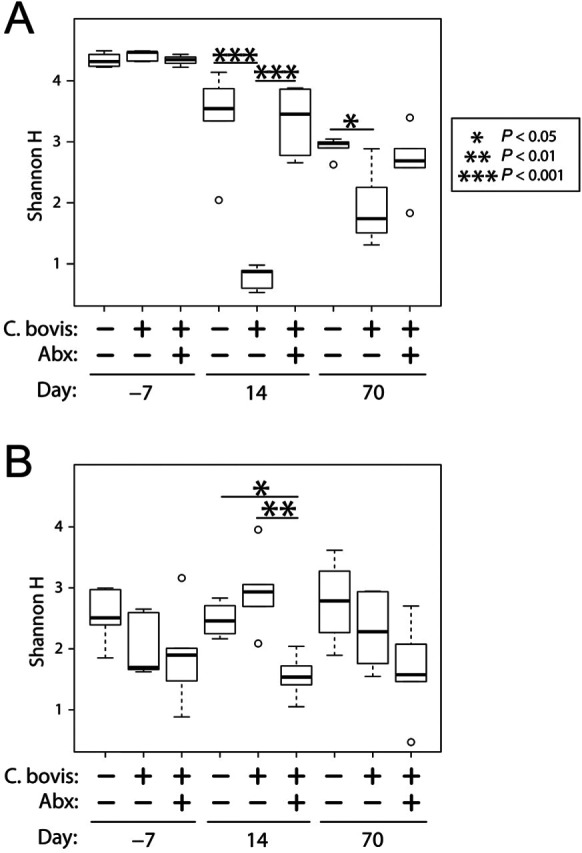
Microbial α-diversity of skin microbiota. Boxplots summarize Shannon Diversity (H) scores, stratified according to mouse strain (nude, upper panel; NSG, lower panel), treatment group, and time point. Significance was tested across all 3 treatment arms for each time point by using ANOVA followed by Tukey Honest Significant Difference tests of pairwise comparisons. Each bar represents 5 mice for all groups, except for NSG Cb−Abx− (n = 4).
Effect of C. bovis on resident skin bacteria.
For Cb+Abx− nude mice, the composition of skin microbiota differed significantly (P < 0.01) from both the Cb−Abx− and Cb+Abx+ groups at both 14 and 70 d after inoculation (Figure 3). After C. bovis inoculation, Corynebacterium spp. were the most prominent population among Cb+Abx− mice, accounting for 90% of the bacteria on the skin at 14 d and 60% on 70 d (P < 0.001 as compared with Cb−Abx− mice at both time points). This dramatic increase in Corynebacterium spp. in the Cb+Abx− group at 14 d after inoculation resulted in a significant (P < 0.001) reduction in α-diversity as compared with Cb−Abx− (Figure 4), with less significant (P < 0.05) differences as compared with Cb−Abx− mice at 70 d (Figure 4).
Among the Cb+Abx− NSG mice, only approximately 10% of the SM was composed of Corynebacterium spp. at 14 d, with no significant difference compared with Cb−Abx− mice at that time (P = 0.099, Figure 3). The abundance of Corynebacterium spp. increased to 45% by 70 d after inoculation (P = 0.0028 as compared with Cb−Abx−). Due to this increase in Corynebacterium spp., the composition of the SM at 70 d approached statistical significance (P < 0.1) as compared with the negative control group. For both nude and NSG mice, visualization of differences in microbiota by principal coordinates analysis confirmed the distinct segregation of C. bovis–positive control group at 14 and 70 d after inoculation (Figure 5).
Figure 5.
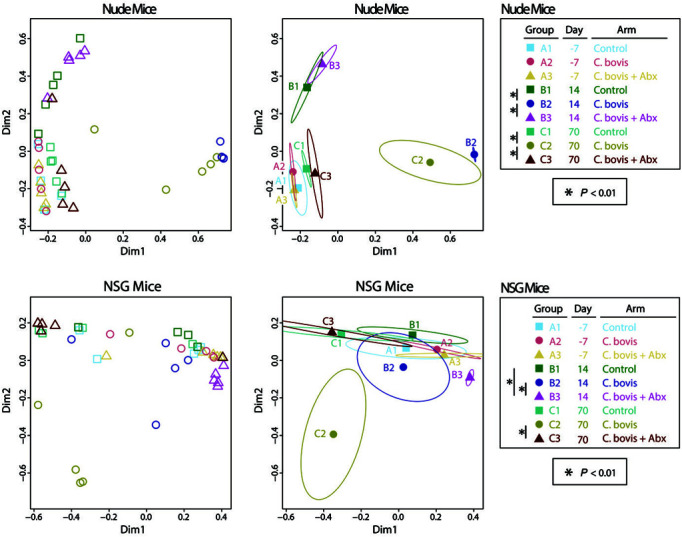
Principal coordinates analysis of skin microbiota. Panels display plots of individual samples from either nude (upper panels) or NSG (lower panels) mice. Left panels show individual samples (open symbols), whereas right panels use ellipses to define a 1 SD spread in each group about the within-group centroids (filled symbols). Treatment groups and time points are indicated by different symbols and color-coding, as summarized in the key. Significance was assessed by between-group PERMANOVA tests and reflect the same statistical findings as in Figure 3.
Effects of systemic antibiotics and C. bovis on resident skin bacteria.
The administration of oral amoxicillin-clavulanic acid beginning immediately after C. bovis inoculation and continuing for 14 d prevented colonization and expansion of Corynebacterium spp. on nude mice. At 14 d after inoculation, the Cb+Abx+ group did not differ significantly from the Cb−Abx− group in either SM composition (P < 0.1; Figure 3) or diversity (P = 0.98; Figure 4). At 14 d after inoculation, the Cb+Abx+ differed significantly in both composition (P < 0.01; Figure 3) and α-diversity (P < 0.001; Figure 4) from the Cb+Abx− group. At 70 d after inoculation, the Cb+Abx+ did not differ significantly from the Cb−Abx− group in either composition or α-diversity.
In contrast to the situation in nude mice, oral amoxicillin-clavulanic acid did not prevent an initial increase in Corynebacterium spp. on the skin of NSG after 14 d of treatment as compared with Cb+Abx− mice (Figure 3). At 14 d after inoculation, Corynebacterium spp. growth and amoxicillin-clavulanic acid treatment significantly (P = 0.0079) altered the composition of the SM between Cb−Abx− and Cb+Abx+ mice. This difference was characterized by a marked increase in Staphylococcus spp. and marked decreases in Lactobacillus spp., Clostridium spp., Methylobacterium spp., Bacteroidales S24-7, and nonclostridial Lachnospiraceae, all of which contributed to the decrease in microbial α-diversity (P = 0.040; Figure 4). At 70 d after inoculation, the genus Corynebacterium was still present in the SM of the Cb+Abx+ group, albeit at significantly (P = 0.0054) lower relative abundance as compared with the Cb+Abx− group and with little difference relative to the Cb−Abx− group (P = 0.25, Figure 3). When the overall SM composition of the Cb+Abx+ group was compared with that of the Cb+Abx− group, the marked reduction in Corynebacterium spp. and elimination of C. bovis due to prophylactic antibiotics easily achieved statistical significance (P < 0.01, Figure 3). The SM composition of the Cb+Abx+ group was not significantly different from that of the Cb−Abx− group (P = 0.45) at 70 d after inoculation, but a decrease in Shannon diversity between these groups approached statistical significance (P = 0.1, Figure 4). For both strains of mice, principal coordinate analysis suggested clustering of the Cb+Abx+ groups with their respective Cb−Abx− groups by 70 d after inoculation (Figure 5).
Histopathology assessment of skin.
Antemortem clinical signs of C. bovis infection were not observed in any experimental group of either strain during the study. Cross-sections of the ear pinnae and skin of the dorsal head and neck were reviewed histologically for hyperkeratosis, acanthosis, and inflammatory infiltrate of the dermis. In all groups of nude mice, multifocal areas of mild hyperkeratosis with occasional keratohyaline granules and neutrophils and lymphocytes surrounding hair follicles were present (data not shown). For NSG mice, mild acanthosis and focal areas of hyperkeratosis were observed in the Cb+Abx− group but were not apparent in either the Cb−Abx− or Cb+Abx+ groups. No marked increase in inflammatory infiltrate was observed in any treatment group or strain (data not shown).
Given the lack of clinical signs and the minimal pathology associated with the NSG Cb+Abx− group only, histologic characterization of the skin sections was not pursued further.
Discussion
We recently demonstrated that metaphylactic antibiotics can be used to prevent the transmission of C. bovis from infected NSG mouse breeding pairs to their neonatal offspring.47 In the current study, prophylactic antibiotics prevented the establishment of a perpetuating C. bovis infection in both nude and NSG mice. This benefit was most evident for nude mice, in which the skin was free of C. bovis DNA at 7 d after inoculation and all subsequent time points based on qPCR analysis. Although prophylactic antibiotics successfully prevented infection of NSG mice by the end of the study, qPCR data did not clearly characterize their infection status at earlier time points. At 7 d after inoculation, only 1 of 10 (10%) NSG mice inoculated with C. bovis had detectable C. bovis DNA on their hair coat. This finding suggests that at a minimum, residual DNA from the bacterial inoculum had been groomed off of the majority of NSG mice within 7 d. However, by 15 d after inoculation, 8 of 10 (80%) inoculated NSG mice had C. bovis DNA on their hair coat, independent of prophylactic treatment. In addition, the C. bovis DNA copy number detected on the skin of Cb+Abx+ mice approximated the copy number obtained from the skin of the Cb+Abx− mice on the day after antibiotic discontinuation (15 d after inoculation). This finding suggests equivalent dermal colonization despite prophylactic antibiotics treatment between 7 and 15 d after inoculation. Nevertheless, prophylactic antibiotics prevented the establishment of a perpetuating infection on NSG mice by 42 d after inoculation. These data suggest that NSG mice require at least 14 d of prophylactic antibiotic to prevent the establishment of an infection. However, additional studies are needed to investigate the early kinetics of C. bovis infection in NSG mice, with and without the presence of prophylactic antibiotics, before the duration of treatment can be titrated further for infection prevention.
The SM is resilient and does not shift significantly after treatment with oral antibiotics commonly used to disrupt the gut microbiome (GM).44,62,67 Ampicillin is commonly used in antibiotic cocktails to disrupt or deplete the GM of mice.59 With prolonged administration of oral ampicillin to mice, skin dysbiosis was not observed.10 Similarly in our current experiments, 14 d of oral amoxicillin-clavulanic acid did not lead to any significant difference in bacterial abundance or α-diversity at either 15 or 70 d after inoculation of Cb+Abx+ nude mice. In contrast, the SM for Cb+Abx+ NSG mice significantly shifted in both bacterial abundance and α-diversity by the final day of treatment. However, drawing a direct correlation between oral antibiotics and these significant changes in the SM for NSG mice is difficult because an antibiotic-only treatment group was not part of the study design. In addition, C. bovis was detected on the skin at 15 d after inoculation and may have contributed to these significant changes. Nevertheless, by 70 d after inoculation (56 d after antibiotic discontinuation), our data demonstrate a complete elimination of C. bovis and recovery of the composition of the SM for NSG mice as determined by qPCR and 16S microbiome analysis.
In the absence of prophylactic antibiotics after inoculation, C. bovis became highly abundant on the skin of both nude and NSG mice but at very different rates (Figure 2). At 15 d after inoculation, Corynebacterium spp. accounted for 90% and 10% of the skin bacteria of nude and NSG mice, respectively. By 70 d after inoculation, the abundance rates in nude and NSG mice (60% and 45%, respectively) more closely approximated each other (Figure 3). But on 70 d after inoculation, infected nude mice had 5.5-fold more C. bovis DNA on their skin than did NSG mice (Figure 2). These results demonstrate that C. bovis grew more rapidly and in greater numbers on the skin of nude mice as compared with NSG mice. The mechanism or inherent physiology that contributes to this difference is currently unknown.
The use of prophylactic antibiotics to prevent C. bovis infection has been attempted previously, without success.5 In that study, 2 groups of naïve nude mice were housed in a room with enzootically C. bovis–infected nude mice. For the first 14 d, one group received amoxicillin-impregnated chow (200 mg/kg), whereas the other group received no treatment. After 14 d of prophylaxis, antibiotics were discontinued, but the housing conditions remained the same. According to that study, the time to C. bovis infection and clinical disease were not significantly different between groups.5 However, the time of exposure to an infectious dose was not defined.5 In the current study, mice were inoculated acutely with a known infectious dose of C. bovis and then were strictly maintained to prevent further C. bovis exposure during and after antibiotic treatment. Because the time of inoculation and environmental contamination were controlled in our experiment, our design gives credence to the conclusion that prophylactic antibiotics prevent C. bovis infection only when mice are maintained in a C. bovis–free environment during and after antibiotic withdrawal.
As a tool for the management of C. bovis–free colonies, prophylactic antibiotics may be practical only in situations where C. bovis exposure is identified quickly or is imminent. C. bovis exposure can be anticipated during the implantation of potentially or known C. bovis–contaminated tumor tissue. Recently we demonstrated that contaminated patient-derived xenograft (PDX) tumor tissue could transmit C. bovis infection to immunodeficient recipient mice.37 In addition, 2 independent diagnostic laboratories have reported that C. bovis is a common contaminant of eukaryotic cell lines and tumor tissues submitted for diagnostic analysis.37 At our institution, we have addressed this concern by providing prophylactic antibiotics to naïve recipient mice scheduled to receive PDX tumor tissue that is suspected of being contaminated, received from another institution at which the C. bovis status is unknown, or had questionable aseptic harvest of the donor tumor tissue.37 The extrapolation of our data for the use of prophylactic antibiotics to prevent C. bovis transmission from ‘high-risk’ PDX tissue has been successful in our hands. Since we initiated this practice 9 y ago, C. bovis infection after PDX implantation has not occurred.
A variety of clinically relevant antibiotics, including doxycycline, macrolides such as erythromycin, and the fluoroquinolone ciprofloxacin have shown detrimental effects on cancer cell growth either in vitro or in vivo.11,25,33,48,66 In addition, several other less clinically relevant antibiotics and antifungal compounds have shown similar effects when their specific mechanism of action interferes with a eukaryotic cellular function.1,51,57 Amoxicillin is in the aminopenicillin class of bicyclic β-lactam antibiotics. Bicyclic β-lactams are specific to and inhibit peptidoglycan crosslinking during bacterial cell wall remodeling or division. The failure of this peptidoglycan matrix, not present in eukaryotic cell walls, results in bacterial cytolysis. Because of its specific mechanism of action, the β-lactam antibiotic penicillin has been used for decades to prevent bacterial growth in eukaryotic cell cultures and is recommended by American Type Tissue Collection when antibiotics are used.2 Although modification of the β-lactam structure into a monocyclic form has been investigated for tumor cell-killing ability, bicyclic β-lactams are not known to be a viable anticancer candidate.31
Antibiotics have been shown to significantly alter the GM of rodents.14 When amoxicillin is administered orally to mice at a dose and for a duration that approximates those in our study, diversity in the GM decreased. The GM that remains demonstrated a decrease in the genus Bifidobacterium and was highly enriched in the Proteobacteria phylum (mainly Escherichia-Shigella and Klebsiella), which is a microbial signature of dysbiosis.35 In addition, oral amoxicillin increases the abundance and diversity of antibiotic-resistance genes, β-lactam ring resistance genes, in surviving bacteria.35 Similarly, mice treated with the combination of amoxicillin and clavulanic acid show a decrease in the relative abundance of the phylum Firmicutes (mainly Clostridiales) and a concurrent expansion of the phylum Proteobacteria.3 Rats treated with oral amoxicillin likewise show a reduction in GM diversity, a decrease in Firmicutes, and an increase in the relative abundance of Bacteroidetes and Proteobacteria.24,61 Due to the consistent findings of these previous reports on the impact of oral amoxicillin on the GM of rodents, with and without clavulanic acid, we elected not to assess the GM in the current study.
The direct influence of antibiotics on the GM can indirectly affect the innate and adaptive immune systems of mice. Through a sequence of direct and indirect actions, antibiotics can increase or decrease tumor growth and metastasis in syngeneic or allograft tumors in immunocompetent mice.26,56 The exact mechanism remains elusive to date, but what is known has been reviewed elsewhere.26,27,36 The vast majority of this literature involves immunocompetent mice, and data on the gut-immune-cancer axis in immunocompromised mice is minimal. However, several studies have shown that antibiotics administered to immunodeficient mice can promote growth and metastasis,7 inhibit or delay growth,58 or have negligible effects in a variety of xenograft tumor models.56 Independent of immune status of the mouse model used, some evidence also indicates a direct interplay between tumor cells and the GM. For colorectal and pancreatic cancers specifically, the presence of bacteria either in the tumor23,32 or in direct contact with tumors cells9,53 can increase xenograft tumor growth. In turn, the elimination of the intratumor bacteria by antibiotic therapy potentially can alter tumor sensitivity to chemotherapeutics.23 Similarly, an antibiotic-induced decrease in the amount and complexity of the GM likewise can alter tumor cytokine expression phenotype9 or even reduce xenograft tumor growth.4 Thus moving forward, reluctance to use prophylactic antibiotics to prevent C. bovis–infection by contaminated tumor tissue should not be based on the mechanism of action of the antibiotic on tumor cells, but rather on the off-target effects antibiotics can have on intratumor bacteria or the GM. Nevertheless, this risk must be weighed against the complications induced by C. bovis infection,37 the negative effect of C. bovis on cancer models,63 the high likelihood of spread of the infection within immunodeficient mouse colonies,6 and proven difficulty in eliminating this agent from research institutions.5,39,42
Motivated by our institution’s goal to eliminate endemic opportunistic pathogen C. bovis from our immunodeficient mouse colonies, this project investigated the potential use of prophylactic antibiotics to prevent C. bovis infection. A 14 d course of oral amoxicillin-clavulanic acid in nude and NSG mice rendered C. bovis unable to establish a perpetuating infection and prevented concurrent infection from further exposure. This study also allowed us to determine the composition of the SM of these strains of mice and the influence of C. bovis on skin microbial communities. Our findings also show the significant effect of C. bovis and the antimicrobial treatment on skin commensals and document the potential benefit of prophylactic antibiotics to prevent C. bovis infection.
Acknowledgments
This project was funded with a grant to CAM from the American College of Laboratory Animal Medicine (ACLAM) Foundation. DI, CER, and DNF were supported in part by funding from the Colorado University Denver GI and Liver Innate Immunity Program.
References
- 1. Alibek K, Bekmurzayeva A, Mussabekova A, Sultankulov B. 2012. Using antimicrobial adjuvant therapy in cancer treatment: a review. Infect Agent Cancer 7: 33. 10.1186/1750-9378-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. American Type Culture Collection. [Internet]. 2020. Adding antibiotics or antimycotics to cell culture media. [Cited 16 Aug 2020]. Available at: https://www.atcc.org/support/faqs/216ac/Adding%20antibiotics%20or%20antimycotics%20to%20cell%20culture%20medium%E2%80%9379.aspx.
- 3. Benakis C, Brea D, Caballero S, Faraco G, Moore J, Murphy M, Sita G, Racchumi G, Ling L, Pamer EG, Iadecola C, Anrather J. 2016. Commensal microbiota affects ischemic stroke outcome by regulating intestinal γδ T cells. Nat Med 22: 516– 523. 10.1038/nm.4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D, Neuberg D, Huang K, Guevara F, Nelson T, Chipashvili O, Hagan T, Walker M, Ramachandran A, Diosdado B, Serna G, Mulet N, Landolfi S, Ramon YCS, Fasani R, Aguirre AJ, Ng K, Élez E, Ogino S, Tabernero J, Fuchs CS, Hahn WC, Nuciforo P, Meyerson M. 2017. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 358: 1443– 1448. 10.1126/science.aal5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Burr HN, Lipman NS, White JR, Zheng J, Wolf FR. 2011. Strategies to prevent, treat, and provoke corynebacterium-associated hyperkeratosis in athymic nude mice. J Am Assoc Lab Anim Sci 50: 378– 388. [PMC free article] [PubMed] [Google Scholar]
- 6. Burr HN, Wolf FR, Lipman NS. 2012. Corynebacterium bovis: epizootiologic features and environmental contamination in an enzootically infected rodent room. J Am Assoc Lab Anim Sci 51: 189– 198. [PMC free article] [PubMed] [Google Scholar]
- 7. Cheung MK, Yue GGL, Tsui KY, Gomes AJ, Kwan HS, Chiu PWY, Lau CBS. 2020. Discovery of an interplay between the gut microbiota and esophageal squamous cell carcinoma in mice. Am J Cancer Res 10: 2409– 2427. [PMC free article] [PubMed] [Google Scholar]
- 8. Clifford CB, Walton BJ, Reed TH, Coyle MB, White WJ, Amyx HL. 1995. Hyperkeratosis in athymic nude mice caused by a coryneform bacterium: microbiology, transmission, clinical signs, and pathology. Lab Anim Sci 45: 131– 139. [PubMed] [Google Scholar]
- 9. Cremonesi E, Governa V, Garzon JFG, Mele V, Amicarella F, Muraro MG, Trella E, Galati-Fournier V, Oertli D, Däster SR, Droeser RA, Weixler B, Bolli M, Rosso R, Nitsche U, Khanna N, Egli A, Keck S, Slotta-Huspenina J, Terracciano LM, Zajac P, Spagnoli GC, Eppenberger-Castori S, Janssen KP, Borsig L, Iezzi G. 2018. Gut microbiota modulate T cell trafficking into human colorectal cancer. Gut 67: 1984– 1994. 10.1136/gutjnl-2016-313498. [DOI] [PubMed] [Google Scholar]
- 10. Dellacecca ER, Cosgrove C, Mukhatayev Z, Akhtar S, Engelhard VH, Rademaker AW, Knight KL, Le Poole IC. 2020. Antibiotics drive microbial imbalance and vitiligo development in mice. J Invest Dermatol 140: 676– 687.e6. 10.1016/j.jid.2019.08.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Duivenvoorden WCM, Popović SV, Lhoták Š, Seidlitz E, Hirte HW, Tozer RG, Singh G. 2002. Doxycycline decreases tumor burden in a bone metastasis model of human breast cancer. Cancer Res 62: 1588– 1591. [PubMed] [Google Scholar]
- 12. Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27: 2194– 2200. 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ericsson AC, Davis J, Spollen W, Bivens N, Givan S, Hagan C, McIntosh M, Franklin CL. 2015. Effects of vendor and genetic background on the composition of the fecal microbiota of inbred mice. PLoS One 10: e0116704. 10.1371/journal.pone.0116704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ericsson AC, Franklin CL. 2015. Manipulating the gut microbiota: methods and challenges. ILAR J 56: 205– 217. 10.1093/ilar/ilv021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ewing B, Green P. 1998. Base-calling of automated sequencer traces using phred. II. error probabilities. Genome Res 8: 186– 194. 10.1101/gr.8.3.186. [DOI] [PubMed] [Google Scholar]
- 16. Ewing B, Hillier L, Wendl MC, Green P. 1998. Base-calling of automated sequencer traces using phred. I. accuracy assessment. Genome Res 8: 175– 185. 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 17. Fagre AC, Pugazhenthi U, Cheleuitte-Nieves C, Crim MJ, Henderson KS, Fong DL, Leszczynski JK, Schurr MJ, Daniels JB, Manuel CA. 2021. Antimicrobial susceptiblity of corynebacterium bovis isolates from immunodeficient rodents. Comp Med 71: 210– 214. 10.30802/AALAS-CM-20-000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Findley K, Grice EA. 2014. The skin microbiome: A focus on pathogens and their association with skin disease. PLoS Pathog 10: e1004436. 10.1371/journal.ppat.1004436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Frank DN. 2009. BARCRAWL and BARTAB: software tools for the design and implementation of barcoded primers for highly multiplexed DNA sequencing. BMC Bioinformatics 10: 362. 10.1186/1471-2105-10-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Frank DN, Giese APJ, Hafren L, Bootpetch TC, Yarza TKL, Steritz MJ, Pedro M, Labra PJ, Daly KA, Tantoco MLC, Szeremeta W, Reyes-Quintos MRT, Ahankoob N. 2021. Otitis media susceptibility and shifts in the head and neck microbiome due to Safter inoculationNK5 variants. J Med Genet 58: 442– 452. 10.1136/jmedgenet-2020-106844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Frank JA, Reich CI, Sharma S, Weisbaum JS, Wilson BA, Olsen GJ. 2008. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl Environ Microbiol 74: 2461– 2470. 10.1128/AEM.02272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Geddes A, Gould I. Penicillins and related drugs: ampicllin, amoxicillin, and other ampicillin-like penicillins, p 55- 93. In: Lindsay MG, editor. Kucers’ the use of antibiotics sixth ed: a clinical review of antibacterial, antifungal, antiparasitic and antiviral drugs. Boca Raton (FL): CRC Press. [Google Scholar]
- 23. Geller LT, Barzily-Rokni M, Danino T, Jonas OH, Shental N, Nejman D, Gavert N, Zwang Y, Cooper ZA, Shee K, Thaiss CA, Reuben A, Livny J, Avraham R, Frederick DT, Ligorio M, Chatman K, Johnston SE, Mosher CM, Brandis A, Fuks G, Gurbatri C, Gopalakrishnan V, Kim M, Hurd MW, Katz M, Fleming J, Maitra A, Smith DA, Skalak M, Bu J, Michaud M, Trauger SA, Barshack I, Golan T, Sandbank J, Flaherty KT, Mandinova A, Garrett WS, Thayer SP, Ferrone CR, Huttenhower C, Bhatia SN, Gevers D, Wargo JA, Golub TR, Straussman R. 2017. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science 357: 1156– 1160. 10.1126/science.aah5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Graversen KB, Bahl MI, Larsen JM, Ballegaard A-SR, Licht TR, Bøgh KL. 2020. Short-term amoxicillin-induced perturbation of the gut microbiota promotes acute intestinal immune regulation in Brown Norway rats. Front Microbiol 11: 496. 10.3389/fmicb.2020.00496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Herold C, Ocker M, Ganslmayer M, Gerauer H, Hahn EG, Schuppan D. 2002. Ciprofloxacin induces apoptosis and inhibits proliferation of human colorectal carcinoma cells. Br J Cancer 86: 443– 448. 10.1038/sj.bjc.6600079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jain T, Sharma P, Are AC, Vickers SM, Dudeja V. 2021. New insights into the cancer–microbiome–immune axis: decrypting a decade of discoveries. Front Immunol 12: 622064. 10.3389/fimmu.2021.622064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kim J, Lee HK. 2021. The role of gut microbiota in modulating tumor growth and anticancer agent efficacy. Mol Cells 44: 356– 362. 10.14348/molcells.2021.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim Y, Lee Y-S, Yang J-Y, Lee S-H, Park Y-Y, Kweon M-N. 2017. The resident pathobiont Staphylococcus xylosus in Nfkbiz-deficient skin accelerates spontaneous skin inflammation. Sci Rep 7: 6348. 10.1038/s41598-017-05740-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kobayashi T, Glatz M, Horiuchi K, Kawasaki H, Akiyama H, Kaplan Daniel H, Kong Heidi H, Amagai M, Nagao K. 2015. Dysbiosis and staphylococcus aureus colonization drives inflammation in atopic dermatitis. Immunity 42: 756– 766. 10.1016/j.immuni.2015.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Korte SW, Franklin CL, Dorfmeyer RA, Ericsson AC. 2018. Effects of fenbendazole-impregnated feed and topical moxidectin during quarantine on the gut microbiota of C57BL/6 mice. J Am Assoc Lab Anim Sci 57: 229– 235. [PMC free article] [PubMed] [Google Scholar]
- 31. Kuhn D, Coates C, Daniel K, Chen D, Bhuiyan M, Kazi A, Turos E, Dou QP. 2004. Beta-lactams and their potential use as novel anticancer chemotherapeutics drugs. Front Biosci 9: 2605– 2617. 10.2741/1420. [DOI] [PubMed] [Google Scholar]
- 32. Kumar R, Herold JL, Schady D, Davis J, Kopetz S, Martinez-Moczygemba M, Murray BE, Han F, Li Y, Callaway E, Chapkin RS, Dashwood W-M, Dashwood RH, Berry T, Mackenzie C, Xu Y. 2017. Streptococcus gallolyticus subsp. gallolyticus promotes colorectal tumor development. PLoS Pathog 13: e1006440. 10.1371/journal.ppat.1006440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lamb R, Ozsvari B, Lisanti CL, Tanowitz HB, Howell A, Martinez-Outschoorn UE, Sotgia F, Lisanti MP. 2015. Antibiotics that target mitochondria effectively eradicate cancer stem cells, across multiple tumor types: Treating cancer like an infectious disease. Oncotarget 6: 4569– 4584. 10.18632/oncotarget.3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lane DJ. 1991. 16S/23S rRNA sequencing, p. 115– 175. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York (NY): Wiley. [Google Scholar]
- 35. Lin H, Wang Q, Yuan M, Liu L, Chen Z, Zhao Y, Das R, Duan Y, Xu X, Xue Y, Luo Y, Mao D. 2020. The prolonged disruption of a single-course amoxicillin on mice gut microbiota and resistome, and recovery by inulin, Bifidobacterium longum and fecal microbiota transplantation. Environ Pollut 265: 114651. 10.1016/j.envpol.2020.114651. [DOI] [PubMed] [Google Scholar]
- 36. Liu X, Chen Y, Zhang S, Dong L. 2021. Gut microbiota-mediated immunomodulation in tumor. J Exp Clin Cancer Res 40: 221. 10.1186/s13046-021-01983-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Manuel CA, Bagby SM, Reisinger JA, Pugazhenthi U, Pitts TM, Keysar SB, Arcaroli JJ, Leszczynski JK. 2017. Procedure for horizontal transfer of patient-derived xenograft tumors to eliminate Corynebacterium bovis. J Am Assoc Lab Anim Sci 56: 166– 172. [PMC free article] [PubMed] [Google Scholar]
- 38. Manuel CA, Pugazhenthi U, Leszczynski J. 2016. Surveillance of a ventilated rack system for corynebacterium bovis by sampling exhaust air manifolds. J Am Assoc Lab Anim Sci 55: 58– 65. [PMC free article] [PubMed] [Google Scholar]
- 39. Manuel CA, Pugazhenthi U, Spiegel SP, Leszczynski JK. 2017. Detection and elimination of Corynebacterium bovis from barrier rooms with an environmental sampling surveillance program. J Am Assoc Lab Anim Sci 56: 202– 209. [PMC free article] [PubMed] [Google Scholar]
- 40. Marx JO, Vudathala D, Murphy L, Rankin S, Hankenson FC. 2014. Antibiotic administration in the drinking water of mice. J Am Assoc Lab Anim Sci 53: 301– 306. [PMC free article] [PubMed] [Google Scholar]
- 41. McIntyre AR, Lipman NS. 2007. Amoxicillin-clavulanic acid and trimethoprim-sulfamethoxazole in rodent feed and water: effects of compounding on antibiotic stability. J Am Assoc Lab Anim Sci 46: 26– 32. [PubMed] [Google Scholar]
- 42. Miedel EL, Ragland NH, Engelman RW. 2018. Facility-wide eradication of Corynebacterium bovis by using PCR-validated vaporized hydrogen peroxide. J Am Assoc Lab Anim Sci 57: 465– 476. 10.30802/AALAS-JAALAS-17-000135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Miedel EL, Ragland NH, Slate AR, Engelman RW. 2020. Persistent Corynebacterium bovis infectious hyperkeratotic dermatitis in immunocompetent epidermal-mutant dep/dep mice. Vet Pathol 57: 586– 589. 10.1177/0300985820922219. [DOI] [PubMed] [Google Scholar]
- 44. Naik S, Bouladoux N, Wilhelm C, Molloy MJ, Salcedo R, Kastenmuller W, Deming C, Quinones M, Koo L, Conlan S, Spencer S, Hall JA, Dzutsev A, Kong H, Campbell DJ, Trinchieri G, Segre JA, Belkaid Y. 2012. Compartmentalized control of skin immunity by resident commensals. Science 337: 1115– 1119. 10.1126/science.1225152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Oksanen J, Blanchet G, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MH, Szoecs E, Wagner H. [Internet]. 2013. Vegan: community ecology package. R package version 2. 5– 7. [Cited 12 Dec 2021]. Available at: http://vegan.r-forge.r-project.org.
- 46. Olli KE, Rapp C, O’Connell L, Collins CB, McNamee EN, Jensen O, Jedlicka P, Allison KC, Goldberg MS, Gerich ME, Frank DN, Ir D, Robertson CE, Evans CM, Aherne CM. 2020. Muc5ac expression protects the colonic barrier in experimental colitis. Inflamm Bowel Dis 26: 1353– 1367. 10.1093/ibd/izaa064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pearson EC, Pugazhenthi U, Fong DL, Smith DE, Nicklawsky AG, Habenicht LM, Fink MK, Leszczynski JK, Schurr MJ, Manuel CA. 2020. Metaphylactic antibiotic treatment to prevent Corynebacterium bovis transmission to immunocompromised mouse offspring. J Am Assoc Lab Anim Sci 59: 712– 718. 10.30802/AALAS-JAALAS-20-000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pillozzi S, D’Amico M, Petroni G, Veltroni M, Favre C, Becchetti A, Basso G, Arcangeli A. 2016. Macrolide antibiotics by modulating the autophagic flux exert an antileukemic effect through the involvement of hERG1 potassium channel. Blood 128: 3953. 10.1182/blood.V128.22.3953.3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pruesse E, Peplies J, Glockner FO. 2012. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28: 1823– 1829. 10.1093/bioinformatics/bts252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41 D1: D590– D596. 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Raynal NJ-M, Lee JT, Wang Y, Beaudry A, Madireddi P, Garriga J, Malouf GG, Dumont S, Dettman EJ, Gharibyan V, Ahmed S, Chung W, Childers WE, Abou-Gharbia M, Henry RA, Andrews AJ, Jelinek J, Cui Y, Baylin SB, Gill DL, Issa J-PJ. 2016. Targeting calcium signaling induces epigenetic reactivation of tumor suppressor genes in cancer. Cancer Res 76: 1494– 1505. 10.1158/0008-5472.CAN-14-2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Robertson CE, Harris JK, Wagner BD, Granger D, Browne K, Tatem B, Feazel LM, Park K, Pace NR, Frank DN. 2013. Explicet: graphical user interface software for metadata-driven management, analysis and visualization of microbiome data. Bioinformatics 29: 3100– 3101. 10.1093/bioinformatics/btt526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. 2013. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe 14: 195– 206. 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Scanziani E, Gobbi A, Crippa L, Giusti AM, Pesenti E, Cavalletti E, Luini M. 1998. Hyperkeratosis-associated coryneform infection in severe combined immunodeficient mice. Lab Anim 32: 330– 336. 10.1258/002367798780559239. [DOI] [PubMed] [Google Scholar]
- 55. Schloss PD, Westcott SL. 2011. Assessing and improving methods used in operational taxonomic unit-based approaches for 16S rRNA gene sequence analysis. Appl Environ Microbiol 77: 3219– 3226. 10.1128/AEM.02810-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sethi V, Kurtom S, Tarique M, Lavania S, Malchiodi Z, Hellmund L, Zhang L, Sharma U, Giri B, Garg B, Ferrantella A, Vickers SM, Banerjee S, Dawra R, Roy S, Ramakrishnan S, Saluja A, Dudeja V. 2018. Gut microbiota promotes tumor growth in mice by modulating immune response. Gastroenterology 155: 33– 37.e6. 10.1053/j.gastro.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Solit DB, Zheng FF, Drobnjak M, Münster PN, Higgins B, Verbel D, Heller G, Tong W, Cordon-Cardo C, Agus DB, Scher HI, Rosen N. 2002. 17-Allylamino-17-demethoxygeldanamycin induces the degradation of androgen receptor and HER-2/neu and inhibits the growth of prostate cancer xenografts. Clin Cancer Res 8: 986– 993. [PubMed] [Google Scholar]
- 58. Thomas RM, Gharaibeh RZ, Gauthier J, Beveridge M, Pope JL, Guijarro MV, Yu Q, He Z, Ohland C, Newsome R, Trevino J, Hughes SJ, Reinhard M, Winglee K, Fodor AA, Zajac-Kaye M, Jobin C. 2018. Intestinal microbiota enhances pancreatic carcinogenesis in preclinical models. Carcinogenesis 39: 1068– 1078. 10.1093/carcin/bgy073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tirelle P, Breton J, Riou G, Déchelotte P, Coëffier M, Ribet D. 2020. Comparison of different modes of antibiotic delivery on gut microbiota depletion efficiency and body composition in mouse. BMC Microbiol 20: 340. 10.1186/s12866-020-02018-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Towne JW, Wagner AM, Griffin KJ, Buntzman AS, Frelinger JA, Besselsen DG. 2014. Elimination of Pasteurella pneumotropica from a mouse barrier facility by using a modified enrofloxacin treatment regimen. J Am Assoc Lab Anim Sci 53: 517– 522. [PMC free article] [PubMed] [Google Scholar]
- 61. Tulstrup MV-L, Christensen EG, Carvalho V, Linninge C, Ahrné S, Højberg O, Licht TR, Bahl MI. 2015. Antibiotic treatment affects intestinal permeability and gut microbial composition in Wistar Rats dependent on antibiotic class. PLoS One 10: e0144854. 10.1371/journal.pone.0144854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Uberoi A, Bartow-McKenney C, Zheng Q, Flowers L, Campbell A, Knight SAB, Chan N, Wei M, Lovins V, Bugayev J, Horwinski J, Bradley C, Meyer J, Crumrine D, Sutter CH, Elias P, Mauldin E, Sutter TR, Grice EA. 2021. Commensal microbiota regulates skin barrier function and repair via signaling through the aryl hydrocarbon receptor. Cell Host Microbe 29: 1235– 1248.e8. 10.1016/j.chom.2021.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Vedder AR, Miedel E, Ragland N, Balasis M, Letson C, Engelman RW, Padron E. 2019. Effects of Corynebacterium bovis on engraftment of patient-derived chronic myelomonocytic leukemia cells in NSGS mice. Comp Med 69: 276– 282. 10.30802/AALAS-CM-18-000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Watts JL, Rossbach S. 2000. Susceptibilities of Corynebacterium bovis and Corynebacterium amylocolatum isolates from bovine mammary glands to 15 antimicrobial agents. Antimicrob Agents Chemother 44: 3476– 3477. 10.1128/AAC.44.12.3476-3477.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wheatley EG, Curtis BJ, Hulsebus HJ, Boe DM, Najarro K, Ir D, Robertson CE, Choudhry MA, Frank DN, Kovacs EJ. 2020. Advanced age impairs intestinal antimicrobial peptide response and worsens fecal microbiome dysbiosis following burn injury in mice. Shock 53: 71– 77. 10.1097/SHK.0000000000001321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Yatsunami J, Fukuno Y, Nagata M, Tominaga M, Aoki S, Tsuruta N, Kawashima M, Taniguchi S, Hayashi S. 1999. Antiangiogenic and antitumor effects of 14-membered ring macrolides on mouse B16 melanoma cells. Clin Exp Metastasis 17: 359– 365. 10.1023/A:1006605725619. [DOI] [PubMed] [Google Scholar]
- 67. Zhang M, Jiang Z, Li D, Jiang D, Wu Y, Ren H, Peng H, Lai Y. 2015. Oral antibiotic treatment induces skin microbiota dysbiosis and influences wound healing. Microb Ecol 69: 415– 421. 10.1007/s00248-014-0504-4. [DOI] [PubMed] [Google Scholar]


