Abstract
The spread of antibiotic resistance among pathogenic bacteria is a serious threat to humans and animals. Therefore, unnecessary use should be minimized, and new antimicrobial agents with novel mechanisms of action are needed. We have developed an efficient method for measuring the action of antibiotics which is applied to a gram-positive strain, Staphylococcus aureus RN4220. The method utilizes the firefly luciferase reporter gene coupled to the metal-inducible cadA promoter in a plasmid, pTOO24. Correctly timed induction by micromolar concentrations of antimonite rapidly triggers the luciferase gene transcription and translation. This sensitizes the detection system to the action of antibiotics, and especially for transcriptional and translational inhibitors. We show the results for 11 model antibiotics with the present approach and compare them to an analytical setup with a strain where luciferase expression is under the regulation of a constitutive promoter giving only a report of metabolic inhibition. The measurement of light emission from intact living cells is shown to correlate extremely well (r = 0.99) with the conventional overnight growth inhibition measurement. Four of the antibiotics were within a 20% concentration range and four were within a 60% concentration range of the drugs tested. This approach shortens the assay time needed, and it can be performed in 1 to 4 h, depending on the sensitivity needed. Furthermore, the assay can be automatized for high-throughput screening by the pharmaceutical industry.
The gram-positive pathogenic staphylococcus Staphylococcus aureus is considered a threat to human health due to its capacity to efficiently infect hospital patients with weakened immune status. The methicillin-resistant S. aureus (MRSA) has been found in serious hospital outbreaks that have proved difficult to treat. Together with methicillin resistance, MRSA has also obtained other resistances through evolution, thereby making the situation even worse (for a review, see reference 7). The recent development of MRSA outbreaks and the emergence of resistant strains of Mycobacterium tuberculosis among others have led to a new interest in the pharmaceutical industry in searching for new antimicrobial agents. High-throughput screening (HTS) technologies applied to high-diversity combinatorial chemistry molecular libraries make it possible to find lead molecules with new modes of action as antimicrobial agents.
Luciferases are a heterologous group of intriguing proteins that produce visible light, i.e., bioluminescence. There do not appear to exist any evolutionary relationships between different luciferases in a wide variety of species capable of bioluminescence. Molecular oxygen is the only common factor needed for emission of light by the luciferases. Other substrates and cofactors show wide chemical varieties (15, 17). The most well-known bioluminescence phenomenon is from the North American firefly, Photinus pyralis. The enzymatic reaction is as follows: luciferase ATP + d-luciferin + O2 ⇒ AMP + oxyluciferin + PPi + CO2 + light (∼560 nm)
The firefly luciferase gene is by far the reporter of choice, not only due to its very sensitive detection limit of 0.05 amol/sample (10) but also because it has been used in commercial detection reagents and vectors for long time. As a consequence of all this, it has been expressed in a very wide variety of cell types of procaryotic and eucaryotic origin (3). Expression of the firefly luciferase gene inserted under the control of a regulatory element to be studied allows the quantitation of factors affecting this regulation specifically in real time. Furthermore, various regulatory circuits and their control, such as signal transduction via G-protein-coupled receptors, can be studied by simple measurement of light emission from intact living cells (reviewed in reference 8).
Recently, the bacterial luciferase operon modified for expression in gram-positive organisms was used to study the infection process of S. aureus in living mice (2). This system does not necessitate the external addition of any substrates for bioluminescence. On the other hand, the firefly luciferase gene has been expressed in S. aureus by members of our group and others (11, 12). These studies showed that detection of light emission from intact living cells can be done after the addition of d-luciferin. In our study we used the firefly luciferase gene as a sensitive reporter of heavy metal contamination by inserting the gene under a metal-responsive genetic element, cadA, of the staphylococcal plasmid pI258 (12). Here we have applied this system for the measurement of action against the S. aureus RN4220 reporter strain, and we show that the system is sensitized to different antimicrobial agents. The sensor can be triggered to produce luciferase protein by inducing the encoding gene with various metal ions, such as Cd2+, Pb2+, Hg2+, Zn2+, and antimonite ions (SbO2−). Cadmium ions are the best inducers, but instead, in this study, we used less toxic antimonite ions (1.0 μM) to induce the luciferase synthesis. Since the machinery is rapidly turned on, the action of transcriptional and translational inhibitors will especially have an effect on light production. Bioluminescence measurements can be performed from the beginning of the assay till the end in microtitration plates, and hence the strain can be used to screen large chemical libraries. We show that the results obtained closely correlate with those from conventional overnight cultivation experiments.
MATERIALS AND METHODS
Materials.
All the antibiotics were from Sigma except for chloramphenicol and erythromycin, which were from Serva Feinchemie GmbH&Co, (Heidelberg, Germany), and ciprofloxacin-HCl, which was from Bayer. The antibiotic stock solutions were stored frozen at −20°C at a concentration of 10 or 40 mg/ml. The cultivation media were from Difco. d-luciferin was from BioOrbit Oy (Turku, Finland). Antimonite (C4H4KO7Sb) was of purum grade (≥99%) from Fluka. The buffer chemicals were from Sigma. All stocks and dilutions of chemicals were made into ultra-pure commercial infusion grade water (Pharmacia, Uppsala, Sweden).
Bacterial strains.
The bacterial strains S. aureus RN4220/pCSS810 and S. aureus RN4220/pTOO24 used in this study have been characterized previously (12). The strain S. aureus RN4220/pCSS810 produces firefly luciferase constitutively. In strain S. aureus RN4220/pTOO24, the firefly luciferase production is under the control of the cadA promoter and cadC regulatory protein of the staphylococcal plasmid pI258 (9, 18).
Cultivation of bacteria.
Both of the strains were cultivated in Luria-Bertani (LB) medium containing kanamycin (30 μg/ml) at 30°C. The cells were grown to an optical density at 600 nm of 1.5 and subjected to an ice bath after a 1-to-20 dilution with the medium until further use.
Fifty-percent-inhibitory-concentration (IC50) measurements by cultivation.
The diluted cells (1 ml) were added to 14-ml Falcon tubes containing 0.5 ml of different antibiotic dilutions in LB and 0.5 ml of broth (LB plus kanamycin [30 μg/ml]). The cells were grown for 12 h at 30°C with shaking (250 rpm), after which the optical density was measured at 600 nm with from 1 to 10 dilutions.
Antibiotic treatments and measurement of light emission.
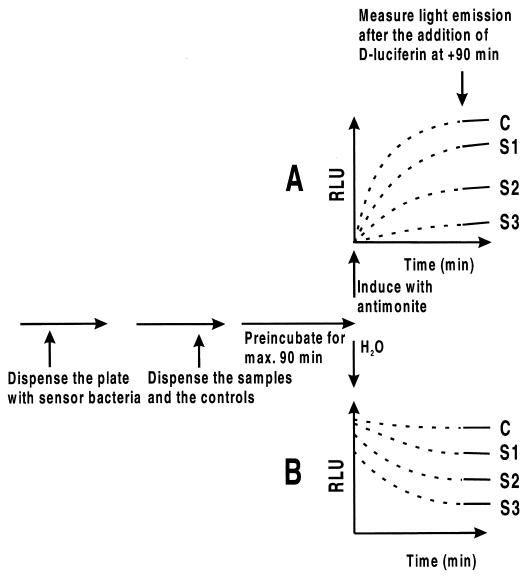
A schematic presentation of the pipetting order and typical light emission pattern are shown in Fig. 1 to visualize the approaches. A suitable dilution of the cells, either RN4220/pTOO24 or RN4220/pCSS810 (50 μl), was dispensed into white 96-well microtitration plate wells and preincubated with different antibiotics, labeled as samples S1, S2 and S3 (25 μl diluted into H20) at 30°C. For Fig. 1A, the firefly luciferase reporter synthesis was triggered by adding the inducer antimonite (25 μl) to a 1 μM concentration, and incubation was continued. For Fig. 1B, water instead of the inducer was pipetted to the constitutive expression system. Light emission was obtained after the addition of the luciferase substrate, d-luciferin, which was added after 90 min at concentration of 0.5 mM (100 μl) in an appropriate buffer (see below). The bioluminescence was measured immediately after the dispensing of the substrate using a Labsystems Luminoskan microtitration plate luminometer (Helsinki, Finland) with an integral type of measurement for 5 s. In some cases the measurement reading was repeated after 30 min.
FIG. 1.
Flow chart and a schematic representation of the response curves for inducible- and constitutive-expression-based methods. Diagrams A and B show how the light emission changes in the two methods during the assay, as if the substrate for d-luciferin were already present at the beginning. In reality, the substrate is added at 90 min (dotted line changes to solid one) as calculated from the end of the preincubation period. The curves in diagram A (the inducible system) indicate that the amount of luciferase protein, shown as RLU, at time point zero min is negligible, and it start to accumulate only after the inducer is added as a function of drug samples S1 to S3 affecting the system in comparison to water control (C). On the other hand, in the constitutive expression method B, a large amount of luciferase protein is already present in the sensor cells, the activity of which is being affected more or less by samples S1 to S3 and is compared to the control (C), unaffected measurement.
Measurement of pH dependence.
For the study of the optimal pH for the d-luciferin substrate penetration inside the reporter cells, 50 mM Na-citrate, morpholineethanesulfonic acid (MES) and morpholinepropanesulfonic acid (MOPS) buffers were used. The cells were preincubated for 90 min at room temperature together with different concentrations of the test antibiotic chloramphenicol, after which they were challenged by adding the antimonite inducer to a 1 μM concentration. After an induction period of 120 min, 100 μl of 1 mM d-luciferin (final concentration, 0.5 mM in the measurement cuvette) in different buffers was added by using a manual dispenser and measured for the immediate bioluminescence emission. The measurements were done in three parallel determinations, and they were repeated twice.
RESULTS
The experimental setup.
The metal-inducible reporter system utilizing bioluminescent S. aureus RN4220/pTOO24 for the measurement of the presence of certain heavy metals has been described earlier (12). In this study the reporter system is applied for a different purpose, and emphasis was put on optimizing the sensor system for the measurement of the effects of various antimicrobial agents. The synthesis of the reporter enzyme, firefly luciferase, was turned on by using antimonite ions as inducing molecules. We studied the optimal preincubation time from −120 min to zero by incubating a suitable dilution of freshly cultivated sensor cells together with different concentrations of a model antibiotic, chloramphenicol. It was found that 90 min is enough to obtain a full response, i.e., maximal test drug penetration through the staphylococcal cell membrane (data not shown). It should be noted that in the fishing of leads from combinatorial libraries, the preincubation period plays a major role. If one wants to screen for well-penetrating compounds, then the preincubation period should be decreased to a few minutes rather than 90 min, shown in Fig. 1. A schematic presentation as a flow chart and response curves is shown in Fig. 1. Figure 1A shows the inducible system, and panel B shows response curves for the constitutive expression system. In the metal-inducible system, the luciferase production starts as the inducer is added. If toxic antibiotics are present, the level of synthesis will be lower than that for the control nontreated sample. In the constitutive system the reporter cells already contain a certain amount of the luciferase enzyme at the beginning of the experiment, and addition of antibiotic samples results mainly in metabolic inhibition, such as depletion of the intracellular ATP pool.
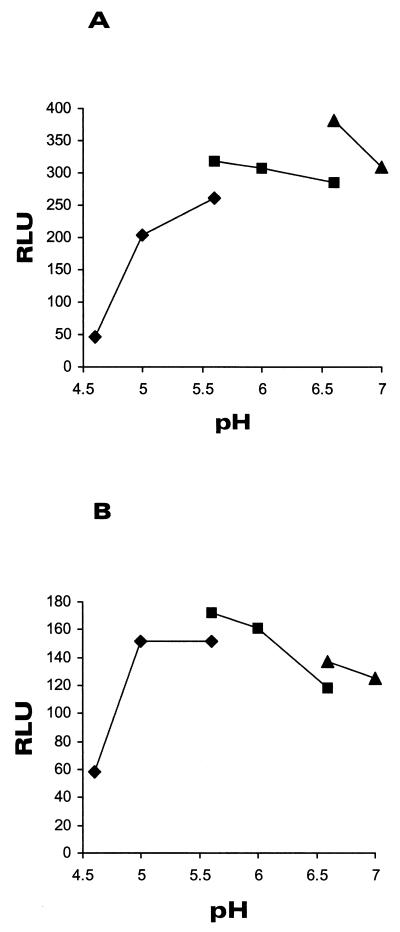
The dependence of bioluminescence on pH.
Different buffers were used to study the transport of the substrate for firefly luciferase, d-luciferin, inside the S. aureus/pTOO24 cells. The pH profile from 4.6 to 7.0 using Na citrate, MES, and MOPS buffers is shown in Fig. 2. We studied two different parameters simultaneously, namely, the functionality of the cells as measured by the induction capacity (induction factor) and responsiveness to the action of different concentrations of chloramphenicol. In the optimum pH of 6.6 (the inducible system), the light emission signal (Fig. 2A) and induction factor were the highest, and the inhibitory effect of chloramphenicol could be clearly quantitated (data not shown). The control experiment with constitutive light production (Fig. 2B) gave similar results. From this experiment with light emission, measurements were done with MOPS buffer at pH 6.6.
FIG. 2.
The pH dependency of light production of S. aureus RN4220/pTOO24 and RN4220/pCSS810 cells. The assay was performed in citrate-phosphate buffer (pH range, 4.6 to 5.6 [⧫]), MES (pH range, 5.6 to 6.6 [▪]) and MOPS (pH range, 6.6 to 7.0 [▴]) as described in Materials and Methods. Panel A shows the pH dependency of d-luciferin penetration into the indicator RN4220/pTOO24 cells, and Panel B shows results for strain RN4220/pCSS810.
The bioluminescence response of S. aureus towards model antibiotics.
Selected model antibiotics belonging to different molecular families and also having different mechanisms of action were tested for effects against RN4220/pTOO24 (inducible system) and against RN4220/pCSS810 (constitutive production of luciferase). The cells were kept together with different dilutions of antibiotics at room temperature for a 90-min preincubation period at pH 7.0. The results of this experiment are given in Table 1, showing a comparison in selected low and high concentrations of antibiotics. Both the inducible and the constitutive system contain a chloramphenicol acetyltransferase gene (cat) in the reporter plasmid downstream of the luciferase gene (12). In the constitutive system, the artificial operon is under a strong T5 promoter (5), whereas in the inducible system it is under the control of a relatively weak cadA promoter. As is evident from the table when comparing the chloramphenicol inhibition data, the cat gene does not work in the inducible system, and one could also speculate that the expression of cat in the constitutive system is not optimal for S. aureus in this experimental setup. Furthermore, the inducible system is in all but one case (that of sulfadiazine) more sensitive than the constitutive system. The power of the induction system is especially well seen with rifampin and tetracycline, where extremely low concentrations (a few nanograms per milliliter) make a clear-cut difference when comparing inducible and constitutive systems with low and high concentrations. Please note that the concentrations shown in Table 1 are different in the low and high range in the inducible and constitutive system for some of the antibiotics to obtain reasonable numbers (those that differ considerably from 0%). The coefficients of variation in the inducible system were 2 to 6% as measured with three replicas from original relative light units (RLUs). The coefficients of variation in the constitutive system were 2 to 7% as measured with three replicas from original RLUs.
TABLE 1.
Comparison between the bioluminescent inducible and constitutive expression systems for the detection of antimicrobial action powera
| Antibiotic | Low concn (ng/ml) | % from water blank | High concn (ng/ml) | % from water blank |
|---|---|---|---|---|
| RN4220/pTOO24 | ||||
| Rifampin | 1.9 | 40.0 | 64.0 | 0.2 |
| Tetracycline | 25.6 | 35.9 | 64.0 | 3.3 |
| Chloramphenicol | 640.0 | 36.2 | 3200.0 | 5.2 |
| Ampicillin | 128.0 | 98.0 | 640.0 | 15.3 |
| Cefotaxime | 128.0 | 75.0 | 640.0 | 19.5 |
| Norfloxacin | 128.0 | 85.3 | 3200.0 | 8.3 |
| Ciprofloxacin | 128.0 | 74.4 | 3200.0 | 29.2 |
| Spiramycin | 128.0 | 91.7 | 3200.0 | 26.4 |
| Erythromycin | 128.0 | 37.1 | 640.0 | 1.4 |
| Trimethoprim | 128.0 | 81.0 | 3200.0 | 22.3 |
| Sulfadiazine | 128.0 | 80.6 | 3200.0 | 96.7 |
| RN4220/pCSS810 | ||||
| Rifampin | 64.0 | 14.5 | 640.0 | 11.9 |
| Tetracycline | 64.0 | 21.7 | 640.0 | 8.4 |
| Chloramphenicol | 640.0 | 81.9 | 3200.0 | 61.7 |
| Ampicillin | 128.0 | 100.8 | 640.0 | 76.3 |
| Cefotaxime | 128.0 | 90.6 | 640.0 | 65.1 |
| Norfloxacin | 128.0 | 85.2 | 3200.0 | 39.8 |
| Ciprofloxacin | 128.0 | 70.3 | 3200.0 | 44.7 |
| Spiramycin | 128.0 | 85.9 | 3200.0 | 49.3 |
| Erythromycin | 128.0 | 56.4 | 640.0 | 19.2 |
| Trimethoprim | 128.0 | 90.1 | 3200.0 | 33.7 |
| Sulfadiazine | 128.0 | 45.4 | 3200.0 | 51.3 |
The S. aureus indicator strains RN4220/pTOO24 and RN4200/pCSS810 were freshly cultivated for the measurement and diluted. The cells were added on the wells of a white microtitration plate, preincubated with antibiotic for 90 min at room temperature, induced by 1 μM antimonite and measured for their bioluminescence immediately after the dispension of d-luciferin at +90 min (and repeated after 30 min, resulting in a total assay time of 210 min). The table shows results from low and high concentrations of each particular antibiotic. The results shown are means of three parallel measurements, and the coefficients of variation are between 2 and 7%.
Correlation between rapid bioluminescence measurement and conventional growth inhibition.
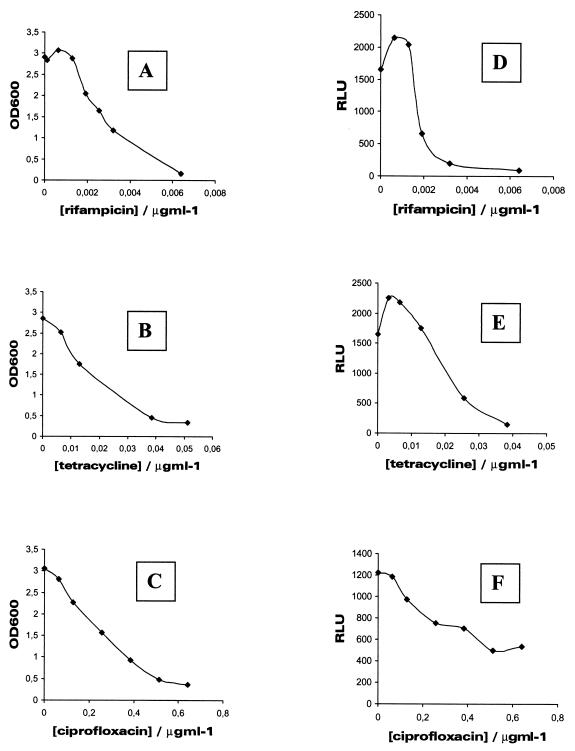
Three typical dose-response curves for the inducible system and conventional cultivation are shown in Fig. 3 to visualize how similar the two measurement methods are. We sometimes encounter situations when very small, nanogram amounts of certain antimicrobial agents cause minor activation of bioluminescence. This is seen here especially in the case of rifampin, which causes a 40% activation with 1-ng/ml concentration in the assay. Similar activation was found with ampicillin (100 ng/ml), cefotaxime (10 ng/ml), norfloxacin (100 ng/ml), spiramycin (100 ng/ml), and tetracycline (2 ng/ml). These slight activation phenomena with low doses may be results of small changes in consumption of cellular ATP pools that are increases in partial inhibition of metabolic routes by the antibiotics. Activation was also seen in the growth inhibition assay in the case of rifampin, which increased the viability by 15% at a 1-ng/ml concentration.
FIG. 3.
Typical growth inhibition and luminescence inhibition curves for three antibiotics. S. aureus RN4220/pTOO24 cells were treated with various concentrations of rifampin (A and D), tetracycline (B and E), and ciprofloxacin (C and F). The conventional overnight cultivation is shown in panels A, B, and C, and luminescence inhibition is shown in panels D, E, and F. The assays were performed as described in Materials and Methods.
Table 2 shows the IC50 values for all antibiotics for the strain RN4220/pTOO24 as measured by the inducible bioluminescence system and a conventional growth inhibition assay. Sulfadiazine and trimethoprim were omitted from this table since IC50 values could not be counted. The correlation factor between the two methods was an r value of 0.99. Strain RN4220/pCSS810 did not show any differences in conventional growth inhibition experiments (data not shown).
TABLE 2.
IC50 values for nine antibiotics for the strain RN4220/pTOO24 as measured by an inducible bioluminescence system and a conventional growth inhibition assay
| Antibiotic | IC50 (μg/ml) with method
|
|
|---|---|---|
| Bioluminescence | Cultivation | |
| Rifampin | 0.0018 | 0.003 |
| Tetracycline | 0.0195 | 0.02 |
| Chloramphenicol | 0.408 | 0.906 |
| Ampicillin | 0.350 | 0.211 |
| Cefotaxime | 0.077 | 0.080 |
| Norfloxacin | 0.604 | 0.529 |
| Ciprofloxacin | 0.349 | 0.247 |
| Spiramycin | 5.105 | 4.486 |
| Erythromycin | 0.030 | 0.225 |
DISCUSSION
The search for new antimicrobial drugs is currently very intensive and should be done more effectively now, since combinatorial chemistry approaches have been available for a decade (reviewed in reference 1). The generation of tens of thousands of lead compounds calls for efficient screening technologies (HTS or ultra-high-throughput screening). Long-term cultivation is an end-point method that answers the question of whether the indicator bacteria are living or not. It does not tell anything of the mode of action of the antibiotic tested. Certain groups of lead candidates with antimicrobial action may be more easily identified in a false screening procedure, and this predomination results in biased hit-scoring from a chemical library. It may happen that only agents that affect membranes can be fished out of the chemicals pool. Also, if a library is initially screened by pooling the compounds to decrease the number of measurements, this biased situation may result in the loss of valuable gold nuggets in the panning process. Therefore, new tools should be generated which allow a more targeted search, emphasizing the possibility of finding compounds with predetermined characteristics.
We have approached this question by making several improvements to existing functional microbial methods. First, we used the luciferase gene, the action of which can be monitored in real time from living cells. This resulted in a considerable savings in assay time, since results are obtained in tens of minutes rather than in several hours or even days. This also opens the possibility of screening a vast amount of compounds, since 96- or 384-well plates can be used. Second, we have inserted the luciferase gene under an inducible promoter, which resulted in specific amplification of the effects of those agents that affect transcription and translation in particular. This resulted in a further time savings, since induction of luciferase synthesis can be triggered at a predetermined time point. Third, we have generated a system that works in a nonpathogenic type strain of S. aureus, RN4220, (4), which works as a model system for its pathogenic relative, MRSA. Finally, we used the firefly luciferase as a marker gene and protein. Since one of the substrates is ATP, an essential energy source of each living cell, any disturbances in the intracellular level are directly reflected in light emission levels measured from the reporter cells. Therefore, minor events happening before the bacteria are killed are detected as a decreased emission of light.
One of the commonly proposed pitfalls for insect luciferase reporter genes is their lack of intrinsic luminescence capability and the need to disrupt the cells prior to d-luciferin addition and measurement. However, for a long time it has been known that measurement from intact living Escherichia coli cells carrying the firefly luciferase gene works perfectly well when d-luciferin is being incorporated inside the cells in a protonated form at pH 5.0 (16). In a recent study members of our group showed that it is actually wiser to measure luciferase reporter activity from intact living cells using the pH method in order to obtain more reproducible results than with disrupted E. coli cells and in vitro activity measurement (13). Various bacterial strains behave somewhat differently with regard to d-luciferin penetration inside cells. We have previously shown that Streptococcus mutans has an optimum pH of 6.0, indicating the effect of the different molecular structure of a gram-positive cellular membrane (6). Likewise, here we have shown that another gram-positive organism, S. aureus, has an even more neutral optimum pH for d-luciferin incorporation, with the optimum at pH 6.6 (Fig. 2). One could speculate that such a pH might allow one to incubate S. aureus cells in the presence of the substrate for a luciferase reaction from the beginning of the assay, making it possible to continually monitor the effects of different antibiotics. It remains to be seen whether this kind of approach could be valid and whether it could be applied to homogenous HTS applications in the future.
Members of our group have previously shown a similar amplification approach, described in this study, to be valid with E. coli, where certain model analytes were tested with a system consisting of a strong and inducible λ pL promoter controlling expression of various luciferase genes (5). In that study the effects of translational inhibitors, for instance, were seen in as much as 10,000-fold-lower concentrations than with the constitutive approach. It was also noticed that metabolic inhibitors affected both systems rather similarly during the short incubation period used. The fact that the effects of certain antibiotics were much greater than in this study is because the phage λ pL promoter is one of the strongest ones reported. This results in extremely high amplification of the effects of antimicrobial agents. In this study we have shown that the previous experience with E. coli is also applicable to a gram-positive human pathogen, S. aureus. We have also previously freeze-dried several strains of light-emitting bacteria (with either a constitutive or an inducible system) and shown that such cells, once reconstituted, are fully functional and behave as though they are freshly cultivated (5, 12–14). This fact creates an opportunity to use these indicator cells as ordinary reagents that can be taken from the shelf for direct use without any need for cultivation or complicated incubations whose operations do not fit with the tight time schedules of HRA.
Recently an engineered bacterial luciferase operon that was optimized to work in gram-positive organisms was described (2). The construction was expressed in S. aureus and used for monitoring the infection process in living mice utilizing a sensitive charge-coupled-device camera and digital image processing. The bacterial infection was experimentally shown to be cured by amoxicillin treatment as judged by the disappearance of in vivo light emission and correlation with CFU counting. This study shows how powerful bioluminescence technologies are in studies concerning antibiotic action against microbial cells. In this study we have generated a system for amplifying the effects of antibiotics and characterized it with model compounds of different modes of action. The approach was shown to correlate extremely well with the conventional growth inhibition method, and the amplification resulted in a clear improvement in sensitive detection of antibiotic action compared to the constitutive expression system. The system should be readily scaled up for HTS purposes.
ACKNOWLEDGMENTS
We thank Marko Virta and Jussi Kurittu for fruitful discussions.
REFERENCES
- 1.Birnbaum S, Mosbach K. Peptide screening. Curr Opin Biotechnol. 1992;3:49–54. doi: 10.1016/0958-1669(92)90125-3. [DOI] [PubMed] [Google Scholar]
- 2.Francis K P, Joh D, Bellinger-Kawahara C, Hawkinson M J, Purchio T F, Contag P R. Monitoring bioluminescent Staphylococcus aureus infections in living mice using a novel luxABCDEconstruct. Infect Immun. 2000;68:3594–3600. doi: 10.1128/iai.68.6.3594-3600.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gelmini S, Pinzani P, Pazzagli M. Luciferase gene as reporter: comparison with the CAT gene and use in transfection and microinjection of mammalian cells. Methods Enzymol. 2000;305:557–576. doi: 10.1016/s0076-6879(00)05513-0. [DOI] [PubMed] [Google Scholar]
- 4.Kreiswirth B N, Lofdahl S, Betley M J, O'Reilly M, Schlievert P M, Bergdoll M S, Novick R P. The toxic shock syndrome exotoxin structural gene is not detectable transmitted by a prophage. Nature. 1983;305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- 5.Lampinen J, Virta M, Karp M. Use of controlled luciferase expression to monitor chemicals affecting protein synthesis. Appl Environ Microbiol. 1995;61:2981–2989. doi: 10.1128/aem.61.8.2981-2989.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loimaranta V, Tenovuo J, Koivisto L, Karp M. Generation of bioluminescent Streptococcus mutansand its usage in rapid analysis of antimicrobial compounds. Antimicrob Agents Chemother. 1998;42:1906–1910. doi: 10.1128/aac.42.8.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mazel D, Davies J. Antibiotic resistance in microbes. Cell Mol Life Sci. 1999;56:742–754. doi: 10.1007/s000180050021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Milligan G, Rees S. Chimaeric G proteins: their potential use in drug discovery. Trends Pharmacol Sci. 1999;20:118–124. doi: 10.1016/s0165-6147(99)01320-6. [DOI] [PubMed] [Google Scholar]
- 9.Nucifora G, Chu L, Misra T K, Silver S. Cadmium resistance from Staphylococcus aureus plasmid pI258 cadAgene results from cadmium-efflux ATPase. Proc Natl Acad Sci USA. 1989;86:3544–3548. doi: 10.1073/pnas.86.10.3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pazzagli M, Devine J H, Peterson D O, Baldwin T O. Use of bacterial and firefly luciferases as reporter genes in DEAE-dextran-mediated transfection of mammalian cells. Anal Biochem. 1992;204:315–323. doi: 10.1016/0003-2697(92)90245-3. [DOI] [PubMed] [Google Scholar]
- 11.Steidler L, Yu W, Fiers W, Remaut E. The expression of Photinus pyralis gene in Staphylococcus aureusCowan I allows the development of a live amplifiable tool for immunoprotection. Appl Environ Microbiol. 1996;62:2356–2359. doi: 10.1128/aem.62.7.2356-2359.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tauriainen S, Karp M, Chang W, Virta M. Luminescent bacterial sensor for cadmium and lead. Biosens Bioelectronics. 1998;13:931–938. doi: 10.1016/s0956-5663(98)00027-x. [DOI] [PubMed] [Google Scholar]
- 13.Tauriainen S, Virta M, Chang W, Karp M. Measurement of firefly luciferase reporter gene activity from cells and lysates using Escherichia coliarsenite and mercury sensors. Anal Biochem. 1999;272:191–198. doi: 10.1006/abio.1999.4193. [DOI] [PubMed] [Google Scholar]
- 14.Tauriainen S, Virta M, Karp M. Detecting bioavailable toxic metals and metalloids from natural water samples using luminescent sensor bacteria. Water Res. 2000;34:2661–2666. [Google Scholar]
- 15.Wilson T, Hastings J W. Bioluminescence. Annu Rev Cell Dev Biol. 1998;14:197–230. doi: 10.1146/annurev.cellbio.14.1.197. [DOI] [PubMed] [Google Scholar]
- 16.Wood K V, DeLuca M. Photographic detection of luminescence in Escherichia colicontaining the gene for firefly luciferase. Anal Biochem. 1987;161:501–507. doi: 10.1016/0003-2697(87)90480-5. [DOI] [PubMed] [Google Scholar]
- 17.Wood K V. The chemical mechanism and evolutionary development of beetle bioluminescence. Photochem Photobiol. 1995;62:662–673. [Google Scholar]
- 18.Yoon K P, Silver S. A second gene in the Staphylococcus aureus cadAcadmium resistance determinant of plasmid pI258. J Bacteriol. 1991;173:7636–7642. doi: 10.1128/jb.173.23.7636-7642.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]