Abstract
Objective
Metagenomic next-generation sequencing (mNGS) has the potential to overcome the shortcomings of traditional culture methods. This study aimed to assess the diagnostic value of mNGS in patients with lower respiratory tract infections (LRTIs).
Methods
This retrospective observational study sequentially enrolled 47 patients with LRTIs admitted to Shenzhen Hospital of Southern Medical University between February 2019 and November 2020. Pathogens in bronchoalveolar lavage fluid (BALF) samples were investigated to compare diagnoses by mNGS with culture methods.
Results
Compared with culture methods, mNGS had a diagnostic sensitivity of 80% and a specificity of 35.13% with an agreement rate of 44.68% between these two methods. mNGS significantly increased the pathogen detection rate.
Conclusions
mNGS may show some advantages in identifying a wide range of LRTI pathogens, improving the sensitivity for viruses and atypical pathogens. The clinical application of NGS technology is worth looking forward to.
Keywords: Metagenomic next-generation sequencing, bronchoalveolar lavage fluid, lower respiratory tract infection, diagnosis, culture, pathogen
Introduction
Lower respiratory tract infections (LRTIs) include community-acquired pneumonia (CAP), hospital-acquired pneumonia, and ventilator-associated pneumonia. They are the most common diseases worldwide, and their high mortality and morbidity result in them also being the most common cause of death in low-income countries. 1 Many types of pathogen cause LRTIs, including bacteria, viruses, fungi, and atypical pathogens and parasites atypical pathogens such as Mycoplasma pneumoniae. Of those who are hospitalized, around 62% of adult patients with CAP and 25% of children with CAP have an unclear etiology.2,3 The rational and appropriate use of antibiotics is crucial for patients with LRTIs, but failure to obtain a timely etiological diagnosis can lead to heavy antibiotic use and multiple infectious complications, which increase mortality, healthcare costs, and antibiotic resistance.
Culture is currently the first-line method for pathogen detection in LRTIs. However, routine cultures of respiratory samples are time consuming, technically intensive, and error prone. Moreover, pathogens such as viruses, atypical pathogens and parasites, and slow-growing bacteria are not suited to cultivation, which may result in a lower detection rate. For example, the pathogens of 46% of cases of CAP could not be identified by cultivation approaches or targeted PCR. 4
With the development of molecular biology, metagenomic next-generation sequencing (mNGS) technology has been applied in microbial detection. mNGS was originally used to evaluate sterile body fluids such as cerebrospinal fluid and blood,5,6 but more recently it has been used for pathogen detection and the identification of LRTIs.7,8 mNGS allows the simultaneous and high-throughput identification of infectious agents, and its characteristics of accuracy and a relatively rapid turnaround time offer advantages over culture methods.
In this study, the mNGS data of 47 bronchoalveolar lavage fluid (BALF) samples from 47 patients with LRTIs were summarized, and the diagnostic value of mNGS was compared with culture in pathogen detection.
Methods
Patients and sample collection
This retrospective study consecutively enrolled 47 patients with LRTIs who were admitted to Shenzhen Hospital (Southern Medical University, Shenzhen, China) between February 2019 and November 2020. The diagnosis of LRTIs was made based on a composite reference standard, including clinical presentation, microbiological tests, radiography, and laboratory findings. The reporting of this study conforms to STROBE guidelines. 9
Patients provided their written informed consent to undergo bronchoscopy. Ethical approval was not sought because this was a retrospective study and patients remained anonymous. BALF samples from all 47 patients were sent to the Shenzhen Hospital microbiological laboratory for bacterial and fungal culture, and the BGI-Huada Genomics Institute (Shenzhen, China) for mNGS. Laboratory criteria for calling pathogens in bronchoalveolar lavage culture were that a bacterial culture colony count >10,000 cfu/mL was considered pathogenic, otherwise it was regarded as contamination.
mNGS and analysis
DNA extraction
A total of 3 mL BALF was inactivated at 65°C for 30 minutes immediately after collection. Next, 500 µL BALF was placed in a 1.5-mL microcentrifuge tube with 1 g 0.5-mm glass beads. Tubes were agitated vigorously at 2800–3200 rpm for 30 minutes on a vortex mixer attached to a horizontal platform. DNA was extracted from 300-μL samples using the TIANamp micro DNA kit (TIANGEN Biotech, Beijing, China) according to the manufacturer’s instructions.
Construction of DNA libraries
DNA libraries were constructed by DNA fragmentation, end-repair, adaptor ligation, and PCR amplification using a PMseq™ high-throughput detection kit for infectious pathogens (BGI Genomics, Wuhan, China) with the following conditions: 98°C for 2 minutes, followed by 12 cycles of 98°C for 15 s, 56°C for 15 s, and 72°C for 30 s, with a final extension at 72°C for 5 minutes. The Agilent 2100 Bioanalyzer system (Agilent, Santa Clara, CA, USA) was used for quality control of the library fragment size to around 300 bp. The Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific Inc., Waltham, MA, USA) was used to control the DNA library concentration, and the library was pooled according to the detected concentration then circularized to form a single-chain circular structure. DNB nanospheres were then generated by rolling circle replication, loaded into the sequencing chip and sequenced. MGISEQ-2000 quality control was performed using the Agilent 2100 Bioanalyzer system, and qualified libraries were sequenced on the BGISEQ 50 platform.
Sequencing and bioinformatic analysis
Low-quality reads shorter than 35 bp were removed to maintain high-quality sequencing data. Human host sequences were computed and deleted, then mapped to the human reference genome (hg19) using the Burrows–Wheeler alignment tool to remove sequence reads from the human host. After erasing low-complexity reads, the remaining data were compared with the four Microbial Genome Database for Comparative Analysis, which includes bacteria, viruses, fungi, and parasites. 8 The classification reference database was extracted from NCBI (ftp://ftp.ncbi.nlm.nih.Gov/genomes/), and includes 4152 whole-genome sequences of viral taxa, 3446 bacterial genomes or scaffolds, 206 genomes of fungi associated with human infection, and 140 parasites associated with human diseases.
The criteria for a positive mNGS result in the BGI-Huada Genomics Institute are provided in supplementary information. Pathogens detected by mNGS were classified into three categories: (1) definite, in which a BALF mNGS result was concordant with results from microbiologic tests (BALF culture, nucleic acid-based testing, and pathological examination); (2) probable, in which a BALF mNGS-based pathogen was the likely cause of LRTI based on clinical, radiologic, or laboratory findings; or (3) possible, in which a BALF mNGS-based pathogen had pathogenic potential but was not consistent with the clinical presentation. 7 Pathogens detected by mNGS that met either definite or probable criteria were judged to be causative pathogens.
Statistical analysis
Statistical analyses were performed using SPSS 22.0 statistical software (IBM Corp, Armonk, NY, USA). Continuous variables are presented as means ± standard deviation. The paired McNemar chi-square test for a matched fourfold table was used to compare the diagnostic efficiency of mNGS and traditional culture. P < 0.05 was considered statistically significant.
Results
Of the 47 patients with LRTIs, 26 were men. The mean age was 48.87 ± 18.83 years. Detailed demographic information is shown in Table 1. A total of 33 patients had CAP, of which 12 had severe pneumonia and nine had tuberculosis. Regarding comorbidity, eight patients had malignancies or were immunocompromised, and 10 patients had hypoproteinemia.
Table 1.
Demographic information.
| Characteristic | Patients, n |
|---|---|
| Age, years, mean ± standard deviation | 48.87 ± 18.83 |
| Male, n (%) | 26 (55.32) |
| Infection types | |
| CAP | 33 |
| Severe pneumonia | 12 |
| AECOPD | 2 |
| AEBX | 1 |
| Lung abscess | 2 |
| Tuberculosis | 9 |
| Comorbidity | |
| COPD | 4 |
| Diabetes | 4 |
| Hypoproteinemia | 10 |
| Malignancy or Immunocompromised | 8 |
| Hypertension | 4 |
| Cardiovascular | 6 |
| Chronic liver diseases | 9 |
| Renal diseases | 4 |
AEXB: acute exacerbation of bronchiectasis; CAP: community-acquired pneumonia; COPD: chronic obstructive pulmonary disease; AECOPD: acute exacerbation chronic obstructive pulmonary disease.
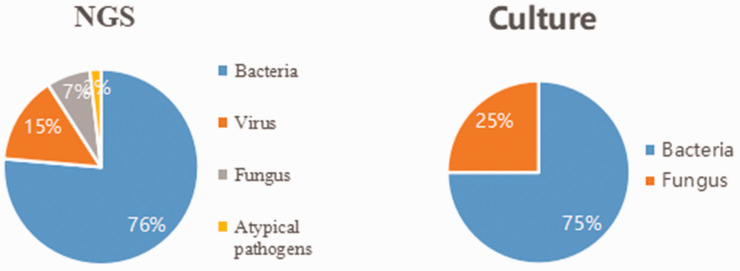
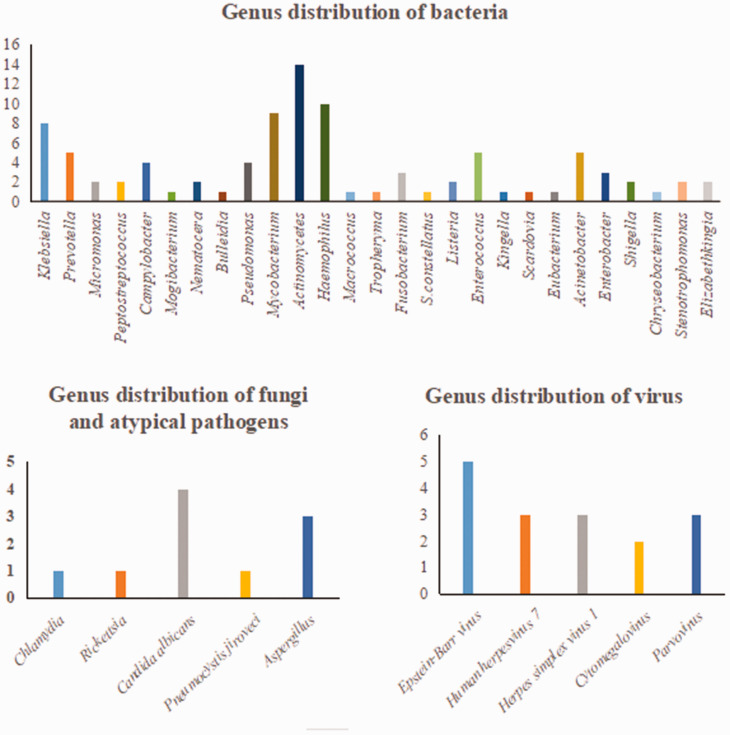
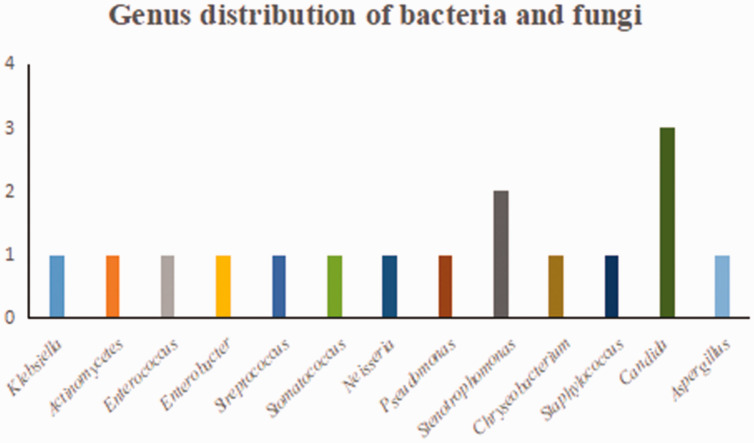
Detailed results of pathogen detection are shown in Supplementary Table 1. Most pathogens detected by the two methods were bacteria (mNGS, 76%; culture, 75%) (Figure 1). A total of 84 bacteria, eight fungi, 16 viruses, and two atypical pathogens were identified by mNGS. By contrast, the culture method only identified 12 bacteria and four fungi (Table 2). Figure 2 shows the genus distribution of bacteria, fungi, atypical pathogens, and viruses identified by mNGS, among which Actinomycetes, Haemophilus, Mycobacterium, and Klebsiella were the most common. Epstein–Barr virus was the most common virus identified by mNGS, and the most common fungus was Candida albicans. Stenotrophomonas was the most common genus of bacteria identified by culture methods (Figure 3). mNGS detected a wider spectrum of pathogens than the culture method, and was also able to detect viruses and atypical pathogens.
Figure 1.
Comparison of culture and mNGS identification with respect to pathogen classification categories.
mNGS, metagenomic next-generation sequencing.
Table 2.
Detectable pathogen count by mNGS and culture methods.
| Detection method |
|||
|---|---|---|---|
| Pathogen | mNGS | Culture PCR | Total |
| Bacteria (+) | 84 | 12 | 96 |
| Mtc (+) | 7 | 0 | 14 |
| Viruses (+) | 16 | 0 | 17 |
| Fungi (+) | 8 | 4 | 12 |
| Atypical pathogens (+) | 2 | 0 | 2 |
Mtc: Mycobacterium tuberculosis complex.
Figure 2.
Genus distribution of bacteria, fungi, atypical pathogens, and viruses by mNGS.
mNGS, metagenomic next-generation sequencing.
Figure 3.
Genus distribution of bacteria and fungi by culture.
The LRTI detection rate was 68.09% (32/47) by mNGS and 21.28% (10/47) by culture (Table 3). Compared with the culture method, mNGS had a diagnostic sensitivity of 80% and a specificity of 35.13%, with an agreement rate of 44.68%. There was a significant difference between mNGS and culture findings (P < 0.01).
Table 3.
Comparison of mNGS and traditional culture analyses of bronchoalveolar lavage fluid samples.
| mNGS | Culture | Total | |
|---|---|---|---|
| + | – | ||
| + | 8 | 24 | 32 |
| – | 2 | 13 | 15 |
| Total | 10 | 37 | 47 |
Discussion
With the development of metagenomic sequencing, increasing numbers of researchers are exploring its application in pathogen diagnosis.10,11 Compared with traditional culture methods, mNGS offers obvious advantages in the pathogenic diagnosis of LRTIs. In the present study, detectability of pathogens by mNGS was notably higher than by traditional culture methods, especially for viruses, atypical pathogens, and Mycobacterium tuberculosis, which could not be cultivated. However, mNGS results should be combined with epidemiological and clinical characteristics in the identification of pathogenic microorganisms.
The accurate and timely identification of pathogens is essential for the treatment of LRTIs, especially those involving infections with atypical pathogens or in critically ill patients. In our study, mNGS significantly increased the positive pathogenic diagnosis rate for LRTIs with a high sensitivity (80%) compared with traditional culture methods. Chen et al previously explored the mNGS diagnosis of clinical samples from 20 bacterial and fungal infections, and observed higher detection rates with mNGS than culture methods. 12 However, a diagnostic study of BALF mNGS in 32 critically ill patients conducted by Li et al found that mNGS did not increase the positive diagnosis rate but improved the sensitivity of pathogen detection. 8 These differences might reflect variations in populations studied. Of the 47 patients included in our study, nine had tuberculosis and eight had malignant diseases or immunodeficiencies, which made it difficult for pathogens to be cultivated. Additionally, different culture conditions in laboratories could lead to variations in positive rates of pathogens.
We also found an increase in the number of types of pathogens and the diagnosis rate of mNGS compared with the culture method, especially for pathogens not easily cultivated by traditional methods such as viruses, atypical pathogens, and tuberculosis; this is consistent with the results of previous studies. For instance, Chen et al found that mNGS improved the diagnosis rate of Chlamydia psittaci through a retrospective analysis of nine cases of psittacosis pneumonia, 13 while Jin et al showed that the overall sensitivity of mNGS was superior to that of culture (49.6% vs 35.2%, respectively) in the diagnosis of 125 cases of active tuberculosis. 14 mNGS was also reported to improve the diagnosis rate of viral infections in plasma samples, and to identify dengue virus 1 and Ebola virus, 15 while in our study it identified Orientia tsutsugamushi. Overall, mNGS can broaden the pathogen detection spectrum of LRTIs, which is helpful for the targeted therapy of patients.
Actinomycosis is a rare infectious disease caused by the opportunistic facultative anaerobic bacteria actinomycetes. 16 Culture conditions are complex and the bacteria demonstrate slow growth, resulting in a poor culture rate, 17 so a diagnosis of actinomycosis mainly depends on histopathology. 18 However, our study identified actinomycetes as the most common type of bacteria in BALF specimens of LRTIs. Thus, mNGS could be helpful in making a diagnosis of actinomycetes, although because actinomycetes normally colonize the human mouth and digestive tract, a positive test may reflect contamination.
Another advantage of mNGS is that it detects multiple pathogens simultaneously, which helps shorten the turnaround time. It also aids good antibacterial management of LRTIs, such as by providing evidence of the presence or absence of particular bacterial pathogens, enabling the unnecessary use of antibiotics to be stopped. However, mNGS has a relatively poor level of specificity, and does not distinguish whether a pathogen is a colonizing or pathogenic bacterium and whether it is alive or dead. For example, a high percentage of C. albicans was identified in our study, but they were considered colonizers or contaminants, not pathogens. The low observed prevalence of Candida pneumonia is consistent with that reported in previous studies.19,20 Therefore, it is important to rationally and objectively utilize mNGS results, and to note that they currently cannot fully replace conventional detection methods. Future diagnoses and treatments of infectious diseases should combine multiple disciplines in diagnosis, thereby enabling precise treatments to be carried out.
Our study had the following shortcomings. First, it was limited by non-random patient selection methods and a relatively small sample size. Second, we could not compare mNGS with other non-laboratory culture methods, such as PCR and pathogen antibodies because of sample size limitations. Third, the proportion of critically ill patients or those difficult to diagnose was relatively high in our study. Therefore, antibacterial drugs had often been used before BALF sample collection, which may have reduced the culture detection rate although it did not affect mNGS detection. Future studies of larger sample sizes are needed to fully investigate and compare the pathogen diagnosis rate between mNGS and traditional methods.
In conclusion, mNGS appears to show some advantages over culture methods in pathogen identification and the diagnosis rate of LRTIs, and is expected to become a powerful tool for clinical diagnosis.
Supplemental Material
Supplemental material, sj-pdf-1-imr-10.1177_03000605221089795 for Application of metagenomic next-generation sequencing for bronchoalveolar lavage diagnostics in patients with lower respiratory tract infections by Dandan Zhang, Xue Yang, Junli Wang, Jian Xu and Mengyi Wang in Journal of International Medical Research
Supplemental material, sj-pdf-2-imr-10.1177_03000605221089795 for Application of metagenomic next-generation sequencing for bronchoalveolar lavage diagnostics in patients with lower respiratory tract infections by Dandan Zhang, Xue Yang, Junli Wang, Jian Xu and Mengyi Wang in Journal of International Medical Research
Acknowledgements
The authors are very grateful to the patients for participating in this study.
Footnotes
Declaration of conflicting interests: The authors declare no competing interests.
Funding: This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
ORCID iD: Dandan Zhang https://orcid.org/0000-0001-5777-9826
Supplemental material
Supplementary 1 Criteria for a positive mNGS result.
Supplementary Table 1 Detailed results of LRTI patients, BALF samples detected by NGS and culture method.
References
- 1.Wunderink RG, Waterer G. Advances in the causes and management of community acquired pneumonia in adults. Bmj 2017; 358: j2471. [DOI] [PubMed] [Google Scholar]
- 2.Jain S, Self WH, Wunderink RG, et al. Community-acquired pneumonia requiring hospitalization among U.S. adults. N Engl J Med 2015; 373: 415–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jain S, Williams DJ, Arnold SR, et al. Community-acquired pneumonia requiring hospitalization among U.S. children. N Engl J Med 2015; 372: 835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Musher DM, Roig IL, Cazares G, et al. Can an etiologic agent be identified in adults who are hospitalized for community-acquired pneumonia: Results of a one-year study. J Infect 2013; 67: 11–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilson MR, Naccache SN, Samayoa E, et al. Actionable diagnosis of neuroleptospirosis by next-generation sequencing. N Engl J Med 2014; 370: 2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blauwkamp TA, Thair S, Rosen MJ, et al. Analytical and clinical validation of a microbial cell-free DNA sequencing test for infectious disease. Nat Microbiol 2019; 4: 663–674. [DOI] [PubMed] [Google Scholar]
- 7.Chen H, Yin Y, Gao H, et al. Clinical utility of in-house metagenomic next-generation sequencing for the diagnosis of lower respiratory tract infections and analysis of the host immune response. Clin Infect Dis 2020; 71: S416–S426. [DOI] [PubMed] [Google Scholar]
- 8.Li Y, Sun B, Tang X, et al. Application of metagenomic next-generation sequencing for bronchoalveolar lavage diagnostics in critically ill patients. Eur J Clin Microbiol Infect Dis 2020; 39: 369–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Von Elm E, Altman DG, Egger M, et al. The strengthening the Reporting of Observational studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. Ann Intern Med 2007; 147: 573–577. [DOI] [PubMed] [Google Scholar]
- 10.Yu G, Zhao W, Shen Y, et al. Metagenomic next generation sequencing for the diagnosis of tuberculosis meningitis: A systematic review and meta-analysis. PLoS One 2020; 15: e0243161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gu W, Deng X, Lee M, et al. Rapid pathogen detection by metagenomic next-generation sequencing of infected body fluids. Nat Med 2021; 27: 115–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen P, Sun W, He Y. Comparison of the next-generation sequencing (NGS) technology with culture methods in the diagnosis of bacterial and fungal infections. J Thorac Dis 2020; 12: 4924–4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen X, Cao K, Wei Y, et al. Metagenomic next-generation sequencing in the diagnosis of severe pneumonias caused by Chlamydia psittaci. Infection 2020; 48: 535–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin W, Pan J, Miao Q, et al. Diagnostic accuracy of metagenomic next-generation sequencing for active tuberculosis in clinical practice at a tertiary general hospital. Ann Transl Med 2020; 8: 1065–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jerome H, Taylor C, Sreenu V, et al. Metagenomic next-generation sequencing aids the diagnosis of viral infections in febrile returning travellers. J Infect 2019; 79: 383–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang W, Ren D, Xu C, et al. Pulmonary actinomycosis diagnosed by radial endobronchial ultrasound coupled with microbiological next-generation sequencing: a case report and brief literature review. Int J Infect Dis 2020; 100: 379–381. [DOI] [PubMed] [Google Scholar]
- 17.Tristan F, Florent V, Judith K, et al. Actinomycosis: etiology, clinical features, diagnosis, treatment, and management. Infection & Drug Resistance 2014; 2014: 183–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Himeji D, Hara S, Kawaguchi T, et al. Pulmonary Actinomyces graevenitzii infection diagnosed by bronchoscopy using endobronchial ultrasonography with a guide sheath. Intern Med 2018; 57: 2547–2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang L, Song J, Wang Y, et al. Metagenomic next-generation sequencing for pulmonary fungal infection diagnosis: lung biopsy versus bronchoalveolar lavage fluid. Infect Drug Resist 2021; 14: 4333–4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao Z, Song J, Yang C, et al. Prevalence of fungal and bacterial co-infection in pulmonary fungal infections: a metagenomic next generation sequencing-based study. Front Cell Infect Microbiol 2021; 11: 749905. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-imr-10.1177_03000605221089795 for Application of metagenomic next-generation sequencing for bronchoalveolar lavage diagnostics in patients with lower respiratory tract infections by Dandan Zhang, Xue Yang, Junli Wang, Jian Xu and Mengyi Wang in Journal of International Medical Research
Supplemental material, sj-pdf-2-imr-10.1177_03000605221089795 for Application of metagenomic next-generation sequencing for bronchoalveolar lavage diagnostics in patients with lower respiratory tract infections by Dandan Zhang, Xue Yang, Junli Wang, Jian Xu and Mengyi Wang in Journal of International Medical Research