Abstract
Stenotrophomonas maltophilia is an emerging nosocomial pathogen that displays high-level intrinsic resistance to a variety of structurally unrelated antimicrobial agents. Efflux mechanisms are known to contribute to acquired multidrug resistance in this organism, and indeed, one such multidrug efflux system, SmeDEF, was recently identified. Still, the importance of SmeDEF to intrinsic antibiotic resistance in S. maltophilia had not yet been determined. Reverse transcription-PCR confirmed expression of the smeDEF genes in wild-type S. maltophilia, and deletion of smeE or smeF in wild-type strains rendered the mutants hypersusceptible to several antimicrobials, suggesting that SmeDEF contributes to intrinsic antimicrobial resistance in this organism. Expression of smeDEF was also enhanced in an in vitro-selected multidrug-resistant mutant, although deletion of smeF but not of smeE in these mutants compromised antimicrobial resistance. Apparently, hyperexpressed SmeF is capable of functioning with additional multidrug efflux components to promote multidrug resistance in S. maltophilia.
Stenotrophomonas maltophilia is an aerobic, nonfermentative, gram-negative bacterium ubiquitous in nature (24). This organism has increasingly emerged as a nosocomial pathogen, particularly for immunocompromised patients, although little is known about the virulence mechanisms of and risk factors for S. maltophilia (5, 8, 19, 21). S. maltophilia is characterized by its high-level intrinsic resistance to a variety of structurally unrelated antimicrobials, including β-lactams, quinolones, and aminoglycosides (7). Combinations of antimicrobial agents are often needed for the therapy of S. maltophilia infections, which remain a challenge to physicians. Multiple mechanisms are involved in the high-level antimicrobial resistance of S. maltophilia. Constitutive (and inducible) production of the L1 and L2 β-lactamases is the major determinant for β-lactam (including carbapenem) resistance in this organism (25, 31, 34). Aminoglycoside-modifying enzymes such as O-nucleotidyltransferases and N-acetyltransferases (AAC) in S. maltophilia have been reported (12, 14, 32). Indeed, a good correlation between expression of the acetyltransferase AAC (6′)-Iz gene and the level of resistance to an aminoglycoside, tobramycin, was recently demonstrated in this organism (X.-Z. Li, L. Zhang, and K. Poole, submitted for publication).
Multiple antibiotic resistance in S. maltophilia, like in other gram-negative bacteria, is also attributable in part to limited outer membrane permeability (20) and active antibiotic extrusion (2, 3, 26, 34), although these mechanisms are poorly characterized to date. The outer membrane limits access of drugs to their bacterial targets (22), while multidrug efflux pumps actively remove drugs from the cell (23, 26). Several multidrug efflux systems have been identified in S. maltophilia to date, including SmeABC (X.-Z. Li, L. Zhang, and K. Poole, submitted for publication; GenBank accession number AF173226) and SmeDEF (3). Multidrug-resistant (MDR) strains hyperexpressing SmeM, a homologue of the outer membrane components of multidrug efflux systems in several gram-negative bacteria (17, 26–28, 34) have also been reported (34), and these strains were thought to hyperexpress yet a third multidrug efflux system. These efflux systems utilize transporters of the resistance-nodulation-cell division family (29) and are homologous to the major multidrug efflux systems, MexAB-OprM, MexCD-OprJ, MexEF-OprN, and MexXY-OprM, of Pseudomonas aeruginosa (1, 13, 15, 26–28). SmeABC appears not to contribute to intrinsic resistance in S. maltophilia, although SmeC does contribute to the acquired multidrug resistance of certain MDR strains (Li et al., submitted). Similarly, SmeDEF hyperexpression is associated with the multidrug resistance of certain in vitro-selected (3) and clinical (3a) MDR strains, although the significance of this efflux system vis-à-vis intrinsic resistance remains unknown. In this report, we assessed the contribution of SmeDEF in intrinsic and acquired antimicrobial resistance in S. maltophilia.
MATERIALS AND METHODS
Bacterial strains, plasmids and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Luria-Bertani (LB) broth (1% [wt/vol] Difco tryptone, 0.5% [wt/vol] Difco yeast extract, and 0.5% [wt/vol] NaCl) and agar (LB broth containing 1.5% [wt/vol] agar) were used as the growth media throughout, and bacterial cells were cultivated at 37°C. Plasmid pEX18Tc (11) and its derivatives were maintained in Escherichia coli with 10 μg of tetracycline per ml.
TABLE 1.
S. maltophila strains and plasmids used in this studya
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| ULA-511 | Wild type | 9 |
| K1765 | ULA-511 ΔsmeE | This study |
| K1793 | ULA-511 ΔsmeF | This study |
| K1449 | L1 and L2 β-lactamase-deficient double mutant of ULA-511 | 34 |
| K1766 | K1449 ΔsmeE | This study |
| K1795 | K1449 ΔsmeF | This study |
| K1385 | ULA-511 MDR mutant, overproduces SmeF (previously called SmeM) | 34 |
| K1767 | K1385 ΔsmeE | This study |
| K1794 | K1385 ΔsmeF | This study |
| K1439 | ULA-511 MDR mutant | 34 |
| K1768 | K1439 ΔsmeE | This study |
| K1796 | K1439 ΔsmeF | This study |
| Plasmids | ||
| pEX18Tc | Broad-host-range gene replacement vector; 6.35 kb; sacB; tetracycline resistant | 11 |
| pLZ706 | pEX18Tc::ΔsmeE | This study |
| pLZ755 | pEX18Tc::ΔsmeF | This study |
With the exception of strain ULA-511 (the reference strain), all strains listed are laboratory isolates.
DNA methodology.
Basic DNA procedures, including restriction endonuclease digestions, ligations, transformations and agarose gel electrophoresis, were performed as described previously (30). The alkaline lysis method (30) or a plasmid midi kit (Qiagen, Inc.) was used to isolate plasmids from E. coli DH5α and S. maltophilia. The genomic DNA of S. maltophilia was extracted by the method of Barcak et al. (6). DNA fragments used in cloning were extracted from agarose gels using Prep-A-Gene (Bio-Rad, Richmond, Calif.) as per the manufacturer's instructions. Nucleotide sequencing of plasmid-borne DNA was carried out by Cortec DNA Services, Inc. (Queen's University) using universal or custom primers.
Construction of ΔsmeE mutants.
To construct ΔsmeE mutants, two separate PCRs were performed to amplify two DNA fragments of ca. 0.85 and 0.76 kb, corresponding to the regions upstream and downstream, respectively, of the smeE gene sequence to be deleted. Sequences 5′ to the deletion were amplified from genomic DNA of S. maltophilia ULA-511 using primers smee5xz (5′-TGCAGAATTCCTACTTCTCCTCCAACAG-3′; anneals 228 to 245 bp downstream of the smeE start codon [GenBank accession number AJ252200]; HindIII site underlined) and smee2xz (5′ AGCGTCTAGAGCAGGAACAGGTACATCAC-3′; anneals 1,060 to 1,078 bp downstream of the smeE start codon; XbaI site underlined), while sequences 3′ to the deletion were amplified using primers smee3xz (5′-AGCATCTAGAAGTTCCGCATCGACATCGAC-3′; anneals 921 to 940 bp upstream of the smeE stop codon; XbaI site underlined) and smee4xz (5′-TAGCAAGCTTAAGGCGAGCGAGGTCATCA-3′; anneals 175 to 194 bp downstream of the smeE stop codon; EcoRI site underlined). The PCR mixture contained 50 ng of S. maltophilia chromosomal DNA, 40 pmol of each primer, 0.2 mM deoxynucleoside triphosphate, 2 mM MgSO4, and 10% (vol/vol) dimethyl sulfoxide in 1× thermoreaction buffer (New England Biolabs, Mississauga, Ontario, Canada) and was heated for 5 min at 94°C before the addition of 2 U of Vent DNA polymerase (New England Biolabs) per reaction. The reaction was then processed for 30 cycles of 1 min at 94°C, 40 s at 56°C, and 40 s at 72°C, before finishing with 10 min at 72°C. The two smeE-containing PCR products were purified using a Qiaquick PCR purification kit (Qiagen, Inc.) and were digested by HindIII-XbaI and XbaI-EcoRI, respectively. Initially, the XbaI-EcoRI-digested 3′ fragment was cloned into XbaI-EcoRI-restricted pEX18Tc, yielding pLZ689, and then the HindIII-XbaI-digested 5′ fragment was cloned into HindIII-XbaI-restricted pLZ689, yielding pLZ706. This latter plasmid carried a 1,104-bp in-frame deletion of the smeE gene as confirmed by nucleotide sequencing. Plasmid pLZ706 was subsequently used to transform E. coli S17-1, from which it was mobilized into strains ULA-511, K1385, K1439, and K1449 via conjugation as described earlier (34). Transconjugants carrying pLZ706 in the chromosome were selected on LB agar containing tetracycline (40 μg/ml) and norfloxacin (2.5 μg/ml; for counterselection). Transconjugants were then streaked onto LB agar containing 10% (wt/vol) sucrose, and sucrose-resistant colonies arising after overnight incubation at 37°C were screened for the presence of the smeE deletion using PCR with primer pair smee5xz and smee4xz.
Construction of ΔsmeF mutants.
To construct ΔsmeF mutants, two separate PCRs were performed to amplify two DNA fragments of ca. 0.92 and 0.85 kb, corresponding to the regions upstream and downstream, respectively, of the smeF gene sequence to be deleted. Sequences 5′ to the deletion were amplified from genomic DNA of S. maltophilia ULA-511 using primers smef1xz (5′-TGACGAATTCTGGTCCGTGAAGAACGACAA-3′; anneals 845 to 826 bp upstream of the smeF gene; EcoRI site underlined) and smef2xz (5′-AGCGTCTAGATGGCGGCAATGGAGAGGAAC-3′; anneals 51 to 32 bp downstream of the smeF start site; XbaI site underlined), while sequences 3′ to the deletion were amplified with primers smef3xz (5′-AGACTCTAGATGTCACCGAGCAGTTCAG-3′; anneals 362 to 379 bp downstream of the smeF start site; XbaI site underlined) and smef4xz (5′-TAGCAAGCTTCATCCAGGCTGACATTCAAC-3′; anneals 224 to 243 bp upstream of the smeF stop site; HindIII site underlined). The PCRs were carried out as described above for the smeE deletion construct, and the products were similarly purified using the Qiaquick PCR purification kit. The PCR products were digested with EcoRI and XbaI or XbaI and HindIII, as appropriate, and were cloned separately into appropriately restricted pEX18Tc, yielding plasmids pLZ753 (5′ upstream fragment) and pLZ754 (3′ downstream fragment). Following nucleotide sequencing to ensure that no errors had been introduced as a result of the PCR, the 3′ downstream fragment was liberated from pLZ754 by digestion with XbaI-HindIII and was cloned into XbaI-HindIII-restricted pLZ753. The resulting plasmid (pLZ755), carrying the smeF gene with an internal 310-bp deletion, was introduced into E. coli S17-1 and mobilized into S. maltophilia strains ULA-511, K1385, K1439, and K1449 as described above. Transconjugants carrying pLZ755 in the chromosome were selected on LB agar containing tetracycline (25 μg/ml, ULA-511 and K1449; and 40 μg/ml, K1385 and K1439) and norfloxacin (2.5 μg/ml; for counterselection). Sucrose-resistant colonies were then recovered as described above and were screened for the presence of a chromosomal smeF deletion using PCR with primers smef1xz and smef4xz. Reaction mixtures were formulated as described above using the same parameters, with the exception of the 72°C incubation, which was for 80 s.
RT-PCR.
Total bacterial RNA was isolated from LB-grown, late-log-phase (A600 = ca 1.0) cultures (1 ml) of S. maltophilia strains using the Qiagen RNeasy Mini Kit (Qiagen, Inc.). Following treatment with RNase-free DNase (2 U of enzyme/μg of RNA for 60 min at 37°C; Promega, Madison, Wis.), the RNA was repurified using the same kit. Samples (0.005, 0.05, and 0.5 μg) of DNase-treated RNA were then used as the template for reverse transcription-PCR (RT-PCR) with the Qiagen OneStep RT-PCR kit (Qiagen, Inc.) according to a protocol supplied by the manufacturer. Primer pairs used were specific for smeD (smed3xz, 5′-CCAAGAGCCTTTCCGTCAT-3′; and smed4xz, 5′-TCTCGGACTTCAGCGTGAC-3′), smeE (smee5xz [see sequence above] and smee2xz [see sequence above]); smeF (smef1xz [see sequence above] and smef2xz [see sequence above]), and blaL2 (sml6xz, 5′-CGTCGCCGATTCCTGCAGTT-3′; and sml7xz, 5′-CGGTGTTGTCGCTGGTGATG-3′). Thirty picomoles of each primer was used per reaction (final volume of 50 μl), which involved a 30-min incubation at 50°C, followed by 15 min at 95°C, and by 30 cycles of 30 s at 94°C, 30 s at 55°C, and 30 s at 72°C, before finishing with 10 min at 72°C. A 15-μl sample of each reaction product was analyzed by agarose (1.4% [wt/vol]) gel electrophoresis for the expected 140- (smeD), 871-(smeE), 917- (smeF), or 410-bp (blaL2) RT-PCR product. To assess the influence of growth on smeDEF expression, RNA was extracted from log-phase (A600 = 0.8) and stationary-phase (i.e., overnight; A600 > 2.0) cultures and was subjected to RT-PCR as described above. To control for DNA contamination of RNA samples, non-RT reactions (i.e., PCRs) were carried out on 0.5 μg of RNA. In no instance was a product obtained in the absence of a RT reaction.
Antimicrobial susceptibility assay.
Susceptibility testing was carried out in LB medium by the twofold serial dilution method with an inoculum of 5 × 105 cells/ml. Data were reported as MICs, which reflected the lowest concentration of antibiotic inhibiting visible cell growth after an overnight incubation at 37°C. Most antimicrobials were purchased from Sigma-Aldrich Canada Ltd. (Oakville, Ontario, Canada). Others were kindly provided by the following sources: moxifloxacin and BAYy3118 (a monofluorinated quinolone) from Bayer AG (Leverkusen, Germany); clinafloxacin from Parke-Davis Pharmaceutical Research (Ann Arbor, Mich.); trovafloxacin from Pfizer Inc. (Groton, Conn.); gemifloxacin (SB-265805) from SmithKline Beecham (Frythe, Welwyn, United Kingdom); cefpirome from Roussel UCLAF (Paris, France); pirazmonam and cefepime from the Squibb Institute (Princeton, N.J.); imipenem from Merck Sharp Dohme Canada (Montreal, Canada); azithromycin from Pfizer Canada Inc. (Kirkland, Quebec, Canada); and tigilcycline (GAR-936; a glycycline) from Wyeth-Ayerst (Pearl River, N.Y.).
Membrane isolation and SDS-polyacrylamide gel electrophoresis.
Outer membranes were prepared as Sarkosyl (1.5% [wt/vol])-insoluble cell envelopes as described previously (34). Fifty micrograms of outer membrane protein was then loaded onto sodium dodecyl sulfate (SDS)–12% polyacrylamide gels and electrophoresed as described earlier (34).
Organic-solvent tolerance assay.
Two approaches were employed to assess the organic-solvent tolerance of S. maltophilia (16, 18). The first involved assessment of cell growth by measuring the increase in optical density at 600 nm (OD600). Briefly, stationary-phase cells were diluted into 30 ml of prewarmed (37°C) LB broth and were incubated (with shaking) for 2 h at 37°C. At the early exponential phase of growth (OD600 = 0.2), n-hexane was added at a final concentration of 2 to 5% (vol/vol), and growth was monitored for 5 h. The second approach involved overlaying solvent onto LB agar plates inoculated with bacteria. Briefly, stationary-phase LB broth cultures were diluted into the same medium to yield a suspension of approximately 107 cells/ml. A 5-μl aliquot of the cell suspension was placed in duplicate on LB agar and allowed to dry before n-hexane (1 ml) was overlaid onto LB agar plates, and the plates were incubated overnight at 37°C.
RESULTS AND DISCUSSION
SmeDEF contributes to intrinsic resistance.
Multidrug efflux systems play an important role in intrinsic as well as mutationally acquired multidrug resistance in gram-negative bacteria (26). Recently, S. maltophilia was shown to possess multiple efflux systems, including SmeABC (Li et al., submitted) and SmeDEF (3), both of which play a role in acquired multidrug resistance. To assess the role of the SmeDEF multidrug efflux system in intrinsic antibiotic resistance, RT-PCR was employed initially to assess expression of the smeDEF genes in wild-type S. maltophilia. As shown in Fig. 1A to C, expression of all three genes was observed in wild-type ULA-511 cells (lanes 1 to 3), although expression of both smeE and smeF was weak and observable only at the higher concentrations of RNA template (Fig. 1, lanes 1 and 2). Recovery of smeE signal at least could be enhanced in strain ULA-511 by increasing the number of cycles used in the RT-PCR (data not shown). The substantial expression of smeD seen in ULA-511 stands in contrast to the very weak signal obtained for wild-type S. maltophilia in Northern blots with an smeD probe (3). Despite the apparently weaker expression seen here for smeE and smeF, it is unlikely that these are expressed at levels markedly below those for smeD. Rather, the differences observed likely reflect differences in the efficiency of the individual gene-specific RT-PCRs.
FIG. 1.
smeDEF expression (A to C) in S. maltophilia measured by RT-PCR of RNA isolated from strains ULA-511 (lane 2, 0.5 μg of RNA amplified; lane 3, 0.05 μg of RNA; and lane 4, 0.005 μg of RNA), K1385 (lane 5, 0.5 μg of RNA amplified; lane 6, 0.05 μg of RNA; and lane 7, 0.005 μg of RNA), and K1439 (lane 8, 0.5 μg of RNA amplified; lane 9, 0.05 μg of RNA; and lane 10, 0.005 μg of RNA). RT-PCR using primers for the blaL2 gene (D) is included as a control (lane designations as above for smeDEF). Lane 1, DNA size markers (100-bp ladder).
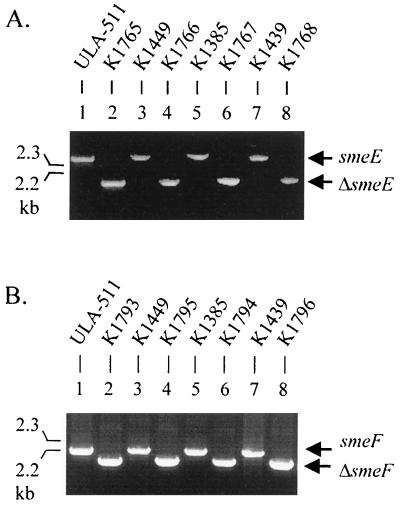
To assess the contribution of SmeDEF to intrinsic resistance in strain ULA-511, markerless chromosomal smeE and smeF deletion derivatives of this strain were constructed (Fig. 2) by using a homologous recombination procedure as described in Materials and Methods. As shown in Table 2, the ΔsmeE and ΔsmeF derivatives of ULA-511 (designated strains K1765 and K1793, respectively) showed increased susceptibility to a variety of structurally unrelated antimicrobials, including quinolones, tetracyclines, macrolides, chloramphenicol, and novobiocin (two to eightfold decrease in MICs), indicating that the SmeDEF efflux system contributed to intrinsic resistance to these agents. The mutants did not show changes in susceptibility to aminoglycoside antibiotics, including amikacin, gentamicn, kanamycin, tobramycin, and streptomycin (Table 2 and data not shown), suggesting that this class of antibiotics was not a substrate for the SmeDEF multidrug efflux system. Because of the high-level production of the L1 and L2 β-lactamases in wild-type S. maltophilia, the role of SmeDEF in β-lactam efflux was investigated in ΔsmeE and ΔsmeF derivatives (designated K1766 and K1795, respectively) of the L1 and L2 β-lactamase-deficient mutant K1449. As with the sme deletion derivatives of wild-type strain ULA-511, strains K1766 and K1795 showed similar hypersusceptibility to multiple non-β-lactam antimicrobials (Table 2). This strain generally did not, however, show any change in susceptibility to most β-lactam compounds (including penicillins, cephalosporins, and carbapenems) compared with its parent strain K1449, although slightly increased susceptibility to carbenicillin, piperacillin, cefoperazone, cefepime, and cefpirome was observed (twofold decrease in MICs). Thus, β-lactams are generally not good substrates for the SmeDEF efflux system, and the pattern of resistance provided by SmeDEF in wild-type cells is very reminiscent of that seen for this efflux system in SmeDEF-hyperexpressing MDR mutants (3).
FIG. 2.
Confirmation of smeE and smeF deletions by PCR amplification of the smeE (A) and smeF (B) genes of S. maltophilia strains using genomic DNA as templates and primer pairs smee5xz and smee4xz (smeE) and smef1xz and smef4xz (smeF). Strain designations are indicated above the lanes. DNA size markers are shown at left.
TABLE 2.
Effect of smeE and smeF deletions on antimicrobial susceptibility of S. maltophilia
| Antimicrobial agent | MIC (μg/ml) fora
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ULA-511 (wild type) | K1765 (ULA-511 ΔsmeE) | K1793 (ULA-511 ΔsmeF) | K1449 (ULA-511 ΔL1/ΔL2) | K1766 (K1449 ΔsmeE) | K1795 (K1449 ΔsmeF) | K1385 (ULA-511 MDR) | K1767 (K1385 ΔsmeE) | K1794 (K1385 ΔsmeF) | K1439 (ULA-511 MDR) | K1768 (K1439 ΔsmeE) | K1796 (K1439 ΔsmeF) | |
| Quinolones | ||||||||||||
| Nalidixic acid | 16 | 4 | 4 | 8 | 4 | 4 | 64 | 64 | 4 | 64 | 64 | 64 |
| Norfloxacin | 16 | 4 | 8 | 16 | 4 | 4 | 128 | 128 | 16 | 128 | 32 | 64 |
| Ciprofloxacin | 8 | 2 | 2 | 4 | 1 | 1 | 32 | 32 | 4 | 64 | 32 | 32 |
| BAYy3118 | 0.06 | 0.015 | 0.015 | 0.06 | 0.015 | 0.015 | 1 | 0.5 | 0.06 | 1 | 1 | 1 |
| Clinafloxacin | 0.125 | 0.032 | 0.064 | 0.125 | 0.032 | 0.032 | 1 | 0.5 | 0.125 | 2 | 2 | 2 |
| Gemifloxacin | 1 | 0.25 | 0.5 | 1 | 0.25 | 0.25 | 8 | 4 | 1 | 8 | 8 | 8 |
| Moxifloxacin | 0.5 | 0.125 | 0.25 | 0.5 | 0.06 | 0.125 | 4 | 2 | 1 | 8 | 8 | 8 |
| Trovafloxacin | 0.25 | 0.125 | 0.25 | 0.25 | 0.125 | 0.125 | 8 | 8 | 0.5 | 8 | 8 | 8 |
| Tetracyclines | ||||||||||||
| Tetracycline | 8 | 2 | 4 | 8 | 2 | 2 | 16 | 16 | 4 | 8 | 4 | 4 |
| Doxycycline | 0.5 | 0.125 | 0.25 | 0.5 | 0.25 | 0.25 | 4 | 4 | 0.5 | 2 | 2 | 2 |
| Minocycline | 0.125 | 0.06 | 0.06 | 0.125 | 0.06 | 0.06 | 1 | 1 | 0.25 | 0.25 | 0.25 | 0.125 |
| Tigilcycline | 0.5 | 0.125 | 0.25 | 0.5 | 0.125 | 0.25 | 2 | 2 | 0.25 | 0.5 | 0.25 | 0.125 |
| Macrolides | ||||||||||||
| Erythromycin | 512 | 128 | 256 | 512 | 128 | 128 | 1,024 | 1,024 | 512 | 512 | 128 | 128 |
| Azithromycin | 256 | 128 | 128 | 128 | 128 | 128 | 512 | 512 | 256 | 128 | 64 | 64 |
| β-Lactams | ||||||||||||
| Penicillin G | NDb | ND | ND | 1 | 1 | 0.5 | ND | ND | ND | ND | ND | ND |
| Ampicillin | ND | ND | ND | 2 | 2 | 0.5 | ND | ND | ND | ND | ND | ND |
| Carbenicillin | ND | ND | ND | 1 | 0.5 | 0.5 | ND | ND | ND | ND | ND | ND |
| Piperacillin | ND | ND | ND | 2 | 1 | 1 | ND | ND | ND | ND | ND | ND |
| Cefuslodin | ND | ND | ND | 2 | 2 | 1 | ND | ND | ND | ND | ND | ND |
| Cefotaxime | ND | ND | ND | 1 | 1 | 0.5 | ND | ND | ND | ND | ND | ND |
| Cefoperazone | ND | ND | ND | 2 | 1 | 1 | ND | ND | ND | ND | ND | ND |
| Cefepime | ND | ND | ND | 0.5 | 0.25 | 0.25 | ND | ND | ND | ND | ND | ND |
| Cefpirome | ND | ND | ND | 0.5 | 0.25 | 0.25 | ND | ND | ND | ND | ND | ND |
| Imipenem | ND | ND | ND | 0.25 | 0.25 | 0.25 | ND | ND | ND | ND | ND | ND |
| Meropenem | ND | ND | ND | 0.125 | 0.06 | 0.06 | ND | ND | ND | ND | ND | ND |
| Pirazmonam | ND | ND | ND | 0.5 | 0.5 | 0.5 | ND | ND | ND | ND | ND | ND |
| Other antibiotics | ||||||||||||
| Chloramphenicol | 8 | 4 | 4 | 8 | 4 | 4 | 64 | 64 | 16 | 64 | 64 | 64 |
| Kanamycin | 1,024 | 1,024 | 1,024 | 1,024 | 1,024 | 512 | 256 | 256 | 128 | 1,024 | 1,024 | 1,024 |
| Novobiocin | 2,560 | 1,280 | 1,280 | 2,560 | 1,280 | 1,280 | 5,120 | 5,120 | 2,560 | 5,120 | 5,120 | 2,560 |
| Rifampin | 8 | 8 | 8 | 8 | 8 | 8 | 16 | 16 | 8 | 16 | 16 | 16 |
| D-Cycloserine | 512 | 512 | 512 | 512 | 512 | 256 | 512 | 512 | 256 | 1,024 | 1,024 | 512 |
| Trimethoprim | 16 | 16 | 8 | 16 | 16 | 8 | 128 | 128 | 64 | 128 | 128 | 128 |
| Toxicants | ||||||||||||
| SDS | 3,200 | 200 | 400 | 3,200 | 200 | 400 | 3,200 | 3,200 | 400 | 3,200 | 1,600 | 800 |
| Crystal violet | 16 | 8 | 8 | 16 | 4 | 4 | 32 | 32 | 16 | 32 | 32 | 16 |
| Acriflavine | 256 | 64 | 128 | 256 | 32 | 32 | >256 | >256 | 256 | 256 | 128 | 256 |
| Proflavine | >256 | 64 | 128 | >256 | 64 | 128 | >256 | >256 | 256 | >256 | 256 | >256 |
| Ethidium bromide | 512 | 64 | 128 | 512 | 64 | 64 | 1,024 | 512 | 256 | 1,024 | 512 | 512 |
MICs for isogenic pairs of SmeE+ and SmeE− strains are shown. Relevant phenotypes are highlighted below strains.
ND, not determined due to the production of L1 and L2 β-lactamases in these strains.
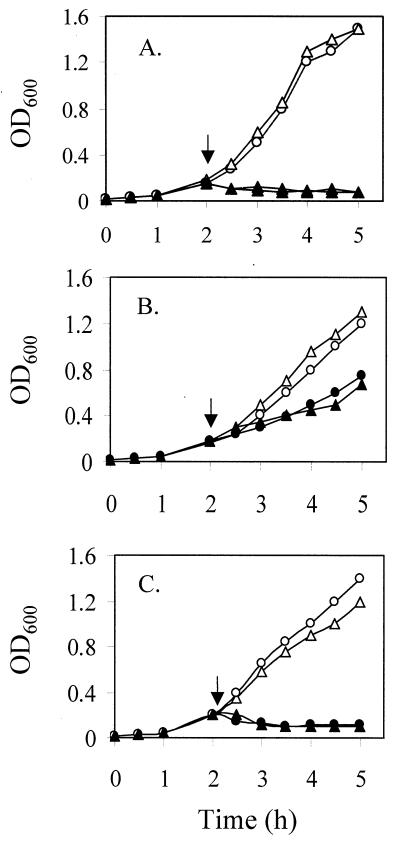
By using the ΔsmeE and ΔsmeF mutants, the substrate range of SmeDEF was also assessed with nonantibiotic, toxic compounds, including dyes, detergents, and organic solvents, given the contribution of related efflux systems to, e.g., solvent tolerance in P. aeruginosa (16, 18) and E. coli (33). Again, the sme deletion mutants displayed enhanced susceptibility to several agents, including SDS, crystal violet, acriflavine, proflavine, and ethidium bromide (4- to 16-fold decrease in MICs) (Table 2), although no detectable change in tolerance to n-hexane on LB agar plates was observed (see Fig. 4A; data not shown). Thus, SmeDEF appears to accommodate dyes and detergents (not previously tested) but is unable to export solvents. These data highlight the broad substrate specificity of the SmeDEF system, which, like other multidrug efflux systems in gram-negative bacteria (26), accommodates a wide range of antimicrobials.
FIG. 4.
Effect of smeE deletion on organic solvent tolerance of S. maltophilia ULA-511 and its MDR mutants K1385 and K1439. Cells of ULA-511 (▵, ▴) and K1765 (ULA-511 ΔsmeE; ○, ●) (A), K1385 (▵, ▴) and K1767 (K1385 ΔsmeE; ○, ●) (B), and K1439 (▵, ▴) and K1768 (K1439 ΔsmeE; ○, ●) (C) were grown in LB broth at 37°C to the early exponential phase, at which time (arrow) n-hexane at 2% (vol/vol) was added (▴, ●) and growth was determined by monitoring OD600. Control cultures (▵, ○) received no supplementation.
Hyperexpression of SmeDEF in mutants with acquired multidrug resistance.
By using the available smeDEF-specific primers, RT-PCR was employed to assess the expression levels of smeDEF in two previously reported MDR strains of S. maltophilia, K1385 and K1439 (34). MDR strain K1385 (but not K1439) clearly hyperexpressed the smeDEF genes (Fig. 1A to C, compare lanes 5 to 7 to lanes 2 to 4), although, in the case of the smeD gene, this was most evident when the lowest concentration of RNA as template in RT-PCR was used (compare, lanes 4 and 7 in Fig. 1A). Intriguingly, however, deletion of smeE in this strain failed to compromise the increased multidrug resistance of this mutant (K1767 in Table 2). In contrast, loss of smeF markedly enhanced antimicrobial susceptibility in this MDR strain, specifically to SmeDEF substrate antimicrobials (K1794 in Table 2). This is reminiscent of MDR mutants hyperexpressing SmeABC, where elimination of SmeC (a SmeF homologue) but not of SmeB (a SmeE homologue) also compromised the enhanced multidrug resistance of this mutant (Li et al., submitted). In the previous instance, it was suggested that SmeC was functioning independently of SmeAB as the outer membrane constituent of an unidentified multidrug efflux system (i.e., SmeABC itself was not a multidrug efflux system) that was responsible for the multidrug resistance of the mutant. Still, it was also possible that SmeABC itself functioned as a multidrug transporter but that, upon loss of SmeB, SmeC could function with yet another efflux system to maintain the resistance of the original SmeABC-hyperexpressing mutant. Here then, too, it may be that SmeDEF hyperexpression is responsible for the multidrug resistance of K1385 but that, upon elimination of SmeE, SmeF then functions with yet another efflux system that maintains the resistance of this mutant. Still, it is also possible that another efflux system dependent upon SmeF is responsible for the multidrug resistance of K1385 and that SmeDEF hyperexpression serves only to provide sufficient SmeF for this other system. In any case, the smeE deletion is clearly present in the K1385 strain (K1767 in Fig. 2, lane 6), and, thus, the retained multidrug resistance of K1767 is not attributable to SmeDEF. As expected, deletion of smeE or smeF had no impact on the resistance profile of MDR strain K1439 (K1768 and K1796 in Table 2), which apparently hyperexpresses an as-yet-unidentified multidrug efflux system.
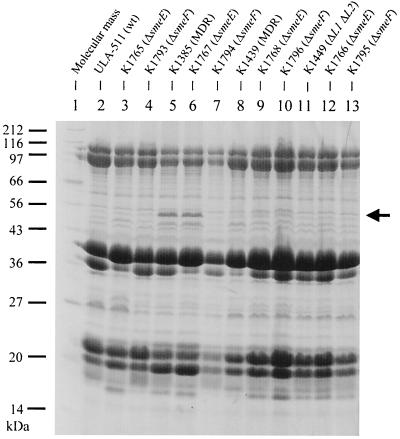
MDR strain K1385 was previously shown to hyperproduce an outer membrane protein that was designated SmeM (2) (Fig. 3, compare lanes 2 and 5). In light of data presented here showing that K1385 hyperexpresses smeDEF, however, it is likely that this protein is, in fact, SmeF. Indeed, introduction of a smeF deletion into K1385 eliminated this protein (Fig. 3, lane 7), although an in-frame deletion of smeE did not (Fig. 3, lane 6). Previously, comparison of the available amino acid sequences of SmeM (derived from amino acid sequencing of a CNBr-generated peptide) and SmeF (deduced from the nucleotide sequence of the corresponding gene) revealed two differences out of a 24-amino-acid stretch (3, 34). This variation may be due to strain differences (the partial SmeM amino acid sequence was derived from strain ULA-511 [34], while the deduced SmeF sequence came from strain D457 [3]). Indeed, S. maltophilia displays great strain-to-strain variation (10), as seen, for example, with the L1 and L2 β-lactamases, which demonstrate substantial variation in amino acid sequences (4, 31). Again, as expected, deletion of smeE or smeF had no impact on the outer membrane profiles of the MDR strain K1439 (Fig. 3, compare lanes 9 and 10 with lane 8).
FIG. 3.
Outer membrane protein profiles of smeE and smeF deletion derivatives of wild-type and MDR S. maltophilia. Strain designations are provided on top and molecular mass markers on the left. The SmeM protein is indicated with an arrow.
Intriguingly, while solvent tolerance in K1385, as in ULA-511 (see above), was not affected by the smeE deletion (Fig. 4B), overexpression of SmeM itself in K1385 was associated with enhanced solvent tolerance relative to the ULA-511 parental strain, as assayed in LB broth (compare Fig. 4A and B) or on LB agar plates (data not shown). Elimination of smeF in K1385, however, did have a modest impact on solvent tolerance as seen on LB agar plates (data not shown). Still, as with the smeE deletion in ULA-511, deletion of smeF in this wild-type strain did not have any influence on solvent susceptibility (data not shown). Thus, while SmeDEF may not accommodate solvents, some system associated with SmeF apparently does. Finally, in contrast to K1385, MDR strain K1439 failed to display any enhancement in solvent tolerance relative to its parent (Fig. 4C; data not shown), indicating that whatever efflux system that might be expressed in this mutant does not accommodate solvents.
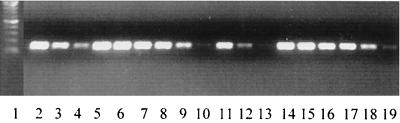
A previous report highlighted growth phase regulation of smeDEF expression, although, owing to a paucity of detectable (with Northern blotting) smeDEF-specific transcripts, this was very obvious only for an MDR mutant hyperexpressing SmeDEF, where expression was maximal in early log phase and undetectable in stationary phase (3). To assess the influence of growth on smeDEF expression in wild-type cells then, RT-PCR with smeD-specific primers (which readily identified message in wild-type S. maltophilia [Fig. 1A]) was used to examine smeD message (as a measure of smeDEF expression) in log-phase (Fig. 5, lanes 2 to 4) and stationary-phase (Fig. 5 lanes 11 to 13) cells of ULA-511. As indicated in the previous report, expression of smeDEF does decline in stationary-versus log-phase cells (Fig. 5, compare lanes 11 to 13 with lanes 2 to 4), though it is certainly detectable in stationary-phase cells (Fig. 5, lanes 11 and 12), in contrast to the previous report (3). A decline in smeD message in the stationary phase, however, is not evident in the SmeDEF-hyperexpressing strain K1385 (Fig. 5, compare lanes 14 to 16 with lanes 5 to 7), and substantial message is detectable in the stationary phase (Fig. 5, lanes 14 to 16), again in contrast to the previous report (3). A recent report on clinical strains of S. maltophilia overproducing SmeDEF did, however, note one mutant which also expressed smeD (using RT-PCR) at high level in both the log and stationary phases (3a). Still, most of the SmeDEF-producing clinical strains did show a marked decline in smeD message with growth, being undetectable at stationary phase (3a). Nonetheless, these clinical isolates undoubtedly carried mutations responsible for smeDEF overexpression, and it is unclear what impact these might have on growth phase regulation. It is certainly clear from the data presented here that smeDEF is expressed in the stationary phase in wild-type cells, and one can only surmise that the earlier Northern blot experiments were insufficiently sensitive to detect this. That the more recent study by the same authors using RT-PCR also failed to detect smeDEF message in apparently wild-type S. maltophilia (3a) is puzzling and is possibly explained by a preexisting mutation that compromised smeDEF expression. Interestingly, smeD levels do not appear to be affected by the growth phase in MDR strain K1439 (which does not hyperexpress SmeDEF) (Fig. 5, compare lanes 17 to 19 with lanes 8 to 10). This may result from some impact of the MDR determinant of this mutant on smeDEF expression.
FIG. 5.
Growth phase influence on expression of smeD. RNA was isolated from log-phase (lanes 2 to 10) and stationary-phase (lanes 11 to 19) cultures of S. maltophilia ULA-511 (lanes 2 and 11, 0.5 μg of RNA amplified; lanes 3 and 12, 0.05 μg of RNA; and lanes 4 and 13, 0.005 μg of RNA), K1385 (lanes 5 and 14, 0.5 μg of RNA amplified; lanes 6 and 15, 0.05 μg of RNA; and lanes 7 and 16, 0.005 μg of RNA), and K1439 (lanes 8 and 17, 0.5 μg of RNA amplified; lanes 9 and 18, 0.05 μg of RNA; and lanes 10 and 19, 0.005 μg of RNA). Lane 1, molecular weight markers (100-bp ladder).
ACKNOWLEDGMENTS
This work was supported by funding from the Canadian Bacterial Diseases Network (one of the Networks of Centers of Excellence). K.P. is a Canadian Cystic Fibrosis Foundation Scholar.
REFERENCES
- 1.Aires J R, Kohler T, Nikaido H, Plesiat P. Involvement of an active efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob Agents Chemother. 1999;43:2624–2628. doi: 10.1128/aac.43.11.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alonso A, Martinez J L. Multiple antibiotic resistance in Stenotrophomonas maltophilia. Antimicrob Agents Chemother. 1997;41:1140–1142. doi: 10.1128/aac.41.5.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alonso A, Martinez J L. Cloning and characterization of SmeDEF, a novel multidrug efflux pump from Stenotrophomonas maltophilia. Antimicrob Agents Chemother. 2000;44:3079–3086. doi: 10.1128/aac.44.11.3079-3086.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Alonso A, Martinez J L. Expression of multidrug efflux pump SmeDEF by clinical isolates of Stenotrophomonas maltophilia. Antimicrob Agents Chemother. 2001;45:1879–1881. doi: 10.1128/AAC.45.6.1879-1881.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avison M B, Higgins C S, von Heldreich C J, Bennett P M, Walsh T R. Plasmid location and molecular heterogeneity of the L1 and L2 β-lactamase genes of Stenotrophomonas maltophilia. Antimicrob Agents Chemother. 2001;45:413–419. doi: 10.1128/AAC.45.2.413-419.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ballestero S, Virseda I, Escobar H, Suarez L, Baquero F. Stenotrophomonas maltophilia in cystic fibrosis patients. Eur J Clin Microbiol Infect Dis. 1995;14:728–729. doi: 10.1007/BF01690887. [DOI] [PubMed] [Google Scholar]
- 6.Barcak G J, Chandler M S, Redfield R J, Tomb J-F. Genetic systems in Haemophilus influenzae. Methods Enzymol. 1991;204:321–337. doi: 10.1016/0076-6879(91)04016-h. [DOI] [PubMed] [Google Scholar]
- 7.Denton M, Kerr K G. Microbiological and clinical aspects of infections associated with Stenotrophomonas maltophilia. Clin Microbiol Rev. 1988;11:57–80. doi: 10.1128/cmr.11.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denton M, Todd N J, Kerr K G, Hawkey P M, Littlewood J M. Molecular epidemiology of Stenotrophomonas maltophilia isolated from clinical specimens from patients with cystic fibrosis and associated environmental samples. J Clin Microbiol. 1998;36:1953–1958. doi: 10.1128/jcm.36.7.1953-1958.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Felici A, Amicosante G, Oratore A, Strom R, Ledent P, Joris B, Fanuel L, Frere J-M. An overview of the kinetic parameters of class B β-lactamases. Biochem J. 1993;291:151–155. doi: 10.1042/bj2910151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hauben L, Vauterin L, Moore E R, Hoste B, Swings J. Genomic diversity of the genus Stenotrophomonas. Int J Syst Bacteriol. 1999;49:1749–1760. doi: 10.1099/00207713-49-4-1749. [DOI] [PubMed] [Google Scholar]
- 11.Hoang T T, Karkhoff-Schweizer R R, Kutchma A J, Schweizer H P. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene. 1998;212:77–86. doi: 10.1016/s0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 12.King B A, Shannon K P, Phillips I. Aminoglycoside 6′-N acetyltransferase production by an isolate of Pseudomonas maltophilia. J Antimicrob Chemother. 1978;4:467–468. doi: 10.1093/jac/4.5.467-a. [DOI] [PubMed] [Google Scholar]
- 13.Köhler T, Michea-Hamzehpour M, Henze U, Gotoh N, Curty L K, Pechere J-C. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol Microbiol. 1997;23:345–354. doi: 10.1046/j.1365-2958.1997.2281594.x. [DOI] [PubMed] [Google Scholar]
- 14.Lambert T, Ploy M-C, Denis F, Courvalin P. Characterization of the chromosomal aac(6′)-Iz gene of Stenotrophomonas maltophilia. Antimicrob Agents Chemother. 1999;43:2366–2371. doi: 10.1128/aac.43.10.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li X-Z, Nikaido H, Poole K. Role of mexA-mexB-oprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:1948–1953. doi: 10.1128/aac.39.9.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li X-Z, Poole K. Organic solvent-tolerant mutants of Pseudomonas aeruginosa display multiple antibiotic resistance. Can J Microbiol. 1999;45:17–22. doi: 10.1139/cjm-45-1-18. [DOI] [PubMed] [Google Scholar]
- 17.Li X-Z, Poole K. Mutational analysis of the OprM outer membrane component of the MexA-MexB-OprM multidrug efflux system of Pseudomonas aeruginosa. J Bacteriol. 2001;183:12–27. doi: 10.1128/JB.183.1.12-27.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li X-Z, Zhang L, Poole K. Role of the multidrug efflux systems of Pseudomonas aeruginosa in organic solvent tolerance. J Bacteriol. 1998;180:2987–2991. doi: 10.1128/jb.180.11.2987-2991.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marshall W F, Keating M R, Anhalt J P, Steekelberg J M. Xanthomonas maltophilia: an emergng nosocomial pathogen. Mayo Clin Proc. 1989;64:1097–1104. doi: 10.1016/s0025-6196(12)64979-9. [DOI] [PubMed] [Google Scholar]
- 20.Mett H, Rosta S, Schacher B, Frei R. Outer membrane permeability and β-lactamase content in Pseudomonas maltophilia clinical isolates and laboratory mutants. Rev Infect Dis. 1988;10:765–769. doi: 10.1093/clinids/10.4.765. [DOI] [PubMed] [Google Scholar]
- 21.Micozzi A, Venditti M, Monaco M, Friedrich A, Taglietti F, Santilli S, Martino P. Bacteremia due to Stenotrophomonas maltophilia in patients with hematologic malignancies. Clin Infect Dis. 2000;31:705–711. doi: 10.1086/314043. [DOI] [PubMed] [Google Scholar]
- 22.Nikaido H. Prevention of drug access to bacterial targets: role of permeability barrier and active efflux. Science. 1994;264:382–388. doi: 10.1126/science.8153625. [DOI] [PubMed] [Google Scholar]
- 23.Nikaido H. Antibiotic resistance caused by gram-negative multidrug efflux pumps. Clin Infect Dis. 1998;27:S32–S41. doi: 10.1086/514920. [DOI] [PubMed] [Google Scholar]
- 24.Palleroni N J, Bradbury J F. Stenotrophomonas, a new bacterial genus for Xanthomonas maltophilia. Int J Syst Bacteriol. 1993;43:606–609. doi: 10.1099/00207713-43-3-606. [DOI] [PubMed] [Google Scholar]
- 25.Payne D J, Cramp R, Batson J H, Neal J, Knowles D. Rapid identification of metallo- and serine β-lactamases. Antimicrob Agents Chemother. 1994;38:991–996. doi: 10.1128/aac.38.5.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poole K. Efflux-mediated resistance to fluoroquinolones in gram-negative bacteria. Antimicrob Agents Chemother. 2000;44:2233–2241. doi: 10.1128/aac.44.9.2233-2241.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poole K, Heinrichs D E, Neshat S. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J Bacteriol. 1993;175:7363–7372. doi: 10.1128/jb.175.22.7363-7372.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poole K, Gotoh N, Tsujimoto H, Zhao Q, Wada A, Yamasaki T, Neshat S, Yamagishi J-I, Li X-Z, Nishino T. Overexpression of the mexC-mexD-oprJ efflux operon in nfxB-type multidrug-resistant strains of Pseudomonas aeruginosa. Mol Microbiol. 1996;21:713–724. doi: 10.1046/j.1365-2958.1996.281397.x. [DOI] [PubMed] [Google Scholar]
- 29.Saier M H., Jr A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol Mol Biol Rev. 2000;64:354–411. doi: 10.1128/mmbr.64.2.354-411.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 31.Sanschagrin F, Dufresne J, Levesque R C. Molecular heterogeneity of the L-1 metallo-β-lactamase family from Stenotrophomonas maltophilia. Antimicrob Agents Chemother. 1998;42:1245–1248. doi: 10.1128/aac.42.5.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vanhoof R, Sonck P, Hannecart-Pokorni E. The role of lipopolysaccharide anionic binding sites in aminoglycoside uptake in Stenotrophomonas (Xanthomonas) maltophilia. J Antimicrob Chemother. 1995;35:167–171. doi: 10.1093/jac/35.1.167. [DOI] [PubMed] [Google Scholar]
- 33.White D G, Goldman J D, Demple B, Levy S B. Role of the acrAB locus in organic solvent tolerance mediated by expression of marA, soxS, or robA in Escherichia coli. J Bacteriol. 1997;179:6122–6126. doi: 10.1128/jb.179.19.6122-6126.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang L, Li X-Z, Poole K. Multiple antibiotic resistance in Stenotrophomonas maltophilia: involvement of a multidrug efflux system. Antimicrob Agents Chemother. 2000;44:287–293. doi: 10.1128/aac.44.2.287-293.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]