Abstract
Caspofungin acetate (MK-0991) is an antifungal antibiotic that inhibits the synthesis of 1,3-β-d-glucan, an essential component of the cell wall of several pathogenic fungi. Caspofungin acetate was recently approved for the treatment of invasive aspergillosis in patients who are refractory to or intolerant of other therapies. The activity of 1,3-β-d-glucan synthesis inhibitors against Aspergillus fumigatus has been evaluated in animal models of pulmonary or disseminated disease by using prolongation of survival or reduction in tissue CFU as assay endpoints. Because these methods suffer from limited sensitivity or poor correlation with fungal growth, we have developed a quantitative PCR-based (qPCR) (TaqMan) assay to monitor disease progression and measure drug efficacy. A. fumigatus added to naïve, uninfected kidneys as either ungerminated conidia or small germlings yielded a linear qPCR response over at least 4 orders of magnitude. In a murine model of disseminated aspergillosis, a burden of A. fumigatus was detected in each of five different organs at 4 days postinfection by the qPCR assay, and the mean fungal load in these organs was 1.2 to 3.5 log10 units greater than mean values determined by CFU measurement. When used to monitor disease progression in infected mice, the qPCR assay detected an increase of nearly 4 log10 conidial equivalents/g of kidney between days 1 and 4 following infection, with a peak fungal burden that coincided with the onset of significant mortality. Traditional CFU methodology detected only a marginal increase in fungal load in the same tissues. In contrast, when mice were infected with Candida albicans, which does not form true mycelia in tissues, quantitation of kidney burden by both qPCR and CFU assays was strongly correlated as the infection progressed. Finally, treatment of mice with induced disseminated aspergillosis with either caspofungin or amphotericin B reduced the A. fumigatus burden in infected kidneys to the limit of detection for the qPCR assay. Because of its much larger dynamic range, the qPCR assay is superior to traditional CFU determination for monitoring the progression of disseminated aspergillosis and evaluating the activity of antifungal antibiotics against A. fumigatus.
Life-threatening fungal infections have become more prevalent as the population of immunocompromised patients has increased (6, 7). The value of existing therapies is tempered by such factors as a lack of sufficient spectrum, toxic side effects, or emerging resistance (13, 31, 37). Currently there are several antifungal agents in clinical development, including novel triazoles and members of a new class of compounds that inhibit the synthesis of 1,3-β-d-glucan, an essential polysaccharide of the fungal cell wall. Caspofungin acetate (MK-0991) is the first compound of this class to be approved for therapeutic use. Caspofungin acetate, indicated for the treatment of invasive aspergillosis in patients who are refractory to or intolerant of other therapies, has shown excellent safety, tolerability, and efficacy in completed clinical studies (J. Maertens, I. Raad, C. A. Sable, A. Ngai, R. Berman, T. F. Patterson, D. Denning, and T. F. Walsh, Abstr. 40th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1103, 2000).
The development of caspofungin for treatment of aspergillosis has been supported by results from animal models of disseminated or pulmonary disease. The efficacy of caspofungin in these animal models was measured by the use of survival as an endpoint (2; E. M. Bernard, T. Ishimaur, and D. Armstrong, Abstr. 36th Intersci. Conf. Antimicrob. Agents Chemother., abstr. F39, 1996). Evaluation of other 1,3-β-d-glucan synthesis (GS) inhibitors (LY303366 and FK463) has been described in models of aspergillosis that employed survival or CFU quantitation in selected organs as an endpoint (28, 29, 34). However, CFU counts do not accurately reflect the number of viable cells for filamentous fungi such as Aspergillus fumigatus. Due to the filamentous nature of these organisms, a large fungal mass is often indistinguishable from single-cell conidial forms when spread on agar plates, since both will usually yield one colony (24). Accordingly, results from experiments employing CFU measurement to quantify in vivo efficacy of GS inhibitors against A. fumigatus have been mixed. One study exploring the efficacy of LY303366 found no significant difference in fungal CFU burden in the lungs of infected rabbits in a model of pulmonary aspergillosis, despite significant improvements in several parameters of therapeutic efficacy (29). Verweij et al. (34) have shown that LY303366 at concentrations ≥2.5 mg/kg of body weight/day significantly reduced the A. fumigatus fungal kidney burden compared to that of vehicle-treated mice in a model of disseminated aspergillosis. A third study, with a murine model of pulmonary aspergillosis and constant infusion of FK463, reported a significant reduction in the CFU count at concentrations of FK463 in plasma of ≥0.55 μg/ml (28).
In an effort to overcome the inherent disadvantages of CFU determination for measuring the load of A. fumigatus in tissues of infected animals, we have developed a method that uses quantitative PCR (qPCR) (TaqMan) analysis (16) to amplify and quantitate an A. fumigatus sequence in complex DNA samples prepared from tissue. The qPCR assay is an adaptation of standard PCR that allows for detection and real-time quantitation of specific amplification products. In qPCR assay, a target DNA sequence is selectively amplified by using sequence-specific sense and antisense primers and Taq DNA polymerase. Also present in reaction mixtures is a dual fluorochrome oligonucleotide hybridization probe (labeled with a reporter such as 6-carboxyfluorescein [FAM] and a quencher such as 6-carboxytetramethyl rhodamine [TAMRA]), which is nonfluorescent due to the physical proximity of the reporter and quencher. During the annealing phase of the PCR, the probe specifically hybridizes to the accumulating product, and during the amplification phase, exonucleolytic activity of Taq DNA polymerase liberates the reporter-labeled nucleotide from the 5′ end of the probe. Release of the reporter nucleotide physically removes it from the environment of the quenching dye, resulting in a fluorescent signal. Reactions are characterized by the time during cycling when a threshold of baseline fluorescence (CT) is exceeded. With increasing amounts of target DNA in the experimental samples, the threshold is exceeded at an earlier cycle of amplification. Samples with no target DNA show no fluorescence and do not exceed the threshold at any point during the standard 40 cycles of qPCR amplification.
PCR has been described in the literature as a tool for detecting and identifying Aspergillus and other pathogenic fungi in clinical samples (25). TaqMan and other qPCR methods have been used to detect and quantitate bacterial (26) or viral (36) burden in other animal models of human infection. Here we describe a qPCR assay that can detect and quantitate A. fumigatus DNA in mouse tissues. We first evaluated this assay by adding either A. fumigatus conidia or mycelium to tissues, extracting total DNA, and demonstrating a linear response over 4 orders of magnitude with a low limit of detection. The primers and probes we chose showed no cross-reactivity with mouse genomic DNA or with DNA from the pathogenic fungus Candida albicans. With tissue samples derived from infected animals, the A. fumigatus DNA burden steadily increased with longer time following infection, reaching a peak that correlated with the onset of mortality. Finally, we show that this assay can quantitate differences in A. fumigatus fungal burden between drug-treated and nontreated control mice, despite minimal changes in CFU determinations from the same experimental samples.
MATERIALS AND METHODS
Drugs.
Caspofungin acetate (MK-0991) was solubilized in sterile distilled water and tested at the dose specified below. Amphotericin B (AmB [Fungizone]; Bristol Myers Squibb, Princeton, N.J.) was reconstituted according to the manufacturer's instructions in sterile distilled water, diluted, and tested at the dose specified below.
Animals.
Conventionally reared, female, DBA/2J (C5′ complement deficient) mice (Jackson Laboratory) with an average weight of 18 to 22 g were used. Mice were housed (10 per cage) in presterilized microisolator cages and provided with sterile bedding, feed, and water. Organs for use in these experiments were removed aseptically from euthanized animals. All procedures were performed in accordance with the highest standards for the humane handling, care, and treatment of research animals and were approved by the Merck Institutional Animal Care and Use Committee. Procedures for the care and use of research animals at Merck meet or exceed all applicable local, national, and international laws and regulations.
Conidia or mycelium added to naïve, uninfected organs.
A glycerol stock of conidia from A. fumigatus strain MF5668 (ATCC 13073) was prepared from a sporulated culture grown on Difco Yeast Malt Extract-Trace Element agar (32) (Difco, Detroit, Mich.) in several cotton-plugged baffled flasks for 5 days at 37°C. Thirty to 35% of the surface of the agar was covered with sterile 4-mm-diameter glass beads, 5 ml of sterile saline was added, and each flask was shaken for 15 min at 175 rpm on an orbital shaker (model 416; Forma Scientific, Marietta, Ohio). The liquid was removed from each flask, pooled, and subjected to ultrasonic treatment for 10 to 15 min at 4°C in a bath sonicator (Branson Ultrasonics Corp., Danbury, Conn.) to further separate spores from mycelia. The suspension was filtered through a sterile glass wool filter loosely packed in a plastic syringe body and concentrated by centrifugation at 3,200 × g for 10 min. The pellet was suspended in 50% glycerol and stored at −80°C. The number of conidia was determined by direct counting with a hemocytometer. Seven serial dilutions were made in 50% glycerol from a stock containing 1.26 × 1010 conidia/ml, and 40-μl aliquots containing known numbers of conidia were added to naïve uninfected organs.
Mycelium used in kidney spiking experiments was prepared by inoculating conidia (105 CFU/ml) from a glycerol suspension into RPMI/Junlon medium {RPMI 1640 medium with l-glutamine (Life Technologies, Rockville, Md.), buffered with 0.165 M MOPS [3-(N-morpholino) propanesulfonic acid; pH 7.0] (Fisher Scientific, Pittsburgh, Pa.) containing 0.15% (wt/vol) Junlon PW110 (Mitsui and Co., Inc., Specialty Chemicals Dept., New York, N.Y.)}. After 20 h at 37°C with shaking (220 rpm), the culture consisted of small dispersed germlings. One-milliliter aliquots of the culture were removed by using wide-bore pipette tips (Rainin Instruments, Woburn, Mass.), and the germlings were collected by centrifugation at 800 × g for 5 min at 25°C. The supernatant was aspirated, and the mean weight of mycelium trapped in the Junlon layer (31.99 mg; n = 5) was determined by subtracting the mean weight of the Junlon layer alone (determined from aliquots of uninoculated media; n = 5). Germlings in the Junlon layer were serially diluted in RPMI/Junlon medium and added in duplicate to naïve uninfected kidneys.
Preparation of inoculum for infection.
A. fumigatus MF5668 was grown on Sabouraud dextrose agar (SDA) slants (BBL, Cockeysville, Md.) at 35°C for 6 to 7 days. Conidia were washed from the surface of agar slants with sterile saline containing 0.01% Tween 20 and quantified by counting with a hemocytometer. Viable counts were determined by serially diluting the conidial suspension, spreading it on SDA plates (BBL), and counting the colonies after growth.
Infection and treatment.
Disseminated aspergillosis was initiated by intravenous (i.v.) injection of 0.2 ml of a suspension of A. fumigatus MF5668 conidia into the lateral tail vein. In the therapy study, mice were treated with a 1.0-mg/kg dose of caspofungin or a 0.5-mg/kg dose of AmB, administered intraperitoneally (i.p.) twice daily (b.i.d.) for a total of 5 days. The first dose was administered immediately after infection. An infected, nontreated control group was included in the therapy study. There were 30 mice per therapy group and 60 mice in the nontreated control group.
Tissue homogenization.
Organ samples were removed from euthanized animals and placed in weighed sterile Whirl Pak bags (Fisher Scientific). Samples designated for qPCR analysis were frozen at −20°C until use. Tissue weights were determined, and primary homogenates were prepared in saline by direct pressure (35). For CFU determination, 5 ml of saline (unless indicated otherwise) was used in preparation of the primary homogenate, and aliquots were serially diluted, spread onto plates (either SDA or YPAD [1% yeast extract, 2% peptone, 2% dextrose, 4 mg of adenine per ml] agar, as indicated), and incubated at 35°C for 24 to 48 h before A. fumigatus colonies were counted. For qPCR analysis, primary homogenates were prepared by adding 3.6 volumes of sterile saline per g of tissue. After applying pressure, 1.0 to 1.5 ml of the primary homogenate was transferred to a sterile 2-ml screw-cap microcentrifuge tube (Sarstedt, Newton, N.C.), and 0.5-mm-diameter glass beads (Biospec, Bartlesville, Okla.) were added. Tissue and hyphae were mechanically disrupted by vigorous agitation in a Bead Beater homogenizer (Biospec) with three bursts of 30 s each at 5,000 rpm with incubation on ice between bursts. This secondary homogenate was collected by centrifugation at 800 × g at 4°C for 5 min and stored at −20°C.
DNA extraction.
DNA was extracted from secondary homogenates with the DNeasy tissue kit (Qiagen, Valencia, Calif.) according to the manufacturer's instructions. Ninety microliters of buffer ATL and 20 μl of proteinase K solution (included in the kit) were added to 90 μl (19.6-mg equivalents) of secondary homogenate. For spleen samples only, 45 μl of homogenate (9.8-mg equivalents) was used; volumes were brought to 90 μl by addition of 45 μl of sterile saline. Samples were incubated overnight at 55°C with shaking (800 rpm). Proteinase K-digested samples were processed according to the manufacturer's instructions and applied to DNeasy columns, and DNA was recovered in 200 μl of elution buffer and stored at −20°C until analysis.
qPCR primers, probes, reactions, and calculations.
Oligonucleotide amplification primers and a dual-labeled fluorogenic oligonucleotide hybridization probe complementary to sequence from the A. fumigatus 18S rRNA gene (GenBank accession no. AB008401) were designed with Primer Express software version 1.5 (Applied Biosystems, Foster City, Calif.). The sequences of these oligonucleotides are as follows: (i) sense amplification primer, 5′-GGCCCTTAAATAGCCCGGT-3′; (ii) antisense amplification primer, 5′-TGAGCCGATAGTCCCCCTAA-3′; and (iii) hybridization probe, 5′-FAM-AGCCAGCGGCCCGCAAATG-TAMRA-3′. All three oligonucleotides were synthesized by Applied Biosystems.
DNA samples were analyzed in triplicate by using the ABI PRISM 7700 sequence detection system (Applied Biosystems). Each 50-μl qPCR consisted of 5 μl of sample DNA, 900 nM sense and antisense primers, 200 nM probe, and TaqMan Universal PCR Master Mix (Applied Biosystems) containing Taq DNA polymerase and the requisite buffers. Reactions were performed in MicroAmp optical 96-well reaction plates (Applied Biosystems) sealed with MicroAmp optical caps (Applied Biosystems). qPCR assays were run according to the manufacturer's directions, and results were analyzed with Sequence Detection System software (version 1.7; Applied Biosystems). Each sample was assigned a CT value, which identifies the cycle number during PCR when fluorescence exceeds a threshold value determined by the software. Differences in DNA recovery between samples were normalized by determining the total DNA concentration (see below) of each sample ([DNA]sample), comparing them to DNA concentration values from naïve, uninfected tissue ([DNA]naïve), and adjusting each sample CT value according to the formula CT(adjusted) = CT + x, where x = log2 ([DNA]sample/[DNA]naïve). A ΔCT value was calculated for each sample as the difference between sample CT(adjusted) and CT values from reaction mixtures containing DNA derived from naïve, uninfected tissue. ΔCT values were used to calculate conidial equivalents (CE) from a standard curve generated from DNA samples prepared from known numbers of conidia added to naïve organs. Samples for the conidial standard curve and from naïve, uninfected tissues were prepared in parallel with the experimental samples and analyzed in triplicate on each 96-well qPCR assay plate. All qPCR results for samples from infected tissues are expressed as CE per gram equivalent of tissue.
For the modified qPCR assay, 200-μl DNA samples (19.6-mg tissue equivalents) were assayed in their entirety as 20 replicate 10-μl aliquots. The sum of all fractional CE was used to calculate log10 CE per gram of kidney. Samples used for standard curves in the modified assay were also analyzed as 10-μl aliquots. Normalization for DNA recovery in the modified qPCR assay was based upon the formula described above; a single DNA adjuster value determined for each homogenate was used for both modified and standard assay calculations.
Determination of DNA concentration.
Mouse genomic DNA (Clontech, Palo Alto, Calif.) was diluted to 2 μg/ml in 10 mM Tris-HCl (pH 8.0)–1 mM EDTA (TE), and 200 μl was placed in the first well of a 96-well black Optiplate (Packard Instruments, Meriden, Conn.). Serial twofold dilutions were made in TE across the plate and used to formulate a standard curve. Experimental DNA samples were diluted 1:100, 1:200, and 1:400 in TE and 100 μl of each of these dilutions was assayed in duplicate. An equal volume (100 μl) of PicoGreen double-stranded DNA (dsDNA) quantitation reagent (Molecular Probes, Eugene, Oreg.), diluted according to the manufacturer's specifications, was added to all wells. Fluorescence was measured with a Fluoroskan II spectrofluorometer (Labsystems, Helsinki, Finland) at excitation/emission wavelengths of 485/538 nm, respectively. DNA was quantitated in the experimental samples based on fluorescence relative to the standard curve.
Candida albicans qPCR analysis.
In this study, treatment of animals, infection, tissue homogenization, DNA extraction, qPCR assays and calculations, and determination of DNA concentration were performed as described for A. fumigatus, with the following modifications. Candida albicans strain MY1055 (Merck, Rahway, N.J.) was grown on SDA at 35°C for 24 h. Growth on the plate was resuspended in sterile saline, the cell concentration was determined by counting with a hemocytometer, and the inoculum was adjusted to 3.75 × 105 cells/ml. The viable cell count, as determined by serially diluting the cell suspension 10-fold, spreading it out on SDA plates, and counting the colonies after overnight incubation at 35°C, was 2 × 105 CFU/ml. The final infectious challenge was 4.0 × 104 CFU/mouse, and vehicle was given i.p. once daily for 4 days beginning immediately after infection. Ten mice were monitored for survival for 21 days postinfection. Kidneys from a second vehicle-treated group (n = 25) were removed from euthanized animals at 0.25, 1, 2, 3, 4, 7, and 14 days postinfection (three per group) and sectioned such that one-half of the left kidney and one-half of the right kidney were combined and used for CFU determination, while the other halves were combined and used for qPCR analysis.
The oligonucleotide primers and probe used for qPCR assays were complementary to sequence from the C. albicans 18S rRNA gene (GenBank accession no. AF114470), and the probe was labeled at the 5′ end with VIC (Applied Biosystems). The sequences are as follows: (i) sense amplification primer, 5′-GGACCCAGCCGAGCCTT-3′; (ii) antisense amplification primer, 5′-AAGTAAAAGTCCTGGTTCGCCA-3′; and (iii) hybridization probe, 5′-VIC-CTTCTGGGTAGCCATTT-TAMRA-3′.
For the standard curve relating infected tissue ΔCT values to cell number, DNA samples were prepared from naïve kidneys spiked with cells from a mid-logarithmic-phase culture of C. albicans MY1055 grown at 30°C in RPMI 1640 medium with l-glutamine, buffered with 0.165 M MOPS (pH 7.0). The cell density of the undiluted culture, determined by direct counting with a hemocytometer, was 1.93 × 107 cells/ml. The culture was serially diluted in RPMI medium containing 25% glycerol, and aliquots were stored frozen at −20°C until use.
Genomic DNA.
Genomic DNA for use in the qPCR specificity studies was prepared from C. albicans strain MY1055 or A. fumigatus strain MF5668 as described previously (18).
Statistical analysis.
Analysis of variance was used to statistically assess differences with respect to the CFU endpoint. All tests and comparisons were deemed significant at the 5% level unless noted otherwise. Percent survival was calculated by the Kaplan-Meier technique (20) with animals from either a separate survival group or from the qPCR sample group (censored for euthanatized animals).
RESULTS
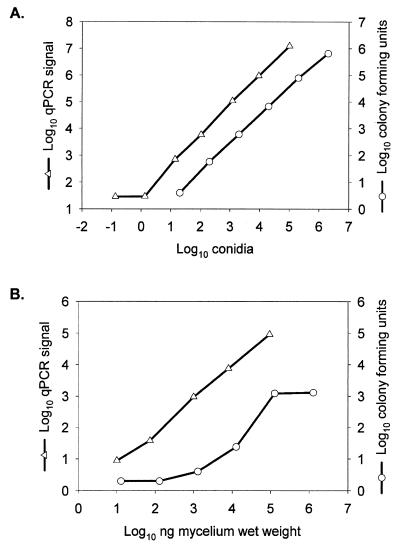
The rRNA gene locus is frequently chosen as the PCR amplicon for both speciation and quantitation of organisms (12, 21, 30). The A. fumigatus 18S rRNA gene was analyzed with ABI Primer Express software to select amplification primers and a hybridization probe suitable for establishing a qPCR assay. By using the BLAST algorithm (4), we performed database searches to demonstrate that the selected primer-probe combinations were specific for Aspergillus spp. (17). The results showed that the oligonucleotides (sense primer, antisense primer, and probe) are identical to the 18S rRNA gene of A. fumigatus and 9 other Aspergillus species in the database, as well as 11 other filamentous fungi. The non-Aspergillus fungi are all ascomycetes and are closely related to members of the genus Aspergillus (data not shown). In order to evaluate the utility of this primer-probe set, defined numbers of single-cell mononucleate A. fumigatus conidia (11) were mixed in a titration experiment with noninfected mouse kidneys. Genomic DNA prepared from these samples was used as a template to establish the qPCR assay. The signal from these reactions was linear over a range of nearly 5 orders of magnitude in this titration series, and the limit of detection was routinely at or near a single conidium (Fig. 1A). No signal was detected when the following templates were tested as negative or specificity controls: (i) a commercial source of mouse genomic DNA, (ii) genomic DNA prepared from kidneys of uninfected mice, or (iii) C. albicans genomic DNA. Parallel samples from the titration of conidia in kidneys were evaluated for CFU (Fig. 1A). The results from this assay were also linear over the same range, and the lowest dilution we tested contained approximately 10 conidia.
FIG. 1.
Quantitation of A. fumigatus conidia or mycelium in mouse kidneys by the CFU and qPCR methodologies. (A) Conidia spiking. (B) Mycelia spiking. Kidney homogenates containing either conidia or mycelia were prepared with 2.5 mL of saline added (CFU; ○) or with 3.6 volumes of saline added per gram of kidney (qPCR; ▵). The qPCR signal represents 2ΔCT (A) or CE per gram of kidney (B). CFU are expressed per gram of kidney in both panels.
A similar titration series was carried out with filamentous A. fumigatus germlings isolated from a liquid culture and added to noninfected mouse kidneys. Analysis of these samples by using the qPCR assay revealed a limit of detection of approximately 10 ng of mycelial wet weight and a linear range that spanned 4 orders of magnitude (Fig. 1B). However, mycelium-spiked kidneys that were evaluated for CFU counts on YPAD plates displayed a distinct lack of linearity across a broad range of hyphal mass, with essentially no change in CFU above 105 ng of mycelial wet weight. In fact, serial dilutions from 104 to 101 ng of mycelium wet weight produced only a 10-fold decrease in the number of CFU (Fig. 1B). In these titration experiments, 2.5 ng of mycelial wet weight corresponded to a single conidial genome equivalent.
To assess whether this qPCR assay might be suitable for measuring A. fumigatus burden in an animal model of disseminated aspergillosis, mice were euthanized 4 days after i.v. infection with A. fumigatus conidia, and the brain, liver, spleen, lungs, and kidneys were removed for analysis. Organs from six animals were assayed by the qPCR method, and tissues from another five animals were evaluated for fungal burden by CFU determination. A. fumigatus was detected in nearly every tissue from each animal (Table 1). Results from the CFU assay indicated that lungs had the lightest A. fumigatus load, while kidneys had the heaviest burden in this mouse model. In the qPCR assay, liver tissue had the lowest A. fumigatus burden, and kidneys had the heaviest load. The absolute fungal load determined by the qPCR assay was significantly larger (in kidney, brain, and lung by more than 3 log10 units) than that determined from CFU measurements of tissues from animals from the same infected group. Standard curves generated from conidia added to naïve uninfected tissue were nearly superimposable for the five tissues surveyed (data not shown). Unlike some reports in the literature in which inhibitors of PCR have been noted in certain tissues (5), the method of sample preparation developed for this assay generated a template that could be amplified regardless of tissue type.
TABLE 1.
Fungal burden in tissues of A. fumigatus-infected mice measured by CFU and qPCR assaysa
| Tissue | Mean log10 CFU/g of tissue | Mean log10 CE/g of tissue |
|---|---|---|
| Kidney | 4.24 ± 0.04 | 7.72 ± 0.38 |
| Spleen | 4.22 ± 0.07 | 5.68 ± 0.14 |
| Brain | 3.04 ± 0.20 | 6.37 ± 0.25 |
| Liver | 3.03 ± 0.19 | 4.48 ± 0.23 |
| Lung | 1.79 ± 0.16b | 5.03 ± 0.39b |
DBA/2J mice were infected with A. fumigatus conidia (1.2 × 106 CFU) and euthanized 4 days later. Fungal burdens were determined by measuring CFU (five animals) or CE (six animals). Values represent the mean ± standard error.
The lungs from one mouse in each group were below the limit of detection.
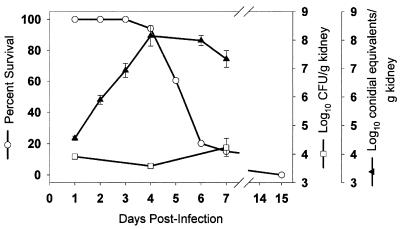
The progression of an A. fumigatus infection in DBA/2J mice was evaluated to compare traditional CFU counts and the qPCR assay in kidney samples and relate the results to survival (Fig. 2). To ensure appropriate sampling when both assays were performed, the right and left kidneys were sectioned, and organ halves were combined for parallel analysis by the CFU and qPCR assays (35). Mortality was first observed at day 4 and continued to increase until day 7 postinfection, after which time none of the animals survived. The A. fumigatus burden in the kidneys of infected animals, when measured at 4 and 7 days following infection by CFU counts, did not change significantly from the initial value determined at day 1. However, the CE per gram of kidney, determined by the qPCR assay, increased nearly 10,000-fold in samples prepared from the same kidneys. Fungal burden increased from 4.55 ± 0.07 log10 CE/g of kidney at day 1 to a peak of 8.15 ± 0.36 log10 CE/g of kidney at day 4 postinfection, when mortality first occurred.
FIG. 2.
Disease progression in A. fumigatus-infected DBA/2J mice monitored by percent survival, CFU, and qPCR analysis. Mice infected with 9.8 × 105 CFU of A. fumigatus were monitored for percent survival (○; n = 60, 23 censored animals), log10 CFU per gram of kidney (□), or log10 CE per gram of kidney (▴). SDA plates were used for CFU enumeration. CFU and CE are the mean ± standard error of three mice, with the exception of CE for days 4 and 6 (n = 5).
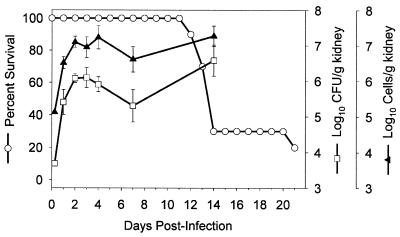
A comparison of CFU and qPCR methodologies was also performed in C. albicans-infected animals. In this study, among a group of 10 mice infected with C. albicans strain MY1055, 30% survived to 14 days postinfection. By 21 days, only 20% of the animals survived (Fig. 3). On selected days up to 14 days postinfection, kidneys from a parallel group of infected mice were assessed both for C. albicans 18S ribosomal DNA (rDNA) content by the qPCR assay and for C. albicans CFU (Fig. 3). The specificity of the oligonucleotides used in this PCR was established in preliminary studies: the primer-probe set failed to give a signal when either A. fumigatus or murine genomic DNA was used as a template (data not shown). Throughout the course of the infection, mean values for log10 CFU or cell equivalents per gram of kidney showed distinctly similar trends (Fig. 3), and in most cases, animals within each group of three had a similar rank order of burden based on values determined by either the CFU or qPCR assay (data not shown). With each animal at all times after infection, the total cell number per gram of kidney estimated by the qPCR assay was consistently higher than the value for CFU per gram of kidney.
FIG. 3.
Progression of infection in mice with disseminated candidiasis. DBA/2J mice were infected with C. albicans and assessed for percent survival (○) by the Kaplan-Meier technique (20) or for kidney burden by both a CFU (□) and a qPCR (▴) assay as described in Materials and Methods. Error bars indicate standard errors.
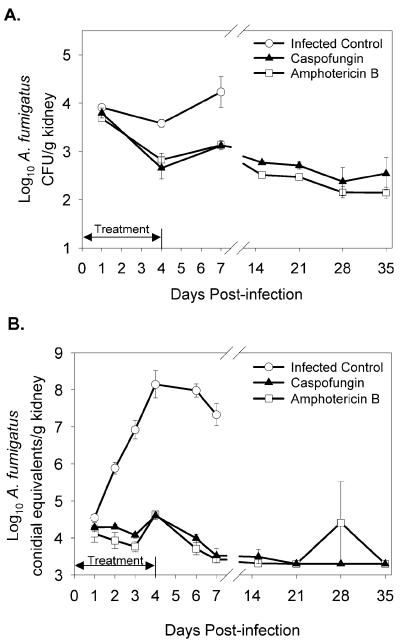
We compared qPCR-based quantitation to CFU measurement in A. fumigatus-infected mice that were given antifungal therapy. The burden in kidneys of caspofungin- or AmB-treated and nontreated mice, determined by CFU counts, is shown in Fig. 4A. The number of CFU per gram of kidney in both of the treated groups of mice was reduced relative to that in untreated controls at days 1, 4, and 7 postinfection. A statistically significant reduction from nontreated animals was observed in the AmB-treated group at all three time points and at days 4 and 7 in the caspofungin-treated group. However, the difference between nontreated and treated groups (for either drug) was relatively small and reached a maximum of ∼1 log10. When the burden in the same animals was measured with the qPCR assay, there was a large difference between treated and nontreated groups (Fig. 4B). In the infected control group, the increase of ∼4 log10 CE seen from day 1 through day 4 postinfection is contrasted with fungal burden in the kidneys of treated mice, which was low during treatment and continued to decrease after the completion of therapy. In a majority of the caspofungin- or AmB-treated animals, the signal in the qPCR assay reached the limit of detection by 15 days postinfection. The efficacies of caspofungin and AmB (1.0 and 0.5 mg/kg/dose, respectively) were equivalent in this model of twice-daily i.p. dosing over a 5-day course of therapy.
FIG. 4.
Effect of caspofungin or AmB on A. fumigatus kidney burden versus that in infected control mice. Animals were infected as described in the legend to Fig. 2. Therapy was administered as described in Materials and Methods. Values (mean ± standard error of three mice unless noted) are log10 CFU per gram of kidney (A) or log10 CE per gram of kidney (B) in the presence of caspofungin therapy (▴), AmB therapy (□), or no therapy (○). The double-headed arrow indicates the duration of the treatment period. On days 4 and 6, five mice from the nontreated group were used for qPCR analysis; on day 35, two mice from the AmB-treated group were used.
The limit of detection for the qPCR assay is based upon the signal produced by a single conidium in a 5-μl sample of DNA (Fig. 1A) derived from a 0.49-mg equivalent of original mouse tissue. Extrapolation to 1 g of tissue produces a limit of detection of ∼2,000 CE, or 3.3 log10 CE/g. Many of the AmB- or caspofungin-treated mice had kidney burdens at or below this limit by 15 days postinfection (Fig. 4B). All of these samples were reevaluated under conditions that lowered the limit of detection to 50 CE, or 1.7 log10 CE per g of tissue (Table 2). The reductions in mean log10 CE/g of kidney from day 1 to day 35 postinfection (the first and last sampling days) were 2.17 and 1.58 log10 CE/g for mice treated with caspofungin or AmB, respectively. This is commensurate with the reduction determined from analysis of the same kidney tissues for mean log10 CFU per gram of kidney (1.42 and 1.54 log10 CFU per g for caspofungin or AmB treatment, respectively) over the same period (Fig. 4A).
TABLE 2.
Reduction of A. fumigatus kidney burden with caspofungin or AmB treatmenta
| No. of days postinfection | Log10 CE/g of kidney
|
|||
|---|---|---|---|---|
| Caspofungin treated
|
AmB treated
|
|||
| Standard assay | Modified assay | Standard assay | Modified assay | |
| 1 | 4.29 ± 0.12 | NDb | 4.12 ± 0.23 | ND |
| 15 | 3.30 ± 0 | 3.13 ± 0.38 | 3.31 ± 0.01 | 3.34c |
| 21 | 3.30 ± 0 | 2.25 ± 0.13 | 3.30 ± 0 | 2.60 ± 0.15 |
| 28 | 3.30 ± 0 | 2.07 ± 0.37d | 4.41 ± 1.11 | 3.38 ± 1.63 |
| 35 | 3.30 ± 0 | 2.12 ± 0.19 | 3.30 ± 0e | 2.54 ± 0.08e |
Values represent mean log10 CE per gram of kidney ± standard error from three animals unless noted otherwise. Standard assay results are also shown in Fig. 4B. The limit of detection for the standard assay is 3.30 log10 CE/g of kidney. In the modified assay, evaluation was performed at a lower limit of assay detection. The limit of detection for the modified assay is 1 conidium per 19.6 mg of tissue, or 1.70 log10 CE/g of kidney.
ND, not determined.
Sufficient homogenate from one of three animals.
Sufficient homogenate from two of three animals.
Only two mice remaining at 35 days postinfection.
DISCUSSION
Filamentous fungi, including A. fumigatus, typically increase their cell mass by apical growth (14). From single-cell uninucleate conidia come germ tubes of growing A. fumigatus hyphae, which are composed of numerous cells bound by septa (15). The quantitation of fungal burden in organs of infected animals has traditionally been accomplished by spreading samples of homogenized tissue on agar plates and enumerating CFU after a period of growth in vitro (35). The method was originally developed for unicellular organisms. While CFU counts generate an accurate representation of the number of A. fumigatus conidia, this method is not adequate for the enumeration of filamentous structures that are not dispersed when spread on agar plates (24). Data from liquid cultures of A. fumigatus show that the number of CFU does not increase over a period of at least 24 h (27), despite obvious increases in hyphal mass. Filamentous hyphae composed of hundreds of cells may only be recorded as a single unit by the traditional CFU methodology.
Caspofungin acetate is a promising new therapeutic agent (Maertens et al., 40th ICAAC) that inhibits fungal 1,3-β-d-glucan synthesis and produces profound morphological changes in A. fumigatus hyphae both in vitro (23) and in vivo (22). A recent evaluation of the in vitro activity of caspofungin against A. fumigatus by using fluorescent indicators of cell viability and death illustrated that cells at the tips and branch points of young germlings were killed preferentially by the drug (C. M. Douglas, J. C. Bowman, G. K. Abruzzo, A. M. Flattery, C. J. Gill, L. Kong, C. Leighton, J. G. Smith, V. B. Pikounis, K. Bartizal, M. B. Kurtz, and H. Rosen, Abstr. 40th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1683, 2000). Whether or not cell death occurs in tissue remains to be determined. Because of the filamentous nature of A. fumigatus growth, CFU measurements would not be expected to accurately quantitate this type of change in mycelial mass. Accordingly, animal model studies to evaluate the efficacy of caspofungin against A. fumigatus have relied on survival rather than CFU enumeration as an endpoint in both pulmonary (Bernard et al., 36th ICAAC) and disseminated (3) models of aspergillosis. However, studies designed to (i) measure the efficacy of caspofungin in combination with other antifungal agents, (ii) model the response to a range of conidial inocula, or (iii) probe the role of various degrees of immunosuppression are not best served by the use of survival as an endpoint.
PCR has been described in several reports as a method with which to detect and speciate organisms in sputum, blood, or other clinical samples from patients with suspected fungal infections (9, 10, 33). A recent paper (25) detailed an assay based on the quantitative PCR Light Cycler System (Roche Diagnostics, Indianapolis, Ind.) for quantification of A. fumigatus DNA in human blood to which known numbers of conidia had been added. The investigators also used the assay to determine fungal burden in a small number of blood samples taken from patients with hematological malignancies. Yamakami and colleagues (38) developed a nested PCR assay to monitor Aspergillus species in human serum samples before and during therapy with AmB. The sample size was small, and two of six patients who had negative PCR results did not respond to therapy and ultimately died.
The results presented in this article provide the first description of a method for real-time PCR-based quantitation of A. fumigatus tissue burden in an animal model of infection. The technical advantage that the qPCR assay exploits is that every cell in a filamentous fungal mass is recognized, independent of other cells within the mass, by the nuclear DNA it contains. When we used this assay to monitor fungal load in a mouse model of infection, the qPCR assay detected a 10,000-fold increase in fungal burden, while the same tissues displayed less than a 10-fold increase in the number of CFU. Significantly, the onset of A. fumigatus-induced mortality coincided with the peak of fungal burden detected by using the qPCR assay. The improved dynamic range of the qPCR method represents an advantage for quantitation of the antifungal efficacy of caspofungin and other therapies.
In the studies reported here, oligonucleotides were designed to specifically amplify a portion of the 18S rRNA gene of A. fumigatus. Since there are approximately 100 copies of the 18S rRNA gene in the Aspergillus genome (17), selection of this target was expected to substantially increase the sensitivity of the method. The A. fumigatus FKS gene (AfFKS; GenBank accession no. U79728), presumed to be single copy in the genome (8), was also evaluated as a potential target sequence for qPCR analysis. The difference in signal intensity between the 18S target and the AfFKS target was nearly 100-fold when purified DNA from A. fumigatus strain MF5668 was used as a template in separate reactions (data not shown).
A number of different homogenization and DNA extraction methods were evaluated in our laboratory to establish the described protocol. These included homogenization by direct pressure alone, disruption with a handheld tissue homogenizer (Polytron; Kinematica AG, Lucerne, Switzerland), and the use of commercial kits and published methods for DNA extraction. We chose the combination of direct pressure and glass bead homogenization because it is efficient, simple, and amenable to processing large numbers of samples. Experiments with tissues spiked with conidia demonstrated that CFU in primary homogenates was reduced by more than 3 log10 following mechanical disruption with glass beads (data not shown). We presume that hyphae were also efficiently broken by this procedure. Direct pressure and glass bead homogenization are also likely to reduce the risk of cross-contamination between samples that may occur with other methods (e.g., physical disruption by Polytron). Finally, the procedure does not rely on enzymatic digestion of the fungal cell wall, which may exhibit changes in physical composition, and therefore susceptibility to digestion, as a function of hyphal age, drug treatment, and/or tissue source.
In measuring A. fumigatus CFU counts from infected organs, roughly 3 mg of homogenized tissue was spread on plates. Making the assumption that a single CFU can be detected in a given sample, mathematical extrapolation predicts that the limit of detection for this assay is approximately 300 CFU per g of tissue, with the exact limit dependent upon actual tissue weight. The 5-μl samples used as a template in the qPCR assay represent 0.49-mg equivalents of the original tissue. Our results suggest that the assay is capable of detecting a single CE in these samples (Fig. 1A); therefore, the limit of detection for the qPCR assay is ∼2,000 CE per g of tissue. If we assay the entire 200-μl DNA sample (derived from 19.6 mg of tissue) in 10-μl aliquots and obtain the sum of the values, the limit is reduced to ∼50 CE per g of tissue (Table 2). However, this is not a practical method when evaluating numerous samples. We are exploring technical improvements that may allow for further reduction of the assay's limit of detection. The dynamic range that the assay provides relative to the CFU method more than compensates for the current difference in the limits of detection.
When comparing the qPCR signal from a sample of infected tissue to a standard curve composed of conidia added to naïve tissue, we have chosen to express the fungal biomass in terms of CE. This is based on the fact that A. fumigatus conidia, like those of other members of the Aspergillus genus, are nearly always uninucleate, while individual cells bounded by septa within a germ tube of growing A. fumigatus hyphae are often multinucleate (11). In fact, apical cells of Aspergillus nidulans have been shown to have as many as 50 nuclei (19). It would therefore not be accurate to extrapolate from CE to cell number.
We found that the yield of total kidney DNA among 94 animals in the caspofungin efficacy study was variable, ranging some 20-fold from a low of 48 to a high of 1,075 μg/ml. There was no significant correlation between DNA yield and time postinfection, between treatment groups, or when comparing treated animals to nontreated animals (data not shown). In a comparison of DNA recovered from naïve tissues with or without added conidia, we determined that the amount of fungal genomic DNA derived from any given tissue sample only represented a fraction of a percent of host genomic DNA. Accordingly, we felt that an adjustment of the CT values to reflect differences in DNA recovery was justified. Despite the broad range of DNA concentrations, the normalization scheme only had a small effect on the final values representing log10 CE per gram of kidney. For example, the largest adjustment factor for the entire study changed the value for a nontreated animal (7 days postinfection) from log10 7.33 to log10 7.66 CE/g of kidney. We are actively pursuing alternate methods to correct for DNA recovery in large groups of samples.
We measured fungal burden by both CFU analysis and real-time qPCR in simple murine models of disseminated aspergillosis or candidiasis. It is interesting that CFU and CE are roughly equal in kidneys of A. fumigatus-infected mice on the first day following infection (Fig. 2), when the fungal burden may consist primarily of small germlings and swollen conidia that would be expected to produce nearly equivalent values by the two methodologies. As the mycelial mass increases during the infection by extension of the filamentous network, fungal burden quantified through CFU counts does not change significantly. By comparison, enumeration of C. albicans burden in kidney tissue from mice with disseminated candidiasis displayed the same trends during disease progression when analyzed by either CFU or qPCR measurement (Fig. 3). Both assays detected an increase in mean C. albicans burden of ca. 2 log10 between 6 h and 2 days postinfection. Previous studies of DBA/2N mice infected with C. albicans MY1055 reported a similar increase in CFU over the same period (1). Peak C. albicans burden measured with the qPCR assay occurred approximately 7 days before the onset of significant mortality, which was unlike the result obtained with A. fumigatus-infected animals (Fig. 3). Relationships between fungal burden and mortality are dependent on pathogenic mechanisms (e.g., production of virulence factors, tissue infiltration and damage, etc.) and would not be expected to be identical for C. albicans and A. fumigatus. We also found that values for C. albicans in log10 cells per gram of kidney determined with the qPCR assay were uniformly higher than values for log10 CFU per gram of kidney. This may reflect the fact that C. albicans growing in tissues generally adopts a pseudohyphal morphology, which could result in fewer CFU than would be expected when the organism is grown in liquid culture as a budding yeast. Alternatively, nonreplicative cells that fail to produce a colony when spread on an agar plate may be detected in the qPCR assay.
Overall, the results from these studies convincingly demonstrate that real-time PCR can be used to monitor the progression of fungal infection in a murine model. For A. fumigatus, CFU determination provides limited insight into the progression of disease and therefore has less utility as an assay to quantitate therapeutic efficacy of novel antifungals. Meanwhile, the qPCR assay detects a significant increase in fungal load, peaking at a time that coincides with the onset of mortality. The assay should be a valuable tool for meaningful quantitative evaluation of the antifungal efficacy of therapies that target A. fumigatus.
ACKNOWLEDGMENTS
We thank Tami Crumley, Patricia Hicks, and Karen Santora for help with preparation of tissue homogenates; Gerald Bills for helpful discussion; and Alex Elbrecht, Hans Zweerink, and Xiaoming Zou for assistance with the qPCR assay.
REFERENCES
- 1.Abruzzo G K, Flattery A M, Gill C J, Kong L, Smith J G, Krupa D, Pikounis V B, Kropp H, Bartizal K. Evaluation of water-soluble pneumocandin analogs L-733560, L-705589, and L-731373 with mouse models of disseminated aspergillosis, candidiasis, and cryptococcosis. Antimicrob Agents Chemother. 1995;39:1077–1081. doi: 10.1128/aac.39.5.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abruzzo G K, Flattery A M, Gill C J, Kong L, Smith J G, Pikounis V B, Balkovec J M, Bouffard A F, Dropinski J F, Rosen H, Kropp H, Bartizal K. Evaluation of the echinocandin antifungal MK-0991 (L-743,782): efficacies in mouse models of disseminated aspergillosis, candidiasis, and cryptococcosis. Antimicrob Agents Chemother. 1997;41:2333–2338. doi: 10.1128/aac.41.11.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abruzzo G K, Gill C J, Flattery A M, Kong L, Leighton C, Smith J G, Pikounis V B, Bartizal K, Rosen H. Efficacy of the echinocandin caspofungin against disseminated aspergillosis and candidiasis in cyclophosphamide-induced immunosuppressed mice. Antimicrob Agents Chemother. 2000;44:2310–2318. doi: 10.1128/aac.44.9.2310-2318.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amicosante M, Richeldi L, Trenti G, Paone G, Campa M, Bisetti A, Saltini C. Inactivation of polymerase inhibitors for Mycobacterium tuberculosis DNA amplification in sputum by using capture resin. J Clin Microbiol. 1995;33:629–630. doi: 10.1128/jcm.33.3.629-630.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anaissie E J, Bodey G P, Rinaldi M G. Emerging fungal pathogens. Eur J Clin Microbiol Infect Dis. 1989;8:323–330. doi: 10.1007/BF01963467. [DOI] [PubMed] [Google Scholar]
- 7.Banerjee S N, Emori T G, Culver D H, Gaynes R P, Jarvis W R, Horan T, Edwards J R, Tolson J, Henderson T, Martone W J. Secular trends in nosocomial primary bloodstream infections in the United States, 1980–1989. National Nosocomial Infections Surveillance. Syst Am J Med. 1991;91(3B):86S–89S. doi: 10.1016/0002-9343(91)90349-3. [DOI] [PubMed] [Google Scholar]
- 8.Beauvais A, Bruneau J M, Mol P C, Buitrago M J, Legrand R, Latgé J P. Glucan synthase complex of Aspergillus fumigatus. J Bacteriol. 2001;183:2273–2279. doi: 10.1128/JB.183.7.2273-2279.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brandt M E, Padhye A A, Mayer L W, Holloway B P. Utility of random amplified polymorphic DNA PCR and TaqMan automated detection in molecular identification of Aspergillus fumigatus. J Clin Microbiol. 1998;36:2057–2062. doi: 10.1128/jcm.36.7.2057-2062.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bretagne S, Costa J M, Marmorat-Khuong A, Poron F, Cordonnier C, Vidaud M, Fleury-Feith J. Detection of Aspergillus species DNA in bronchoalveolar lavage samples by competitive PCR. J Clin Microbiol. 1995;33:1164–1168. doi: 10.1128/jcm.33.5.1164-1168.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burnett J H. Mycogenetics. London, United Kingdom: John Wiley & Sons; 1975. Fungi as organisms for genetic study; pp. 3–22. [Google Scholar]
- 12.Einsele H, Hebart H, Roller G, Loffler J, Rothenhofer I, Muller C A, Bowden R A, van Burik J-A, Engelhard D, Kanz L, Schumacher U. Detection and identification of fungal pathogens in blood by using molecular probes. J Clin Microbiol. 1997;35:1353–1360. doi: 10.1128/jcm.35.6.1353-1360.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghannoum M A, Rice L B. Antifungal agents: mode of action, mechanisms of resistance, and correlation of these mechanisms with bacterial resistance. Clin Microbiol Rev. 1999;12:501–517. doi: 10.1128/cmr.12.4.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griffin D H. Fungal physiology. New York, N.Y: John Wiley and Sons; 1981. pp. 102–130. [Google Scholar]
- 15.Gull K. Form and function of septa in filamentous fungi. Vol. 3. New York, N.Y: John Wiley and Sons; 1978. [Google Scholar]
- 16.Heid C, Stevens J, Livak K J, Williams P M. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 17.Henry T, Iwen P C, Hinrichs S H. Identification of Aspergillus species using internal transcribed spacer regions 1 and 2. J Clin Microbiol. 2000;38:1510–1515. doi: 10.1128/jcm.38.4.1510-1515.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holm C, Meeks-Wagner D W, Fangman W L, Botstein D. A rapid, efficient method for isolating DNA from yeast. Gene. 1986;42:169–173. doi: 10.1016/0378-1119(86)90293-3. [DOI] [PubMed] [Google Scholar]
- 19.Kaminskyj S G, Hamer J E. hyp loci control cell pattern formation in the vegetative mycelium of Aspergillus nidulans. Genetics. 1998;148:669–680. doi: 10.1093/genetics/148.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaplan E L, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 1958;53:457–481. [Google Scholar]
- 21.Kawamura S, Maesaki S, Noda T, Hirakata Y, Tomono K, Tashiro T, Kohno S. Comparison between PCR and detection of antigen in sera for diagnosis of pulmonary aspergillosis. J Clin Microbiol. 1999;37:218–220. doi: 10.1128/jcm.37.1.218-220.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurtz M B, Bernard E M, Edwards F F, Marrinan J A, Dropinski J, Douglas C M, Armstrong D. Aerosol and parenteral pneumocandins are effective in a rat model of pulmonary aspergillosis. Antimicrob Agents Chemother. 1995;39:1784–1789. doi: 10.1128/aac.39.8.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kurtz M B, Heath I B, Marrinan J, Dreikorn S, Onishi J, Douglas C. Morphological effects of lipopeptides against Aspergillus fumigatus correlate with activities against (1,3)-β-d-glucan synthase. Antimicrob Agents Chemother. 1994;38:1480–1489. doi: 10.1128/aac.38.7.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Latgé J-P. Aspergillus fumigatus and aspergillosis. Clin Microbiol Rev. 1999;12:310–350. doi: 10.1128/cmr.12.2.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loeffler J, Henke N, Hebart H, Schmidt D, Hagmeyer L, Schumacher U, Einsele H. Quantification of fungal DNA by using fluorescence resonance energy transfer and the Light Cycler system. J Clin Microbiol. 2000;38:586–590. doi: 10.1128/jcm.38.2.586-590.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lyons S R, Griffen A L, Leys E J. Quantitative real-time PCR for Porphyromonas gingivalis and total bacteria. J Clin Microbiol. 2000;38:2362–2365. doi: 10.1128/jcm.38.6.2362-2365.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manavathu E K, Cutright J L, Chandrasekar P H. Organism-dependent fungicidal activities of azoles. Antimicrob Agents Chemother. 1998;42:3018–3021. doi: 10.1128/aac.42.11.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsumoto S, Wakai Y, Nakai T, Hatano K, Ushitani T, Ikeda F, Tawara S, Goto T, Matsumoto F, Kuwahara S. Efficacy of FK463, a new lipopeptide antifungal agent, in mouse models of pulmonary aspergillosis. Antimicrob Agents Chemother. 2000;44:619–621. doi: 10.1128/aac.44.3.619-621.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petraitis V, Petraitiene R, Groll A H, Bell A, Callender D P, Sein T, Schaufele R L, McMillian C L, Bacher J, Walsh T J. Antifungal efficacy, safety, and single-dose pharmacokinetics of LY303366, a novel echinocandin B, in experimental pulmonary aspergillosis in persistently neutropenic rabbits. Antimicrob Agents Chemother. 1998;42:2898–2905. doi: 10.1128/aac.42.11.2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reiss E, Tanaka K, Bruker G, Chazalet V, Coleman D, Debeaupuis J P, Hanazawa R, Latge J P, Lortholary J, Makimura K, Morrison C J, Murayama S Y, Naoe S, Paris S, Sarfati J, Shibuya K, Sullivan D, Uchida K, Yamaguchi H. Molecular diagnosis and epidemiology of fungal infections. Med Mycol. 1998;36(Suppl. 1):249–257. [PubMed] [Google Scholar]
- 31.Rex J H, Rinaldi M G, Pfaller M A. Resistance of Candida species to fluconazole. Antimicrob Agents Chemother. 1995;39:1–8. doi: 10.1128/aac.39.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schulman M D, Valentino D, Nallin M, Kaplan L. Avermectin B2 O-methyltransferase activity in “Streptomyces avermitilis”mutants that produce increased amounts of the avermectins. Antimicrob Agents Chemother. 1986;29:620–624. doi: 10.1128/aac.29.4.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang C M, Holden D W, Aufauvre-Brown A, Cohen J. The detection of Aspergillus spp. by the polymerase chain reaction and its evaluation in bronchoalveolar lavage fluid. Am Rev Respir Dis. 1993;148:1313–1317. doi: 10.1164/ajrccm/148.5.1313. [DOI] [PubMed] [Google Scholar]
- 34.Verweij P E, Oakley K L, Morrissey J, Morrissey G, Denning D W. Efficacy of LY303366 against amphotericin B-susceptible and -resistant Aspergillus fumigatus in a murine model of invasive aspergillosis. Antimicrob Agents Chemother. 1998;42:873–878. doi: 10.1128/aac.42.4.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walsh T J, McEntee C, Dixon D M. Tissue homogenization with sterile reinforced polyethylene bags for quantitative culture of Candida albicans. J Clin Microbiol. 1987;25:931–932. doi: 10.1128/jcm.25.5.931-932.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weinberger K M, Wiedenmann E, Bohm S, Jilg W. Sensitive and accurate quantitation of hepatitis B virus DNA using a kinetic fluorescence detection system (TaqMan PCR) J Virol Methods. 2000;85:75–82. doi: 10.1016/s0166-0934(99)00154-8. [DOI] [PubMed] [Google Scholar]
- 37.White T C, Marr K A, Bowden R A. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin Microbiol Rev. 1998;11:382–402. doi: 10.1128/cmr.11.2.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamakami Y, Hashimoto A, Yamagata E, Kamberi P, Karashima R, Nagai H, Nasu M. Evaluation of PCR for detection of DNA specific for Aspergillus species in sera of patients with various forms of pulmonary aspergillosis. J Clin Microbiol. 1998;36:3619–3623. doi: 10.1128/jcm.36.12.3619-3623.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]