Abstract
Plant aerial development relies on meristem activity which ensures main body plant axis development during plant life. While the shoot apical meristem (SAM) formed in the embryo only contributes to the main stem, the branched structure observed in many plants relies on axillary meristems (AMs) formed post-embryonically. These AMs initiate from a few cells of the leaf axil that retain meristematic characteristics, increase in number, and finally organize into a structure similar to the SAM. In this review, we will discuss recent findings on de novo establishment of a stem cell population and its regulatory niche, a key step essential for the indeterminate fate of AMs. We stress that de novo stem cell formation is a progressive process, which starts with a transient regulatory network promoting stem cell formation and that is different from the one acting in functional meristems. This transient stage can be called premeristems and we discuss whether this concept can be extended to the formation of meristems other than AMs.
Keywords: Arabidopsis thaliana, rice, stem cells, CLAVATA3, WUSCHEL, HAIRY MERISTEM, axillary meristem
Introduction
Plants are characterized by continuous organogenesis and growth throughout their life by the action of meristems. These structures are formed by a few hundreds or thousands of cells, depending on the species, that are maintained undifferentiated and proliferating by the combined action of meristematic genes such as the KNOTTED1-LIKE Homeobox encoding gene SHOOT MERISTEMLESS (STM) and an hormonal balance of high cytokinin (CK) to low gibberellin (Shani et al., 2006; Hay and Tsiantis, 2010; Maugarny-Calès and Laufs, 2018; Shi and Vernoux, 2022). Within this population of STM-expressing meristematic cells lies a specific subpopulation of semi-permanent stem cells that can be recognized by the expression of the CLV3 gene (Fletcher et al., 1999). These stem cells divide infrequently to replenish themselves while producing cells contributing to the meristem organogenetic activity. These stem cells are maintained by the activity of a stem cell niche that provides a cellular environment regulating stem cell division and preventing their differentiation (Bäurle and Laux, 2003; Dinneny and Benfey, 2008). While meristem multicellularity and complexity of the regulatory interactions between its different functional domains may be an advantage for the robustness of established meristems, they become, however, challenges to overcome when it comes to producing new meristems. Such new meristems are nevertheless repetitively produced during the plant life, turning for instance into axillary meristems (AMs) that increase the branching pattern of the plant. Here, we will discuss current knowledge and hypothesis of how an organized meristem emerges from a small group of meristematic cells, concentrating mostly on the formation of the stem cell population and its regulatory niche.
How Are Stem Cells Regulated in an Arabidopsis Meristem?
In the apical part of the shoot apical meristem (SAM) lies a group of semi-permanent stem cells maintained in their undifferentiated and pluripotent state by an underlying organizing center (OC) that contributes to the stem cell niche function (Laux et al., 1996). The WUSCHEL (WUS) gene is a meristematic stem cell fate regulator expressed in the OC and encoding a HOMEOBOX-like transcription factor that moves through plasmodesmata to promote CLAVATA3 (CLV) expression in the stem cell pool (Laux et al., 1996; Mayer et al., 1998; Yadav et al., 2011; Daum et al., 2014). To ensure its function in establishing the stem cell niche, WUS proteins form homodimers but also act through monomers or heterodimers with STM (Perales et al., 2016; Rodriguez et al., 2016; Sloan et al., 2020; Su et al., 2020). In turn, the small secreted peptide CLV3 inhibits WUS expression upon binding to receptor kinases such as CLV1, CLV2, CORYNE (CRN), or BARELY ANY MERISTEM 1–3 (BAM1-3; Clark, 2001; Brand et al., 2002; DeYoung et al., 2006; DeYoung and Clark, 2008; Müller et al., 2008; Ogawa et al., 2008; Schlegel et al., 2021). This core regulatory network contributes to the maintenance of the stem cell population while additional interacting regulators such as the HAIRY MERISTEM (HAM) transcription factors contribute to the positioning of the stem cells. WUS can bind and form heterodimers with HAM proteins to define the expression domain of CLV3 (Zhou et al., 2015, 2018). HAM1 and HAM2 are expressed in domains partially overlapping WUS domains. Specifically, both genes are expressed in the medullar and peripheral zones of the apical meristem but not in the L1 or L2 layers of the central zone (Schulze et al., 2010; Zhou et al., 2018; Han et al., 2020a). Expression and modeling data suggest that an apical-basal gradient of HAM1/2 genes is established in the apical meristem to define CLV3 expression pattern in the SAM. The establishment of the HAM gradient is in part mediated by miR171 that target the HAM genes and the transcription factor ARABIDOPSIS THALIANA MERISTEM LAYER 1 (ATML1) that promotes miR171 expression in the epidermis (Wang et al., 2010; Takanashi et al., 2018; Han et al., 2020b). Furthermore, an epidermal signal of CK is associated with the CLV-WUS genetic network. Indeed, modeling suggests that a combination of long and short range epidermal signals that could be CK production and response, respectively, act as positional cues for patterning the WUS domain (Leibfried et al., 2005; Chickarmane et al., 2012; Gruel et al., 2016). Finally, auxin signaling is locally controlled to maintain a low level of auxin response in the stem cell niche because the presence of a high level of auxin signaling could induce organ emergence from the center of the meristem and severely disturb meristem integrity (Shi et al., 2018; Ma et al., 2019; Galvan-Ampudia et al., 2020). Altogether, this network contributes to maintenance of stem cell homeostasis via a balance between their loss and renewal and proper spatial positioning of the stem cell and stem cell niche to allow meristem activity to respond to environmental signals (Yoshida et al., 2011; Pfeiffer et al., 2016; Landrein et al., 2018).
How Is an Arabidopsis Axillary Meristem Initiated?
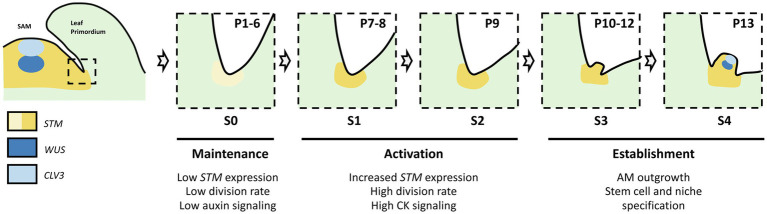
The initiation of an AM during the vegetative phase in the axils of the rosette leaves of Arabidopsis has been extensively studied and several stages have been defined based on cellular and morphological features (Long and Barton, 2000; Xin et al., 2017; Figure 1). Thus, axils of leaf primordia 1–6 (P1-6, counted from the SAM) show no morphological or cellular evidence of AM initiation (stage S0). At S1, P7–8 leaf axils show cell divisions which intensify at S2 in P9 axils. Then, at S3, a bulge forms in P10-12 axils and becomes a dome-shaped structure at S4 in P13 axils, and primordia start to emerge from the S5 AM at ≥ P14.
Figure 1.
Steps of axillary meristem formation in the axils of Arabidopsis thaliana rosette leaves. Developmental stages (S0 to S4) are shown relative to leaf age (defined in plastochrones, P1 to P13).
Thus, AM initiation can be divided into two phase, a maintenance phase during which some meristematic cells remain latent in the leaf axils (stage S0) and an activation phase in which the division of these cells leads to the formation of an AM (S1 to S4; Grbić and Bleecker, 2000; Long and Barton, 2000). Live imaging showed that there are only a very limited number of cell divisions in the latent axillary meristem, which contributes to limiting the risk of somatic mutations that could be propagated in the axillary branches (Burian et al., 2016). During the maintenance phase, STM remains expressed at a low level in the leaf axil and keeps cells in an undifferentiated state in contrast to their neighboring cells which undergo differentiation. Such maintenance of STM expression requires depletion of auxin from the axillary region and involves at the molecular level a self-activation loop facilitated by a permissive epigenetic environment (Wang et al., 2014a,b; Cao et al., 2020). During the activation phase, STM expression increases to induce division of these cells and the bulging out of the initiating AM (Shi et al., 2016; Xin et al., 2017). The importance of STM in AM formation is illustrated by the absence of AM formation in a high proportion of leaf axils in the weak allele of STM, stm-bumpershoot1 (Shi et al., 2016). A local pulse of CK response is required to stimulate STM expression and promote AM formation, possibly through a mutual positive feedback loop between STM and CK (Wang et al., 2014b). Multiple factors, such as CUP-SHAPED COTYLEDON 1–3 (CUC1-3), LATERAL SUPPRESSOR (LAS), REVOLUTA (REV), DORNRÖSCHEN (DRN) and DORNRÖSCHEN LIKE (DRNL), REGULATOR OF AXILLARY MERISTEMS 1–3 (RAX1-3), REGULATOR OF AXILLARY MERISTEM FORMATION (ROX), and ARGONAUTE 10 (AGO10), provide spatial and temporal cues for the local activation of STM expression, and thus control the pattern of AM formation (Greb et al., 2003; Hibara et al., 2006; Keller et al., 2006; Müller et al., 2006; Raman et al., 2008; Yang et al., 2012; Shi et al., 2016; Zhang et al., 2018, 2020).
How Is a New Stem Cell Niche Formed in an Arabidopsis Axillary Meristem?
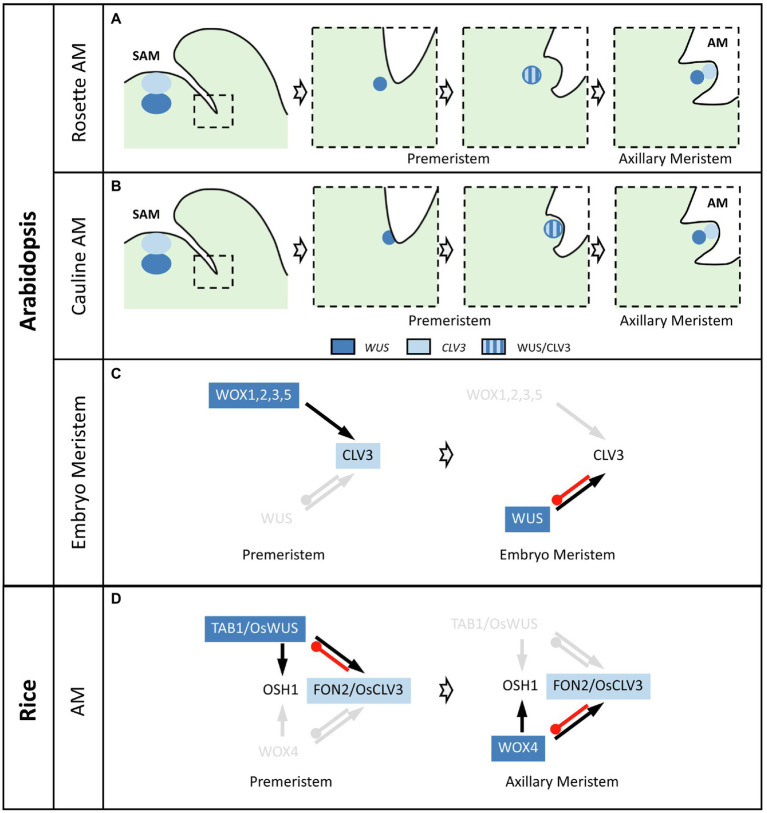
The observation that AMs result from the division of a few cells maintaining STM expression in the axils is in agreement with the so-called “detached meristem” concept for AM formation (Steeves and Sussex, 1989). In this “detached meristem” model, meristematic cells “detached” from the SAM are maintained in the axil of the leaf before being amplified to generate the new AM (Long and Barton, 2000). This model is opposed to the “de novo origin of AM” model, in which differentiated cells regain a meristematic fate to form a new AM (McConnell and Barton, 1998). However, while cells of the “detached meristem” express meristem markers such as STM, they express neither the stem cell marker CLV3 nor WUS, clearly indicating that they are not yet organized in a properly structured meristem (Figure 2A). In fact, WUS becomes activated only at S1 after the increase in STM expression and WUS activates CLV3 expression at S2 (Wang et al., 2017; Xin et al., 2017). A similar temporal succession of WUS and CLV3 activation is also observed during AM establishment in cauline leaf axils of Arabidopsis (Nicolas et al., 2022). Interestingly, in both rosette and cauline AM, WUS and CLV3 are initially expressed in overlapping domains and not in mostly separated domains as in the SAM. More precisely, in cauline AMs, WUS and CLV3 are initially expressed in overlapping apical domains while later WUS expression shifts down from the apical to a central domain (Nicolas et al., 2022; Figure 2B). The opposite dynamic is observed in rosette AMs, as WUS and CLV3 are co-expressed in the center of S4 rosette AM while later on, CLV3 expression domain shifts upwards and relocates to an apical position (Figure 2A; Xin et al., 2017). This spatial rearrangement of CLV3 expression in rosette AMs is mainly ensured by the establishment of an apico-basal gradient of HAM gene expression. The role of the HAM genes during AM formation is demonstrated by the phenotype of the triple mutant ham1 ham2 ham3 in which CLV3 expression does not shift from a central to an apical domain and AM formation is compromised. Complementation of the triple ham1 ham2 ham3 mutant with each of the HAM genes showed that HAM1 and 2 play the most prominent role for CLV3 expression pattern dynamics during AM formation. Interestingly, the apico-basal gradient of HAM gene expression is established progressively during AM formation. Indeed, the HAM genes are first expressed uniformly at S2 and S3 and it is only from S4 and S5 onwards that a gradient of expression is formed. Such a gradient is in part a consequence of the epidermis-specific expression of their negative regulators miR171 (Zhou et al., 2018; Han et al., 2020a,b).
Figure 2.
Regulatory dynamics driving de novo stem cell formation during meristem formation. (A) Stem cell formation in Arabidopsis thaliana rosette AMs. Neither WUS nor CLV3 are expressed in the axils of young rosette leaf primordia from which the AM will initiate (first panel). WUS expression becomes expressed in the inner part of the axil (second panel) and later activates CLV3 in an inner, overlapping domain (third panel). These stages can be defined as premeristem. Finally, WUS and CLV3 expressions resolve in two separate domains, in the inner and apical part of the rosette AM, respectively (last panel). (B) Stem cell formation in Arabidopsis thaliana cauline AMs. Neither WUS nor CLV3 are expressed in the axils of young cauline leaf primordia from which the AM will initiate (first panel). WUS expression becomes expressed in the apical part of the axil (second panel) and later activates CLV3 in an apical, overlapping domain (third panel). These stages can be defined as premeristem. Finally, WUS and CLV3 expressions resolve in two separate domains, in the inner and apical part of the cauline AM, respectively (last panel). (C) Stem cell formation in Arabidopsis thaliana embryo meristems. CLV3 expression is activated by the WOX1, 2, 3, and 5 genes at the premeristem stage, while in established and active meristems WUS activates CLV3 expression. (D) Stem cell formation in rice AMs. FON2 (the CLV3 ortholog) expression is activated by TAB1 (the WUS ortholog) at the premeristem stage, while in established and active meristems WOX4 activates FON2 expression. TAB1 or WOX4 promotes OSH1 (the STM ortholog) expression in premeristems and meristems, respectively. In (C) and (D), black arrows mean expression activation while red lines mean repression. Genes and interactions indicated in light gray are not present at the described stage.
What drives the formation of a new stem cell population and its niche is not fully understood yet. WUS is a central actor in the process as the wus mutant has a very strong AM initiation defect in contrast to the clv3-2 mutant which has only very weak AM initiation defects (Wang et al., 2017; Xin et al., 2017). De novo WUS expression in AMs is promoted by CK, through binding of the type-B Arabidopsis response regulator proteins (ARRs), which mediate the transcriptional response to CK, to the WUS promoter (Wang et al., 2017). Thus, CK signaling could be a link between the AM activation phase where it forms a positive feedback loop with STM and the AM establishment phase where it promotes WUS expression. Another link between the early phases of meristem initiation and its establishment is provided by the CUC genes. As described before, CUC genes are required for AM initiation (Hibara et al., 2006; Raman et al., 2008), likely by preventing cell differentiation and maintaining cells in a meristematic fate. However, recent results show that expression of those boundary genes has to be downregulated from the initiating meristem to proceed to the establishment phase and allow stem cell formation (Nicolas et al., 2022). CUC ectopic expression in the developing AM perturbs its growth and prevents stem cell establishment as shown by delayed expression of WUS and CLV3. Repression of the CUC genes from the initiating meristem results from the redundant action of two NGATHA-LIKE (NGAL) transcription factors, DPA4 and SOD7. Thus, the boundary identity needs to be repressed in order to allow de novo establishment of stem cells in newly formed AM and permit proper expression of WUS and CLV3 (Nicolas et al., 2022).
Altogether, Arabidopsis AM establishment appears as being a gradual process. Although AMs derive from boundary cells that maintain a meristematic fate, a prerequisite for AM establishment is the repression of boundary fate from the developing AM. CK initiates de novo stem cell niche establishment by activating WUS expression. In parallel, an apico-basal gradient of HAM activity is established which contributes to the dynamic (re)positioning of the stem cells and stem cell niche and the formation of an active meristem.
How Is a New Stem Cell Niche Formed in a Rice Axillary Meristem?
Development of the rice AMs during the vegetative phase leading to rice tillering provides another example of a dynamic reorganization of the regulatory network leading to de novo stem cell niche establishment. In rice, the axillary bud is formed on the side of the main stem (also called culm in rice) and is composed by a protective, modified leaf, the prophyll, enclosing a few leaf primordia and the AM. The first sign of AM initiation is a small group of small, dense cells that readably forms a bulge growing out from the stem in P3 axils, a transient state called “premeristem” by Tanaka et al. (2015). Cells of the premeristem are expressing OSH1, a marker for meristematic cells and orthologue of Arabidopsis STM (Oikawa and Kyozuka, 2009; Tanaka et al., 2015). In P4, the developing AM grows through cell division forming a cone-like structure initiating first the prophyll and later true leaf primordia. LAX PANICLE1 (LAX1, orthologous to the bHLH transcription factor ROX of Arabidopsis thaliana), LAX PANICLE2 (a nuclear protein interacting with LAX1), and MONOCULM1 (orthologous to the GRAS transcription factor LATERAL SUPPRESSOR of Arabidopsis thaliana) synergistically promote OSH1 expression and are required for AM formation (Oikawa and Kyozuka, 2009; Tabuchi et al., 2011).
Apparition of the stem cells in rice AM can be followed by the expression of FLORAL ORGAN NUMBER2 (FON2), that like its Arabidopsis orthologue CLV3 marks the stem cells in meristems (Suzaki et al., 2006). This revealed an early specification of the stem cells in rice developing AMs as FON2 expression is detected in a central, apical subset of the premeristem (Tanaka and Hirano, 2020). fon2 mutants show larger developing AMs with enlarged FON2 expressing domains, while conversely in FON2 overexpressors AM have a reduced size, are often flat, and show reduced OSH1 expression that is not maintained, suggesting that FON2 is required for AM maintenance (Tanaka and Hirano, 2020).
TAB1 (TILLERS ABSENT 1, also called MONOCULM3), the rice orthologue of the WUS gene is required for AM formation in rice (Lu et al., 2015; Tanaka et al., 2015; Shao et al., 2019). TAB1 is expressed early on in the developing AM, in a central, apical domain of the premeristem that overlaps with the FON2 expression domain (Tanaka et al., 2015; Tanaka and Hirano, 2020). Like in Arabidopsis, TAB1 expression is induced by CKs and mutation of TAB1 leads to the absence of a functional AM, with only the development of the prophyll in some cases (Lu et al., 2015; Tanaka et al., 2015). Accordingly, expression of OSH1 is strongly reduced in tab1 mutants, suggesting TAB1 promotes AM formation through promoting OSH1 expression to prevent cell differentiation (Figure 2D). In contrast, expression of LAX1 and MOC1 is not modified in tab1 mutants, suggesting that these genes act upstream or in parallel to TAB1. However, in contrast to what occurs in Arabidopsis, TAB1 expression is not maintained in rice AM once the prophyll is initiated (Tanaka et al., 2015). Instead, WOX4, the closest TAB1 paralog becomes expressed in the AM. AM defects of the tab1 mutant can be partially rescued by expressing WOX4 under the control of the TAB1 promoter, showing a strong functional conservation between WOX4 and TAB1 proteins (Tanaka and Hirano, 2020). Furthermore, tab1 AM defects are also partially rescued in the tab1 fon2 double mutant, in which a precocious expression of WOX4 is observed (Tanaka and Hirano, 2020). Hence, while TAB1 is required for the formation of the premeristem, WOX4 is required for the later function of the established AM, which is in agreement with WOX4 being required for SAM maintenance during rice vegetative development and TAB1 being not required (Ohmori et al., 2013; Tanaka et al., 2015).
Altogether these observations indicate that AM formation in rice involves a transient phase, called premeristem, and that the stem cell promoting role of WUS during the premeristem stage is later taken over by WOX4 in functional meristems (Figure 2D).
Can the Concept of Premeristem Be Extended to All Axillary Meristems?
The concept of premeristem was coined by Tanaka et al. (2015) to describe a transient stage when rice AMs are formed by a group of cells with meristematic features but are not yet alike functional meristems. In rice, a difference between axillary premeristem and established AM is the involvement of the WUS ortholog TAB1 instead of WOX4. In contrast, in the case of Arabidopsis, expression and genetic data indicate that the same gene, WUS, is acting in establishing and active AMs. However, the expression patterns of WUS or CLV3 are reorganized during Arabidopsis AM formation, possibly as a result of HAM gene dynamics. We therefore suggest to extend the concept of premeristems to all developing AM that are formed by a group of meristematic cells and in which the regulatory networks are not yet similar to the ones acting in established meristems either because the actors are different as in rice or because the actors are expressed in different domains, as in Arabidopsis. Whether this concept may apply to developing AMs in other species awaits precise molecular and genetic deciphering of the processes at play.
Can the Concept of Premeristem Be Extended to All Newly Formed Meristems?
Beside AMs, plants form new meristems in other contexts. Depending on the species, floral meristems (FM) have different origins and they initiate a new stem cell population, which is not permanently maintained in relation with the determinate fate of flowers. In the case of Arabidopsis, FMs arise at the flank of the inflorescence SAM. In fact, Arabidopsis FMs are thought as modified AMs, with the subtending leaf being reduced to a cryptic bract which growth can be derepressed in some mutant backgrounds (Long and Barton, 2000; Ohno et al., 2004). WUS expression starts in FM at late stage 1 and becomes stronger at stage 2 (Mayer et al., 1998; Grégoire et al., 2018; Nicolas et al., 2022) when it induces CLV3 expression, which thus begins to be expressed at stage 2 (Seeliger et al., 2016; Prunet et al., 2017; Nicolas et al., 2022). Like in the AMs, increased CK signaling is observed in young FMs (Yoshida et al., 2011) and could contribute to WUS activation. More recently, it has been shown that the meristem patterning gene REVOLUTA together with the main floral determinant LEAFY, in part through its target RAX1, contribute to WUS activation in a partially redundant manner (Denay et al., 2018). Finally, as in AMs, DPA4 and SOD7 are required for efficient de novo stem cell formation in FMs (Nicolas et al., 2022). Despite these findings, our understanding of the mechanisms at play during stem cell formation in FM lacks the detail required to unambiguously recognize or exclude a transient stage as defined for the premeristem.
The first meristem formed is the embryonic meristem. In embryos, WUS is expressed early from the 16-cell stage onwards (Mayer et al., 1998). However, it is not required for the induction of CLV3, which starts to be expressed from the transition stage and at the heart stage in the subepidermal layer of the meristem, then extends into the epidermis leading to the formation of the apical meristem (Zhang et al., 2017b). Instead of WUS, other genes from the same family, the WUSCHEL-LIKE HOMEBOX 1 (WOX1), WOX2, WOX3, and WOX5 genes, are required for stem cell establishment via activation and spatial rearrangement of CLV3 expression (Zhang et al., 2017b). Therefore, formation of the embryonic meristem involves a transient stage during which both the actors (WOX1,2,3,5 instead of WUS) and the expression pattern of the actors (CLV3 starting with an expression in the inner layers) are different from what is observed in the established SAM (Figure 2C). This transient phase during which WOX genes activate CLV3 expression could be seen as a premeristem stage.
Finally, new meristems can also be formed during in vitro culture through different ways (Ikeuchi et al., 2019). In the most widely used protocol, tissue explants are first cultured on an auxin-rich callus-inducing medium and then switched to a shoot-inducing medium characterized by a high CK level that promotes the formation of shoot meristems. CK-mediated activation of WUS expression by ARR proteins is a key step of shoot meristem formation, as it can be by-passed by forced WUS expression (Meng et al., 2017; Zhang et al., 2017a). Indeed, WUS activation results from two successive steps: first, cell divisions erase repressive epigenetic marks on the WUS locus, and, second, ARR proteins interact with the class III homeodomain-leucine zipper (HD-ZIP III) transcription factors on the WUS locus to promote its expression. This interaction between ARRs and HD-ZIP III allows limiting the expression pattern of WUS, as CK-mediated ARR activity is widely present in the regenerating callus while HD-ZIP III gene expression is more localized (Zhang et al., 2017a). However, initial expression pattern of WUS is reorganized during regeneration to reach the pattern typical of the OC in regenerated meristems (Meng et al., 2017; Zhang et al., 2017a). Shoot meristems can also be formed by direct conversion of lateral root meristems without the formation of an intermediate callus (Rosspopoff et al., 2017). Here again, formation of shoot meristems goes along with the induction of WUS expression as a response to CK treatments. Interestingly, during their activation phase, WUS and CLV3 show largely overlapping expression domains that only later resolve into patterns characteristic of bona fide meristem (Rosspopoff et al., 2017).
Conclusion and Perspectives
Formation of new meristems occurs repeatedly during plant development, starting from the embryonic meristem to AMs and FMs. Such structures are formed by meristematic cells that contain at their center a group of stem cells and stem cell niche. To form them, one could envisage two scenarios: the formation of a group of meristematic cells in a first step and in a second step, the definition of a functional stem cells and niche or, alternatively, a progressive increase of the pool of meristematic cells with the successive definition of stem cells niche and stem cells in a dynamic way. The observations discussed above all plead for the second scenario and reveal a transient, dynamic phase which can be called premeristem leading to the formation of functional meristem. Notably, the formation of this premeristem is marked by a transient regulatory network that leads to the regulatory network active in established AM but differs from it, either by identity or by the patterns of the actors involved. Furthermore, some factors such as the CUC or RAX1 genes or CK may contribute to the coordination between the increase of the meristematic cells and the formation of a stem cell niche. In addition, the role of other factors such as the oxidative status or mechanical signals that have been recently shown to regulate the stem cell or its niche could be investigated for their role during meristem formation (Weits et al., 2019; Bhattacharya et al., 2022).
Author Contributions
AN and PL wrote the review. All authors contributed to the article and approved the submitted version.
Funding
The IJPB benefits from the support of Saclay Plant Sciences-SPS (ANR-17-EUR-0007).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank N. Arnaud, L. Chelysheva, and J. C. Palauqui for helpful comments on the manuscript.
References
- Bäurle I., Laux T. (2003). Apical meristems: the plant’s fountain of youth. BioEssays 25, 961–970. doi: 10.1002/bies.10341, PMID: [DOI] [PubMed] [Google Scholar]
- Bhattacharya S., Wolfenson H., Shivdasani R., Shalom-Feuerstein R. (2022). Stem cell responses to stretch and strain. Trends Cell Biol. 32, 4–7. doi: 10.1016/j.tcb.2021.10.007, PMID: [DOI] [PubMed] [Google Scholar]
- Brand U., Grünewald M., Hobe M., Simon R., Grunewald M., Hobe M., et al. (2002). Regulation of CLV3 expression by two homeobox genes in Arabidopsis. Plant Physiol. 129, 565–575. doi: 10.1104/pp.001867, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burian A., Barbier de Reuille P., Kuhlemeier C. (2016). Patterns of stem cell divisions contribute to plant longevity. Curr. Biol. 26, 1385–1394. doi: 10.1016/j.cub.2016.03.067, PMID: [DOI] [PubMed] [Google Scholar]
- Cao X., Wang J., Xiong Y., Yang H., Yang M., Ye P., et al. (2020). A self-activation loop maintains Meristematic cell fate for branching. Curr. Biol. 30, 1893.e4–1904.e4. doi: 10.1016/j.cub.2020.03.031, PMID: [DOI] [PubMed] [Google Scholar]
- Chickarmane V. S. V. S., Gordon S. P. S. P., Tarr P. T. P. T., Heisler M. G. M. G., Meyerowitz E. M. E. M. (2012). Cytokinin signaling as a positional cue for patterning the apical-basal axis of the growing Arabidopsis shoot meristem. Proc. Natl. Acad. Sci. U. S. A. 109, 4002–4007. doi: 10.1073/pnas.1200636109, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark S. E. (2001). Cell signalling at the shoot meristem. Nat. Rev. Mol. Cell Biol. 2, 276–284. doi: 10.1038/35067079 [DOI] [PubMed] [Google Scholar]
- Daum G., Medzihradszky A., Suzaki T., Lohmann J. U. (2014). A mechanistic framework for noncell autonomous stem cell induction in Arabidopsis. Proc. Natl. Acad. Sci. 111, 14619–14624. doi: 10.1073/pnas.1406446111, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denay G., Tichtinsky G., Le Masson M., Chahtane H., Huguet S., Lopez-Vidriero I., et al. (2018). Control of stem-cell niche establishment in Arabidopsis flowers by REVOLUTA and the LEAFY-RAX1 module. bioRxiv [Preprint]. 1–37. doi: 10.1101/488114 [DOI] [Google Scholar]
- DeYoung B. J., Bickle K. L., Schrage K. J., Muskett P., Patel K., Clark S. E. (2006). The CLAVATA1-related BAM1, BAM2 and BAM3 receptor kinase-like proteins are required for meristem function in Arabidopsis. Plant J. 45, 1–16. doi: 10.1111/j.1365-313X.2005.02592.x, PMID: [DOI] [PubMed] [Google Scholar]
- DeYoung B. J., Clark S. E. (2008). BAM receptors regulate stem cell specification and organ development Through complex interactions With CLAVATA signaling. Genetics 180, 895–904. doi: 10.1534/genetics.108.091108, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinneny J. R., Benfey P. N. (2008). Plant stem cell niches: standing the test of time. Cell 132, 553–557. doi: 10.1016/j.cell.2008.02.001, PMID: [DOI] [PubMed] [Google Scholar]
- Fletcher J. C., Brand U., Running M. P., Simon R., Meyerowitz E. M. (1999). Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283, 1911–1914. doi: 10.1126/science.283.5409.1911, PMID: [DOI] [PubMed] [Google Scholar]
- Galvan-Ampudia C. S., Cerutti G., Legrand J., Brunoud G., Martin-Arevalillo R., Azais R., et al. (2020). Temporal integration of auxin information for the regulation of patterning. elife 9, 1–65. doi: 10.7554/eLife.55832, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grbić V., Bleecker A. B. (2000). Axillary meristem development in Arabidopsis thaliana. Plant J. 21, 215–223. doi: 10.1046/j.1365-313x.2000.00670.x [DOI] [PubMed] [Google Scholar]
- Greb T., Clarenz O., Schäfer E., Müller D., Herrero R., Schmitz G., et al. (2003). Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev. 17, 1175–1187. doi: 10.1101/gad.260703, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruel J., Landrein B., Tarr P., Schuster C., Refahi Y., Sampathkumar A., et al. (2016). An epidermis-driven mechanism positions and scales stem cell niches in plants. Sci. Adv. 2, e1500989–e1500924. doi: 10.1126/sciadv.1500989, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H., Geng Y., Guo L., Yan A., Meyerowitz E. M., Liu X., et al. (2020a). The overlapping and distinct roles of HAM family genes in Arabidopsis shoot meristems. Front. Plant Sci. 11, 541968. doi: 10.3389/fpls.2020.541968, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H., Yan A., Li L., Zhu Y., Feng B., Liu X., et al. (2020b). A signal cascade originated from epidermis defines apical-basal patterning of Arabidopsis shoot apical meristems. Nat. Commun. 11:1214. doi: 10.1038/s41467-020-14989-4, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay A., Tsiantis M. (2010). KNOX genes: versatile regulators of plant development and diversity. Development 137, 3153–3165. doi: 10.1242/dev.030049, PMID: [DOI] [PubMed] [Google Scholar]
- Hibara K., Karim M. R., Takada S., Taoka K., Furutani M., Aida M., et al. (2006). Arabidopsis CUP-SHAPED COTYLEDON3 regulates postembryonic shoot meristem and organ boundary formation. Plant Cell 18, 2946–2957. doi: 10.1105/tpc.106.045716, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi M., Favero D. S., Sakamoto Y., Iwase A., Coleman D., Rymen B., et al. (2019). Molecular mechanisms of plant regeneration. Annu. Rev. Plant Biol. 70, 377–406. doi: 10.1146/annurev-arplant-050718-100434 [DOI] [PubMed] [Google Scholar]
- Keller T., Abbott J., Moritz T., Doerner P. (2006). Arabidopsis REGULATOR OF AXILLARY MERISTEMS1 controls a leaf axil stem cell niche and modulates vegetative development. Plant Cell 18, 598–611. doi: 10.1105/tpc.105.038588, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrein B., Formosa-Jordan P., Malivert A., Schuster C., Melnyk C. W., Yang W., et al. (2018). Nitrate modulates stem cell dynamics in Arabidopsis shoot meristems through cytokinins. Proc. Natl. Acad. Sci. U. S. A. 115, 1382–1387. doi: 10.1073/pnas.1718670115, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laux T., Mayer K. F., Berger J., Jurgens G. (1996). The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122, 87–96. doi: 10.1242/dev.122.1.87, PMID: [DOI] [PubMed] [Google Scholar]
- Leibfried A., To J. P. C., Busch W., Stehling S., Kehle A., Demar M., et al. (2005). WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438, 1172–1175. doi: 10.1038/nature04270, PMID: [DOI] [PubMed] [Google Scholar]
- Long J., Barton M. K. (2000). Initiation of axillary and floral meristems in Arabidopsis. Dev. Biol. 218, 341–353. doi: 10.1006/dbio.1999.9572, PMID: [DOI] [PubMed] [Google Scholar]
- Lu Z., Shao G., Xiong J., Jiao Y., Wang J., Liu G., et al. (2015). MONOCULM 3, an Ortholog of WUSCHEL in Rice, is required for tiller bud formation. J. Genet. Genomics 42, 71–78. doi: 10.1016/j.jgg.2014.12.005, PMID: [DOI] [PubMed] [Google Scholar]
- Ma Y., Miotk A., Šutiković Z., Ermakova O., Wenzl C., Medzihradszky A., et al. (2019). WUSCHEL acts as an auxin response rheostat to maintain apical stem cells in Arabidopsis. Nat. Commun. 10:5093. doi: 10.1038/s41467-019-13074-9, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maugarny-Calès A., Laufs P. (2018). Getting leaves into shape: a molecular, cellular, environmental and evolutionary view. Dev. 145:dev161646. doi: 10.1242/dev.161646, PMID: [DOI] [PubMed] [Google Scholar]
- Mayer K. F. K. F., Schoof H., Haecker A., Lenhard M., Jürgens G., Laux T. (1998). Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95, 805–815. doi: 10.1016/S0092-8674(00)81703-1, PMID: [DOI] [PubMed] [Google Scholar]
- McConnell J. R., Barton M. K. (1998). Leaf polarity and meristem formation in Arabidopsis. Development 125, 2935–2942. doi: 10.1242/dev.125.15.2935 [DOI] [PubMed] [Google Scholar]
- Meng W. J., Cheng Z. J., Sang Y. L., Zhang M. M., Rong X. F., Wang Z. W., et al. (2017). Type-B ARABIDOPSIS RESPONSE REGULATORs specify the shoot stem cell niche by dual regulation of WUSCHEL. Plant Cell 29, 1357–1372. doi: 10.1105/tpc.16.00640, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller R., Bleckmann A., Simon R. (2008). The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell 20, 934–946. doi: 10.1105/tpc.107.057547, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller D., Schmitz G., Theres K., Muller D., Schmitz G., Theres K., et al. (2006). Blind homologous R2R3 Myb genes control the pattern of lateral meristem initiation in Arabidopsis. Plant Cell 18, 586–597. doi: 10.1105/tpc.105.038745, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas A., Maugarny-Calès A., Adroher B., Chelysheva L., Li Y. (2022). De novo stem cell establishment in meristems requires repression of organ boundary cell fate. bioRxiv [Preprint]. 1–63. doi: 10.1101/2022.02.03.478990 [DOI] [PMC free article] [PubMed]
- Ogawa M., Shinohara H., Sakagami Y., Matsubayash Y. (2008). Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science 319:294. doi: 10.1126/science.1150083, PMID: [DOI] [PubMed] [Google Scholar]
- Ohmori Y., Tanaka W., Kojima M., Sakakibara H., Hirano H.-Y. (2013). WUSCHEL-RELATED HOMEOBOX4 is involved in meristem maintenance and is negatively regulated by the CLE gene FCP1 in Rice. Plant Cell 25, 229–241. doi: 10.1105/tpc.112.103432, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno C. K., Reddy G. V., Heisler M. G. B., Meyerowitz E. M. (2004). The Arabidopsis JAGGED gene encodes a zinc finger protein that promotes leaf tissue development. Development 131, 1111–1122. doi: 10.1242/dev.00991, PMID: [DOI] [PubMed] [Google Scholar]
- Oikawa T., Kyozuka J. (2009). Two-step regulation of LAX PANICLE1 protein accumulation in axillary meristem formation in Rice. Plant Cell 21, 1095–1108. doi: 10.1105/tpc.108.065425, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales M., Rodriguez K., Snipes S., Yadav R. K., Diaz-Mendoza M., Reddy G. V. (2016). Threshold-dependent transcriptional discrimination underlies stem cell homeostasis. Proc. Natl. Acad. Sci. 113, E6298–E6306. doi: 10.1073/pnas.1607669113, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer A., Janocha D., Dong Y., Medzihradszky A., Schöne S., Daum G., et al. (2016). Integration of light and metabolic signals for stem cell activation at the shoot apical meristem. elife 5, 1–21. doi: 10.7554/eLife.17023, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunet N., Yang W., Das P., Meyerowitz E. M., Jack T. P. (2017). SUPERMAN prevents class B gene expression and promotes stem cell termination in the fourth whorl of Arabidopsis thaliana flowers. Proc. Natl. Acad. Sci. 114, 7166–7171. doi: 10.1073/pnas.1705977114, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman S., Greb T., Peaucelle A., Blein T., Laufs P., Theres K. (2008). Interplay of miR164, CUP-SHAPED COTYLEDON genes and LATERAL SUPPRESSOR controls axillary meristem formation in Arabidopsis thaliana. Plant J. 55, 65–76. doi: 10.1111/j.1365-313X.2008.03483.x, PMID: [DOI] [PubMed] [Google Scholar]
- Rodriguez K., Perales M., Snipes S., Yadav R. K., Diaz-Mendoza M., Reddy G. V. (2016). DNA-dependent homodimerization, sub-cellular partitioning, and protein destabilization control WUSCHEL levels and spatial patterning. Proc. Natl. Acad. Sci. 113, E6307–E6315. doi: 10.1073/pnas.1607673113, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosspopoff O., Chelysheva L., Saffar J., Lecorgne L., Gey D., Caillieux E., et al. (2017). Direct conversion of root primordium into shoot meristem relies on timing of stem cell niche development. Development 144, 1187–1200. doi: 10.1242/dev.142570, PMID: [DOI] [PubMed] [Google Scholar]
- Schlegel J., Denay G., Wink R., Pinto K. G., Stahl Y., Schmid J., et al. (2021). Control of Arabidopsis shoot stem cell homeostasis by two antagonistic CLE peptide signalling pathways. elife 10, 1–30. doi: 10.7554/eLife.70934, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze S., Schafer B. N., Parizotto E. A., Voinnet O., Theres K., Schäfer B. N., et al. (2010). LOST MERISTEMS genes regulate cell differentiation of central zone descendants in Arabidopsis shoot meristems. Plant J. 64, 668–678. doi: 10.1111/j.1365-313X.2010.04359.x, PMID: [DOI] [PubMed] [Google Scholar]
- Seeliger I., Frerichs A., Glowa D., Velo L., Comelli P., Chandler J. W., et al. (2016). The AP2-type transcription factors DORNRÖSCHEN and DORNRÖSCHEN-LIKE promote G1/S transition. Mol. Gen. Genomics. 291, 1835–1849. doi: 10.1007/s00438-016-1224-x, PMID: [DOI] [PubMed] [Google Scholar]
- Shani E., Yanai O., Ori N. (2006). The role of hormones in shoot apical meristem function. Curr. Opin. Plant Biol. 9, 484–489. doi: 10.1016/j.pbi.2006.07.008 [DOI] [PubMed] [Google Scholar]
- Shao G., Lu Z., Xiong J., Wang B., Jing Y., Meng X., et al. (2019). Tiller bud formation regulators MOC1 and MOC3 cooperatively promote tiller bud outgrowth by activating FON1 expression in Rice. Mol. Plant 12, 1090–1102. doi: 10.1016/j.molp.2019.04.008, PMID: [DOI] [PubMed] [Google Scholar]
- Shi B., Guo X., Wang Y., Xiong Y., Wang J., Hayashi K. i., et al. (2018). Feedback from lateral organs controls shoot apical meristem growth by modulating Auxin transport. Dev. Cell 44, 204.e6–216.e6. doi: 10.1016/j.devcel.2017.12.021, PMID: [DOI] [PubMed] [Google Scholar]
- Shi B., Vernoux T. (2022). Hormonal control of cell identity and growth in the shoot apical meristem. Curr. Opin. Plant Biol. 65:102111. doi: 10.1016/j.pbi.2021.102111, PMID: [DOI] [PubMed] [Google Scholar]
- Shi B., Zhang C., Tian C., Wang J., Wang Q., Xu T., et al. (2016). Two-step regulation of a Meristematic cell population acting in shoot branching in Arabidopsis. PLoS Genet. 12, e1006168–e1006120. doi: 10.1371/journal.pgen.1006168, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan J., Hakenjos J. P., Gebert M., Ermakova O., Gumiero A., Stier G., et al. (2020). Structural basis for the complex DNA binding behavior of the plant stem cell regulator WUSCHEL. Nat. Commun. 11:2223. doi: 10.1038/s41467-020-16024-y, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeves T. A., Sussex I. M. A. (1989). Patterns in Plant Development. New York: Cambridge University Press. [Google Scholar]
- Su Y. H., Zhou C., Li Y. J., Yu Y., Tang L. P., Zhang W. J., et al. (2020). Integration of pluripotency pathways regulates stem cell maintenance in the arabidopsis shoot meristem. Proc. Natl. Acad. Sci. U. S. A. 117, 22561–22571. doi: 10.1073/pnas.2015248117, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzaki T., Toriba T., Fujimoto M., Tsutsumi N., Kitano H., Hirano H.-Y. (2006). Conservation and diversification of meristem maintenance mechanism in Oryza sativa: function of the FLORAL ORGAN NUMBER2 gene. Plant Cell Physiol. 47, 1591–1602. doi: 10.1093/pcp/pcl025, PMID: [DOI] [PubMed] [Google Scholar]
- Tabuchi H., Zhang Y., Hattori S., Omae M., Shimizu-Sato S., Oikawa T., et al. (2011). LAX PANICLE2 of Rice encodes a novel nuclear protein and regulates the formation of axillary meristems. Plant Cell 23, 3276–3287. doi: 10.1105/tpc.111.088765, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takanashi H., Sumiyoshi H., Mogi M., Hayashi Y., Ohnishi T., Tsutsumi N. (2018). miRNAs control HAM1 functions at the single-cell-layer level and are essential for normal embryogenesis in Arabidopsis. Plant Mol. Biol. 96, 627–640. doi: 10.1007/s11103-018-0719-8, PMID: [DOI] [PubMed] [Google Scholar]
- Tanaka W., Hirano H. (2020). Antagonistic action of TILLERS ABSENT1 and FLORAL ORGAN NUMBER2 regulates stem cell maintenance during axillary meristem development in rice. New Phytol. 225, 974–984. doi: 10.1111/nph.16163, PMID: [DOI] [PubMed] [Google Scholar]
- Tanaka W., Ohmori Y., Ushijima T., Matsusaka H., Matsushita T., Kumamaru T., et al. (2015). Axillary meristem formation in Rice requires the WUSCHEL Ortholog TILLERS ABSENT1. Plant Cell 27, 1173–1184. doi: 10.1105/tpc.15.00074, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Kohlen W., Rossmann S., Vernoux T., Theres K. (2014a). Auxin depletion from the leaf axil conditions competence for axillary meristem formation in Arabidopsis and tomato. Plant Cell 26, 2068–2079. doi: 10.1105/tpc.114.123059, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Mai Y. X., Zhang Y. C., Luo Q., Yang H. Q. (2010). MicroRNA171c-targeted SCL6-II, SCL6-III, and SCL6-IV genes regulate shoot branching in arabidopsis. Mol. Plant 3, 794–806. doi: 10.1093/mp/ssq042, PMID: [DOI] [PubMed] [Google Scholar]
- Wang J., Tian C., Zhang C., Shi B., Cao X., Zhang T. Q., et al. (2017). Cytokinin signaling activates WUSCHEL expression during axillary meristem initiation. Plant Cell 29, 1373–1387. doi: 10.1105/tpc.16.00579, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wang J., Shi B., Yu T., Qi J., Meyerowitz E. M., et al. (2014b). The stem cell niche in leaf axils is established by auxin and cytokinin in Arabidopsis. Plant Cell 26, 2055–2067. doi: 10.1105/tpc.114.123083, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weits D. A., Kunkowska A. B., Kamps N. C. W., Portz K. M. S., Packbier N. K., Nemec Venza Z., et al. (2019). An apical hypoxic niche sets the pace of shoot meristem activity. Nature 569, 714–717. doi: 10.1038/s41586-019-1203-6, PMID: [DOI] [PubMed] [Google Scholar]
- Xin W., Wang Z., Liang Y., Wang Y., Hu Y. (2017). Dynamic expression reveals a two-step patterning of WUS and CLV3 during axillary shoot meristem formation in Arabidopsis. J. Plant Physiol. 214, 1–6. doi: 10.1016/j.jplph.2017.03.017, PMID: [DOI] [PubMed] [Google Scholar]
- Yadav R. K. R. K., Perales M., Gruel J., Girke T., Jonsson H., Reddy G. V., et al. (2011). WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes Dev. 25, 2025–2030. doi: 10.1101/gad.17258511, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Wang Q., Schmitz G., Müller D., Theres K. (2012). The bHLH protein ROX acts in concert with RAX1 and LAS to modulate axillary meristem formation in Arabidopsis. Plant J. 71, 61–70. doi: 10.1111/j.1365-313X.2012.04970.x, PMID: [DOI] [PubMed] [Google Scholar]
- Yoshida S., Mandel T., Kuhlemeier C. (2011). Stem cell activation by light guides plant organogenesis. Genes Dev. 25, 1439–1450. doi: 10.1101/gad.631211, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Fan L., Le B. H., Ye P., Mo B., Chen X. (2020). Regulation of ARGONAUTE10 expression enables temporal and spatial precision in axillary meristem initiation in Arabidopsis. Dev. Cell 55, 603.e5–616.e5. doi: 10.1016/j.devcel.2020.10.019, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T.-Q., Lian H., Zhou C.-M., Xu L., Jiao Y., Wang J.-W. (2017a). A two-step model for de novo activation of WUSCHEL during plant shoot regeneration. Plant Cell 29, 1073–1087. doi: 10.1105/tpc.16.00863, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Tucker E., Hermann M., Laux T. (2017b). A molecular framework for the embryonic initiation of shoot meristem stem cells. Dev. Cell 40, 264.e4–277.e4. doi: 10.1016/j.devcel.2017.01.002, PMID: [DOI] [PubMed] [Google Scholar]
- Zhang C., Wang J., Wenkel S., Chandler J. W., Werr W., Jiao Y. (2018). Spatiotemporal control of axillary meristem formation by interacting transcriptional regulators. Development 145, 1–10. doi: 10.1242/dev.158352, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Liu X., Engstrom E. M., Nimchuk Z. L., Pruneda-Paz J. L., Tarr P. T., et al. (2015). Control of plant stem cell function by conserved interacting transcriptional regulators. Nature 517, 377–380. doi: 10.1038/nature13853, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Yan A., Han H., Li T., Geng Y., Liu X., et al. (2018). Hairy meristem with wuschel confines clavata3 expression to the outer apical meristem layers. Science 361, 502–506. doi: 10.1126/science.aar8638, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]