Abstract
“Junk DNA” is a popular yet controversial concept that states that organisms carry in their genomes DNA that has no positive impact on their fitness. Nonetheless, biochemical functions have been identified for an increasing fraction of DNA elements traditionally seen as “Junk DNA”. These findings have been interpreted as fundamentally undermining the “Junk DNA” concept. Here, we reinforce previous arguments that this interpretation relies on an inadequate concept of biological function that does not consider the selected effect of a given genomic structure, which is central to the “Junk DNA” concept. Next, we suggest that another (though ignored) confounding factor is that the discussion about biological functions includes two different dimensions: a horizontal, ecological dimension that reflects how a given genomic element affects fitness in a specific time, and a vertical, temporal dimension that reflects how a given genomic element persisted along time. We suggest that “Junk DNA” should be used exclusively relative to the horizontal dimension, while for the vertical dimension, we propose a new term, “Spam DNA”, that reflects the fact that a given genomic element may persist in the genome even if not selected for on their origin. Importantly, these concepts are complementary. An element can be both “Spam DNA” and “Junk DNA”, and “Spam DNA” can also be recruited to perform evolved biological functions, as illustrated in processes of exaptation or constructive neutral evolution.
Keywords: spam DNA, genome evolution, biological function, exaptation, purifying selection, positive selection
Significance.
“Junk DNA” is a popular yet controversial concept that states that organisms carry in their genomes DNA that has no positive impact on fitness. A recent study suggested that genomic elements traditionally seen as “junk” may play key biological roles in Schistosoma mansoni. Here, we criticize this conclusion and highlight that the concept of biological function includes both a current (horizontal) and a historical (vertical) meaning. While the term “Junk DNA” is best suited for the horizontal definition, we propose the term “Spam DNA” to account for the vertical definition of function. Acknowledging these differences is fundamental for a better understanding of genome evolution and how non-adaptive processes may originate proper biological function.
About a decade ago, the ENCODE (Encyclopedia of DNA Elements) consortium used multi-omics data to declare the death of “Junk DNA” (ENCODE Project Consortium 2012). This conclusion was met with much fanfare and was highly publicized by several high-profile scientific journals, such as Nature (Ecker et al. 2012), Science (Pennisi 2012), and The Lancet (2012). Subsequently, several authors raised important conceptual issues that were poorly considered in the ENCODE study, ranging from the definition of biological function to the correct understanding of Ohno’s view of the C-value paradox, ultimately vindicating the validity of the “Junk DNA” concept (Eddy 2012; Graur et al. 2013; Doolittle 2013; Niu and Jiang 2013; Palazzo and Gregory 2014).
Echoing ENCODE’s perspective in regard to the prominence of biochemical function, a recent study published in this journal analyzed new data for a class of genomic elements in S. mansoni to conclude that “it is tempting to speculate that more of the WE ‘junk-DNA’ than expected might be functional and relevant” (Stitz et al. 2021). An accompanying opinion piece highlights the redemption of repetitive elements from their “Junk DNA” status into “vital sources of [genomic] variation” (McGrath 2021), which fits in the view that “the days of ‘Junk DNA’ are over” (Stitz et al. 2021).
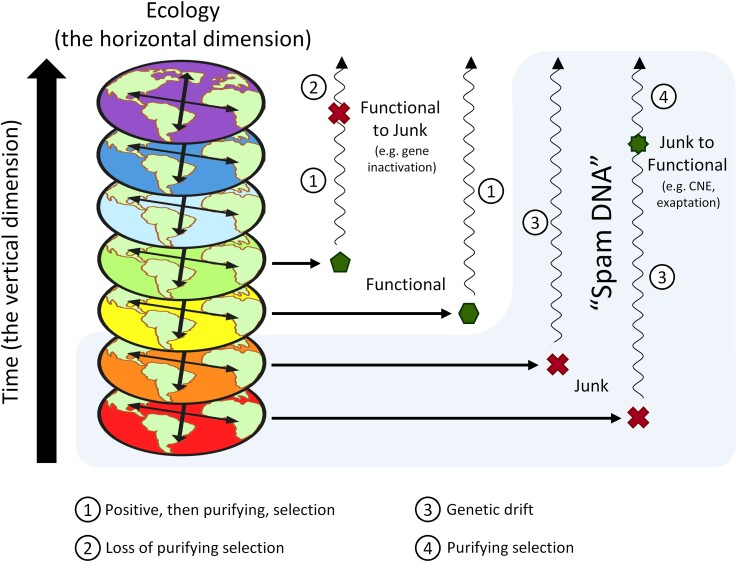
That such an important concept in genome evolution such as “Junk DNA” has been buried alive twice over a decade is worrying, as substantial evidence supports the notion that “Junk DNA” is common in many genomes (see e.g., Eddy 2012; Doolittle 2013; Graur et al. 2013; Niu and Jiang 2013; Palazzo and Gregory 2014; Palazzo and Lee 2015; Koonin 2016). One reason for the ongoing conundrum around “Junk DNA” is the unresolved debate over alternative definitions of biological function. Another and less appreciated issue is the complex discussion about the current versus past function of a genomic element, which, as we argue, should be seen as representing a horizontal and a vertical dimension of the evolutionary process, respectively. One way out of this problem is having different terms for each dimension. We propose that while the horizontal dimension is well captured by “Junk DNA”, a new concept, “Spam DNA”, could account for the vertical dimension (fig. 1). Hopefully, this distinction will help to clarify the current debate about function in genome evolution.
Fig. 1.
The horizontal and vertical dimensions in genome evolution, and the possible fates of new genomic elements. The vertical dimension can be thought of as a succession of (horizontal) time slices where the ecological relations are always changing. Depending on the impact of selection, a new genomic element can originate as “Functional” (green polygons) or “Junk” (red crosses) in a specific time slice. Along different time slices, “Junk” elements may be converted into “Functional” ones (if they acquire “maintenance functions”) and vice-versa (if they are no longer maintained by selection). On the other hand, “Spam DNA”, highlighted in the blue shade, depends on neutral processes causing the persistence of this element along the time, irrespective of their future function.
W-elements in Schistosoma, “Junk DNA,” and Function
Stitz et al. (2021) used bioinformatics and omics data to characterize 19 W-element families (WEFs) in the genome of S. mansoni. Differently from the initial observation that W-elements (WEs) were restricted to the W chromosome, they found homologs of all WEFs in the autosomes (though only four WEFs had full-length copies). They suggested that WEs could be a class of transposable elements and that their similarities with noncoding RNAs (ncRNAs) indicated a potential effect on gene expression, in line with the observation of differential expression among samples and biological replicates. Finally, they showed that some WEFs contained sequences similar to the hammerhead (HHD) class of ribozymes and that some of these were able to perform the expected self-cleavage reaction. The hypothesis raised by the authors is that WEs increase genome plasticity by transposition and by altering gene regulation. Should WEFs be considered “Junk DNA”?
Susumo Ohno coined the term “Junk DNA” (Ohno 1972) in the wake of the discussions about genome size and the lack of any obvious correlation with biological complexity—“genomes carry some fraction of DNA that has little or no adaptive advantage for the organism at all” (Eddy 2012). In Ohno’s own words, “is it a wonder that our genome too is filled with the remains of extinct genes?” (Ohno 1972). While it may be possible that some WEs in S. mansoni play regulatory roles that are important for the organism survival (and therefore are no longer “Junk DNA”), a large fraction of WEs is constituted by partial sequences or have no associated ncRNA motifs (Stitz et al. 2021). As suggested by others, “Junk DNA” should be considered as a null hypothesis in genomics (Niu and Jiang 2013; Koonin 2016). Is it reasonable to assume that these elements play an adaptive role for the organism?
The previous question is important because if the days of “Junk DNA” were over, one would have to generalize the findings obtained for specific (putatively functional) WEs onto WEs as a whole and then onto transposable elements as a whole, etc. Otherwise, the genome of S. mansoni would still harbor a significant amount of “junk.” A similar point has been made by Graur et al. (2013) against ENCODE estimates of functional elements in the human genome. Perhaps more important, however, is the discussion about how we recognize biological function and what we mean by it. There is good evidence that WEs are present in the transcriptome in diverse conditions and that some elements with the predicted HHD motif have a ribozyme action (Stitz et al. 2021). Aside from that, the functional predictions “are mainly based on bioinformatics analyses and have to be substantiated by functional analyses” (McGrath 2021). Thus, we do not know if the predicted ncRNAs do affect gene expression nor (and this is the most important) if they have any effect on fitness. Differential WE expression across life stages could simply reflect the expression of distinct genomic regions, while variation between replicates could suggest transcriptional noise.
Finally, what do we mean by biological function? Should we use the selected effect (SE) or the causal role (CR) definition for it (Graur et al. 2013; Doolittle and Brunet 2017; Doolittle 2018)? In short, CR aims at answering “what it does,” while for SE, the true question is “why it is there” (Doolittle 2018). Commonly used examples for the inadequacy of the CR definition in biology are that 1) it is not the function of the heart to make sound (even if it does) and that 2) it is not the function of trinucleotide repeats in the HTT gene to cause Huntington’s disease (even if it does). Conversely, an element has SE function if it contributes to organism fitness. As noted by others, the discussion about “Junk DNA” only makes sense under the SE definition (Doolittle 2013; Graur et al. 2013). Stitz et al. (2021) test for function under a CR rationale, but this can only give us candidates for evolved (SE) biological functions (Thomas 2017). The days of “Junk DNA” are not over.
Beyond SE, and the Multiple Origins of Functional Elements
Even if we agree that SE better represents “proper” biological function, it is much harder to infer biological function under the SE definition (compared with CR). In principle, sequence conservation is a proxy for function because purifying (or negative) selection eliminates deleterious mutations that affect fitness (Graur et al. 2013). However, sequence conservation is less relevant in the case of 4-fold degenerated codon positions, which are clearly not “Junk DNA” (Palazzo and Lee 2015), and a similar argument can be made for DNA performing structural functions, which have also been called “nucleoskeletal” (Cavalier-Smith 1978) and “nucleotypic” (Gregory 2001).
There is, nonetheless, another difficulty with the SE definition. When trying to answer the “why” question, there is a horizontal dimension that reflects how a given genomic element currently affects fitness, but there is also a vertical dimension that reflects how a given genomic element originated and has been maintained in the genome. In some sense, this is obvious, as “nothing in biology makes sense except in the light of evolution” (Dobzhansky 1973). However, what if different evolutionary processes contribute differently to these dimensions?
Doolittle and Brunet (2017) propose that three questions must be made for assigning proper biological function to a DNA segment, which can be briefly summarized as follows:
Is it (this DNA segment) expressed in the phenotype?
Does such expression make a positive contribution to organismal fitness?
Is it present in the genome due to past positive selection related to such expression?
It should be clear that questions 1 and 2 are “horizontal,” while question 3 is “vertical.” These authors conclude that only when we answer “yes” to all three questions a DNA segment can earn a true SE status (and thus be called functional). This has been called the “strong SE” approach (SSE). Therefore, under the SSE definition, DNA segments representing exaptations (Gould and Vrba 1982) or resulting from constructive neutral evolution (CNE) (Stoltzfus 1999), that represent a “yes” for the first two questions, but a clear “no” for the third one should not be considered functional, as they originated by processes unrelated to positive natural selection.
Different research groups have been debating whether the third question should be a sine qua non for function. For example, Linquist et al. (2020) proposed to distinguish between maintenance functions (if conditions 1 and 2 above are satisfied) and origin functions (if condition 3 is satisfied). A different view is held by Brzović and Šustar (2020), who put the emphasis on the action of purifying selection (similar to the idea of maintenance functions), calling it the “weak etiological monism” (WEM) principle, which would be more appropriate than SSE. More recently, Brunet et al. (2021), who favors the SSE definition for biological function but recognizes the role of exaptation and CNE in genome evolution, argued that the WEM suffers from two major drawbacks. First, it could lead to a non-Darwinian account of biology, and second, it would allow that several traits that arose neutrally be considered functional even though they did not originate “for” any adaptive role. The abovementioned discussion is fundamental to the “Junk DNA” debate because the latter is defined by the lack of (evolved) biological function. Paradoxically as it may seem, under the SSE definition, elements that contribute positively to fitness and are maintained by purifying selection would still count as “junk” only because they did not originate as an adaptation.
While we agree that positive selection is the fundamental process by which novelties arise under the Darwinian canon, WEM does emphasize fitness and selection, which are central to Darwinian thought. Commenting on the impact of reading Malthus’ essay, Darwin wrote “it at once struck me that under these circumstances favorable variations would tend to be preserved, and unfavorable ones to be destroyed” (our emphasis) (Darwin, 1887). Furthermore, the notion of positive and purifying selection is much associated with the fitness impact of new mutations (Eyre-Walker and Keightley 2007). However, because Darwin had no mechanism for the origin of variation, stating that only positive selection is genuinely “Darwinian” is, in our opinion, inadequate. Perhaps a stronger argument in support of WEM is that modern evolutionary theory has no problem in recognizing that other evolutionary processes can lead to evolutionary adaptive novelties (Koonin 2011; Brosius 2019; Muñoz-Gómez et al. 2021).
Defining Function and “Junk DNA”—The Horizontal Perspective
Suppose we were able to delete from the genome an element that originated via exaptation (or CNE) and that this leads to the death of the organism. Is it reasonable to say that this genomic element is not functional (in the SE sense)? In our opinion, the answer is no. As this example illustrates, the SEE principle may result in a paradox, at least regarding any notion of “function” minimally grounded on biological intuition. Therefore, if a given genomic element maintains the adaptive value of the organism, it fully qualifies as functional. In other words, “maintenance function” is sufficient to ascribe proper biological function (fig. 1).
Returning to “Junk DNA”, when Ohno came up with this concept he was trying to explain an observation: Why organisms have DNA that does not contribute to fitness. This is a “horizontal” question that can be fully accounted for using the WEM criterium for biological function. Even if we concede that the origin of elements constituting “Junk DNA” was also relevant in Ohno’s formulation, it was not the major pattern begging for an explanation. Thus, we submit that “Junk DNA” is represented by all genomic elements, informational or structural, that do not contribute to the fitness of the organism. In other words, that are currently devoid of “maintenance function.”
“Spam DNA”—The Vertical Perspective
If “Junk DNA” is to be restricted to the horizontal dimension, some solution must be provided to deal with the origins of genomic elements. When a given genomic element originates (by mutation, recombination, transposition, horizontal transfer, etc.) there are two possibilities, either it is adaptive (i.e., it confers an evolutionary advantage to its possessor due to informational or structural roles played by it) or it is nonadaptive (neutral or deleterious). If the element is adaptive, positive selection is the evolutionary process responsible for its persistence. On the other hand, when nonadaptive elements persist in the genome, they do so despite not being selected for (from the organismal perspective). Even if slightly deleterious, nonadaptive DNA may persist in genomes of organisms having low effective population size (Lynch 2007). Transposable elements and other selfish DNA (Doolittle and Sapienza 1980; Orgel and Crick 1980; Ågren and Clark 2018), for example, are bona fide nonadaptive DNA.
As previously discussed, nonadaptive DNA can be exapted for novel functions or can be involved in CNE (Brosius 2019; Muñoz-Gómez et al. 2021). On the horizontal perspective, this marks the transition “from Junk to Function,” but they remain nonadaptive in their origin (fig. 1). A similar, though more radical alternative, is the “Genome Balance Hypothesis,” that suggests that blooms of selfish elements become ultimately selected for maintaining a balanced transcription of networked genes (Freeling et al. 2015). For such cases, distinguishing among the horizontal and the vertical perspectives is crucial. We propose that “Spam DNA” may be a useful term to account for the vertical dimension (persistence). Similar to what happens in our email boxes, “Spam DNA” accumulates in the genome despite not being selected for, neither it accumulates to serve a future purpose. “Spam DNA” represents every genomic element which has not been selected for during its origin in the genome, even if it currently participates in relevant biological functions. Importantly, “Spam DNA” is not a more inclusive term that encompasses all “Junk DNA,” but truly a historical definition (fig. 1). For example, some cases of gene inactivation may originate “Junk DNA” from “non-Spam” precursors. During the process of nonorthologous gene displacement, nonorthologous genes become responsible for the same essential function in different organisms, which may lead to subsequent gene inactivation (in which case it persists as “Junk DNA”) and loss (Koonin et al. 1996, Koonin 2011).
Conclusions
In summary, we propose “Spam DNA” as a companion to “Junk DNA” to account for the origin of genome elements that persisted in the genome despite no positive selection (fig. 1). Even though considered derogatory by many scientists (Brosius and Gould 1992; Makalowski 2003), the term “Junk DNA” caught on. While we cannot know if “Spam DNA” will be that long-lived, we believe that it adds a layer of complexity to the discussion of genome evolution while being of easy understanding. Current molecular evolution theory suggests that genomic parasites are an expected feature of cellular organisms (Iranzo et al. 2016). Therefore, “Junk DNA” and “Spam DNA” are not meant to be only “catchy” terms but relevant concepts in genome evolution. It would be possible to test if a given genomic element constitutes “Spam DNA” by investigating signatures of positive selection associated with this element on the phylogenetic branch in which it originates, even though this may be challenging.
“Junk DNA” (and we hope “Spam DNA” as well) is a very good term to caution against a pan-adaptationist view of evolution (Gould and Lewontin 1979; Koonin 2016), taking in full account the possibility of tinkering (Jacob 1977), which is illustrated by the argument that “Junk DNA” (which is kept) is different from “Garbage DNA” (which is thrown away) (Graur et al. 2013). In this sense, it is interesting to note that while other terms such as “Spandrel DNA” or “Tinker DNA” could have been proposed instead of “Spam DNA,” they could give the wrong impression that nonadaptive elements persist in a genome with the purpose of playing some beneficial role in the future (Makalowski 2003), which is clearly not the case.
Finally, it is interesting to note that Brunet et al. (2021) defended the SSE definition for biological function as a means of avoiding the perils of pan-adaptationism, on the one hand, and of intelligent design, on the other. We have the opposite view. Explicitly allowing proper biological functions to arise neutrally and/or by tinkering represents an even stronger case for the lack of “design” in the genomes of living organisms, including our own (Avise 2010).
Acknowledgments
We thank Diogo Meyer, Guillermo Reales, Rodrigo Ligabue-Braun and three anonymous reviewers for comments on the first version of the manuscript. We thank Coordenação de Aperfeiçoamento de Pessoal de Nível Superior for a scholarship awarded to R.B-M., and Conselho Nacional de Desenvolvimento Científico e Tecnológico for scholarships awarded to P.I.C.C.F., M.V., and A.L.S.Z.
Contributor Information
Nelson J.R. Fagundes, Postgraduate Program in Genetics and Molecular Biology, Institute of Biosciences, Federal University of Rio Grande do Sul, Porto Alegre, Brazil Postgraduate Program in Animal Biology, Institute of Biosciences, Federal University of Rio Grande do Sul, Porto Alegre, Brazil.
Rafael Bisso-Machado, Postgraduate Program in Genetics and Molecular Biology, Institute of Biosciences, Federal University of Rio Grande do Sul, Porto Alegre, Brazil.
Pedro I.C.C. Figueiredo, Postgraduate Program in Genetics and Molecular Biology, Institute of Biosciences, Federal University of Rio Grande do Sul, Porto Alegre, Brazil
Maikel Varal, Postgraduate Program in Genetics and Molecular Biology, Institute of Biosciences, Federal University of Rio Grande do Sul, Porto Alegre, Brazil.
André L.S. Zani, Postgraduate Program in Genetics and Molecular Biology, Institute of Biosciences, Federal University of Rio Grande do Sul, Porto Alegre, Brazil
Literature Cited
- The Lancet . 2012. Cracking ENCODE. Lancet 380:950. [DOI] [PubMed] [Google Scholar]
- ENCODE Project Consortium . 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ågren JA, Clark AG. 2018. Selfish genetic elements. PLoS Genet. 14:e1007700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avise JC. 2010. Footprints of nonsentient design inside the human genome. Proc Natl Acad Sci U S A. 107:8969–8976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J. 2019. Exaptation at the molecular genetic level. Sci China Life Sci. 62:437–452. [DOI] [PubMed] [Google Scholar]
- Brosius J, Gould SJ. 1992. On “genomenclature”: a comprehensive (and respectful) taxonomy for pseudogenes and other “junk DNA”. Proc Natl Acad Sci U S A. 89:10706–10710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet TDP, Doolittle WF, Bielawski JP. 2021. The role of purifying selection in the origin and maintenance of complex function. Stud Hist Philos Sci. 87:125–135. [DOI] [PubMed] [Google Scholar]
- Brzović Z, Šustar P. 2020. Postgenomics function monism. Stud Hist Philos Biol Biomed Sci. 80:101243. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. 1978. Nuclear volume control by nucleoskeletal DNA, selection for cell volume and cell growth rate, and the solution of the DNA C-value paradox. J Cell Sci. 34:247–278. [DOI] [PubMed] [Google Scholar]
- Darwin F. 1887, editors. The life and letters of Charles Darwin, including an autobiographical chapter. London: (UK: ): John Murray. [Google Scholar]
- Dobzhansky T. 1973. Nothing in biology makes sense except in the light of evolution. Am Biol Teach. 35:125–129. [Google Scholar]
- Doolittle WF. 2013. Is junk DNA bunk? A critique of ENCODE. Proc Natl Acad Sci U S A. 110:5294–5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle WF. 2018. We simply cannot go on being so vague about ‘function’. Genome Biol. 19:223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle WF, Brunet TDP. 2017. On causal roles and selected effects: our genome is mostly junk. BMC Biol. 15:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle WF, Sapienza C. 1980. Selfish genes, the phenotype paradigm and genome evolution. Nature 284:601–603. [DOI] [PubMed] [Google Scholar]
- Ecker J, et al. 2012. ENCODE explained. Nature 489:52–54. [DOI] [PubMed] [Google Scholar]
- Eddy SR. 2012. The C-value paradox, junk DNA and ENCODE. Curr Biol. 22:R898–R899. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A, Keightley PD. 2007. The distribution of fitness effects of new mutations. Nat Rev Genet. 8:610–618. [DOI] [PubMed] [Google Scholar]
- Freeling M, Xu J, Woodhouse M, Lisch D. 2015. A solution to the C-value paradox and the function of junk DNA: the genome balance hypothesis. Mol Plant. 8:899–910. [DOI] [PubMed] [Google Scholar]
- Gould SJ, Lewontin RC. 1979. The spandrels of San Marco and the Panglossian paradigm: a critique of the adaptationist programme. Proc R Soc Lond B Biol Sci. 205:581–598. [DOI] [PubMed] [Google Scholar]
- Gould SJ, Vrba E. 1982. Exaptation—a missing term in the science of form. Paleobiology 8:4–15. [Google Scholar]
- Graur D, et al. 2013. On the immortality of television sets: “function” in the human genome according to the evolution-free gospel of ENCODE. Genome Biol Evol. 5:578–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory TR. 2001. The bigger the C-value, the larger the cell: genome size and red blood cell size in vertebrates. Blood Cells Mol Dis. 27:830–843. [DOI] [PubMed] [Google Scholar]
- Iranzo J, Puigbò P, Lobkovsky AE, Wolf YI, Koonin EV. 2016. Inevitability of genetic parasites. Genome Biol Evol. 8:2856–2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F. 1977. Evolution and tinkering. Science 196:1161–1166. [DOI] [PubMed] [Google Scholar]
- Koonin EV. 2011. The logic of chance: the nature and origin of biological evolution. Upper Saddle River (NJ): FT Press. [Google Scholar]
- Koonin EV. 2016. Splendor and misery of adaptation, or the importance of neutral null for understanding evolution. BMC Biol. 14:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Mushegian AR, Bork P. 1996. Non-orthologous gene displacement. Trends Genet. 12:334–336. [PubMed] [Google Scholar]
- Linquist S, Doolittle WF, Palazzo AF. 2020. Getting clear about the F-word in genomics. PLoS Genet. 16:e1008702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. 2007. The frailty of adaptive hypotheses for the origins of organismal complexity. Proc Natl Acad Sci U S A. 104:8597–8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makalowski W. 2003. Genomics. Not junk after all. Science 300:1246–1247. [DOI] [PubMed] [Google Scholar]
- McGrath C. 2021. Highlight – “junk DNA” no more: repetitive elements as vital sources of flatworm variation. Genome Biol Evol. 13:evab217. [Google Scholar]
- Muñoz-Gómez SA, Bilolikar G, Wideman JG, Geiler-Samerotte K. 2021. Constructive neutral evolution 20 years later. J Mol Evol. 89:172–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu D-K, Jiang L. 2013. Can ENCODE tell us how much junk DNA we carry in our genome? Biochem Biophys Res Commun. 430:1340–1343. [DOI] [PubMed] [Google Scholar]
- Ohno S. 1972. So much “junk” DNA in our genome. Brookhaven Symp Biol. 23:366–370. [PubMed] [Google Scholar]
- Orgel LE, Crick FH. 1980. Selfish DNA: the ultimate parasite. Nature 284:604–607. [DOI] [PubMed] [Google Scholar]
- Palazzo AF, Gregory TR. 2014. The case for junk DNA. PLoS Genet. 10:e1004351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzo AF, Lee ES. 2015. Non-coding RNA: what is functional and what is junk? Front Genet. 6:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennisi E. 2012. Genomics. ENCODE project writes eulogy for junk DNA. Science 337:1159, 1161. [DOI] [PubMed] [Google Scholar]
- Stitz M, et al. 2021. Satellite-like W-elements: repetitive, transcribed, and putative mobile genetic factors with potential roles for biology and evolution of Schistosoma mansoni. Genome Biol Evol. 13:evab204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltzfus A. 1999. On the possibility of constructive neutral evolution. J Mol Evol. 49:169–181. [DOI] [PubMed] [Google Scholar]
- Thomas PD. 2017. The gene ontology and the meaning of biological function. In: Dessimoz C, Škunca N, editors. The gene ontology handbook, methods in molecular biology. Vol. 1446. New York (NY): Humana Press. p. 15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]