Abstract
Conventional methods for quantifying and phenotyping antigen‐specific lymphocytes can rapidly deplete irreplaceable specimens. This is due to the fact that antigen‐specific T and B cells have historically been analyzed in independent assays each requiring millions of cells. A technique that facilitates the simultaneous detection of antigen‐specific T and B cells would allow for more thorough immune profiling with significantly reduced sample requirements. To this end, we developed the B and T cell tandem lymphocyte evaluation (BATTLE) assay, which allows for the simultaneous identification of SARS‐CoV‐2 Spike reactive T and B cells using an activation induced marker (AIM) T cell assay and dual‐color B cell antigen probes. Using this assay, we demonstrate that antigen‐specific B and T cell subsets can be identified simultaneously using conventional flow cytometry platforms and provide insight into the differential effects of mRNA vaccination on B and T cell populations following natural SARS‐CoV‐2 infection.
Keywords: antigen‐specific, B cells, natural infection, SARS‐CoV‐2, spike protein, T cells, vaccination
1. INTRODUCTION
The identification, quantification, and characterization of antigen‐specific lymphocytes is a critical prerequisite for understanding the breadth, magnitude, and functional potential of vaccine and/or natural pathogen‐elicited immunity. Multiple distinct methodologic approaches have been developed that allow for the identification of antigen‐specific T and B cells. For the identification of antigen‐specific T cells, these include quantification of functional cytokine‐producing T cells by ELISPOT, flow cytometric detection of intracellular cytokine production following peptide stimulation, staining with peptide/HLA probes, and upregulation of activation‐induced markers following antigen engagement [1, 2, 3, 4, 5, 6]. Analogous approaches have also been developed for the identification of antigen specific B cells including, but not limited to, B cell ELISPOT assays and the use of fluorescently conjugated protein probes to stain antigen‐specific B cells [7, 8, 9, 10]. However, a major limitation of the approaches developed thus far is that they interrogate rare B and T cell subsets in isolation. An approach capable of identifying and phenotyping antigen‐specific B cells, CD4+ T cells, and CD8+ T cells simultaneously would preserve valuable samples by performing multiple assays in parallel, making the assessment of B and T cells that recognize the same antigen more efficient.
In this study, we endeavored to simultaneously identify and characterize SARS‐CoV‐2 spike protein‐specific B cells, CD4+ T cells, and CD8+ T cells in the same Peripheral Blood Mononuclear Cell (PBMC) sample. We used the combination of an Activation‐Induced Marker (AIM) assay to identify spike reactive T cells following stimulation with overlapping spike peptide pools, and spike reactive B cells by baiting with dual spike trimer fluorescent tetramers. This approach was then used to quantify and longitudinally track the T and B cell populations induced by natural SARS‐CoV‐2 infection and subsequent SARS‐CoV‐2 mRNA vaccination in the same individuals. Flow cytometric analysis of these antigen‐specific populations revealed that while the spike‐specific B cell, and to lesser degree CD8+ T cell responses, were consistently enhanced by mRNA vaccination, a clear boosting effect on spike‐specific CD4+ T cells was only detectable in individuals with low or undetectable T cell responses following natural infection. Furthermore, we validated our observations of SARS‐CoV‐2‐specific B cell functional enhancement upon vaccination with paired serological analysis. Our findings suggest divergent patterns of B and T cell subset mobilization with repeated antigen encounter and highlight the utility of examining these populations in concert.
2. MATERIALS AND METHODS
2.1. Study design
PBMC and sera for this study were obtained from the previously described SUNY Upstate Medical University Convalescent COVID‐19 Plasma Donor protocol [11, 12]. Samples used in this study were collected between April 22, 2020 and June 17, 2021. A subset of 10 donors was selected for focused analysis in the study using the following criteria: (1) blood collection at least 30 days after but not longer than 75 days after their last COVID‐19 symptom, (2) immunization with two doses of a SARS‐CoV‐2 mRNA vaccine at least 9 months after a negative polymerase chain reaction (PCR) test (8/10 Pfizer‐BioNTech, 2/10 Moderna mRNA‐1273), and (3) blood collection at least 30 days, but not longer than 22 weeks following vaccination. One subject was excluded from the final analysis due to immunosuppressive treatment for autoimmune disease at the time of sample collection. Archived sera and PBMC from healthy individuals collected prior to December 2019 were used as negative controls. Sample size was determined based on subject availability and conformity of sample collection to the described timeline.
2.2. Spike tetramer generation
Full‐length SARS‐CoV‐2 Spike (2P‐stabilized, C‐terminal Histidine/Avi‐tagged) was obtained from BEI resources (Manassas, VA, Cat. NR53524) and biotinylated using a BirA ubiquitin ligase (Avidity, Aurora, CO, Cat. Bir500A) following the manufacturer's recommended protocol. Biotinylated spike protein was purified using a 40 K molecular weight cut‐off (MWCO) 2 ml Pierce Zeba™ desalting column (Thermo Fisher Scientific, Waltham, MA, Cat. 87768), and mixed with streptavidin BV421 (BD, Cat. 563259) or streptavidin allophycocyanin (APC) (BD Cat. 554067) separately at a 20:1 ratio (~6:1 molar ratio) as previously described [3]. Tetramerized Spike probes were stored at 4°C until use. Just prior to probe staining, spike protein probes were added one‐by‐one to FACS wash buffer (1x PBS, 2% fetal bovine serum) containing 5 μM free d‐biotin (Avidity, Cat. Bir500A). Both streptavidin‐fluor conjugates were used to stain dimethyl sulfoxide (DMSO) control samples to further verify the absence of significant frequencies of nonspecific streptavidin‐binding B cells.
2.3. Activation induced marker (AIM) assay
Spike‐specific T cells were quantitated as a frequency of AIM+ (OX40+CD69+) CD4+ T or (CD25+CD69+) CD8+ T cells among PBMC following 24 h of stimulation with overlapping peptide pools covering the length of SARS‐CoV‐2 spike peptide (BEI, Cat. NR52402), as previously described [11]. Cryopreserved PBMC were thawed, washed twice, and cultured in complete cell culture media: RPMI 1640 medium (Corning, Tewksbury, MA, Cat. 10‐040‐CV) supplemented with 10% heat‐inactivated fetal bovine serum (Corning, Cat. 35‐010‐CV), 1% L‐glutamine (Lonza, Basel, Switzerland, Cat. BW17‐605E), and 1% penicillin/streptomycin (Gibco, Waltham, MA, Cat. 15140122). Cellular viability was assessed by trypan blue exclusion and cells were resuspended at a concentration of 5 × 106 cells/ml. Next, spike peptide pool cocktail was added to each well, diluted to a final concentration of 1 μg/ml/peptide (DMSO concentration 0.5%). Incubation with an equimolar amount of DMSO (0.5%) was used as a negative control, or monoclonal antihuman CD3 (Mabtech, Sweden, Cat. 3420‐2AST‐10) at a 1:1000 dilution for positive control wells. Cells were cultured for 24 h at 37°C, 5% CO2 in polystyrene plates.
2.4. Staining and flow cytometry
Stimulated PBMC were washed twice in warm complete culture media and transferred to polypropylene 96‐well plates for staining in the dark with 50 μl of spike tetramer probe cocktail containing 100 ng Spike per probe (total 200 ng) at 4°C for 1 h prior to surface staining in FACS wash buffer with the antibodies listed in Table S1 at 4°C for 30 min. Cells were washed with 1× PBS to avoid binding of antibodies to viability dye and stained with the LIVE/DEAD Fixable Aqua Stain Kit (Thermo, Cat. L34957) in 1× PBS at 4°C for 30 min. to exclude dead cells. The cells were washed again with FACS buffer to quench remaining dye. Single‐color compensation controls were prepared using Onecomp eBeads™ compensation beads (Thermo, Cat. 01111142) and Arc™ Amine Reactive Compensation Bead Kit (Thermo, Cat. A10628) according to manufacturer's instructions. For streptavidin‐fluorophore compensation, biotin antihuman CD19 (BioLegend, San Diego, CA, Cat. 302203) was used as a primary stain. Stained cells and compensation controls were acquired on a BD Fortessa or Aria II and analyzed using FlowJo (BD), version 10.8.0. No significant differences were observed between flow instruments. The antibody panel utilized in all experiments is shown in Table S1. Validation was performed between batches of spike tetramer probes on the same sample to confirm consistency of binding and fluorescence properties. This was measured as the frequency and number of spike+ B cells of total B cells between the same cells stained with the old or new batch.
The frequency of spike‐specific B cells was expressed as a percentage of total B cells (CD19+, CD3/CD14/CD56/LIVE/DEAD−, lymphocytes), or as number per 106 PBMC. Spike‐specific CD4+ and CD8+ T cell quantities were calculated as background (DMSO) subtracted data. Flow cytometry data has been submitted under ID FR‐FCM‐Z4PB and annotated at http://flowrepository.org.
2.5. ELISA
SARS‐CoV‐2 Spike/receptor binding domain (RBD) and nucleocapsid (N) antibody titers were quantified using a sandwich ELISA protocol. In brief, 96 well NUNC MaxSorb flat‐bottom plates were coated with 3.99 μg/ml recombinant SARS‐CoV‐2 Wuhan‐Hu‐1 spike RBD protein (Sino Biological, Beijing, PR China, Cat. 40592‐V08B), 1 μg/ml SARS‐CoV‐2 Wuhan‐Hu‐1 nucleocapsid protein (Sino, Cat. 40588‐V08B), 0.15 μg/ml SARS‐CoV‐2 alpha variant spike RBD protein (BEI, Cat. NR‐54004), or 0.15 μg/ml beta variant spike RBD protein (BEI, Cat. NR‐55278) diluted in sterile 1× phosphate buffered saline (PBS). Plates were washed and blocked for 30 min. at RT with 0.25% bovine serum albumin (BSA) + 1% normal goat serum in 1× PBS + 0.1% Tween (0.1% PBST) after overnight incubation 4°C. Serum samples were heat‐inactivated for 30 min. at 56°C and serially diluted fourfold eight times in singlet starting at 1:200 prior to incubation for 2 h at RT on the blocked plates. Plates were washed and antigen‐specific antibody binding was detected using antihuman IgG HRP (Horseradish Peroxidase) (MilliporeSigma, St. Louis, MO, Cat. SAB3701362), or antihuman IgM HRP (SeraCare, Milford, MA, Cat. 5220‐0328). Secondary antibody binding was quantified using the Pierce 3,3′,5,5′‐Tetramethylbenzidine (TMB) Substrate Kit (Thermo, Cat. 34021) and Synergy HT plate reader (BioTek, Winooski, VT). Antibody binding data were analyzed by nonlinear regression (One site specific binding with Hill slope) to determine EC50 titers, reported as equilibrium dissociation constant (Kd) values, in GraphPad Prism 9.1.0 (GraphPad Software, La Jolla, CA). Samples with a calculated EC50 of greater than the highest serum dilution used in the assay (1:200) were assigned a value of >1:200. Each ELISA run compared experimental samples to the same known SARS‐CoV‐2‐positive and SARS‐CoV‐2 naïve serum samples.
2.6. Statistical analyses
Statistical analyses were performed using Prism version 9.1.0 (GraphPad), except for correlations. Spearman's rho rank correlation coefficients were computed in R (version 1.3.1093) using the rcorr function in the Hmisc package (version 4.4‐1). Statistical significance was represented as shown: ns: p > 0.05, *: p < 0.05, **: p < 0.01, ***: p < 0.001, and ****: p < 0.0001. Individual analyses performed and sample size (n) are indicated in the figure legend. Statistical tests were chosen based on descriptive statistics for the specified dataset. The column in each column plot indicates the arithmetic mean of the data set.
2.7. Study approval
All participants provided written informed consent prior to participation in the study, which was performed according to protocols approved by the Institutional Review Board (IRB) of the SUNY Upstate Medical University under IRB number 1587400 for experimental samples and 1839296‐1 for pre‐pandemic control samples. All clinical investigation was conducted according to Declaration of Helsinki principles.
2.8. Graphics
Study design and assay workflow schematics were created with BioRender.com.
3. RESULTS
3.1. BATTLE assay
We developed an assay workflow (Figure 1A) for the simultaneous detection of SARS‐CoV‐2 spike‐specific T and B cells from cryopreserved PBMC. Cryopreserved PBMC were thawed and stimulated for 24 h with overlapping peptide pools spanning SARS‐CoV‐2 spike to stimulate SARS‐CoV‐2 spike protein‐reactive T cells, followed by incubation with fluorescent spike trimer tetramers to bait spike‐binding B cells. These steps were followed by staining with fluorescent antibodies and viability dye prior to interrogation by flow cytometry or sorting. The gating strategy for flow cytometry is illustrated in Figure S1. Using this approach, we were able to identify and quantify SARS‐CoV‐2 spike‐reactive B cells (Figure 1B) in tandem with conventional activation‐induced marker (AIM) T cell stimulation using as few as 3 × 106 PBMC per sample from SARS‐CoV‐2 mRNA vaccine recipients. Notably, overnight culture or stimulation with overlapping peptide pools did not significantly interfere with the detection of antigen‐specific B cells using fluorescent spike B cell probes, although a modest but statistically insignificant reduction was observed between cells stained fresh from cryopreservation and overnight culture with DMSO or peptide pools (Figure 1C). Our results demonstrate that B and t cell tandem lymphocyte evaluation (BATTLE) is an efficient assay capable of simultaneously quantifying and isolating antigen‐specific B and T cells from a single PBMC sample.
FIGURE 1.
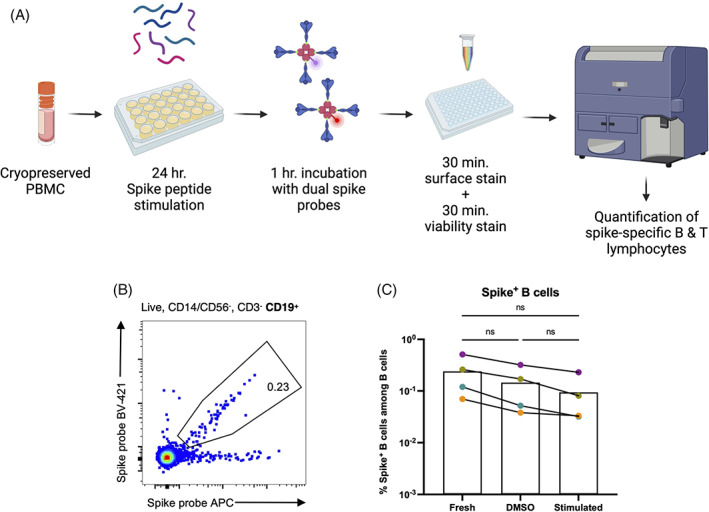
BATTLE: B and T tandem lymphocyte evaluation. (A) Schematic of BATTLE assay workflow. (B) Representative flow cytometry plot of SARS‐CoV‐2 spike protein‐specific B cells detected from the PBMC of a vaccinated individual using the BATTLE assay. Pre‐gating is shown above the plot. The frequency of cells within the double‐positive gate among B cells is shown inside the gate. (C) Total PBMC from SARS‐CoV‐2 mRNA vaccinated individuals were stimulated with overlapping spike peptide pools or DMSO alone and baited with dual spike BCR probes. An aliquot of each sample was also stained with the probes directly following thawing from cryopreservation (“Fresh”) for comparison. The frequency among B cells of B cells positive for both probes are shown, with each dot representing an individual. Matched fresh, DMSO control and peptide pool‐stimulated samples from the same individual are connected by a solid line. Matched groups were analyzed by Repeated Measures one‐way ANOVA, with the Geisser–Greenhouse correction (n = 4, exact p = 0.0682), and Tukey's multiple comparison's test, with individual variances computed for each comparison (n = 4, adjusted p = 0.1311, p = 0.1392, and p = 0.2038 for fresh vs. DMSO, fresh vs. stim., and DMSO vs. stim., respectively). [Color figure can be viewed at wileyonlinelibrary.com]
3.2. Study design and serology
To demonstrate the utility of our new approach, we used the BATTLE assay to examine the impact of SARS‐CoV‐2 mRNA vaccination on spike protein‐specific adaptive immunity following natural infection. We selected a cohort of subjects from a large study of convalescent plasma donors who experienced symptomatic but mild infection and had a peripheral blood draw between 30 and 75 days of symptom resolution (Figure 2A). This window was selected to capture the profile of antigen‐specific memory lymphocytes proximally to natural infection, but following resolution of acute inflammatory responses to infection. We included subjects who were vaccinated with two doses of mRNA vaccine at least 9 months following a negative SARS‐CoV‐2 PCR test and had a subsequent blood draw more than 30 days, but no more than 22 weeks following vaccination. These time constraints were chosen to ensure that immune memory was stable prior to vaccination, and to capture the profile of antigen‐specific memory lymphocytes proximally to vaccination, but following resolution of acute vaccine‐driven inflammation.
FIGURE 2.
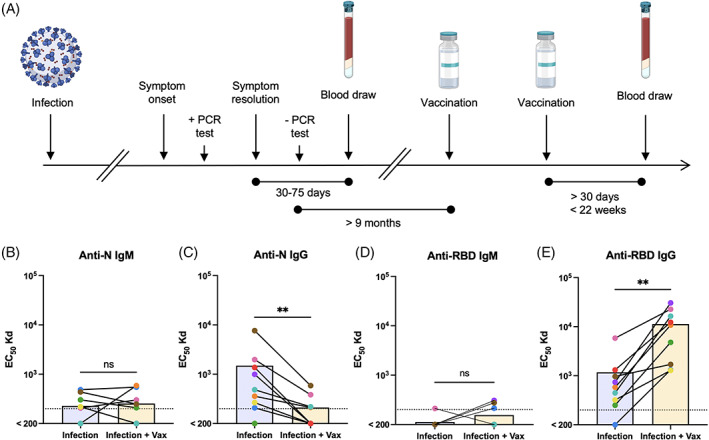
Study design and serology. (A) Timeline of SARS‐CoV‐2 convalescent study and sample collection. (B–E) Paired sera following natural infection and after subsequent vaccination from the same individual were assessed for IgM and IgG binding to SARS‐CoV‐2 nucleocapsid (B, C) or spike RBD (D, E) proteins. Each dot color represents an individual and a solid line connects pairs. Differences between infection and infection + vaccination groups were analyzed by Wilcoxon matched‐pairs signed rank test (n = 9, exact two‐tailed p > 0.9999 (B), p = 0.0078, 0.2500, 0.0039 [C–E]). The LOD is represented as a dotted line [Color figure can be viewed at wileyonlinelibrary.com]
To validate and extend the clinical utility of our findings, we evaluated the serological profiles of each donor from the same visits used in the BATTLE assay. As expected, IgG against the SARS‐CoV‐2 nucleocapsid protein, which is induced by natural infection but not mRNA vaccination, declined between the first and postvaccination samples, while anti‐N IgM was largely below the level of reliable detection (Figure 2B,C). Serum anti‐spike receptor‐binding domain (RBD) IgG, but not IgM, responses also were enhanced by mRNA vaccination (Figure 2D,E). Additionally, IgG, but not IgM, raised against the spike RBD from SARS‐CoV‐2 variant strains alpha and beta was significantly enhanced following mRNA vaccination, although not all subjects had equally improved variant reactivity (Figure S2).
3.3. SARS‐CoV‐2 mRNA vaccination significantly increases the number and frequency of spike‐reactive B cells among peripheral blood B cells following natural infection
To determine if SARS‐CoV‐2 mRNA vaccination had an impact on the B cell response targeting spike protein, we examined spike‐reactive B cells within postinfection and postinfection plus vaccination PBMC samples. We observed a significant increase in both the frequency and number of spike tetramer double‐positive B cells in postvaccination samples, compared to natural infection alone (Figures 3A,D and S3a). Importantly, spike‐specific B cells were virtually absent in PBMC samples collected prior to the onset of the SARS‐CoV‐2 pandemic, demonstrating the specificity of the assay.
FIGURE 3.
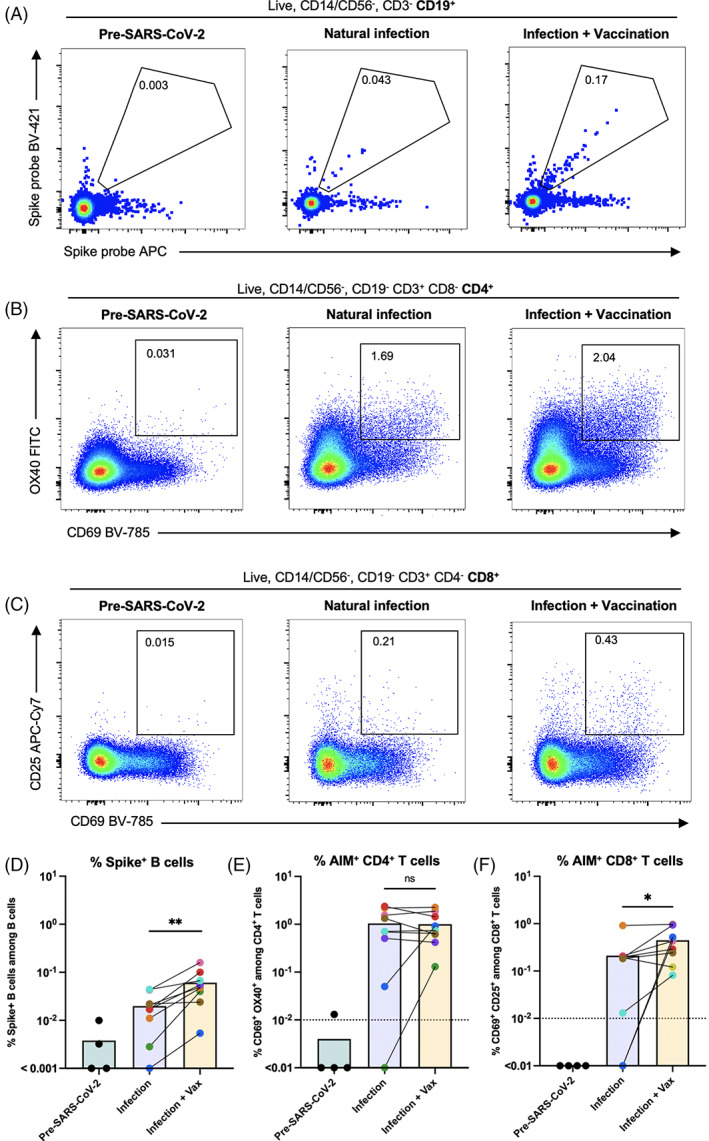
Spike‐specific lymphocyte responses following natural infection and subsequent vaccination. (A–C) Representative flow cytometry plots of SARS‐CoV‐2 spike protein‐specific B cells (A), OX40/CD69+ CD4+ (B) and CD25/CD69+ CD8+ (C) T cells detected from PBMC using the BATTLE assay from a sample collected independently prior to the onset of the pandemic, and following natural infection and subsequent vaccination in the same individual (left to right, respectively). Pre‐gating is shown above each panel. The frequency of cells within the double‐positive gate among B cells, CD4+ T cells, and CD8+ T cells is shown inside the gates, respectively. (D) The frequency among B cells of cells positive for both probes are shown, with each dot or dot pair representing an individual. Paired postinfection and postinfection plus vaccination samples from the same individual are connected by a solid line. Paired groups were analyzed by Wilcoxon matched‐pairs signed rank test (n = 9, exact two‐tailed p = 0.0039). (E) The frequency among CD4+ T cells of cells positive for activation‐induced markers OX40 and CD69 are shown, following subtraction of DMSO control values. Each dot or dot pair represents an individual. Paired postinfection and postinfection plus vaccination samples from the same individual are connected by a solid line. Paired groups were analyzed by Wilcoxon matched‐pairs signed rank test (n = 9, exact two‐tailed p > 0.9999). The LOD is represented as a dotted line. (F) The frequency among CD8+T cells of cells positive for activation‐induced markers CD25 and CD69 are shown, following subtraction of DMSO control values. Each dot or dot pair represents an individual. Paired postinfection and postinfection plus vaccination samples from the same individual are connected by a solid line. Paired groups were analyzed by Wilcoxon matched‐pairs signed rank test (n = 9, exact two‐tailed p = 0.0234). The LOD is represented as a dotted line. [Color figure can be viewed at wileyonlinelibrary.com]
Taken together, our data are in alignment with other studies in which an enhancement of spike‐reactive adaptive B cell responses was observed in individuals who received SARS‐CoV‐2 mRNA vaccination following natural infection [13].
3.4. SARS‐CoV‐2 mRNA vaccination differentially impacts the spike‐specific peripheral CD4+ and CD8+ T cell compartments following natural infection
We next sought to determine whether spike‐reactive CD4+ and CD8+ T cells within the same PBMC vial were similarly enhanced by mRNA vaccination after natural SARS‐CoV‐2 infection. We quantified the expression of activation‐induced markers on these T cell subsets and found that, unlike for B cells targeting spike protein, the frequency of CD4+ spike peptide‐activated T cells was not significantly enhanced following mRNA vaccination compared with infection alone (Figure 3B,E). Although significant changes were not observed for the full cohort, the frequency and number of AIM+ CD4+ T cells was increased following vaccination for some individuals, particularly for those with a lower initial spike‐specific adaptive response. The same trend was observed for subjects with low AIM+ CD8+ T cell frequencies following natural infection, but unlike the spike‐specific CD4+ T cell compartment, CD8+ T cells reactive to spike were marginally enhanced in frequency following mRNA vaccination (Figure 3C,F). The same patterns were present when adjusting for the number of cells used as input for the assay, although for AIM+ CD8+ T cells differences between the groups no longer reached statistical significance (Figure S3b,c). As observed for spike‐specific B cells, few AIM+ T cells were present in PBMC samples collected prior to the onset of the SARS‐CoV‐2 pandemic, and unlike T cells from exposed subjects, those from pre‐SARS‐CoV‐2 samples were largely unresponsive to spike peptide pool stimulation (Figure S3b,d).
Correlative analysis of pooled data from both infection and infection plus vaccination samples revealed a positive correlation between the frequency of SARS‐CoV‐2 spike‐specific B cells and serum IgG titers against the spike RBD from the original Wuhan strain, as well as the alpha and beta variants (Figure S3f). The frequency of AIM+ CD8+ T cells also positively correlated with anti‐RBD IgG, but this was only significant for the original SARS‐CoV‐2 strain. These results propose a coordinated response to SARS‐CoV‐2 between B cells and CD8+ T cells, potentially without sustained synchronization with the CD4+ T cell compartment. Taken together, these data suggest that mRNA vaccination of previously infected individuals influences T cell subsets asymmetrically, and may allow those who mount a sub‐optimal response to infection to achieve an immunological set point not reached via natural infection.
4. DISCUSSION
Here we demonstrate that B cells and CD4+ and CD8+ T cells specific for the same antigen can be simultaneously identified and quantified in a single PBMC sample. Furthermore, our results suggest that the ability for SARS‐CoV‐2 spike‐specific B and T cells to be enhanced by mRNA vaccination following natural SARS‐CoV‐2 infection may not be equal. Other studies have demonstrated that the spike‐specific B and T cell responses are highly heterogeneous within the human population [3, 14, 15, 16], findings which are echoed here. Our work suggests that despite this heterogeneity, trends in the patterns of antigen‐specific B and T cell expansion and contraction can be detected even within small cohorts. Future work will examine whether, like quantity, phenotypic characteristics such as lineage subset markers, integrin and chemokine expression, and effector function‐associated markers are also heterogeneous within the spike‐specific lymphocyte populations identified. Complementary transcriptional analysis of the sorted cells via bulk or single cell RNA sequencing will also provide wider insight into the features of antigen‐specific cells, as well as the ability to interrogate B and T cell receptor diversity.
Although our study was limited to the examination of PBMC, it is possible that these observations extend to other sources of lymphocytes, such as lymph node aspirates, bronchioalveolar lavage fluid, and bone marrow. Antigen‐specific lymphocytes have been identified within these specimens in humans and non‐human primates [17, 18, 19], suggesting that BATTLE can be applied to maximize the yield of information gathered from these valuable samples. Although AIM assays can be performed with as few as 1 × 106 PBMC per well (3 × 106 including controls) [3, 20], the detection of antigen‐specific B cells can utilize 1–1.2 × 107 PBMC per sample [3, 21], with this number varying considerably based on the expected frequency of the population. Thus, BATTLE can reduce the number of PBMC required to detect both antigen‐specific B and T cells by up to 12 million PBMC.
Unlike other studies, we did not detect substantial frequencies or numbers of SARS‐CoV‐2 spike‐binding T cells in nonexposed individuals [22, 23]. We also did not detect SARS‐CoV‐2 cross‐reactive B cells at appreciable frequencies in pre‐pandemic controls, as has been suggested by serological studies from other groups [24, 25]. We attribute these disparities between our observations and others' potentially to demographic variability between our sampling region and populations and those sampled in other studies. It is also possible that the sensitivity of our assay was lower, although we consider this unlikely to be the case, as the quantities of antigen‐specific T cells we observed postinfection and postvaccination were well‐aligned with those in the literature [3, 26].
The observation of increased quantities of spike‐specific B cells following mRNA vaccination after natural infection supports the idea that mRNA vaccination is beneficial even after natural adaptive immunity has developed. Additionally, our results suggest that spike‐reactive B cells may possess a capacity for vaccine‐elicited expansion beyond that of spike‐reactive CD4+ T cells. The mechanism by which this process may occur is still an active area of investigation, but an attractive hypothesis is that B and T cell populations each achieve an independent immunological set point following resolution of acute infection or vaccine‐induced activation. As the epitopes recognized by B cells, CD4+ T cells, and CD8+ T cells, the timing, and settings in which these events occur, and the environmental signals these population receive can differ considerably, it is tempting to speculate that uncoordinated dynamic expansion and contraction of B and T cells can lead to the generation of immunological memory in a compartmentalized manner. This interpretation would be supported with recent work describing coordinated and timely B and T cell responses that lead to the generation of superior adaptive immunity, and importantly that the lack thereof can have pathological consequences [27, 28, 29]. BATTLE is an ideal approach that can be harnessed to explore these concepts.
There are some important limitations to the use of BATTLE worth additional consideration. Our study did not exclude the possibility that coculture during T cell stimulation and/or BCR baiting has an impact on each cell population at the phenotypic, population subset, or transcriptional level. Indeed, it will be important to examine the transcriptional signatures of T and B cells isolated via BATTLE compared to traditional assays to explore this possibility. Likewise, future users of BATTLE would be advised to carefully assess whether the population dynamics of B and T cell subsets in specific contexts are altered by the assay, particularly if activated and effector lymphocytes are of interest. If the co‐stimulatory conditions do indeed cause activation or exhaustion of B or T cells, we envision the adaptation of this assay to study the outcome of any resulting cognate recognition‐driven processes. Alternatively, the duration of co‐stimulation may be modified to optimize the assay to the needs of particular research questions. Importantly, our development of BATTLE exclusively assessed PBMC following cryopreservation for quantification of antigen‐specific lymphocytes relative to those of other cryopreserved samples.
Given the paucity of antigen‐specific memory lymphocytes following infection and vaccination, the findings here have the potential to increase the breadth of diagnostic and experimental protocols which rely on interrogation of these populations. Thus, our studies have potential implications beyond the study of SARS‐CoV‐2 and mRNA vaccination. We plan to adapt BATTLE to examine adaptive responses to additional antigens in the future.
AUTHOR CONTRIBUTIONS
Krista L Newell: Conceptualization (equal); data curation (lead); formal analysis (lead); investigation (lead); methodology (equal); validation (lead); visualization (lead); writing – original draft (lead); writing – review and editing (lead). Mitchell J Waldran: Investigation (supporting); writing – review and editing (supporting). Stephen J Thomas: Funding acquisition (supporting); project administration (supporting); supervision (supporting); writing – review and editing (supporting). Timothy P Endy: Funding acquisition (supporting); project administration (supporting); supervision (supporting); writing – review and editing (supporting). Adam Waickman: Conceptualization (equal); funding acquisition (lead); methodology (equal); project administration (lead); supervision (lead); writing – original draft (supporting); writing – review and editing (supporting).
FUNDING INFORMATION
This work was supported by the State of New York with funds awarded to Adam T. Waickman.
CONFLICT OF INTEREST
Stephen J. Thomas reports compensation from Pfizer, during the conduct of the study; personal fees from Merck, Sanofi, Takeda, Themisbio, and Janssen, outside the submitted work. All other authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supporting information
MIFlowCyt MIFlowCyt Item Checklist.
Supplementary Figure S1 BATTLE assay gating strategy.
The flow cytometry gating strategy used to identify SARS‐CoV‐2 spike protein‐specific B and T cells is shown using a representative sample.
Supplementary Figure 2: SARS‐CoV‐2 variant spike RBD serology.
A‐D: Paired sera following natural infection and after subsequent vaccination from the same individuals were assessed for IgM and IgG binding to SARS‐CoV‐2 alpha (A, B) or beta (C, D) variant spike RBD proteins. Each dot color represents an individual and pairs are connected by a solid line. Differences between infection and infection + vaccination groups were analyzed by Wilcoxon matched‐pairs signed rank test (n = 9, exact two‐tailed p = 0.6250, 0.0195, 0.3125, 0.0469 [A–D]).
Supplementary Figure 3: AIM + T cell analysis and correlative analysis.
A: The number of B cells positive for both probes per 1 × 106 cells plated for stimulation is shown, with each dot or dot pair representing an individual. Paired postinfection and postinfection plus vaccination samples from the same individual are connected by a solid line. Paired groups were analyzed by Wilcoxon matched‐pairs signed rank test (n = 9, exact two‐tailed p = 0.0039).
B: The number of CD4+ T cells positive for activation‐induced markers OX40 and CD69 per 1 × 106 cells plated for stimulation is shown, following subtraction of DMSO control values. Each dot or dot pair represents an individual. Paired postinfection and postinfection plus vaccination samples from the same individual are connected by a solid line. Paired groups were analyzed by Wilcoxon matched‐pairs signed rank test (n = 9, exact two‐tailed p = 0.5703).
C: The number of CD8+ T cells of cells positive for activation‐induced markers CD25 and CD69 are shown, following subtraction of DMSO control values. Each dot or dot pair represents an individual. Paired postinfection and postinfection plus vaccination samples from the same individual are connected by a solid line. Paired groups were analyzed by Wilcoxon matched‐pairs signed rank test (n = 9, exact two‐tailed p = 0.2500).
D. The frequency of AIM+ CD4+ T cells among total CD4+ T cells is shown with each pair of dots representing an individual. Paired DMSO control and peptide pool‐stimulated samples from the same individual are connected by a solid line (n = 4 pre‐SARS‐CoV‐2, 20 post‐SARS‐CoV‐2). Subjects exposed to SARS‐CoV‐2 mRNA vaccination only (turquoise), natural infection only (purple), and infection plus vaccination (orange) are shown.
E. The frequency of AIM+ CD8+ T cells among total CD8+ T cells is shown with each pair of dots representing an individual. Paired DMSO control and peptide pool‐stimulated samples from the same individual are connected by a solid line (n = 4 pre‐SARS‐CoV‐2, 20 post‐SARS‐CoV‐2). Subjects exposed to SARS‐CoV‐2 mRNA vaccination only (turquoise), natural infection only (purple), and infection plus vaccination (orange) are shown.
F. Spearman's rho rank correlation coefficients are graphed by circle color according to the scale on the right of the panel. The size of the circle indicates increasing statistical significance. Correlation matrix was generated from data as presented in the previous figures representing the combined SARS‐CoV‐2 infection and vaccination‐elicited immune response.
Supplementary Table 1: Fluorescent antibodies used in BATTLE assay.
Supporting Information
ACKNOWLEDGMENTS
We gratefully acknowledge excellent technical assistance provided by the Upstate Medical University Flow Cytometry Core. We thank L. Phelps and M. Miller for their technical support. We thank Drs. J. Wilmore, H. Friberg, J. Currier, G. Gromowski, and A. Wegman for their helpful comments. The following reagents were obtained through BEI Resources, NIAID, NIH: Spike Glycoprotein (Stabilized) from SARS‐Related Coronavirus 2, Wuhan‐Hu‐1 with C‐Terminal Histidine and Avi Tags, Recombinant from HEK293F Cells, NR‐53524, Peptide Array, SARS‐Related Coronavirus 2 Spike (S) Glycoprotein, NR‐52402, Spike Glycoprotein RBD from SARS‐Related Coronavirus 2, United Kingdom Variant with C‐Terminal Histidine Tag, Recombinant from HEK293 Cells, NR‐54004, and Spike Glycoprotein RBD from SARS Related Coronavirus 2, Beta Variant with C‐Terminal Histidine Tag, Recombinant from HEK293 Cells, NR‐55278.
Newell KL, Waldran MJ, Thomas SJ, Endy TP, Waickman AT. Simultaneous analysis of antigen‐specific B and T cells after SARS‐CoV‐2 infection and vaccination. Cytometry. 2022;101(6):474–482. 10.1002/cyto.a.24563
Funding information The State of New York
DATA AVAILABILITY STATEMENT
Flow cytometry data has been submitted under ID FR‐FCM‐Z4PB and annotated at http://flowrepository.org. All other data are available upon request.
REFERENCES
- 1. Altman JD, Moss PA, Goulder PJ, Barouch DH, McHeyzer‐Williams MG, Bell JI, et al. Phenotypic analysis of antigen‐specific T lymphocytes. Science. 1996;274:94–6. [PubMed] [Google Scholar]
- 2. Dan JM, Lindestam Arlehamn CS, Weiskopf D, da Silva Antunes R, Havenar‐Daughton C, Reiss SM, et al. A cytokine‐independent approach to identify antigen‐specific human germinal center T follicular helper cells and rare antigen‐specific CD4+ T cells in blood. J Immunol. 2016;1950(197):983–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dan JM, Mateus J, Kato Y, Hastie KM, Yu ED, Faliti CE, et al. Immunological memory to SARS‐CoV‐2 assessed for up to 8 months after infection. Science. 2021;371:eabf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Frentsch M, Arbach O, Kirchhoff D, Moewes B, Worm M, Rothe M, et al. Direct access to CD4+ T cells specific for defined antigens according to CD154 expression. Nat Med. 2005;11:1118–24. [DOI] [PubMed] [Google Scholar]
- 5. Kern F, Faulhaber N, Frömmel C, Khatamzas E, Prösch S, Schönemann C, et al. Analysis of CD8 T cell reactivity to cytomegalovirus using protein‐spanning pools of overlapping pentadecapeptides. Eur J Immunol. 2000;30:1676–82. [DOI] [PubMed] [Google Scholar]
- 6. Maecker HT, Dunn HS, Suni MA, Khatamzas E, Pitcher CJ, Bunde T, et al. Use of overlapping peptide mixtures as antigens for cytokine flow cytometry. J Immunol Methods. 2001;255:27–40. [DOI] [PubMed] [Google Scholar]
- 7. Boonyaratanakornkit J, Taylor JJ. Techniques to study antigen‐specific B cell responses. Front Immunol. 2019;10:1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Czerkinsky CC, Nilsson LA, Nygren H, Ouchterlony O, Tarkowski A. A solid‐phase enzyme‐linked immunospot (ELISPOT) assay for enumeration of specific antibody‐secreting cells. J Immunol Methods. 1983;65:109–21. [DOI] [PubMed] [Google Scholar]
- 9. Julius MH, Masuda T, Herzenberg LA. Demonstration that antigen‐binding cells are precursors of antibody‐producing cells after purification with a fluorescence‐activated cell sorter. Proc Natl Acad Sci U S A. 1972;69:1934–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Moody MA, Haynes BF. Antigen‐specific B cell detection reagents: use and quality control. Cytom. Part J Int Soc Anal Cytol. 2008;73:1086–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fang H, Wegman AD, Ripich K, Friberg H, Currier JR, Thomas SJ, et al. Persistent COVID‐19 symptoms minimally impact the development of SARS‐CoV‐2‐specific T cell immunity. Viruses. 2021;13:916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Newell KL, Clemmer DC, Cox JB, Kayode YI, Zoccoli‐Rodriguez V, Taylor HE, et al. Switched and unswitched memory B cells detected during SARS‐CoV‐2 convalescence correlate with limited symptom duration. PLoS One. 2021;16:e0244855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang Z, Muecksch F, Schaefer‐Babajew D, Finkin S, Viant C, Gaebler C, et al. Naturally enhanced neutralizing breadth against SARS‐CoV‐2 one year after infection. Nature. 2021;595:426–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rodda LB, Netland J, Shehata L, Pruner KB, Morawski PA, Thouvenel CD, et al. Functional SARS‐CoV‐2‐specific immune memory persists after mild COVID‐19. Cell. 2020;184:169–183.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sette A, Crotty S. Adaptive immunity to SARS‐CoV‐2 and COVID‐19. Cell. 2021;184:861–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Weiskopf D, Schmitz KS, Raadsen MP, Grifoni A, Okba NMA, Endeman H, et al. Phenotype and kinetics of SARS‐CoV‐2‐specific T cells in COVID‐19 patients with acute respiratory distress syndrome. Sci Immunol. 2020;5:eabd2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Corbett KS, Nason MC, Flach B, Gagne M, O'Connell S, Johnston TS, et al. Immune correlates of protection by mRNA‐1273 vaccine against SARS‐CoV‐2 in nonhuman primates. Science. 2021;373:eabj0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kaneko N, Kuo H‐H, Boucau J, Farmer JR, Allard‐Chamard H, Mahajan VS, et al. Loss of Bcl‐6‐expressing T follicular helper cells and germinal centers in COVID‐19. Cell. 2020;183:143–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Turner JS, Kim W, Kalaidina E, Goss CW, Rauseo AM, Schmitz AJ, et al. SARS‐CoV‐2 infection induces long‐lived bone marrow plasma cells in humans. Nature. 2021;595:421–5. [DOI] [PubMed] [Google Scholar]
- 20. GeurtsvanKessel CH, Geers D, Schmitz KS, Mykytyn AZ, Lamers MM, Bogers S, et al. Divergent SARS CoV‐2 omicron‐reactive T‐ and B cell responses in COVID‐19 vaccine recipients. Sci Immunol. 2022;7:eabo2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hartley GE, Edwards ESJ, Aui PM, Varese N, Stojanovic S, McMahon J, et al. Rapid generation of durable B cell memory to SARS‐CoV‐2 spike and nucleocapsid proteins in COVID‐19 and convalescence. Sci Immunol. 2020;5:eabf8891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher CR, et al. Targets of T cell responses to SARS‐CoV‐2 coronavirus in humans with COVID‐19 disease and unexposed individuals. Cell. 2020;181:1489–1501.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mateus J, Grifoni A, Tarke A, Sidney J, Ramirez SI, Dan JM, et al. Selective and cross‐reactive SARS‐CoV‐2 T cell epitopes in unexposed humans. Science. 2020;370:89–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ng KW, Faulkner N, Cornish GH, Rosa A, Harvey R, Hussain S, et al. Preexisting and de novo humoral immunity to SARS‐CoV‐2 in humans. Science. 2020;370:1339–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wec AZ, Wrapp D, Herbert AS, Maurer DP, Haslwanter D, Sakharkar M, et al. Broad neutralization of SARS‐related viruses by human monoclonal antibodies. Science. 2020;369:731–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rydyznski Moderbacher C, Ramirez SI, Dan JM, Grifoni A, Hastie KM, Weiskopf D, et al. Antigen‐specific adaptive immunity to SARS‐CoV‐2 in acute COVID‐19 and associations with age and disease severity. Cell. 2020;183:996–1012.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lucas C, Klein J, Sundaram ME, Liu F, Wong P, Silva J, et al. Delayed production of neutralizing antibodies correlates with fatal COVID‐19. Nat Med. 2021;27:1178–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Painter MM, Mathew D, Goel RR, Apostolidis SA, Pattekar A, Kuthuru O, et al. Rapid induction of antigen‐specific CD4+ T cells is associated with coordinated humoral and cellular immunity to SARS‐CoV‐2 mRNA vaccination. Immunity. 2021;54:2133–2142.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zohar T, Loos C, Fischinger S, Atyeo C, Wang C, Slein MD, et al. Compromised humoral functional evolution tracks with SARS‐CoV‐2 mortality. Cell. 2020;183:1508–1519.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
MIFlowCyt MIFlowCyt Item Checklist.
Supplementary Figure S1 BATTLE assay gating strategy.
The flow cytometry gating strategy used to identify SARS‐CoV‐2 spike protein‐specific B and T cells is shown using a representative sample.
Supplementary Figure 2: SARS‐CoV‐2 variant spike RBD serology.
A‐D: Paired sera following natural infection and after subsequent vaccination from the same individuals were assessed for IgM and IgG binding to SARS‐CoV‐2 alpha (A, B) or beta (C, D) variant spike RBD proteins. Each dot color represents an individual and pairs are connected by a solid line. Differences between infection and infection + vaccination groups were analyzed by Wilcoxon matched‐pairs signed rank test (n = 9, exact two‐tailed p = 0.6250, 0.0195, 0.3125, 0.0469 [A–D]).
Supplementary Figure 3: AIM + T cell analysis and correlative analysis.
A: The number of B cells positive for both probes per 1 × 106 cells plated for stimulation is shown, with each dot or dot pair representing an individual. Paired postinfection and postinfection plus vaccination samples from the same individual are connected by a solid line. Paired groups were analyzed by Wilcoxon matched‐pairs signed rank test (n = 9, exact two‐tailed p = 0.0039).
B: The number of CD4+ T cells positive for activation‐induced markers OX40 and CD69 per 1 × 106 cells plated for stimulation is shown, following subtraction of DMSO control values. Each dot or dot pair represents an individual. Paired postinfection and postinfection plus vaccination samples from the same individual are connected by a solid line. Paired groups were analyzed by Wilcoxon matched‐pairs signed rank test (n = 9, exact two‐tailed p = 0.5703).
C: The number of CD8+ T cells of cells positive for activation‐induced markers CD25 and CD69 are shown, following subtraction of DMSO control values. Each dot or dot pair represents an individual. Paired postinfection and postinfection plus vaccination samples from the same individual are connected by a solid line. Paired groups were analyzed by Wilcoxon matched‐pairs signed rank test (n = 9, exact two‐tailed p = 0.2500).
D. The frequency of AIM+ CD4+ T cells among total CD4+ T cells is shown with each pair of dots representing an individual. Paired DMSO control and peptide pool‐stimulated samples from the same individual are connected by a solid line (n = 4 pre‐SARS‐CoV‐2, 20 post‐SARS‐CoV‐2). Subjects exposed to SARS‐CoV‐2 mRNA vaccination only (turquoise), natural infection only (purple), and infection plus vaccination (orange) are shown.
E. The frequency of AIM+ CD8+ T cells among total CD8+ T cells is shown with each pair of dots representing an individual. Paired DMSO control and peptide pool‐stimulated samples from the same individual are connected by a solid line (n = 4 pre‐SARS‐CoV‐2, 20 post‐SARS‐CoV‐2). Subjects exposed to SARS‐CoV‐2 mRNA vaccination only (turquoise), natural infection only (purple), and infection plus vaccination (orange) are shown.
F. Spearman's rho rank correlation coefficients are graphed by circle color according to the scale on the right of the panel. The size of the circle indicates increasing statistical significance. Correlation matrix was generated from data as presented in the previous figures representing the combined SARS‐CoV‐2 infection and vaccination‐elicited immune response.
Supplementary Table 1: Fluorescent antibodies used in BATTLE assay.
Supporting Information
Data Availability Statement
Flow cytometry data has been submitted under ID FR‐FCM‐Z4PB and annotated at http://flowrepository.org. All other data are available upon request.


