Abstract
Basic neuroscience research employs numerous forms of antibodies as key reagents in diverse applications. While the predominant use of antibodies is as immunolabeling reagents, neuroscientists are making increased use of intracellular antibodies or intrabodies. Intrabodies are recombinant antibodies genetically encoded for expression within neurons. These can be used to target various cargo (fluorescent proteins, reporters, enzymes, etc.) to specific molecules and subcellular domains to report on and manipulate neuronal function with high precision. Intrabodies have the advantages inherent in all genetically encoded recombinant antibodies but represent a distinct subclass in that their structure allows for their expression and function within cells. The high precision afforded by the ability to direct their expression to specific cell types, and the selective binding of intrabodies to targets within these allows intrabodies to offer unique advantages for neuroscience research, given the tremendous molecular, cellular and morphological complexity of brain neurons. Intrabodies expressed within neurons have been used for a variety of purposes in basic neuroscience research. Here I provide a general background to intrabodies and their development, and examples of their emerging utility as valuable basic neuroscience research tools.
Keywords: Intrabody, brain, neuron, scFv, nanobody, FingR, recombinant, plasmid, live cell imaging
1. Introduction
Antibodies play a critical role in many areas of basic neuroscience research. The predominant use of antibodies is as immunolabeling reagents, for example in immunohistochemistry, immunocytochemistry, and immunoblotting (“Western blotting”), in which antibodies have access to their binding site or epitope on their target protein regardless of whether it is inside the cell, on the cell surface, or is a secreted molecule. However, as antibodies cannot cross the plasma membrane, their use is limited to those that bind to secreted proteins or to extracellular domains of cell surface proteins when used in vivo or in in vitro in cellular preparations containing intact cells. Intracellular antibodies or intrabodies are antibodies present in the cytoplasm of cells. Historically, to introduce antibodies into cells conventional IgG antibodies were microinjected into the cytoplasm. Examples of this approach include numerous studies in which antibodies were microinjected into the cytoplasm of neurons in primary culture [e.g., [1–7]], and studies in which antibodies were passively infused/dialyzed into cells with a recording electrode/patch pipet during electrophysiological analyses [e.g., [8–11]]. However, due to the need to microinject or patch onto individual neurons to introduce conventional IgG antibodies into their cytoplasm, studies using this approach were limited to small numbers of neurons in culture. There are numerous reports of conventional IgG antibodies against intracellular proteins being taken up into neurons in vivo and in vitro. These include those against against tau (e.g., [12–14]), α-synuclein (e.g., [15]), TDP-43 (e.g., [16]), and RAN (e.g., [17]). However, this uptake can be relatively inefficient and sporadic, such that researchers are not able to reliably control the levels of cytoplasmic antibody in specific cell types.
A major advance in employing intracellular antibodies in neuroscience research has come from the development of recombinant antibodies. Recombinant antibodies are antibody (immunoglobulin) proteins whose nucleic acid coding regions, or fragments thereof, have been cloned into expression plasmids. Such plasmids are typically used to direct expression of secreted recombinant antibody that can be used as one would use conventional antibodies (e.g., as an immunolabeling reagent for immunohistochemistry, immunocytochemistry, immunoblots, for microinjection/dialysis into neurons, etc.), although recombinant antibodies have numerous advantages over conventional antibodies [18–20]. However, recombinant antibody expression plasmids and/or recombinant viruses can also be engineered to direct expression of the encoded protein as intrabodies in the cytoplasm of neurons. This allows for reliable expression, and when used in conjunction with genetic elements that yield inducible and/or selective expression in specific classes of neurons provides genetically encoded intrabodies substantial advantages for utility as tools for basic neuroscience research.
2. Different forms of intrabodies used in neuroscience research
2.1. Requirements for use of different forms of antibodies as intrabodies
While it is possible to use intact IgG antibodies in approaches employing microinjection or infusion/dialysis of antibodies through a patch pipet to introduce antibodies into neurons, it is not possible to effectively express this form of antibody inside cells as an intrabody. Intact IgG antibodies are formed as a heterotetramer comprising two identical heavy (H) and two identical light (L) chains (Figure 1). Both polypeptide chains contain leader sequences that direct their translation to endoplasmic reticulum (ER)-bound ribosomes and their translocation across the ER membrane into the ER lumen, in which the H and L chains coassemble into the functional heterotetramer. The association of the single H and L chains within each of the two identical heterodimers, as well as the association of these heterodimers with one another to form the intact IgG occur via extensive non-covalent interactions within both their variable and constant domains and via covalent disulfide bonds. Disulfide bonds form and are retained in oxidizing environments, such as the ER lumen and the extracellular environment. They can also be retained in intact IgG molecules introduced into the reducing environment of the cytoplasm via microinjection, etc. However, de novo assembly of cytoplasmically expressed H and L chains into an IgG in the reducing environment of the cytoplasm is inefficient such that this form of antibody cannot typically be used as an intrabody. As such, it is necessary to engineer alternate forms of single chain antibodies for use as intrabodies, two prominent forms being single chain variable fragments or scFvs and nanobodies. While these IgG-derived single chain intrabodies still contain intrachain disulfide bonds, it is possible to develop scFvs and nanobodies that effectively fold and function when expressed cytoplasmically. Alternate forms of intrabodies have been developed from non-antibody binders that lack disulfide bonds and have other attributes that enhance their folding and stability.
Figure 1. Comparison of typical mammalian IgG and camelid heavy-chain only IgG and their derivatives.
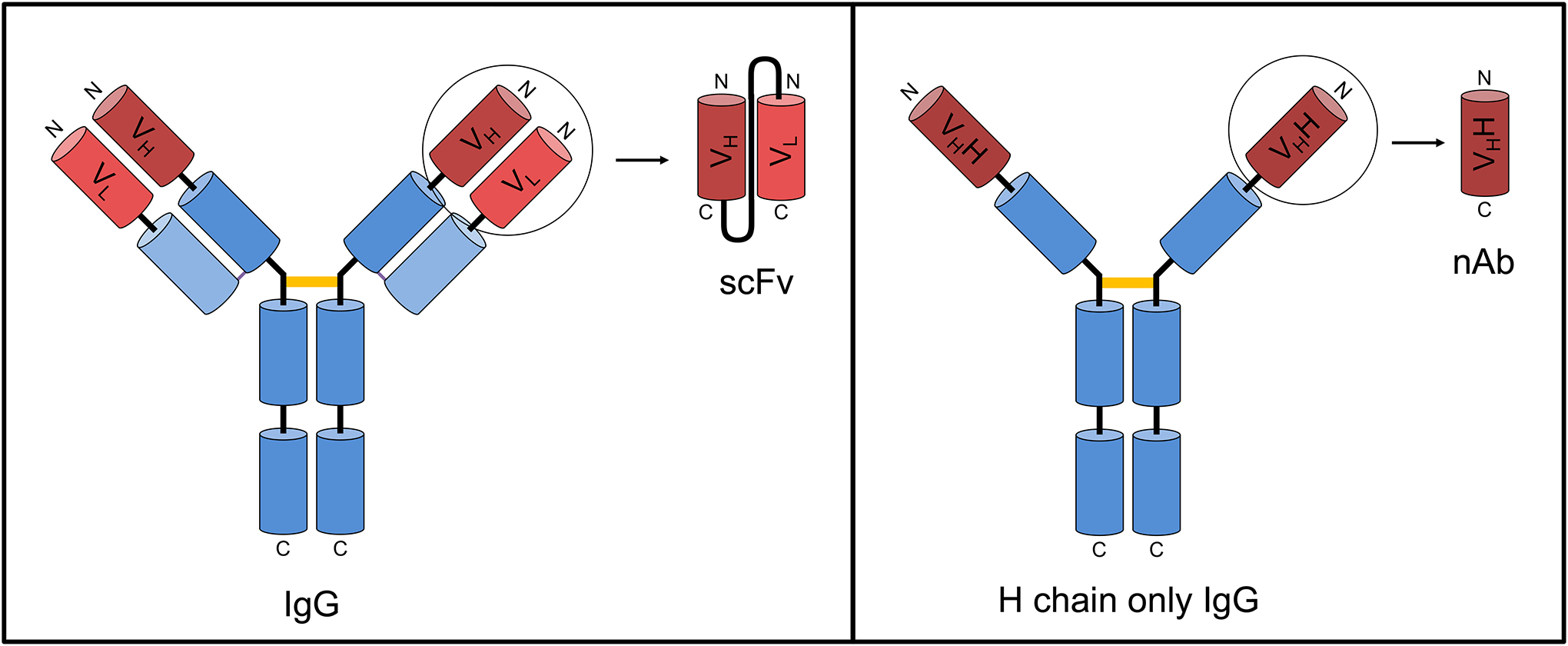
Left. A typical mammalian IgG molecule is a heterotetramer comprising two heavy and two light chains. Both chains contain a variable domain or FV region as shown in red, and one or more constant domains as shown in blue. The antigen binding site or paratope is formed by noncovalent association of the VH and VL domains. The primary region of covalent disulfide bond linkage of the two identical H + L chain heterodimers is shown by an orange bar. Typical mammalian H + L chain IgGs can be miniaturized to various forms including scFvs as shown. Right. Camelid heavy chain-only IgGs lack light chains and exist as a homodimer of two identical H chains. In this case the antigen binding site or paratope is formed by a single VHH domain, which can function autonomously as a nAb. The primary region of covalent disulfide bond linkage of the two identical H chain monomers is shown by an orange bar.
2.2. Single chain variable fragments (scFvs)
Single chain variable fragments or scFvs are a widely used form of single chain polypeptide recombinant antibody fragments. Derived from conventional IgGs, scFvs are monomeric and generated by tethering a VH and VL region to one another to generate using a flexible linker [21, 22]. Since their development in the 1980s, scFvs have had widespread use, primarily as therapeutics and as probes for diagnostic imaging. The small size of scFvs facilitates their penetration into tissue, which enhances their efficacy in these applications. They have also been used widely as intrabodies, the latter term being coined to describe intracellularly expressed scFvs [23]. The conceptual design of scFvs is to replicate in a single polypeptide the antigen binding surface formed upon the coassembly of the VH and VL regions of the separate H and L chains of an intact IgG (Figure 1, left). This concept is complicated in that the VH and VL regions are aligned with their respective free N-termini and C-termini in parallel (Figure 1, left). This is not possible to replicate in a single polypeptide format, a problem that the design attempts to circumvent by using a flexible linker, of sufficient length to allow for V regions to align in parallel when the C-terminus of the leading V region is fused to the N-terminus of the lagging V region to create a scFv (Figure 1, left). This format results in N-terminus of the leading V region being free as it is in the intact IgG, but that of the lagging V region fused to the C-terminus of the flexible linker sequence. In some cases, this unnatural configuration leads to reduced efficiency of folding and coassembly of the leading and lagging V regions, resulting in misfolded and/or non-functional scFvs. There is an extensive literature on these and other challenges associated with the successful generation of scFvs using V regions derived from IgGs, and numerous strategies for overcoming these problems should they occur [e.g., [24–28] and many others]. The extensive research in this area has led to basic design guidelines that can often yield the generation of a functional scFv that recapitulates the utility of the IgG from which the V region sequences were derived without further engineering. We have employed as a first pass a design employing a VH-linker-VL orientation and a flexible (GGGGS)4 linker, followed by expression in mammalian cells, regardless of the primary sequence characteristics of the component VH and VL regions. While our experience is limited, to date we have had an ≈60% (16/27) first pass success rate using this approach to convert mAbs into VH-linker-VL form of scFvs without any further engineering.
2.3. Camelid VHH domain fragments or Nanobodies (nAbs)
Among mammals, camelids (llamas, alpacas, dromedaries, etc.) are unique in producing as part of their humoral immune repertoire fully functional IgG antibodies that contain only a heavy chain [29] (Figure 1, right). It is possible to use the variable or VHH domain from these heavy chain only antibodies, termed a “nanobody” (nAb), as the smallest form of a functional antibody fragment (≈15 kD), which is ≈1/10 the size of a conventional IgG antibody. nAbs have numerous advantages over conventional IgG antibodies for use as intrabodies. Unlike scFvs, the VHH domain is synthesized, folds and functions as a single polypeptide chain, such that they are typically more easy to produce and are more stable than scFvs, making them valuable for use as intrabodies when expressed in mammalian cells including neurons.
2.4. Antibody mimetics-FingRs
There are numerous forms of recombinant antibody mimetics that can be developed to exhibit high affinity and specific binding to their cognate target protein comparable to antibodies. These genetically encoded molecules typically comprise a stable backbone scaffold upon which is built a huge variety of flexible binding surface sequences to generate high complexity libraries from which target-specific binders can be isolated. Monobodies are based on a backbone of stable fibronectin repeats [30]. Those developed for use in neuroscience research were isolated through mRNA display, hence their name FingRs (for Fibronectin intrabodies generated with mRNA display) and were subsequently extensively validated for efficacy and specificity as intrabodies [31, 32]. One aspect of their design that enhances their utility as intrabodies as they were also engineered to be expressed with an innovative transcriptional control system to limit FingR expression levels to those which saturate target protein binding, with any unbound FingRs acting as transcriptional repressors to inhibit their further expression, preventing accumulation of unbound intrabody.
3. Applications of intrabodies in neuroscience research
The use of intrabodies in neuroscience research is relatively recent. Intrabodies offer unique advantages in neuroscience research in being genetically encoded reagents whose expression can be directed to specific cell types, within which target binding can yield accumulation at specific sites. Intrabodies have gained their most substantial use in primary cultures of brain neurons, which can be used to study many aspects of neuronal function in a system readily accessible to introduction of genetically-encoded tools such as intrabodies by transfection, and to other manipulations [33]. While less commonly used than dissociated neuronal cultures, organotypic slice cultures can be made from different brain regions and are also accessible to introduction of plasmid DNA by conventional or biolistic transfection or by electroporation, or by introduction of recombinant viral vectors, and offer enhanced preservation of cytoarchitecture and cell populations [34]. Intrabodies can also be introduced into brain neurons in vivo by in utero electroporation [35, 36]. In this method, plasmid DNA is injected into the ventricular system of fetal mice or rats within a surgically exposed uterus. A series of electric shocks are used to transiently destabilize cell membranes, allowing for uptake of the plasmid DNA. The uterus is returned to the abdominal cavity and after completion of in utero development and birth the pups are used in experiments at different ages, as dictated by the nature of the experiment. While it is possible to obtain highly reliable expression of plasmid encoded proteins including intrabodies in cerebral cortex by this method, reliable expression in other brain regions is more difficult, although technical developments continue to expand the utility of this technique [e.g., [37, 38]]. Viral-based delivery systems offer advantages of broader and more efficient expression [39] but with the additional requirement to develop recombinant viruses for each intrabody. Each of these experimental systems has been used to express intrabodies in brain neurons, allowing for novel and valuable insights into distinct aspects of neuronal structure and function. However, it remains that the use of intrabodies in certain areas of neuroscience research remains a challenge due to the relative inaccessibility of cells within the brain, and the need to keep the structure of the brain intact for studies at the level of the complex circuits that underlie brain function.
3.1. Examples of uses of scFvs as intrabodies
scFvs employed as genetically encoded intrabodies have played a key role in numerous basic neuroscience research studies. One application of scFv intrabodies has been to knockdown protein expression. scFvs against the inhibitory synapse cytoplasmic scaffolding protein gephyrin were tagged with a nuclear localization signal and when expressed as intrabodies in cultured hippocampal neurons led to loss of endogenous gephyrin expression at inhibitory synapses [40]. This led to a reduction in the amplitude of whole cell ionic currents arising from inhibitory glycine receptors, consistent with a corresponding knockdown of these gephyrin-associated receptors at inhibitory synapses. However, the amplitude excitatory currents from AMPA-type glutamate receptors, with which gephyrin does not associate were unaffected. This supported the utility of these scFv intrabodies to selectively knockdown inhibitory synaptic function [40]. These same scFvs were subsequently used to define the role of gephyrin to support distinct aspects of inhibitory synaptic signaling as mediated through gephyrin-associated GABAA receptors [41]. scFv-mediated gephyrin knockdown in cultured rat hippocampal neurons led to a reduction in synaptic GABAA receptor immunolabeling, and a decrease in the overall number of GABAergic synapses as revealed by immunolabeling for the versicular GABA transporter VGAT, a marker of inhibitory presynaptic terminals. Electrophysiological analyses showed associated changes in the functional characteristics of GABAA receptor-mediated synaptic currents. Lastly, these studies revealed that gephyrin, in its role as an organizer of GABAA receptors at inhibitory synapses, supports both phasic and tonic forms of GABAergic inhibition [41]. Studies employing these same scFv intrabodies reveled that the impact of gephyrin-knockdown could be rescued by neuroligin 2, a gephyrin binding protein that mediates transynaptic adhesion between pre- and post-synaptic membranes and development and maintenance of GABAergic synapses [42].
scFvs have also been expressed as intrabodies in neuronal cells to examine the impact of knocking down expression of the neurotrophin receptor p75NTR [43]. The scFvs were not used as intrabodies in the classical sense, as they were developed to bind to extracellular domains of the neurotrophin receptor. These intrabodies contained an N-terminal leader sequence such that they were translated from ER-bound ribosomes and translocated into the lumen of the ER, as would occur for a secreted scFv. However, these scFvs also contained a C-terminal KDEL ER retrieval sequence, such that they were not secreted but accumulated in the ER. When expressed in PC12 cells and in mouse motor neuron neuroblastoma NSC19 cells they reduced cell surface expression of P75NTR, presumably by preventing receptor trafficking to the cell surface. Importantly, the knockdown effect could be maintained up to eight days without activating the ER unfolded protein response, which can occur from accumulation of unfolded proteins in the lumen of the ER. Expression of these scFv intrabodies in PC12 cells also inhibited NGF-induced neurite outgrowth, reinforcing the important role of p75NTR in mediating signaling by this important neuronal growth factor [43].
scFvs expressed as intrabodies have also been used to track the dynamic of proteins in neuronal cells. One study employed an scFv developed from the widely used conventional mAb 12CA5 that recognizes a defined epitope on the influenza hemagglutinin protein [44] that is now routinely used as the HA epitope tag. The published sequences of the 12CA5 VH and VL domains were used to generate a conventional VH-linker-VL scFv, but this failed to fold and function as an intrabody [45]. A chimeric anti-HA scFv was subsequently engineered by introducing the crucial sequences from 12CA5 onto a stable scFv scaffold yielding a functional intrabody the authors termed a Frankenbody or FB [46]. A FB-GFP fusion was expressed as an intrabody in neurons expressing the HA-tagged plasma membrane ion channel Kv2.1, which is highly clustered in neurons [47], FB-GFP yielded highly specific labeling of plasma membrane Kv2.1 clusters that could be maintained up to 8 days posttransfection. The FB scFv intrabody coupled to a photoactivatable probe was an effective reporter for single particle tracking of HA-tagged proteins, including newly synthesized proteins with HA tags at their N-termini. This allowed for real-time imaging of sites of local translation that occurs in dendrites of neurons, sites which were found to have much higher mobility than sites of translation imaged in a non-neuronal cell line [46]. A set of Frankenbody scFv plasmids with various FP tags for expression in mammalian cells as intrabodies are available from the open-source plasmid repository Addgene.
An scFv intrabody was also used to define the activity-dependent palmitoylation of the important synaptic scaffolding protein PSD-95 in neurons. A palmitoylation-state specific scFv fused to GFP was found to be an effective reporter of the palmitoylated form of PSD-95 when expressed as an intrabody in living neurons [48]. Superresolution STED live cell imaging of this reporter revealed that palmitoylated PSD-95 was localized in specific nanodomains within the postsynaptic density (Figure 2A). Moreover, PSD-95 palmitoylation at these sites was dynamic and regulated by neuronal activity, contributing to the activity-dependent plasticity in synaptic structure that underlies dynamic changes in excitatory synapse function [48]. This same scFv intrabody was subsequently used to show that the palmitoylated form of PSD-95 colocalizes in synaptic nanodomains with both the AMPA and NMDA subtypes of synaptic glutamate receptors, and how changes in the expression of enzymes impacting palmitoylation state modulated PSD-95 palmitoylation and localization [49]. These studies underscore the utility of using state-specific scFvs as intrabodies to gain insights into specific subsets of target proteins that could not otherwise be visualized in live neurons.
Figure 2. Examples of intrabodies in neurons.
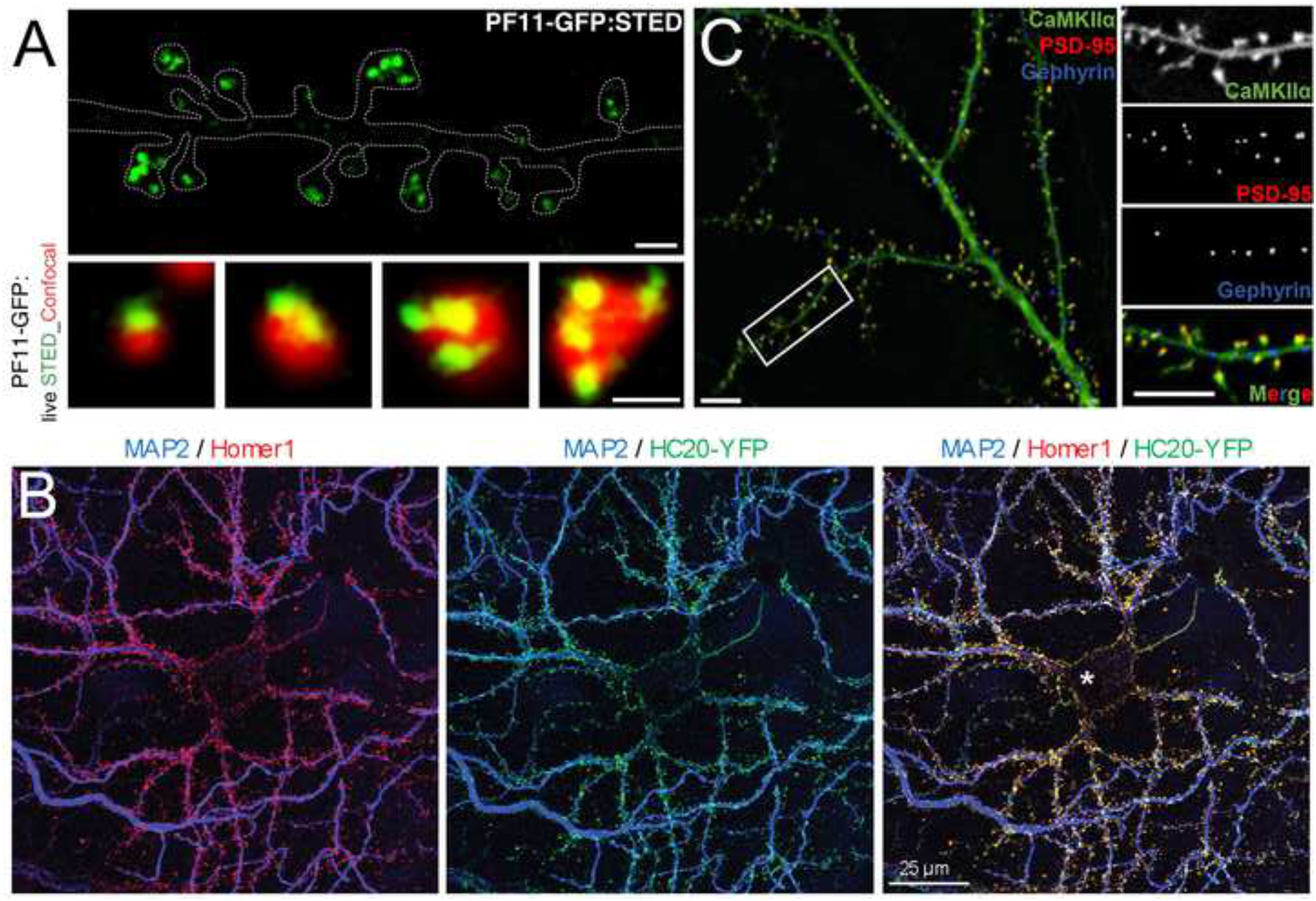
A. scFv intrabody against palmitoylated PSD-95. Live cell superresolution imaging (green) and conventional imaging (red) of PF11-GFP intrabody localization in cultured hippocampal neurons showing nanodomains of palmitoylated PSD-95 within dendritic spines visible with superresolution imaging (see outline of dendrite in top panel). From reference [42]. B. nAb intrabody against Homer1. Confocal imaging of cultured hippocampal neurons showing precise localization of the nAb-YFP intrabody (green) nine days after infection with nAb-encoding lentivirus, fixed and immunolabeled for Homer 1 (red) and the dendritic marker MAP2. From reference [60]. C. FingRs against PSD-95, gephyrin and CamKIIα. Simultaneous multiplex confocal imaging of cultured hippocampal neurons showing localization of FingRs against PSD-95 (red), Gephyrin (blue) and CamKIIα (green), demonstrating the utility of FingRs to visualize the precise localization of these endogenous proteins. Used with permission from reference [69].
3.2. Examples of using nanobodies as intrabodies
3.2.1. Examples of using anti-GFP nanobodies as intrabodies
Anti-GFP nAbs have been used as intrabodies for a variety of purposes in neuroscience research, ranging from circuit-specific transcriptional profiling, cell-specific manipulation of gene expression, to reporting on specific neuronal subcellular compartments. Anti-GFP nAbs have been used in in an innovative approach to selectively capture actively translated transcripts in brain neurons [50]. Transgenic mice that express an anti-GFP nAb [51] fused to a ribosomal protein (Rpl10a) were developed. Using a retrogradely transported virus microinjected into specific brain regions, GFP was expressed in brain neurons in a circuit-specific manner. Beads coated with an anti-GFP mAb that binds to an epitope distinct from the nAb was used to capture ribosomes with nAb-bound GFP. These represent ribosomes from neurons whose axons project to the brain region into which the retrograde virus was microinjected. Identification of transcripts that were being actively translated on these captured ribosomes allowed for a determination of circuit-specific gene expression in different brain regions. In a subsequent study, this innovative intrabody-based approach was used to define genes expressed in a distinct subset of midbrain neurons that participate in reward circuitry [52].
The transgenic mice expressing an anti-GFP nAb fused to the Rpl10a ribosomal protein that were used in the initial intrabody-based ribosome profiling study [50] were subsequently employed in studies showing it is possible to use this intrabody to enhance the fluorescent signal in GFP expressing neurons [53]. The binding of certain anti-GFP nAbs had been previously shown to enhance GFP fluorescence in vitro [54, 55]. Expressing GFP in mouse brain neurons mice expressing the nAb-Rpl10a intrabody led to enhanced GFP fluorescence in vivo. Moreover, instead of being broadly localized in expressing neurons, including throughout the axon and the extensive dendritic arbor, the GFP was highly concentrated in the cell body through its capture by the ribosome localized anti-GFP intrabody. This strategy, or others employing intrabodies specific for soma-localized targets, could be used as an alternate approach to those employing trafficking signals to localize and concentrate proteins such as optogenetic reporters and actuators to the soma to enhance their efficacy and utility [e.g., [56–58]], which have the potential drawback of competing with endogenous proteins for access to trafficking machinery.
Previously developed anti-GFP nAbs [54] were also employed as intrabodies in an innovative strategy to manipulate gene expression [59]. Many transcription factors are composed of separable autonomous DNA binding and activation domains, which when brought together reconstitute transcription factor activity, the basis of the yeast two hybrid system [60]). A GFP-dependent transcriptional activation system was developed employing two distinct anti-GFP nAbs, each fused to a different transcription factor domain, whereby their simultaneous binding results in transcription factor activation. This was used to manipulate expression of both endogenous genes and optogenetic tools such as channelrhodopsin-2 in a cell-specific manner in mouse retina and in brain. The authors extended this technique to then generate mutations in nAbs that lead to their conditional destabilization in the absence of target binding [61], leading to selective accumulation and function of nAb-cargo fusions in only those cells that express the target. Destabilized nAbs against multiple target proteins could be used to manipulate gene expression in diverse cell types including neurons in mouse brain. In theory it should be possible to tap into the growing collection of publicly available nAb-based intrabodies directed against neuronal targets with restricted cellular expression to manipulate gene expression in distinct types of brain neurons.
Anti-GFP nAbs employed as intrabodies have been used as reporters of neuronal plasma membrane domains enriched in lipid signaling molecules. Anti-GFP nAbs tagged with mCherry were used to target a GFP-tagged protein containing the pleckstrin homology (PH) domain of the δ isoform of phospholipase C (PH-PLCδ), which exhibits selective high affinity binding to the important lipid signaling molecule PtdIns(4,5)P2 or PIP2. When coexpressed in neuronal PC12 cells, the mCherry-tagged nAb intrabody colocalized with the PH-PLCδ reporter at the inner face of the plasma membrane [62]. The authors then replaced the mCherry on the nAb with the photoconvertible protein mEos2, which can be stochastically photoconverted from green to red emission to allow for its use in single particle imaging. This study also employed nAbs against endogenous proteins in PC12 cells in a similar manner.
3.2.2. Examples of using nanobodies against endogenous brain targets as intrabodies
Nanobodies have been employed to knockdown cell surface expression of endogenous neuronal proteins. A nAb against the cytoplasmic Cavβ auxiliary subunit of voltage-activated Ca2+ channels was fused to a ubiquitin E3-ligase and used to selectively knockdown plasma membrane expression and function of the subset of voltage-gated Ca2+ channels containing Cavβ subunits in dorsal route ganglion neurons, as well as cardiomyocytes and pancreatic beta cells [63]. This study underscores the utility of employing intrabodies as effective mechanism to deliver cargo such as functional enzymes to specific molecular and/or subcellular targets to impact cell function.
Nanobodies have been used to track single particle dynamics of endogenous GPCRs in neuronal cells. Conformationally-specific nAbs against the active and inactive states of the β2-adrenergic receptor (β2-AR) and the guanine nucleotide-free form of the αs G protein subunit were used previously to track different states of receptors in living cells [64, 65]. They were subsequently used to track single-particle dynamics of exogenous and endogenous β2-ARs in neuronal PC12 cells to interrogate the distinct state-specific dynamics of these important components of the GPCR signalosome at a level of resolution not possible by studying the FP-tagged proteins themselves [62]. This includes identifying distinct highly immobile states of activated β2-ARs that form transient nanoclusters and that could represent signaling platforms for binding to other components of the β2-AR signalosome [62]. These studies underscore the utility of using state-specific nAbs to gain insights into specific subsets of target proteins that could not otherwise be visualized in live neuronal cells.
Nanobodies against a series of endogenous neuronal proteins were developed to target fluorescent reporters to specific sites in neurons [66]. Specific targeting of these nAbs to distinct subcellular sites in neurons (Figure 2B) could be maintained up to 9 days when lentiviral expression systems were employed [66], suggesting that these nAbs against different endogenous targets could be used to direct delivery of other cargo to diverse subcellular sites in neurons to report on or manipulate the local environment. Plasmids encoding these originally developed nAbs as and many others are available from the open-source plasmid repository Addgene.
3.3. Examples of using FingRs as intrabodies
The most widely used intrabodies recognizing endogenous neuronal proteins are the non-antibody “FingRs” developed by Arnold, Roberts and colleagues targeting the synaptic scaffolding proteins PSD-95 and gephyrin [31], and CamKIIα [32], a protein kinase that is a critical mediator of synaptic plasticity. When used as intrabodies these FingRs bind to and accumulate at sites of target protein expression without impacting the target proteins themselves [31, 32]. These have primarily been used as intrabody fusions with fluorescent proteins in imaging studies defining the subcellular localization and its dynamic regulation of their targets,. However, FingRs have also been fused to proteins with other functionalities, for example to an E3 ubiquitin ligase, to knockdown their synaptic target protein gephyrin and disrupt inhibitory synapses [67]. These have an advantage over methods employing fluorescent protein-tagged synaptic proteins to image synapses as the FingRs minimally impact their target protein expression and function [31, 32]. Plasmids encoding the originally developed FingRs as well as numerous derivatives are available from Addgene.
GFP-tagged anti-gephyrin FingR intrabodies were employed to visualize the impact of visual deprivation on GABAergic inhibition on layer 2/3 neurons in mouse primary visual cortex [68]. The authors employed in utero electroporation to introduce plasmids encoding the GFP-tagged anti-gephyrin FingR into brain neurons, and GFP was imaged in postnatal day 28 (P28) brain. The imaging revealed an increased density of gephyrin puncta corresponding to inhibitory synapses in layer 2/3 neurons in response to visual deprivation, as a likely mechanism underlying the enhanced GABAergic inhibition as measured in electrophysiological recordings. GFP-tagged anti-gephyrin FingR intrabodies were similarly used to image inhibitory synapses in brain slices prepared from P14 mouse brain, again after employing in utero electroporation to introduce the intrabody plasmids into brain [69]. After establishing that the fluorescent intrabody puncta corresponded to functional inhibitory synapses, the authors used live cell imaging to define the subcellular localization of these synapses and found that in both layer 1 and layer 2/3 dendrites, the bulk (≈80%) were on dendritic shafts, and that very few spines (3%) contained inhibitory synapses [69]. GFP-tagged anti-PSD-95 FingR intrabodies were used in a similar manner to map the distribution and size of excitatory synapses along dendritic arbors of neurons in brain slices [70]. GFP-positive puncta corresponding to the postsynaptic density or PSD of dendritic spines were quantified as to their number and size at various locations throughout the dendritic arbor of hippocampal CA1 pyramidal neurons. Parallel measurements of NMDA receptor-driven calcium signals allowed for a determination of how synaptic function related to structure. The results showed that the thinner distal dendrites had smaller spines but larger NMDA receptor-driven calcium signals, while the converse was seen in thicker proximal dendrites. The authors concluded that dendritic location is an important determinant governing synapse structure and function [70]. More recently a set of recombinant adeno-associated virus (AAV) and retrovirus vectors were developed that allow for constitutive and Cre-dependent expression of fluorescent protein tagged anti-PSD-95 and anti-gephyrin FingRs as intrabodies in brain neurons [39]. The availability of these viral vectors from the open-source plasmid repository Addgene will greatly expand the use of these intrabodies in vitro and in vivo well beyond that possible with plasmid transfection or electroporation.
Fusing the anti-gephyrin FingR an E3 ligase was shown to be an effective approach to knock down gephyrin expression and disrupt inhibitory synapse function by dispersing synaptic GABAA receptors [67]. A recent study generated an AAV vector to express this intrabody in mouse brain followed by generation of hippocampal slices [71]. While different degrees of disruption of inhibitory synaptic function was seen across different CA1 neurons, the authors found that the impact on a specific form of excitatory synaptic plasticity termed cumulative LTP positively correlated with the degree of inhibitory synapses disruption. This led to an overall conclusion that inhibitory synapses are necessary for suppression of this form of excitatory synaptic plasticity [71].
An innovative use of the anti-PSD-95 FingR was in studies employing the optical dimerizer CRY2/CIB1 to reversibly manipulate the composition of the postsynaptic compartment in a light-dependent manner [72]. Separate proteins tagged with either CRY2 or CIB1 will reversibly dimerize when exposed to blue light. A set of synaptic scaffolding proteins fused to CRY2 were exogenously expressed in cultured neurons. To circumvent any artifacts that could come from overexpression of these synaptic proteins, the authors also employed the anti-PSD-95 FingR, which unlike overexpression of the synaptic proteins, accumulates at the PSD through its binding to endogenous PSD-95 but does not detectably alter the molecular composition of the synapse [31]. The anti-PSD-95 FingR fused to CRY2, which then reversibly dimerizes with and recruits CIB1-tagged proteins to the postsynaptic density or PSD upon blue light exposure. The authors fused AMPA type glutamate receptor GluA1 subunits to CIB1, such that they were able to reversibly manipulate the levels of AMPA receptors in the postsynaptic membrane, mimicking what occurs during activity-dependent plasticity. The authors found that while this would activate synapses that had few receptors to begin with, this in itself did not alter synaptic strength of existing functional synapses, supporting a model that additional events are needed to strengthen these existing synapses and providing novel insights through the innovative use of this intrabody-based platform [72].
The Bayer lab has made extensive use of the GFP-tagged anti-CamKIIα FingR [32] as an intrabody for live cell imaging of endogenous CamKIIα in neurons. Live imaging of cultured hippocampal neurons transfected with a plasmid encoding the GFP-tagged anti-CamKIIα FingR allowed for studies of the dynamic aggregation of endogenous CamKIIα [73]. CamKIIα plays a crucial role in regulating the function and plasticity of excitatory synapses. This includes the enhanced glutamate release that occurs under ischemic conditions that leads to excitotoxic death of neurons, and triggers extrasynaptic aggregation of CamKIIα. However, while CamKIIα aggregation had been studied using immunolabeling of endogenous CamKIIα in fixed neurons, and by live cell imaging of overexpressed GFP- CamKIIα, the dynamics of endogenous CamKIIα aggregation in living neurons had not been visualized. By employing the GFP-tagged anti-CamKIIα FingR as an intrabody for live cell imaging the authors were able to determine that CamKIIα aggregation does not require the enzymatic activity of this protein kinase [73]. A similar live cell imaging approach employing the GFP-tagged anti-CamKIIα intrabody was used to show that a specific protein kinase, DAPK1, plays a crucial role during long-term depression or LTD by suppressing CamKIIα synaptic accumulation and binding to the NMDA receptor subunit GluN2B as occurs during LTP [74]. By employing FingRs with different fluorescent protein tags, the anti-CamKIIα and anti-PSD-95 intrabodies were used in conjunction to define the population of CamKIIα at excitatory synapses in live cell imaging experiments [74]. A subsequent study employed all three available FingRs, each tagged with a different fluorescent protein, in conjunction with simultaneous multiplex live cell imaging experiments in transfected cultured hippocampal neurons [75]. These studies focused on the dynamic impact of soluble amyloid-β peptide oligomers (Aβ), which numerous studies have shown blocks hippocampal LTP while enhancing LTD. Employing these intrabodies with distinct fluorescent protein tags allowed for simultaneous multiplex imaging (Figure 2C) of the dynamic response of endogenous PSD-95, gephyrin and CamKIIα proteins to Aβ treatment. The authors found that Aβ treatment blocks endogenous CamKIIα accumulation at excitatory synapses during LTP, but not its accumulation at inhibitory synapses during LTD. Employing this real-time, intrabody based live cell imaging approach, the authors also defined the specific parameters (requirement for CaMKII activity, time and dose dependence, and synapse-specific requirement for Aβ) underlying the pathological impact of Aβ on synaptic plasticity. A recent study employed these same three intrabodies in determining the distinct mechanistic requirements for translocation of CamKIIα to excitatory versus inhibitory synapses [76]. The authors found that a complex code of CamKIIα autophosphorylation at distinct sites and on the different subunits within the oligomeric CamKIIα complex underlies whether CamKIIα translocates to excitatory synapses to yield LTP, or to inhibitory synapses to yield LTD.
4. Conclusions
The increasing availability of genetically-encoded intrabodies that can be expressed in neurons has led to their use in a wide variety of basic neuroscience research applications. This should only increase as techniques for gene transfer into neurons in both ex vivo and in vivo environments and plasmids and recombinant viruses encoding intrabodies become more widely accessible to neuroscientists. Development and open availability of a larger toolbox of the various forms of recombinant antibodies (scFvs and nAbs) and antibody mimetics (FingRs and others) that can be used as intrabodies to bind to endogenous neuronal proteins in living neurons will fuel further expansion of the use of this powerful approach to report on and manipulate neuronal function. Enhancing the utility of this expanded toolbox with emerging technologies for regulating the function of intrabodies already present in cells, represents an attractive path for future advances. As one example, methods have been developed to express intrabodies in a non-functional form and once they have accumulated activating them with light. These modified intrabodies, termed optobodies, have been developed using approaches employing distinct photoswitchable proteins. These include expressing split N- and C-terminal fragments of nAbs, with each fragment fused to a photoswitchable dimerization domain that lead to the reconstitution of the entire nAb structure and function in response to light [77]. Another approach is to generate improperly folded nAbs fused to a photoswitchable light–oxygen–voltage (LOV) domain that allosterically drives the correct light-induced folding of the nanobody and recovery of function [78]. Incorporation of photocaged amino acids into nAbs has also been used to generate optobodies [79]. An alternate approach is to use chemical or light stimulation to induce the coupling a specific functional protein, in this case an E3 ubiquitin ligase, to an intrabody to yield stimulus-dependent target protein degradation [80]. Employing such approaches to generate inducible intrabodies against endogenous neuronal proteins such as those described here represents a powerful approach for future neuroscience research advances. These may also provide additional routes for the development of intrabodies with enhanced potential for therapeutic use.
Acknowledgements
Our work developing recombinant antibodies for neuroscience research, including for use as intrabodies, is supported by National Institutes of Health Grant U24NS109113 to J. S. Trimmer. This funding source was not involved in any aspect of the writing or submission of this article.
Footnotes
Conflict of Interest
The author has no conflict of interest to declare
References
- [1].Andersson F, Jakobsson J, Low P, Shupliakov O, Brodin L, Perturbation of syndapin/PACSIN impairs synaptic vesicle recycling evoked by intense stimulation, J Neurosci 28(15) (2008) 3925–33 10.1523/JNEUROSCI.1754-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Theiss C, Napirei M, Meller K, Impairment of anterograde and retrograde neurofilament transport after anti-kinesin and anti-dynein antibody microinjection in chicken dorsal root ganglia, Eur J Cell Biol 84(1) (2005) 29–43 10.1016/j.ejcb.2004.09.001. [DOI] [PubMed] [Google Scholar]
- [3].Tint I, Fischer I, Black M, Acute inactivation of MAP1b in growing sympathetic neurons destabilizes axonal microtubules, Cell Motil Cytoskeleton 60(1) (2005) 48–65 10.1002/cm.20045. [DOI] [PubMed] [Google Scholar]
- [4].Evergren E, Marcucci M, Tomilin N, Low P, Slepnev V, Andersson F, Gad H, Brodin L, De Camilli P, Shupliakov O, Amphiphysin is a component of clathrin coats formed during synaptic vesicle recycling at the lamprey giant synapse, Traffic 5(7) (2004) 514–28 10.1111/j.1398-9219.2004.00198.x. [DOI] [PubMed] [Google Scholar]
- [5].Humeau Y, Doussau F, Vitiello F, Greengard P, Benfenati F, Poulain B, Synapsin controls both reserve and releasable synaptic vesicle pools during neuronal activity and short-term plasticity in Aplysia, J Neurosci 21(12) (2001) 4195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Gad H, Ringstad N, Low P, Kjaerulff O, Gustafsson J, Wenk M, Di Paolo G, Nemoto Y, Crun J, Ellisman MH, De Camilli P, Shupliakov O, Brodin L, Fission and uncoating of synaptic clathrin-coated vesicles are perturbed by disruption of interactions with the SH3 domain of endophilin, Neuron 27(2) (2000) 301–12 10.1016/s0896-6273(00)00038-6. [DOI] [PubMed] [Google Scholar]
- [7].Neame SJ, Rubin LL, Philpott KL, Blocking cytochrome c activity within intact neurons inhibits apoptosis, J Cell Biol 142(6) (1998) 1583–93 10.1083/jcb.142.6.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Vassilev P, Scheuer T, Catterall WA, Inhibition of inactivation of single sodium channels by a site-directed antibody, Proc Natl Acad Sci U S A 86(20) (1989) 8147–51 10.1073/pnas.86.20.8147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Murakoshi H, Trimmer JS, Identification of the Kv2.1 K+ channel as a major component of the delayed rectifier K+ current in rat hippocampal neurons, J Neurosci 19(5) (1999) 1728–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Anderson D, Mehaffey WH, Iftinca M, Rehak R, Engbers JD, Hameed S, Zamponi GW, Turner RW, Regulation of neuronal activity by Cav3-Kv4 channel signaling complexes, Nat Neurosci 13(3) (2010) 333–7 10.1038/nn.2493. [DOI] [PubMed] [Google Scholar]
- [11].Wang K, Lin MT, Adelman JP, Maylie J, Distinct Ca2+ sources in dendritic spines of hippocampal CA1 neurons couple to SK and Kv4 channels, Neuron 81(2) (2014) 379–87 10.1016/j.neuron.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Shamir DB, Deng Y, Wu Q, Modak S, Congdon EE, Sigurdsson EM, Dynamics of internalization and intracellular interaction of Tau antibodies and human pathological Tau protein in a human neuron-like model, Front Neurol 11 (2020) 602292 10.3389/fneur.2020.602292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Asuni AA, Boutajangout A, Quartermain D, Sigurdsson EM, Immunotherapy targeting pathological tau conformers in a tangle mouse model reduces brain pathology with associated functional improvements, J Neurosci 27(34) (2007) 9115–29 10.1523/JNEUROSCI.2361-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Gu J, Congdon EE, Sigurdsson EM, Two novel Tau antibodies targeting the 396/404 region are primarily taken up by neurons and reduce Tau protein pathology, J Biol Chem 288(46) (2013) 33081–95 10.1074/jbc.M113.494922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Gustafsson G, Eriksson F, Moller C, da Fonseca TL, Outeiro TF, Lannfelt L, Bergstrom J, Ingelsson M, Cellular uptake of alpha-Synuclein oligomer-selective antibodies is enhanced by the extracellular presence of alpha-Synuclein and mediated via Fcgamma receptors, Cell Mol Neurobiol 37(1) (2017) 121–131 10.1007/s10571-016-0352-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Pozzi S, Codron P, Soucy G, Renaud L, Cordeau PJ, Dutta K, Bareil C, Julien JP, Monoclonal full-length antibody against TAR DNA binding protein 43 reduces related proteinopathy in neurons, JCI Insight 5(21) (2020) 10.1172/jci.insight.140420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Nguyen L, Montrasio F, Pattamatta A, Tusi SK, Bardhi O, Meyer KD, Hayes L, Nakamura K, Banez-Coronel M, Coyne A, Guo S, Laboissonniere LA, Gu Y, Narayanan S, Smith B, Nitsch RM, Kankel MW, Rushe M, Rothstein J, Zu T, Grimm J, Ranum LPW, Antibody therapy targeting RAN proteins rescues C9 ALS/FTD phenotypes in C9orf72 mouse model, Neuron 105(4) (2020) 645–662 e11 10.1016/j.neuron.2019.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bradbury AR, Pluckthun A, Getting to reproducible antibodies: the rationale for sequenced recombinant characterized reagents, Protein Eng Des Sel 28(10) (2015) 303–5 10.1093/protein/gzv051. [DOI] [PubMed] [Google Scholar]
- [19].Bradbury A, Pluckthun A, Reproducibility: Standardize antibodies used in research, Nature 518(7537) (2015) 27–9 10.1038/518027a. [DOI] [PubMed] [Google Scholar]
- [20].Trimmer JS, Recombinant antibodies in basic neuroscience research, Curr Protoc Neurosci 94(1) (2020) e106 10.1002/cpns.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Bird RE, Hardman KD, Jacobson JW, Johnson S, Kaufman BM, Lee SM, Lee T, Pope SH, Riordan GS, Whitlow M, Single-chain antigen-binding proteins, Science 242(4877) (1988) 423–6. [DOI] [PubMed] [Google Scholar]
- [22].Huston JS, Levinson D, Mudgett-Hunter M, Tai MS, Novotny J, Margolies MN, Ridge RJ, Bruccoleri RE, Haber E, Crea R, et al. , Protein engineering of antibody binding sites: recovery of specific activity in an anti-digoxin single-chain Fv analogue produced in Escherichia coli, Proc Natl Acad Sci U S A 85(16) (1988) 5879–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Chen SY, Bagley J, Marasco WA, Intracellular antibodies as a new class of therapeutic molecules for gene therapy, Hum Gene Ther 5(5) (1994) 595–601 10.1089/hum.1994.5.5-595. [DOI] [PubMed] [Google Scholar]
- [24].Unkauf T, Hust M, Frenzel A, Antibody affinity and stability maturation by error-prone PCR, Methods Mol Biol 1701 (2018) 393–407 10.1007/978-1-4939-7447-4_22. [DOI] [PubMed] [Google Scholar]
- [25].Murphy C, Stack E, Krivelo S, Breheny M, Ma H, O’Kennedy R, Enhancing recombinant antibody performance by optimally engineering its format, J Immunol Methods 463 (2018) 127–133 10.1016/j.jim.2018.10.005. [DOI] [PubMed] [Google Scholar]
- [26].Schaefer JV, Honegger A, Pluckthun A, Construction of scFv Fragments from hybridoma or spleen cells by PCR assembly, in: Kontermann R, Dubel S (Eds.), Antibody Engineering, Springer Verlag, Heidelberg, Germany, 2010, pp. 21–44. [Google Scholar]
- [27].Chiu ML, Goulet DR, Teplyakov A, Gilliland GL, Antibody structure and function: the basis for engineering therapeutics, Antibodies (Basel) 8(4) (2019) 10.3390/antib8040055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Tiller KE, Tessier PM, Advances in antibody design, Annu Rev Biomed Eng 17 (2015) 191–216 10.1146/annurev-bioeng-071114-040733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Muyldermans S, Nanobodies: natural single-domain antibodies, Annu Rev Biochem 82 (2013) 775–97 10.1146/annurev-biochem-063011-092449. [DOI] [PubMed] [Google Scholar]
- [30].Koide A, Bailey CW, Huang X, Koide S, The fibronectin type III domain as a scaffold for novel binding proteins, J Mol Biol 284(4) (1998) 1141–51 10.1006/jmbi.1998.2238. [DOI] [PubMed] [Google Scholar]
- [31].Gross GG, Junge JA, Mora RJ, Kwon HB, Olson CA, Takahashi TT, Liman ER, Ellis-Davies GC, McGee AW, Sabatini BL, Roberts RW, Arnold DB, Recombinant probes for visualizing endogenous synaptic proteins in living neurons, Neuron 78(6) (2013) 971–85 10.1016/j.neuron.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Mora RJ, Roberts RW, Arnold DB, Recombinant probes reveal dynamic localization of CaMKIIalpha within somata of cortical neurons, J Neurosci 33(36) (2013) 14579–90 10.1523/JNEUROSCI.2108-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kaech S, Banker G, Culturing hippocampal neurons, Nat Protoc 1(5) (2006) 2406–15 10.1038/nprot.2006.356. [DOI] [PubMed] [Google Scholar]
- [34].Humpel C, Organotypic brain slice cultures: A review, Neuroscience 305 (2015) 86–98 10.1016/j.neuroscience.2015.07.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Saito T, Nakatsuji N, Efficient gene transfer into the embryonic mouse brain using in vivo electroporation, Dev Biol 240(1) (2001) 237–46 10.1006/dbio.2001.0439. [DOI] [PubMed] [Google Scholar]
- [36].Meyer-Dilhet G, Courchet J, In utero cortical electroporation of plasmids in the mouse embryo, STAR Protoc 1(1) (2020) 100027 10.1016/j.xpro.2020.100027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].dal Maschio M, Ghezzi D, Bony G, Alabastri A, Deidda G, Brondi M, Sato SS, Zaccaria RP, Di Fabrizio E, Ratto GM, Cancedda L, High-performance and site-directed in utero electroporation by a triple-electrode probe, Nat Commun 3 (2012) 960 10.1038/ncomms1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Baumgart J, Baumgart N, Cortex-, hippocampus-, thalamus-, hypothalamus-, lateral septal nucleus- and striatum-specific in utero electroporation in the C57BL/6 mouse, J Vis Exp (107) (2016) e53303 10.3791/53303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Bensussen S, Shankar S, Ching KH, Zemel D, Ta TL, Mount RA, Shroff SN, Gritton HJ, Fabris P, Vanbenschoten H, Beck C, Man HY, Han X, A viral toolbox of genetically encoded fluorescent synaptic tags, iScience 23(7) (2020) 101330 10.1016/j.isci.2020.101330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Zacchi P, Dreosti E, Visintin M, Moretto-Zita M, Marchionni I, Cannistraci I, Kasap Z, Betz H, Cattaneo A, Cherubini E, Gephyrin selective intrabodies as a new strategy for studying inhibitory receptor clustering, J Mol Neurosci 34(2) (2008) 141–8 10.1007/s12031-007-9018-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Marchionni I, Kasap Z, Mozrzymas JW, Sieghart W, Cherubini E, Zacchi P, New insights on the role of gephyrin in regulating both phasic and tonic GABAergic inhibition in rat hippocampal neurons in culture, Neuroscience 164(2) (2009) 552–62 10.1016/j.neuroscience.2009.07.063. [DOI] [PubMed] [Google Scholar]
- [42].Varley ZK, Pizzarelli R, Antonelli R, Stancheva SH, Kneussel M, Cherubini E, Zacchi P, Gephyrin regulates GABAergic and glutamatergic synaptic transmission in hippocampal cell cultures, J Biol Chem 286(23) (2011) 20942–51 10.1074/jbc.M111.234641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Zhang C, Helmsing S, Zagrebelsky M, Schirrmann T, Marschall AL, Schungel M, Korte M, Hust M, Dubel S, Suppression of p75 neurotrophin receptor surface expression with intrabodies influences Bcl-xL mRNA expression and neurite outgrowth in PC12 cells, PLoS One 7(1) (2012) e30684 10.1371/journal.pone.0030684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Wilson IA, Niman HL, Houghten RA, Cherenson AR, Connolly ML, Lerner RA, The structure of an antigenic determinant in a protein, Cell 37(3) (1984) 767–78 10.1016/0092-8674(84)90412-4. [DOI] [PubMed] [Google Scholar]
- [45].Wongso D, Dong J, Ueda H, Kitaguchi T, Flashbody: a next generation fluobody with fluorescence intensity enhanced by antigen binding, Anal Chem 89(12) (2017) 6719–6725 10.1021/acs.analchem.7b00959. [DOI] [PubMed] [Google Scholar]
- [46].Zhao N, Kamijo K, Fox PD, Oda H, Morisaki T, Sato Y, Kimura H, Stasevich TJ, A genetically encoded probe for imaging nascent and mature HA-tagged proteins in vivo, Nat Commun 10(1) (2019) 2947 10.1038/s41467-019-10846-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Lim ST, Antonucci DE, Scannevin RH, Trimmer JS, A novel targeting signal for proximal clustering of the Kv2.1 K+ channel in hippocampal neurons, Neuron 25(2) (2000) 385–97 S0896-6273(00)80902-2 [pii]. [DOI] [PubMed] [Google Scholar]
- [48].Fukata Y, Dimitrov A, Boncompain G, Vielemeyer O, Perez F, Fukata M, Local palmitoylation cycles define activity-regulated postsynaptic subdomains, J Cell Biol 202(1) (2013) 145–61 10.1083/jcb.201302071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Jeyifous O, Lin EI, Chen X, Antinone SE, Mastro R, Drisdel R, Reese TS, Green WN, Palmitoylation regulates glutamate receptor distributions in postsynaptic densities through control of PSD95 conformation and orientation, Proc Natl Acad Sci U S A 113(52) (2016) E8482–E8491 10.1073/pnas.1612963113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Ekstrand MI, Nectow AR, Knight ZA, Latcha KN, Pomeranz LE, Friedman JM, Molecular profiling of neurons based on connectivity, Cell 157(5) (2014) 1230–42 10.1016/j.cell.2014.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Rothbauer U, Zolghadr K, Tillib S, Nowak D, Schermelleh L, Gahl A, Backmann N, Conrath K, Muyldermans S, Cardoso MC, Leonhardt H, Targeting and tracing antigens in live cells with fluorescent nanobodies, Nat Methods 3(11) (2006) 887–9 10.1038/nmeth953. [DOI] [PubMed] [Google Scholar]
- [52].Pomeranz LE, Ekstrand MI, Latcha KN, Smith GA, Enquist LW, Friedman JM, Gene expression profiling with Cre-conditional pseudorabies virus reveals a subset of midbrain neurons that participate in reward circuitry, J Neurosci 37(15) (2017) 4128–4144 10.1523/JNEUROSCI.3193-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Chen Y, Jang H, Spratt PWE, Kosar S, Taylor DE, Essner RA, Bai L, Leib DE, Kuo TW, Lin YC, Patel M, Subkhangulova A, Kato S, Feinberg EH, Bender KJ, Knight ZA, Garrison JL, Soma-targeted imaging of neural circuits by ribosome tethering, Neuron 107(3) (2020) 454–469 e6 10.1016/j.neuron.2020.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Kirchhofer A, Helma J, Schmidthals K, Frauer C, Cui S, Karcher A, Pellis M, Muyldermans S, Casas-Delucchi CS, Cardoso MC, Leonhardt H, Hopfner KP, Rothbauer U, Modulation of protein properties in living cells using nanobodies, Nat Struct Mol Biol 17(1) (2010) 133–8 10.1038/nsmb.1727. [DOI] [PubMed] [Google Scholar]
- [55].Kubala MH, Kovtun O, Alexandrov K, Collins BM, Structural and thermodynamic analysis of the GFP:GFP-nanobody complex, Protein Sci 19(12) (2010) 2389–401 10.1002/pro.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Baker CA, Elyada YM, Parra A, Bolton MM, Cellular resolution circuit mapping with temporal-focused excitation of soma-targeted channelrhodopsin, Elife 5 (2016) 10.7554/eLife.14193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Mahn M, Gibor L, Patil P, Cohen-Kashi Malina K, Oring S, Printz Y, Levy R, Lampl I, Yizhar O, High-efficiency optogenetic silencing with soma-targeted anion-conducting channelrhodopsins, Nat Commun 9(1) (2018) 4125 10.1038/s41467-018-06511-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Shemesh OA, Linghu C, Piatkevich KD, Goodwin D, Celiker OT, Gritton HJ, Romano MF, Gao R, Yu CJ, Tseng HA, Bensussen S, Narayan S, Yang CT, Freifeld L, Siciliano CA, Gupta I, Wang J, Pak N, Yoon YG, Ullmann JFP, Guner-Ataman B, Noamany H, Sheinkopf ZR, Park WM, Asano S, Keating AE, Trimmer JS, Reimer J, Tolias AS, Bear MF, Tye KM, Han X, Ahrens MB, Boyden ES, Precision calcium imaging of dense neural populations via a cell-body-targeted calcium indicator, Neuron 107(3) (2020) 470–486 e11 10.1016/j.neuron.2020.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Tang JC, Szikra T, Kozorovitskiy Y, Teixiera M, Sabatini BL, Roska B, Cepko CL, A nanobody-based system using fluorescent proteins as scaffolds for cell-specific gene manipulation, Cell 154(4) (2013) 928–39 10.1016/j.cell.2013.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Fields S, Song O, A novel genetic system to detect protein-protein interactions, Nature 340(6230) (1989) 245–6 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- [61].Tang JC, Drokhlyansky E, Etemad B, Rudolph S, Guo B, Wang S, Ellis EG, Li JZ, Cepko CL, Detection and manipulation of live antigen-expressing cells using conditionally stable nanobodies, Elife 5 (2016) 10.7554/eLife.15312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Gormal RS, Padmanabhan P, Kasula R, Bademosi AT, Coakley S, Giacomotto J, Blum A, Joensuu M, Wallis TP, Lo HP, Budnar S, Rae J, Ferguson C, Bastiani M, Thomas WG, Pardon E, Steyaert J, Yap AS, Goodhill GJ, Hilliard MA, Parton RG, Meunier FA, Modular transient nanoclustering of activated beta2-adrenergic receptors revealed by single-molecule tracking of conformation-specific nanobodies, Proc Natl Acad Sci U S A 117(48) (2020) 30476–30487 10.1073/pnas.2007443117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Morgenstern TJ, Park J, Fan QR, Colecraft HM, A potent voltage-gated calcium channel inhibitor engineered from a nanobody targeted to auxiliary CaVbeta subunits, Elife 8 (2019) 10.7554/eLife.49253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Irannejad R, Tomshine JC, Tomshine JR, Chevalier M, Mahoney JP, Steyaert J, Rasmussen SG, Sunahara RK, El-Samad H, Huang B, von Zastrow M, Conformational biosensors reveal GPCR signalling from endosomes, Nature 495(7442) (2013) 534–8 10.1038/nature12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Staus DP, Strachan RT, Manglik A, Pani B, Kahsai AW, Kim TH, Wingler LM, Ahn S, Chatterjee A, Masoudi A, Kruse AC, Pardon E, Steyaert J, Weis WI, Prosser RS, Kobilka BK, Costa T, Lefkowitz RJ, Allosteric nanobodies reveal the dynamic range and diverse mechanisms of G-protein-coupled receptor activation, Nature 535(7612) (2016) 448–52 10.1038/nature18636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Dong JX, Lee Y, Kirmiz M, Palacio S, Dumitras C, Moreno CM, Sando R, Santana LF, Sudhof TC, Gong B, Murray KD, Trimmer JS, A toolbox of nanobodies developed and validated for use as intrabodies and nanoscale immunolabels in mammalian brain neurons, Elife 8 (2019) e48750 10.7554/eLife.48750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Gross GG, Straub C, Perez-Sanchez J, Dempsey WP, Junge JA, Roberts RW, Trinh le A, Fraser SE, De Koninck Y, De Koninck P, Sabatini BL, Arnold DB, An E3-ligase-based method for ablating inhibitory synapses, Nat Methods 13(8) (2016) 673–8 10.1038/nmeth.3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Kannan M, Gross GG, Arnold DB, Higley MJ, Visual deprivation during the critical period enhances layer 2/3 GABAergic inhibition in mouse V1, J Neurosci 36(22) (2016) 5914–9 10.1523/JNEUROSCI.0051-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Kwon T, Merchan-Perez A, Rial Verde EM, Rodriguez JR, DeFelipe J, Yuste R, Ultrastructural, molecular and functional mapping of GABAergic synapses on dendritic spines and shafts of neocortical pyramidal neurons, Cereb Cortex 29(7) (2019) 2771–2781 10.1093/cercor/bhy143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Walker AS, Neves G, Grillo F, Jackson RE, Rigby M, O’Donnell C, Lowe AS, Vizcay-Barrena G, Fleck RA, Burrone J, Distance-dependent gradient in NMDAR-driven spine calcium signals along tapering dendrites, Proc Natl Acad Sci U S A 114(10) (2017) E1986–E1995 10.1073/pnas.1607462114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Davenport CM, Rajappa R, Katchan L, Taylor CR, Tsai MC, Smith CM, de Jong JW, Arnold DB, Lammel S, Kramer RH, Relocation of an extrasynaptic GABAA receptor to inhibitory synapses freezes excitatory synaptic strength and preserves memory, Neuron 109(1) (2021) 123–134 e4 10.1016/j.neuron.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Sinnen BL, Bowen AB, Forte JS, Hiester BG, Crosby KC, Gibson ES, Dell’Acqua ML, Kennedy MJ, Optogenetic control of synaptic composition and function, Neuron 93(3) (2017) 646–660 e5 10.1016/j.neuron.2016.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Barcomb K, Goodell DJ, Arnold DB, Bayer KU, Live imaging of endogenous Ca(2+)/calmodulin-dependent protein kinase II in neurons reveals that ischemia-related aggregation does not require kinase activity, J Neurochem 135(4) (2015) 666–73 10.1111/jnc.13263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Goodell DJ, Zaegel V, Coultrap SJ, Hell JW, Bayer KU, DAPK1 mediates LTD by making CaMKII/GluN2B binding LTP specific, Cell Rep 19(11) (2017) 2231–2243 10.1016/j.celrep.2017.05.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Cook SG, Goodell DJ, Restrepo S, Arnold DB, Bayer KU, Simultaneouslive imaging of multiple endogenous proteins reveals a mechanism for Alzheimer’s-related plasticity impairment, Cell Rep 27(3) (2019) 658–665 e4 10.1016/j.celrep.2019.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Cook SG, Buonarati OR, Coultrap SJ, Bayer KU, CaMKII holoenzyme mechanisms that govern the LTP versus LTD decision, Sci Adv 7(16) (2021) 10.1126/sciadv.abe2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Yu D, Lee H, Hong J, Jung H, Jo Y, Oh BH, Park BO, Heo WD, Optogenetic activation of intracellular antibodies for direct modulation of endogenous proteins, Nat Methods 16(11) (2019) 1095–1100 10.1038/s41592-019-0592-7. [DOI] [PubMed] [Google Scholar]
- [78].Gil AA, Carrasco-Lopez C, Zhu L, Zhao EM, Ravindran PT, Wilson MZ, Goglia AG, Avalos JL, Toettcher JE, Optogenetic control of protein binding using light-switchable nanobodies, Nat Commun 11(1) (2020) 4044 10.1038/s41467-020-17836-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Joest EF, Winter C, Wesalo JS, Deiters A, Tampé R, Light-guided intrabodies for on-demand in situ target recognition in human cells, Chemical Science 12(16) (2021) 5787–5795 10.1039/D1SC01331A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Deng W, Bates JA, Wei H, Bartoschek MD, Conradt B, Leonhardt H, Tunable light and drug induced depletion of target proteins, Nat Commun 11(1) (2020) 304 10.1038/s41467-019-14160-8. [DOI] [PMC free article] [PubMed] [Google Scholar]


