Abstract
Accurate detection of early COVID-19 cases is crucial to reduce infections and deaths, however, it remains a challenge. Here, we used the results from a seroprevalence study in 50 US states to apply our Retrospective Methodology to Estimate Daily Infections from Deaths (REMEDID) with the aim of analyzing the initial spread of SARS-CoV-2 infections across the US. Our analysis revealed that the virus likely entered the country through California on December 28, 2019, which corresponds to 16 days prior to the officially recognized entry date established by the Centers of Disease Control and Prevention. Furthermore, the REMEDID algorithm provides evidence that SARS-CoV-2 entered, on average, a month earlier than previously reflected in official data for each US state. Collectively, our mathematical modeling provides more accurate estimates of the initial COVID-19 cases in the US, and has the ability to be extrapolated to other countries and used to retrospectively track the progress of the pandemic. The use of approaches such as REMEDID are highly recommended to better understand the early stages of an outbreak, which will enable health authorities to improve mitigation and preventive measures in the future.
Keywords: COVID-19, Experimental retrospective analysis, REMEDID, Epidemics, Pandemic, First infection
1. Introduction
SARS-CoV-2 was probably circulating in Hubei province, China, between mid-October and mid-November 2019 (Pekar et al., 2021), and it was detected for the first time in Wuhan, China, in December 2019 (WHO, 2020) subsequently spreading rapidly throughout the world. The details from early confirmed cases were described by Worobey (2021). In the United States of America (US), according to data aggregated by USAFacts (USAFacts, 2021) from the Centers for Disease Control and Prevention (CDC), state- and local-level public health agencies, the first documented cases emerged in Washington state on January 22, 2020, followed by Illinois (on January 24), and California and Arizona (both on January 26). These were isolated cases, since the second/third report of cases in these states only took place 38/39, 7 /37, 1/3, and 40/41 days later (Kujawski et al., 2020). However, the dissemination of the virus may have been even faster than previously appreciated.
Identifying the very first case of a pandemic is an arduous task, which has been further emphasized in the context of COVID-19 due to the high proportion of asymptomatic and mildly symptomatic individuals, and underreported cases (Bajema et al., 2020; Basavaraju et al., 2021; Havers et al., 2020; Li et al., 2020; Pollán et al., 2020; Barber et al. 2021; Irons and Raftery, 2021; Noh and Danuser, 2021). Several attempts have been made to this end. In China, 100 cases were retrospectively confirmed in December 2019 (WHO, 2021). In France, a retrospective analysis of respiratory samples of an individual hospitalized on December 27, 2019, was positive for SARS-CoV-2, which is around a month before the first case had been reported (Deslandes et al., 2020). In Italy, the retrospective analysis of wastewater samples found that the virus was already circulating on 18 December 2019 in Milan and Turin (LaRosa et al., 2021). Besides, a retrospective computational analysis suggested that the first infection in Italy was in late November 2019 (Fochesato et al., 2021). In the US, retrospective analysis of blood samples identified virus introduction earlier than reported in Illinois, Massachusetts, Wisconsin, Pennsylvania, and Mississippi (Althoff et al., 2021), and even between December 13-16, 2019, in California, Oregon, and Washington (Basavaraju et al., 2021). The objective of this study is to provide insights into the early stages of the COVID-19 outbreak in the US from a likelihood-based estimation procedure.
We report the results from an independent retrospective data analysis to reconstruct the daily infections time series at the beginning of the pandemic. These new time series reconcile reported deaths, clinical information of the illness, and the results of a seroprevalence study (Bajema et al., 2020), unlike official records, which present a general underestimate of cases, and then an overestimate of the Case Fatality Ratio (CFR). Besides, official data usually refers to the diagnosis date and not to the date of the infection, which is relevant for modeling purposes. Finally, the first infection of each reconstructed time series is identified for each state, providing information about where and when the virus was introduced in the US, which provides valuable information about the early spread dynamics of the virus.
2. Methods
Overall, COVID-19 deaths have been more thoroughly documented than infections, and we would like to transfer that thoroughness from death records to the infection records. The reason being, is that the number of infections at the beginning of the pandemic were generally of poor quality, either because no one was looking for them yet or because there were not enough diagnosis tests. Therefore, it can be useful to apply our algorithm Retrospective Methodology to Estimate Daily Infections from Deaths (REMEDID) (García-García et al., 2021) to reconstruct the time series of new infections, as it was done in Spain. To do so, some information about COVID-19 is needed.
Given that an individual died due to COVID-19, the question of when they got infected remains. The period from infection to death is the addition of the incubation period (IP) and the illness onset to death (IOD) period. Then, as far as IP and IOD are known, the date of infection can be inferred by subtracting the IP+IOD from the date of death. However, IP and IOD are not fixed values, but random variables that can be approximated by probability distributions. The convolution of their probability density functions (PDF) defines the PDF of the period from infection to death. Let f(t) be such PDF, where t represents time since infection. As data are usually given daily, let F(n) be a discrete approximation to f(t) representing the probability of death n days after infection. Then, given a COVID-19 death on a certain day n, the probability of having contracted the disease 1 day before is F(1); 2 days before is F(2), and so on. If more than one death was produced on day n, say x(n) deaths, the associated infections can be dated as follows: x(n)⋅F(1) infections were produced 1 day before; x(n)⋅F(2) were produced 2 days before, and so on.
If the CFR is known, the total infections can be inferred from deaths. Following the previous reasoning from the opposite point of view, the infections on day n that ended in death, y(n), can be inferred as the addition of deaths on day n+1 that were infected on day n, x(n+1)⋅F(1); deaths on day n+2 that were infected on day n, x(n+2)⋅F(2); and so on. Then,
| (1) |
For each infection that ends in death, it can therefore be assumed that there were 100/CFR infections. So, given a time series of deaths produced by the illness, x(n), the infections can be inferred as
| (2) |
To make sense of the inferred infections they have been rounded to the nearest integer (positive) number. Then, the first non-null element defines the date of the first infection.
All the computations in this study were implemented in Matlab R2019b, while graphics were made in R software with the packages usmap_0.5.2, viridis_0.6.2, and ggplot2_3.3.4. The nature of the data is public and anonymous, hence, no ethical approval was required for this study.
3. Data
We used the IP and IOD distributions estimated by Linton et al. 2020 from initial cases in Wuhan, China. The IP was approximated by a lognormal distribution with mean=5.6 days and median=5 days, while the IOD was also by a lognormal distribution with mean=14.5 days and median=13.2 days.
An accurate estimate of CFR is needed as a further input parameter for the REMEDID algorithm. Nevertheless, even if deaths are accurately estimated, the CFR cannot be estimated if the number of infections are unknown or inaccurate. So, here we have a circular reasoning because CFR is needed to infer infection time series, but infections are needed to estimate the CFR. However, the circular reasoning can be broken thanks to seroprevalence studies, which determine the accumulated infections up to a certain date. We used the seroprevalence study developed by Bajema et al. (2020), which was carried out at the following four different time periods in 2020: from July 27 to August 13; from August 10 to 17; from August 24 to September 10; and from September 8 to 24. The accumulated infections detected for each period are associated to a specific date for each state. Although the number of accumulated infections in a given period should be larger than those from any given previous period, this is not always the case when dealing with a relatively low number of cases per time interval. Therefore, for each state, we consider the averaged infections for the four periods in relation to the average date from such time periods. The accumulated deaths from USAFacts up to those dates, plus the proportional deaths detected subsequently according to the convolution of IP and IOP distributions, are used to estimate a mean CFR for each state.
4. Results
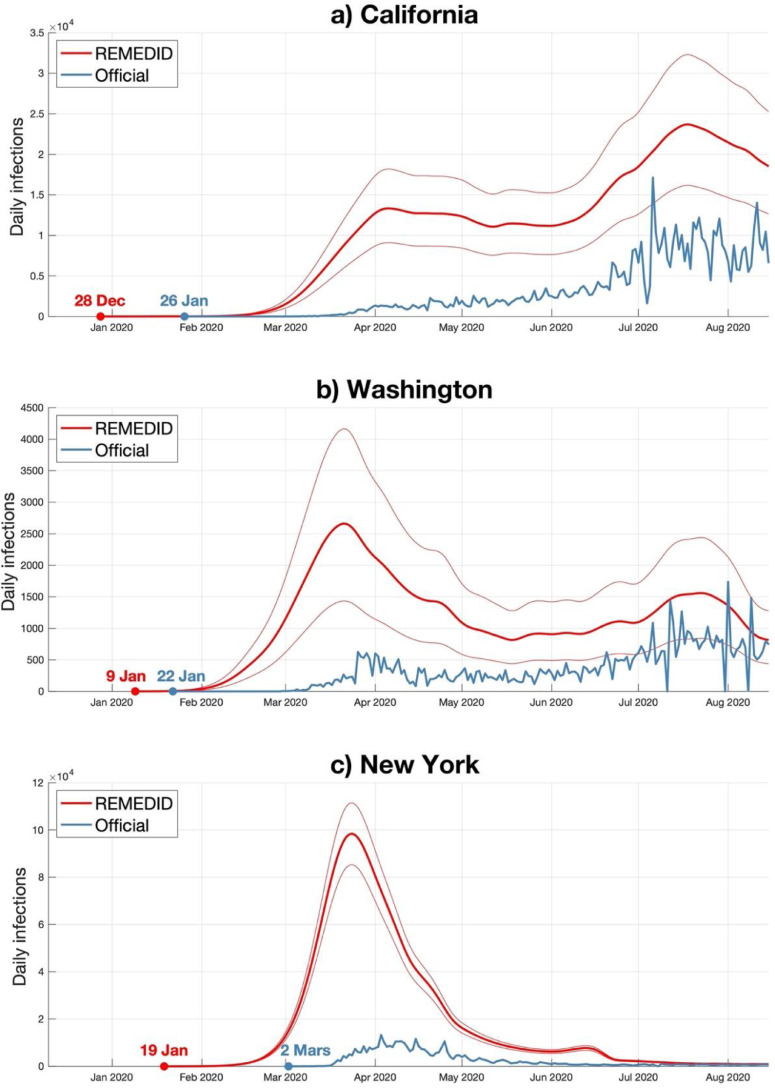
The daily infections time series have been estimated by applying the REMEDID algorithm in each state, and they will be referred to as IR. Similarly, daily infections from official records will be referred to as IO. As an example, Fig. 1 shows the IR and IO for the states of California, Washington, and New York (the rest of the states are shown in the Supplementary Material). The error band for the IR is derived from the 95% confidence interval (CI) estimated in the seroprevalence study. We interpret that the first day presenting at least one infection in IR is the day when COVID-19 entered each state for the first time. The dates of such first day is shown in Fig. 2 a and column 2 of Table 1 , for each state. Similarly, Fig. 2b and column 3 of Table 1 show the dates of the first officially documented cases, that is for the first case in IO. Note that there is no data for the US territories. The first IR case in the US was located in California on December 28, 2019, that is 16 days before the first officially documented case, and 3 days before the Wuhan Municipal Health first reported a cluster of pneumonia cases of unknown origin (ECDC, 2020).
Fig. 1.
Daily infections from official data (blue) and estimated from REMEDID (red) for three states: a) California; b) Washington; and c) New York. Dots represent the 1st infection. The error band for the IR is derived from the 95% confidence interval (CI) estimated in the seroprevalence study.
Fig. 2.
First COVID-19 cases recorded for each state from: a) REMEDID infections, and b) officially documented infections. The common color bar scale ranges from December 2019 to March 2020.
Table 1.
Dates corresponding to the first COVID-19 cases for each state within the US based on both our REMEDID modeling and officially reported data, and the differences in days between them. Dates format is dd/mm/yy.
| State | Date of 1st REMEDID case | Date of 1st documented case | Difference in days |
|---|---|---|---|
| Alabama, AL | 3/2/20 | 13/3/20 | 39 |
| Alaska, AK | 10/2/20 | 12/3/20 | 31 |
| Arizona, AZ | 2/2/20 | 26/1/20 | -7 |
| Arkansas, AR | 3/2/20 | 11/3/20 | 37 |
| California, CA | 28/12/19 | 26/1/20 | 29 |
| Colorado, CO | 28/1/20 | 6/3/20 | 38 |
| Connecticut, CT | 2/2/20 | 9/3/20 | 36 |
| Delaware, DE | 7/2/20 | 12/3/20 | 34 |
| Florida, FL | 22/1/20 | 2/3/20 | 40 |
| Georgia, GA | 22/1/20 | 3/3/20 | 41 |
| Hawaii, HI | 12/2/20 | 7/3/20 | 24 |
| Idaho, ID | 3/2/20 | 14/3/20 | 40 |
| Illinois, IL | 28/1/20 | 24/1/20 | -4 |
| Indiana, IN | 28/1/20 | 6/3/20 | 38 |
| Iowa, IA | 3/2/20 | 9/3/20 | 35 |
| Kansas, KS | 30/1/20 | 8/3/20 | 38 |
| Kentucky, KY | 30/1/20 | 9/3/20 | 39 |
| Louisiana, LA | 23/1/20 | 9/3/20 | 46 |
| Maine, ME | 11/2/20 | 12/3/20 | 30 |
| Maryland, MD | 31/1/20 | 6/3/20 | 35 |
| Massachusetts, MA | 1/2/20 | 1/2/20 | 0 |
| Michigan, MI | 27/1/20 | 10/3/20 | 43 |
| Minnesota, MN | 1/2/20 | 6/3/20 | 34 |
| Mississippi, MS | 2/2/20 | 12/3/20 | 39 |
| Missouri, MO | 31/1/20 | 8/3/20 | 37 |
| Montana, MT | 12/2/20 | 13/3/20 | 30 |
| Nebraska, NE | 5/2/20 | 6/3/20 | 30 |
| Nevada, NV | 29/1/20 | 5/3/20 | 36 |
| New Hampshire, NH | 15/2/20 | 2/3/20 | 16 |
| New Jersey, NJ | 24/1/20 | 5/3/20 | 41 |
| New Mexico, NM | 11/2/20 | 11/3/20 | 29 |
| New York, NY | 19/1/20 | 2/3/20 | 43 |
| North Carolina, NC | 3/2/20 | 3/3/20 | 29 |
| North Dakota, ND | 13/2/20 | 12/3/20 | 28 |
| Ohio, OH | 30/1/20 | 9/3/20 | 39 |
| Oklahoma, OK | 29/1/20 | 7/3/20 | 38 |
| Oregon, OR | 26/1/20 | 29/2/20 | 34 |
| Pennsylvania, PA | 27/1/20 | 6/3/20 | 39 |
| Rhode Island, RI | 12/2/20 | 1/3/20 | 18 |
| South Carolina, SC | 30/1/20 | 6/3/20 | 36 |
| South Dakota, SD | 8/2/20 | 9/3/20 | 30 |
| Tennessee, TN | 30/1/20 | 5/3/20 | 35 |
| Texas, TX | 26/1/20 | 5/3/20 | 39 |
| Utah, UT | 2/2/20 | 7/3/20 | 34 |
| Vermont, VT | 4/2/20 | 8/3/20 | 33 |
| Virginia, VA | 29/1/20 | 8/3/20 | 39 |
| Washington, WA | 9/1/20 | 22/1/20 | 13 |
| West Virginia, WV | 15/2/20 | 17/3/20 | 31 |
| Wisconsin, WI | 30/1/20 | 9/3/20 | 39 |
| Wyoming, WY | 28/2/20 | 12/3/20 | 13 |
The IR data reports that initial COVID-19 cases in the US occurred earlier than previously recorded in IO for 96% of the states. On average, the first IR cases occurred 32 days prior to the IO case count. The fourth column in Table 1 corresponds to the difference in the number of days between the estimated first infection date of IR and IO, where a positive (negative) value means that the first IR case was earlier (later) than the first IO case.
5. Discussion
From daily deaths during the first wave and seroprevalence studies along with theoretical knowledge about the COVID-19, an alternative daily infections data for each US state has been estimated. The new data allows us to revise, independently from official infection records, the dynamic of the beginning of the pandemic in the US. In general, the daily infections were underestimated during the first wave and the official infections were delayed between one or two weeks. It is especially evident in New York state, where IR reached a maximum of 98,454 (95% CI = 85,364–111,520) on March 24, 2020, while IO reported a maximum of only 13,262 infections on April 3 (Fig. 1c), that is less than the 15% of the total infections and with a week and a half delay. Another major result is that the first official cases in IO are quite delayed with respect to those in IR. We will focus on the dates of the first infections.
Although the dates of the first infections have been estimated in studies based on sample repositories (Deslandes et al., 2020; Althoff et al., 2021; Basavaraju et al., 2021; LaRosa et al., 2021; Valenti et al., 2021; WHO, 2021), they have not been reported in other studies based on retrospective models (Barber et al. 2021; Irons and Raftery, 2021; Noh and Danuser, 2021). Our approach allows to model daily infections and estimate the date of first infection. However, the IR does not necessarily show the earliest ever estimated case. For example, the earliest REMEDID case is observed in California (December 28, 2019) around 2 weeks later than those retrospectively reported by Basavaraju et al. (2021) from serologic testing of blood donation specimens from an existing repository. The blood collected in California was donated on December 13-16, 2019. An explanation of this discrepancy with REMEDID data may be that early infections were produced in an above-average proportion of individuals with a low risk of death.
The flight connections with China, and specially with Wuhan, may explain the spatial distribution of the earliest cases in the US. The first and second states presenting REMEDID infections were California and Washington, respectively, both on the West Coast of the US. The third state was New York. These results are consistent with the fact that California and New York received the largest number of flight connections from China. In December 2019, the only two direct flights from Wuhan airport to the US were to San Francisco in California (8071 passengers), and New York (5849 passengers), while other Chinese airports sent 299,278 passengers to California, 97,897 to New York, 38,149 to Washington state, and 266,273 to other 7 states (data.transportation.gov). Therefore, it makes sense that California had the first case because this was the state that received the highest number of travelers directly from Wuhan. The first and second documented cases in the US were a man and a woman traveling from Wuhan to Washington and Illinois states with arrival dates on January 15 and 13, 2020, respectively (CDC, 2020; Holshue et al., 2020).
The high percentage of mild and asymptomatic cases hindered the detection of cases at the beginning of the pandemic. The first documented case of Illinois did not lead to a local outbreak since it was rapidly isolated. Apparently only the patient's husband was infected, accounting for the first documented secondary transmission of COVID-19 in the US. However, the Illinois case was not the only one, since Althoff et al. (2021) retrospectively reported a case on January 7, 2020, from blood specimens belonging to the All of Us Research Program. It makes sense to think that there were more cases since the two earliest documented cases were detected because the hosts presented symptoms and went to the hospital, which happens in a low portion of infections (Rippinger et al., 2021). For example, the Spanish seroprevalence study reported a third of completely asymptomatic infections during the first wave (Pollán et al., 2020); and in Italy, another seroprevalence study in a random sample of blood donors exposed many more infections in Milan, Italy, than was initially detected at the beginning of the pandemic in February 2020 (Valenti et al., 2021).
REMEDID algorithm estimates infections of mild and asymptomatic (thus undetected) cases, which produces remarkable differences compared to official records. The application of the REMEDID algorithm assumes that the proportion of mild and asymptomatic cases was similar at the beginning of the epidemic and during the period covered by the seroprevalence study. This scenario is plausible since there was not any new virus variant becoming dominant till the alpha variant (B.1.1.7 lineage), which was first detected in England in September 2020 (PHE, 2020). Differences between IO and IR regarding the early spread are significant. For example, Illinois dropped from the 2nd to 13th position using our REMEDID infection score. The first IR cases are dated around a month earlier than in the IO ones, revealing that: (i) it was more likely that SARS-CoV-2 spread to US states a month earlier on average than previously reported in official records; (ii) there was a generalized underdetection of cases during the beginning of the pandemic. Only Arizona and Illinois showed earlier first cases in documented infections than in our REMEDID analysis. Finally, West Virginia was the last state to report a COVID-19 infection (on March 17, 2020), contrary to our REMEDID analysis that identified Wyoming as the last state on its ranking (on February 28, 2020).
The REMEDID algorithm provides information about the early stage of the pandemic when official records are expected to be of lower quality, although it has pros and cons. For example, it presents some advantages with respect to other retrospective analyses that rely on sample repositories (Deslandes et al., 2020; Althoff et al., 2021; Basavaraju et al., 2021; LaRosa et al., 2021; Valenti et al., 2021; WHO, 2021) that may or may not exist. If they do not exist when the health crisis breaks out, these retrospective studies will no longer be feasible. On the contrary, the REMEDID algorithm is based on seroprevalence studies that can be planned and carried out after the illness outbreak took place. In fact, it is highly recommended to apply the algorithm to all regions with available seroprevalence studies to estimate daily infections, and infer their first infections. However, this dependence has a counterpart that limits the algorithm application, since it can only be applied to regions where seroprevalence studies are available. A second advantage is that the REMEDID reconstruction of daily infections allows to infer the first infection date, which is not the case in other approaches that are also reconstructing infections from deaths (Irons and Raftery, 2021). Another limitation comes from the IP and IOD distributions, which were estimated in China and may differ for the US. The IP is known to show geographical differences (Cheng et al., 2021), and some differences are expected for the IOD as far as it partially depends on the health system of each country. However, the study can easily be redone as soon as an IP and IOD distribution will be available for the US. Finally, results depend on the quality of the daily deaths time series. Iuliano et al. (2021) reported that 24% of deaths attributable to COVID-19 in the US were undocumented from March 8, 2020, to May 29, 2021. However, if deaths were underreported homogeneously throughout the studied period, the CFR would also be underreported but not the result of the REMEDID algorithm. This is because deaths and CFR are inversely related in Eq. 2.
The calculation of infections using REMEDID algorithm present several advantages with respect to official records since they are compatible with: (i) the stochastic information available about the COVID-19, such as IP and IOD distributions; (ii) the seroprevalence studies, then providing a realistic total amount of infections; and (iii) daily death time series. Besides, the infections estimated from REMEDID can be relevant to understanding the viral spread and provide substantial evidence that COVID-19 transmission occurs more rapidly than previously observed through official recorded data. This is underscored by the observation that SARS-CoV-2 arrived in the US before it was even reported by the Wuhan authorities in China. The situation was similar in Spain where, during the first COVID-19 wave, the 1st official case was detected on February 20, 2020, in contrast to our REMEDID model, which identified the 1st case 43 days earlier on January 8 (García-García et al., 2021). Our results using a mathematical modeling approach reveal a generalized and significant delay in the detection of the first viral cases in the US, which may extend to numerous other countries around the globe.
6. Conclusion
Although the delayed detection of early cases of COVID-19 has been generalized in all countries, our methodology allows their quantification from daily deaths and seroprevalence studies. In the US, the virus was introduced around a month earlier than officially reported by most of the states. The results presented in this study are important to improve our understanding of the early spread of the virus, which is crucial to prevent or mitigate future epidemics.
Declarations
Funding
This work was supported by the University of Alicante [COVID-19 2020-41.30.6P.0016 to CB] and the Montó-Dénia Research Station (Agreement Ajuntament de Dénia-O.A. Parques Nacionales-Conselleria de Agricultura, Desarrollo Rural, Emergencia Climática y Transición Ecológica, Generalitat Valenciana) [2020-41.30.6O.00.01 to CB].
Cesar de la Fuente-Nunez holds a Presidential Professorship at the University of Pennsylvania, is a recipient of the Langer Prize by the AIChE Foundation and acknowledges funding from the Institute for Diabetes, Obesity, and Metabolism, the Penn Mental Health AIDS Research Center of the University of Pennsylvania, the Nemirovsky Prize, the Dean's Innovation Fund from the Perelman School of Medicine at the University of Pennsylvania, the National Institute of General Medical Sciences of the National Institutes of Health under award number R35GM138201, and the Defense Threat Reduction Agency (DTRA; HDTRA11810041 and HDTRA1-21-1-0014).
Availability of data and material
All data are available in this article and the references cited.
Code availability
MATLAB REMEDID code described in García-García et al. 2021, and available in GitHub https://github.com/isavig/REMEDID.
Authors' contributions
D.G., C.F.N. and C.B. designed the hypothesis. D.G., E.M. and I.V. performed the data analysis. All authors wrote and reviewed the manuscript. D.G. and C.B. coordinated the work.
Declaration of Competing Interest
Not applicable
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.sste.2022.100517.
Appendix. Supplementary materials
References
- Althoff KN, Schlueter DJ, Anton-Culver H, Cherry J, Denny JC, Thomsen I, et al. Antibodies to Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) in All of Us Research Program Participants. Clin. Infect. Dis. 2021 doi: 10.1093/cid/ciab519. 2 January to 18 March 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajema KL, Wiegand RE, Cuffe K, Patel S V, Iachan R, Lim T, et al. Estimated SARS-CoV-2 Seroprevalence in the US as of September 2020 Invited Commentary Supplemental content. JAMA Intern. Med. 2020 doi: 10.1001/jamainternmed.2020.7976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber RM, Sorensen RJD, Pigott DM, Bisignano C, Carter A, Amlag JO, et al. Estimating global, regional, and national daily and cumulative infections with SARS-CoV-2 through Nov 14, 2021: a statistical analysis. Lancet. 2022 doi: 10.1016/S0140-6736(22)00484-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basavaraju S.V., Patton ME, Grimm K, Rasheed MAU, Lester S, Mills L, et al. Serologic testing of us blood donations to identify severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-reactive antibodies: December 2019-January 2020. Clin. Infect. Dis. 2021;72:E1004–E1009. doi: 10.1093/cid/ciaa1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC. Second travel-related Case of 2019 Novel Coronavirus Detected in United States. Press release. 2020. https://www.cdc.gov/media/releases/2020/p0124-second-travel-coronavirus.html Accessed 10 June 2021.
- Cheng C, Zhang DD, Dang D, Geng J, Zhu P, Yuan M, et al. The incubation period of COVID-19: a global meta-analysis of 53 studies and a Chinese observation study of 11 545 patients. Infect. Dis. Poverty. 2021;10:119. doi: 10.1186/s40249-021-00901-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes A, Berti V, Tandjaoui-Lambotte Y, Alloui C, Carbonnelle E, Zahar JR, et al. SARS-CoV-2 was already spreading in France in late December 2019. Int. J. Antimicrob. Agents. 2020;55 doi: 10.1016/j.ijantimicag.2020.106006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ECDC. Cluster of pneumonia cases caused by a novel coronavirus, Wuhan, China. 2020. https://www.ecdc.europa.eu/sites/default/files/documents/Risk assessment - pneumonia Wuhan China 17 Jan 2020.pdf. Accessed 10th June 2021.
- Fochesato A, Simoni G, Reali F, Giordano G, Domenici E, Marchetti L. A retrospective analysis of the COVID-19 pandemic evolution in Italy. Biology (Basel) 2021;10:311. doi: 10.3390/biology10040311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-García D, Vigo MI, Fonfría ES, Herrador Z, Navarro M, Bordehore C. Retrospective methodology to estimate daily infections from deaths (REMEDID) in COVID-19: the Spain case study. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-90051-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havers FP, Reed C, Lim T, et al. Seroprevalence of antibodies to SARS-CoV-2 in 10 sites in the United States, March 23–May 12, 2020. JAMA Intern. Med. 2020;180:1576–1586. doi: 10.1001/jamainternmed.2020.4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holshue ML, DeBolt C, Lindquist S, Lofy KH, Wiesman J, Bruce H, et al. First Case of 2019 Novel Coronavirus in the United States. N. Engl. J. Med. 2020;382:929–936. doi: 10.1056/nejmoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irons NJ, Raftery AE. Estimating SARS-CoV-2 infections from deaths, confirmed cases, tests, and random surveys. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2103272118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuliano AD, Chang HH, Patel NH, Threlkel R, Kniss K, Reich J, et al. Estimating under-recognized COVID-19 deaths, United States, march 2020-may 2021 using an excess mortality modelling approach. Lancet Regional Health – Americas. 2021;1 doi: 10.1016/j.lana.2021.100019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujawski SA, Wong KK, Collins JP, Epstein L, Killerby ME, Midgley CM, et al. Clinical and virologic characteristics of the first 12 patients with coronavirus disease 2019 (COVID-19) in the United States. Nat. Med. 2020;26:861–868. doi: 10.1038/s41591-020-0877-5. [DOI] [PubMed] [Google Scholar]
- La Rosa G, Mancini P, Bonanno Ferraro G, Veneri C, Iaconelli M, Bonadonna L, et al. SARS-CoV-2 has been circulating in northern Italy since December 2019: Evidence from environmental monitoring. Sci. Total Environ. 2021;750 doi: 10.1016/j.scitotenv.2020.141711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Pei S, Chen B, Song Y, Zhang T, Yang W, et al. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV2) Science. 2020;368:489–493. doi: 10.1126/science.abb3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linton NM, Kobayashi T, Yang Y, Hayashi K, Akhmetzhanov AR, Jung SM, et al. Incubation period and other epidemiological characteristics of 2019 novel coronavirus infections with right truncation: a statistical analysis of publicly available case data. J. Clin. Med. 2020;9 doi: 10.3390/jcm9020538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh J, Danuser G. Estimation of the fraction of COVID-19 infected people in U.S. states and countries worldwide. PLoS One. 2021;16 doi: 10.1371/journal.pone.0246772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekar J, Worobey M, Moshiri N, Scheffler K, Wertheim JO. Timing the SARS-CoV-2 index case in Hubei province. Science. 2021;372(80-):412–417. doi: 10.1126/science.abf8003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PHE. Investigation of novel SARS-COV-2 variant. Variant of Concercn 202012/01. Technical briefing 3. 2020. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/959360/Variant_of_Concern_VOC_202012_01_Technical_Briefing_3.pdf. Accessed 27th May 2021.
- Pollán M, Pérez-Gómez B, Pastor-Barriuso R, Oteo J, Hernán MA, Pérez-Olmeda M, et al. Prevalence of SARS-CoV-2 in Spain (ENE-COVID): a nationwide, population-based seroepidemiological study. Lancet. 2020;396:535–544. doi: 10.1016/S0140-6736(20)31483-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippinger C, Bicher M, Urach C, Brunmeir D, Weibrecht N, Zauner G, et al. Evaluation of undetected cases during the COVID-19 epidemic in Austria. BMC Infect. Dis. 2021;21 doi: 10.1186/s12879-020-05737-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- USAFacts. Data accessed on March 15, 2021 from https://usafacts.org/visualizations/coronavirus-covid-19-spread-map/.
- Valenti L, Bergna A, Pelusi S, Facciotti F, Lai A, Tarkowski M, et al. SARS-CoV-2 seroprevalence trends in healthy blood donors during the COVID-19 outbreak in Milan. Blood Transfus. 2021;19:181–189. doi: 10.2450/2021.0324-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. WHO-convened global study of origins of SARS-COV-2: China part. 2021. https://www.who.int/publications/i/item/who-convened-global-study-of-origins-of-sars-cov-2-china-part. Accessed 14th April 2021.
- WHO. Coronavirus disease 2019 (COVID-19). Situation Report-94. 2020. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200423-sitrep-94-covid-19.pdf?sfvrsn=b8304bf0_4. Accessed 26th February 2021.
- Worobey M. Dissecting the early COVID-19 cases in Wuhan. Science. 2021;374:eabm4454. doi: 10.1126/science.abm4454. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in this article and the references cited.