Abstract
Due to controversial reports concerning antibacterial activity of different graphene based materials it is very important to investigate their antibacterial action on a wide range of Gram-positive and Gram-negative bacteria. In this paper we have investigated the structure induced phototoxic antibacterial activity of four types of graphene based materials: graphene oxide (GO), graphene quantum dots (GQDs), carbon quantum dots (CQDs) and nitrogen doped carbon quantum dots (N-CQDs). Antibacterial activity was tested on 19 types of bacteria. It is found that nanometer-size CQDs and N-CQDs are the most potent agents whereas micrometer-size GO has very poor antibacterial activity. Electron paramagnetic resonance measurements confirmed photodynamic production of singlet oxygen for all types of used quantum dots. Detailed analysis has shown that N-CQDs are an excellent photodynamic antibacterial agent for treatment of bacterial infections induced by Enterobacter aerogenes (E. aerogenes), Proteus mirabilis (P. mirabilis), Staphylococcus saprophyticus (S. saprophyticus), Listeria monocytogenes (L. monocytogenes), Salmonella typhimurium (S. typhimurium) and Klebsiella pneumoniae.
Antibacterial potentials of four graphene based nanomaterials are tested on a wide range of bacteria.
1. Introduction
In the last decade, graphene based materials especially graphene oxide, graphene quantum dots and carbon quantum dots have been the subject of investigation of diverse groups of researchers due to their unique properties. Bio-application of graphene based materials is of particular interest especially in the area of bio-imaging, drug delivery, cancer treatment or antibacterial activity.1–4
Many types of bacterial strains have a crucial role in different bacterial infections. Of particular interest is the bacteria that cause infections in healthcare facilities. Some of the infections result from specific antibiotic treatments, venous catheter insertions, and/or surgical procedures.5 Because of that, it is important to develop a new class of antibacterial agents for partial or complete removal of bacteria to improve human health.6–10 One of the major problem for effective bacterial release is their growing resistance to applied drugs.
The usage of antimicrobial photodynamic therapy (APDT) is one of the possibilities to minimize the spread of multidrug resistant bacterial infections.3 APDT involves the use of visible light, combined with a photosensitizer (light sensitive dye) and oxygen present in and around cells.2 In this way, this therapy is excellent for the treatment of skin, wound and burn infections. Skin, wounds and burns are easily accessible for PDT.11 It has been observed earlier that the degree of antibacterial activity of graphene based materials is affected by their shape, surface functionalization, size, stability and size distribution.12–16
GO is an atom thick layer of carbon atoms arranged in a honeycomb structure and bonded together by σ bonds. Apart from intrinsic corrugations and topological defects (i.e. pentagons, heptagons or their combination), GO can have other types of defects such as vacancies, adatoms, edges/cracks, adsorbed impurities.17 The average height of GO sheet is about 0.8 nm due to many functional groups distributed over the surface and edges of graphene sheets. There are many reports concerning antibacterial activity of GO.12,18–20 Some of them claim that GO has significant antibacterial effect especially to Escherischia coli (E. coli) whereas the other emphasized completely different role. Main mechanisms of bacteria death caused by GO are: membrane rupture and production of reactive oxygen species (ROS). Significant roughness or peaks with high aspect ratios can cause bacterial membrane to rupture under its own weight upon contact.21 Faria et al. found that GO is also composed of one additional layer (oxidative debris) which increases toxicity of GO significantly.22
GQDs are a 0-dimensional graphene based material produced by top-down methods, with lateral dimension up to 100 nm and large surface area, good water solubility, tunable fluorescence, high photostability, excellent biocompatibility and low toxicity.23–26 GQDs are derived from the sp2 carbon network, and their photoluminescence property can be tuned by size, edge structure, shape, functional groups, defects, and heteroatom doping. At ambient light conditions GQDs show relative low in vivo and in vitro toxicity.27 But under blue light irradiation they become photoactive and show significant toxicity toward U251 human glioma cells.28 The cell death induced by photoexcited GQDs displayed morphological and/or biochemical characteristics of both apoptosis and autophagy. Under blue light irradiation GQDs induce bacterial death (Staphylococcus aureus-S. aureus and E. coli) as well29 and neither GQDs nor light exposure alone were able to cause oxidative stress and reduce the bacteria viability. Barneck et al. found earlier that blue light with wavelength longer than 405 nm did not kill bacteria itself.30 Chong et al. showed that GQDs exhibit significant phototoxicity through increasing intracellular ROS levels and reducing cell viability upon blue light irradiation.31 Those groups confirmed that light-induced formation of ROS originates from the electron–hole pair and, more importantly, reveal that singlet oxygen is generated by photoexcited GQDs via both energy-transfer and electron-transfer pathways. Hui et al. claim that GQDs prepared by rupturing C60 cage (i.e., C60-GQD) effectively kills S. aureus, including its antibiotic-tolerant persisters, but not Bacillus subtilis (B. subtilis), E. coli, or Pseudomonas aeruginosa (P. aeruginosa) whereas diverse GQDs with basal planes similar to those of graphene oxide sheets (i.e., GO-GQDs) lack antibacterial property.32
CQDs represents small carbon nanoparticles produced by bottom-up methods with sizes below 10 nm. They have outstanding features such as high chemical stability, environmental friendliness, broadband optical absorption, low toxicity, strong photoluminescence emission and optical properties and can be easily produce at large-scale with low cost.33,34 CQDs can be doped by different techniques to improve their properties especially photoluminescence and production of ROS.35–37 Meziani et al. reports that CQDs were highly effective in bacteria-killing with visible-light illumination.38 In fact, the inhibition effect could be observed even simply under ambient light conditions. Thakur et al. claim that Cipro@C-dots conjugate also showed enhanced antimicrobial activity against both model Gram-positive and Gram-negative microorganisms.39 Liu et al. demonstrated that carbon nanodots obtained from metronidazole at 250 °C for 8 h can only inhibit the growth of obligate anaerobes, such as Porphyromonas gingivalis (P. gingivalis) directly.40 Li et al. investigated antibacterial activity of spermidine-capped fluorescent carbon quantum dots (Spd-CQDs) toward to nonmultidrug-resistant E. coli, S. aureus, B. subtilis, and P. aeruginosa bacteria and also multidrug-resistant bacteria, methicillin-resistant S. aureus (MRSA).41 They claim that the minimal inhibitory concentration of Spd–CQDs is much lower (>25 000-fold) than that of spermidine, indicating their promising antibacterial characteristics.
In this work, we have investigated structural induced antibacterial potentials of four graphene based materials: GO, GQDs, CQDs and N-CQDs on a wide range of Gram-positive and Gram-negative bacteria (19 types of bacteria strains) cultured in laboratory conditions. We have used different techniques to characterize surface morphology, surface functionalization, photoluminescence, potentials to generate ROS and antibacterial activity of GO and quantum dots. Electron paramagnetic measurements (EPR) have been especially used to investigate the potentials of these species to generate not only singlet oxygen but also superoxide and hydroxyl radicals in dark, ambient light (AL) and blue light (BL) at 470 nm irradiation. We have also studied the possible mechanism of bacteria killing by these carbon based species.
2. Experimental
2.1. Sample preparation
GO was synthesized by the oxidation of graphite powder (Z-346, Timrex KS6 graphite, Bodio, Switzerland) using improved Hummers method without NaNO3 in reaction mixture described in detail in ref. 42 GQDs have been prepared by electrochemical procedure as described in our previous work.24 CQDs and N-CQDs are prepared as follows: typically, citric acid (10 g) or mixture of citric acid (4 g) with 4 mL of ammonium hydroxide are dissolved in distilled water (10 mL) with stirring for 30 min, respectively.43,44 Then, the prepared solution is transferred into a porcelain boat and then heated in air at 210 °C for 1 h with a heating rate of 10 °C min−1. The obtained carbogenic product is dissolved in acetone (40 mL) by use of ultrasonic tip. The supernatants containing carbon dots are collected after centrifugation at 8.000 rpm for 15 min. Then, the acetone is removed by rotary evaporation, and solid-state carbon dots (CQD and N-CQD) were redispersed in water, filtered through 50 nm filter and characterized. The pH values of CQD and N-CQD colloids were tuned to 7.
All samples are designated as follows: graphene oxide-GO, graphene quantum dots-GQD, CQDs prepared from thermal decomposition of citric acid-CQDCA and N-CQDs as CQDNH.
2.2. Sample characterization
Surface morphology and the lateral dimension of all samples were observed by transmission electron microscopy (TEM-JEOL JEM-1400 operated at 120 kV) and atomic force microscopy (AFM- Bruker, Germany). For TEM, GO samples were drop-casted on holey carbon grids while GQD, CQDCA and CQDNH samples were drop-casted on amorpous carbon grids. All samples for AFM were deposited by spin-coating on freshly cleaved mica. The AFM measurements were performed at room temperature in air. Gwyddion software was used to measure lateral dimension and height size of carbon based nanoparticles.24
Zeta potential (ζ-potential) of all samples were assessed by DLS using Zetasizer Nano-ZS (Malvern Instruments, UK) equipped with a 4 mW helium/neon laser (λ = 633 nm) and thermo-electric temperature controller. All measurements were performed at 25 °C. Values of zeta potential were averaged from five measurements.
XPS was performed using a Thermo Scientific K-Alpha XPS system (Thermo Fisher Scientific, UK) equipped with a micro-focused, monochromatic Al Kα X-ray source.
The micro attenuated total reflection (ATR) FTIR spectra of all samples deposited on aluminum foil were measured at room temperature in the spectral range from 400 to 4000 cm−1 on a Nicolet 8700 spectrometer. The spectral resolution was 4 cm−1.
UV-Vis spectra of all samples were recorded by a Shimadzu UV-Vis-NIR SolidSpec-3700 spectrophotometer in the range of 200–800 nm at room temperature.
The PL spectra of all samples were recorded on a RF-5301PC spectrofluorophotometer (Shimadzu, Japan) in 1 cm cuvette at the right angle arrangement. The experimental set-up for detecting the fluorescence lifetimes was based on a time-correlated single-photon counting (TCSPC) set-up, similar to that described in ref. 45 The fluorescence decays were measured with a 50 ns time base and sampled using 1024 temporal channels. The curves were fitted using SPCImage software (Becker & Hickl, Germany).
For EPR measurements, a Spectrometer MiniScope 300, Magnettech, Berlin, Germany was used. Measurements were performed at the temperature of 37 °C. The microwave power was 1 mW (microwave attenuation of 20 dB), with a modulation amplitude of 0.2 mT. The instrument was operating at a nominal frequency of 9.5 GHz.
All samples were treated with different irradiation treatment: one set of samples was kept in dark; the second set was exposed to AL while the third set was exposed to BL irradiation at 470 nm.
To follow the formation of singlet oxygen (1O2), 2,2,6,6-tetramethylpiperidine (TEMP) was used as a spin trap. With 1O2, TEMP molecules quickly react and form stable, EPR active product, TEMP-1O2 (TEMPO). All samples in the concentration of 0.2 wt%, were mixed with TEMP solution in ethanol, at a final concentration of 30 mM. Prepared mixtures were exposed to three different irradiation treatments at the temperature of 37 °C in the air-saturated environment for 24 h. EPR measurements were recorded during 24 h.
The concentration of generated singlet oxygen was estimated based on TEMPO produced in the photochemical reaction in solutions. We calculated the integrated intensity of EPR spectrum of 2,2-diphenyl-1-picrylhydrazyl (DPPH) solution (at a concentration of 432 μM). This compound is a stable free radical. The integrated intensity of TEMPO signals in each spectrum was calculated and divided by the value obtained for DPPH solution.
Additionally, the production of hydroxyl (HO˙) and superoxide (O2−˙) radicals was investigated using 5,5-dimethyl-1-pyrroline-N-oxide (DMPO) as a spin trap. The molecules of DMPO react with both HO˙ and O2−˙ to form stable, radical products with distinct EPR spectra. These reactions allow following the production of these oxygen containing radical species in carbon nanomaterial dispersions. The mixtures of sample and DMPO (concentration of 15 mM) were exposed also to three different irradiation treatments at the temperature of 37 °C in the air-saturated environment for 24 h.
2.3. Bacteria strains and culture conditions
The Gram-negative bacteria Escherichia coli (E. coli ATCC 25922), Salmonella enterica subsp. Enterica (S. enteritidas ATCC 13076), Enterobacter aerogenes (E. aerogenes ATCC 13048), Citrobacter freundii (C. freundi ATCC 43864), Salmonella typhmurium (S. typhimurium ATCC 14028), Pseudomonas aeruginosa (P. aeruginosa ATCC 27853), Proteus mirabilis (P. mirabilis ATCC 35659), Klebsiella pneumoniae subsp. pneumoniae (Klebsiella pneumoniae ATCC 13883) and Proteus vulgaris (Proteus vulgaris ATCC 13315) as well as the Gram-positive bacteria Staphylococcus saprophyticus subsp. saprophyticus (S. saprophiticus ATCC 15035), Staphylococcus aureus (S. aureus ATCC 25923 and S. aureus ATCC 25923), Listeria ivanovii (L. ivanovii ATCC 19119), Listeria innocua (L. inocun ATCC 33090), Enterococcus faecalis (E. faccalis ATCC 2912), Listeria monocytogenes (L. monocytogenes ATCC 19112), Bacillus subtilis subsp. Spizizenii (B. spieizeneii ATCC 6633) and Enterococcus faecium (E. faccium ATCC 6057) were used. Selected species of bacteria originate from ATCC (American Type Culture Collection, Rockville, Maryland). All tests were performed in Muller–Hinton broth (MHB).
2.4. Antibacterial activity of GO, GQD, CQDCA and CQDNH
The GO stock solutions of 10 mg mL−1 and the GQD, CQDCA and CQDNH stock solutions of 5 mg mL−1 were sterilized by gamma irradiation at a dose of 10 kGy and used to examine the kinetics of bacterial growth using 96-well plates. The pH of prepared colloids was adjusted to value 7. By microdilution method in 96 multi-well microtiter plates,46 minimum inhibitory concentration (MIC) of the samples against the test bacteria was determined. A volume of 100 μL stock solutions of samples (in 10% DMSO, 2 mg mL−1) was pipetted into the first row of the plate. Fifty μL of Mueller Hinton was added to the other wells. A volume of 50 μL from the first test wells was pipetted into the second well of each microtiter line, and then 50 μL of scalar dilution were transferred from the second to the twelfth well. Ten μL of resazurin indicator solution (prepared by dissolution of a 270-mg tablet in 40 mL of sterile distilled water) and 30 μL of nutrient broth were added to each well. Finally, 10 μL of bacterial suspension (106 CFU/mL−1) was added to each well. The growth conditions and the sterility of the medium were checked, for each strain. To control the sensitivity of the tested bacteria was used standard antibiotic amracin (A). Plates were wrapped loosely with transparent cling film to ensure that bacteria did not become dehydrated and prepared in triplicate, and then they were placed in an incubator at 37 °C for 24 h for the bacteria and color change were assessed visually. Well plates were irradiated by blue light (LED, 465–470 nm, 10 W) at a distance of 20 cm from the light source. Any color change from purple to pink or colorless was recorded as positive. The lowest concentration at which color change occurred was taken as the MIC value. The average of 3 values was calculated, and the obtained value was taken as the MIC for the tested compounds and standard drug.
After the MIC assay, 100 μL of the medium in the wells with no visible growth was aspirated and spread on Mueller–Hinton agar. The agar plates were incubated at 37 °C overnight to check the viability of the bacteria. Minimum bactericidal concentration (MBC) was recorded as the lowest concentration of the compound that killed 99.9% of the initially inoculated bacteria.
2.5. Loss of 260-nm-absorbing material
The extracellular 260-nm-absorbing material released by the cells was determined using slightly modified method described by Carson et al.47 Bacterial suspensions (pretreatment sample) of approximately 105–106 CFU/mL−1 were taken, diluted in a ratio 1 : 100, and filtered through a 0.22-μm pore size filter (Sartorius, Germany). The GO, GQD, CQDCA and CQDNH were added in 1 mL of E. coli suspensions to reach final concentration of 0.1 mg mL−1. Cells without samples were used as a control. All the samples were incubated at 37 °C, and additional aliquots of control and treated cell suspensions were removed after 24 h, diluted and filtered as described above and dyalized. The release of UV-absorbing material was measured using a Shimadzu UV-1800 UV-VIS Spectrophotometer. The obtained results of the measurements at 260 nm at each time were expressed as a proportion of the initial optical density 260 (OD260) value. An OD260 is defined as the amount of light at a 260 nm wavelength which will be absorbed by an oligo if it is resuspended in 1 mL water and the concentration is read in a 1 cm quartz cuvette. The assay was carried out in triplicates.
3. Results and discussion
3.1. Surface morphology of GO, GQD, CQDCA and CQDNH
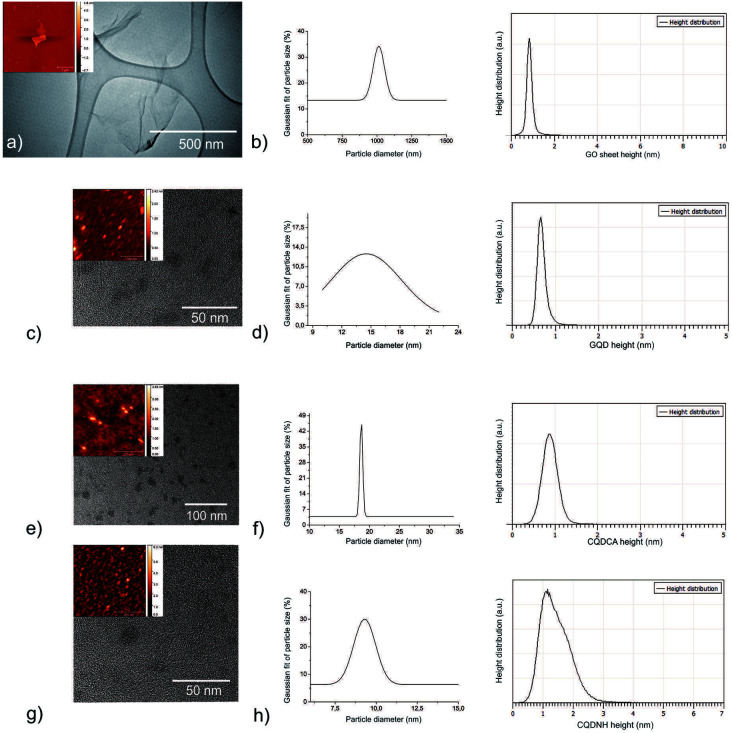
TEM and AFM were used to visualize surface morphology, particle size and height distributions of all samples calculated from more than 100 dots and 50 GO sheets-Fig. 1(a–h). Fig. 1a and b presents surface morphology and the Gaussian fit of lateral size and height distributions of GO. More than 30% of GO sheets has lateral dimension of about 1 micron whereas their average height is about 1 nm. The height value indicates the existence of functional groups over whole surface of GO sheets related to the fact that typical height value of single layer graphene sheet is about 0.33 nm.48Fig. 1(c and d) presents surface morphology and Gaussian fit of particle size and height distributions of GQD. It can be seen from these figures that GQD has disc-like shape and very wide distribution of particle size with average diameter of 14 ± 2 nm-Fig. 1d. Based on statistical calculations conducting on several AFM images, 90% GQD has height values of 0.85 nm. Fig. 1(e and f) presents surface morphology and particle size and height distributions of CQDCA. CQDCA have disc-like shape and very narrow distribution of particle size with average diameter of 22.5 ± 2.5 nm whereas their average heights are less than 1 nm. Fig. 1g and h presents surface morphology and particle size and height distributions of CQDNH sample. It can be seen from these figures that the CQDNH have disc-like shape and average diameter of 12.5 ± 2.5 nm whereas their heights are predominantly 1.2 nm.
Fig. 1. TEM micrographs, top view AFM images (insets), Gaussian fit of particle size and height distributions of the: (a and b) GO; (c and d) GQDs; (e and f) CQDCA and (g and h) CQDNH samples. The particle size and height of all dots were calculated for set of 100 dots whereas for the GO sample set had 50 GO sheets.
3.2. Zeta potential of GO, GQD, CQDCA and CQDNH
In Table S1 (ESI†), the average zeta potential of graphene based nanoparticles obtained from five measurements are presented. Based on data presented in Table S1,† we concluded that colloids of all samples (GO, GQD, CQDCA, CQDNH) are stable, electrostatically stabilized and negatively charged. The pH values of all colloids are tuned to 7. GQD and CQDNH colloids are less negatively charged than CQDCA colloid according to values of zeta potential. This happens due to protonation of acidic and amino groups (C–OH, COOH, NH2) which decreases the negative charge on the sheets.
3.3. XPS and FTIR measurements of GO, GQD, CQDCA and CQDNH
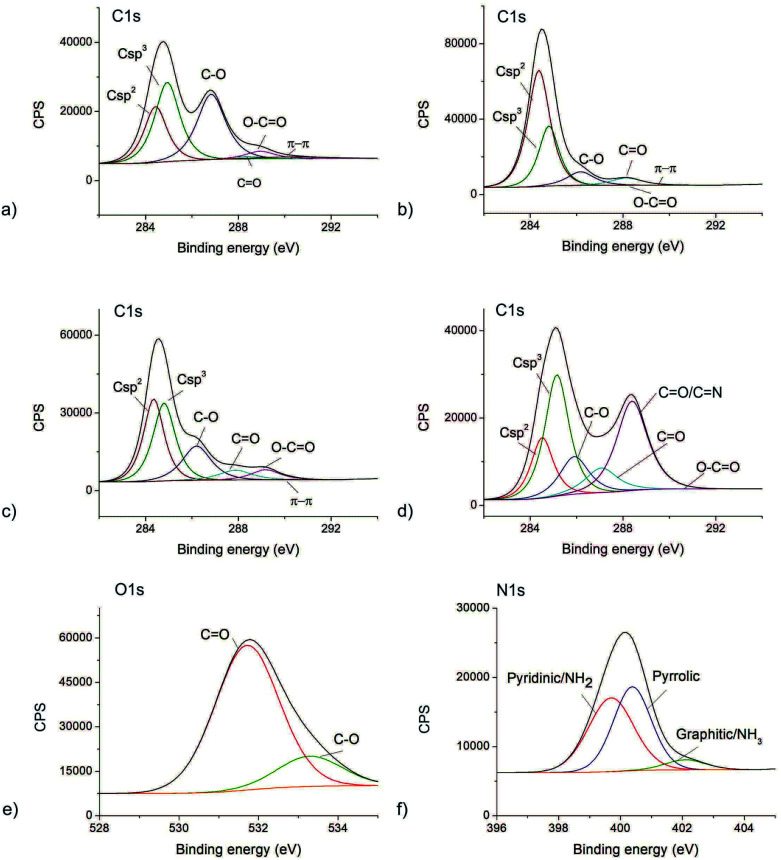
XPS method was used to determine the elemental content of the GO, GQDs, CQDCA and CQDNH samples. The content of elements detected in all samples is presented in Table 1. The nitrogen can be detected in all samples. The highest amount of this element was detected in the CQDNH sample (12.9%).
XPS data of elemental analysis of all samples in atomic% (at%).
| Name | GO | GQDs | CQDCA | CQDNH |
|---|---|---|---|---|
| C1s | 48.1 | 84.7 | 76.2 | 63.1 |
| O1s | 50.8 | 15.2 | 23.3 | 24.0 |
| N1s | 0.7 | 0.2 | 0.5 | 12.9 |
| S2p | 0.4 | — | — | — |
The sulfur is detected only in the GO sample as a consequence of the production method. All XPS spectra are fitted to study different bonds in all samples. The results are presented in Fig. 2 and Table S2.† The high-resolution C1s XPS spectrum of all samples can be deconvoluted in 6 peaks-Fig. 2a–d. Fig. 2e shows O1s XPS spectrum of the CQDNH sample and can be deconvoluted in 2 peaks at 531.71 (C O) and 533.28 eV (C–O) indicating the existence of C O and C–O, respectively.49Fig. 2f represents N1s XPS spectrum of the CQDNH sample and can be deconvoluted in 3 peaks at 399.7 (pyridinic/NH2), 400.38 (pyrrolic) and 402.1 eV (graphitic/NH3). The peak at 399.7 eV could be attributed either from pyridinic or NH2 groups. The pyridinic N is in the range of 398.0–399.3 eV whereas amino groups are at 399.2 eV.50,51
Fig. 2. The deconvoluted XPS spectra of: (a) C1s GO; (b) C1s GQDs; (c) C1s CQDCA, (d) C1s CQDNH and (e) O1s CQDNH and (f) N1s CQDNH samples.
In Table S2,† the values of the characteristic bonds detected in all samples are presented.
Based on data presented in Table S2† it can be observed that there is a huge structural difference among the GO, GQD, CQDCA and CQDNH samples. The GO sample shows less crystallinity and the most defective structure (the contents of sp3 and C–O bonds) than other samples. Oxygen functional groups are dominantly distributed over the GO sheet surface (the amounts of C–O–33.7 vs. C O–1.4 at%). GQD shows more crystallinity and the lowest content of sp3 bonds compared to other samples. It means that they have the best ordered sp2 structure compared to other samples. Oxygen functional groups are uniformly distributed on the basal planes and along edges of graphene plane. As for the CQDCA sample it has the similar contents of sp2 and sp3 bonds and almost three times higher content of C–O vs. C O bonds.
FTIR spectra of all samples are presented in Fig. S1.† As for the GO sample (curve 1), the sharp peaks at 1600, 1427, 1134, 1071 and 1004 cm−1 can be attributed to C C stretching mode, C–OH stretching mode and C–O, respectively whereas broad band at 3300 cm−1 corresponds to a strong stretching mode of OH group.52
As for the GQD (curve 2), peaks near 1579 and 1698 cm−1 can be assigned to C C stretching of graphite. Peak at 1004 cm−1 stems from C–O bonds in carboxyl groups and peak at 1388 cm−1 stems from symmetric stretching of carboxyl groups. A peak at 1076 cm−1 originates from C–O stretching vibrations of alkoxy groups. Two peaks at 2890 cm−1 and 2930 cm−1 stem from C–H stretching vibrations. The broad band at 3423 cm−1 is associated with the O–H stretching vibrations. These results display good hydrophilic properties of GQD.53 As for the CQDCA sample (curve 3), the peaks at 923 and 1414 cm−1 correspond to C–H bending vibrations whereas a peak at 1200 cm−1 can be attributed to C–O vibrations; the peaks at 1707 and 1765 cm−1 can be assigned to C O groups whereas broad band at 3200 cm−1 stem from O–H stretching vibrations.54
As for the CQDNH sample (curve 4), the peaks at 1291, 1359 and 1418 cm−1 correspond to C–H bending vibrations, a peak at 1200 cm−1 stem from C–O vibrations whereas a peak at 1596 cm−1 originates from N–H bending vibrations. A peak at 1706 cm−1 can be attributed to carbonyl groups whereas the peaks between 3100 and 3500 cm−1 correspond to primary amine (NH2) groups.54 The obtained FTIR results agree with XPS results.
3.4. UV-Vis and PL measurements of GO, GQD, CQDCA and CQDNH
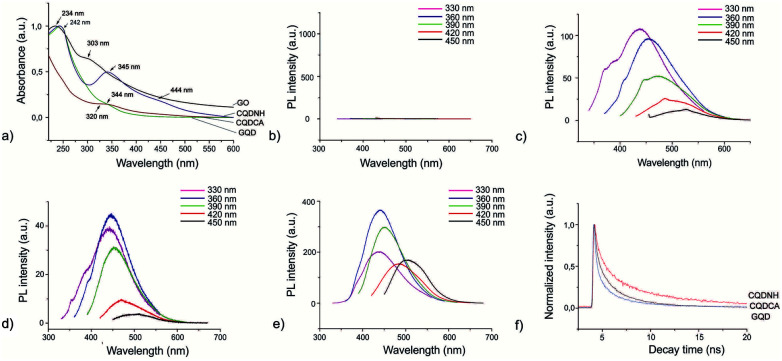
UV-Vis spectra of all samples are presented in Fig. 3a. The absorption curve of the GO sample has one peak at 234 nm which can be attributed to π–π* transitions of C C bonds and a shoulder at 303 nm which is attributed to n–π* transitions of C O bonds whereas the absorption curve of the GQD sample has a shoulder at 320 nm.26,55,56 The absorption spectrum of CQDCA sample has a peak at 344 nm due to n–π* transitions of C O bonds.57,58 The CQDNH sample has three absorption peaks: at 242, 345 and 444 nm. The former peak is due to π–π*transitions of C C bonds and the two last are from the trapping of excited state energy of the surface states, which can lead to strong fluorescence.45,59
Fig. 3. (a) UV-Vis spectra of GO (black curve), GQD (red curve), CQDCA (green curve) and CQDCH (blue curve); (b) PL spectra of GO; (c) PL spectra of GQD; (d) PL spectra of CQDCA; (e) PL spectra of CQDNH at different excitation wavelengths; (f) PL decay time for GQD (blue curve), CQDCA (black curve) and CQDNH (red curve).
Fig. 3(b–e) presents PL spectra of all specimens at different excitation wavelengths (330, 360, 390, 420 and 450 nm). As can be seen from Fig. 3b, the GO sample does not show photoluminescence at any excitation wavelengths. All other samples show photoluminescence property. But there are significant PL up-shifts between excitation and emission wavelengths. These PL up-shifts are from 50–111 nm. The highest PL up-shift (111 nm) is for the CQDCA sample at excitation wavelength of 330 nm whereas the lowest (50 nm) is for the same sample under two excitation wavelengths (420 and 450 nm). Based on the recorded PL spectra we can conclude that all samples emit blue light under excitation of 330, 360 and 390 nm whereas under excitation of 420 nm emit blue-green light and under excitation of 450 nm they emit green light. In the case of GQD sample PL intensity decreases with the increase of excitation wavelength. In the case of the CQDCA and CQDNH samples, the highest PL intensity is during excitation of 390 nm.
The main reasons for PL of all samples excluded GO are electron–hole recombination, zig-zag edges as well as surface defects.60 Based on the obtained results (TEM, AFM, XPS and PL) we concluded that excitation-dependent PL behavior is related to the different surface states of the GQD, CQDCA and CQDNH. From XPS studies, there are multiple C–, N– and O– containing functional groups on the surface of the produced quantum dots, resulting in various surface states with different energy levels and thus a series of emissive traps. Under different excitation wavelengths, the corresponding surface state emissive trap will be dominant, giving excitation-dependent PL.
The time-resolved fluorescence decay curves of all samples measured by time-correlated single photon counting method are illustrated in Fig. 3f and S2.† The decay curve can be fitted to a triple-exponential function-Fig. S2.† All experimental curves are fitted toward the following relation:
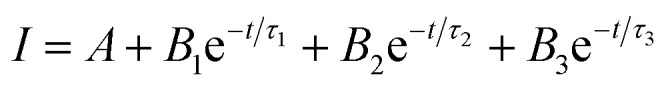 |
1 |
where I is the intensity, A, B1, B2 and B3 are constants, τ1, τ2, and τ3 are lifetimes of quantum dots.
At the emission wavelength of 450 nm, the observed lifetimes of the GQD are τ1 = 0.2 ns, τ2 = 1.7 ns, τ3 = 6.6 ns, whereas for the CQDCA τ1 = 0.3 ns, τ2 = 1.8 ns, τ3 = 5.5 ns and for the CQDNH τ1 = 1.05 ns, τ2 = 4.5 ns, τ3 = 9.7 ns. Average lifetimes of the GQD, CQDCA and CQDNH are 0.84, 1.56 and 3.1 ns, respectively under emission wavelength of 450 nm.
3.5. EPR measurements of GO, GQD, CQDCA and CQDNH induced ROS production
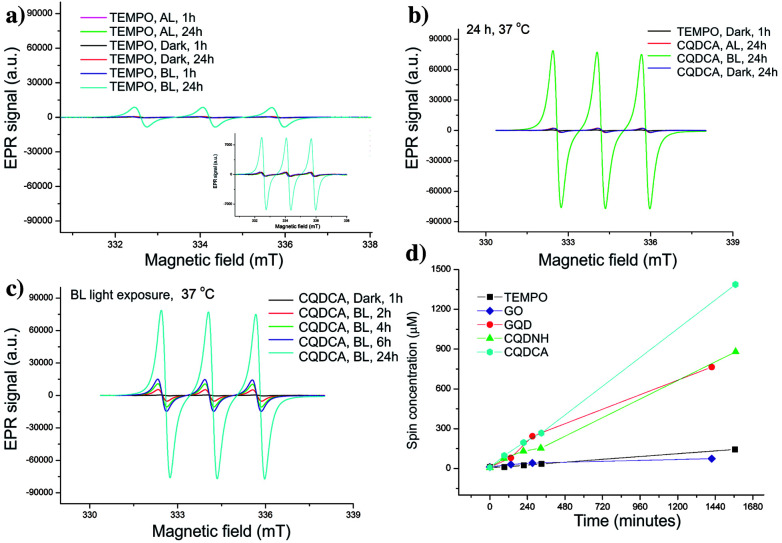
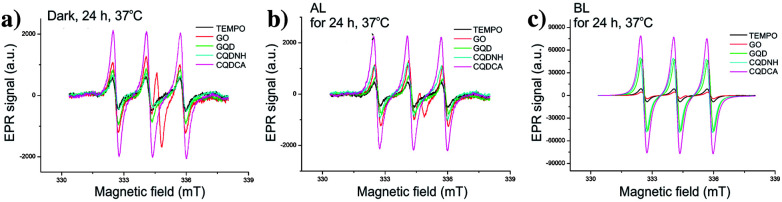
EPR technique has been used to investigate the ROS production of all samples. At first, trap solution was tested under the same experimental conditions as all samples- Fig. 4a. These spectra indicate the presence of TEMPO in the dark, under the AL and upon BL irradiation for 1 h. The obtained results showed that certain amount of TEMPO was present in the starting trap solution. The higher increase of TEMPO signal was observed after 24 h of BL irradiation.
Fig. 4. EPR spectra of TEMP at different conditions (a), sample CQDCA after 24 h of irradiation and in dark (b), sample CQDCA after the different time of BL irradiation (c) and spin concentration for all samples after the different time of BL irradiation (d).
Further, the singlet oxygen production of the GO, GQD, CQDCA and CQDNH samples has been studied. In the Fig. 4b, the EPR spectrum of the CQDCA sample after 24 h exposure at different light conditions are presented. We can see that intensities of characteristic EPR signals are low after 24 h irradiation with AL as for the samples kept in the dark. The signal intensity increases significantly only under BL exposure. These results suggest that only BL induces large singlet oxygen production in the solution containing the CQDCA. By investigating the time effect on 1O2 production upon BL exposure (Fig. 4c), we noticed that the production of 1O2 was increased by time.
By comparing signal intensities from different samples (Fig. 5a–c), we observe that GO induces the lowest change of the TEMPO signal intensity at all studied experimental conditions. By contrary, the highest signal intensities are observed for the CQDCA, whereas the GQD and the CQDNH cause similar enhancements of TEMPO signal in the EPR spectra. All analysed samples cause the highest production of TEMPO and thus singlet oxygen upon BL irradiation. The samples which were exposed to AL and kept in the dark showed a low 1O2 production.
Fig. 5. EPR spectra of GO, GQD, CQDCA and CQDNH with TEMP as a trap, after 24 h at 37 °C in the dark (a), under AL (b) and upon BL exposure (c).
The EPR spectra of GO and quantum dots irradiated by BL were used for calculation of spin concentration and these results are present in the Fig. 4d. The spin concentration of TEMP and GO solutions shows a very small increase over 24 h of BL irradiation. Even in the first 4 h of BL irradiation, the concentration of spin was higher in the GQD than for the CQDCA. In the following period dynamics of free radicals formation was changed which resulted in the similar values of spin concentration formed in these two samples after 24 h. The highest spin concentration was measured in the CQDCA sample and it was 57% higher than spin concentration produced by the CQDNH and GQD.
Based on presented results we concluded that GO and quantum dots samples produce a small amount of singlet oxygen under AL. Similar, very low production was observed when samples were kept in dark. But under BL irradiation, the GQD, CQDCA and CQDNH samples produce singlet oxygen intensively.32 The highest concentration of singlet oxygen production was measured in the CQDCA sample.
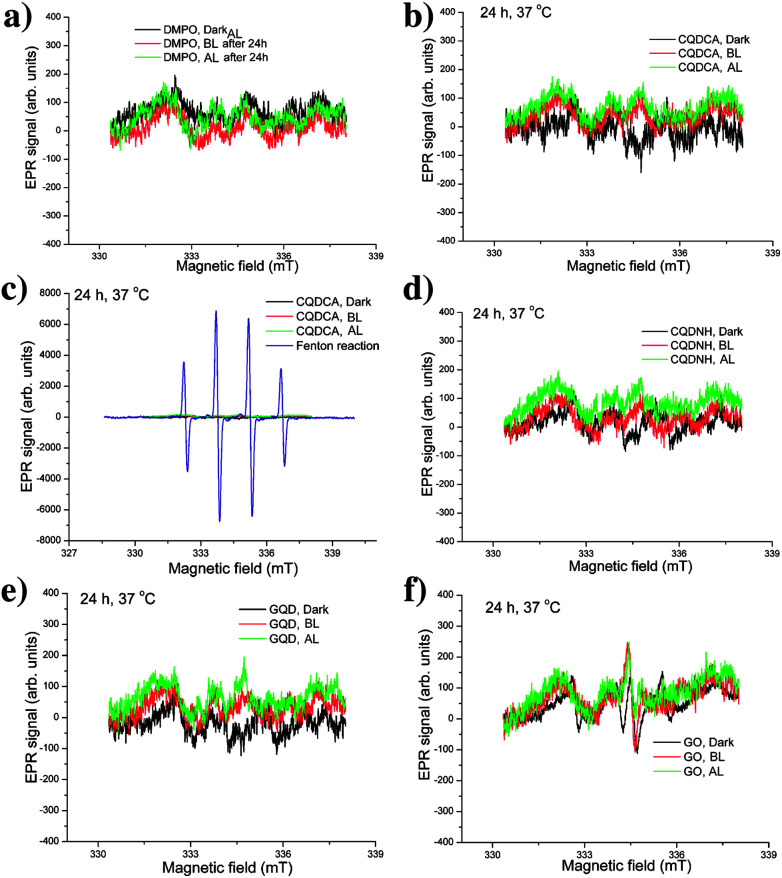
We have also analysed the production of hydroxyl and superoxide radicals upon different irradiation conditions using DMPO as a spin trap specific for both of these radicals. Firstly, we conducted the control experiment to study the effect of irradiation for 24 h on the EPR spectra of DMPO in water. These experiments are presented in the Fig. 6a. As can be observed, signals with very low intensities are detected for DMPO left in dark, irradiated with BL and AL. These signals can be considered as insignificant and thus the presented measurements indicate that selected experimental conditions do not have effects on DMPO. In the presence of the CQDCA sample, the intensity of EPR signals did not change, Fig. 6b. By comparing these EPR spectra with spectrum obtained from Fenton reaction in the presence of DMPO (Fig. 6c), it can be clearly observed that under condition of high hydroxyl radical production, four high-intensity signals can be observed. Considering spectra of the referent sample (Fig. 6a), Fenton reaction (Fig. 6c) and the CQDCA (Fig. 6b and c), it can be concluded that the CQDCA sample is not capable for generation of hydroxyl and superoxide radicals. Similar low-intensity signals were noticed for the CQDNH, GQD and GO samples (Fig. 6d–f). These results confirm that the CQDNH, GQD and GO samples under the studied conditions also do not induce the formation of hydroxyl and superoxide radicals.
Fig. 6. EPR spectra of DMPO at different conditions (a), CQDCA with DMPO after 24 h of irradiation and in the dark (b), Fenton reaction and the CQDCA sample at different conditions (c), EPR spectra of CQDNH (d), GO (e) and GQD (f) with DMPO as a trap, after 24 h at 37 °C in dark, under AL and upon BL exposure.
Taking into account the comprehensive EPR analysis, we concluded that the CQDCA, CQDNH and GQD produce significant amount of single oxygen only under BL irradiation. Additionally, investigated samples are not capable of generating hydroxyl and superoxide radicals.
Ge et al. found that GQD generate singlet oxygen through energy transfer to molecular oxygen.61 Chong et al. claimed that superoxide anion is involved in the generation of singlet oxygen, implying that electron transfer is an intermediate step for generation of singlet oxygen by photoexcited GQD.31 Our results indicate that energy transfer has a crucial role in singlet oxygen production. Our experiments did not confirm any production of superoxide or hydroxyl radicals by investigated samples. The singlet oxygen generation by CQDNH is on the same level as GQD as and twice less than that by CQDCA under BL irradiation. We suppose that CQDCA generates singlet oxygen at the highest level due to lack of agglomeration. Namely, results of zeta potential measurements indicate that this sample has the highest value of zeta potential compared to GQD and CQDNH (Table S2†).
3.6. Antibacterial activity of GO, GQD, CQDCA and CQDNH
In Tables 2 and S3† are listed the values of MIC and MBC of the GO, GQD, CQDCA and CQDNH samples. During AL and BL exposure alone all bacteria strains were able to grow. So we concluded that light exposure alone does not affect the bacteria at all. But upon BL exposure of bacteria in the presence of GO, GQD, CQDCA and CQDNH, all samples have shown some antibacterial activity toward used bacterial strains. As for the GO sample, it shows poor activity against to all tested bacteria with MIC in the range of 500–4000 μg mL−1 and MBC of 1000–8000 μg mL−1. The GO inhibits growth of the following bacteria best: E. coli, E. aerogenes, P. aeruginosa, Klebsiella pneumoniae and Bacillus subtilis (MIC-500 μg mL−1; MBC-1000 μg mL−1). All tested quantum dots samples (GQD, CQDCA and CQDNH) show activity against all tested bacteria with MIC from 3.905–250 μg mL−1 and MBC from 7.81–500 μg mL−1. Two bacteria strains, E. aerogenes and P. Mirabilis, are the most sensitive bacteria strains to the CQDNH sample (3.905 μg mL−1). The effects of the GQD and CQDCA on these bacteria strains are very effective but not as good as the effect of the CQDNH. S. saprophyticus, L. monocytogenes, S. typhimurium, Klebsiella pneumoniae bacteria strains are slightly less sensitive to the GQD but equally sensitive to the CQDNH and CQDCA. E. faccalis is the most resistive bacteria to all used samples. Comparing obtained results to the MIC/MBC of applied antibiotics we can conclude that amracin acts more effective than tested nanoparticles samples.
The values of MIC (μg mL−1) of the GO, GQD, CQDCA and CQDNH samples.
| Bacteria strains | MIC value | (μL mL−1) | |||
|---|---|---|---|---|---|
| GO | GQD | CQDCA | CQDNH | A* | |
| S. saprophyticus ATCC 15035 | >1000.0 | 15.62 ± 0.49 | 7.81 ± 0.24 | 7.81 ± 0.24 | 0.24 ± 0.02 |
| S. aureus ATCC 25923 | >2000.0 | 125.00 ± 0.97 | 62.50 ± 0.49 | 62.50 ± 0.49 | 0.97 ± 0.03 |
| L. ivanovii ATCC 19119 | >3000.0 | 62.50 ± 0.97 | 62.50 ± 0.49 | 31.25 ± 0.24 | 0.49 ± 0.02 |
| L. inocun ATCC 33090 | >4000.0 | 125.00 ± 0.49 | 125.00 ± 0.97 | 62.5 ± 0.24 | 0.97 ± 0.03 |
| E. faccalis ATCC 2912 | >1000.0 | 250.00 ± 0.24 | 250.00 ± 0.24 | 125.00 ± 0.49 | 0.49 ± 0.02 |
| L. monocytogenes ATCC 19112 | >3000.0 | 15.62 ± 0.24 | 7.81 ± 0.49 | 7.81 ± 0.24 | 0.49 ± 0.02 |
| B. spieizeneii ATCC 6633 | >1000.0 | 125.00 ± 0.97 | 125.00 ± 0.24 | 62.50 ± 0.97 | 0.24 ± 0.02 |
| E. faccium ATCC 6057 | >4000.0 | 62.50 ± 0.49 | 62.50 ± 0.24 | 31.25 ± 0.24 | 0.97 ± 0.03 |
| E. coli ATCC 25922 | >500.0 | 62.50 ± 0.49 | 31.25 ± 0.97 | 31.25 ± 0.49 | 0.49 ± 0.02 |
| S. enteritidas ATCC 13076 | >1000.0 | 250.00 ± 0.24 | 125.00 ± 0.97 | 125.00 ± 0.24 | 0.97 ± 0.02 |
| E. aerogenes ATCC 13048 | >500.0 | 7.81 ± 0.97 | 7.81 ± 0.24 | 3.905 ± 0.97 | 0.49 ± 0.02 |
| C. freundi ATCC 43864 | >3000.0 | 62.50 ± 0.24 | 62.50 ± 0.24 | 31.25 ± 0.49 | 0.49 ± 0.02 |
| S. typhimurium ATCC 14028 | >4000.0 | 15.62 ± 0.49 | 7.81 ± 0.24 | 7.81 ± 0.97 | 0.24 ± 0.02 |
| P. aeroginosa ATCC 27853 | >500.0 | 62.50 ± 0.97 | 62.50 ± 0.24 | 31.25 ± 0.49 | 0.97 ± 0.03 |
| P. mirabilis ATCC 35659 | >1000.0 | 7.81 ± 0.49 | 7.81 ± 0.49 | 3.905 ± 0.97 | 0.49 ± 0.02 |
| S. aureus ATCC 25923 | >2000.0 | 125.00 ± 0.97 | 125.00 ± 0.49 | 62.50 ± 0.24 | 0.97 ± 0.03 |
| Klebsiella pneumoniae ATCC 13883 | >500.0 | 15.62 ± 0.49 | 7.81 ± 0.24 | 7.81 ± 0.49 | 0.49 ± 0.02 |
| Proteus vulgaris ATCC 13315 | >1000.0 | 125.00 ± 0.24 | 62.50 ± 0.49 | 62.50 ± 0.97 | 0.49 ± 0.02 |
| Bacillus subtilis ATCC 6633 | >500.0 | 62.50 ± 0.97 | 31.25 ± 0.24 | 31.25 ± 0.49 | 0.49 ± 0.02 |
3.7. Loss of 260-nm-absorbing material
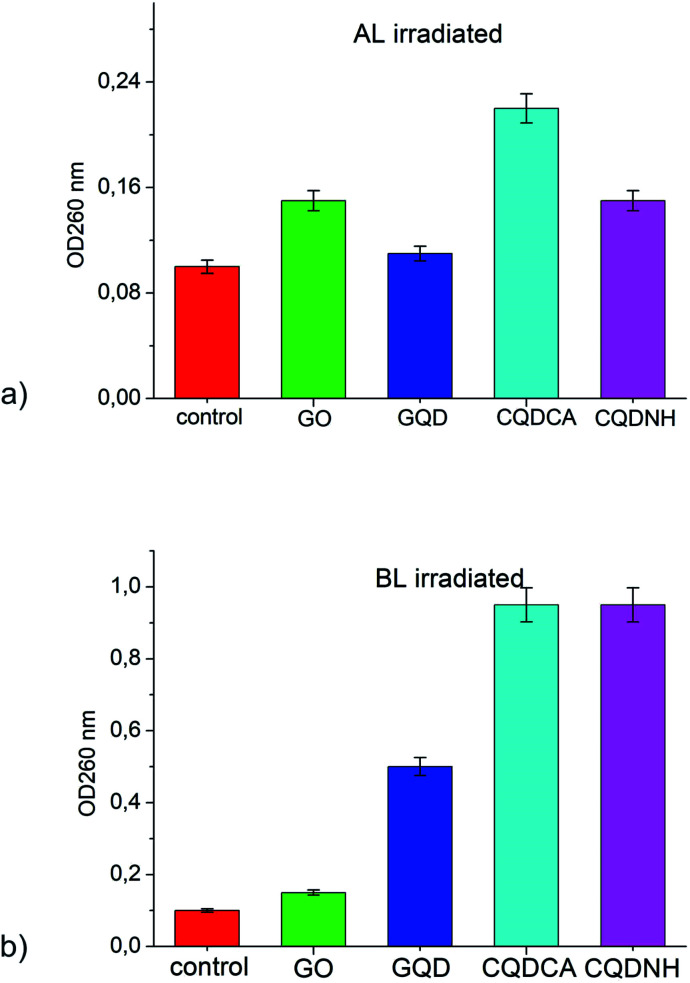
The presence of materials, in cell free medium, that absorb at 260 nm indicate that large molecules (e.g. nucleic acids) have been lost from the cell interior and that major membrane damage has occurred. The OD260s values of the filtrates from E. coli control suspensions remained approximately the same after 24 h under Al and BL. The OD260s of the filtrates from E. coli suspensions treated with 1 mg mL−1 of GO has remained almost the same during the whole assay period under AL and BL-Fig. 7(a and b). All other samples (GQD, CQDCA and CQDNH) have shown the same absorbance during 24 h under AL irradiation-Fig. 7a. But, under BL irradiation, the OD260s of the filtrates from E. coli suspensions treated with 0.1 mg mL−1 of CQDNH has shown very high absorbance-Fig. 7b. Obtained results indicate loss of absorbing material, i.e. leakage of interior content of E. coli.
Fig. 7. Presence of 260-nm-absorbing material in the filtrates of: (a) E. coli after treatment with GO, GQD, CQDCA and CQDNH at 24 h, compared to E. coli control suspension under AL and (b) E. coli after treatment with GO, GQD, CQDCA and CQDNH at 24 h, compared to E. coli control suspension under BL. The mean ± SD for three replicates are illustrated.
4. Discussion
Conducted structural and antibacterial research of different graphene based materials has shown that various aspects of used materials can affect their antibacterial affinity: chemical composition, shape, size, surface functionalization and ROS production. Diameters of the investigated samples are varying in the following way: CQDNH < GQD < CQDCA < GO. All samples except GO produce singlet oxygen predominantly under the BL. We detected by EPR that all samples including GO produce singlet oxygen even at AL conditions (light and dark). None of them produce hydroxyl or superoxide radicals at any conditions. Generated singlet oxygen attacks bacteria membrane wall. Due to increased porosity of bacteria membrane wall singlet oxygen enters inside bacteria, damages cytoplasmic membrane and causes its lipid peroxidation.62 By comparing antibacterial activity of GQD and CQDCA we found that antibacterial efficacy of GQD is weaker toward some bacterial strains (S. saprophyticus, L. monocytogenes, E. coli, S. typhimurium, Klebsiella pneumoniae, Proteus vulgaris and Bacillus subtilis). But MIC values of GQD toward E. coli, S. aureus, B. subtilis and P. aeruginosa are lower 4 and 8 times compared to MIC values of GQDs produced by conversion of multi-walled carbon nanotubes to GQDs.63
MIC values of CQDNH toward E. coli and B. subtilis are similar with results of other authors.13 Since CQDNH has been shown as the most potent antibacterial agent we assumed that the presence of amino groups on the basal plane of these particles (proved by XPS and FTIR measurements) can contribute to the bacterial death as well. Amino groups are known to adsorb onto the bacterial membrane wall, which provides the molecules bearing this functional groups are diffusing into the cell interior, where the disruption of the cytoplasmic membrane finally leads to the bacterial cell destruction.13,64 Therefore, the action of CQDNH on bacterial membrane wall can be twofold: generated singlet oxygen penetrates membrane wall whereas amino groups adsorb on membrane wall thus enabling CQDNH to enter bacteria cell and cause oxidative stress.
Despite different reports related to antibacterial activity of GO, we have found poor antibacterial activity of this material compared to other three used materials. Their MIC and MBC are 2 orders of magnitude higher than that of the CQDCA and especially the CQDNH. Castrillon et al. found that interaction between GO-E. coli membrane walls are predominantly repulsive due to lipopolysaccharide bridging.65
Barbolina et al.66 have shown that GO with sulfur content over 1.7% has antibacterial effect contrary to GO with smaller amount of sulfur. In our experiment the content of sulfur in the GO sample was 0.4%. Therefore, sulfur impurity did not affect the antibacterial activity of this material. Other important parameters (lateral size, shape, surface roughness, and surface functionalization) did not improve its antibacterial potential significantly. In our previous report, we analyzed antibacterial potential of exfoliated graphene67 and found that this material had very poor antibacterial activity, i.e. it had only bacteriostatic not bactericidal activity at certain conditions. Further, we found that its antibacterial potential has been improved by curcumin functionalization.68
5. Conclusions
In this work we have compared structural properties as well as antibacterial potentials of four different graphene based materials: GO, GQDs, CQDs and N-CQDs. Used quantum dots and GO have different diameters, lateral size, and chemical composition. It was found that all BL triggered samples except GO produce singlet oxygen. Compared to other tested nanoparticles GO has poor antibacterial action. However, GO is capable to inhibit the growth of E. coli, E. aerogenus, P. aeroginosa, Klebsiella pneumoniae and B. subtilis. It was demonstrated that N-CQDs have shown the most potent antibacterial activity against E. aerogenes, P. mirabilis, S. saprophyticus, L. monocytogenes, S. typhimurium and Klebsiella pneumoniae. The GQD and CQDCA have shown double less efficiency against E. aerogenes and P. mirabilis. The CQDCA has shown very good antibacterial activity toward S. saprophyticus, L. monocytogenes, S. typhimurium and Klebsiella pneumoniae as well. But MIC values of CQDCA are double less than those of CQDNH. Oxidative stress of bacteria is the main reason for bacteria death after the action of BL photoexcited quantum dots. The obtained results enable possible usage of GQDs and CQDs as antibacterial agents in the future.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This research was supported by the SASPRO Programme project 1237/02/02-b. The research leading to these results has received funding from the People Programme (Marie Curie Actions) European Union's Seventh Framework Programme under REA grant agreement No. 609427. Research has been further co-funded by the Slovak Academy of Sciences. Research was also supported by the Ministry of Education, Science and Technological Development of the Republic of Serbia (project no. 172003) and multilateral scientific and technological cooperation in the Danube region (DS021). We are gratefully acknowledged to Mrs Marcela Kimličkova for performing zeta potential measurements.
Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra04664f
Notes and references
- Albert K. Hsu H. Y. Molecules. 2016;21:1585. doi: 10.3390/molecules21111585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabish T. A. Zhang S. Winyard P. G. Redox Biol. 2018;15:34. doi: 10.1016/j.redox.2017.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karahan H. E. Wiraja C. Xu C. Wei J. Wang Y. Wang L. Liu F. Chen Y. Adv. Healthcare Mater. 2018;7:e1701406. doi: 10.1002/adhm.201701406. [DOI] [PubMed] [Google Scholar]
- Szunerits S. Boukherroub R. J. Mater. Chem. B. 2016;4:6892. doi: 10.1039/C6TB01647B. [DOI] [PubMed] [Google Scholar]
- Fair R. J. Tor Y. Perspect. Med. Chem. 2014;6:25. doi: 10.4137/PMC.S14459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q. Li J. Zhong X. Dai Z. Lu Z. Yang H. Chen R. Adv. Powder Technol. 2018;29:2082. doi: 10.1016/j.apt.2018.05.015. [DOI] [Google Scholar]
- Wei D. Tian F. Lu Z. Yang H. Chen R. RSC Adv. 2016;6:52264. doi: 10.1039/C6RA05754C. [DOI] [Google Scholar]
- Zhao Y. Ye C. Liu W. Chen R. Jiang X. Angew. Chem., Int. Ed. Engl. 2014;53:8127. doi: 10.1002/anie.201401035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z. Rong K. Li J. Yang H. Chen R. J. Mater. Sci.: Mater. Med. 2013;24:1465. doi: 10.1007/s10856-013-4894-5. [DOI] [PubMed] [Google Scholar]
- Li J. Rong K. Zhao H. Li F. Li Z. Chen R. J. Nanosci. Nanotechnol. 2013;13:6806. doi: 10.1166/jnn.2013.7781. [DOI] [PubMed] [Google Scholar]
- Dai T. Tegos G. P. Lu Z. Huang L. Zhiyentayev T. Franklin M. J. Baer D. G. Hamblin M. R. Antimicrob. Agents Chemother. 2009;53:3929. doi: 10.1128/AAC.00027-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegab H. M. ElMekawy A. Zou L. Mulcahy D. Saint C. P. Ginic-Markovic M. Carbon. 2016;105:362. doi: 10.1016/j.carbon.2016.04.046. [DOI] [Google Scholar]
- Travlou N. A. Giannakoudakis D. A. Algarra M. Labella A. M. Rodríguez-Castellón E. Bandosz T. J. Carbon. 2018;135:104. doi: 10.1016/j.carbon.2018.04.018. [DOI] [Google Scholar]
- Liu S. B. Hu M. Yeng T. H. Wu R. Jiang R. R. Wei J. Wang L. Kong J. Chen Y. Langmuir. 2012;28:12364. doi: 10.1021/la3023908. [DOI] [PubMed] [Google Scholar]
- Perreault F. Fonseca de Faria A. Nejati S. Elimelech M. ACS Nano. 2015;9:7226. doi: 10.1021/acsnano.5b02067. [DOI] [PubMed] [Google Scholar]
- Moritz M. Geszke-Moritz M. Chem. Eng. J. 2013;228:596. doi: 10.1016/j.cej.2013.05.046. [DOI] [Google Scholar]
- Zhu Y. Murali S. Cai W. Li X. Suk J. W. Potts J. R. Ruoff R. S. Adv. Mater. 2010;22:3906. doi: 10.1002/adma.201001068. [DOI] [PubMed] [Google Scholar]
- Guo X. Mei N. J. Food Drug Anal. 2014;22:105. doi: 10.1016/j.jfda.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S. B. Zeng T. H. Hofmann M. Burcombe E. Wei J. Jiang R. R. Kong J. Chen Y. ACS Nano. 2011;5:6971. doi: 10.1021/nn202451x. [DOI] [PubMed] [Google Scholar]
- Chong Y. Ge C. Fang G. Wu R. Zhang H. Chai Z. Chen C. Yin J. J. Environ. Sci. Technol. 2017;51:10154. doi: 10.1021/acs.est.7b00663. [DOI] [PubMed] [Google Scholar]
- Ivanova E. P. Hasan J. Webb H. K. Truong V. K. Watson G. S. Watson J. A. Baulin V. A. Pogodin S. Wang J. Y. Tobin M. J. Löbbe C. Crawford R. J. Small. 2012;20:2489. doi: 10.1002/smll.201200528. [DOI] [PubMed] [Google Scholar]
- Faria F. A. Perreault F. Elimelech M. ACS Appl. Nano Mater. 2018;1:1164. doi: 10.1021/acsanm.7b00332. [DOI] [Google Scholar]
- Zhou S. Xu H. Gan W. Yuan Q. RSC Adv. 2016;6:110775. doi: 10.1039/C6RA24349E. [DOI] [Google Scholar]
- Jovanović S. P. Syrgiannis Z. Marković Z. M. Bonasera A. Kepić D. P. Budimir M. D. Milivojević D. D. Spasojević V. D. Dramićanin M. D. Pavlović V. B. Todorović Marković B. M. ACS Appl. Mater. Interfaces. 2015;7:25865. doi: 10.1021/acsami.5b08226. [DOI] [PubMed] [Google Scholar]
- Jovanović S. Marković Z. Budimir M. Spitalsky Z. Vidoeski B. Todorović Marković B. Opt. Quantum Electron. 2016;48:259. doi: 10.1007/s11082-016-0516-z. [DOI] [Google Scholar]
- Zhu S. Zhang J. Qiao C. Tang S. Li Y. Yuan W. Li B. Tian L. Liu F. Hu R. Gao H. Wei H. Zhang H. Sun H. Yang B. Chem. Commun. 2011;47:6858. doi: 10.1039/C1CC11122A. [DOI] [PubMed] [Google Scholar]
- Wang S. Cole I. S. Li Q. RSC Adv. 2016;6:89867. doi: 10.1039/C6RA16516H. [DOI] [Google Scholar]
- Marković Z. Ristić B. Arsikin K. Klisić D. Harhaji-Trajković Lj. Todorović-Marković B. Kepić D. Kravić-Stevović T. Jovanović S. Milenković M. Milivojević D. Bumbaširević V. Dramićanin M. Trajković V. Biomaterials. 2012;33:7084. doi: 10.1016/j.biomaterials.2012.06.060. [DOI] [PubMed] [Google Scholar]
- Ristić B. Z. Milenković M. M. Dakić I. R. Todorović-Marković B. M. Milosavljević M. S. Budimir M. D. Paunović V. G. Dramićanin M. D. Marković Z. M. Trajković V. S. Biomaterials. 2014;35:4428. doi: 10.1016/j.biomaterials.2014.02.014. [DOI] [PubMed] [Google Scholar]
- Barneck M. D. Rhodes N. L. De la Presa M. Allen J. P. Poursaid A. E. Nourian M. M. Firpo M. A. Langell J. T. J. Surg. Res. 2016;206:316. doi: 10.1016/j.jss.2016.08.006. [DOI] [PubMed] [Google Scholar]
- Chong Y. Ge C. Fang G. Tian X. Ma X. Wen T. Wamer W. G. Chen C. Chai Z. Yin J. J. ACS Nano. 2016;10:8690. doi: 10.1021/acsnano.6b04061. [DOI] [PubMed] [Google Scholar]
- Hui L. Huang J. Chen G. Zhu Y. Yang L. ACS Appl. Mater. Interfaces. 2016;8:20. doi: 10.1021/acsami.5b10132. [DOI] [PubMed] [Google Scholar]
- Namdari P. Negahdari B. Eatemadi A. Biomed. Pharmacother. 2017;87:209. doi: 10.1016/j.biopha.2016.12.108. [DOI] [PubMed] [Google Scholar]
- Wang Y. Hu A. J. Mater. Chem. C. 2014;2:6921. doi: 10.1039/C4TC00988F. [DOI] [Google Scholar]
- Tan J. Zou R. Zhang J. Li W. Zhang L. Yue D. Nanoscale. 2016;8:4742. doi: 10.1039/C5NR08516K. [DOI] [PubMed] [Google Scholar]
- Zhang Y. Wang Y. Feng X. Zhang F. Yang Y. Liua X. Appl. Surf. Sci. 2016;387:1236. doi: 10.1016/j.apsusc.2016.07.048. [DOI] [Google Scholar]
- Song Z. Quan F. Xu Y. Liu M. Cui L. Liu J. Carbon. 2016;104:169. doi: 10.1016/j.carbon.2016.04.003. [DOI] [Google Scholar]
- Meziani M. J. Dong X. Zhu L. Jones L. P. LeCroy G. E. Yang F. Wang S. Wang P. Zhao Y. Yang L. Tripp R. A. Sun Y. P. ACS Appl. Mater. Interfaces. 2016;8:10761. doi: 10.1021/acsami.6b01765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur M. Pandey S. Mewada A. Patil V. Khade M. Goshi E. Sharon M. J. Drug Delivery. 2014;2014:282193. doi: 10.1155/2014/282193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. Lu S. Tang Q. Zhang K. Yu W. Sun H. Yang B. Nanoscale. 2017;9:7135. doi: 10.1039/C7NR02128C. [DOI] [PubMed] [Google Scholar]
- Li Y. J. Harroun S. G. Su Y. C. Huang C. F. Unnikrishnan B. Lin H. J. Lin C. H. Huang C. C. Adv. Healthcare Mater. 2016;19:2545. doi: 10.1002/adhm.201600297. [DOI] [PubMed] [Google Scholar]
- Chen J. Yao B. Li C. Shi G. Carbon. 2013;64:225. doi: 10.1016/j.carbon.2013.07.055. [DOI] [Google Scholar]
- Wang S. Chen Z. G. Cole I. Li Q. Carbon. 2015;82:304. doi: 10.1016/j.carbon.2014.10.075. [DOI] [Google Scholar]
- Wang H. Sun P. Cong S. Wu J. Gao L. Wang Y. Dai X. Yi Q. Zou G. Nanoscale Res. Lett. 2016;11:27. doi: 10.1186/s11671-016-1231-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chorvát Jr D. Chorvátová A. Eur. Biophys. J. 2006;36:73. doi: 10.1007/s00249-006-0104-4. [DOI] [PubMed] [Google Scholar]
- Satyajit D. Sarker L. N. Kumarasamy Y. Methods. 2007;42:321. doi: 10.1016/j.ymeth.2007.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson C. F. Mee B. J. Riley T. V. Antimicrob. Agents Chemother. 2002;46:1914. doi: 10.1128/AAC.46.6.1914-1920.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatamie S. Akhavan O. Sadrnezhaada S. K. Ahadian M. M. Shirolkar M. M. Wang H. Q. Mater. Sci. Eng., C. 2015;55:482. doi: 10.1016/j.msec.2015.05.077. [DOI] [PubMed] [Google Scholar]
- Lin L. Rong M. Liu S. Song X. Zhong Y. Yan J. Wang Y. Chen X. Nanoscale. 2015;7:1872. doi: 10.1039/C4NR06365A. [DOI] [PubMed] [Google Scholar]
- Daems N. Sheng X. Vankelecom I. F. J. Pescarmona P. P. J. Mater. Chem. A. 2014;2:4085. doi: 10.1039/C3TA14043A. [DOI] [Google Scholar]
- http://www.xpsfitting.com/search/label/Nitrogen. http://www.xpsfitting.com/search/label/Nitrogen
- Jana M. Saha S. Khanra P. Murmu N. C. Srivastava S. K. Kuilaa T. Lee J. H. J. Mater. Sci. Eng. B. 2014;186:33. doi: 10.1016/j.mseb.2014.03.004. [DOI] [Google Scholar]
- Tosić D. Marković Z. Jovanović S. Prekodravac J. Budimir M. Kepić D. Holclajtner-Antunović I. Dramićanin M. Todorović-Marković B. Synth. Met. 2016;218:43. doi: 10.1016/j.synthmet.2016.04.016. [DOI] [Google Scholar]
- Zhang C. Hao R. Liao H. Hou Y. Nano Energy. 2013;2:88. doi: 10.1016/j.nanoen.2012.07.021. [DOI] [Google Scholar]
- Lai Q. Zhu S. Luo X. Zou M. Huang S. AIP Adv. 2012;2:032146. doi: 10.1063/1.4747817. [DOI] [Google Scholar]
- Luo Z. Lu Y. Somers L. A. Johnson A. T. C. J. Am. Chem. Soc. 2009;131:898. doi: 10.1021/ja807934n. [DOI] [PubMed] [Google Scholar]
- Zhu S. Meng Q. Wang L. Zhang J. Song Y. Jin H. Zhang K. Angew. Chem., Int. Ed. 2013;52:3953. doi: 10.1002/anie.201300519. [DOI] [PubMed] [Google Scholar]
- Zhang F. Feng X. Zhang Y. Yan L. Yang Y. Liu X. Nanoscale. 2016;8:8618. doi: 10.1039/C5NR08838K. [DOI] [PubMed] [Google Scholar]
- Zhang R. Chen W. Biosens. Bioelectron. 2014;55:83. doi: 10.1016/j.bios.2013.11.074. [DOI] [PubMed] [Google Scholar]
- Jovanović S. P. Marković Z. M. Syrgiannis Z. Dramićanin M. D. Arcudi F. La Parola V. Budimir M. Todorović Marković B. M. Mater. Res. Bull. 2017;93:183. doi: 10.1016/j.materresbull.2017.04.052. [DOI] [Google Scholar]
- Ge J. Lan M. Zhou B. Liu W. Guo L. Wang H. Jia Q. e Niu G. Huang X. Zhou H. Meng X. Wang P. Lee C. S. Zhang W. Han X. Nat. Commun. 2014;5:4596. doi: 10.1038/ncomms5596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker T. Canales M. Noimark S. Page K. Parkin I. Faull J. Bhatti M. Ciric L. Sci. Rep. 2017;7:15298. doi: 10.1038/s41598-017-15565-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas A. A. Khandelwal P. Das R. Salunke G. Alam A. Ghorai S. Chattopadhyay S. Poddar P. J. Mater. Chem. B. 2017;5:785. doi: 10.1039/C6TB02446G. [DOI] [PubMed] [Google Scholar]
- Reichel V., Functionalization of cellulose acetate surfaces for removal the of endocrine disruption compounds, Diploma thesis, Department of Colloid Science: University of Graz, 2012 [Google Scholar]
- Vargas Castrillon S. R. Perreault F. Fonseca de Faria A. Environ. Sci. Technol. Lett. 2015;2:112. doi: 10.1021/acs.estlett.5b00066. [DOI] [Google Scholar]
- Barbolina I. I. Woods C. R. Lozano N. Kostarelos K. Novoselov K. S. Roberts I. S. 2D Materials. 2016;3:1. doi: 10.1088/2053-1583/3/2/025025. [DOI] [Google Scholar]
- Marković Z. M. Matijašević D. M. Pavlović V. B. Jovanović S. P. Holclajtner-Antunović I. D. Špitalsky Z. Mičušik M. Dramićanin M. D. Milivojević D. D. Nikšić M. P. Todorović Marković B. M. J. Colloid Interface Sci. 2017;500:30. doi: 10.1016/j.jcis.2017.03.110. [DOI] [PubMed] [Google Scholar]
- Marković Z. M. Kepić D. P. Matijašević D. M. Pavlović V. B. Jovanović S. P. Stanković N. K. Milivojević D. D. Spitalsky Z. Holclajtner-Antunović I. D. Bajuk-Bogdanović D. V. Nikšić M. P. Todorović Marković B. M. RSC Adv. 2017;7:36081. doi: 10.1039/C7RA05027E. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.