Abstract
Background/Aim: Cholangiocarcinoma (CCA) is a stem cell-based cancer. The in vivo tumor microenvironment is not present in two-dimensional (2D) cultures, which is one of the limitations in cancer stem cell (CSC) research. Thus, we aimed to establish three-dimensional (3D) culture mimicking extracellular matrix (ECM) that could serve as a niche for CSC enrichment in CCA.
Materials and Methods: Silk fibroin-gelatin/hyaluronic acid/heparan sulfate (SF-GHHs) scaffolds were fabricated by lyophilization in various ratios and compared to silk fibroin (SF) scaffold. The physical and biological characteristics of the scaffolds were investigated.
Results: The SF-GHHs 1:2 scaffold with pore size of 350±102 μm harbored optimal porosity, good water uptake, and stable beta-sheet that supported the increase in KKU-213A cell proliferation and aggregation. The CSC and the epithelial-mesenchymal transition (EMT) markers were significantly upregulated in this scaffold compared to 2D. Moreover, drug sensitivity against cisplatin and gemcitabine in 3D culture was significantly higher than that in 2D culture.
Conclusion: The SF-GHHs 1:2 scaffold could simulate ECM that may serve as a CSC niche of CCA, and reinforce stemness and EMT properties, suggesting its suitability for 3D CCA model, which supports CSC and new targeting drug research in CCA.
Keywords: Silk fibroin-based scaffold, three-dimensional culture, cancer stem cell, epithelial-mesenchymal transition, cholangiocarcinoma
Cholangiocarcinoma (CCA) is an aggressive cancer that arises from the epithelium of bile ducts with the highest incidence in Northeastern Thailand where liver flukes are endemic (1). The only curative treatment is surgery but is not effective in patients with late-stage cancer (2). Many lines of evidence indicated the implication of cancer stem cells (CSCs) in CCA (3-9). CSCs are a tumor cell subpopulation that is capable of self-renewal and differentiation. CSCs are related to cancer initiation, progression, and resistance to chemo- and radio-therapies (9). Although the traditional two-dimensional (2D) culture has long been used in cancer research, it cannot mimic tumor microenvironment, which plays crucial roles in cell-cell and cell-matrix interactions giving rise to the differences in cancer morphology, proliferation, invasion, metastasis, signaling pathways and other biological functions when compared to in vivo conditions (10,11). Three-dimensional (3D) cell culture was established to mimic tumor-like in vivo conditions providing more predictive data for in vivo tests of CSCs in cancer research (12). There are two main types of 3D cancer models for CSC enrichment including scaffold-free and scaffold-based methods (12).
Most 3D CCA models are scaffold-free methods and organoid models (13,14). However, the tumor microenvironment is not represented in scaffold-free applications, whereas organoid generation is difficult, expensive, and time-consuming (14). The 3D porous scaffold provides a favorable tumor microenvironment for cell proliferation and adhesion, resulting in differences in drug response, and increased CSC and epithelial-mesenchymal transition (EMT)-related gene expression when compared to 2D models (15–19). Moreover, a previous study showed that chitosan-alginate scaffolds promoted CSC proliferation and enrichment in glioblastoma, prostate, liver, and breast cancers (16).
Silk-fibroin (SF) extracted from silkworm cocoon is widely used biomaterial for engineering 3D scaffolds because of non-cytotoxicity and low antigenicity (20–22). It can be combined with various additives such as chitosan, gelatin, and hyaluronic acid (23). SF has been successfully used in 3D cancer models such as lung, liver, and breast cancers (17,19,24). Gelatin, a collagen derivative, is a water-soluble natural polymer containing cell adhesion motifs, including the arginine–glycine–aspartic acid (RGD) motif (25,26). Hyaluronic acid (HA) is a non-immunogenic and non-toxic glycosaminoglycan (27). It binds CD44 specific receptor to promote tumor cell proliferation, adhesion, and migration (28). Elevated levels of HA have been reported in CCA (29). Heparan sulfate (HS) is a linear polysaccharide that can be found either unconjugated or conjugated to proteins as heparan sulfate proteoglycan (HSPG) (30). HSPGs play a role in cell growth, adhesion, and mobility (31). HSPG is highly expressed in CCA (31,32). HS and HSPGs can act as coreceptors for mitogenic growth factor receptors, thus enhancing their signaling (30).
This study aimed to fabricate scaffolds for 3D culture models of CCA cells by incorporating SF with gelatin, hyaluronic acid, and heparan sulfate (SF-GHHs), which could enrich CSCs and reflect tumor behavior better than the 2D system. The SF-GHHs scaffolds were characterized for their physical properties and tested for their biological properties using a CCA cell line and examining cell proliferation and morphology in comparison with the SF scaffold. The expression of genes related to CSCs and EMT, and drug sensitivity were investigated in comparison with the 2D culture system.
Materials and Methods
Cell line and cell culture. The CCA cell line KKU-213A was kindly provided by the Cholangiocarcinoma Research Institute, Khon Kaen University, Thailand. In 2D and 3D cell cultures, KKU-213A was cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin (all from Thermo Fisher Scientific, Waltham, MA, USA). Cell cultures were maintained at 37˚C with 5% CO2, and the medium was replaced every 2 to 3 days.
Preparation of silk fibroin (SF) solution. The SF solution was prepared by cutting Bombyx mori silk cocoons (Nangnoi-Srisaket 1), which then were boiled in 0.02 M Na2CO3 for 1 h to remove sericin, washed with warm distilled water, and dried overnight in hot air oven. The degummed silk was dissolved in CaCl2/CH3CH2OH/H2O solution (molar ratio 1:2:8) followed by dialysis against distilled water for 3 days. The SF solution was adjusted with distilled water to a 3% (W/V) final concentration.
Preparation of silk fibroin (SF) and silk fibroin-gelatin/hyaluronic acid/heparan sulfate (SF-GHHs) scaffolds. The scaffolds were fabricated by a freeze-drying technique. To prepare GHHs, 3% Gelatin type A, isolated from porcine skin (Sigma-Aldrich, St. Louis, MO, USA), 0.03% hyaluronic acid sodium salt, Streptococcus equi 91% (Alfa Aesar, Ward Hill, MA, USA), and 10 μg/ml heparan sulfate sodium salt from bovine kidney (Sigma-Aldrich) were dissolved in distilled water at 40˚C. Then, 3% SF solution and GHHs solution were blended at the ratios 2:1, 1:1, and 1:2 (V/V). All scaffolds were cross-linked with 1% N-(3-Dimethylaminopropyl)-N’-ethylcarbodiimide hydrochloride (EDC, Sigma-Aldrich)/0.5% N-Hydroxysuccinimide (NHS, Sigma-Aldrich) at room temperature for 15 min. Subsequently, 3% SF solution or blended SF-GHHs solution was added into molds. All scaffolds were frozen initially at –20˚C for 20 min and subsequently –80˚C for 24 h and lyophilized for 48 h. The scaffolds were soaked in 90% methanol for 1 h, washed by distilled water then cross-linked in 1% EDC/0.5% NHS for 30 min and lyophilized for 24 h. All scaffolds were stored in a desiccator until further use.
Pore size. The structure and morphology of scaffolds were analyzed by scanning electron microscope (SEM, Jeol, JSM-IT200 InTouchScope™, Tokyo, Japan). The scaffolds were cut and placed onto the sample stage with adhesive tape followed by vacuum spray of gold films. Pore size was observed under SEM. Pore diameters of scaffolds were determined from SEM images using Image J software (U. S. National Institutes of Health, Bethesda, MD, USA).
Porosity. The porosity of scaffolds was determined using hexane by a liquid displacement method (33). Scaffolds were placed in a graduated cylinder with a known volume of the displacement liquid to fill the pore of scaffolds. The porosity was calculated using the following formula:
% Porosity=((V1-V3)/(V2-V3))×100
V1=known volume of hexane that was used to submerge the scaffold; V2=volume of the hexane and hexane-impregnated scaffold; V3=volume of remaining hexane after removing the hexane-impregnated scaffold.
Water uptake property. The water uptake property was measured by weighting scaffolds. The scaffolds were placed in phosphate buffer saline (PBS) at 37˚C for 24 h and weighed to measure the weight of scaffolds (Ws). Then, the scaffolds were dried and weighed to measure the dry weight of scaffolds (Wd). The water uptake of scaffold was calculated according to the following formula (34):
% Water uptake=(Ws-Wd)/Ws)×100%
Attenuated total reflection–Fourier transform infrared (ATR-FTIR) spectroscopy: spectral acquisition and spectral analysis. Three individual scaffolds of SF, GHHs, and blended SF:GHHs at ratios 2:1, 1:1, and 1:2 lyophilized in a 48-well plate mold were analyzed by portable Agilent ATR-FTIR spectrometer 4500 series (Agilent Technologies, Santa Clara, CA, USA). The parameters for spectral acquisition were 64 co-added scans in both background and sample, 4 cm–1 spectral resolution and 4,000-650 cm–1 spectral range in 5 replications. Acquired spectra derived from each group were averaged and performed a spectral pre-processing under 15 spectral smoothing point using S-Golay with third polynomial orders and standard normal varied (SNV) in 1,800-900 cm–1 spectral region.
Cell seeding and culture. The scaffolds were cut into small pieces (5 mm diameter×3 mm height), sterilized with 70% ethanol, exposed to UV light for 1 h, and evaporated for 30 min. The scaffolds were rinsed with PBS three times, soaked in DMEM and incubated at 37˚C under 5% CO2 for 4 h, after which were seeded with the CCA cell line KKU-213A at a density of 5×104 cells per scaffold (10 μl) and placed in a 48-well plate. After 4 h of initial cell attachment, 500 μl of DMEM was added to each well. The cell-seeded scaffolds were cultured at 37˚C under 5% CO2. The medium was changed every 2 to 3 days until harvested.
Cell proliferation assay. The 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium) (MTS) assay is a colorimetric method which determines cell metabolic activity. Cell-seeded scaffolds were determined for cell proliferation after cultivation on days 0, 3, 5, and 10 using CellTiter 96® AQueous One Solution Cell Proliferation Assay (Promega, Madison, WI, USA). Cell-free scaffolds were used to obtain the baseline absorbance at each specific time point. On the day of assay, the medium was removed, then 40 μl of MTS reagent and 200 μl of DMEM were added, and scaffolds were incubated at 37˚C in a 5% CO2 atmosphere for 4 h. Then, 100 μl of the solution was transferred to a 96-well plate before the absorbance was determined at 490 nm using an RT-2100C Microplate Reader (Rayto Life and Analytical Sciences, Shenzhen, PR China).
Hematoxylin and eosin (H&E) staining. For histological examination, the cell-seeded scaffolds were removed from a 48-well plate after 10 days of culture and immediately fixed in 10% buffered formalin. Then, the scaffolds were dehydrated through a graded ethanol series and embedded in paraffin. Embedded samples were sectioned and stained with hematoxylin and eosin (H&E) for histological analysis.
Cell morphology. The morphology of KKU-213A cells cultured in scaffolds was studied by SEM. After 10 days of culture, cell-seeded scaffolds were fixed with 4% paraformaldehyde overnight, then dehydrated by gradient concentration of ethanol and air-dried. Samples were cut and placed onto the sample stage with adhesive tape followed by vacuum spray of gold films and SEM analysis.
RNA extraction, cDNA synthesis, and quantitative reverse transcription polymerase chain reaction (qRT-PCR). Total RNA was extracted from cells cultured in 3D and 2D systems on day 3 using TRIzol® reagent (Thermo Fisher Scientific) and transcribed into cDNA using the ImProm-II™ Reverse Transcription System (Promega) according to the manufacturer’s protocols. The primer sequences of stemness- and EMT-related genes are shown in Table I. The housekeeping gene, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), was used as an internal control. qRT-PCR was performed in triplicate on an Exicycler™ 96 Real-Time Quantitative Thermal Block (Bioneer, Daejeon, Korea). PCR products were quantitated by AccuPower® GreenStar™ qPCR PreMix (Bioneer). The relative expression quantification was calculated by the 2–ΔΔCt method, where ΔCt=(Cttarget-Ctreference) (35).
Table I. List of primer sequences.
Nanog: Nanog homeobox; Sox2: sex determining region Y-Box 2; Snail1: snail family transcriptional repressor 1; Snail2: snail family transcriptional repressor 2; Twist1: twist family bHLH transcription factor 1; Zeb1: zinc finger E-box binding homeobox 1; MMP2: matrix metalloproteinase-2; MMP9: matrix metalloproteinase-9; GAPDH: glyceraldehyde 3-phosphate dehydrogenase.
Cytotoxicity assay. In 2D culture system, 100 μl of KKU-213A (2,000 cells/well) was seeded in triplicate into 96-well plates and cultured for 24 h before being treated with different concentrations of cisplatin (Kemoplat, Fresenius Kabi, New Delhi, India) or gemcitabine (Gemita, Fresenius Kabi) for another 48 h or 72 h, respectively. Cell viability was performed using MTS assay. Cells were incubated with MTS reagent at 37˚C for 4 h and absorbances were detected using an RT-2100C Microplate Reader (Rayto Life and Analytical Sciences) at a wavelength of 490 nm. The inhibitory concentration 50 (IC50) was then determined. In 3D culture system, cell-seeded scaffolds (5×104 cells/scaffold) were cultured in triplicate for 3 days before being treated with different concentrations of cisplatin or gemcitabine for another 48 h or 72 h, respectively. The IC50 was determined using the MTS assay as mentioned above.
Statistical analysis. The statistical analysis was performed using SPSS version 26 software (IBM, Armonk, NY, USA). Graph and IC50 were generated by GraphPad prism 8 for Windows (GraphPad Software, La Jolla, CA, USA). All normal distribution data are presented as mean±SD. Differences between groups were analyzed using one-way analysis of variance (ANOVA) with Tukey’s post-hoc comparisons test. For porosity test, nonparametric data were analyzed by Kruskal–Wallis One-way ANOVA and are presented as median. Differences between gene expression data were determined using Student’s t-test. A value of p<0.05 was considered as statistically significant.
Results
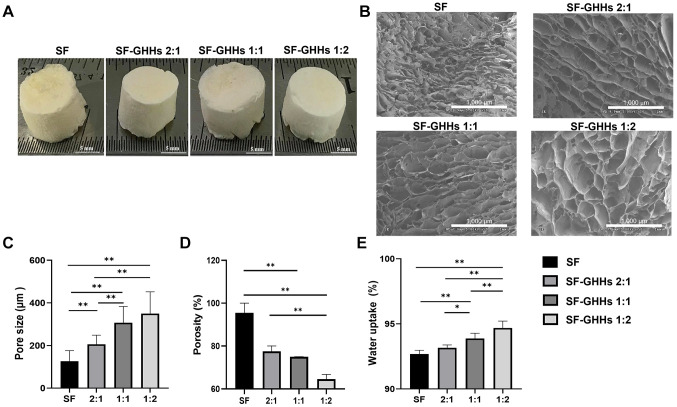
Scaffold fabrication and characterization. Macroscopic features of SF and SF-GHHs scaffolds with different blending ratios (2:1, 1:1, and 1:2) fabricated by a freeze-drying technique are shown in Figure 1A. The scaffolds used for physical characterization was cut into 12 mm diameter×1 mm height pieces. The porous structure of scaffolds was observed by SEM (Figure 1B). The pore size was determined from SEM images. As shown in Figure 1C, the SF scaffold possessed the smallest pore size with the average diameter of 127±49 μm. The average pore diameters of SF-GHHs scaffolds with different blending ratios (2:1, 1:1, and 1:2) were 206±43 μm, 307±47 μm, and 350±102 μm, respectively. The increase in pore size of SF-GHHs scaffolds was related to the increased GHHs content. By contrast, the SF scaffold showed the highest porosity 95.5% (88.2-100%) when compared to that of SF-GHHs scaffolds; 77.5% (75-80%), 75% (70-75%), and 64.6% (62.5-66.7%) for 2:1, 1:1, and 1:2, respectively. The porosity percentage of scaffolds was decreased when GHHs content was increased as shown in Figure 1D. The water uptake percentages of SF, SF-GHHs 2:1, 1:1, and 1:2 were 92.69±0.28%, 93.17±0.21%, 93.88±0.40%, and 94.69±0.53%, respectively. The water uptake percentage of the SF-GHHs 1:2 scaffold was significantly higher than those of other scaffolds, which was related to its pore size (Figure 1E).
Figure 1. Scaffold fabrication and characterization. (A) Macroscopic appearance of silk fibroin (SF), silk fibroin-gelatin/hyaluronic acid/heparan sulfate (SF-GHHs) with different blending ratios; SF-GHHs 2:1, SF-GHHs 1:1, and SF-GHHs 1:2 scaffolds (Diameter=12 mm, height=7 mm). Scale bar indicates 5 mm. (B) Scanning electron microscope images of SF, SF-GHHs 2:1, SF-GHHs 1:1, and SF-GHHs 1:2. Scale bar indicates 1,000 μm. Comparison of physical characteristics among scaffolds including (C) pore size, (D) porosity percentage, and (E) water uptake percentage. Statistically significant differences between groups at: *p<0.05 and **p<0.01.
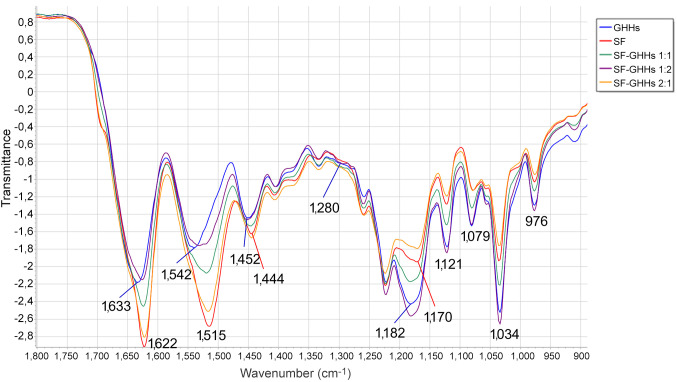
ATR-FTIR spectral analysis was performed to confirm the chemical structures of SF, GHHs, and blended SF-GHHs 2:1, 1:1, and 1:2 scaffolds. Amide I and amide II are two major bands of IR spectrum used for protein structural analysis. The amide I region (1,600-1,700 cm–1) is mainly involved in the C=O stretching of peptide bonds (nearly 80%) and is related to secondary structural elements of proteins. The amide II presented peaks at 1,480-1,575 cm–1 indicating CN stretching and NH bending (36). The SF scaffold showed peaks at 1,622 cm–1 (amide I) and 1,515 cm–1 (amide II), whereas the GHHs scaffold exhibited peaks at 1,633 cm–1, and 1,542 cm–1 of amide I and amide II, respectively. The amide I of blended SF-GHHs scaffold shifted from 1,633 cm–1 to 1,622 cm–1 and amide II was revealed by peak shifting from 1,542 cm–1 to 1,515 cm–1. The peaks at 1,622 cm–1 indicated beta-sheet formation in the scaffolds (37). An increased peak intensity was related to SF content in the mixture of SF-GHHs scaffold. Moreover, SF-GHHs scaffolds demonstrated a peak at 1,280 cm–1 representing hydroxyproline, which indicated the structure of gelatin (38), peaks at 1,182 cm–1, 1,121 cm–1 and 1,079 cm–1 indicated the structure of glycosaminoglycans in HS (39,40), and the O-CH3 (976 cm–1) indicated the structure of HS (41). SF-GHHs scaffolds also showed a peak of the C-O-C linkage in saccharide units of HA at 1,034 cm–1 (42) (Figure 2). Therefore, the ATR-FTIR spectroscopy analysis demonstrated the incorporation of GHHs with SF in SF-GHHs scaffolds and conformation of beta-sheet secondary structure, which is a stable form.
Figure 2. Attenuated total reflection–Fourier transform infrared (ATR-FTIR) transmittance spectra of scaffolds. Silk fibroin (SF) (red line), silk fibroin-gelatin/hyaluronic acid/heparan sulfate (SF-GHHs) 2:1 (orange line), SF-GHHs 1:1 (green line), SF-GHHs 1:2 (purple line) and gelatin/hyaluronic acid/heparan sulfate (GHHs) scaffolds (blue line) in 1,800-900 cm–1 spectral range. The numbers represent the absorption peaks of scaffolds.
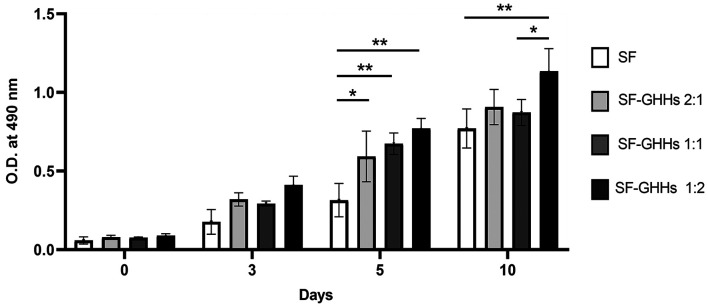
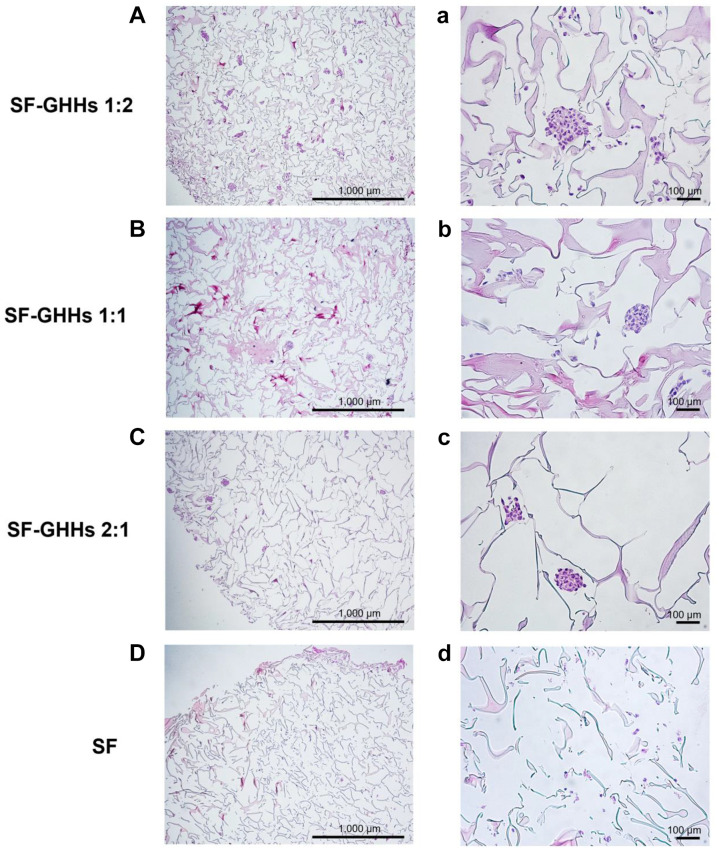
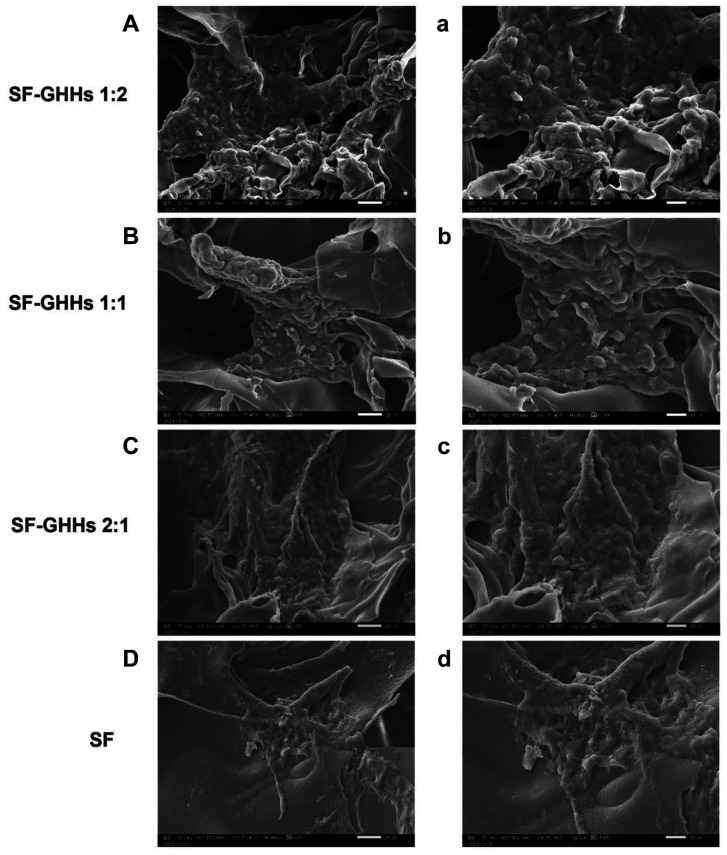
Cell proliferation, morphology and distribution. All scaffolds were non-toxic and could support KKU-213A cell proliferation. Interestingly, the GHHs components in SF-GHHs significantly enhanced the proliferation of KKU-213A cells compared to SF after 5 days in culture. The highest proliferation rate was observed in SF-GHHs 1:2 on day 10 and remained significantly different from SF while the other SF-GHHs did not (Figure 3). Histological examination of KKU-213A cell line after 10 days in culture in scaffolds is shown in Figure 4. Scaffolds were stained pink and cell nuclei were stained deep purple. KKU-213A cells could infiltrate into scaffolds. H&E images showed clusters of enlarged dense cells in the pores and along the edges of all SF-GHHs scaffolds but were higher in SF-GHHs 1:2, whereas scattered cells were observed in the SF scaffold. SEM images of KKU-213A cell morphology after 10 days in culture in scaffolds are shown in Figure 5. Aggregates of enlarged spherical cells were found in SF-GHHs 1:2 more than in other SF-GHHs scaffolds, whereas an elongated cell morphology was noticed in the SF scaffold. The results indicated that the GHHs components in SF-GHHs also promoted cell aggregation and spheroid formation, in particular, the SF-GHHs 1:2 scaffold. Based on its superior physical and biological characteristics compared to other scaffolds, the SF-GHHs 1:2 was selected for further experiments in comparison with 2D cultures.
Figure 3. The proliferation rates of KKU-213A cells cultured in scaffolds. Cell proliferation rates were compared among scaffolds including silk fibroin (SF) and silk fibroin-gelatin/hyaluronic acid/heparan sulfate (SF-GHHs) with different blending ratios (2:1, 1:1, and 1:2). Cell proliferation was determined on days 0, 3, 5, and 10 of cultivation using the 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2Htetrazolium) assay. Values are expressed as mean±SD (n=3). Statistically significant differences between groups at: *p<0.05 and **p<0.01.
Figure 4. Histological analysis of KKU-213A cells by hematoxylin and eosin staining. Cells were seeded on silk fibroin-gelatin/hyaluronic acid/heparan sulfate (SF-GHHs) 1:2 (A, a), SF-GHHs 1:1 (B, b), SF-GHHs 2:1 (C, c), and silk fibroin (SF) scaffolds (D, d) and cultured for 10 days. Clusters of enlarged dense cells were observed in all SF-GHHs scaffolds but higher in SF-GHHs 1:2. Cells were scattered in the SF scaffold. Scale bars=1,000 μm, with 4× magnification (A-D); scale bars=100 μm, with 20× magnification (a-d).
Figure 5. Scanning electron microscope images of KKU-213A cells. Cells were cultured for 10 days in the silk fibroin-gelatin/hyaluronic acid/heparan sulfate (SF-GHHs) 1:2 (A, a), SF-GHHs 1:1 (B, b), SF-GHHs 2:1 (C, c), and silk fibroin (SF) scaffolds (D, d). Distinct spheroids were discerned in SF-GHHs 1:2 compared to other SF-GHHs scaffolds. An elongated cell morphology was observed in the SF scaffold. Scale bars=20 μm, with 600´ magnification (A-D); scale bars=10 μm, with 1,000´ magnification (a-d).
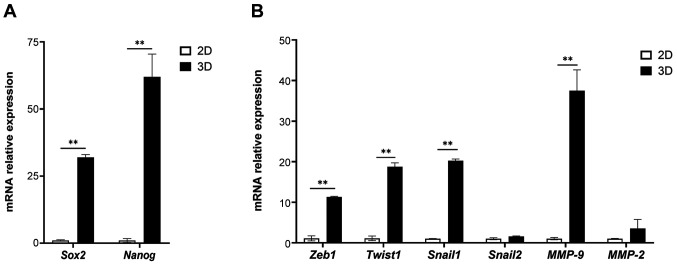
Upregulation of stemness- and EMT-related genes in 3D culture. After 3 days in culture, the expression of stemness-related genes (Sox2 and Nanog) and EMT-related genes (Zeb1, Twist1, Snail-1, and MMP-9) in KKU-213A cells was significantly upregulated in the 3D culture compared to the monolayer 2D condition (Figure 6). EMT and CSCs are closely related to each other, and play a key role in metastasis (43). Our findings indicated the drawback of 2D culture that could not support the growth of CSCs and EMT process, whereas the scaffold-based 3D model overcame these pitfalls.
Figure 6. Comparison of relative expression of stemness- and epithelial-mesenchymal transition (EMT)-related genes between 2D and 3D models. mRNA expression of stemness- (Sox2 and Nanog) (A) and EMT-related genes (Zeb1, Twist1, Snail1, Snail2, MMP-9 and MMP-2) (B) derived from 2D and 3D models was determined by quantitative reverse transcription polymerase chain reaction. Values are presented as mean±SD (n=3). Statistically significant differences between 2D and 3D at: **p<0.01.
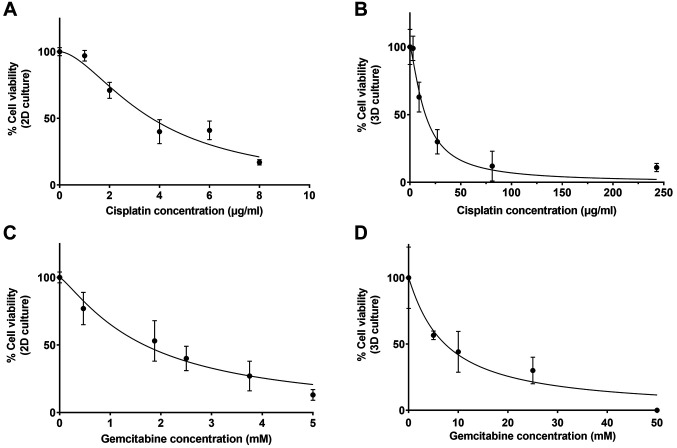
Effect of 2D and 3D cell cultures on treatment response. To compare anti-cancer drug response between 2D and 3D cell culture systems, KKU-213A cells were treated with different concentrations of cisplatin and gemcitabine as shown in Figure 7. For cisplatin, the IC50 was 15.19 μg/ml in 3D culture, which is 4.1-fold higher than that in 2D culture (IC50=3.68 μg/ml). For gemcitabine, the IC50 was 7.34 mM in 3D culture, which is 4.4-fold higher than that in 2D culture (IC50=1.67 mM). Therefore, KKU-213A cells grown in the SF-GHHs 1:2 scaffold showed increased resistance to both cisplatin and gemcitabine when compared to 2D culture.
Figure 7. Comparison of anti-cancer drug response between 2D and 3D culture models. KKU-213A cells grown in 2D (A, C) and 3D (B, D) systems were treated with cisplatin or gemcitabine, and cell viability was measured by the 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium) assay. All measurements were performed in triplicates and the data are presented as mean± SD.
Discussion
The study of Cardinale et al. on CSCs in CCA subtypes indicated that CSCs represented more than 30% of the CCA tumor mass and implicated in CCA carcinogenesis (3). In the simulation of the stem cell niche, a 3D model was generated to mimic the native ECM of CCA in this study. Collagen is a major structural ECM protein in portal tracts of the liver, one component of which is the bile duct (44). Moreover, high levels of HA and HSPG have been observed in CCA (29,32). In this study, SF-GHHs composite scaffolds for 3D CCA culture comprising silk fibroin (SF), gelatin (G), hyaluronic acid (HA) and heparan sulfate (HS) with different ratios (2:1, 1:1, and 1:2) were constructed and compared to SF scaffold. The constructed scaffolds showed stable beta-sheet conformation (45). It was found that high SF content was associated with increased intensity of beta-sheet conformation, high porosity and decreased pore size. Previous studies showed that high SF content led to high porosity (46,47). The increased GHHs levels in our scaffolds resulted in increased pore size and water uptake percentage. Dondajewska et al. showed that SF scaffolds with a pore size of 250-500 μm improved cancer cell infiltration and proliferation (17), supporting our finding that the SF-GHHs 1:2 scaffold with a pore size of 350±102 μm is suitable for promoting CCA cell proliferation. Porous scaffolds are important to enhance cell attachment and infiltration. The study of Dhamecha et al. indicated that the porous poly lactide co-glycolide microspheres (PPMS) with a porosity of 45.5% facilitated attachment and growth of A549 lung tumor cells (48). Thus, the SF-GHHs 1:2 scaffold with a porosity of 64.6% should provide sufficient porous structures for cell infiltration and growth. The highest water uptake of 94.69% found in the SF-GHHs 1:2 scaffold resulted from high levels of negatively charged HA and HS, allowing them to attract and retain water (49). Therefore, the physical characteristics of the SF-GHHs 1:2 scaffold showed a stable form with high water uptake, optimal pore size and porosity, which are suitable for CCA culture.
Regarding the biological properties, the SF-GHHs 1:2 scaffold promoted cell proliferation, attachment and aggregation resulting in spheroid formation and suggesting the contribution of natural polymers as an ECM for cancer cells. Gelatin contains the arginine–glycine–aspartic acid (RGD) motifs, which enhance cell adhesion through the interaction with integrin (26). The interaction between HA and its receptors, CD44 and receptor for HA-mediated motility (RHAMM) designated as CD168, plays pivotal roles in cell proliferation, migration and invasion (28). Heparan sulfate binds fibroblast growth factor (FGF) ligands and receptors, leading to FGF receptor (FGFR) dimerization, which promoted oncogenic signaling resulting in tumor growth (30), suggesting the role of HS in supporting cell growth in the 3D model. Our finding indicated the biological functions of the SF-GHHs 1:2 scaffold in providing a tumor microenvironment that supported CCA cell aggregation and expansion. The study of Tit-oon et al. showed that CCA cells aggregated and expanded in the collagen scaffold, whereas a flat monolayer was observed in 2D culture (50).
Sex determining region Y-Box 2 (Sox2) and nanog homeobox (Nanog) are transcription factors that maintain pluripotency and self-renewal in embryonic stem cells and have been used as markers for CSCs (51). The upregulation of Sox2 and Nanog was associated with poor prognosis in intrahepatic CCA (7,8).The upregulation of Sox2 and Nanog in KKU-213A cells grown on the SF-GHHs 1:2 scaffold, compared to 2D cell culture, suggested that the optimal ratio of natural polymers in the scaffold might serve as a CSC niche that promotes CSC proliferation and enrichment. Our finding was consistent with a previous study on 3D tumor spheroids of head and neck cancer in which increased expression of the CSC markers, including Sox2 and Nanog, was observed (52). Moreover, the upregulation of CSC-related genes was elucidated in the 3D cancer models compared to the 2D systems (16,53,54).
EMT is a reversible process in which epithelial cells transform into mesenchymal cells (55). Many of the EMT-inducing pathways control transcription factors of snail family transcriptional repressors (SNAIL), zinc finger E-box binding homeobox (ZEB), and twist family bHLH transcription factor (TWIST), which act as transcriptional repressors of E-cadherin leading to motility of epithelial cells (55). Matrix metalloproteinase (MMP)-2 and MMP-9 are gelatinase subgroups of matrix metalloproteinases (MMPs) that have been implicated in invasion and metastasis (56). EMT and CSCs are closely related to each other. During cancer metastasis, the EMT process enables cancer cell dissemination, by which disseminated cancer cells acquire self-renewal capability, similar to CSCs, which express markers associated with EMT (57). Moreover, in many types of cancer, only cancer cells within the CSC-enriched subpopulation show EMT activation (58). The upregulation of Snail-1, Twist1, Zeb1 and MMP-9 was observed in the SF-GHHs 1:2 scaffold compared to 2D system. This finding is in agreement with previous studies on primary CCA, where expression of Zeb1, Twist and Snail was found to be upregulated and associated with tumor metastasis, progression, poor prognosis and short patients’ survival (59–61). The upregulation of Snail-1, Twist, Zeb1 and MMP was demonstrated in 3D collagen scaffolds compared to 2D culture (15,53).
The potential link between EMT and CSCs has been illustrated in many types of human cancer, both of which are involved in drug resistance (58). A combination of gemcitabine and cisplatin is a standard first-line regimen for the treatment of advanced CCA (62). Cisplatin and gemcitabine were selected for drug sensitivity tests in 2D and 3D cancer models. The KKU-213A cells in the 3D culture were more resistant to both anticancer drugs than in the 2D system. There are many factors involved in increased drug resistance in 3D models including reduced penetration of anticancer drugs, increased cell compaction and adhesion, enhanced pro-survival signaling and upregulated drug resistance genes (19,63). Our results were in agreement with previous studies, which showed that liver and breast cancer cells grown in 3D silk scaffolds had higher chemoresistance than those grown in 2D culture (24,63,64).
In conclusion, a novel 3D SF-GHHs 1:2 scaffold provides an optimal tumor microenvironment that supports cell adhesion, enhances cell proliferation, and allows spheroid formation. This 3D scaffold also enhances the expression of CSC markers and EMT-related genes, and anti-cancer drug resistance. Thus, the 3D SF-GHHs 1:2 scaffold may be useful for studying CSCs and anti-cancer therapeutics in vitro.
Conflicts of Interest
The Authors declare no competing interests in relation to this study.
Authors’ Contributions
O.B. and T.L. conceived the project and designed the experiments. O.B. performed the experiments and statistical analyses of the data. M.W. assisted with data analysis and interpretation. J.D. interpreted the data. O.B. wrote the original manuscript. T.L. supervised the whole project, edited and reviewed, and approved final manuscript.
Acknowledgements
This project received financial support from the Centre for Research and Development of Medical Diagnostic Laboratories, Khon Kaen University, Khon Kean, Thailand. The Authors would like to thank the Cholangiocarcinoma Research Institute, Khon Kaen University for kindly providing the CCA cell line; and the SEM core facility, Faculty of Medicine, Khon Kaen University for providing SEM services. Gene expression was investigated by Real-time PCR at Srinagarind Hospital Excellence Laboratory, Clinical Laboratory Unit, Srinagarind Hospital, Faculty of Medicine, Khon Kean University, Khon Kaen, Thailand.
References
- 1.Sripa B, Pairojkul C. Cholangiocarcinoma: lessons from Thailand. Curr Opin Gastroenterol. 2008;24(3):349–356. doi: 10.1097/MOG.0b013e3282fbf9b3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartolini I, Risaliti M, Fortuna L, Agostini C, Ringressi MN, Taddei A, Muiesan P. Current management of intrahepatic cholangiocarcinoma: from resection to palliative treatments. Radiol Oncol. 2020;54(3):263–271. doi: 10.2478/raon-2020-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cardinale V, Renzi A, Carpino G, Torrice A, Bragazzi MC, Giuliante F, DeRose AM, Fraveto A, Onori P, Napoletano C, Franchitto A, Cantafora A, Grazi G, Caporaso N, D’Argenio G, Alpini G, Reid LM, Gaudio E, Alvaro D. Profiles of cancer stem cell subpopulations in cholangiocarcinomas. Am J Pathol. 2015;185(6):1724–1739. doi: 10.1016/j.ajpath.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iwahashi S, Utsunomiya T, Shimada M, Saito Y, Morine Y, Imura S, Ikemoto T, Mori H, Hanaoka J, Bando Y. High expression of cancer stem cell markers in cholangiolocellular carcinoma. Surg Today. 2013;43(6):654–660. doi: 10.1007/s00595-012-0437-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shimada M, Sugimoto K, Iwahashi S, Utsunomiya T, Morine Y, Imura S, Ikemoto T. CD133 expression is a potential prognostic indicator in intrahepatic cholangiocarcinoma. J Gastroenterol. 2010;45(8):896–902. doi: 10.1007/s00535-010-0235-3. [DOI] [PubMed] [Google Scholar]
- 6.Suwannakul N, Ma N, Thanan R, Pinlaor S, Ungarreevittaya P, Midorikawa K, Hiraku Y, Oikawa S, Kawanishi S, Murata M. Overexpression of CD44 variant 9: a novel cancer stem cell marker in human cholangiocarcinoma in relation to inflammation. Mediators Inflamm. 2018;2018:4867234. doi: 10.1155/2018/4867234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang MX, Gan W, Jing CY, Zheng SS, Yi Y, Zhang J, Xu X, Lin JJ, Zhang BH, Qiu SJ. High expression of Oct4 and Nanog predict poor prognosis in intrahepatic cholangiocarcinoma patients after curative resection. J Cancer. 2019;10(5):1313–1324. doi: 10.7150/jca.28349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gu MJ, Jang BI. Clinicopathologic significance of Sox2, CD44 and CD44v6 expression in intrahepatic cholangiocarcinoma. Pathol Oncol Res. 2014;20(3):655–660. doi: 10.1007/s12253-014-9745-2. [DOI] [PubMed] [Google Scholar]
- 9.Wu HJ, Chu PY. Role of cancer stem cells in cholangiocarcinoma and therapeutic implications. Int J Mol Sci. 2019;20(17):4154. doi: 10.3390/ijms20174154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hutmacher DW, Loessner D, Rizzi S, Kaplan DL, Mooney DJ, Clements JA. Can tissue engineering concepts advance tumor biology research. Trends Biotechnol. 2010;28(3):125–133. doi: 10.1016/j.tibtech.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 11.Chaicharoenaudomrung N, Kunhorm P, Noisa P. Three-dimensional cell culture systems as an in vitro platform for cancer and stem cell modeling. World J Stem Cells. 2019;11(12):1065–1083. doi: 10.4252/wjsc.v11.i12.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang C, Yang Z, Dong DL, Jang TS, Knowles JC, Kim HW, Jin GZ, Xuan Y. 3D culture technologies of cancer stem cells: promising ex vivo tumor models. J Tissue Eng. 2020;11:2041731420933407. doi: 10.1177/2041731420933407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vicent S, Lieshout R, Saborowski A, Verstegen MMA, Raggi C, Recalcati S, Invernizzi P, van der Laan LJW, Alvaro D, Calvisi DF, Cardinale V. Experimental models to unravel the molecular pathogenesis, cell of origin and stem cell properties of cholangiocarcinoma. Liver Int. 2019;39 Suppl 1:79–97. doi: 10.1111/liv.14094. [DOI] [PubMed] [Google Scholar]
- 14.Massa A, Varamo C, Vita F, Tavolari S, Peraldo-Neia C, Brandi G, Rizzo A, Cavalloni G, Aglietta M. Evolution of the experimental models of cholangiocarcinoma. Cancers (Basel) 2020;12(8):2308. doi: 10.3390/cancers12082308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu LJ, Zhang J, Xiao ZF, Dai B, Sun MY, Chen L, Chen B. Three-dimensional collagen scaffold enhances the human adenoid cystic carcinoma cancer stem cell and epithelial-mesenchymal transition properties. J Biomed Mater Res B Appl Biomater. 2014;102(4):772–780. doi: 10.1002/jbm.b.33058. [DOI] [PubMed] [Google Scholar]
- 16.Florczyk SJ, Kievit FM, Wang K, Erickson AE, Ellenbogen RG, Zhang M. 3D porous chitosan-alginate scaffolds promote proliferation and enrichment of cancer stem-like cells. J Mater Chem B. 2016;4(38):6326–6334. doi: 10.1039/C6TB01713D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dondajewska E, Juzwa W, Mackiewicz A, Dams-Kozlowska H. Heterotypic breast cancer model based on a silk fibroin scaffold to study the tumor microenvironment. Oncotarget. 2017;9(4):4935–4950. doi: 10.18632/oncotarget.23574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leung M, Kievit FM, Florczyk SJ, Veiseh O, Wu J, Park JO, Zhang M. Chitosan-alginate scaffold culture system for hepatocellular carcinoma increases malignancy and drug resistance. Pharm Res. 2010;27(9):1939–1948. doi: 10.1007/s11095-010-0198-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li J, Zhou Y, Chen W, Yuan Z, You B, Liu Y, Yang S, Li F, Qu C, Zhang X. A novel 3D in vitro tumor model based on silk fibroin/chitosan scaffolds to mimic the tumor microenvironment. ACS Appl Mater Interfaces. 2018;10(43):36641–36651. doi: 10.1021/acsami.8b10679. [DOI] [PubMed] [Google Scholar]
- 20.Acharya C, Ghosh SK, Kundu SC. Silk fibroin protein from mulberry and non-mulberry silkworms: cytotoxicity, biocompatibility and kinetics of L929 murine fibroblast adhesion. J Mater Sci Mater Med. 2008;19(8):2827–2836. doi: 10.1007/s10856-008-3408-3. [DOI] [PubMed] [Google Scholar]
- 21.Janani G, Nandi SK, Mandal BB. Functional hepatocyte clusters on bioactive blend silk matrices towards generating bioartificial liver constructs. Acta Biomater. 2018;67:167–182. doi: 10.1016/j.actbio.2017.11.053. [DOI] [PubMed] [Google Scholar]
- 22.Choi JH, Kim DK, Song JE, Oliveira JM, Reis RL, Khang G. Silk fibroin-based scaffold for bone tissue engineering. Adv Exp Med Biol. 2018;1077:371–387. doi: 10.1007/978-981-13-0947-2_20. [DOI] [PubMed] [Google Scholar]
- 23.Li Z, Ji S, Wang Y, Shen X, Liang H. Silk fibroin-based scaffolds for tissue engineering. Frontiers of Materials Science. 2019;7(3):237–247. doi: 10.1007/s11706-013-0214-8. [DOI] [Google Scholar]
- 24.Kundu B, Saha P, Datta K, Kundu SC. A silk fibroin based hepatocarcinoma model and the assessment of the drug response in hyaluronan-binding protein 1 overexpressed HepG2 cells. Biomaterials. 2013;34(37):9462–9474. doi: 10.1016/j.biomaterials.2013.08.047. [DOI] [PubMed] [Google Scholar]
- 25.Nii T, Makino K, Tabata Y. Three-dimensional culture system of cancer cells combined with biomaterials for drug screening. Cancers (Basel) 2020;12(10):2754. doi: 10.3390/cancers12102754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klimek K, Ginalska G. Proteins and peptides as important modifiers of the polymer scaffolds for tissue engineering applications-a review. Polymers (Basel) 2020;12(4):844. doi: 10.3390/polym12040844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shuborna NS, Chaiyasamut T, Sakdajeyont W, Vorakulpipat C, Rojvanakarn M, Wongsirichat N. Generation of novel hyaluronic acid biomaterials for study of pain in third molar intervention: a review. J Dent Anesth Pain Med. 2019;19(1):11–19. doi: 10.17245/jdapm.2019.19.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Misra S, Hascall VC, Markwald RR, Ghatak S. Interactions between hyaluronan and its receptors (CD44, RHAMM) regulate the activities of inflammation and cancer. Front Immunol. 2015;6:201. doi: 10.3389/fimmu.2015.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lv H, Yu G, Sun L, Zhang Z, Zhao X, Chai W. Elevate level of glycosaminoglycans and altered sulfation pattern of chondroitin sulfate are associated with differentiation status and histological type of human primary hepatic carcinoma. Oncology. 2007;72(5-6):347–356. doi: 10.1159/000113145. [DOI] [PubMed] [Google Scholar]
- 30.Nagarajan A, Malvi P, Wajapeyee N. Heparan sulfate and heparan sulfate proteoglycans in cancer initiation and progression. Front Endocrinol (Lausanne) 2018;9:483. doi: 10.3389/fendo.2018.00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sabit H, Tsuneyama K, Shimonishi T, Harada K, Cheng J, Ida H, Saku T, Saito K, Nakanuma Y. Enhanced expression of basement-membrane-type heparan sulfate proteoglycan in tumor fibro-myxoid stroma of intrahepatic cholangiocarcinoma. Pathol Int. 2001;51(4):248–256. doi: 10.1046/j.1440-1827.2001.01201.x. [DOI] [PubMed] [Google Scholar]
- 32.Batmunkh E, Tátrai P, Szabó E, Lódi C, Holczbauer A, Páska C, Kupcsulik P, Kiss A, Schaff Z, Kovalszky I. Comparison of the expression of agrin, a basement membrane heparan sulfate proteoglycan, in cholangiocarcinoma and hepatocellular carcinoma. Hum Pathol. 2007;38(10):1508–1515. doi: 10.1016/j.humpath.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 33.Kim UJ, Park J, Kim HJ, Wada M, Kaplan DL. Three-dimensional aqueous-derived biomaterial scaffolds from silk fibroin. Biomaterials. 2005;26(15):2775–2785. doi: 10.1016/j.biomaterials.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 34.Park HJ, Lee OJ, Lee MC, Moon BM, Ju HW, Lee Jm, Kim JH, Kim DW, Park CH. Fabrication of 3D porous silk scaffolds by particulate (salt/sucrose) leaching for bone tissue reconstruction. Int J Biol Macromol. 2015;78:215–223. doi: 10.1016/j.ijbiomac.2015.03.064. [DOI] [PubMed] [Google Scholar]
- 35.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 36.Kong J, Yu S. Fourier transform infrared spectroscopic analysis of protein secondary structures. Acta Biochim Biophys Sin (Shanghai) 2007;39(8):549–559. doi: 10.1111/j.1745-7270.2007.00320.x. [DOI] [PubMed] [Google Scholar]
- 37.DeBari MK, Abbott RD. Microscopic considerations for optimizing silk biomaterials. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2019;11(2):e1534. doi: 10.1002/wnan.1534. [DOI] [PubMed] [Google Scholar]
- 38.Stelling AL, Toher D, Uckermann O, Tavkin J, Leipnitz E, Schweizer J, Cramm H, Steiner G, Geiger KD, Kirsch M. Infrared spectroscopic studies of cells and tissues: triple helix proteins as a potential biomarker for tumors. PLoS One. 2013;8(3):e58332. doi: 10.1371/journal.pone.0058332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh R, Sahu S, Thangaraj M, Karthikeyan V. Anticoagulant potential of marine polychaete (Nereis species) Journal of Biological & Scientific Opinion. 2013;1(4):337–340. doi: 10.7897/2321-6328.01412. [DOI] [Google Scholar]
- 40.Sanden KW, Kohler A, Afseth NK, Böcker U, Rønning SB, Liland KH, Pedersen ME. The use of Fourier-transform infrared spectroscopy to characterize connective tissue components in skeletal muscle of Atlantic cod (Gadus morhua L.) J Biophotonics. 2019;12(9):e201800436. doi: 10.1002/jbio.201800436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shetty G, Kendall C, Shepherd N, Stone N, Barr H. Raman spectroscopy: elucidation of biochemical changes in carcinogenesis of oesophagus. Br J Cancer. 2006;94(10):1460–1464. doi: 10.1038/sj.bjc.6603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vasi AM, Popa MI, Butnaru M, Dodi G, Verestiuc L. Chemical functionalization of hyaluronic acid for drug delivery applications. Mater Sci Eng C Mater Biol Appl. 2014;38:177–185. doi: 10.1016/j.msec.2014.01.052. [DOI] [PubMed] [Google Scholar]
- 43.Babaei G, Aziz SG, Jaghi NZZ. EMT, cancer stem cells and autophagy; The three main axes of metastasis. Biomed Pharmacother. 2021;133:110909. doi: 10.1016/j.biopha.2020.110909. [DOI] [PubMed] [Google Scholar]
- 44.Bedossa P, Paradis V. Liver extracellular matrix in health and disease. J Pathol. 2003;200(4):504–515. doi: 10.1002/path.1397. [DOI] [PubMed] [Google Scholar]
- 45.Tsukada M, Gotoh Y, Nagura M, Minoura N, Kasai N, Freddi G. Structural changes of silk fibroin membranes induced by immersion in methanol aqueous solutions. Journal of Polymer Science Part B: Polymer Physics. 2021;32(5):961–968. doi: 10.1002/polb.1994.090320519. [DOI] [Google Scholar]
- 46.Sawatjui N, Damrongrungruang T, Leeanansaksiri W, Jearanaikoon P, Limpaiboon T. Fabrication and characterization of silk fibroin-gelatin/chondroitin sulfate/hyaluronic acid scaffold for biomedical applications. Materials Letters. 2019;126:207–210. doi: 10.1016/j.matlet.2014.04.018. [DOI] [Google Scholar]
- 47.Gil ES, Kluge JA, Rockwood DN, Rajkhowa R, Wang L, Wang X, Kaplan DL. Mechanical improvements to reinforced porous silk scaffolds. J Biomed Mater Res A. 2011;99(1):16–28. doi: 10.1002/jbm.a.33158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dhamecha D, Le D, Movsas R, Gonsalves A, Menon JU. Porous polymeric microspheres with controllable pore diameters for tissue engineered lung tumor model development. Front Bioeng Biotechnol. 2020;8:799. doi: 10.3389/fbioe.2020.00799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Habanjar O, Diab-Assaf M, Caldefie-Chezet F, Delort L. 3D cell culture systems: tumor application, advantages, and disadvantages. Int J Mol Sci. 2021;22(22):12200. doi: 10.3390/ijms222212200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tit-Oon P, Chokchaichamnankit D, Khongmanee A, Sawangareetrakul P, Svasti J, Srisomsap C. Comparative secretome analysis of cholangiocarcinoma cell line in three-dimensional culture. Int J Oncol. 2014;45(5):2108–2116. doi: 10.3892/ijo.2014.2636. [DOI] [PubMed] [Google Scholar]
- 51.Mcgrath NA, Fu J, Gu SZ, Xie C. Targeting cancer stem cells in cholangiocarcinoma (Review) Int J Oncol. 2020;57(2):397–408. doi: 10.3892/ijo.2020.5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Melissaridou S, Wiechec E, Magan M, Jain MV, Chung MK, Farnebo L, Roberg K. The effect of 2D and 3D cell cultures on treatment response, EMT profile and stem cell features in head and neck cancer. Cancer Cell Int. 2019;19:16. doi: 10.1186/s12935-019-0733-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen L, Xiao Z, Meng Y, Zhao Y, Han J, Su G, Chen B, Dai J. The enhancement of cancer stem cell properties of MCF-7 cells in 3D collagen scaffolds for modeling of cancer and anti-cancer drugs. Biomaterials. 2012;33(5):1437–1444. doi: 10.1016/j.biomaterials.2011.10.056. [DOI] [PubMed] [Google Scholar]
- 54.Gheytanchi E, Naseri M, Karimi-Busheri F, Atyabi F, Mirsharif ES, Bozorgmehr M, Ghods R, Madjd Z. Morphological and molecular characteristics of spheroid formation in HT-29 and Caco-2 colorectal cancer cell lines. Cancer Cell Int. 2021;21(1):204. doi: 10.1186/s12935-021-01898-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vaquero J, Guedj N, Clapéron A, Nguyen Ho-Bouldoires TH, Paradis V, Fouassier L. Epithelial-mesenchymal transition in cholangiocarcinoma: From clinical evidence to regulatory networks. J Hepatol. 2017;66(2):424–441. doi: 10.1016/j.jhep.2016.09.010. [DOI] [PubMed] [Google Scholar]
- 56.Roomi MW, Monterrey JC, Kalinovsky T, Rath M, Niedzwiecki A. Patterns of MMP-2 and MMP-9 expression in human cancer cell lines. Oncol Rep. 2009;21(5):1323–1333. doi: 10.3892/or_00000358. [DOI] [PubMed] [Google Scholar]
- 57.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, Campbell LL, Polyak K, Brisken C, Yang J, Weinberg RA. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133(4):704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shibue T, Weinberg RA. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol. 2017;14(10):611–629. doi: 10.1038/nrclinonc.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Duangkumpha K, Techasen A, Loilome W, Namwat N, Thanan R, Khuntikeo N, Yongvanit P. BMP-7 blocks the effects of TGF-β-induced EMT in cholangiocarcinoma. Tumour Biol. 2014;35(10):9667–9676. doi: 10.1007/s13277-014-2246-9. [DOI] [PubMed] [Google Scholar]
- 60.Kong D, Liang J, Li R, Liu S, Wang J, Zhang K, Chen D. Prognostic significance of snail expression in hilar cholangiocarcinoma. Braz J Med Biol Res. 2012;45(7):617–624. doi: 10.1590/s0100-879x2012007500070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Terashita K, Chuma M, Hatanaka Y, Hatanaka K, Mitsuhashi T, Yokoo H, Ohmura T, Ishizu H, Muraoka S, Nagasaka A, Tsuji T, Yamamoto Y, Kurauchi N, Shimoyama N, Toyoda H, Kumada T, Kaneoka Y, Maeda A, Ogawa K, Natsuizaka M, Kamachi H, Kakisaka T, Kamiyama T, Taketomi A, Matsuno Y, Sakamoto N. ZEB1 expression is associated with prognosis of intrahepatic cholangiocarcinoma. J Clin Pathol. 2016;69(7):593–599. doi: 10.1136/jclinpath-2015-203115. [DOI] [PubMed] [Google Scholar]
- 62.Guo X, Shen W. Latest evidence on immunotherapy for cholangiocarcinoma. Oncol Lett. 2020;20(6):381. doi: 10.3892/ol.2020.12244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Talukdar S, Kundu S. A non-mulberry silk fibroin protein based 3D in vitro tumor model for evaluation of anticancer drug activity. Advanced Functional Materials. 2021;22(22):4778–4788. doi: 10.1002/adfm.201200375. [DOI] [Google Scholar]
- 64.Mishra A, Mukhopadhyay SK, Dey S. Evaluation of cyclosaplin efficacy using a silk based 3D tumor model. Biomolecules. 2019;9(4):123. doi: 10.3390/biom9040123. [DOI] [PMC free article] [PubMed] [Google Scholar]