Abstract
Polygenic risk scores (PRSs) of common genetic variants have shown promise in prostate cancer risk stratification, but their validity across populations has yet to be confirmed. We evaluated a multiethnic PRS model based on 269 germline genetic risk variants (261 were available for analysis) using an independent population of 13 628 US men. The PRS was strongly associated with prostate cancer but not with any other disease. Comparing men in the top PRS decile with those at average risk (40%-60%), the odds ratio of prostate cancer was 3.89 (95% confidence interval = 3.24 to 4.68) for men of European ancestry and 3.81 (95% confidence interval = 1.48 to 10.19) for men of African ancestry. By age 85 years, the cumulative incidence of prostate cancer for European American men was 7.1% in the bottom decile and 54.1% in the top decile. This suggests that the PRS can be used to identify a substantial proportion of men at high risk for prostate cancer.
Genome-wide association studies (GWAS) have identified over 260 germline genetic variants independently associated with prostate cancer risk (1–3). Although individual variants contribute to risk only modestly, combining information from multiple variants into a polygenic risk score (PRS) has shown promise to aid in stratification of future prostate cancer risk (1,3-5). In a multiethnic GWAS of 107 247 prostate cancer cases and 127 006 controls, a PRS including 269 genetic variants effectively stratified prostate cancer risk across ethnic groups (3). In the replication sample, the odds ratio (OR) comparing men in the top decile with men at average genetic risk in the 40th-60th percentile of the PRS ranged between 3.53 for men of African ancestry and 4.17 for men of European ancestry. The estimated lifetime risk of prostate cancer in the top decile of the PRS was just below 40% for men in the European and African ancestry groups.
Before widespread integration of a PRS into clinical practice, the external validity of findings across populations and settings should be assessed. Although the association between the PRS from the multiethnic GWAS and prostate cancer was confirmed in the replication samples, it is unrealistic to assume that genetic variants influence the risk of prostate cancer equally in all populations (6). Furthermore, a PRS model should be validated in populations and settings in which it is intended to be used. To address this, we evaluated this multiethnic PRS in an independent study population of men included in the Mass General Brigham Biobank (7). Individuals in the biobank were patients seen at affiliated hospitals in the greater Boston area and were not recruited for any specific disease.
We first designed a case-control study of men age 40 years or older recruited between 2010 and 2018. Cases were defined as having a biopsy-confirmed prostate cancer documented by a pathology report (82.5%) or at least 2 prostate cancer–related billing codes. The case-control population consisted of 13 628 men (1643 cases and 11 985 controls), of whom 12 473 (91.5%) were of European, 524 (3.8%) of African, 402 (2.9%) of admixed American, and 229 (1.7%) of Asian ancestry (based on genetically defined ancestry). We also designed a cohort study of men initially free from prostate cancer, restricted to men of European ancestry due to the small number of men with non-European ancestries. Of the 11 908 men included in the cohort analysis, 847 presented with prostate cancer during a median of 7 (interquartile range = 5-9) years of follow-up. To evaluate if the PRS was specific to prostate cancer, we also conducted a phenome-wide association study (8) of 1093 International Classification of Diseases (ICD)-9 and ICD-10 categories in men of European ancestry. Detailed information on materials and methods can be found in the Supplementary Materials (available online). All participants provided written informed consent, and the study was approved by the Partners Human Research Institutional Review Board (2019P002655). All statistical tests were 2-sided.
In the case-control analysis, the PRS was strongly associated with prostate cancer in the European and African ancestry groups, with an 11-fold gradient in odds of disease. Compared with men at average risk (40th-60th percentile), the odds ratio of prostate cancer for men of European ancestry was 0.34 (95% confidence interval [CI] = 0.25 to 0.46) in the bottom decile and 3.89 (95% CI = 3.24 to 4.68) in the top decile (Table 1). In men of African ancestry, the odds ratio was 0.15 (95% CI = 0.01 to 0.92) in the bottom decile and 3.81 (95% CI = 1.48 to 10.19) in the top decile. The analysis was underpowered for the other ancestry groups due to few cases (Supplementary Table 1, available online).
Table 1.
Odds ratio with 95% confidence interval for the association between PRS and prostate cancer from the case-control analysis
| PRS category | European ancestry |
African ancestry |
||
|---|---|---|---|---|
| Cases/controls | ORa (95% CI) | Cases/controls | ORb (95% CI) | |
| (1554/10 918) | (67/457) | |||
| By quartiles | ||||
| 0%-25% | 177/2943 | 0.45 (0.38 to 0.54) | 8/123 | 0.59 (0.24 to 1.35) |
| 25%-75% | 700/5536 | 1.00 (Ref.) | 28/234 | 1.00 (Ref.) |
| 75%-100% | 677/2440 | 2.46 (2.18 to 2.78) | 31/100 | 2.66 (1.44 to 4.98) |
| By deciles | ||||
| 0%-10% | 55/1193 | 0.34 (0.25 to 0.46) | 1/52 | 0.15 (0.01 to 0.92) |
| 10%-20% | 79/1168 | 0.51 (0.39 to 0.67) | 6/46 | 1.21 (0.37 to 3.70) |
| 20%-30% | 98/1149 | 0.66 (0.52 to 0.85) | 5/48 | 0.73 (0.21 to 2.32) |
| 30%-40% | 100/1148 | 0.68 (0.53 to 0.87) | 5/47 | 0.59 (0.16 to 1.93) |
| 40%-60% | 285/2210 | 1.00 (Ref.) | 10/94 | 1.00 (Ref.) |
| 60%-70% | 166/1080 | 1.21 (0.98 to 1.50) | 5/48 | 0.77 (0.21 to 2.49) |
| 70%-80% | 183/1064 | 1.42 (1.15 to 1.74) | 7/45 | 1.31 (0.42 to 3.90) |
| 80%-90% | 223/1024 | 1.87 (1.54 to 2.29) | 13/39 | 2.27 (0.85 to 6.16) |
| 90%-100% | 365/883 | 3.89 (3.24 to 4.68) | 15/38 | 3.81 (1.48 to 10.19) |
Adjusted for age at blood collection, genotyping platform, and the first 10 principal components. CI = confidence interval; OR = odds ratio; PRS = polygenic risk score.
Adjusted for age at blood collection, genotyping platform, and the first 2 principal components.
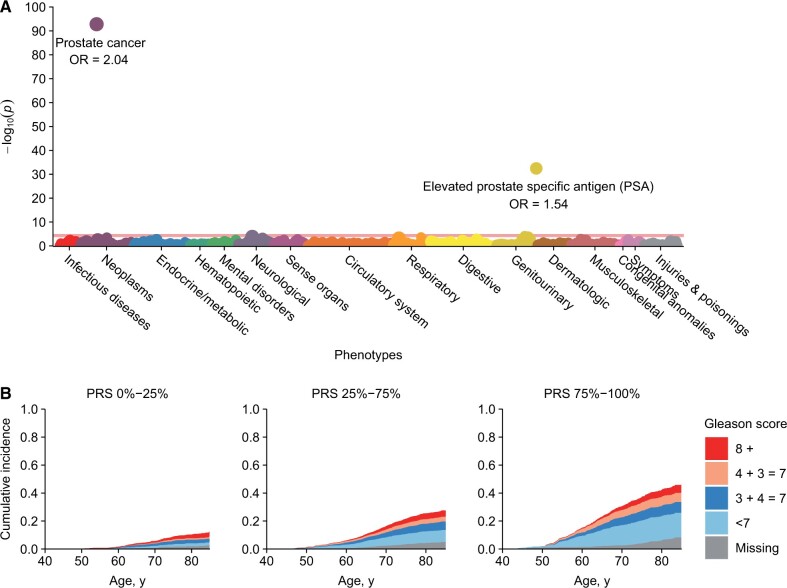
The PRS was highly specific for prostate cancer in men of European ancestry. In the phenome-wide association study of over 1000 disease categories, only 2 yielded statistically significant associations with the PRS after correction for multiple testing: prostate cancer and presence of elevated prostate-specific antigen (Figure 1, A;Supplementary Table 2, available online).
Figure 1.
Polygenic risk score (PRS) phenome-wide association results and absolute lifetime risk of prostate cancer in men of European ancestry. (A) Manhattan plot for phenome-wide association study (PheWAS) of 1093 disease categories (based on billing codes) with the prostate cancer PRS in men of European ancestry. The horizontal line indicates phenome-wide–level significance (2-sided P = 4.6 × 10−5) using Bonferroni correction. Adjustments were made for age at blood collection, genotyping platform, and the first 10 principal components. Odds ratios (ORs) are reported as per 1-SD increase in the PRS. (B) Cumulative incidence of prostate cancer (overall and by Gleason score) for men of European ancestry included in the cohort analysis, stratified by PRS quartile.
In the cohort analysis, the cumulative incidence of prostate cancer in men of European ancestry by age 85 years was 42.9% for men in the top quartile of the PRS compared with 10.3% in the bottom quartile (by deciles: 54.1% in the top decile compared with 7.1% in the bottom decile) (Figure 1, B;Supplementary Table 3, available online). The PRS did not demonstrate a difference for high-, intermediate-, and low-grade prostate cancer. However, because the PRS was strongly associated with an increased risk of overall prostate cancer, the absolute risk of a Gleason score 7 or higher tumor was highest in the highest PRS quartile. By age 85 years, it was 19.9% in the top quartile compared with 7.0% in the bottom quartile.
Our results were largely in agreement with the prior multiethnic GWAS. We observed a slightly weaker association between the PRS and prostate cancer in men of European ancestry—but not in men of African ancestry—compared with both the main analysis and the replication sample, which may be related to differences in population structures, prostate cancer screening, or the hospital-based design. The studied population may be enriched with men seeking health care for prostate cancer and may not represent the general population in terms of PRS distribution. Although the trend was similar, we observed higher lifetime absolute risk estimates for men of European ancestry across all PRS categories compared with those estimated in the multiethnic GWAS.
Our findings support the use of a PRS as part of risk stratification for prostate cancer in men of European or African ancestry. An advantage of a multiethnic PRS is a single tool that can be used in all individuals. Although the PRS on its own is not specific to risk of aggressive disease (3,9), it provides an opportunity to integrate markers with other serum, urine, or imaging diagnostic tests (10–12) that are specific for aggressive disease. The PRS can narrow the number of men at high risk of prostate cancer and enable a more focused assessment for aggressive disease. This PRS could potentially be used to identify men at increased risk of prostate cancer who would then undergo targeted screening and prophylaxis. Meanwhile, men in the lowest risk of the PRS could safely avoid future screenings. The evaluated PRS may provide more refined risk estimation than current clinical tools allow.
Funding
This work was supported by the DiNovi Family Foundation, the National Cancer Institute at the National Institutes of Health (P30 CA006516 and CA 014089), William Casey, the Swedish Society for Medical Research (P19-0017), and the Prostate Cancer Foundation.
Notes
Role of the funder: The funder had no role in the design of the study; the collection, analysis, and interpretation of the data; the writing of the manuscript; and the decision to submit the manuscript for publication.
Disclosures: The authors report no conflict of interest.
Author contributions: AP: conceptualization, data curation, formal analysis, investigation, methodology, software, visualization, writing—original draft. KLP: conceptualization, investigation, validation, writing—review and editing. SK: conceptualization, investigation, validation, writing—review and editing. PK: conceptualization, methodology, investigation, validation, writing—review and editing. DVC: investigation, validation, writing—review and editing. CH: investigation, validation, writing—review and editing. LAM: conceptualization, investigation, validation, writing—review and editing. ASK: conceptualization, funding acquisition, methodology, project administration, supervision, investigation, validation, writing—review and editing.
Prior presentations: Preliminary result from this study has been presented at the Thirteenth Annual Prostate Cancer Program Retreat, Fort Lauderdale, Florida March 3, 2020.
Data Availability
Variants and weights used to generate the PRS can be found under: https://www.pgscatalog.org/publication/PGP000122/. Individual-level genotype and clinical data can be obtained from the Mass General Brigham Biobank (https://personalizedmedicine.partners.org/biobank/), but restrictions apply to the availability of these data and data are not publicly available. Data are available for all Mass General Brigham investigators and their external affiliates, including academic and commercial affiliates, provided a protocol from the Institutional Review Board (IRB) and a data use agreement. Samples and data shared with an external entity must be de-identified. Investigators can contact the corresponding author or biobank@partners.org for more information.
Supplementary Material
References
- 1. Schumacher FR, Al Olama AA, Berndt SI, et al. ; The Profile Study. Association analyses of more than 140,000 men identify 63 new prostate cancer susceptibility loci. Nat Genet. 2018;50(7):928–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Benafif S, Kote-Jarai Z, Eeles RA; PRACTICAL Consortium. A review of prostate cancer genome-wide association studies (GWAS). Cancer Epidemiol Biomarkers Prev. 2018;27(8):845–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Conti DV, Darst BF, Moss LC, et al. Trans-ancestry genome-wide association meta-analysis of prostate cancer identifies new susceptibility loci and informs genetic risk prediction. Nat Genet. 2021;53(1):65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gronberg H, Adolfsson J, Aly M, et al. Prostate cancer screening in men aged 50-69 years (STHLM3): a prospective population-based diagnostic study. Lancet Oncol. 2015;16(16):1667–1676. [DOI] [PubMed] [Google Scholar]
- 5. Kader AK, Sun J, Reck BH, et al. Potential impact of adding genetic markers to clinical parameters in predicting prostate biopsy outcomes in men following an initial negative biopsy: findings from the REDUCE trial. Eur Urol. 2012;62(6):953–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Janssens A. Validity of polygenic risk scores: are we measuring what we think we are? Hum Mol Genet. 2019;28(R2):R143–R50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Karlson EW, Boutin NT, Hoffnagle AG, Allen NL.. Building the partners HealthCare Biobank at partners personalized medicine: informed consent, return of research results, recruitment lessons and operational considerations. J Pers Med. 2016;6(1):2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carroll RJ, Bastarache L, Denny JC.. R PheWAS: data analysis and plotting tools for phenome-wide association studies in the R environment. Bioinformatics. 2014;30(16):2375–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fantus RJ, Helfand BT.. Germline genetics of prostate cancer: time to incorporate genetics into early detection tools. Clin Chem. 2019;65(1):74–79. [DOI] [PubMed] [Google Scholar]
- 10. Ahmed HU, El-Shater Bosaily A, Brown LC, et al. Diagnostic accuracy of multi-parametric MRI and TRUS biopsy in prostate cancer (PROMIS): a paired validating confirmatory study. Lancet. 2017;389(10071):815–822. [DOI] [PubMed] [Google Scholar]
- 11. Kasivisvanathan V, Rannikko AS, Borghi M, et al. MRI-targeted or standard biopsy for prostate-cancer diagnosis. N Engl J Med. 2018;378(19):1767–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Preston MA, Batista JL, Wilson KM, et al. Baseline prostate-specific antigen levels in midlife predict lethal prostate cancer. J Clin Oncol. 2016;34(23):2705–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Variants and weights used to generate the PRS can be found under: https://www.pgscatalog.org/publication/PGP000122/. Individual-level genotype and clinical data can be obtained from the Mass General Brigham Biobank (https://personalizedmedicine.partners.org/biobank/), but restrictions apply to the availability of these data and data are not publicly available. Data are available for all Mass General Brigham investigators and their external affiliates, including academic and commercial affiliates, provided a protocol from the Institutional Review Board (IRB) and a data use agreement. Samples and data shared with an external entity must be de-identified. Investigators can contact the corresponding author or biobank@partners.org for more information.