Abstract
Surfactin is a cyclic lipopeptide biosurfactant. Transposon mutagenesis was performed in Bacillus subtilis strain 168, and a surfactin-susceptible mutant, strain 801, was isolated. Analysis of the region of insertion revealed that yerP was the determinant of surfactin self-resistance. YerP had homology with the resistance, nodulation, and cell division (RND) family proton motive force-dependent efflux pumps only characterized in gram-negative strains. The yerP-deficient strain 802, in which the internal region of the yerP gene of B. subtilis strain 168 was deleted, showed susceptibility to acriflavine and ethidium bromide. When strain 802 was converted to a surfactin producer by introducing a functional sfp which encodes a 4′-phosphopantetheinyl transferase and is mutated in B. subtilis strain 168, this yerP-deficient strain produced surfactin, although surfactin production was significantly reduced. The expression of yerP was at its maximum at the end of the logarithmic growth phase and was not induced by surfactin. yerP is the first RND-like gene characterized in gram-positive strains and is supposed to be involved in the efflux of surfactin.
Certain strains of Bacillus subtilis produce surfactin, a cyclic lipopeptide biosurfactant. Surfactin is composed of one β-hydroxy fatty acid, which has a long fatty acid moiety, and seven amino acids, three of which have d configuration. The β-hydroxy fatty acid links with a heptapeptide to form a lactone ring (2, 14, 30). Surfactin is synthesized nonribosomally by large template enzymes. The genes required for surfactin synthesis are identified to date as the srfA operon and sfp. The srfA operon encodes the large template enzymes (5), and sfp encodes a 4′-phosphopantetheinyl transferase that posttranslationally modifies template enzymes to their functional forms (19).
Since surfactin is a strong surfactant that reduces the surface tension of water from 72 to 27 mN/m at a concentration of 20 μM (2), studies on surfactin are focused on properties such as antitumor activity (15), activity against enveloped viruses (44), and activity against the protoplast of Bacillus megaterium (43) and against Mycoplasma (4, 45). In particular, regarding its antiviral and anti-Mycoplasma activities, surfactin is thought to disrupt or disintegrate membranes via physicochemical interaction with the membranes, and its biotechnological and pharmaceutical applications are thus of interest (44, 45).
Together with the increasing knowledge of the special biological activity of surfactin, the question arises as to whether surfactin is toxic to the producing strain. In general, production of antibiotics is closely associated with resistance of the producing microorganisms because these microorganisms must avoid the adverse effects of their own metabolisms. Strategies for acquisition of antibiotic resistance include elimination of the target site of the antibiotic by modification, chemical modification of the antibiotic, and efflux of the antibiotic in the cells (6). However, to date, there is no information on the mechanism of surfactin resistance in B. subtilis.
In our investigation of the gene(s) responsible for lipopeptide production in B. subtilis, we are interested in self-resistance to surfactin, especially in efflux of the product in the environment of the cells. In this study, we used B. subtilis strain 168, which cannot produce surfactin because of a mutation in the sfp gene (referred to as sfp0) (25). By transposon mutagenesis of this strain, we obtained a surfactin-susceptible mutant from which yerP was identified to be the determinant of susceptibility. Moreover, we examined the relationship of yerP deficiency with surfactin production or drug resistance.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in this study are listed in Table 1. Transposon-carrying plasmid pHV1249 (29) was obtained from the Bacillus Genetic Stock Center (Columbus, Ohio). Plasmid pIS284, constructed by I. Smith of The Public Health Research Institute (17, 22) was obtained from T. Tanaka of Tokai University. Escherichia coli DH5α (34) was used for constructing the recombinant plasmids. L medium contained (per liter) 10 g of polypepton (Nippon Pharmaceutical Co., Ltd., Tokyo, Japan), 5 g of yeast extract (Oriental Yeast Co., Ltd., Tokyo, Japan), and 5 g of NaCl and was adjusted to pH 7.2 with 1 N NaOH. An L-agar plate is L medium solidified with 2% (wt/vol) agar. When necessary, antibiotics were added at the following concentrations: ampicillin, 50 μg/ml; chloramphenicol, 5 μg/ml; erythromycin, 10 μg/ml; tetracycline, 20 μg/ml; and neomycin, 20 μg/ml. No. 3S medium contained (per liter) 30 g of polypepton S (Nippon Pharmaceutical Co., Ltd.), 10 g of glucose, 1 g of KH2PO4, and 0.5 g of MgSO4 · 7H2O and was adjusted to pH 7.0 with 2 N NaOH.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Characteristic(s)a | Reference(s) or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | supE44 lacU169(φ80 lacZ M15)hsdR17 recA1 endA1 gyrA96 thi-1 relA | 34 |
| B. subtilis | ||
| 168 | trpC2 | Laboratory stock |
| 801 | trpC2 yerP::mini-Tn10 Cmr | This study |
| 802 | trpC2 yerP::Nmr | This study |
| 803 | trpC2 amyE::[yerP′-′lacZ (Cmr)] | This study |
| 804 | trpC2 amyE::[yerO′-′lacZ (Cmr)] | This study |
| 805 | trpC2 amyE::[′lacZ (Cmr)] | This study |
| 806 | trpC2 yerP::NmramyE::[yerP′-′lacZ (Cmr)] | This study |
| 807 | trpC2 yerP::NmramyE::[yerO′-′lacZ (Cmr)] | This study |
| 808 | trpC2 yerP::NmramyE::[′lacZ (Cmr)] | This study |
| 168::sfp | 168 sfp+ | This study |
| 802::sfp | 802 sfp+ | This study |
| 802::sfp(pTB522-yerP) | 802::sfp with plasmid pTB522-yerP | This study |
| 802::sfp(pTB522) | 802::sfp with plasmid pTB522 | This study |
| MI112 | recA4 leuB8 arg-15 SP βR(hsdR-cotA+-purB+)202-5 (rrnH-rrnG) | 40 |
| Plasmids | ||
| pHV1249 | mini-Tn10 Apr Emr Cmr | 29 |
| pTB522 | Tcr | 13 |
| pTB522-yerP::Tn | pTB522 with 5.5-kb BglII fragment containing yerP::mini-Tn10 from strain 801 | This study |
| pTB522-yerP | pTB522 with 3.9-kb BglII fragment containing intact yerP | This study |
| pUCN1 | Nmr | 42 |
| pUC18 | Cloning vector, Apr | 34 |
| pUC18-yerP::Tn | pUC18 with 5.5-kb BglII fragment containing yerP::mini-Tn10 from strain 801, Apr Cmr | This study |
| pUC18-yerP::Nmr | EcoRV-BamHI fragment of pUC18-yerP::Tn was replaced by the Nmr cassette from pUCN1, Apr Nmr | This study |
| pIS284 | Promoterless lacZ integrated into amyE, constructed by exchanging the small EcoRI-SacI fragment of pAC5 for the small EcoRI-SacI fragment from pTKlac, Apr Cmr | 17, 22 |
| pISPyerP | pIS284 with 0.5-kb BamHI-BglII fragment containing the yerP-yerO intercoding region (yerP′-′lacZ), Apr Cmr | This study |
| pISPyerO | pIS284 with 0.5-kb BamHI-BglII fragment containing the yerP-yerO intercoding region (yerO′-′lacZ), Apr Cmr | This study |
Apr, ampicillin resistant; Cmr, chloramphenicol resistant; Nmr, neomycin resistant; Tcr, tetracycline resistant.
Source of surfactin.
Surfactin was purified from a solid-state culture of B. subtilis strain MI113(pC115) as reported previously (26). Analytical high-performance liquid chromatography (HPLC) indicated that the purity of original surfactin and its fatty acid derivatives was higher than 80% (wt/wt). We used commercial surfactin (Wako Pure Chemical Co., Ltd., Osaka, Japan) as a control.
Serial dilution experiments.
Bacteria were grown in L medium at 37°C for 6 or 24 h, and the suspensions were serially diluted with 150 mM NaCl. Four microliters of each dilution was spotted onto L-agar-surfactin (100 or 10,000 μg/ml) plates or L-agar-drug (acriflavine [0.25, 0.5, 1, 2.5, 5, or 10 μg/ml], ethidium bromide [1, 2.5, 5, or 10 μg/ml], tetracycline [1, 2.5, 5, or 10 μg/ml], Triton X-100 [50, 100, 250, or 500 μg/ml], and sodium dodecyl sulfate [SDS] [50, 100, 250, or 500 μg/ml]) plates and incubated at 37°C for 12 h.
Evaluation of surfactin susceptibility.
One hundred microliters of a 12-h culture of B. subtilis strain 168 and its derivatives was inoculated into 5 ml of L medium and cultivated at 37°C at 120 strokes per minute. After 6 and 24 h, 100 μl of 105 diluted cultures with 150 mM NaCl was spread on L-agar plates containing 0, 5, 10, 25, 50, or 100 μg of surfactin/ml. These plates were incubated at 37°C for 24 h, and the numbers of viable cells were counted.
Transformation, DNA manipulation, and transposon mutagenesis.
DNA transformation of B. subtilis was performed according to the method of Anagnostopoulos and Spizizen (1) as described previously (41). E. coli transformation was carried out using the CaCl2 method as described previously (41). Transposon mini-Tn10 mutagenesis was carried out as described previously (42).
Cloning of yerP.
Cloning of yerP was performed by the gap repair method. First, the chromosome of the mutant strain 801 was digested by BglII, which does not cut intact yerP, and then cloned into the BamHI site of pUC18. This library was transformed into E. coli DH5α and screened for both ampicillin and chloramphenicol resistance. The plasmid obtained, pUC18-yerP::Tn, harbored yerP::mini-Tn10. The SmaI-XbaI fragment of pUC18-yerP::Tn was blunt ended and ligated to the blunted HindIII site of pTB522, which autonomously replicates in B. subtilis (13). The ligation mixture was transformed into competent cells of the recA mutant MI112 to avoid integration into the recipient chromosome (16, 40). The resulting pTB522-yerP::mini-Tn10 was transformed into B. subtilis strain 168. Finally, a chloramphenicol-sensitive and tetracycline-resistant strain, which had lost mini-Tn10 by transformation-mediated gap repair and harbored pTB522-yerP, was selected. Transposon elimination was confirmed by sequencing.
Deletion of yerP.
Construction of a yerP-deficient strain was carried out as follows. pUC18-yerP::Tn was digested with BamHI and EcoRV and ligated with a BamHI-SmaI fragment of a neomycin-resistant cassette from pUCN1. The resulting plasmid, pUC18-yerP::Nmr, was linearized with ScaI and transformed into strain 168. Deletion of yerP was confirmed by Southern analysis using digoxigenin labeling and a detection kit (Roche) as described previously (42). A probe for yerP was prepared as follows. The yerP coding sequence was amplified by PCR with primers yerPF (5′-CGCAGATCTGGAGGATATGATGAACCACG-3′) and yerPR (5′-GGCTCTAGATTACTCTTCTTCCGTTCCCG-3′) and then labeled with digoxigenin. Then, the yerP-deficient strain 802 was produced.
Conversion of yerP-deficient strain to a surfactin producer.
To convert the yerP-deficient strain 802 to a potential surfactin producer, the sfp0 gene on the chromosome was exchanged with sfp as follows. First, an sfp0-deficient strain, which has a chloramphenicol resistance gene instead of the deleted segment, was obtained by a double crossover of plasmid pECΔ1, which was previously designed for lpa-8 disruption (41). The resulting strain was transformed with two DNAs by congression: one from plasmid pMMN6 that has an intact sfp gene and the other from plasmid pTB522 that is able to autonomously replicate and has a tetracycline resistance gene. Tetracycline-resistant transformants, which are candidates for sfp-containing strains, were assayed for chloramphenicol resistance by plate streaking. A chloramphenicol-sensitive strain that had exchanged its chloramphenicol resistance gene with sfp by homologous recombination was selected. Finally, this strain was cultured without tetracycline in L medium for 10 generations and spread on an L-agar plate, and then a tetracycline-sensitive strain was selected. Thus, we obtained 802::sfp, which exchanged its intrinsic sfp0 with functional sfp. The control strain, 168::sfp, was also obtained by using the same method.
Primer extension analysis.
Strain 168 was cultured in L medium at 37°C. When cell growth reached an optical density at 600 nm (OD600) of 1, 200 ml of culture was subjected to preparation of mRNA by the procedure of Igo and Losick (12). For primer extension analysis, the IRD41-labeled primer (5′-GGCTGCCGTTACAATAATCGTCATC-3′) complementary to the sequence located 51 to 75 nucleotides downstream from the putative start codon of the yerP was obtained from Nisshinbo Co., Ltd., Atsugi, Japan. Total RNA was dissolved in 20 μl of hybridization buffer {80 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)] buffer (pH 6.4), 2 mM EDTA, 800 mM NaCl, 50% formamide}, added to 1.8 μl of primer (1 pmol/μl), denatured at 80°C for 15 min, and then gradually cooled to 30°C. After ethanol precipitation, the pellet was dissolved in extension buffer (4 μl of Moloney murine leukemia virus reverse transcriptase [TOYOBO], 8 μl of 5× buffer for Moloney murine leukemia virus reverse transcriptase, 16 μl of 2.5 mM deoxynucleoside triphosphate, 10 μl of H2O, and 2 μl of RNase inhibitor) and then incubated at 42°C for 1 h. The resulting cDNA was subjected to sequencing by the Li-cor dNA automated DNA sequencer.
Construction of lacZ fusions with promoters of yerP and yerO
To monitor the expression patterns of yerP and yerO, lacZ fusions with the promoters of yerP and yerO were constructed using transcriptional fusion vector pIS284, which has promoterless lacZ inside amyE. The BglII-BamHI fragment, containing the yerO-yerP intergenetic region, was cloned in the BamHI site of the pUB18 polylinker site of pIS284. The resultant plasmids, pISPyerP (which contains yerP-lacZ) and pISPyerO (which contains yerO-lacZ), were linearized by digestion with SacI and transformed into strains 168 and 802. Transcriptional fusions of yerP′-′lacZ and yerO′-′lacZ, which localized at amyE, were thus constructed.
β-Galactosidase assay.
Strains of B. subtilis harboring transcriptional lacZ fusions were inoculated into 50 ml of L medium, and growth was monitored by measuring the OD600. During the early logarithmic phase (OD600 of 0.2 to 0.4), 25 ml of the culture was transferred to a different flask and surfactin was added to a final concentration of 500 μg/ml. At each sampling time, aliquots of cell culture were subjected to a β-galactosidase assay using the method of Nagami and Tanaka (24). β-Galactosidase activity was expressed in Miller units (23).
Quantitative analysis of surfactin.
Forty milliliters of B. subtilis culture grown in No. 3S medium was acidified to pH 2.0 with 12 N HCl. The precipitate was collected by centrifugation and extracted with methanol. The extract was filtered through a 0.2-μm-pore-size polytetrafluoroethylene membrane (Advantec, Tokyo, Japan), and the concentration of surfactin was determined by reversed-phase HPLC as described previously (41).
Nucleotide sequencing analysis.
Double-stranded DNA cloned in pUC18 was sequenced using a Li-cor dNA model 4000 DNA sequencer with the Thermo Sequenase cycle sequencing kit (Amersham) and IRD41 labeled primers (Nisshinbo). The BLAST program was used for a homology search of the DNA Data Bank of Japan database.
RESULTS
Screening of surfactin-susceptible strains.
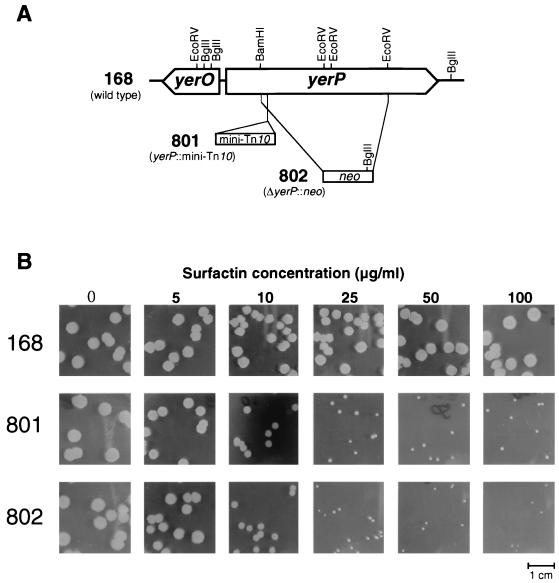
B. subtilis strain 168 was transformed with transposon-carrying pHV1249, and mini-Tn10 insertion mutants were selected using chloramphenicol. About 6,500 mutants were replicated on both L-agar plates and L-agar-surfactin plates containing 500 μg of surfactin/ml. One strain, named strain 801, showed significantly slower growth on the L-agar-surfactin plate than on the L-agar plate. To identify the region associated with self-resistance to surfactin, we constructed a plasmid library of strain 801 in E. coli and recovered the mini-Tn10-inserted region using the chloramphenicol resistance gene of mini-Tn10. Sequencing of this region revealed that mini-Tn10 was inserted into the yerP region (http://www.pasteur.fr/Bio/SubtiList/) (Fig. 1A). To confirm that yerP was a determinant of growth inhibition by surfactin, yerP was deleted from strain 168. The yerP-deficient strain, designated strain 802, and strain 801 exhibited slow growth in the presence of surfactin (Fig. 1B). Based on these results, we concluded that yerP was important for normal growth in the presence of surfactin.
FIG. 1.
(A) Physical and genetic map of the yerP-yerO region of yerP-deficient strains derived from B. subtilis strain 168. The locations of the yerO and yerP coding regions (Z99107) are indicated by arrows pointing in the direction of transcription. The positions of the relevant restriction sites are indicated. (B) Photographs of growth inhibition of 6-h cultures of strains 801, 802, and 168 on L-agar-surfactin plates with various surfactin concentrations after a 12-h incubation in an incubator at 37°C.
Sequence analysis of the yerP gene.
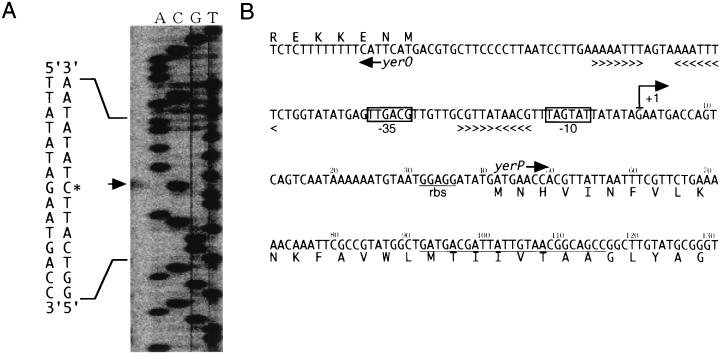
Primer extension analysis of total RNA isolated from the cells of B. subtilis strain 168 grown in L medium showed that the starting point of yerP transcription corresponds to a G residue at position +1, as shown in Fig. 2. From this result, the ribosome binding site of yerP was predicted to be GGAGG, 32 bp downstream of +1, and therefore, the start codon was thought to be ATG, 42 bp downstream of +1. The deduced amino acid length of YerP is 1,052 amino acids (mass, 114 kDa), which is 13 amino acids shorter than the YerP registered in the database (http://www.pasteur.fr/Bio/SubtiList/) (GenBank, EMBL, and DDBJ accession number Z99107). YerP had homology with the resistance, nodulation, and cell division (RND) family of the proton-dependent multidrug efflux system as follows: 36% identical to the putative protein (AF105976) of Staphylococcus aureus, 23% identical to AcrB (U00734) of E. coli, 22% identical to MexB (L11616) of Pseudomonas aeruginosa, 23% identical to MtrD (U60099) of Neisseria gonorrhoeae, 21% identical to CzcA (M26073) of Alcaligenes eutrophus, and 24% identical to NolGHI (X58632) of Rhizobium meliloti.
FIG. 2.
(A) Determination of the transcriptional start site of yerP by primer extension. The arrow indicates the band corresponding to the yerP-specific primer extension product. (B) The nucleotide sequence of the intergenetic yerO-yerP region, the nontranscribed yerO strand and the transcribed yerP strand, is shown in the 5′-to-3′ direction. The deduced primary structure of the polypeptide encoded by yerP is shown in the single-letter code below the nucleotide sequence. The yerP transcriptional start site (+1) defined by the primer extension method, the −35 and −10 regions of the promoter, and the putative ribosome binding site are indicated below the nucleotide sequence. > < indicates an inverted repeat sequence. The complementary sequence of the primer, used in the primer extension method, is underlined.
According to SubtiList (http://www.pasteur.fr/Bio/SubtiList/), there is a putative transcriptional regulator gene, yerO, upstream from yerP and oriented in a divergent direction, as shown in Fig. 1 and 2.
Phenotypic characterization of yerP
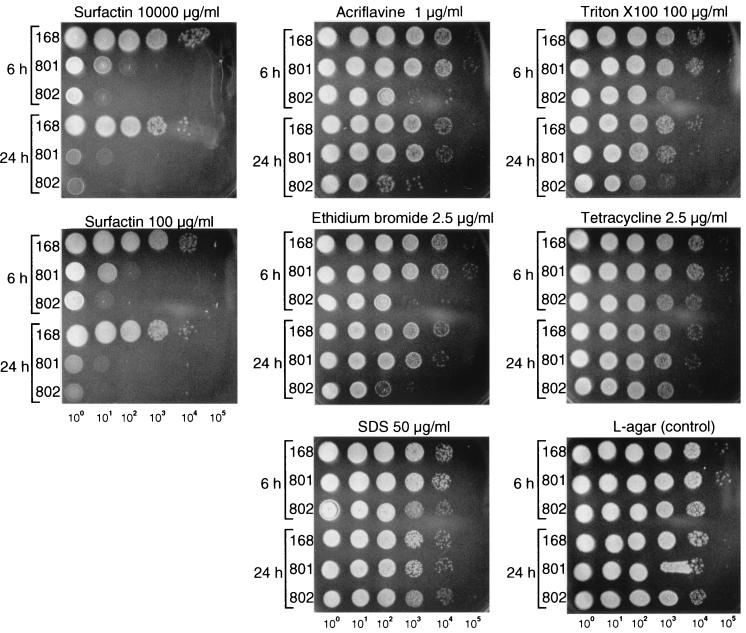
The drug and surfactin susceptibilities of yerP-deficient mutants were quantitatively assayed in serial dilution experiments. We chose SDS, Triton X-100, tetracycline, ethidium bromide, and acriflavine as the drugs. Two different growth phase cultures (a 6-h culture in the early stationary phase and a 24-h culture in the late stationary phase) were serially diluted and spotted on L-agar-drug plates and L-agar-surfactin plates. After 12 h of incubation at 37°C, the colony formations of the mutant strains were compared with that of strain 168. Figure 3 shows representative results of the serial dilution experiments. Strains 801 and 168 showed similar susceptibilities to all five drugs, while strain 802 was more sensitive to acriflavine and ethidium bromide. Apparently, both strains 801 and 802 indicated severe susceptibility to 100 μg of surfactin/ml, with strain 802 being more sensitive to 100 μg of surfactin/ml than strain 801. However, 10,000 μg of surfactin/ml, a concentration 10 times higher than that produced by strain 168::sfp (1,283 μg of surfactin/ml), had an effect similar to that of 100 μg of surfactin/ml on the three strains.
FIG. 3.
Drug and surfactin susceptibility of yerP-deficient strains 801 and 802. Bacteria were grown in L medium for 6 or 24 h. Four microliters of 100 through 105 serial dilutions were spotted on L-agar-drug and L-agar-surfactin plates and incubated at 37°C for 12 h. The initial numbers of bacteria were as follows: strain 168 (6 h), 7.0 × 108 CFU/ml; strain 801 (6 h), 6.1 × 108 CFU/ml; strain 802 (6 h), 5.4 × 108 CFU/ml; strain 168 (24 h), 2.4 × 108 CFU/ml; strain 801 (24 h), 2.0 × 108 CFU/ml; and strain 802 (24 h), 3.7 × 108 CFU/ml. In the 100 spot (nondilution spot) of all strains, no colony was observed at concentrations higher than following: acriflavine, 5 μg/ml; ethidium bromide, 10 μg/ml; tetracycline, 10 μg/ml; Triton X-100, 500 μg/ml; and SDS, 250 μg/ml. Only one result for each drug is shown.
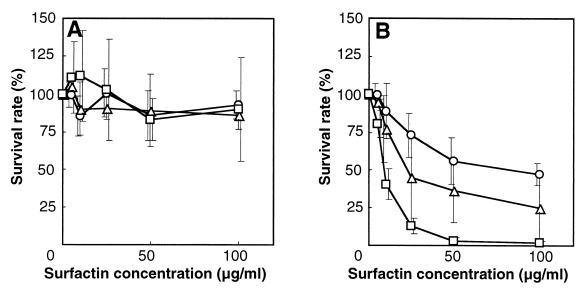
To evaluate the surfactin susceptibility of strains 801 and 802, their survival rates on L-agar-surfactin plates containing different concentrations of surfactin were measured (Fig. 4). Two different growth phase cultures (a 6-h culture in the early stationary phase and a 24-h culture in the late stationary phase) of strains 168, 801, and 802 were spread on L-agar-surfactin plates, and after 24 h of incubation at 37°C, the numbers of viable cells were counted. None of the strains of the 6-h culture showed a significant reduction in survival rate (expressed as colony number) (Fig. 4A), whereas an apparently severe growth inhibition of colony formation was observed as shown in Fig. 1. However, for strains in the 24-h culture (Fig. 4B), survival rates were reduced as surfactin concentration increased. The reduction in the number of cells of strains 801 and 802 was more significant than that of strain 168, and strain 802 was more sensitive to surfactin than strain 801.
FIG. 4.
Surfactin susceptibility of strains 801 (▵), 802 (□), and 168 (○). The 6-h (A) and 24-h (B) cultures were plated on L agar-surfactin plates containing different surfactin concentrations, and the survival rates were measured after 24 h of incubation. Error bars represent standard deviations of the means (n = 3).
Expression and regulation of yerP′-′lacZ transcriptional fusion.
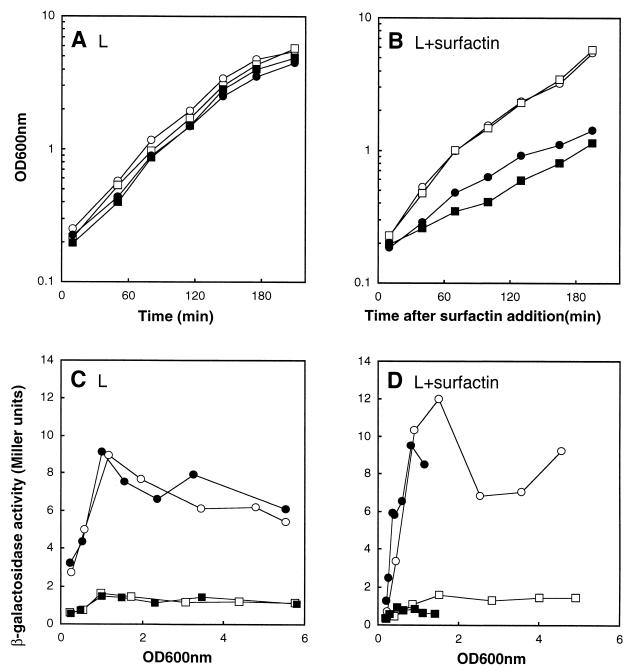
To study the function of the yerP promoter, we constructed a transcriptional yerP′-′lacZ fusion at amyE in strains 168 (yerP+) and 802 (yerP) and then monitored β-galactosidase activity in the presence or absence of surfactin (Fig. 5). In the absence of surfactin, lacZ activity reached its maximum at the end of the logarithmic phase in both the yerP+ (strain 803) and yerP (strain 806) backgrounds (Fig. 5A and C). In the presence of 500 μg of surfactin/ml, significantly slower growth was observed for the yerP strain 806 background but no change in growth rate was observed for the yerP+ (strain 803) background (Fig. 5A and B). Although there was a significant growth difference between strains 806 and 803, the expression pattern of yerP (expressed in Miller units as a function of OD600) was almost unchanged (Fig. 5D). Furthermore, the specific induction of yerP by surfactin was not observed.
FIG. 5.
Growth (A and B) and expression (C and D) of yerP-lacZ and yerO-lacZ in L medium alone (A and C) or supplemented with surfactin (500 μg/ml) (B and D). The change in expression of yerP-lacZ and yerO-lacZ was determined by β-galactosidase specific activity and calculated in Miller units and is shown plotted against the OD600. ○, strain 803 (yerP+/yerP-lacZ); □, strain 804 (yerP+/yerO-lacZ); ●, strain 806 (ΔyerP/yerP-lacZ); ▪, strain 807 (ΔyerP/yerO-lacZ). The expression levels of the control strains inserted promoterless lacZ into the amyE gene of both the yerP+ (strain 805) and ΔyerP (strain 808) backgrounds were lower than 2 Miller units at each measured point (data not shown).
We also constructed a transcriptional yerO′-′lacZ fusion and monitored its expression pattern in a similar manner. In all experiments, lacZ activity was very low and we could not observe the effect of surfactin on the gene.
Effect of yerP deficiency on surfactin production.
To investigate the effect of yerP deficiency on surfactin production, B. subtilis strain 802 was converted to a potential surfactin producer by introducing the functional sfp gene which encodes a 4′-phosphopantetheinyl transferase (19, 25). The resultant strain, 802::sfp, was cultivated in No. 3S medium at 30°C for 72 h, and the amount of secreted surfactin was measured quantitatively by HPLC. Surfactin production by 802::sfp was observed, although the production was approximately 6-fold lower than that by 168::sfp (224 μg of surfactin/ml for 802::sfp versus 1,283 μg of surfactin/ml for 168::sfp). However, some of the single colonies formed on a plate from the 72-h culture of strain 802::sfp could not produce surfactin (data not shown). A complementation experiment on the yerP deficiency of 802::sfp by introducing pTB522-yerP, which has an intact yerP, showed almost complete recovery of surfactin production [115 μg of surfactin/ml for 802::sfp(pTB522) versus 1,098 μg of surfactin/ml for 802::sfp(pTB522-yerP)]. These results indicate that yerP is involved in effective surfactin production but is not essential for the production of surfactin.
DISCUSSION
We found that yerP encodes a protein that is involved in surfactin resistance in B. subtilis strain 168. The yerP-deficient strains showed severer growth inhibition as surfactin concentration increased up to 100 μg/ml (Fig. 1 and 4). However, at a concentration of 10,000 μg/ml, growth inhibition was similar to that at 100 μg/ml (Fig. 3). This suggests that most of the surfactin molecules at a concentration of 10,000 μg/ml form micelles because the critical micellar concentration of surfactin is known to be 220 μM (or 228 μg/ml) (30), and the toxicity of micellar surfactin molecules is weak.
The deduced amino acid sequence of YerP has homology with those of the RND family, which is associated with multidrug resistance (9, 10, 20, 21, 32, 33), heavy metal ion export (27, 28, 36), transport of oligosaccharides (3), and solvent tolerance (18). Most of these RNDs play a role in resistance to a wide range of noxious compounds, and some RNDs are thought to be associated with metabolite secretion by cells. P. aeruginosa MexB is known to transport not only various drugs but also siderophore and pyoverdine (31), and R. meliloti NolGHI is involved in secreting oligosaccharides that act as nodulation signals (3). In this study, the susceptibility of strain 802 to acriflavine and ethidium bromide was observed, and thus the relationship between YerP and multidrug resistance in B. subtilis was suggested (Fig. 3). There is no direct evidence of the effluxion of surfactin by YerP; however, the observation that surfactin, which is an inherent metabolite of B. subtilis, severely inhibited the growth of the yerP-deficient strain indicated that susceptibility of the mutant to surfactin was caused by the lack of active efflux that plays a major role in secreting newly synthesized surfactin into the medium. Although these functions have been characterized in gram-negative bacteria, this is the first report demonstrating an RND-like gene in gram-positive bacteria.
The expression of yerP, which was not induced by surfactin, was at its maximum level when cell growth reached the end of the logarithmic growth phase (Fig. 5). As shown in Fig. 1, the genetic organization of yerP and yerO indicates the possibility that yerO encodes the regulator that controls the expression level of yerP during growth. However, no activation of the transcription of yerO was observed (Fig. 5), indicating that the expression of yerP is associated with the cell growth phase and is not dependent on external surfactin. A similar growth-phase-dependent expression of the efflux pump was also observed in MexA-MexB-OprM in P. aeruginosa (8).
To examine whether yerP is responsible for surfactin production in B. subtilis, the yerP-deficient strain was converted to a potential surfactin producer by the introduction of functional sfp. The production of surfactin in the yerP-deficient strain was observed, although it was significantly low (224 μg/ml). The findings that surfactin was produced and that yerP-deficient strains persistently survived at surfactin concentrations higher than 10,000 μg/ml (Fig. 3) suggest the existence of other mechanisms that act to cause the effluxion of surfactin. Since some colonies from the culture of strain 802::sfp could not produce surfactin (data not shown), the possible existence of a non-surfactin-producing strain in the culture due to suppression mutation of strain 802::sfp could not be excluded. However, due to the potentially high susceptibility of the yerP-deficient strain, as shown in Fig. 3, we could not evaluate the non-surfactin-producing population.
Certain strains of B. subtilis are coproducers of surfactin and other lipopeptides (11, 30, 35, 41). We showed that B. subtilis strain 168 is intrinsically equipped with synthetases for lipopeptides of surfactin and the antifungal plipastatin (42). On the other hand, the in vivo production of engineered surfactin derivatives with novel structures has been successfully performed by recombining template enzyme genes (7, 37, 38, 39). Therefore, it is of interest whether or not yerP is associated with the efflux of these nonsurfactin lipopeptides.
ACKNOWLEDGMENTS
We thank T. Ano for valuable suggestions.
This study was supported by a grant-in-aid for scientific research from the Ministry of Education, Sports, Science, and Technology in Japan.
REFERENCES
- 1.Anagnostopoulos C, Spizizen J. Requirements for transformation in Bacillus subtilis. J Bacteriol. 1961;81:741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arima K, Kakinuma A, Tamura G. Surfactin, a crystalline peptide-lipid surfactant produced by Bacillus subtilis: isolation, characterization and its inhibition of fibrin clot formation. Biochim Biophys Res Commun. 1968;31:488–494. doi: 10.1016/0006-291x(68)90503-2. [DOI] [PubMed] [Google Scholar]
- 3.Baev N, Endre G, Petrovics G, Banfalvi Z, Kondorosi A. Six nodulation genes of nod box locus 4 in Rhizobium meliloti are involved in nodulation signal production: nodM codes for D-glucosamine synthetase. Mol Gen Genet. 1991;228:113–124. doi: 10.1007/BF00282455. [DOI] [PubMed] [Google Scholar]
- 4.Beven L, Wroblewski H. Effect of natural amphipathic peptides on viability, membrane potential, cell shape and motility of mollicutes. Res Microbiol. 1997;148:163–175. doi: 10.1016/S0923-2508(97)87647-4. [DOI] [PubMed] [Google Scholar]
- 5.Cosmina P, Rodriguez F, de Ferra F, Grandi G, Perego M, Venema G, van Sinderen D. Sequence and analysis of the genetic locus responsible for surfactin synthesis in Bacillus subtilis. Mol Microbiol. 1993;8:821–831. doi: 10.1111/j.1365-2958.1993.tb01629.x. [DOI] [PubMed] [Google Scholar]
- 6.Cundliffe E. How antibiotic-producing organisms avoid suicide. Annu Rev Microbiol. 1989;43:207–233. doi: 10.1146/annurev.mi.43.100189.001231. [DOI] [PubMed] [Google Scholar]
- 7.de Ferra F, Rodriguez F, Tortora O, Tosi C, Grandi G. Engineering of peptide synthetase. Key role of the thioesterase-like domain for efficient production of recombinant peptides. J Biol Chem. 1997;272:25304–25309. doi: 10.1074/jbc.272.40.25304. [DOI] [PubMed] [Google Scholar]
- 8.Evans K, Poole K. The MexA-MexB-OprM multidrug efflux system of Pseudomonas aeruginosa is growth-phase regulated. FEMS Microbiol Lett. 1999;173:35–39. doi: 10.1111/j.1574-6968.1999.tb13481.x. [DOI] [PubMed] [Google Scholar]
- 9.Hagman K E, Pan W, Spratt B G, Balthazar J T, Judd R C, Shafer W M. Resistance of Neisseria gonorrhoeae to antimicrobial hydrophobic agents is modulated by the mtrRCDE efflux system. Microbiology. 1995;141:611–622. doi: 10.1099/13500872-141-3-611. [DOI] [PubMed] [Google Scholar]
- 10.Hagman K E, Lucas C E, Balthazar J T, Snyder L, Nilles M, Judd R C, Shafer W M. The MtrD protein of Neisseria gonorrhoeae is a member of the resistance/nodulation/division protein family constituting part of an efflux system. Microbiology. 1997;143:2117–2125. doi: 10.1099/00221287-143-7-2117. [DOI] [PubMed] [Google Scholar]
- 11.Hiraoka H, Asaka O, Ano T, Shoda M. Characterization of Bacillus subtilis RB14, coproducer of peptide antibiotics iturin A and surfactin. J Gen Appl Microbiol. 1992;38:635–640. [Google Scholar]
- 12.Igo M M, Losick R. Regulation of a promoter that is utilized by minor forms of RNA polymerase holoenzyme in Bacillus subtilis. J Mol Biol. 1986;191:615–624. doi: 10.1016/0022-2836(86)90449-3. [DOI] [PubMed] [Google Scholar]
- 13.Imanaka T, Himeno T, Aiba S. Effect of in vitro DNA rearrangement in the NH2-terminal region of the penicillinase gene from Bacillus licheniformis on the mode of expression in Bacillus subtilis. J Gen Microbiol. 1985;131:1753–1763. doi: 10.1099/00221287-131-7-1753. [DOI] [PubMed] [Google Scholar]
- 14.Kakinuma A, Ouchida A, Shima T, Sugio H, Isono M, Tamura G, Arima K. Confirmation of the structure of surfactin by mass spectrometry. Agric Biol Chem. 1969;33:1669–1671. [Google Scholar]
- 15.Kameda Y, Ouhira M, Matsui K, Kanamoto S, Hase T, Atsusaka T. Antitumor activity of Bacillus natto. V. Isolation and characterization of surfactin in the culture medium of Bacillus natto KMD 2331. Chem Pharm Bull (Tokyo) 1974;22:938–944. doi: 10.1248/cpb.22.938. [DOI] [PubMed] [Google Scholar]
- 16.Keggins K M, Lovett P S, Duvall E J. Molecular cloning of genetically active fragments of Bacillus DNA in Bacillus subtilis and properties of the vector plasmid pUB110. Proc Natl Acad Sci USA. 1978;75:1423–1427. doi: 10.1073/pnas.75.3.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kenney T J, Moran C P., Jr Genetic evidence for interaction of ςA with two promoters in Bacillus subtilis. J Bacteriol. 1991;173:3282–3290. doi: 10.1128/jb.173.11.3282-3290.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kieboom J, Dennis J J, de Bont J A M, Zylstra G J. Identification and molecular characterization of an efflux pump involved in Pseudomonas putida S12 solvent tolerance. J Biol Chem. 1998;273:85–91. doi: 10.1074/jbc.273.1.85. [DOI] [PubMed] [Google Scholar]
- 19.Lambalot R H, Gehring A M, Flugel R S, Zuber P, LaCelle M, Marahiel M A, Reid R, Khosla C, Walsh C T. A new enzyme superfamily—the phosphopantetheinyl transferases. Chem Biol. 1996;3:923–936. doi: 10.1016/s1074-5521(96)90181-7. [DOI] [PubMed] [Google Scholar]
- 20.Ma D, Cook D N, Alberti M, Pon N G, Nikaido H, Hearst J E. Molecular cloning and characterization of acrA and acrE genes of Escherichia coli. J Bacteriol. 1993;175:6299–6313. doi: 10.1128/jb.175.19.6299-6313.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma D, Cook D N, Alberti M, Pon N G, Nikaido H, Hearst J E. Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol Microbiol. 1995;16:45–55. doi: 10.1111/j.1365-2958.1995.tb02390.x. [DOI] [PubMed] [Google Scholar]
- 22.Martin-Verstraete I, Débarbouillé M, Klier A, Rapoport G. Mutagenesis of the Bacillus subtilis “−12, −24” promoter of the levanase operon and evidence for the existence of an upstream activating sequence. J Mol Biol. 1992;226:85–99. doi: 10.1016/0022-2836(92)90126-5. [DOI] [PubMed] [Google Scholar]
- 23.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. pp. 352–355. [Google Scholar]
- 24.Nagami Y, Tanaka T. Molecular cloning and nucleotide sequence of a DNA fragment from Bacillus natto that enhances production of extracellular proteases and levansucrase in Bacillus subtilis. J Bacteriol. 1986;166:20–28. doi: 10.1128/jb.166.1.20-28.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakano M, Corbell M N, Besson J, Zuber P. Isolation and characterization of sfp: a gene that functions in the production of the lipopeptide biosurfactant, surfactin, in Bacillus subtilis. Mol Gen Genet. 1992;232:313–321. doi: 10.1007/BF00280011. [DOI] [PubMed] [Google Scholar]
- 26.Nakayama S, Takahashi S, Hirai M, Shoda M. Isolation of new variants of surfactin by a recombinant Bacillus subtilis. Appl Microbiol Biotechnol. 1997;48:80–82. [Google Scholar]
- 27.Nies D H, Nies A, Chu L, Silver S. Expression and nucleotide sequence of a plasmid-determined divalent cation efflux system from Alcaligenes eutrophus. Proc Natl Acad Sci USA. 1989;86:7351–7355. doi: 10.1073/pnas.86.19.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nies D H. The cobalt, zinc, and cadmium efflux system CzcABC from Alcaligenes eutrophus functions as a cation-proton antiporter in Escherichia coli. J Bacteriol. 1995;177:2707–2712. doi: 10.1128/jb.177.10.2707-2712.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petit M A, Bruand C, Janniére L, Ehrlich S D. Tn10-derived transposons active in Bacillus subtilis. J Bacteriol. 1990;172:6736–6740. doi: 10.1128/jb.172.12.6736-6740.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peypoux F, Bonmatin J M, Wallach J. Recent trends in the biochemistry of surfactin. Appl Microbiol Biotechnol. 1999;51:553–563. doi: 10.1007/s002530051432. [DOI] [PubMed] [Google Scholar]
- 31.Poole K, Heinrichs D E, Neshat S. Cloning and sequence analysis of an EnvCD homologue in Pseudomonas aeruginosa: regulation by iron and possible involvement in the secretion of the siderophore pyoverdine. Mol Microbiol. 1993;10:529–544. doi: 10.1111/j.1365-2958.1993.tb00925.x. [DOI] [PubMed] [Google Scholar]
- 32.Poole K, Krebes K, McNally C, Neshat S. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J Bacteriol. 1993;175:7363–7372. doi: 10.1128/jb.175.22.7363-7372.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poole K, Gotoh N, Tsujimoto H, Zhao Q, Wada A, Yamasaki T, Neshat S, Yamagishi J, Li X Z, Nishino T. Overexpression of the mexC-mexD-oprJ efflux operon in nfxB-type multidrug-resistant strains of Pseudomonas aeruginosa. Mol Microbiol. 1996;21:713–724. doi: 10.1046/j.1365-2958.1996.281397.x. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 35.Sandrin C, Peypoux F, Michel G. Coproduction of surfactin and iturin A, lipopeptides with surfactant and antifungal properties, by Bacillus subtilis. Biotechnol Appl Biochem. 1990;12:370–375. [PubMed] [Google Scholar]
- 36.Schmidt T, Schlegel H G. Combined nickel-cobalt-cadmium resistance encoded by the ncc locus of Alcaligenes xylosoxidans 31A. J Bacteriol. 1994;176:7045–7054. doi: 10.1128/jb.176.22.7045-7054.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schneider A, Stachelhaus T, Marahiel M A. Targeted alteration of the substrate specificity of peptide synthetases by rational module swapping. Mol Gen Genet. 1998;257:308–318. doi: 10.1007/s004380050652. [DOI] [PubMed] [Google Scholar]
- 38.Stachelhaus T, Schneider A, Marahiel M A. Rational design of peptide antibiotics by targeted replacement of bacterial and fungal domains. Science. 1995;269:69–72. doi: 10.1126/science.7604280. [DOI] [PubMed] [Google Scholar]
- 39.Stachelhaus T, Schneider A, Marahiel M A. Engineered biosynthesis of peptide antibiotics. Biochem Pharmacol. 1996;52:177–186. doi: 10.1016/0006-2952(96)00111-6. [DOI] [PubMed] [Google Scholar]
- 40.Tanaka T, Sakaguchi K. Construction of a recombinant plasmid composed of B. subtilis leucine genes and a B. subtilis (natto) plasmid: its use as cloning vehicle in B. subtilis 168. Mol Gen Genet. 1978;165:269–276. doi: 10.1007/BF00332526. [DOI] [PubMed] [Google Scholar]
- 41.Tsuge K, Ano T, Shoda M. Isolation of a gene essential for biosynthesis of the lipopeptide antibiotics plipastatin B1 and surfactin in Bacillus subtilis YB8. Arch Microbiol. 1996;165:243–251. doi: 10.1007/s002030050322. [DOI] [PubMed] [Google Scholar]
- 42.Tsuge K, Ano T, Hirai M, Nakamura Y, Shoda M. The genes degQ, pps, and lpa-8 (sfp) are responsible for conversion of Bacillus subtilis 168 to plipastatin production. Antimicrob Agents Chemother. 1999;43:2183–2192. doi: 10.1128/aac.43.9.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsukagoshi N, Tamura G, Arima K. A novel protoplast-bursting factor (surfactin) obtained from Bacillus subtilis IAM 1213. II. The interaction of surfactin with bacterial membranes and lipids. Biochim Biophys Acta. 1970;196:211–214. doi: 10.1016/0005-2736(70)90008-8. [DOI] [PubMed] [Google Scholar]
- 44.Vollenbroich D, Özel M, Vater J, Kamp R M, Pauli G. Mechanism of inactivation of enveloped viruses by the biosurfactant surfactin from Bacillus subtilis. Biologicals. 1997;25:289–297. doi: 10.1006/biol.1997.0099. [DOI] [PubMed] [Google Scholar]
- 45.Vollenbroich D, Pauli G, Özel M, Vater J. Antimycoplasma properties and application in cell culture of surfactin, a lipopeptide antibiotic from Bacillus subtilis. Appl Environ Microbiol. 1997;63:44–49. doi: 10.1128/aem.63.1.44-49.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]