Abstract
Gamma (γ)-terpinene, a monoterpene compound, which is generally used in the pharmaceutical and cosmetics industries, due to its physical and chemical properties, is expected to become one of the more influential compounds used as an alternative biofuel in the future. It is necessary to seek more sustainable technologies such as microbial engineering for γ-terpinene production. In this study, we metabolically engineered Escherichia coli to produce γ-terpinene by introducing a heterologous mevalonate (MVA) pathway combined with the geranyl diphosphate synthase gene and γ-terpinene synthase gene. Subsequently, the culture medium and process conditions were optimised with a titre of 19.42 mg L−1 obtained. Additionally, in-depth analysis at translation level for the engineered strain and intermediate metabolites were detected for further analysis. Finally, the fed-batch fermentation of γ-terpinene was evaluated, where a maximum concentration of 275.41 mg L−1 with a maintainable feedstock of glycerol was achieved.
A sustainable technology for gamma (γ)-terpinene was constructed in an engineered Escherichia coli. In-depth analysis at translation level for the engineered strain and intermediate metabolites were analyzed. The fed-batch fermentation of γ-terpinene was 275.41 mg L−1.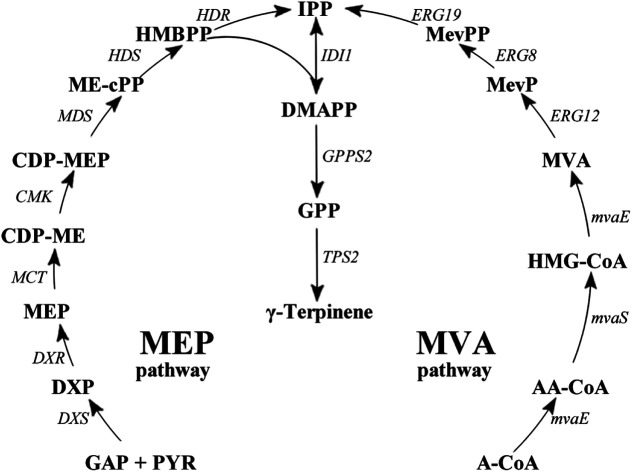
1. Introduction
Monoterpenes (C10H16) are compounds that are part of the terpene class, consisting of two isoprene units. They are known to form a remarkably sophisticated defence system in plants in the fight against predators such as insects and herbivores,1,2 as well as having a number of other production purposes such as a hydrocarbon antioxidants,3–5 in fragrances,6 as fine chemicals and in medicinal products.7–11 Recently, an increased interest in the production of transportation fuels from renewable resources has catalysed many research endeavours focussing on developing microbial systems for production of these natural resources.12–14 In particular, monoterpenes are considered to be the best candidates as precursors in the production of alternative aviation fuels. Previously, it has been reported that the hydrogenated monoterpenes, limonene and pinene, enhanced cold weather performance of jet fuel mixtures. It was shown that limonene could match the volumetric energy, whereas pinene could match the net heat of combustion characteristics of the aviation turbine fuel JP-10.15,16
Gamma (γ)-terpinene (CAS: 99-85-4), is also a known potential biofuel alternative; is synthesised from the cyclisation of the geranyl diphosphate (GPP);17–22 and is produced from either the methylerythritol 4-phosphate pathway (MEP) or the mevalonate (MVA) pathway (Fig. 1).23 The current isolation technique of γ-terpinene from plants and citrus fruit is inefficient, requiring substantial expenditure of natural resources as well as the use of a number of environmentally hazardous chemicals.24,25Escherichia coli has well established metabolic pathways, simple genetic manipulation technology, and its fermentation technology is approaching maturity.26 Therefore, a more sustainable technology using E. coli for the production of γ-terpinene is required to allow it to be considered as a viable bio-based advanced biofuel in the future.
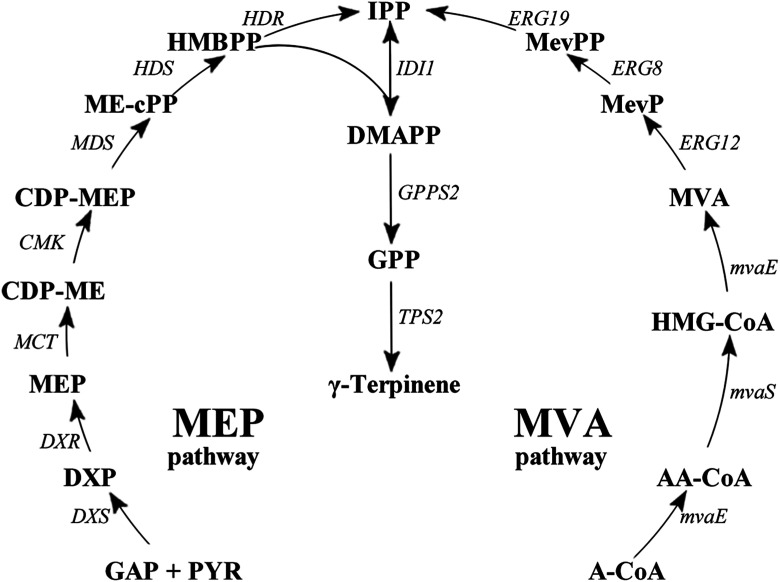
Fig. 1. γ-Terpinene biosynthesis pathway. γ-Terpinene was produced from the MEP pathway and MVA pathway, respectively. Pathway intermediates: GAP, glyceraldehyde 3-phosphate; PYR, pyruvate; DXP, 1-deoxy-d-xylulose 5-phosphate; MEP, 2C-methyl-D-erythritol 4-phosphate; CDP-ME, 4-diphosphocytidyl-2C-methyl-D-erythritol; CDP-MEP, 2-phospho-4-(cytidine-5′-di-phospho)-2C-methyl-D-erythritol; MEcPP, 2C-methyl-D-erythritol 2,4-cyclodiphosphate; HMBPP, 1-hydroxy-2-methyl-2(E)-butenyl 4-diphosphate; IPP, isopentenyl diphosphate; DMAPP, dimethylallyl diphosphate; GPP, geranyl diphosphate; A-CoA, acetyl-CoA; AA-CoA, acetoacetyl-CoA; HMG-CoA, 3-hydroxy-3-methylglutaryl-CoA; MevP, mevalonate 5-phosphate; MevPP, mevalonate 5-diphosphate. Enzymes in MEP pathway: DXS, DXP synthase; DXR, DXP reductoisomerase; MCT, CDP-ME synthase; CMK, CDP-ME kinase; MDS, ME-cPP synthase; HDS, HMBPP synthase; HDR, HMBPP reductase; IDI, IPP isomerase. Enzymes in MVA pathway: MvaE, acetyl-CoA acetyltransferase/HMG-CoA reductase; MvaS, HMG-CoA synthase; ERG12, mevalonate kinase; ERG8, phosphomevalonate kinase; ERG19, mevalonate pyrophosphate decarboxylase; IDI1, IPP isomerase.
γ-Terpinene synthase (TPS) belongs to the monoterpene cyclases family, and is the catalysis responsible for the conversion of GPP into γ-terpinene.27,28 Currently, TPS extraction has been reported from Eucalyptus, Origanum vulgare, Citrus limon, Citrus unshiu, Thymus vulgaris, and Thymus caespititius. Furthermore, it has been shown that TPS extracted specifically from Coriandrum sativum, C. limon, and O. vulgare was able to catalyse the conversion of GPP into γ-terpinene as well as a number of other products, such as α-thujene, myrcene, sabinene, limonene, pinene, linalool, α-phellandrene, ocimene, cymene, α-terpinolene, α-thujene and α-terpinene.29–31 Moreover, the microbial production of GPP was available in E. coli. Therefore, when TPS was introduced into E. coli, the microbial production technique for γ-terpinene was able to be constructed. Additionally, to ensure sustainable production of γ-terpinene by a microorganism like E. coli, the ability to use a renewable carbon source such as glycerol is necessary, along with a more developed and optimised fermentation medium and process conditions.32–34
Based on our previously work,33,34 in this study γ-terpinene was produced by assembling a biosynthetic pathway in an engineered E. coli strain, using the heterologous MVA pathway combined with the addition of the acetyl-CoA acetyltransferase/HMG-CoA reductase gene (mvaE), HMG-CoA synthase gene (mvaS) from Enterococcus faecalis, geranyl diphosphate synthase gene (GPPS2) gene from Abies grandis and TPS2 gene from T. vulgaris, due to its catalytic specificity. Additionally, on the chromosome of this particular E. coli strain the mevalonate kinase gene (MK/ERG12), phosphomevalonate kinase gene (PMK/ERG8), mevalonate pyrophosphate decarboxylase gene (PMD/ERG19) and IPP isomerase gene (idi/IDI1) were present.35 Finally, fed-batch fermentation of γ-terpinene was evaluated using the optimised culture medium and process conditions. This study has begun the necessary foundations needed for a more sustainable future method of γ-terpinene production.
2. Materials and methods
2.1. Plasmid construction and bacterial strain
All bacterial strains used in this study are listed in Table 1. E. coli DH5α (Invitrogen, California, USA) was used as the host to extract all necessary plasmids. E. coli BL21(DE3) (Invitrogen) and E. coli CIBTS1756 (ref. 35) was used as the host strains to over express the necessary proteins and therefore to produce γ-terpinene. All plasmids used in this study (Table 1), were constructed using standard cloning techniques as per manufactures protocols. Antibiotic resistance, PCR and DNA sequencing were utilised for the verification of alleles as well as plasmid constructs.
Plasmids and strains used in this study.
| Name | Relevant characteristics | Reference |
|---|---|---|
| Plasmids | ||
| pACYCDuet-1 | P15A origin. CmR. PT7 | Novagen |
| pTrcHis2B | ColE1 origin. AmpR. Ptrc | Invitrogen |
| pGH | pUC origin. AmpR. PT7 | Generay |
| pTrc-low | ColE1 origin. CmR. Ptrc::ERG12–ERG8–ERG19–IDI1 | 34 |
| pHW1 | P15A origin. CmR. PT7::TPS2 | This work |
| pHW2 | P15A origin. CmR. PT7::GPPS2–TPS2 | This work |
| pHW3 | P15A origin. CmR. PT7::mvaS–mvaE–GPPS2–TPS2 | This work |
| Strains | ||
| E. coli BL21(DE3) | E. coli B dcm ompT hsdS (rB− mB−) gal | Invitrogen |
| E. coli CIBTS1756 | BL21, glmS-pstS::P*L-MKMM-PMKSC-PMDSC-idiSC | 35 |
| E. coli DH5α | deoR, recA1, endA1, hsdR17 (rk−, mk+), phoA, supE44, λ−, thi-1, gyrA96, relA1 | Invitrogen |
| S. cerevisiae | Type strain | ATCC 204508 |
| HW1 | E. coli BL21(DE3) harboring pHW1 | This work |
| HW2 | E. coli BL21(DE3) harboring pHW2 | This work |
| HW3 | E. coli BL21(DE3) harboring pHW3 | This work |
| HW4 | E. coli BL21(DE3) harboring pHW3 and pTrc-low | This work |
| HW5 | E. coli CIBTS1756 harboring pHW3 | This work |
TPS (TPS2, GenBank: KR920616) from T. vulgaris, was analysed using online software (http://www.expasy.org/tools/), optimised to the preferred codon usage of E. coli (http://www.genscript.com/cgi-bin/tools/rare_codon_analysis) and chemically synthesised by the company (Sangon Biotech, Shanghai, China).
2.2. Analytical methods
Cell growth was monitored by measuring the optical density at 600 nm (OD600) with a spectrophotometer (Cary 50 UV-Vis, Varian). All the samples were measured in triple.
Product characterisation was carried out by capillary GC-MS using an Agilent 5975C system chromatograph with the following settings; A HP-INNOWAX capillary column (30 m × 320 μm × 0.25 μm, Agilent, Palo Alto, CA, USA), with helium as the carrier gas at a flow rate of 1 mL min−1, the injector temperature at 260 °C, the ion source temperature at 240 °C, EI 70 eV and the mass range 35–300 m/z. The following oven temperature program was carried out: 75 °C for 1 min with an increase of 10 °C min−1 until 100 °C was reached, where it was held for 5 min. The product γ-terpinene was quantified by gas chromatography with a flame ionisation detector and an Agilent HP-INNOWAX (30 m × 320 μm × 0.25 μm) column. The oven temperature was initially held at 75 °C for 1 min, with increases of 10 °C min−1 until 100 °C was reached, where it was held for 5 min. The temperatures of the infector and the detector were held at 240 °C and 260 °C, respectively. A standard curve of known concentrations of γ-terpinene (Sigma-Aldrich, USA) was used in the conversion of the peak area into γ-terpinene concentration.
To analyse the translation level perturbation for the engineered strain. The cultured bacterial cells were collected from the fermentation culture (5 mL) by centrifugation at 12 000 rpm for 5 min, and then washed with sterile distilled water. The washed pellets were suspended in 500 μL Tris–HCl buffer (pH 8.0) and subject to ultrasonication. The cell lysates were centrifuged and the supernatants obtained were mixed with 2× sodium dodecyl sulfate (SDS) sample buffer, heated at 100 °C for 5 min and then analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) according to standard protocols.
Intermediate metabolites concentrations in the culture supernatant were determined using an Agilent 1200 series high-performance liquid chromatography (HPLC) system equipped with an Aminex HPX-87H (Bio-Rad, Hercules, CA) column (300 × 7.8 mm). All samples were filtered through 0.22 μm syringe filter. Ultrapure water with 5 mM H2SO4 was used as the eluent at a flow rate of 0.5 mL min−1. The oven temperature was maintained at 25 °C, peaks were detected by a refractive index detector (RID). Quantitation of the intermediates were performed using an external standards method.
2.3. Culture media and fed-batch conditions
For all cloning and strain pre-culture purposes Luria–Bertani (LB) medium (10 g L−1 NaCl, 10 g L−1 peptone, 5 g L−1 yeast extract) with appropriate antibiotics was used. For shake-flask fermentation, γ-terpinene was produced in a 600 mL sealed bottle with 50 mL of a pre-established fermentation medium (40 g L−1 glycerol, 5 g L−1 yeast extract, 9.8 g L−1 K2HPO4, 0.3 g L−1 ferric ammonium citrate, 2.1 g L−1 citric acid monohydrate, 0.1 g L−1 MgSO4, 1 mL L−1 trace element solution).36 For fed-batch fermentation, γ-terpinene was detected from off-gas, and grown and produced in 2 L of fermentation medium described above, with ampicillin (100 μg mL−1), chloramphenicol (34 μg mL−1). The fed-batch medium consisted of 350 g L−1 glycerol, 5 g L−1 yeast extract, 9.8 g L−1 K2HPO4, 2.1 mg L−1 ferric ammonium citrate and 0.3 g L−1 citric acid monohydrate. The trace element solution used in both shake-flask and fed-batch fermentation contained 0.37 g L−1 (NH4)6Mo7O24·4H2O, 0.29 g L−1 ZnSO4·7H2O, 2.47 g L−1 H3BO3, 0.25 g L−1 CuSO4·5H2O and 1.58 g L−1 MnCl·4H2O.
2.4. γ-Terpinene production in shake-flask and fed-batch fermentation
The pre-culture was prepared by inoculating a single colony of E. coli from a freshly grown LB agar plate, into 5 mL of LB media (with appropriate antibiotics). The choice of antibiotic was dependent upon the plasmid in the E. coli (Table 1). We use ampicilin and chloramphenicol in the engineering strains containing the pACYCDuet-1 and pTrc-low plasmid, respectively. The culture was left to grow in a shaking bath (IS-RDS3 Incu-Shaker CRYSTAL, USA) operated at 180 rpm and 37 °C for 10 h. For γ-terpinene production in shake-flask experiments, a rotary shaker at 180 rpm and at 37 °C was used. A 600 mL sealed bottle containing 50 mL of fermentation medium described above, was inoculated with 1% pre-culture (with appropriate antibiotics). The initial pH was adjusted to 6.0. When the OD600 reached 0.6–0.8, production of γ-terpinene was induced by the addition of isopropy-β-D-thiogalactoside (IPTG) to the final concentration of 0.1 mM for 24 h. Then, a gas sample were taken from the headspace of the sealed bottle and analysed by previously described gas chromatography methods. In particular, cells were collected for protein analysis at 6 h after induction.
For γ-terpinene fed-batch fermentation, a 250 mL triangle bottle with 50 mL of the fermentation medium described above was used and was inoculated with 1% pre-culture. The culture was grown at 37 °C with shaking (180 rpm) for about 4 h. When the OD600 reached 0.8–1.0, it was then used to inoculate a 5 L fermenter (BIOSTAT Bplus MO 5 L, Sartorius, Germany) containing a further 2 L of fermentation medium. When the OD600 reached 20, the production of γ-terpinene was induced by the addition of IPTG to a final concentration of 0.1 mM. The temperature was initially maintained at 37 °C, and then reduced to 30 °C after induction. The following parameters were maintained throughout the production, pH was sustained at 6.0 via an automated addition of ammonia or 20% H2SO4, the stirring speed was initially 400 rpm and then linked to the dissolved oxygen (DO) concentration to maintain a 20% saturation and the flow velocity of air was controlled at 3 L min−1. During the course of fermentation, the fed-batch medium described above was fed into the system at a controlled flow rate of 3%; 1 mL samples of off-gas and fermentation liquid were taken every 3 h for gas chromatography analysis and OD600 measurements to monitor the production of γ-terpinene and the growth of the bacterium, respectively.
3. Results and discussion
3.1. The effect of different strains
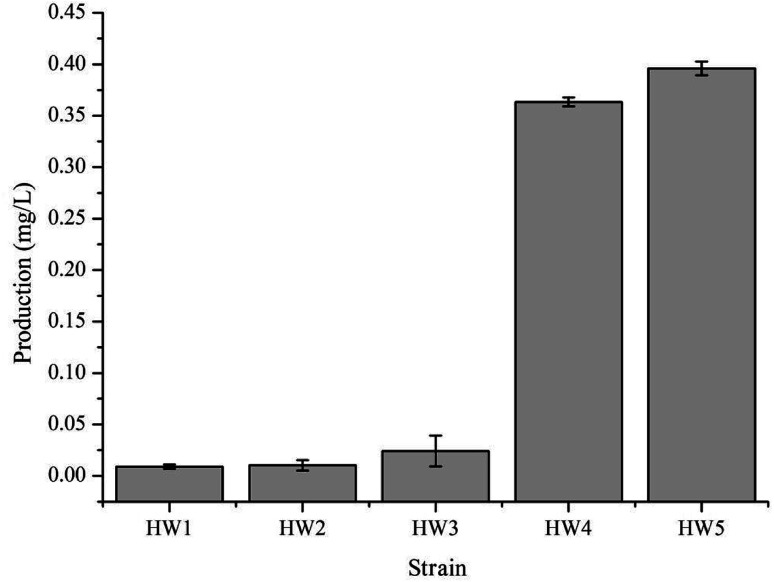
In order to confirm that the observed difference in γ-terpinene production was indeed related to the heterogeneous expression of the TPS2 gene, a number of strains were constructed in this study (Table 1). We used the amino acid sequence of the TPS2 gene from T. vulgaris to optimise the gene for the preferred codon usage of E. coli and had the gene synthesised by Sangon Biotech (Shanghai, China). Initial testing of the pACYC–TPS2 plasmid (pHW1) in the E. coli strain BL21 (DE3) for the production of γ-terpinene only yielded trace amounts with the MEP pathway. In order to begin the improvement of the production of γ-terpinene, the GPPS2 gene from A. grandis was added into the plasmid to improve the efficiency of the conversion of DMAPP to GPP and generate the plasmid pACYC–GPPS2–TPS2 (pHW2) and to form the strain HW2. Following on from this strain, the mvaE and mvaS genes from E. faecalis were added to form pACYC–mvaE–mvaS–GPPS2–TPS2 (pHW3) and to build the strain HW3. HW3 resulted in a 2-fold increase in the production of γ-terpinene when compared with the previous strain, HW2. Subsequently, to generate HW4, the plasmids pHW3 and pTrc-low34 which contains the MVA pathway genes ERG12, ERG8, ERG19, and IDI1 from Saccharomyces cerevisiae were used. The yield of the HW4 strain increased significantly, with it being 15 times higher than that of the HW3 strain. Finally, the E. coli strain CIBTS1756 (ref. 35) was transformed with pHW3 to construct the final strain, HW5. As illustrated in Fig. 2, the production of γ-terpinene in HW4 and HW5 was clearly higher than that of the other strains HW1, HW2, and HW3. Furthermore, the HW4 strain, which also contained the plasmid pTrc-low, increased the production of γ-terpinene more than 15-fold compared with HW3. The pTrc-low plasmid had low stability in the HW4 strain, therefore to avoid the loss of the pTrc-low plasmid and to also reduce the use of the antibiotics, we integrated the pTrc-low plasmid into the E. coli chromosome of the HW5 strain. This integrated strain resulted in a 9.0% increase in γ-terpinene production compared to the previous strains.
Fig. 2. γ-Terpinene production in the varying E. coli strains. HW1, HW2, and HW3 strains contained the plasmid pHW1, pHW2, and pHW3, respectively. HW4 contained two plasmids, pTrc-low and pHW3. HW5 was the CIBTS1756 strain with the plasmid of pHW3. Each strain for γ-terpinene production was cultured in a shake-flask containing 20 g L−1 glycerol, 5 g L−1 yeast extract, 9.8 g L−1 K2HPO4, 0.3 g L−1 ferric ammonium citrate, 2.1 g L−1 citric acid monohydrate, 0.1 g L−1 MgSO4 and 1 mL L−1 trace element solution. Protein production was induced with 0.1 mM IPTG and 100 μg mL−1 ampicillin, 34 μg mL−1 chloramphenicol was added when required.
3.2. The effect of different carbon sources
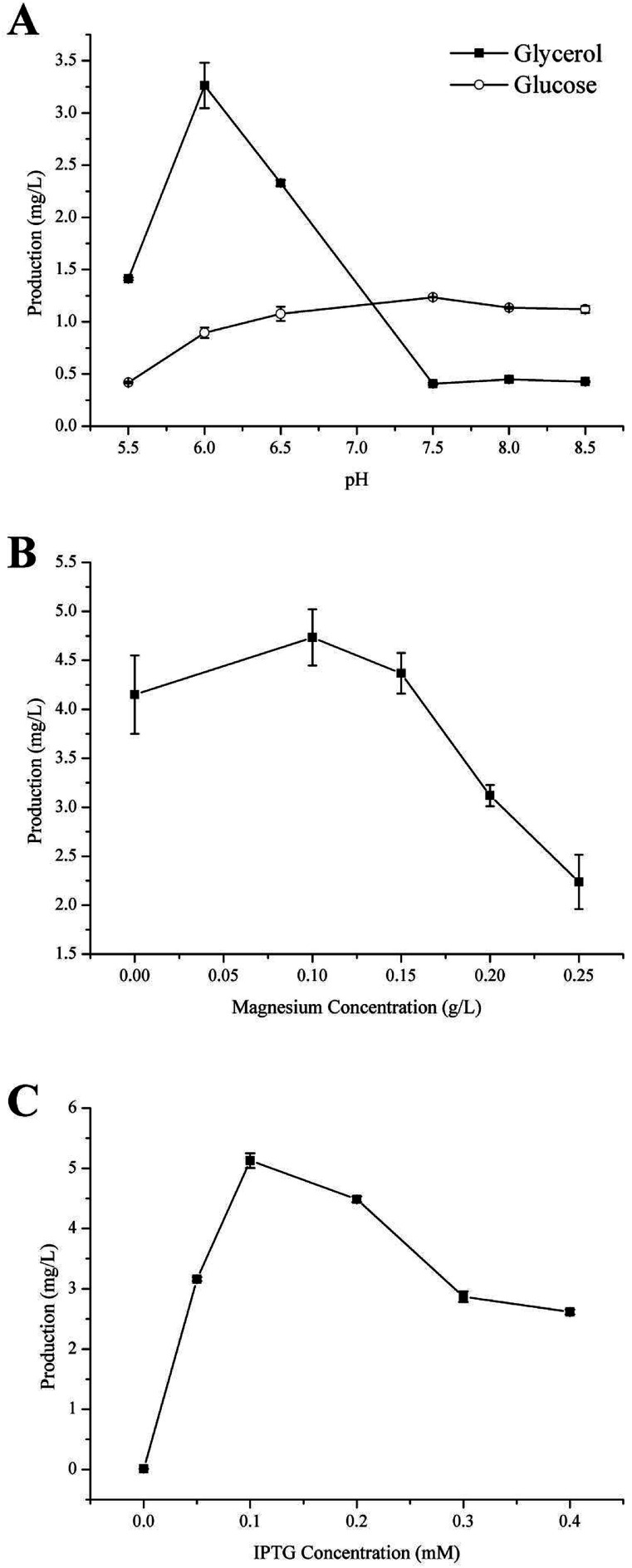
Carbon is the main feedstock in most fermentation media and is known to be quite influential in the process, therefore finding an efficient, as well as relatively sustainable carbon source for the production of γ-terpinene was crucial.37 In this study, we investigated the use of 40 g L−1 glycerol (C3H8O3) or 20 g L−1 glucose (C6H12O6) to culture the strain HW5. Previously, it was been established that the conversion of glycerol to acetoacetyl-CoA is different from that of glucose, with both generating a number of acidic products such as formate, lactate, and acetate during fermentation leading to changes in the pH of the fermentation media.36,38 Therefore, further study to develop a better understanding of the effects of the different carbon sources on the pH during the production of γ-terpinene was required. The effects of the different carbon sources on the pH of the media are shown in Fig. 3A.
Fig. 3. Effects of fermentation source and culture conditions on γ-terpinene production. (A) The effect of varying carbon sources, glycerol and glucose, on both pH and γ-terpinene production in the strain HW5. (B) The effect of varying concentration of magnesium. (C) The effect of the concentration of IPTG on γ-terpinene production. When the OD600 of the culture reached 0.6–0.8, the fermentation broth was induced for 24 h using varying concentrations of IPTG in a shake-flask.
At pH 6.0, the production of γ-terpinene with glycerol as the carbon source during fermentation yielded 3.26 ± 0.02 mg L−1 a 3.64-fold increase on that yielded by fermentation with glucose as the carbon source. However, at pH 7.5, the production of γ-terpinene with glucose as the carbon source during fermentation yielded 1.13 ± 0.01 mg L−1, 3.03-fold higher than that produced with glycerol as the carbon source under the same conditions. These results indicate that the use of glycerol during fermentation is more suitable at the lower pH when compared to glucose and therefore was used in future work. These results clearly highlight the different effects of using glycerol or glucose as the sole carbon source during fermentation.
3.3. The effect of Mg2+
Divalent cations especially magnesium ions are necessary for supporting catalysis of the γ-terpinene synthetase and for also activating several glycolytic enzymes. The appropriate concentration of magnesium is highly important in the activity of the enzyme. In this case, Mg2+ is needed to ensure the conversion of sugar to γ-terpinene during the fermentation process.28,39,40 Moreover, Mg2+ is essential to both physiological and biochemical functions in microorganisms, including growth, cell division, synthesis of essential fatty acids, regulation of cellular ionic levels, and maintaining membrane integrity and permeability.41 To investigate the influence of varying concentrations of Mg2+ on the production of γ-terpinene, various concentrations of Mg2+ were added into the fermentation medium. As shown in Fig. 3B, the production of γ-terpinene was inversely proportional to the Mg concentration, with decreasing yield with increasing concentrations of Mg2+. The most suitable concentration of Mg2+ in the production of γ-terpinene was 70 mg L−1. However, this concentration of Mg2+ was less than what has been established previously. This may be due to having an effect on the fermentation media pH, rather than just affecting microbial cell growth and the activity of the enzymes.42
3.4. The effect of IPTG
Various concentrations of IPTG (0.05 mM, 0.1 mM, 0.2 mM, 0.3 mM, and 0.4 mM) were added into the fermentation media, to develop the most efficient induction of γ-terpinene production. Once the OD600 reached 0.6–0.8, the culture temperature was decreased from 37 °C to 30 °C and incubated for 24 h. A gas sample from the headspace of the sealed cultures was then used in quantification. The induction of γ-terpinene production at the various concentrations of IPTG is shown at Fig. 3C. We found that at low concentrations of IPTG, HW5 was most efficient at producing γ-terpinene, with a yield of 5.13 mg L−1 with an optimised IPTG concentration of 0.1 mM. However, it is known that induction with IPTG can have an impact on the cell biomass, expression yields, as well as affecting plasmid stability,43,44 therefore future work should focus on using a different inducer to lower the stress response of the cell.45
3.5. Toxicity of commercial γ-terpinene to E. coli
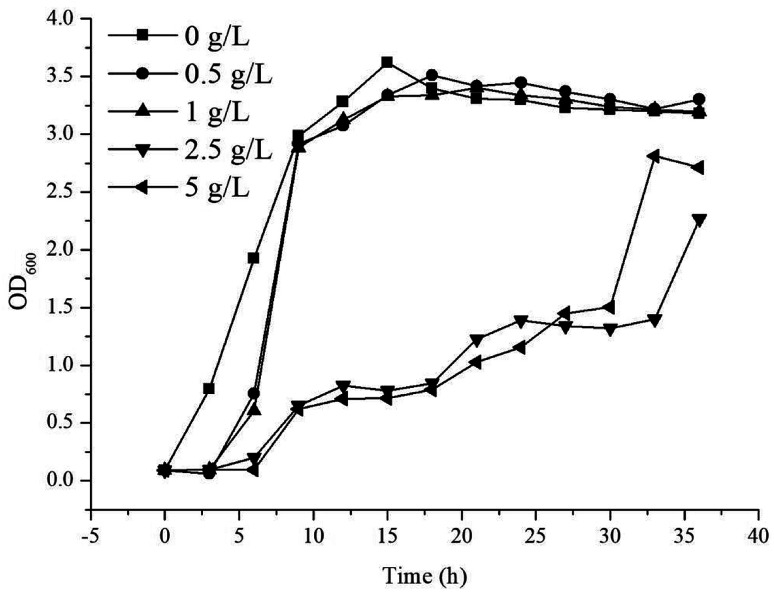
Throughout the induction and subsequent production of γ-terpinene, as its concentration increased, an effect on the growth of the E. coli production strain was noted. In order to understand the effect of the concentration of γ-terpinene on the E. coli production strain further, we measured the OD600 every 3 h for 36 h in 50 mL of LB medium in sealed shake-flask bottles, with various concentrations of commercially available γ-terpinene (0.5 g L−1, 1 g L−1, 2.5 g L−1, and 5 g L−1). As seen in Fig. 4, HW5 was found to grow more slowly at the 2.5 and 5 g L−1 concentrations of commercially available γ-terpinene, with an inhibition rate (IR) at the 12th hour of growth of 74.9% and 78.3%, respectively. These results indicate that γ-terpinene has the ability to slow down and even inhibit the growth of HW5. Therefore, the toxicity caused by the presence of γ-terpinene, at various concentrations on the E. coli production strain is a key factor to address in the future to ensure efficient production.
Fig. 4. The growth of strain HW5 in LB medium with different concentration of commercially available γ-terpinene. The growth (OD600) was monitored every 3 h for 36 h. Concentrations of γ-terpinene were added to the LB medium as follows: 0 g L−1 (■), 0.5 g L−1 (●), 1 g L−1 (▲), 2.5 g L−1 (▼) and 5 g L−1 (◀).
The toxicity of overexpressed proteins has attracted much research attention recently, with it having a major effect on the host biomass as well as the production of certain biochemicals.33,46,47 Research has focussed on increasing product tolerance in the host microbe by understanding membrane fluidity and function,48,49 the function of efflux pumps50,51 and the significance of changed expression of particular genes of those strains that are more tolerant of protein production.52 Furthermore, it may be a better strategy to improve the hydrocarbon biofuels tolerance in the microbial host.53
3.6. Fermentation for γ-terpinene production in both shake-flask and fed-batch cultures
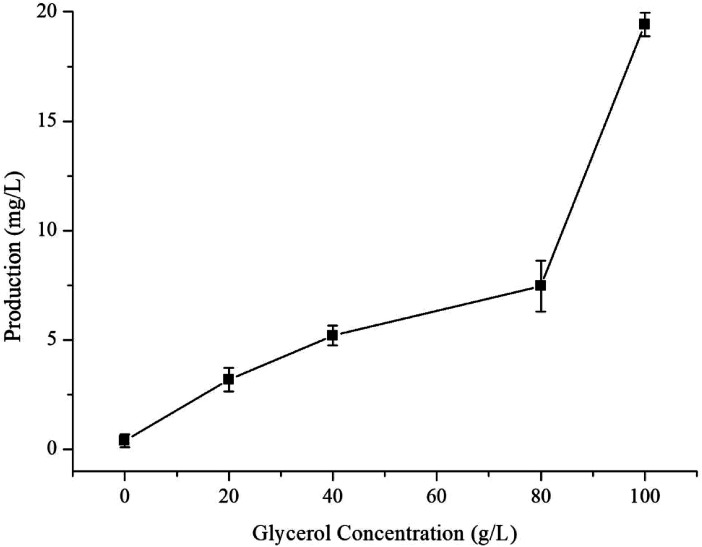
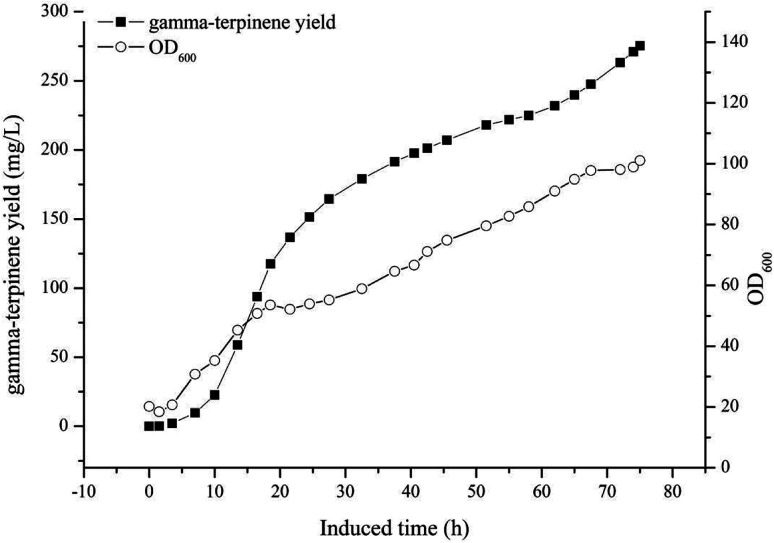
The optimised carbon source, the Mg2+ concentration IPTG and pH culture conditions yielded 5.13 mg L−1 of γ-terpinene. Following on from this, we attempted to control the concentration of glycerol in the fed-batch fermentation (0, 20, 40, 80 and 100 g L−1) to possibly improve the production of γ-terpinene even more so. As we hypothesised, the concentration of glycerol in the culture, had a large effect on the yield, increasing the production of γ-terpinene to 19.42 mg L−1 with 100 g L−1 glycerol (Fig. 5). Due to the positive results obtained in fed-batch fermentation, we performed scaled-up experiments. In a 5 L fermenter with a working volume of 2 L in batch mode, fed-batch medium (containing glycerol) was continuously added at a 3% flow rate once protein production was induced with 0.1 mM IPTG, at an OD600 of 20. The changes in biomass and γ-terpinene production over the course of fermentation are shown in Fig. 6. After 3 days of fermentation, the 2 L culture reached an OD600 of 100, with an accumulated γ-terpinene yield of 275.41 mg L−1.
Fig. 5. The effect of glycerol concentration on γ-terpinene production.
Fig. 6. A time course of γ-terpinene production in the strain HW5 in fed-batch fermentation. A 5 L fermenter containing 2 L of fermentation medium was used, and the temperature was maintained at 37 °C. Once the culture reached an OD600 of 20, protein expression was induced with 0.1 mM IPTG and the temperature was changed to 30 °C. From this process, 275.41 mg L−1 of γ-terpinene was produced.
The HW5 strain enables the microbial synthesis of γ-terpinene, but future work should focus on how to modify the metabolism further and/or improve extraction technologies to get higher production. For example, formate is the main by-product of γ-terpinene glycerol fermentation, therefore if we were to knockout the pfl gene, we would then allow the metabolic flux to favour acetyl-CoA production, meaning less toxic products.35 Also, the yield was found to be proportional to the concentration of glycerol in the fed media, however, the low conversion efficiency of this as the carbon source during the fermentation process was hard to overcome. In future work, it may possible that the glycerol conversion efficiency could be increased with the presence of additional genes in the system including sldAB, gldA, dhaKLM, glpK, and glpD.54–57 Furthermore, further study of glycerol-tolerant strains is needed to improve the balance of glycerol fermentation and yield or protein.58 Finally, the extraction methods used in the isolation of γ-terpinene may be altered to include dodecane or poly-α-olefin in situ extraction technology to improve the yield of γ-terpinene in fermentation production.59,60
3.7. Translation level analysis with SDS-PAGE
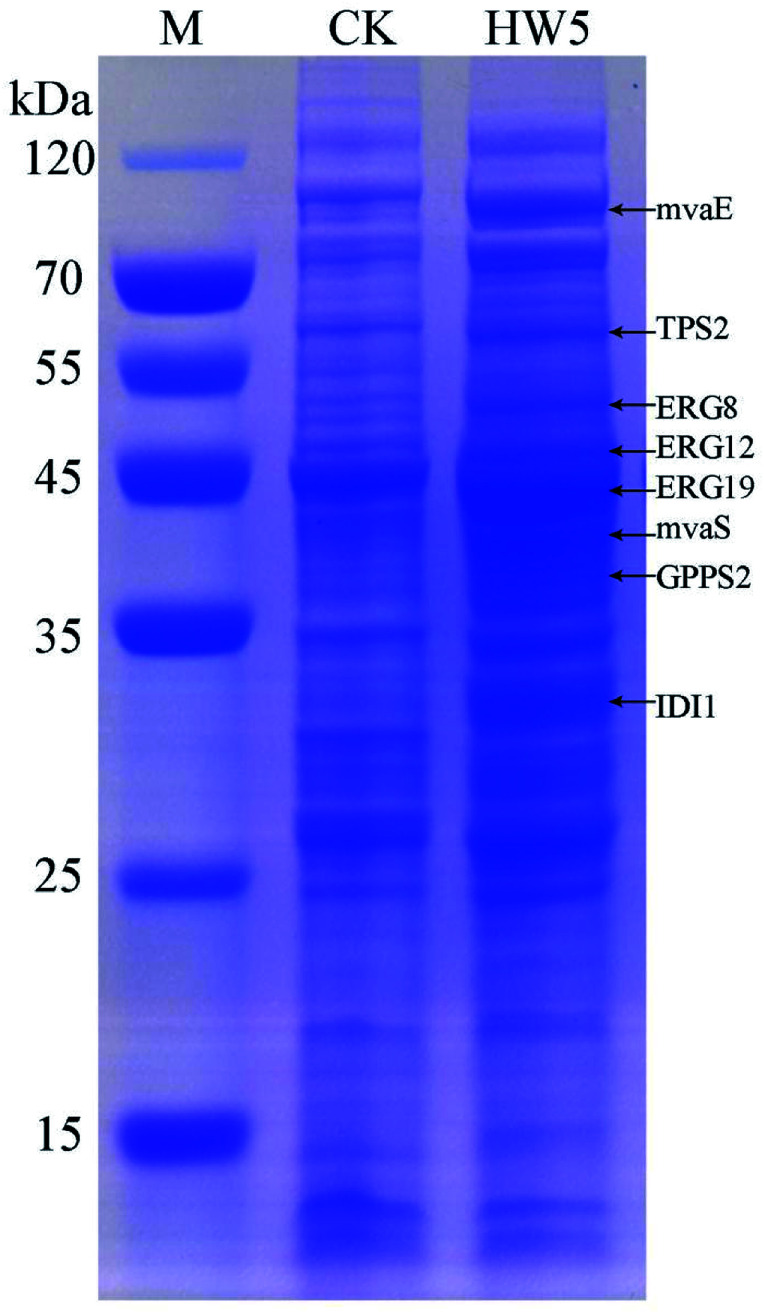
In order to more depth analysis of the perturbation of the engineered strain to provide data for further improvement in the efficiency of our engineered strain for the production of γ-terpinene. SDS-PAGE was chosen to analyze the strain at the protein level. As shown in Fig. 7, the proteins TPS2 (63.3 kDa), GPPS2 (41.3 kDa), mvaE (87.5 kDa), mvaS (43.4 kDa), ERG12 (48.4 kDa), ERG8 (50.5 kDa), ERG19 (44.0 kDa) and IDI1 (33.4 kDa) were successfully expressed. The results indicated that the expression levels of four proteins (ERG12, ERG8, ERG19 and IDI1) in the MVA downstream pathway were relatively high in the HW5 strain. However, protein expression levels of GPPS2 and TPS2 were found to be relatively low. It is known that the high expression of GPPS2 and TPS2 proteins is needed for the efficient production of C10 compound γ-terpinene.61 Therefore, future experiments will focus on optimizing the GPPS2 and TPS2 genes to increase the efficiency of the strain for the production of γ-terpinene.
Fig. 7. SDS-PAGE analysis of protein products. M: the protein marker; CK: negative control BL21 (DE3) with pACYCDuet1; HW5: CIBTS1756 with pACYC–mvaE–mvaS–GPPS2–TPS2.
3.8. Analysis of intermediate metabolites
The monitoring of intermediate metabolites is helpful in further engineering of the engineered strains. In this study, the metabolites of all the engineered strains were detected by HPLC. As shown in Table 2, succinic, lactic, acetic, mevalonic and butanediol were the main by-products. The results indicated that GPPS2 gene in the strain HW2 could cause the increment of lactic and butanediol compared to the strain HW1, which might cause by carbon flowing to the anaerobic respiratory chain. When the two upstream genes (mvaE and mvaS) were introduced into the strain HW2 to form the strain HW3. The mevalonate produced by strain HW3 was nearly five times more than that of strain HW2. The accumulation of mevalonate showed that the two genes are able to play a crucial role in the MVA pathway. Subsequently, the downstream genes of MVA pathway (ERG12, ERG8, ERG19 and IDI1) enhanced (HW4) to convert the accumulated mevalonate into γ-terpinene. Finally, to make the strain more stabile and reduce the use of antibiotics, the four downstream genes were integrated into the E. coli chromosomes (HW5). As shown in Table 2, the accumulation of mevalonate was decreased, meanwhile the butanediol increase notably. Butanediol is a reductive by-product, which indicated that the MVA pathway is more energetically friendly. Just like the production of isoprene in E. coli through MVA pathway, redundant NAD(P)H might accumulate in the cell, which might cause metabolic disturbance to the host.62 Therefore, how to equilibrate the redox balance in E. coli is vitally for further engineering of the engineered strain.
Analysis of intermediate metabolites.
| Succinic acid (mg L−1) | Lactic (mg L−1) | Acetic (mg L−1) | Mevalonate (mg L−1) | Butanediol (mg L−1) | |
|---|---|---|---|---|---|
| HW1 | 437.01 ± 6.62 | 400.67 ± 12.09 | 889.54 ± 35.16 | 125.23 ± 2.63 | 4.11 ± 0.35 |
| HW2 | 397.18 ± 27.09 | 765.06 ± 46.66 | 629.55 ± 84.91 | 122.58 ± 3.41 | 9.25 ± 6.16 |
| HW3 | 324.47 ± 17.13 | 847.58 ± 47.62 | 316.58 ± 10.56 | 603.95 ± 22.44 | 6.46 ± 2.03 |
| HW4 | 11.57 ± 0.48 | 0.00 ± 0.00 | 479.63 ± 20.14 | 119.38 ± 10.32 | 36.30 ± 1.80 |
| HW5 | 63.00 ± 16.93 | 0.00 ± 0.00 | 909.37 ± 36.48 | 119.60 ± 2.53 | 51.25 ± 1.57 |
4. Conclusions
In summary, we have demonstrated the feasibility of producing γ-terpinene in metabolically engineered E. coli in fermentation conditions using glycerol as a carbon source. In this study, γ-terpinene was successfully produced by assembling a biosynthetic pathway using the methylerythritol 4-phosphate or heterologous MVA pathway and by combining the GPPS2 gene and TPS2 gene in E. coli. Subsequently, the culture medium and process conditions were optimised to further enhance the titre of γ-terpinene production, yielding 19.42 mg L−1. Finally, we also evaluated the fed-batch fermentation of γ-terpinene using the optimised culture medium and process conditions, which successfully, when scaled up, reached a total production of 275.1 mg L−1 of γ-terpinene, 1.9 mmol mol−1 glycerol to γ-terpinene. This study has provided a sustainable strategy to produce γ-terpinene using glycerol as carbon source and has laid the foundation for future industrial production of monoterpenes with glycerol fermentation platforms.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This study was supported by National Natural Science Foundation of China (NSF No. 31400084), Hainan's Key Project of Research and Development Plan (No. ZDYF2017155), Taishan Scholars Climbing Program of Shandong (No. TSPD20150210), Youth Innovation Promotion Association CAS (No. 2017252).
References
- Özcan M. Chalchat J. C. Bulg. J. Plant Physiol. 2004;30:68–73. [Google Scholar]
- Tsoukatou M. Tsitsimpikou C. Vagias C. Roussis V. Z. Naturforsch., C: J. Biosci. 2001;56:211–215. doi: 10.1515/znc-2001-3-407. [DOI] [PubMed] [Google Scholar]
- Graßmann J. Vitam. Horm. 2005;72:505–535. doi: 10.1016/S0083-6729(05)72015-X. [DOI] [PubMed] [Google Scholar]
- Hazzit M. Baaliouamer A. Faleiro M. L. Miguel M. G. J. Agric. Food Chem. 2006;54:6314–6321. doi: 10.1021/jf0606104. [DOI] [PubMed] [Google Scholar]
- Mario C. F. Ingold K. U. J. Agric. Food Chem. 2003;51:2758–2765. doi: 10.1021/jf020993f. [DOI] [PubMed] [Google Scholar]
- Lücker J. Schwab W. van Hautum B. Blaas J. van der Plas L. H. W. Bouwmeester H. J. Verhoeven H. A. Plant Physiol. 2004;134:510–519. doi: 10.1104/pp.103.030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aydin S. Basaran A. A. Basaran N. Mutat. Res. 2005;581:43–53. doi: 10.1016/j.mrgentox.2004.10.017. [DOI] [PubMed] [Google Scholar]
- Fukumoto S. Sawasaki E. Okuyama S. Miyake Y. Yokogoshi H. Nutr. Neurosci. 2006;9:73–80. doi: 10.1080/10284150600573660. [DOI] [PubMed] [Google Scholar]
- Hua K. Yang T. Chiu H. Ho C. Nat. Prod. Commun. 2014;9:869–872. [PubMed] [Google Scholar]
- Takahashi Y. Inaba N. Kuwahara S. Kuki W. Biosci., Biotechnol., Biochem. 2003;67:2448–2450. doi: 10.1271/bbb.67.2448. [DOI] [PubMed] [Google Scholar]
- Ramalho T. Oliveira M. Lima A. Bezerra-Santos C. Piuvezam M. Planta Med. 2015;81:1248–1254. doi: 10.1055/s-0035-1546169. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay A. Redding A. M. Rutherford B. J. Keasling J. D. Curr. Opin. Biotechnol. 2008;19:228–234. doi: 10.1016/j.copbio.2008.05.003. [DOI] [PubMed] [Google Scholar]
- Clomburg J. M. Gonzalez R. Appl. Microbiol. Biotechnol. 2010;86:419–434. doi: 10.1007/s00253-010-2446-1. [DOI] [PubMed] [Google Scholar]
- Stephanopoulos G. Science. 2007;315:801–804. doi: 10.1126/science.1139612. [DOI] [PubMed] [Google Scholar]
- Harvey B. G. Wright M. E. Quintana R. L. Energy Fuels. 2010;24:267–273. doi: 10.1021/ef900799c. [DOI] [Google Scholar]
- Tracy N. I. Chen D. Crunkleton D. W. Price G. L. Fuel. 2009;88:2238–2240. doi: 10.1016/j.fuel.2009.02.002. [DOI] [Google Scholar]
- Cheng A. X. Lou Y. G. Mao Y. B. Lu S. Wang L. J. Chen X. Y. J. Integr. Plant Biol. 2007;49:179–186. doi: 10.1111/j.1744-7909.2007.00395.x. [DOI] [Google Scholar]
- McGarvey D. J. Croteao R. Plant Cell. 1995;7:1015–1026. doi: 10.1105/tpc.7.7.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab W. Williams D. C. Davis E. M. Croteau R. Arch. Biochem. Biophys. 2001;392:123–136. doi: 10.1006/abbi.2001.2442. [DOI] [PubMed] [Google Scholar]
- Trapp S. C. Croteau R. B. Genetics. 2001;158:811–832. doi: 10.1093/genetics/158.2.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tholl D. Curr. Opin. Plant Biol. 2006;9:297–304. doi: 10.1016/j.pbi.2006.03.014. [DOI] [PubMed] [Google Scholar]
- Zhang Q. Tiefenbacher K. Nat. Chem. 2015;7:197–202. doi: 10.1038/nchem.2181. [DOI] [PubMed] [Google Scholar]
- Steinbüchel A. Curr. Opin. Microbiol. 2003;6:261–270. doi: 10.1016/S1369-5274(03)00061-4. [DOI] [PubMed] [Google Scholar]
- Bourgou S. Rahali F. Z. Ourghemmi I. Saïdani Tounsi M. Sci. World J. 2012 doi: 10.1100/2012/528593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carasek E. Nardini G. S. Merib J. O. Microchem. J. 2013;109:128–133. doi: 10.1016/j.microc.2012.03.024. [DOI] [Google Scholar]
- Martin V. J. J. Pitera D. J. Withers S. T. Newman J. D. Keasling J. D. Nat. Biotechnol. 2003;21:796–802. doi: 10.1038/nbt833. [DOI] [PubMed] [Google Scholar]
- Alonso W. R. Croteau R. Arch. Biochem. Biophys. 1991;286:511–517. doi: 10.1016/0003-9861(91)90073-R. [DOI] [PubMed] [Google Scholar]
- Rudolph K. Parthier C. Egerer-Sieber C. Geiger D. Muller Y. A. Kreis W. Muller-Uri F. Acta Crystallogr., Sect. F: Struct. Biol. Commun. 2016;72(Pt 1):16–23. doi: 10.1107/S2053230X15023043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocoll C. Asbach J. Novak J. Gershenzon J. Degenhardt J. Plant Mol. Biol. 2010;73:587–603. doi: 10.1007/s11103-010-9636-1. [DOI] [PubMed] [Google Scholar]
- Lücker J. El Tamer M. K. Schwab W. Verstappen F. W. A. van der Plas L. H. W. Bouwmeester H. J. Verhoeven H. A. Eur. J. Biochem. 2002;269:3160–3171. doi: 10.1046/j.1432-1033.2002.02985.x. [DOI] [PubMed] [Google Scholar]
- Galata M. Sarker L. S. Mahmoud S. S. Phytochemicals. 2014;102:64–73. doi: 10.1016/j.phytochem.2014.02.016. [DOI] [PubMed] [Google Scholar]
- Willrodt C. David C. Cornelissen S. Bühler B. Julsing M. K. Schmid A. Biotechnol. J. 2014;9:1000–1012. doi: 10.1002/biot.201400023. [DOI] [PubMed] [Google Scholar]
- Zhang H. Liu C. Cao Y. Feng X. Zheng Y. Zou H. Liu H. Yang J. Xian M. Microb. Cell Fact. 2014;13:20. doi: 10.1186/1475-2859-13-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. Nie Q. Ren M. Feng H. Jiang X. Zheng Y. Liu M. Zhang H. Xian M. Biotechnol. Biofuels. 2013;6:60. doi: 10.1186/1754-6834-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C. Gao X. Jiang Y. Sun B. Gao F. Yang S. Metab. Eng. 2016;37:79–91. doi: 10.1016/j.ymben.2016.05.003. [DOI] [PubMed] [Google Scholar]
- Marzan L. W. Barua R. Akter Y. Arifuzzaman M. Islam M. R. Shimizu K. J. Genet. Eng. Biotechnol. 2017;15:161–168. doi: 10.1016/j.jgeb.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y. H. Kim S. J. Kim H. W. Kim J. Y. Gwak J. S. Chung D. Kim K. H. Park K. Park Y. C. J. Biotechnol. 2017;253:34–39. doi: 10.1016/j.jbiotec.2017.05.014. [DOI] [PubMed] [Google Scholar]
- Kim K. Kim S. K. Park Y. C. Seo J. H. Bioresour. Technol. 2014;156:170–175. doi: 10.1016/j.biortech.2014.01.009. [DOI] [PubMed] [Google Scholar]
- Pejin J. D. Mojović L. V. Pejin D. J. Kocić-Tanackov S. D. Savić D. S. Nikolić S. B. Djukić-Vuković A. P. Fuel. 2015;142:58–64. doi: 10.1016/j.fuel.2014.10.077. [DOI] [Google Scholar]
- Walker G. Nicola R. D. Anthony S. Learmonth R. Enzyme Microb. Technol. 2006;26:678–687. [Google Scholar]
- Cowan J. A. BioMetals. 2002;15:225–235. doi: 10.1023/A:1016022730880. [DOI] [PubMed] [Google Scholar]
- Christensen D. G. Orr J. S. Rao C. V. Wolfe A. J. Appl. Environ. Microbiol. 2017;83:e03034-16. doi: 10.1128/AEM.03034-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan R. Robinson C. Glick B. J. Ind. Microbiol. Biotechnol. 1996;16:145–154. doi: 10.1007/BF01569997. [DOI] [PubMed] [Google Scholar]
- Glick B. R. Biotechnol. Bioeng. 1990;35:668–681. doi: 10.1002/bit.260350704. [DOI] [PubMed] [Google Scholar]
- Piskunova J. Maisonneuve E. Germain E. Gerdes K. Severinov K. Mol. Microbiol. 2017;104:463–471. doi: 10.1111/mmi.13640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlop M. J. Biotechnol. Biofuels. 2011;4:32. doi: 10.1186/1754-6834-4-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X. Zhang H. Yang J. Liu M. Feng H. Liu X. Cao Y. Feng D. Xian M. Appl. Microbiol. Biotechnol. 2013;97:5423–5431. doi: 10.1007/s00253-012-4633-8. [DOI] [PubMed] [Google Scholar]
- Alexandre H. Rousseaux I. Charpentier C. FEMS Microbiol. Lett. 1994;124:17–22. doi: 10.1111/j.1574-6968.1994.tb07255.x. [DOI] [PubMed] [Google Scholar]
- Gustafson J. E. Cox S. D. Liew Y. C. Wyllie S. G. Warmington J. R. Pathology. 2001;33:211–215. doi: 10.1080/00313020123909. [DOI] [PubMed] [Google Scholar]
- Chen B. Ling H. Chang M. W. Biotechnol. Biofuels. 2013;6:21. doi: 10.1186/1754-6834-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling H. Chen B. Kang A. Lee J. M. Chang M. W. Biotechnol. Biofuels. 2013;6:95. doi: 10.1186/1754-6834-6-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi T. Sakai A. Kotani H. Tanabe K. Furusawa C. J. Biotechnol. 2017;255:47–56. doi: 10.1016/j.jbiotec.2017.06.408. [DOI] [PubMed] [Google Scholar]
- Sheng J. Feng X. Front. Microbiol. 2015;6:554. doi: 10.3389/fmicb.2015.00554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gätgens C. Degner U. Bringer-Meyer S. Appl. Microbiol. Biotechnol. 2007;76:553–559. doi: 10.1007/s00253-007-1003-z. [DOI] [PubMed] [Google Scholar]
- Mazumdar S. Clomburg J. M. Gonzalez R. Appl. Environ. Microbiol. 2010;76:4327–4336. doi: 10.1128/AEM.00664-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazdani S. S. Gonzalez R. Metab. Eng. 2008;10:340–351. doi: 10.1016/j.ymben.2008.08.005. [DOI] [PubMed] [Google Scholar]
- Zhang X. Shanmugam K. T. Ingram L. O. Appl. Environ. Microbiol. 2010;76:2397–2401. doi: 10.1128/AEM.02902-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. Wu J. Gao M. J. Zhu L. Zhan X. B. Prep. Biochem. Biotechnol. 2017;47:468–472. doi: 10.1080/10826068.2017.1292288. [DOI] [PubMed] [Google Scholar]
- Meadows A. L. Hawkins K. M. Tsegaye Y. Antipov E. Kim Y. Raetz L. Dahl R. H. Tai A. Mahatdejkul-Meadows T. Xu L. Zhao L. Dasika M. S. Murarka A. Lenihan J. Eng D. Leng J. S. Liu C. L. Wenger J. W. Jiang H. Chao L. Westfall P. Lai J. Ganesan S. Jackson P. Mans R. Platt D. Reeves C. D. Saija P. R. Wichmann G. Holmes V. F. Benjamin K. Hill P. W. Gardner T. S. Tsong A. E. Nature. 2016;537:694–697. doi: 10.1038/nature19769. [DOI] [PubMed] [Google Scholar]
- Tippmann S. Nielsen J. Khoomrung S. Talanta. 2016;146:100–106. doi: 10.1016/j.talanta.2015.08.031. [DOI] [PubMed] [Google Scholar]
- Zhou J. Wang C. Choi E. S. Kim S. W. Enzyme Microb. Technol. 2015;68:50–55. doi: 10.1016/j.enzmictec.2014.10.005. [DOI] [PubMed] [Google Scholar]
- Li M. Nian R. Xian M. Zhang H. Appl. Microbiol. Biotechnol. 2018;102:7725–7738. doi: 10.1007/s00253-018-9200-5. [DOI] [PMC free article] [PubMed] [Google Scholar]