Abstract
Background
SARS-CoV-2 is an RNA virus that primarily causes respiratory disease; however, infection of other tissue has been reported. Evaluation of SARS-CoV-2 in tissue specimens may increase understanding of SARS-CoV-2 pathobiology.
Materials and Methods
A qualitative test for detection of SARS-CoV-2 in formalin-fixed paraffin-embedded (FFPE) tissues was developed and validated using droplet digital PCR (ddPCR), which has a lower limit of detection than reverse transcription (RT)-qPCR. After extraction of total RNA from unstained FFPE tissue, SARS-CoV-2 nucleocapsid (N1, N2) target sequences were amplified and quantified, along with human RPP30 as a control using the Bio-Rad SARS-CoV-2 ddPCR kit.
Results
SARS-CoV-2 was detected in all 21 known positive samples and none of the 16 negative samples. As few as approximately 5 viral copies were reliably detected. Since January 2021, many tissue types have been clinically tested. Of the 195 clinical specimens, the positivity rate was 35% with placenta and fetal tissue showing the highest percentage of positive cases.
Conclusion
This sensitive FFPE-based assay has broad clinical utility with applications as diverse as pregnancy loss and evaluation of liver transplant rejection. This assay will aid in understanding atypical presentations of COVID-19 as well as long-term sequelae.
Keywords: SARS-CoV-2, COVID-19, Viral detection in FFPE, Tissue-based viral detection, Droplet digital PCR, Placental viral infection
1. Introduction
SARS-CoV-2 is a positive-sense, single-stranded RNA virus that causes life-threatening coronavirus disease 2019 (COVID-19) in humans. SARS-CoV-2 infection can result in diverse, multiorgan pathology, the most significant being in the lungs, heart, kidney, central nervous system, liver, lymph nodes, bone marrow, vasculature, intestine, and placenta [1], [2], [3], [4], [5], [6], [7], [8], [9], [10].
SARS-CoV-2 infection is usually identified by the detection of viral RNA using reverse transcriptase polymerase chain reaction (RT-qPCR) on nasopharyngeal or oropharyngeal swabs. Current or prior infection might also be recognized by a serum test for SARS-CoV-2 IgG antibodies. A subset of patients may present without the typical upper respiratory symptoms or with concurrent unusual features that potentially represent long-term sequelae of SARS-CoV-2 [11], [12]. Patients with current or previous SARS-CoV-2 infection who present with “unexplained” acute or acute on chronic respiratory failure or other organ-specific manifestations may undergo tissue biopsy. Furthermore, an autopsy may be performed for individuals with sudden unexplained death. Therefore, a formalin-fixed paraffin-embedded (FFPE)-tissue-based test was needed for evaluation of SARS-CoV-2 for both improved understanding of potential disease manifestations as well as for patient management. Various methods for the identification of SARS-CoV-2 infection in FFPE-tissue specimens have been published including in situ hybridization (ISH), immunohistochemistry (IHC), RT-qPCR, and electron microscopy [1], [6], [11], [13]. Moreover, due to various factors in the preanalytical, analytical, and postanalytical phase, virus detection in tissue biopsy samples presents specific diagnostic challenges. Therefore, potentially more sensitive techniques such as reverse transcription-droplet digital polymerase chain reaction (RT-ddPCR) might enhance detection of SARS-CoV-2 in FFPE tissue or other matrices [11]. In fact, ddPCR has been shown to have a significantly lower limit of detection of SARS-CoV-2 in nasopharyngeal swabs than reverse transcription RT-qPCR [14]. Consequently, detection of SARS-CoV-2 in deceased patients (autopsy tissue) or surgical specimens may confirm a suspected diagnosis among individuals with clinical and/or pathologic manifestations of COVID-19 and may increase understanding of SARS-CoV-2 pathobiology.
The use of ddPCR for nucleotide target analysis has emerged in recent years as a robust and reliable methodology [15], [16], [17]. ddPCR can specifically measure single or multiple nucleic acid targets in a single reaction [15], [16], [18]. With ddPCR, DNA or reverse transcribed cDNA is partitioned into thousands of droplets that are subsequently amplified, and fluorescently-labeled probe signals within each droplet are recorded as either positive or negative, depending on the presence or absence of a nucleotide target. With thousands of individual droplets, a Poisson calculation of target copies per droplet [−ln(1 − p)], where p is the fraction of positive droplets, is applied and the absolute concentration for each nucleotide target is obtained, providing a highly accurate estimation of nucleotide copy number that is crucial for clinical diagnostics [15], [18], [19].
Here the development and performance characteristics are described for a qualitative RT-ddPCR test for detection of SARS-CoV-2 in FFPE tissues, which can be implemented in most clinical molecular diagnostic laboratories.
2. Materials and methods
2.1. Clinical specimens
Thirty-seven tissue samples from 35 autopsy cases (20 positive, 15 negative), a cell block from a cell line infected with SARS-CoV-2, and a cell block from a cell line infected with influenza virus were utilized in this study for accuracy, precision, stability, linearity, and specificity studies. Multiple tissue types were used for test development, including lung, heart, brain, and liver tissue. Slides from autopsy cases of patients who tested positive for COVID-19 (n = 20) by antemortem nasopharyngeal or oropharyngeal swab were reviewed by a pathologist. Blocks with histologic features suggestive of infection were tested by RNA in situ hybridization (ISH) and immunohistochemistry, as previously described, for comparison to the ddPCR assay [11]. Tissue samples from patients who underwent autopsy the year prior to the beginning of the COVID-19 pandemic were included as negative controls. These patients had lung pathology not due to SARS-CoV-2, such as diffuse alveolar damage or influenza virus infection (n = 15). Note that although in the tables the positive samples are represented sequentially by lower numbers and the negative samples are represented sequentially by higher numbers, they were intermixed and the test developers were blinded as to which were positive and negative. They have been re-named and grouped for the purpose of publication.
This assay was launched as a clinically orderable test in January 2021. All specimens analyzed clinically up to November 20, 2021 were included in a retrospective summary describing clinical applications of this test as well as positivity rates.
This study was approved by the Mayo Clinic Institutional Review Board.
2.2. Total RNA extraction and reverse transcriptase ddPCR
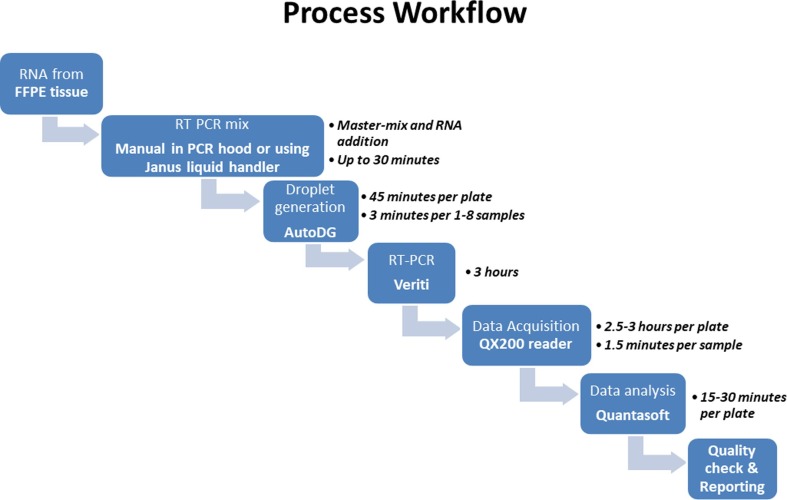
An overview of the RT-ddPCR assay workflow is shown in Fig. 1 . Total RNA, including viral RNA, was extracted from unstained FFPE tissue scrolls cut at 10 µm by either RNeasy® DSP FFPE Kit or miRNeasy FFPE Kit (Qiagen, Hilden, Germany). The RNA was quantified, ranging from 12 to 800 ng/μL, and stored at −80 °C. The assay was performed according to the manufacturer protocol, as outlined in the “Instructions for Use” for the Bio-Rad SARS-CoV-2 ddPCR Test (Bio-Rad Laboratories, CA, USA), which had received Emergency Use Authorization (EUA) from the US Food and Drug Administration (FDA) for nasopharyngeal specimens at the time of development of this tissue-based assay. Briefly, 5.5 uL of RNA eluate (corresponding to a range of 66–800 ng of RNA) was added to 16.5 uL of ddPCR master mix containing 5.5 μL of 4x One Step-RT-ddPCR Supermix, 2.2 μL of reverse transcriptase, 1.1 μL of dithiothreitol, 1.1 μL of the 20x 2019-nCoV CDC ddPCR triplex probe. RNA samples were run both neat and at a 1:100 dilution. After the reaction was partitioned into droplets using the Bio-Rad AutoDG instrument, SARS-CoV-2 nucleocapsid (N1 and N2) target sequences, along with human ribonuclease P/MRP subunit 30 (RPP30) target as a control, were amplified using Veriti thermocyclers (Thermo Fisher Scientific, Inc., Waltham, MA) using the following protocol: RT steps: 25 °C for 3 min and 50 °C for 60 min, followed by the PCR steps: 95 °C for 10 min, then 40 cycles of denaturation at 95 °C for 30 s and annealing/extension at 55 °C for 1 min, and a final enzyme deactivation at 98 °C for 10 min. Droplets were counted as negative or positive for one or more target by the QX200 Droplet Reader and signal data were analyzed using QuantaSoft Analysis Pro software version 1.0.596 (Bio-Rad Laboratories, Hercules, CA, USA). For each run, a low positive for SARS-CoV-2 and a negative sample were included for quality control.
Fig. 1.
Workflow of SARS-Cov-2 analysis by RT-ddPCR in FFPE tissues.
2.3. Assessment of assay performance characteristics
To evaluate the potential impact of the FFPE matrix and establish the limit of detection (LOD), samples with total SARS-CoV-2 RNA copies of 80, 40, 20, 10, and 5 were prepared in triplicate and samples with 0 copies were prepared in pentaplicate using Exact Diagnostics SARS-CoV-2 Standard (Exact Diagnostics LLC, Fort Worth, TX) mixed with extracted RNA from pooled SARS-CoV-2 negative FFPE tissues (523 ng total RNA in each reaction) as a diluent for testing with the RT-ddPCR assay. The LOD was determined based on the lowest signal that could be reproducibly detected and differentiated from background signal observed when no SARS-CoV-2 was present.
All 37 known SARS-CoV-2 positive (n = 21) and negative (n = 16) tissues and cell blocks were utilized to determine accuracy. Intra-assay and inter-assay precision (reproducibility) were evaluated using pooled extracted RNA from two patient FFPE tissue specimens (one negative and one positive, pulmonary derived), one positive quality control (QC) sample derived from a SARS-CoV-2 embedded FFPE cell block, and one negative QC sample derived from an FFPE cell block of tissue infected with influenza A virus. For the intra-assay precision study, each of the four samples were run in triplicate together on the same run. To assess inter-assay precision, each sample was run on three different runs. Concordance of both the qualitative positive or negative result was evaluated, along with the quantitative value of N1 and N2 SARS-CoV-2 targets.
Extensive in silico analyses conducted by the kit manufacturer found no potential cross-reactivity of common respiratory tract pathogens (bacteria, fungi, and viruses including other coronavirus strains), and our laboratory previously identified no cross reactivity with other coronavirus strains when testing nasopharyngeal swab specimens (unpublished data). To further verify the analytical specificity in the FFPE matrix, two blocks of rhesus monkey kidney (RMK) cells infected with Influenza A and Parainfluenza were tested.
2.4. Data analysis
QuantaSoft Analysis Pro software version 1.0.596 (Bio-Rad) was used for droplet cluster classification. The QX200 automated droplet reader counts every acceptable droplet and measures the fluorescence emissions from each droplet using 2 channels (FAM and HEX).
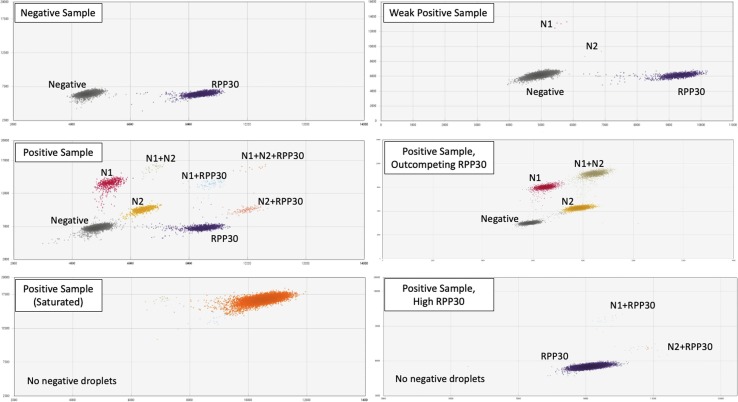
Droplets of different color and intensity were displayed on 2-dimensional plots, allowing counting of negative droplets as well as those positive for N1, N2, RPP30, or a combination of targets (Fig. 2 ). For each fluorophore, the fraction of positive droplets was fitted into a Poisson distribution equation, thereby providing absolute quantification of N1, N2, and RPP30 PCR products per well without a standard curve.
Fig. 2.
Graphical output for 2D plots representing droplets positive for SARS-CoV-2 N1 and N2 targets, colored in red and gold, respectively. Droplets positive for human RPP30 control target are colored in dark blue. Negative droplets are represented in grey. The double positive droplets are tan (N1 + N2), light blue (N1 + RPP30), or salmon (N2 + RPP30), while the triple positive (N1 + N2 + RPP30) droplets are colored orange. Representative plots from a negative sample and a variety of SARS-CoV-2 positive samples are shown.
Standard statistical analyses, including averages, standard deviation (SD), and coefficient of variation (CV), were calculated using Microsoft Excel, version 2102 (Microsoft, Redmond, WA). Rejection criteria for excluding a result included unusual spread of droplets or < 9000 acceptable droplets measured per well.
3. Results
3.1. Multiplex ddPCR
Two droplet clusters and up to eight droplet clusters were observed for a normal (negative) and COVID-19 patient (positive) specimen, respectively, as illustrated in Fig. 2. These clusters included negative droplets and RPP30-positive droplets only with no cluster for N1 or N2 for a negative sample. Positive samples had additional clusters for N1-positive and N2-positive droplets, and may have had clusters for N1 + N2 positive (double positive), N1 + RPP30 positive (double positive), N2 + RPP30 positive (double positive), and N1 + N2 + RPP30 positive (triple positive) droplets. With very high SARS-CoV-2 viral load, the saturated samples showed one or more undefined clusters consisting of N1 and N2 targets, often outcompeting the human RPP30 control target; dilution with 1x TE buffer allowed for separation into the typical cluster pattern. In some cases with strong RPP30 expression, a small number of N1 + RPP30 and N2 + RPP30 double positive droplets were observed without negative or single-positive droplets. The number of droplets generated per reaction ranged from 9000 to 18,299 droplets. Any sample with < 9000 droplets was rejected and repeated.
3.2. Assay performance characteristics
3.2.1. Limit of detection (LOD)
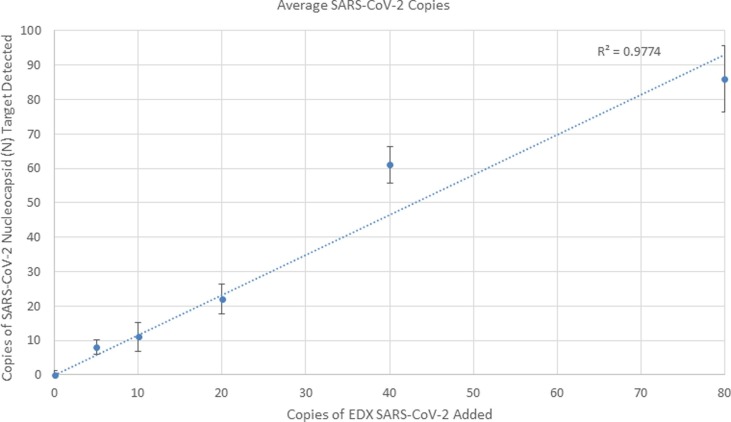
SARS-CoV-2 N1 and N2 targets were detected for all 3 of the triplicate samples prepared with 5 virion copies (Table 1 ). The three replicates had 7, 11, and 12 total positive N1 and N2 droplets, respectively. Two N1-positive droplets were observed in one replicate out of the 5 blank samples prepared with 0 copies of viral RNA, while the remainder had 0 positive N1 or N2 droplets. Additionally, the no template control (NTC) was run in triplicate with no N1 or N2 positive droplets. Therefore, the limit of blank was established at 2 positive droplets and the cut-offs for final calls were set at ≥ 5 N1 and N2 droplets = positive, 0–2 positive droplets = negative, and 3–4 positive droplets = indeterminate. Based on this experiment, 5 copies of viral RNA (equivalent to approximately 5 droplets of N1 and/or N2) can be reliably detected and is expected to be the approximate limit of detection. Additional linear regression analysis of the average observed copies of SARS-CoV-2 RNA detected by the RT-ddPCR assay as compared to the quantity expected in the sample based on the amount added demonstrates that the assay is linear and is shown in Fig. 3 . RPP30 values may differ based on tissue type and size; therefore, a cut-off of 50 positive droplets was selected to ensure that RNA was successfully extracted.
Table 1.
Detection of SARS-CoV-2 targets at different dilutions tested by RT-ddPCR. Average and range of N1 copies, N2 copies, N1 positive droplets, N2 positive droplets, and total droplets for LOD experiments with known SARS-CoV-2 virion copies, as indicated.
| SARS-CoV-2 Copies Input | N1 copies detected /reaction | N2 copies detected /reaction | N1 positive droplets | N2 positive droplets | Total Droplets |
|---|---|---|---|---|---|
| 80 | 80.7 (80–82) | 91.0 (78–101) | 49.7 (39–56) | 57.0 (38–71) | 14,537 (11519–16535) |
| 40 | 58.3 (53–66) | 63.0 (60–66) | 37.7 (34–41) | 40.7 (38–43) | 15,261 (14656–16092) |
| 20 | 24.7 (19–31) | 20.3 (20–21) | 15.3 (11–20) | 12.3 (12–13) | 14,441 (13507–15383) |
| 10 | 9.7 (7–16) | 12.7 (11–14) | 6.0 (4–10) | 8 (7–9) | 14,653 (14377–15103) |
| 5 | 9.3 (7–12) | 7.0 (5–8) | 5.7 (4–7) | 4.3 (3–5) | 14,059 (12848–15280) |
| 0 | 0.8 (0–4) | 0 (0) | 0.4 (0–2) | 0 (0) | 14,821 (12646–16887) |
| NTC | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 16,555 (15492–18412) |
Fig. 3.
Linear regression analysis of the average observed versus expected SARS-CoV-2 RNA copies of the RT-ddPCR assay. Error bars represent the standard deviation.
3.2.2. Accuracy
Total (100%) agreement of qualitative results (detected vs. not detected) was observed for 21 known-positive samples. Near complete agreement (15 of 16 samples; 93.8%) was observed for 16 known-negative samples (Table 2 ). One sample had a total of 4 positive droplets, which would be called indeterminate with the established cut-offs. This sample was heart tissue from autopsy of a patient known to be SARS-CoV-2 positive. As shown in Table 2, the assay was saturated for several positive samples (N1 and N2 copies exceeded upper limits of approximately 20,000,000), but when diluted 1:100 with 1x TE buffer, these samples displayed the expected droplet patterns.
Table 2.
Concordance between ddPCR assay results (run undiluted) and ISH results. CB indicates “cell block” while PA indicates “patient” (which have been deidentified using this code for the purposes of publication). Saturated samples were diluted 1:100 for further analysis (not shown). Samples were called positive, negative, or indeterminate based on the cut-offs established during the LOD studies.
| Sample ID | ISH results | N1 droplets | N2 droplets |
N1 copies/ 20 μL reaction |
N2 copies/ 20 μL reaction |
ddPCR Results |
|---|---|---|---|---|---|---|
| CB-Pos | Positive | 6172 | 5837 | 14,410 | 13,355 | Positive |
| PA-1 | Positive | 12,683 | 12,698 | 158,615 | 168,320 | Positive |
| PA-2 | Positive | 13 | 8 | 21 | 13 | Positive |
| PA-3 | Positive | 1639 | 2262 | 3431 | 4879 | Positive |
| PA-4 | Positive | 13,124 | 13,124 | >20000000 | >20000000 | Positive |
| PA-5 | Positive | 309 | 746 | 580 | 1427 | Positive |
| PA-6 | Positive | 47 | 60 | 65 | 83 | Positive |
| PA-7 | Positive | 18 | 32 | 28 | 50 | Positive |
| PA-8 | Positive | 6 | 5 | 12 | 10 | Positive |
| PA-9 | Positive | 12,632 | 13,002 | 49,153 | 54,618 | Positive |
| PA-10 | Positive | 12,274 | 12,274 | >20000000 | >20000000 | Positive |
| PA-11 | Positive | 11,990 | 11,824 | 35,817 | 34,678 | Positive |
| PA-12 | Positive | 309 | 746 | 580 | 1427 | Positive |
| PA-13 | Positive | 3129 | 3371 | 4884 | 5308 | Positive |
| PA-14 | Positive | 12,874 | 12,895 | 151,061 | 151,061 | Positive |
| PA-15 | Positive | 56 | 54 | 89 | 85 | Positive |
| PA-16 | Positive | 10,663 | 10,909 | 32,292 | 33,947 | Positive |
| PA-17 | Positive | 5 | 10 | 8 | 16 | Positive |
| PA-18 | Positive | 14,760 | 14,760 | 225,876 | 225,876 | Positive |
| PA-19 | Positive | 1900 | 2135 | 3359 | 3810 | Positive |
| PA-20 | Positive | 1187 | 1209 | 1834 | 1870 | Positive |
| CB-Neg | Negative | 0 | 0 | 0 | 0 | Negative |
| PA-22 | Negative | 0 | 0 | 0 | 0 | Negative |
| PA-23 | Negative | 0 | 1 | 0 | 2 | Negative |
| PA-24 | Negative | 0 | 0 | 0 | 0 | Negative |
| PA-25 | Negative | 0 | 0 | 0 | 0 | Negative |
| PA-26 | Negative | 2 | 2 | 3 | 3 | Indeterminate |
| PA-27 | Negative | 1 | 0 | 1 | 0 | Negative |
| PA-28 | Negative | 1 | 0 | 2 | 0 | Negative |
| PA-29 | Negative | 0 | 0 | 0 | 0 | Negative |
| PA-30 | Negative | 0 | 0 | 0 | 0 | Negative |
| PA-31 | Negative | 0 | 0 | 0 | 0 | Negative |
| PA-32 | Negative | 0 | 0 | 0 | 0 | Negative |
| PA-33 | Negative | 1 | 1 | 1 | 1 | Negative |
| PA-34 | Negative | 1 | 1 | 2 | 2 | Negative |
| PA-35 | Negative | 0 | 0 | 0 | 0 | Negative |
| PA-36 | Negative | 0 | 0 | 0 | 0 | Negative |
3.2.3. Reproducibility
Both intra- and inter-assay measurements were 100% concordant in qualitative positive or negative calls among all replicates for each sample. Although this is a qualitative assay, the coefficient of variation (CV) of the number of positive droplets detected and copy number were also evaluated. Across the 3 intra-assay and inter-assay measurements, the negative QC sample had 0 positive N1 and N2 droplets in each replicate, with the exception of 1 N2 droplet (corresponding to 2 copies of N2 in the reaction well) in the intra-assay study. This would have been considered negative in our qualitative assay. Similarly, the negative patient sample had one replicate in the inter-assay study with 1 N2 droplet (corresponding to 2 copies of N2 in the reaction well), and in the intra-assay study there was one replicate with 1 N1 droplet (corresponding to 1 copy of N1) and one replicate with 1 N2 droplet (corresponding to 2 N2 copies in the reaction). Again, each replicate would have been called negative in the qualitative assay. The positive QC sample had a mean of 16,575 copies of N1 per reaction detected, with 10% CV in the inter-assay study and 3% CV in the intra-assay study. This sample had a mean of 15,175 copies of N2 detected per reaction, with 10% CV in the inter-assay study and 3% CV in the intra-assay study. The positive patient sample had a mean of 156,938 copies of N1 detected per reaction, with 3% CV in the inter-assay study and 4% CV in the intra-assay study. This sample had a mean of 174,836 copies of N2 detected per reaction, with 10% CV in the inter-assay study and 8% CV in the intra-assay study. In this qualitative assay, each replicate of both the positive control and positive patient sample would have been called positive.
3.2.4. Analytical specificity
No amplification of either of the nucleocapsid targets was detected (i.e., 0 droplet count for both N1 and N2) for any of the non-SARS-CoV-2 viruses tested.
3.3. Clinical test usage after implementation
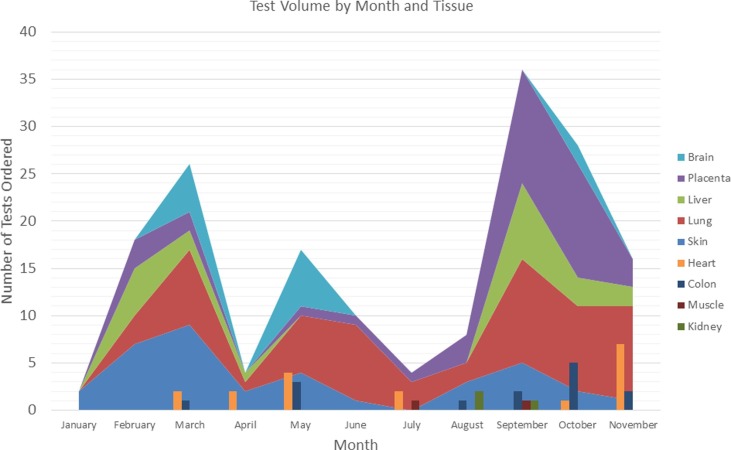
From January 22, 2021, which was the first clinical run using this test, until November 20, 2021, a total of 195 specimens have been run for clinical purposes (Table 3 , Fig. 4 ). Additional testing has been performed on a research basis and not included in this number. Testing has been ordered both by physicians at our institution as well as through our reference laboratory practice. The overall SARS-CoV-2 positivity rate in clinical testing was 35% with placenta and fetal tissue showing the highest percentage of positive cases (Table 3).
Table 3.
SARS-CoV-2 detection in tissues that were analyzed for clinical purposes after the test went live. The total number of test orders, as well as number of positive, negative, indeterminate, and failed cases are provided and are displayed by tissue type. In addition, the percentage of cases that have been positive is provided.
| Tissue Type | Total Tested | Positive | Negative | Indeterminate | Failed | % Positive |
|---|---|---|---|---|---|---|
| Lung | 57 | 25 | 31 | 1 | 0 | 44 |
| Heart or aorta | 13 | 1 | 11 | 0 | 1 | 8.3 |
| Brain (including olfactory bulbs) | 13 | 0 | 13 | 0 |
0 |
0 |
| Colon or small bowel | 14 | 3 | 11 | 0 | 0 | 21 |
| Liver | 20 | 8 | 11 | 1 | 0 | 40 |
| Kidney | 4 | 0 | 4 | 0 | 0 | 0 |
| Skin | 35 | 5 | 29 | 1 | 0 | 14 |
| Placenta and products of conception | 37 | 26 | 9 | 1 | 1 | 72 |
| Muscle | 2 | 1 | 1 | 0 | 0 | 50 |
| Total | 195 | 69 | 120 | 4 | 2 | 35 |
Fig. 4.
Number of clinical tests performed by tissue type over time. Each color represents a tissue type, as defined in the legend. The height of each peak or bar presents the number of that type of tissue, with the combined total of all tissue types is represented by the overall peak height.
Lung tissue was most commonly tested. Among the 25 positive cases were 9 autopsies. Six of these patients were known to be COVID-positive prior to death, though three of those had been positive several weeks prior to death. The remaining three had no known history of a COVID-test. The other positive cases were biopsy specimens, four of which were from patients with a remote history of COVID, while no clinical information was provided for the others. Among the negative lung specimens, 13 were from autopsy cases, two of which had a remote history of COVID – including one who had remained hospitalized for the 3 months since diagnosis. One patient had multiple negative COVID tests prior to death, and no COVID testing was known to have been performed on the remaining patients. The additional negative lung tissue specimens were biopsies and included two patients with a remote history of COVID while no clinical history was provided for the remainder.
4. Discussion
SARS-CoV-2 has been described to result in a variety of pathological manifestations affecting most organ systems. Because formalin fixation can impact the quality and integrity of nucleic acid, published methods for detection of SARS-CoV-2 are either not suitable for testing FFPE tissue specimens or lack sensitivity, and thus might not be helpful or feasible in routine clinical practice [1], [6], [7], [13], [20]. The aim of this study was to develop and validate an easy, reliable, and accurate assay using ddPCR technology for the detection of SARS-CoV-2 in FFPE tissue for routine clinical use. We have developed and validated a qualitative test protocol that directly detects the SARS-CoV-2 virus in human FFPE tissues. Of note, in this assay SARS-CoV-2 detection was possible and reproducible even at low viral load (i.e., 5 copies of viral RNA).
Although many methods are available for detecting SARS-CoV-2 virus in tissues [6], [7], [13], ddPCR accomplishes a level of sensitivity and specificity for SARS-CoV-2 detection unachievable by heavily used RT-qPCR because of inherent technical inconsistencies of this method. For example, PCR inhibitors have a large effect on amplification efficiency, and thus, qPCR assay performance [21], [22]. Concentrating target molecules in smaller reactions, ddPCR can reduce the bias introduced by PCR inhibitors often found in clinical specimens [21], [22], [23]. In addition, the calibration curves needed for qPCR are a major source of assay variability and are difficult to maintain. Consequently, one of the main advantages of ddPCR is the generation of thousands of droplets per sample and this method does not require replicates because it directly counts the number of target molecules rather than relying on a standard curve generated by reference standards or endogenous controls [15], [16], [17], [18]. Therefore, these properties make ddPCR a promising SARS-CoV-2 detection method for samples with low template abundance derived as may be observed in non-respiratory tissues [14].
Since the test became clinically available, it has been ordered for a wide variety of clinical and research purposes by physicians at our institution and across the country. Initially, common indications for clinical testing included evaluating brain or lung tissue, often in the setting of autopsy. Over time, fewer tests were ordered for these indications, and instead testing was used to evaluate lung tissue that showed morphologic features or sequela of an acute or acute on chronic lung injury [24], skin rashes, gastrointestinal tract ulceration and/or inflammation, and other findings in the setting of current or recent COVID-19. In addition, testing has been used in the transplant setting to test organs from COVID-positive donors, as well as the donated organ after transplant in the setting of recipients with SARS-CoV-2 infection to better understand a patient’s clinical course and to distinguish organ damage from SARS-CoV-2 from damage due to rejection. Testing to evaluate placental pathology has also increased over the time the test has been available. Initially testing was often performed to evaluate subtle lesions identified in the placenta, while after the delta variant became wide-spread in the United States, an increase in testing of placenta tissue or products of conception for the indication of fetal demise was noted. Concurrently, although this is not a quantitative test and the raw signal has not been formally evaluated, we noted an increase in the number of placenta samples with a saturated (highly positive) signal after the emergence of the delta variant.
In summary, this work demonstrates the utility and sensitivity of tissue-based testing for detection of SARS-CoV-2 in various FFPE tissue types, autopsy to surgical biopsy samples, in a large group of patients and is a feasible method for use in clinical laboratories. The clinical utility is broad with the assay used for applications as diverse as pregnancy loss and transplant rejection. We showed that ddPCR provides both accuracy and sensitivity in detecting patients with low viral load. SARS-CoV-2 ddPCR testing will also aid in understanding atypical presentations of COVID-19, including in patients who present with “unexplained” symptoms or potential long-term sequelae.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Facchetti F., Bugatti M., Drera E., Tripodo C., Sartori E., Cancila V., Papaccio M., Castellani R., Casola S., Boniotti M.B., Cavadini P., Lavazza A. SARS-CoV2 vertical transmission with adverse effects on the newborn revealed through integrated immunohistochemical, electron microscopy and molecular analyses of Placenta. EBioMedicine. 2020;59 doi: 10.1016/j.ebiom.2020.102951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Falasca L., Nardacci R., Colombo D., Lalle E., Di Caro A., Nicastri E., Antinori A., Petrosillo N., Marchioni L., Biava G., D’Offizi G., Palmieri F., Goletti D., Zumla A., Ippolito G., Piacentini M., Del Nonno F. Postmortem Findings in Italian Patients With COVID-19: A Descriptive Full Autopsy Study of Cases With and Without Comorbidities. J. Infect. Dis. 2020;222(11):1807–1815. doi: 10.1093/infdis/jiaa578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., Liu L., Shan H., Lei C.L., Hui D.S.C., Du B., Li L.J., Zeng G., Yuen K.Y., Chen R.C., Tang C.L., Wang T., Chen P.Y., Xiang J., Li S.Y., Wang J.L., Liang Z.J., Peng Y.X., Wei L., Liu Y., Hu Y.H., Peng P., Wang J.M., Liu J.Y., Chen Z., Li G., Zheng Z.J., Qiu S.Q., Luo J., Ye C.J., Zhu S.Y., Zhong N.S. C. China Medical Treatment Expert Group for, Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harris C.K., Hung Y.P., Nielsen G.P., Stone J.R., Ferry J.A. Bone Marrow and Peripheral Blood Findings in Patients Infected by SARS-CoV-2. Am. J. Clin. Pathol. 2021;155(5):627–637. doi: 10.1093/ajcp/aqaa274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hosier H., Farhadian S.F., Morotti R.A., Deshmukh U., Lu-Culligan A., Campbell K.H., Yasumoto Y., Vogels C.B., Casanovas-Massana A., Vijayakumar P., Geng B., Odio C.D., Fournier J., Brito A.F., Fauver J.R., Liu F., Alpert T., Tal R., Szigeti-Buck K., Perincheri S., Larsen C., Gariepy A.M., Aguilar G., Fardelmann K.L., Harigopal M., Taylor H.S., Pettker C.M., Wyllie A.L., Cruz C.D., Ring A.M., Grubaugh N.D., Ko A.I., Horvath T.L., Iwasaki A., Reddy U.M., Lipkind H.S. SARS-CoV-2 infection of the placenta. J Clin Invest. 2020;130(9):4947–4953. doi: 10.1172/JCI139569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Massoth L.R., Desai N., Szabolcs A., Harris C.K., Neyaz A., Crotty R., Chebib I., Rivera M.N., Sholl L.M., Stone J.R., Ting D.T., Deshpande V. Comparison of RNA In Situ Hybridization and Immunohistochemistry Techniques for the Detection and Localization of SARS-CoV-2 in Human Tissues. Am. J. Surg. Pathol. 2021;45(1):14–24. doi: 10.1097/PAS.0000000000001563. [DOI] [PubMed] [Google Scholar]
- 7.Paniz-Mondolfi A., Bryce C., Grimes Z., Gordon R.E., Reidy J., Lednicky J., Sordillo E.M., Fowkes M. Central nervous system involvement by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) J. Med. Virol. 2020;92(7):699–702. doi: 10.1002/jmv.25915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patane L., Morotti D., Giunta M.R., Sigismondi C., Piccoli M.G., Frigerio L., Mangili G., Arosio M., Cornolti G. Vertical transmission of coronavirus disease 2019: severe acute respiratory syndrome coronavirus 2 RNA on the fetal side of the placenta in pregnancies with coronavirus disease 2019-positive mothers and neonates at birth. Am J Obstet Gynecol MFM. 2020;2(3) doi: 10.1016/j.ajogmf.2020.100145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tabary M., Khanmohammadi S., Araghi F., Dadkhahfar S., Tavangar S.M. Pathologic features of COVID-19: A concise review. Pathol. Res. Pract. 2020;216(9) doi: 10.1016/j.prp.2020.153097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., Guan L., Wei Y., Li H., Wu X., Xu J., Tu S., Zhang Y., Chen H., Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roden A.C., Vrana J.A., Koepplin J.W., Hudson A.E., Norgan A.P., Jenkinson G., Yamaoka S., Ebihara H., Monroe R., Szabolcs M.J., Majumdar R., Moyer A.M., Garcia J.J., Kipp B.R. Comparison of In Situ Hybridization, Immunohistochemistry, and Reverse Transcription-Droplet Digital Polymerase Chain Reaction for Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Testing in Tissue. Arch. Pathol. Lab. Med. 2021;145(7):785–796. doi: 10.5858/arpa.2021-0008-SA. [DOI] [PubMed] [Google Scholar]
- 12.Satterfield B.A., Bhatt D.L., Gersh B.J. Cardiac involvement in the long-term implications of COVID-19. Nat. Rev. Cardiol. 2021 doi: 10.1038/s41569-021-00631-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.von Stillfried S., Boor P. Detection methods for SARS-CoV-2 in tissue. Pathologe. 2021;42(Suppl 1):81–88. doi: 10.1007/s00292-021-00920-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suo T., Liu X., Feng J., Guo M., Hu W., Guo D., Ullah H., Yang Y., Zhang Q., Wang X., Sajid M., Huang Z., Deng L., Chen T., Liu F., Xu K., Liu Y., Zhang Q., Liu Y., Xiong Y., Chen G., Lan K., Chen Y. ddPCR: a more accurate tool for SARS-CoV-2 detection in low viral load specimens. Emerg Microbes Infect. 2020;9(1):1259–1268. doi: 10.1080/22221751.2020.1772678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pinheiro L., Emslie K.R. Basic Concepts and Validation of Digital PCR Measurements. Methods Mol. Biol. 2018;1768:11–24. doi: 10.1007/978-1-4939-7778-9_2. [DOI] [PubMed] [Google Scholar]
- 16.Pinheiro L.B., Coleman V.A., Hindson C.M., Herrmann J., Hindson B.J., Bhat S., Emslie K.R. Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Anal. Chem. 2012;84(2):1003–1011. doi: 10.1021/ac202578x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whale A.S., Huggett J.F., Tzonev S. Fundamentals of multiplexing with digital PCR. Biomol. Detect. Quantif. 2016;10:15–23. doi: 10.1016/j.bdq.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quan P.L., Sauzade M., Brouzes E. dPCR: A Technology Review, Sensors (Basel) 2018;18(4) doi: 10.3390/s18041271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H., Bai R., Zhao Z., Tao L., Ma M., Ji Z., Jian M., Ding Z., Dai X., Bao F., Liu A. Application of droplet digital PCR to detect the pathogens of infectious diseases. Biosci. Rep. 2018;38(6) doi: 10.1042/BSR20181170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Best Rocha A., Stroberg E., Barton L.M., Duval E.J., Mukhopadhyay S., Yarid N., Caza T., Wilson J.D., Kenan D.J., Kuperman M., Sharma S.G., Larsen C.P. Detection of SARS-CoV-2 in formalin-fixed paraffin-embedded tissue sections using commercially available reagents. Lab Invest. 2020;100(11):1485–1489. doi: 10.1038/s41374-020-0464-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nixon G., Garson J.A., Grant P., Nastouli E., Foy C.A., Huggett J.F. Comparative study of sensitivity, linearity, and resistance to inhibition of digital and nondigital polymerase chain reaction and loop mediated isothermal amplification assays for quantification of human cytomegalovirus. Anal. Chem. 2014;86(9):4387–4394. doi: 10.1021/ac500208w. [DOI] [PubMed] [Google Scholar]
- 22.Taylor S.C., Laperriere G., Germain H. Droplet Digital PCR versus qPCR for gene expression analysis with low abundant targets: from variable nonsense to publication quality data. Sci. Rep. 2017;7(1):2409. doi: 10.1038/s41598-017-02217-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dingle T.C., Sedlak R.H., Cook L., Jerome K.R. Tolerance of droplet-digital PCR vs real-time quantitative PCR to inhibitory substances. Clin. Chem. 2013;59(11):1670–1672. doi: 10.1373/clinchem.2013.211045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roden A.C., Boland J.M., Johnson T.F., Aubry M.C., Lo Y.C., Butt Y.M., Maleszewski J.J., Larsen B.T., Tazelaar H.D., Khoor A., Smith M.L., Moua T., Jenkins S.M., Moyer A.M., Yi E.S., Bois M.C. Late Complications of COVID-19: A Morphologic, Imaging, and Droplet Digital Polymerase Chain Reaction Study of Lung Tissue. Arch. Pathol. Lab Med. 2022 doi: 10.5858/arpa.2021-0519-SA. [DOI] [PubMed] [Google Scholar]