Abstract
The ongoing outbreak of the severe acute respiratory syndrome‐coronavirus 2 (SARS‐CoV‐2) has spread globally and poses a threat to public health and National economic development. Rapid and high‐throughput SARS‐CoV‐2 RNA detection without the need of RNA extraction and amplification remain a key challenge. In this study, a new SARS‐CoV‐2 RNA detection strategy using a microfluidic biochip for the rapid and ultrasensitive detection of SARS‐CoV‐2 without RNA extraction and amplification was developed. This new strategy takes advantage of the specific SARS‐CoV‐2 RNA and probe DNA reaction in the microfluidic channel, fluorescence signal regulation by nanomaterials, and accurate sample control by the microfluidic chip. It presents an ultralow limit of detection of 600 copies mL−1 in a large linear detection regime from 1 aM to 100 fM. Fifteen samples were simultaneously detected in 40 min without the need for RNA purification and amplification. The detection accuracy of the strategy was validated through quantitative reverse transcription polymerase chain reaction (qRT‐PCR), with a recovery of 99–113 %. Therefore, the SARS‐CoV‐2 RNA detection strategy proposed in this study can potentially be used for the quantitative diagnosis of viral infectious diseases.
Keywords: amplification-free methods, biosensors, nucleic acids, RNA purification-free methods
Rapid, ultrasensitive, extraction‐free and amplification‐free strategy for the detection of severe acute respiratory syndrome‐coronavirus 2 (SARS‐CoV‐2) RNA using a microfluidic biochip that exhibits an ultralow detection limit (600 copies mL−1) and the ability to simultaneously evaluate 15 RNA samples within 40 min. The proposed RNA detection strategy has prospective applications in SARS‐CoV‐2 detection and COVID‐19 (Corona virus disease 2019) patient diagnosis.
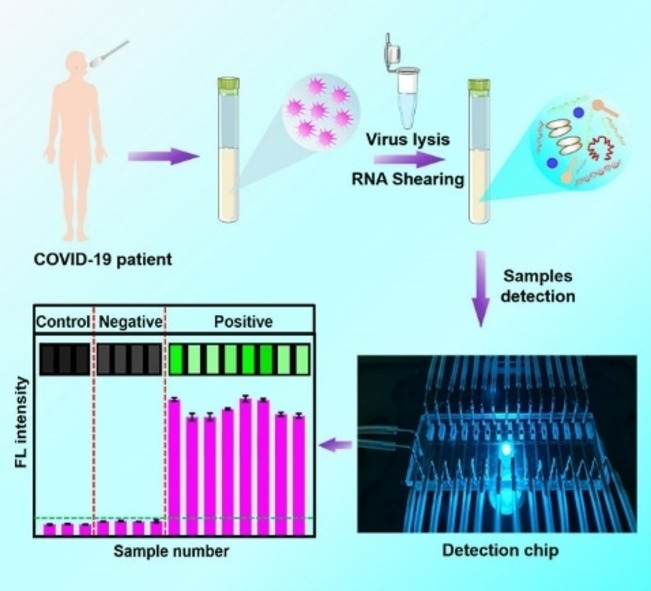
Introduction
Coronavirus disease 2019 (COVID‐19) is a major global health threat, caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2). [1] Development of rapid, accurate, and sensitive molecular diagnostic screening techniques is necessary for a quick and efficient public health response to the emerging viral threat. The currently used detection methods include antibody and nucleic acid detection due to the structural characteristics of SARS‐CoV‐2, with a single positive‐strand RNA genome, containing a spike (S) protein, envelope (E), matrix (M), and a nucleocapsid (N) protein. [2] Antibody detection is used to detect SARS‐specific antibodies produced after infection, such as IgG, IgM and IgA etc. [3] This method can be used for tracking and confirming antibodies after viral infection, recovery, and vaccine injection, but cannot be used to effectively prevent the spread of the disease. [4] The nucleic acid detection method, which accurately targets specific genes, is combined with nucleic acid extraction and amplification techniques for highly sensitive SARS‐CoV‐2 detection. [5] Nucleic acid detection is widely used in the diagnosis and prevention of diseases, and is also a relatively accurate detection method. [6]
Currently, real‐time quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) is the gold standard for diagnosing and confirming the SARS‐CoV‐2 infection. qRT‐PCR has excellent selectivity and sensitivity. However, it is time‐intensive, requires highly qualified technical personnel, and can only be operated in a laboratory‐based hospital with access to large stationary equipment and reagents. [7] To overcome these drawbacks, isothermal nucleic acid amplification assays based on recombinase polymerase amplification, [8] DNA nanoscaffold‐based hybrid chain reaction, [9] loop‐mediated isothermal amplification, [10] and nucleic acid sequence‐based amplification, [11] were employed. However, isothermal amplification may produce false‐positive results when detecting homologous sequences, and the results usually have to be confirmed by agarose gel electrophoresis using SYBR dyes, [12] Clustered regularly interspaced palindromic repeat (CRISPR)‐based nucleic acid detection technology has attracted much attraction as a simply and sensitive SARS‐CoV‐2 RNA detection method. It performs nucleic acid detection by pre‐amplification of the target nucleic acid, followed by CRISPR‐caspase (Cas) enzymes (e. g., Cas9, Cas10, Cas12a, Cas13a, etc) mediated nucleic acid detection, [13] Although CRISPR‐based nucleic acid detection is simple and highly sensitive, it increases the pre‐amplification process, prolongs the detection time, and may cause false‐negative or false‐positive results due to amplification errors. The rapid development of nanoscale science and technology has made field effect transistor (FET)‐based SARS‐CoV‐2 RNA detection possible. FET‐based biosensors[ 3b , 14 ] based on (molybdenum disulfide) MoS2, gold (Au) nanoparticles, graphene, and other nanomaterials can effectively detect the SARS‐CoV‐2 RNA with multiple advantages including the ability to use a small amount of analytes, high sensitivity, and instantaneous measurement. However, FET‐based biosensors have a complicated construction process. Electrochemical biosensors,[ 5b , 6a , 15 ] which convert the signals generated by chemical reactions between different reagents and samples into electrical signals for complete detection, show the advantages of having high sensitivity, low cost, and user‐friendly nature. However, the addition and reaction of different reagents, as well as the extraction and amplification of nucleic acids in the detection process, prolong the overall detection time. The development of microfluidic technology has greatly improved the accuracy of SARS‐CoV‐2 RNA detection. The combination of microfluidic technology with CRISPR‐Cas, FET, and electrochemistry etc. not only shows the advantages of the original techniques, but also enhances the overall accuracy and efficiency of detection.[ 13b , 16 ] However, the extraction and amplification of nucleic acids is still essential, prolonging the overall detection time of these methods. Therefore, there is an urgent need to develop a SARS‐CoV‐2 RNA detection method, without the requirement of nucleic acid extraction and amplification, which could meet the requirements of high throughput screening, low cost, high sensitivity, and ease of operation to effectively detect and block the transmission COVID‐19.
Here, we propose an extraction‐free, amplification‐free, ultrasensitive fluorescence‐based SARS‐CoV‐2 RNA rapid detection method based on a nanomaterial hybrid microfluidic biochip including 15 parallel sensing units. Each sensing unit is independent from each other, avoiding interference between samples during detection. High signal‐to‐noise ratio of the biochip and the high‐precision laser scanner enables accurate detection of targets signal. The hybridization between DNA probe and target RNA presents excellent specificity, and false amplification is greatly reduced because of the ellipsis of target sequence copying. The whole process does not involve any enzyme and expensive regents, and the cost of each test is less than 1$, even cheaper for large scale production. The SARS‐CoV‐2 particles were treated in a lysis solution with ultrasound for 5 min to break the RNA into short RNAs, and then the treated samples were loaded into the microfluidic biochip on which the designed fluorescence‐labeled SARS‐CoV‐2 DNA probes reacted with SARS‐CoV‐2 RNAs in the reaction zone. Fluorescence was detected in the detection zone. SARS‐CoV‐2 pseudovirus was used to demonstrate the capability of the detection strategy. SARS‐CoV‐2 RNA of the 15 samples could be detected in 40 min with a limit of detection (LOD) of 600 copies/mL. Moreover, the accuracy of detection can be improved using multiple designed DNA probes at different sites. Finally, qRT‐PCR was used to verify the rationality and consistency of the chip detection strategy, and at detection recovery of 99–113 % was achieved.
Results and Discussion
Detection principle of SARS‐CoV‐2 RNA without RNA extraction and amplification based on a microfluidic biochip
SARS‐CoV‐2, identified as a single positive‐strand RNA genome [17] is shown in Figure 1a. It includes four specific parts: open reading frame (ORF)‐1a/b, N, E, and S genomes. Detection probes were designed at these sites, and the detailed sequences of the probes and target RNAs are shown in Table S1. The detection process for SARS‐CoV‐2 RNA is shown in Figure 1b. Viruses in the collected throat or nose swab samples were lysed to release the RNA, which is sheared simultaneously during SARS‐CoV‐2 particle lysis by sonication to produce short segments. The target RNA segments were then loaded into the designed DNA probe fixed reaction channel, where the target RNA segments reacted with probes to form a double‐stranded DNA‐RNA complex. The ability of graphene oxide (GO) loaded on glass substrate (Figure S1) to absorb single‐stranded nucleic acids and release double‐stranded nucleic acids enables the double‐stranded DNA‐RNA complex to desorb from the GO nanomaterial surface. The released double‐stranded nucleic acids were pushed into the detection chamber assembled with poly‐L–Lysine (PLL). Then, the DNA‐RNA complex was absorbed by PLL via electrostatic absorption, and the fluorescence intensity from DNA probes enabled the quantitative detection of the target RNA. A laser scanner was used to excite the fluorescent molecules on the DNA probes in the detection area.
Figure 1.
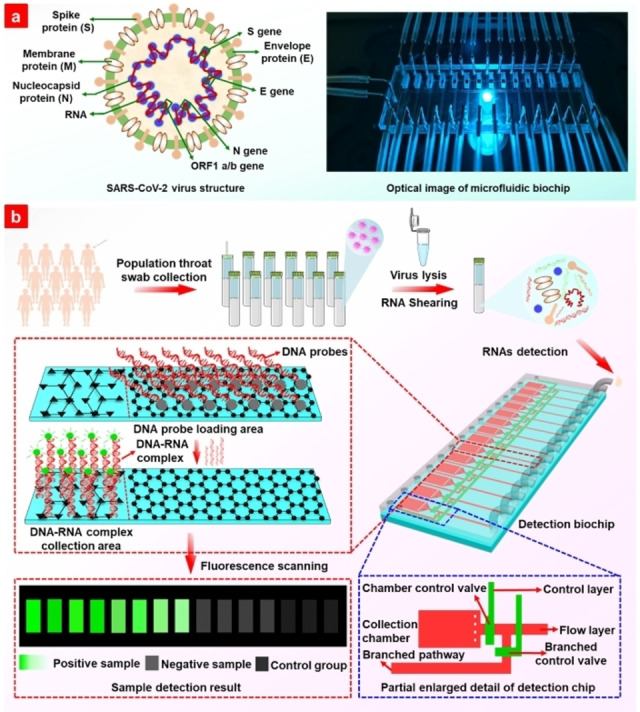
(a) Schematic structure of severe acute respiratory syndrome‐coronavirus 2 (SARS‐CoV‐2) and optical photo of the microfluidic biochip. (b) Schematic procedure for SARS‐CoV‐2 RNA detection without extraction and amplification.
SARS‐CoV‐2 RNA detection performance of the microfluidic biochip
The synthesized RNA segments complementary to the probes were used as targets to demonstrate the basic performance of the biochip. The detected fluorescence signal is the integration of the fluorescence intensity in the entire detection chamber. As shown in Figure S2a, the detected fluorescence signal of 10−9 M target RNA increased when the concentration of probes increased during immobilization with the substrate in the reaction channel. As a result, a maximum probe concentration of 10−4 M was used in subsequent experiments. Figure S2b and c show the shortest probe incubation time with the substrate, and the hybridization time of probe and target RNA was 35 min and 30 min, respectively. The results in Figure S2 indicate that all four pairs of probes and target RNAs were under similar experimental conditions. The detection signal began to decrease when the sample injection rate increased from 1.25 μL/min as shown in Figure 2a, because the rapid loading rate affected the binding efficiency of the DNA probe with the target RNA (a pair of D−E1 and R−E1 was taken as an example). A sample injection rate of 1 μL/min was used to ensure adequate reaction between the probes and target RNAs. Figure 2b shows that a stronger signal was achieved when the loaded sample volume was increased because of the increased total targets, which is extremely important for samples with ultralow targets. The target concentration of 10−18 M in Figure 2b shows similar trends as the target concentrations of 10−13 M and 10−15 M in Figures S3a and b.
Figure 2.
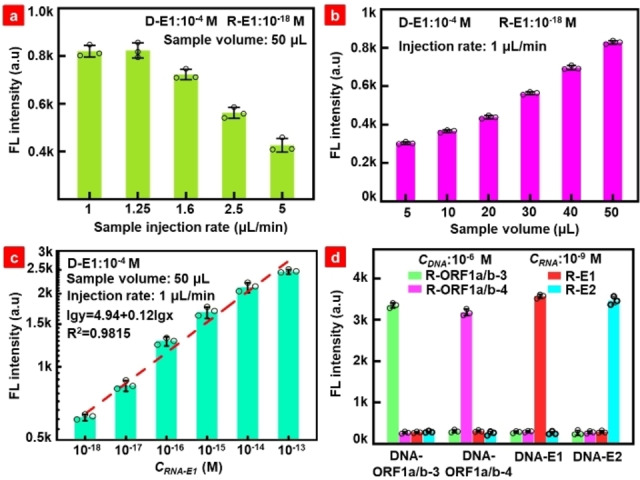
(a) Fluorescence intensity dependence on the sample injection rate with total volume of 50 μL. (b) Fluorescence intensity dependence on the sample volume at the injection rate of 1 μL/min. (c) Quantitative relationship between the target RNA and fluorescence intensity. (d) Detection specificity of the designed DNA probes to SARS‐CoV‐2 RNAs. The circle represents the measured fluorescence values in three repeated experiments.
With the optimized process, the concentration of R−E1 presents a linear relationship with the detected fluorescence intensity on the log scale in the concentration range of 10−13 to 10−18 M (Figure 2c), which is similar to that of the target RNA at another site, ORF1a/b‐3, in Figure S3c. Figure S4 shows the fluorescence scanning map and heat map for different target RNA concentrations, indicating obvious differences in the fluorescence signals. It is worth mentioning that the detected fluorescence signal of the sample is the integration of fluorescence intensity in the entire detection chamber, as shown by the yellow dashed box in Figure S4. LODs of 0.564 aM (338 copies/mL) and 0.643 aM (387 copies/mL) were achieved for R−E1 and R‐ORF1a/b‐3, respectively, using the formula LOD=3Sb1/S. [18] Based on the International Union of Pure and Applied Chemistry (IUPAC) guideline of a 2 : 1 signal to noise ratio, [19] the calculated lower LODs were 0.414 aM (249 copies/mL) and 0.191 aM (115 copies/mL), respectively. A detailed calculation of the LOD is presented in Table S2. The specific reaction results for different pairs of probes and target RNAs are shown in Figure 2d, in which the complementary pairs presented a much stronger signal than the mismatched pairs, indicating the high specificity of the designed probes and the excellent detection performance of the proposed biochip.
SARS‐CoV‐2 pseudovirus RNA detection without RNA extraction and amplification
To simulate the detection of actual samples, pseudoviruses were used to exploit the biochip detection performance. For comparison, RNA purification kit and direct lysis were used to extract the RNA from pseudoviruses, followed by RNA shearing, as shown in Figure 3a. A sonication power of 720 W was used to shear long RNA, and the samples treated using the two approaches presented similar results. The detected signal increased with a sonication time shorter than 5 min. The fluorescence signal tended to be stable when the sonication time increased from 5 min to 10 min; therefore 5 min of sonication was favored for practical applications. We measured the length of RNA by agarose gel electrophoresis after ultrasonication for 5 min, as shown in Figure S5. The result shown that the length of the RNA after the sheraed was approximately 100 bp. For different sonication times, the directly lysed sample showed a higher fluorescence signal than the kit extracted sample, which was also exploited by the scanned fluorescence image and heat map in Figure 3b; the possible reason for this may be the unavoidable RNA loss during the complex manipulation process according to the kit protocol. To further simplify the detection process and reduce the detection time, the lysis process was conducted simultaneously with sonication, which means that sonication started once the lysis buffer was loaded into the sample instead of a separate lysis step followed by sonication. The experimental results in Figure S6 show that both treatments had similar fluorescence signals with 3 min and 5 min sonication, and simultaneous lysis and sonication‐shearing presented slightly higher fluorescence intensity. GO nanomaterials assembled in the reaction channel immobilize the single‐strand probes via π‐π stacking [20] and are not affected by other factors in the lysis buffer. The hybridization of the DNA probe and target RNA weakens the interaction between the probe and GO, enabling the release of the DNA‐RNA complex from the GO substrate and further collection by the PLL chamber. Experiments confirmed that the proposed biochip does not require an RNA purification step, which not only simplifies the sample pretreatment process and reduces treatment time, but also prevents the loss of nucleic acids in samples.
Figure 3.
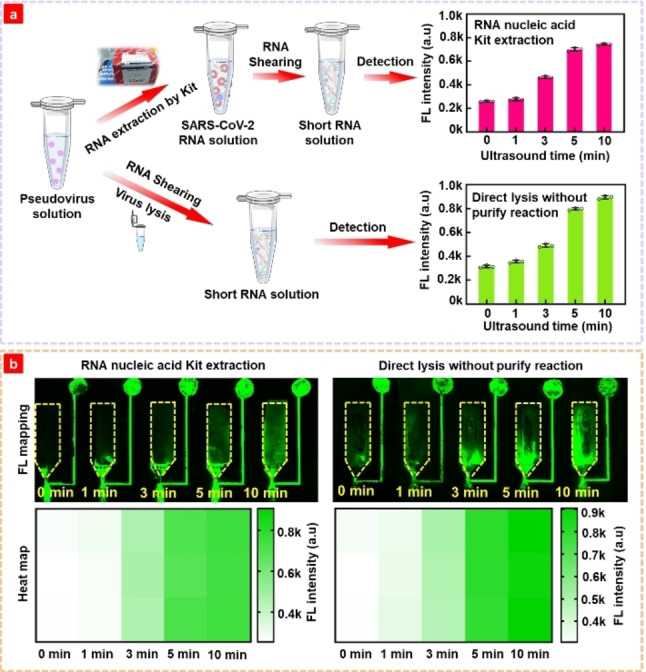
(a) Detection schematic and detected fluorescence of pseudovirus samples obtained by the commercial extraction of SARS‐CoV‐2 RNA using RNeasy Mini Kit and Genomic lysis buffer with different ultrasound time. (b) Fluorescence scanning map and heat map corresponding to the fluorescence intensity result in Figure 3a.
SARS‐CoV‐2 RNA detection using multiple DNA probes without RNA extraction and amplification
During detection, the long RNA is sheared into short segments. More targets can be detected if multiple DNA probes for more genome sites are used. Thus, a total of ten DNA probes at different sites were designed according to SARS‐CoV‐2 RNA genome specificity to test the SARS‐CoV‐2 RNA standard. The SARS‐CoV‐2 pseudovirus particle concentration of 1×10−18 M was used, which had ∼600 copies/mL of the whole RNA sequence. The fluorescence signal increased when DNA probe sites increased for different sample loading rates, as shown in Figure 4. With the increase in the number of designed DNA probes, the possibility of capturing the target RNA segments increases because RNA segments from different sites remain the same after random shearing. The fluorescence mapping in Figure 4b clearly shows the change in fluorescence intensity with the number of DNA probes at different injection rates, and the corresponding heat map was derived by the integration of the fluorescence signal in the whole detection chamber (yellow dashed box). Experimental results indicate that the use of multiple‐site DNA probes is a promising strategy to improve the detection sensitivity. To verify the specificity of the DNA probes with the Middle East respiratory syndrome (MERS), SARS and SARS‐CoV‐2 RNAs, these pseudoviruses were lysed and ultrasound to prepare detection samples. Different pseudovirus samples (the concentrations of all three samples were 600 copies/mL) were loaded into the microchannel to react with the DNA probes, and the fluorescence signal was scanned using a laser scanner. Figure 4c presents at heatmap of the detected fluorescence intensity, in which the color depth represents the fluorescence intensity of each detection. SARS‐CoV‐2 showed much stronger signal intensity than MERS and SARS pseudoviruses, indicating the excellent specificity of the proposed biochip. To further verify the detection interference of multiple DNA probes, the DNA probe E1 (10−4 M×2 μL) used to detect the target RNA segment in pseudovirus samples of different concentration (10−15, 10−16, 10−17, and 10−18 M; 50 μL) to verify the detection sensitivity, and the results are shown in Figure S7. The line fitting formula of fluorescence intensity and concentration is lgy=5.08+0.12lgx. The slope in the fitting formula is consistent with that observed in Figure 2c, indicating that the sensitivity remains consistent with different probes. Furthermore, the research results indicates that multiple probes detection greatly improved the detection sensitivity. Therefore, ten probes were used to detect different concentration pseudovirus (10−17, 10−18, 10−19, and 10−20 M; 50 μL), as shown in Figure S8. The lowest detection concentration of 10−20 M can be achieved. The result indicated that the application of multiple DNA probes greatly improves the detection sensitivity of microfluidic biochip. To verify the performance of the microfluidic biochip in detecting actual samples, we used pseudovirus diluted in throat swab solution to different concentrations (10−17 M, 10−18 M, 10−19 M), and conduct RNA detection. The fluorescence detection results are shown in Figure S9a. The RNA detection deviation from the actual spiked‐concentration is analyzed and shown in Figure S9b. The results showed that the relative deviation range between the detected RNA concentration and the spiked RNA concentration was 1.7 %∼8.8 %, indicating that the microfluidic biochip could accurately detect the specific genes in SARS‐CoV‐2 virus.
Figure 4.
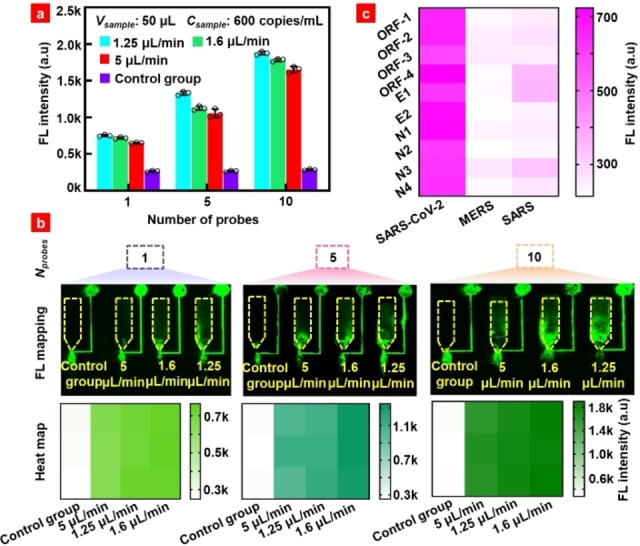
(a) Fluorescence detection of pseudovirus RNA using multiple DNA probes. (b) Fluorescence map and heat map corresponding to the fluorescence intensity in Figure 4a. (c) Specificity heat map of ten DNA probes for SARS‐CoV‐2, MERS, and SARS. Note: One DNA probe is D−E1. The other five DNA probes are D−E1, D‐open reading frame (ORF)‐1a/b‐1, D−N1, D−N2, and D−N4 respectively.
Validation of SARS‐CoV‐2 pseudovirus RNA detection methods by qRT‐PCR
qRT‐PCR, has been widely used as the gold standard method for SARS‐CoV‐2 RNA detection in clinical diagnosis. To validate the detection performance of the proposed microfluidic biochip, qRT‐PCR of SARS, MERS, and SARS‐CoV‐2 pseudovirus RNAs was conducted using the Hifair® III 1st Strand cDNA Synthesis SuperMix and Hieff UNICON® Universal Blue qPCR SYBR Green Master. The RNA sequences of these pseudoviruses are listed in Table S3‐S5. The red parts in Table S3 represent the DNA probe sequences used in the experiment. Figure S10 shows at schematic diagram of qRT‐PCR and the primers used in the experiment. The suitability of the synthetic primers was validated by qRT‐PCR. The primer sequences are shown in Table S6. The amplification and melt curves for SARS‐CoV‐2 pseudovirus RNA are shown in Figure S11. Human glyceraldehyde 3‐phosphate dehydrogenase (GAPDH) endogenous reference gene primers were used as negative controls. Amplification curves show that the four primers in the experimental group had obvious amplification and Ct values, and the difference in Ct values was less than 0.4, while the negative control did not yield a Ct value. There is only one single peak between 80 °C and 90 °C in the melt curves, implying that there is only one product, and the reaction was carried out without any errors. To verify the specificity of the primers, MERS and SARS RNAs were subjected to qRT‐PCR, as shown in Figure S11c and d. There was no obvious peak in the amplification curves, indicating that the four primers have excellent specificity for the detection of SARS‐CoV‐2 RNA. To verify the accuracy of the microfluidic biochip detection results, we performed PCR amplification of different concentrations of RNA using the E primer, as shown in Figure 5a. The cycle at which the line of relative fluorescence unit (RFU) for the sample crosses the set threshold line is called the Ct value. ΔCt value reflects the change in the target gene cycle period relative to the reference gene, and the calculation formula is ΔCt=Ct (target gene) – Ct (reference gene). [21] Ct values appeared in the qPCR curves of samples with different concentrations. SARS and MERS RNAs did not show any significant peak. Figure S12 shows the linear relationship between RNA quantity and ▵Ct value. The value of the standard curve (R2) of 0.9981 and an amplification efficiency of 92.5 % indicated that the test results obtained by qRT‐PCR were reliable. The detection results from the biochip without amplification were compared with those detected by qRT‐PCR, as shown in Figure 5b. Biochip detection had a standard deviation of 2–6.1 % from qRT‐PCR results, with a recovery of 99–113 % (Table S7), which confirmed the detection accuracy of the proposed method.
Figure 5.
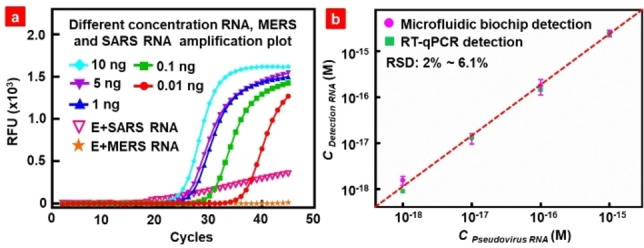
(a) qRT‐PCR results of SARS‐CoV‐2, SARS, and MERS pseudoviruses RNA detection. (b) Correlation between the pseudovirus RNA detection results by the microfluidic biochip and qRT‐PCR results.
For comparison, we summarized the performance details of other SARS‐CoV‐2 RNA detection methods in Table 1. The proposed SARS‐CoV‐2 RNA detection method, without RNA extraction and amplification. It shows pretty low LOD, short pretreatment and detection time, large range of sample volume, and does not need purification and amplification.
Table 1.
Properties of different platforms for the detection of the SARS‐CoV‐2 RNA.
|
Detection methods |
LOD [copies/μL] |
Sample pretreatment and detection time [min] |
Sample volume [μL] |
Purification |
Amplification |
Ref. |
|---|---|---|---|---|---|---|
|
RT‐PCR |
15 |
25/<180 |
10 |
Yes |
Yes |
[7b] |
|
Isothermal ligation |
0.06 |
5/50 |
0.78 |
Yes |
Yes |
[1c] |
|
CRISPR‐Cas12 |
10 |
25/52 |
– |
Yes |
Yes |
[22] |
|
Electrochemical |
6.9 |
60/35 |
2 |
Yes |
Yes |
[15] |
|
FET |
1380 |
35/62 |
10 |
No |
Yes |
[14a] |
|
SPR |
12 |
60/∼10 |
– |
Yes |
Yes |
[23] |
|
Fluorescence polarization |
3 |
25/20 |
2 |
Yes |
Yes |
[24] |
|
Microfluidic biochip |
0.6 |
5/35 |
2–50 |
No |
No |
This work |
Note: The detection time includes the time from sample processing to the reading of the results.
Conclusion
A new rapid, selective, and ultrasensitive SARS‐CoV‐2 RNA detection strategy that does not require the RNA extraction and amplification based on microfluidic biochip platform integrated with graphene nanomaterials and PLL was developed. This strategy has several merits. First, the microfluidic biochip exhibits the characteristic of simultaneous detection of multiple samples due to the appropriate application of GO nanomaterials and PLL biomaterials and the novel structural design of the biochip, which is indicated by the switch from “0” to “signal” when the target is present and the solid‐phase fluorescence signal produced on the chip surface without distributions. It has a low detection limit (∼600 copies/mL) at the optimal injection rate and sample volume. The volume of the loading sample was adjustable, resulting in an ultralow concentration of the detectable target RNA. As more samples are loaded, more DNA‐RNA complexes are loaded into the detection chamber to produce stronger fluorescence signals. Second, by simultaneously using the direct lysis and ultrasound methods to treat the samples, the loss of RNA was avoided, and the need for RNA reverse transcription, purification, and amplification was eliminated. Hybridization between the DNA probe and RNA is feasible even in the lysed solution, shortening the overall detection time. As shown in Figure S13, the whole detection process including sample pretreatment is ∼40 min, which is much shorter than the qRT‐PCR process that takes ∼2 h. Thirdly, multiple DNA probes can detect multiple sites of RNA, which greatly enhances the detection capability of the biochip. Considering all these advantages, the developed microfluidic biochip‐based SARS‐CoV‐2 RNA detection strategy can be used as a rapid and easy‐to‐implement detection method for epidemic diagnostics and other nucleic acid‐based tests.
Conflict of interest
The authors declare no conflict of interest.
1.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Acknowledgements
This work was supported by the National Key R&D Plan of China (Grant No. 2017YFB0405400), Natural Science Foundation for Distinguished Young Scientist of Shandong Province (Grant No. JQ201814), National Science Foundation for Youth Scientist (Grant No. 32001018), the Fundamental Research Funds of Shandong University (2020QNQT001), and Collaborative Innovation Center of Technology and Equipment for Biological Diagnosis and Therapy in Universities of Shandong.
Y. Chu, J. Qiu, Y. Wang, M. Wang, Y. Zhang, L. Han, Chem. Eur. J. 2022, 28, e202104054.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1.
- 1a. Gorbalenya A. E., Baker S. C., Baric R. S., de Groot R. J., Drosten C., Gulyaeva A. A., Haagmans B. L., Lauber C., Leontovich A. M., Neuman B. W., Penzar D., Perlman S., Poon L. L. M., Samborskiy D. V., Sidorov I. A., Sola I., Ziebuhr J., Nat. Micro. 2020, 5, 536–544; [Google Scholar]
- 1b. Kissler S. M., Tedijanto C., Goldstein E., Grad Y. H., Lipsitch M., Science 2020, 368, 860–868; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1c. Woo C. H., Jang S., Shin G., Jung G. Y., Lee J. W., Nat. Biomed. Eng. 2020, 4, 1168–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.
- 2a. Ji T., Liu Z., Wang G., Guo X., Akbar khan S., Lai C., Chen H., Huang S., Xia S., Chen B., Jia H., Chen Y., Zhou Q., Biosens. Bioelectron. 2020, 166, 112455–112472; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2b. Guo L., Ren L., Yang S., Xiao M., Chang D., Yang F., Dela Cruz C. S., Wang Y., Wu C., Xiao Y., Zhang L., Han L., Dang S., Xu Y., Yang Q. W., Xu S. Y., Zhu H. D., Xu Y. C., Jin Q., Sharma L., Wang L., Wang J., Clin. Infect. Dis. 2020, 71, 778–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.
- 3a. Marsh R. A., Orange J. S., Ann. 2019, 123, 444–453; [DOI] [PubMed] [Google Scholar]
- 3b. Seo G., Lee G., Kim M. J., Baek S. H., Choi M., Ku K. B., Lee C. S., Jun S., Park D., Kim H. G., Kim S. J., Lee J. O., Kim B. T., Park E. C., Kim S. I., ACS Nano 2020, 14, 5135–5142; [DOI] [PubMed] [Google Scholar]
- 3c. Li Z., Yi Y., Luo X., Xiong N., Liu Y., Li S., Sun R., Wang Y., Hu B., Chen W., Zhang Y., Wang J., Huang B., Lin Y., Yang J., Cai W., Wang X., Cheng J., Chen Z., Sun K., Pan W., Zhan Z., Chen L., Ye F., J. Med. Virol. 2020, 92, 1518–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.
- 4a. Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., Yu J., Shan S., Zhou B., Song S., Tang X., Yu J., Lan J., Yuan J., Wang H., Zhao J., Zhang S., Wang Y., Shi X., Liu L., Zhao J., Wang X., Zhang Z., Zhang L., Nature 2020, 584, 115–119; [DOI] [PubMed] [Google Scholar]
- 4b. Wang X., Guo X., Xin Q., Pan Y., Hu Y., Li J., Chu Y., Feng Y., Wang Q., Clin. Infect. Dis. 2020, 71, 2688–2694; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4c. Wang C., Wang C., Qiu J., Gao J., Liu H., Zhang Y., Han L., Microchim. Acta 2021, 188, 262–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.
- 5a. Zhou P., Yang X. L., Wang X. G., Hu B., Zhang L., Zhang W., Si H. R., Zhu Y., Li B., Huang C. L., Chen H. D., Chen J., Luo Y., Guo H., Jiang R. D., Liu M. Q., Chen Y., Shen X. R., Wang X., Zheng X. S., Zhao K., Chen Q. J., Deng F., Liu L. L., Yan B., Zhan F. X., Wang Y. Y., Xiao G. F., Shi Z. L., Nature 2020, 579, 270–273; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5b. Zhao H., Liu F., Xie W., Zhou T.-C., OuYang J., Jin L., Li H., Zhao C.-Y., Zhang L., Wei J., Zhang Y.-P., Li C.-P., Sens. Actuators B 2021, 327, 128899–128907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.
- 6a. Chaibun T., Puenpa J., Ngamdee T., Boonapatcharoen N., Athamanolap P., O'Mullane A. P., Vongpunsawad S., Poovorawan Y., Lee S. Y., Lertanantawong B., Nat. Commun. 2021, 12, 802–811; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6b. Thi V. L. D., Herbst K., Boerner K., Meurer M., Kremer L. P., Kirrmaier D., Freistaedter A., Papagiannidis D., Galmozzi C., Stanifer M. L., Boulant S., Klein S., Chlanda P., Khalid D., Miranda I. B., Schnitzler P., Kräusslich H. G., Knop M., Anders S., Sci. Transl. Med. 2020, 12, eabc7075-eabc7087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.
- 7a. Smyrlaki I., Ekman M., Lentini A., de Sousa N. R., Papanicolaou N., Vondracek M., Aarum J., Safari H., Muradrasoli S., Rothfuchs A. G., Albert J., Högberg B., Reinius B., Nat. Commun. 2020, 11, 1–12; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7b. Jung Y., Park G. S., Moon J. H., Ku K., Beak S. H., Lee C. S., Kim S., Park E. C., Park D., Lee J. H., Byeon C. W., Lee J. J., Maeng J. S., Kim S. J., Kim S. I., Kim B. T., Lee M. J., Kim H. G., ACS Infect. Dis. 2020, 6, 2513–2523; [DOI] [PubMed] [Google Scholar]
- 7c. Merindol N., Pépin G., Marchand C., Rheault M., Peterson C., Poirier A., Houle C., Germain H., Danylo A., J. Clin. Virol. 2020, 128, 104423–104426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kim Y., Yaseen A. B., Kishi J. Y., Hong F., Saka S. K., Sheng K., Gopalkrishnan N., Schaus T. E., Yin P., MedRxiv 2020, 21, 1–15. [Google Scholar]
- 9. Jiao J., Duan C., Xue L., Liu Y., Sun W., Xiang Y., Biosens. Bioelectron. 2020, 167, 112479–112485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.
- 10a. Yan C., Cui J., Huang L., Du B., Chen L., Xue G., Li S., Zhang W., Zhao L., Sun Y., Yao H., Li N., Zhao H., Feng Y., Liu S., Zhang Q., Liu D., Yuan J., Clin. Microbiol. Infect. 2020, 26, 773–779; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10b. Huang W. E., Lim B., Hsu C. C., Xiong D., Wu W., Yu Y., Jia H., Wang Y., Zeng Y., Ji M., Chang H., Zhang X., Wang H., Cui Z., Microb. Biotechnol. 2020, 13, 950–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xing W., Liu Y., Wang H., Li S., Lin Y., Chen L., Zhao Y., Chao S., Huang X., Ge S., Deng T., Zhao T., Li B., Wang H., Wang L., Song Y., Jin R., He J., Zhao X., Liu P., Li W., Cheng J., Engineering 2020, 6, 1130–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.
- 12a. Qian J., Boswell S. A., Chidley C., Lu Z. X., Pettit M. E., Gaudio B. L., Fajnzylber J. M., Ingram R. T., Ward R. H., Li J. Z., Springer M., Nat. Commun. 2020, 11, 5920–5929; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12b. Lu R., Wu X., Wan Z., Li Y., Jin X., Zhang C., Int. J. Mol. Sci. 2020, 21, 1–10; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12c. Kashir J., Yaqinuddin A., Med. Hypo. 2020, 141, 109786–109790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.
- 13a. Ooi K. H., Liu M. M., Tay J. W. D., Teo S. Y., Kaewsapsak P., Jin S., Lee C. K., Hou J., Maurer-Stroh S., Lin W., Yan B., Yan G., Gao Y. G., Tan M. H., Nat. Commun. 2021, 12, 1739–1761; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13b. Ding X., Yin K., Li Z., Lalla R. V., Ballesteros E., Sfeir M. M., Liu C., Nat. Commun. 2020, 11, 4711–4720; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13c. Patchsung M., Jantarug K., Pattama A., Aphicho K., Suraritdechachai S., Meesawat P., Sappakhaw K., Leelahakorn N., Ruenkam T., Wongsatit T., Athipanyasilp N., Eiamthong B., Lakkanasirorat B., Phoodokmai T., Niljianskul N., Pakotiprapha D., Chanarat S., Homchan A., Tinikul R., Kamutira P., Phiwkaow K., Soithongcharoen S., Kantiwiriyawanitch C., Pongsupasa V., Trisrivirat D., Jaroensuk J., Wongnate T., Maenpuen S., Chaiyen P., Kamnerdnakta S., Swangsri J., Chuthapisith S., Sirivatanauksorn Y., Chaimayo C., Sutthent R., Kantakamalakul W., Joung J., Ladha A., Jin X., Gootenberg J. S., Abudayyeh O. O., Zhang F., Horthongkham N., Uttamapinant C., Nat. Biomed. Eng. 2020, 4, 1140–1149; [DOI] [PubMed] [Google Scholar]
- 13d. Xiong E., Jiang L., Tian T., Hu M., Yue H., Huang M., Lin W., Jiang Y., Zhu D., Zhou X., Angew. Chem. Int. Ed. 2020, 60, 5307–5315; [DOI] [PubMed] [Google Scholar]
- 13e. Dai Y., Xu W., Somoza R. A., Welter J. F., Caplan A. I., Liu C. C., Angew. Chem. Int. Ed. 2020, 59, 20545–20551; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2020, 132, 20726–20732. [Google Scholar]
- 14.
- 14a. Li J., Wu D., Yu Y., Li T., Li K., Xiao M.-M., Li Y., Zhang Z.-Y., Zhang G.-J., Biosens. Bioelectron. 2021, 183, 113206–113214; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14b. Fathi-Hafshejani P., Azam N., Wang L., Kuroda M. A., Hamilton M. C., Hasim S., Mahjouri-Samani M., ACS Nano 2021, 15, 11461–11469. [DOI] [PubMed] [Google Scholar]
- 15. Alafeef M., Dighe K., Moitra P., Pan D., ACS Nano 2020, 14, 17028–17045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.
- 16a. Ramachandran A., Huyke D. A., Sharma E., Sahoo M. K., Huang C., Banaei N., Pinsky B. A., Santiago J. G., Proc. Nat. Acad. Sci. 2020, 117, 29518–29525; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16b. Donia A., Shahid M. F., Hassan S.-u., Ahmad A., Javed A., Nawaz M., Yaqub T., Bokhari H., BioRxiv 2021, 23, 1–25. [Google Scholar]
- 17. Grifoni A., Sidney J., Zhang Y., Scheuermann R. H., Peters B., Sette A., Cell Host Microbe 2020, 27, 671–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.
- 18a. Lu L., Tu D., Liu Y., Zhou S., Zheng W., Chen X., Nano Res. 2017, 11, 264–273; [Google Scholar]
- 18b. Lee J. U., Nguyen A. H., Sim S. J., Biosens. Bioelectron. 2015, 74, 341–346; [DOI] [PubMed] [Google Scholar]
- 18c. Xia Y., Wang L., Li J., Chen X., Lan J., Yan A., Lei Y., Yang S., Yang H., Chen J., Anal. Chem. 2018, 90, 8969–8976. [DOI] [PubMed] [Google Scholar]
- 19.
- 19a. Xue T., Liang W., Li Y., Sun Y., Xiang Y., Zhang Y., Dai Z., Duo Y., Wu L., Qi K., Shivananju B. N., Zhang L., Cui X., Zhang H., Bao Q., Nat. Commun. 2019, 10, 28–36; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19b. Lim L. K., Ng B. K., Fu C. Y., Tobing L. Y. M., Zhang D. H., Nanotechnology 2017, 28, 235302–235311. [DOI] [PubMed] [Google Scholar]
- 20.
- 20a. Ou X., Hong F., Zhang Z., Cheng Y., Zhao Z., Gao P., Lou X., Xia F., Wang S., Biosens. Bioelectron. 2017, 89, 417–421; [DOI] [PubMed] [Google Scholar]
- 20b. Manohar S., Mantz A. R., Bancroft K. E., Hui C. Y., Jagota A., Vezenov D. V., Nano Lett. 2008, 8, 4365–4372; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20c. Park J. S., Na H.-K., Min D. H., Kim D. E., Analyst 2013, 138, 1745–1749. [DOI] [PubMed] [Google Scholar]
- 21.
- 21a. Morrison T., Hurley J., Garcia J., Yoder K., Katz A., Roberts D., Cho J., Kanigan T., Ilyin S. E., Horowitz D., Dixon J. M., Brenan C. J. H., Nucleic Acids Res. 2006, 34, e123-e123; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21b. Fink L., Seeger W., Ermert L., Hänze J., Stahl U., Grimminger F., Kummer W., Bohle R. M., Nat. Med. 1998, 4, 1329–1333. [DOI] [PubMed] [Google Scholar]
- 22. Broughton J. P., Deng X., Yu G., Fasching C. L., Servellita V., Singh J., Miao X., Streithorst J. A., Granados A., Sotomayor-Gonzalez A., Zorn K., Gopez A., Hsu E., Gu W., Miller S., Pan C. Y., Guevara H., Wadford D. A., Chen J. S., Chiu C. Y., Nat. Biotechnol. 2020, 38, 870–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Moitra P., Alafeef M., Dighe K., Frieman M. B., Pan D., ACS Nano 2020, 14, 7617–7627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee C. Y., Degani I., Cheong J., Lee J. H., Choi H. J., Cheon J., Lee H., Biosens. Bioelectron. 2021, 178, 113049–113055. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


