Abstract
Objective
Respiratory illnesses have information-rich acoustic biomarkers, such as cough, that can potentially play an important role in screening populations for disease risk. To realize that potential, datasets of paired acoustic-clinical samples are needed for the development and validation of acoustic screening models, and protocols for collecting acoustic samples must be efficient and safe. We collected cough acoustic signatures at a high-throughput SARS-CoV-2 testing site on a college campus. Here, we share logistical details and the dataset of acoustic cough signatures paired with the gold standard in SARS-CoV-2 testing of SARS-CoV-2 genomic sequences using qRT-PCR.
Methods
Cough recordings were collected in winter-spring 2021 at a rural residential college (Sewanee, TN, USA), where approximately 2000 students were tested for SARS-CoV-2 on a weekly basis. Cough collection was managed by student volunteers using custom software.
Results
4302 coughs were recorded from 960 participants over 11 weeks. All coughs were COVID-19 negative. Approximately 30 s were required to check-in a participant and collect their cough.
Conclusion
The value of acoustic screening tools depends upon our ability to develop and implement them reliably and quickly. For that to happen, high-quality datasets and logistical insights must be collected and shared on an ongoing basis.
Keywords: COVID-19, SARS-CoV-2, acoustic epidemiology, acoustic screening, machine learning < general, logistics, open source
Introduction
The field of acoustic epidemiology—the objective recording and analysis of sounds’ acoustic features for human disease screening, diagnosis, and surveillance—is rapidly expanding.1–3 Sound has been used for respiratory disease diagnostics for centuries, 4 but recent advances in acoustic modeling and machine learning have compounded what we can learn by listening to patients cough, sneeze, wheeze, breathe, speak, and snore.5–9 Those behaviors all have information-rich acoustic signatures that can be used to screen for the probability of infection or pathology (e.g. TB, 10 COVID-19,11–15 and speech pathology3,16–18). Those sounds can also be aggregated at population level to monitor or predict disease activity. 19 With accurate models, mass acoustic screening could be an important means of early detection, resource triage, cost savings, and epidemic control, particularly in urban centers, where diseases spread rapidly, and in areas where recording devices such as smartphones are more ubiquitous than conventional diagnostic platforms.20–24
Acoustic epidemiology remains a nascent field in which statistical software and their deployment strategies need to be stress-tested, both logistically and analytically. In order to build up a training dataset to develop and evaluate an accurate acoustic classification model, sound samples must be paired with conventional clinical tests, thus compounding the logistical complexity of data collection. Access to such data is a perennial limitation in this work,10,14,25 and its urgency has been highlighted in recent scrutiny of acoustic screening models for SARS-CoV-2 infection.15,25 Sound collection must also be non-intrusive and safe for patients and staff, especially in the context of transmissible diseases.
To facilitate the rapid development of these acoustic screening methods, we present an open-access dataset of 4302 cough recordings, all of which have been confirmed negative for SARS-CoV-2 infection using the highly sensitive RT-PCR assay. All cough recordings were collected from participants providing informed consent for their cough and associated metadata to be released in open-access repositories and used by the research community. The data collection protocols we present here emulate a biosafe mass screening approach which could be deployed in current and future pandemics.
Methods
Screening occurred during the spring academic semester (1 February–12 May, 2021) at the University of the South, a residential liberal arts university in the mid-Cumberland region of Tennessee, USA, where rates of COVID-19 cases were highest in the state's rural sectors. 26 Weekly COVID-19 testing was required of all undergraduate and graduate students (approx. 2000), faculty, and staff were also required to be tested on a monthly basis, yielding 2900 potential participants.
COVID-19 testing occurred on the central campus at the university gymnasium, 10 am to 2 pm on Monday to Thursday, with the capacity to accommodate more than 180 self-collected tests per hour. Participants used a floxed swab to collect from the anterior nares region of the nose and these samples were then assessed for three genes (ORF1ab, N, and S) from the SARS-CoV-2 virus to detect possible COVID-19 disease. Tests were analyzed at a CLIA high complexity clinical laboratory located on campus using the Applied Biosystems TaqPath COVID-19 RT-PCR kit (Cat.# A47817). This COVID-19 testing program was accompanied by standard public health recommended infection control measures including quarantining of PCR-positive cases.
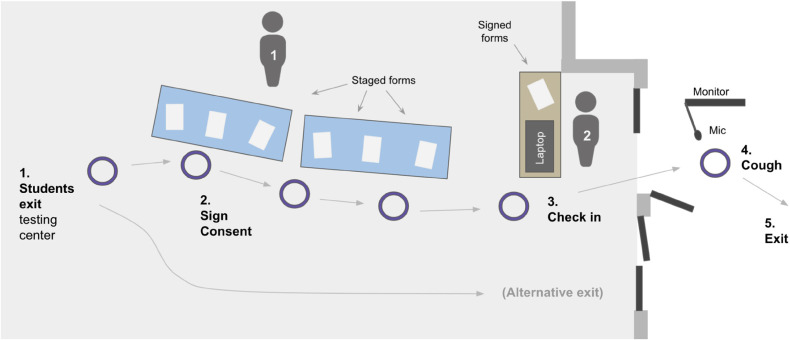
On 21 February, three weeks into the semester, an acoustic screening station was established at the exit of the COVID-19 testing center. This station consisted of a table for participant enrollment, a kiosk for participant sign-in, and a portable recording apparatus that was moved outside during testing hours and placed under an open-air 10-ft × 10-ft tent immediately outside of the exit door (Figure 1). The recording location was outside, well-ventilated by a constant breeze, and away from regular motor traffic.
Figure 1.
Cough collection system at university Sewanee COVID-19 testing site. Staff members (individuals 1 and 2) guide participants from (step 1) the nose-swab testing area to (2) the cough collection station's consent table, then to (3) the sign-in kiosk and (4) cough collection monitor, then to (5) the exit.
The recording apparatus consisted of a computer monitor and microphone (Movo PC-M6, www.movophoto.com) mounted upon a tripod speaker stand (Pyle universal mount, www.pyleaudio.com). The microphone was held in an articulating swivel arm (Movo ASM-5, www.movophoto.com) braced to the speaker stand. The monitor and microphone communicated with the kiosk laptop (Hewlett-Packard laptop running Windows 10) via 15-ft USB and HDMI cables bound together with plastic ties. This apparatus was designed to be high-quality but affordable (<$100 USD, excluding the laptop and monitor).
To raise awareness for this study, the university Public Health Office emailed the student body the week prior to the start of the acoustic screening, with subsequent reminders in the second and third weeks of the study. At the testing center, large-print signs were placed near the exit to direct students to the station. The volunteer at the enrollment table invited students to consider participation before they exited.
At the “cough station,” each participant approached the kiosk and provided their identification information and provided informed consent (Figure 1). A participant-specific URL for a web-based screening app, developed for this study by Hyfe Inc. (www.hyfe.ai), was used to record their cough (a generic version of that app is available at https://hyfe-general.web.app/). Participants were directed out the door to the acoustic recording apparatus to initiate recording, where they were instructed to stand close to the microphone without touching it. The computer monitor, which was mirroring the volunteer's laptop screen, displayed instructions and a countdown specifying when to produce a single cough (mask off), then displayed confirmation that the cough was recorded successfully. If a cough was not recognized, the participant was asked to try again with a maximum of three attempts possible. During this cough collection episode, the screening website recorded a 5-s WAV file (sampling rate 44.1 kHz) to Hyfe's encrypted, HIPAA-compliant servers. Equipment was disinfected after each day of use.
At the sign-in kiosk, a laptop with a custom dashboard application was used to keep pace with the rate of participant traffic (an open-source example of this dashboard, developed in the R environment27–29 is available at https://github.com/hyfe-ai/hyfer). This dashboard referenced a “master” spreadsheet linking the student's identifying information to an anonymized ID code and unique URL. These ID codes protected the medical information of the participants such that only the Principal Investigators could pair the cough recordings with the COVID-19 lab results.
The screening station was attended by two staff members. One was stationed at the participant enrollment table while the other coordinated cough collection at the sign-in kiosk (Figure 1). A coordinator (author EJT) was hired to recruit, train, and manage the staff. This study received approval by Sewanee's Institutional Review Board (IRB, proposal number 16), which was expedited due to the minimal risks to participants.
Results
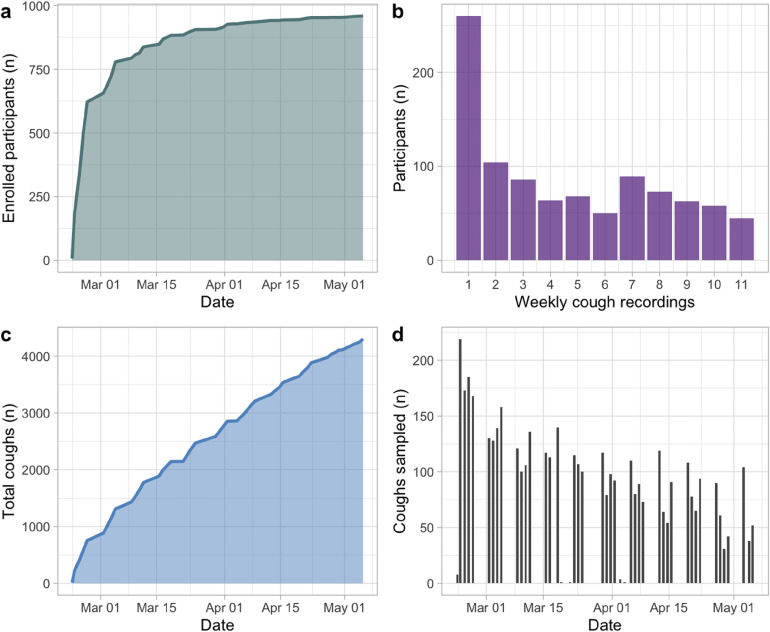
Over the course of 11 weeks (22 February–5 May, 2021), 4302 coughs were recorded from 960 participants (Figure 2a,c). Every cough collected in this study was associated with a negative PCR test for COVID-19. The university detected only 23 positive cases in the entire semester, and none of those cases occurred in participants within two weeks of a visit to the cough collection station.
Figure 2.
Cough collection at COVID-19 testing center in Sewanee, TN, USA, Feb to May 2021. (a) participant enrollment over the course of the study; (b) participant retention, showing the number of participants who visited the cough station in 1, 2, …, 9 weeks of the 11-week project; (c) cumulative coughs sampled; (d) daily breakdown of cough collection.
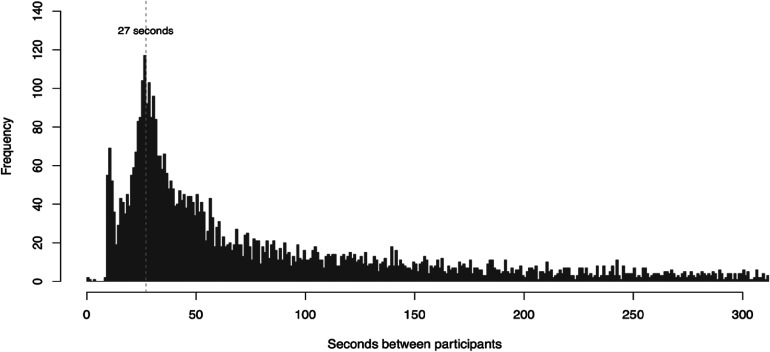
An average of 124 coughs (SD = 68) were sampled per day (Figure 2d), equivalent to 31 coughs per hour. When the testing center was busiest (during lunch hours), the cough collection station was able to check in a student and collect their cough within 30 s (Figure 3).
Figure 3.
Distribution of time intervals between each cough collection at the COVID-19 testing center. During high-traffic periods, in which a line of participants formed at the cough collection station, participants could check in and provide their cough in 27 s.
Sixty-five percent of participants enrolled in the first week, but recruitment continued throughout the study (Figure 2a). Participation rates were highest early in the study, then dwindled into May (Figure 2b). Seventy-five percent of participants participated in the study for at least two weeks, and 47% of participants were retained for at least five weeks. Forty-five participants (5% of cohort) were active for 9 out of the study's 11 weeks. All collected cough sounds were uploaded to a repository at FAIRsharing (https://beta.fairsharing.org/3619) and are now publicly available. 30
Discussion
The COVID-19 pandemic has added urgency to the development of systems for acoustic health screening.11–13,25 Development of acoustic analysis models—as with any machine learning model—depends upon the volume and quality of data used to train them, as well as the availability of external datasets for evaluating their performance. To make such tools easier to develop and evaluate, high-quality datasets must be made publicly available on an ongoing basis.
Other pandemic diseases, such as influenza and tuberculosis, would also benefit from high-performance cough classification software and standard operating procedures for acoustic screening in centers of high population density (e.g. college campuses, prisons, and urban centers) and large event spaces (e.g. airports, stadiums, and concert halls).18,21,23 In contexts like these, robust and affordable acoustic screening could be an important mitigation measure.11,31
For future similar acoustic screening initiatives, either for dataset building or clinical use, we recommend the following improvements to our protocol: (1) incentivize participant retention through loyalty programs; (2) establish identical screening stations at quarantine sites in order to increase the collection of disease-positive coughs; (3) simultaneously collect medical history and health-related habits to enable the development of multivariate models; and (4) consider expanding to settings that are either demographically diverse or homogenous in other ways to further enrich public acoustic datasets. As a result of our remote university setting, most of our participants had a similar profile—young and healthy, in general—which can be a two-edged sword in model training and validation. We encourage acoustic epidemiologists and machine learning experts to use our open-access cough dataset for training and validation purposes in conjunction with other diversified acoustic data sources.
Acknowledgments
The authors thank our team of student volunteers, Sewanee's Public Health Office, Alex Bruce, David Shipps, and Marty Hawkins for supporting this effort.
Footnotes
Conflict of interest: The author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: At the time of data collection, EMK and JB were employees of Hyfe, the company used to collect and store cough sounds. ET, AS, ECS, and SGL have no conflict of interest directly related to this work.
Contributorship: JB, ARS, ECS, and SGL conceived the study. ARS and ECS processed and analyzed COVID-19 test samples. ET managed the cough testing station and its volunteer team. EMK wrote the software, compiled results, and wrote the first draft of the manuscript. SGL, ARS, and ECS provided funding. All authors reviewed and edited the manuscript and approved its final version.
Ethical approval: This study obtained ethical approval from the Sewanee Institutional Review Board.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the Hyfe, Inc., (grant number N/A).
Guarantor: EMK
ORCID iDs: Eric M. Keen https://orcid.org/0000-0002-3053-3612
Simon Grandjean Lapierre https://orcid.org/0000-0003-3646-1573
References
- 1.Hall JI, Lozano M, Estrada-Petrocelli Let al. et al. The present and future of cough counting tools. J Thorac Dis 2020; 12: 5207–5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cho PSP, Birring SS, Fletcher H, et al. Methods of cough assessment. J Allergy Clin Immunol Pract 2019; 7: 1715–1723. [DOI] [PubMed] [Google Scholar]
- 3.Fagherazzi G, Fischer A, Ismael Met al. et al. Voice for health: The use of vocal biomarkers from research to clinical practice. Digital Biomarkers 2021; 5: 78–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collier M. The diagnosis of cough. Lancet 1897; 1645–1646. [Google Scholar]
- 5.Barata F, Kipfer K, Weber Met al. et al. Towards device-agnostic mobile cough detection with convolutional neural networks. In: IEEE International Conference on Healthcare Informatics (ICHI) 2019. [Google Scholar]
- 6.Kvapilova L, Boza V, Dubec PJ, et al. Continuous sound collection using smartphones and machine learning to measure cough. Digit Biomark 2019; 3: 166–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gosh A, Liaquat S, Ahmed S. Healthcare-internet of things (h-iot) can assist and address emerging challenges in healthcare. Int J Sci Innov Res 2020; 01: 2725–3338. [Google Scholar]
- 8.Sriram RD, Subrahmanian E. Transforming health care through digital revolutions. J Indian Inst Sci 2020; 100: 753–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu X, Nemati E, Vatanparvar K, et al. and others. Listen2cough: Leveraging end-to-end deep learning cough detection model to enhance lung health assessment using passively sense audio. Proc ACM Interact Mob Wearable Ubiquitous Technol 2021; 5: 43. [Google Scholar]
- 10.Botha GHR, Theron G, Warren RM, et al. Detection of tuberculosis by automatic cough sound analysis. Physiol Meas 2018; 39.29543189: 1–9. [DOI] [PubMed] [Google Scholar]
- 11.Anthes E. Alexa, do I have COVID-19? Nature 2020; 586: 22–25. [DOI] [PubMed] [Google Scholar]
- 12.Laguarta J, Hueto F, Subirana B. COVID-19 artificial intelligence diagnosis using only cough recordings. IEEE Open J Eng Med Biol 2020; 1: 275–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Imran A, Posophova I, Quershi HN, et al. AI4COVID-19: AI enabled preliminary diagnosis for COVID-19 from cough samples via an app. Inform Med Unlocked 2020; 20: 100378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mouawad P, Dubnov T, Dubon S. Robust detection of COVID-19 in cough sounds. SN Comput Sci 2021; 2: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharma M, Shenoy N, Doshi J, et al. Impact of data-splits on generalization: Identifying COVID-19 from cough and context. ICRL Public Health Workshop 2021; 1: 1–9. [Google Scholar]
- 16.König A, Satt A, Sorin A, et al. Automatic speech analysis for the assessment of patients with predementia and Alzheimer’s disease. Alzheimers Dement 2015; 1: 112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhan A, Mohan S, Tarolli C, et al. Using smartphones and machine learning to quantify Parkinson disease severity: The mobile Parkinson Disease Score. JAMA Neurol 2018; 75: 786–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arora S, Baghai-Ravary L, Tsanas S. Developing a large scale population screening tool for the assessment of Parkinson’s disease using telephone-qualit voice. J Acoustic Soc Am 2019; 145: 2871–2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gabaldon-Figueira JC, Brew J, Doré DHet al. Digital acoustic surveillance for early detection of respiratory disease outbreaks in Spain: A protocol for an observational study. BMJ Open 2021; 11: e051278. PMID: 34215614; PMCID: PMC8257291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Donner J. Research approaches to mobile use in the developing world: A review of the literature. Inform Soc 2008; 24: 140–159. [Google Scholar]
- 21.Spinou A, Birring SS. An update on measurement and monitoring of cough: What are the important study endpoints? J Thorac Dis 2014; 6: S728–S734.25383207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burney P, Jarvis D, Perez-Padilla R. The global burden of chronic respiratory disease in adults. Int J Tuberc Lung Dis 2015; 19: 10–20. [DOI] [PubMed] [Google Scholar]
- 23.Boulet L-P, Coeytaux RP, McCrorry DC, et al. Tools for assessing outcomes in studies of chronic cough: CHEST Guideline and Expert Panel Report. Chest 2015; 147.25522203: 804–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Statista. Smartphone users worldwide 2020-2021. Statista 2021. [Google Scholar]
- 25.Topol EJ. Is my cough COVID-19? Lancet 2020; 396: 1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.TDOH (Tennessee Department of Health). COVID-19 Critical Indicators. 5 November 2021. Accessed 11 November 2021. 2021. https://www.tn.gov/health/cedep/ncov.html
- 27.R Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing, 2020, https://www.R-project.org/. [Google Scholar]
- 28.Winston C, Cheng J, Allaire JJet al. shiny: Web Application Framework for R. R package version 1.6.0. 2021. https://CRAN.R-project.org/package=shiny
- 29.Keen. hyfer: utilities for analyzing cough sounds recorded by Hyfe. R package version 1.0.0. 2021. https://github.com/hyfe-ai/hyfer
- 30.Keen EM, True E, Summers Aet al. et al. Sewanee Cough Study. 2021. https://www.synapse.org/#!Synapse:syn26473927/files/, doi: 10.7303/syn26473927 [DOI]
- 31.Yuen CM, Amanullah F, Dharmadhikari A, et al. Turning off the tap: Stopping tuberculosis transmission through active casefinding and prompt effective treatment. Lancet 2015; 386: 2334–2343. [DOI] [PMC free article] [PubMed] [Google Scholar]