Abstract
In the present study, we integrated liquid chromatography high-resolution mass spectrometry (LC-HRMS) and high-throughput DNA sequencing for prospecting cytotoxic specialized metabolites from Pseudofusicoccum stromaticum, an endophytic fungus associated to the medicinal plant Myracrodruon urundeuva. LC-HRMS profiling allowed identifying putatively eleven compounds in the ethyl acetate extract from P. stromaticum broth. Additionally, a chemical fractionation guided by cytotoxicity combined with spectrometric analysis resulted in the isolation of three compounds: the cyclopeptide cyclo-l-Phe-d-Leu-l-Leu-l-Leu-l-lle along with the known rotenoids rotenolone and tephrosin. MTT assay showed that tephrosin (IC50 0.51 μg mL−1) has strong cytotoxic effect and may be pointed out as the compound responsible for the antiproliferative activity of P. stromaticum. Next Generation Sequencing (NGS) and genome mining of P. stromaticum draft genome revealed 56 contigs codifying specialized metabolites biosynthesis-related enzymes. Nearly half of such genes (44.6%) could be mapped to orphan Biosynthetic Gene Clusters (BGCs) of related plant pathogens belonging to family Botryosphaeriaceae. Also, screening for rotenoids biosynthetic enzymes led to characterization of a putative chalcone isomerase-like (CHI-like) protein. This is the first report of rotenoids biosynthesized by a fungus, unveiling a unique ability of P. stromaticum.
Pseudofusicoccum stromaticum produces cyclopeptides and plant-related rotenoids, which are responsible for its antiproliferative effect.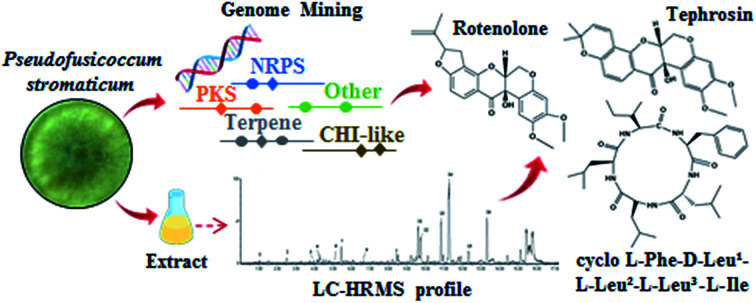
Introduction
The fungal kingdom includes many species with unique and unusual biochemical pathways, which may lead to biosynthesis of diverse metabolites used as pharmaceuticals, as for instance the antibiotic penicillin, the immunosuppressant cyclosporin and the cholesterol-lowering drugs statins.1 In this way, endophytic fungi are particularly attractive not only because they produce their own bioactive substances, but are also able to biosynthesize secondary metabolites from host plants such as the antineoplastic paclitaxel, camptothecin, and podophyllotoxin.2,3 Indeed, medicinal plants are interesting sources for prospecting fungal endophytes with biological potential.4
In order to expedite the discovery of microorganisms producing pharmacologically active compounds, the modern natural products research have relied on an approach that combines the usage of powerful analytical techniques (e.g. liquid chromatography coupled to high-resolution mass spectrometry, LC-HRMS) with genome sequencing technologies (e.g. Next-Generation Sequencing, NGS).5,6
The LC-HRMS analysis is a sensitive method that provides the isotope ratios and accurate masses of quasi-molecular ions leading to only one or few possible elemental compositions. Then, the putative identification of previously reported fungal metabolites may be achieved after searching for these few candidates in natural product databases.7 Nowadays, the annotation of secondary metabolites has been facilitated by surveying powerful public and private databases such as Dictionary of Natural Products, MarinLit Metlin, Massbank, NIST, some of them housing MS spectra of authentic samples,8 besides in silico MS/MS libraries such as CSI:FingerID and MetFRAG,9 and even until a crowdsourced MS/MS platform as Global Natural Products Social.10
Likewise, NGS is combined with bioinformatics tools to reveal biosynthetic gene clusters (BGC's) existing in the genome sequence as well as the enzymes encoded in the gene clusters, enabling the prediction of putative metabolites. Currently, there are many online platforms and databases for mining BGC's such as AntiSMASH and MIBiG.11,12 Therefore, the integrated use of ‘omics’ approaches has been considered the renaissance of natural product research, since it has opened new opportunities among them to engineer genetically organisms for producing molecules of great pharmacological interest with higher yield.13,14
Previous studies have reported highly cytotoxic compounds towards cancer cell lines isolated from endophytic fungi.15,16 In our seeking for natural products with anticancer potential, we found a strain of Pseudofusicoccum stromaticum as endophyte in Myracrodruon urundeuva Fr. All. (Anacardiaceae). P. stromaticum is a fungus belonging to Botryosphaeriaceae family, while M. urundeuva is a medicinal plant with anti-inflammatory and cicatrizing properties widespread in Northeast of Brazil.17 Although the host-plant has been relatively well studied with regard to phytochemical and pharmacological features, no study has been reported for this fungus.
In the present study, we describe the chemical and genomic characterization of a P. stromaticum strain along with its cytotoxicity. LC-HRMS profiling permitted to identify putatively eleven compounds in the ethyl acetate extract from P. stromaticum broth. Additionally, a cytotoxic assay-guided fractionation of this extract resulted in the isolation of three compounds: the new cyclopeptide l-Phe-d-Leu-l-Leu-l-Leu-l-lle (1) besides known rotenoids rotenolone (2) and tephrosin (3). The latter one showed strong cytotoxic effect against a human colorectal cancer cell line (HCT-116). Furthermore, this is the first report of rotenoids biosynthesized by a fungus. Genome mining supported the chemical characterization, revealing 56 contigs codifying secondary metabolites biosynthesis-related enzymes, including nonribosomal peptide-synthetase (NRPS), terpene and type I-polyketide (T1PKS) synthases. Moreover, a putative chalcone isomerase-like protein expected to be enzymatically active and to recognize a chalcanoid substrate other than (2S)-nangerin was retrieved from P. stromaticum genome and may be engaged in rotenoids production in that fungus.
Results and discussion
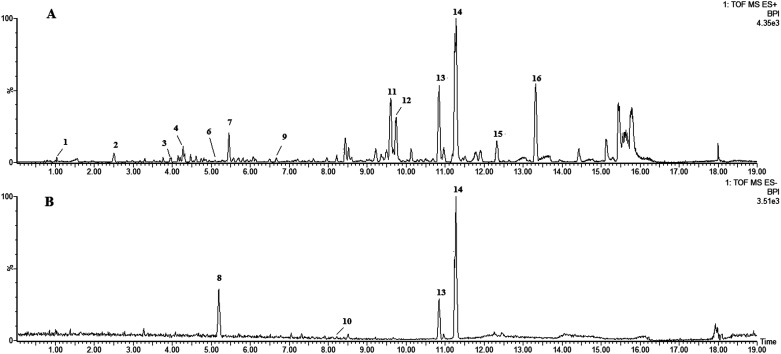
LC-HRMS profiling of P. stromaticum was then performed for the chemical characterization of this fungus (Fig. 1), which was driven by the cytotoxicity evaluation against the HCT-116 cell line of human colon carcinoma. Broth extract presented cytotoxic activity (IC50 = 10.40 μg mL−1), whereas mycelium extract did not show any inhibitory effect on that cell line.
Fig. 1. UPLC-QTOF-MS-MS chromatograms of the ethyl acetate extract from Pseudofusicoccum stromaticum: (A) positive ionization mode and (B) negative mode.
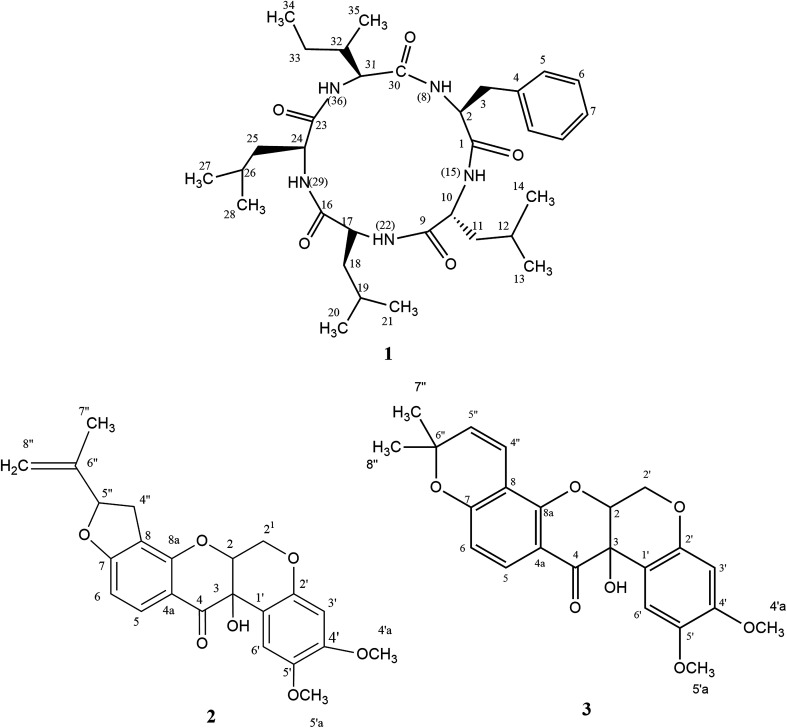
Eleven compounds were putatively identified (Fig. 1 and Table 1) from the broth: choline-O-sulfate, xanthofusin, pestaloficiol W, multicolic acid, 8-(methoxycarbonyl)-1-hydroxy-9-oxo-9H-xanthene-3-carboxylic acid, djalonensone, pestalotiopyrone C, cyclo-Phe-Leu-Val-Leu-Leu, along with compound 1–3 (Fig. 2).
Metabolites of the ethyl acetate extract from Pseudofusicoccum stromaticum characterized by UPLC-q-TOF.
| Peak no. | t R (min) | m/z [M + H]+ | m/z [M − H]− | m/z [M + Na]+ | Molecular formula | m/z calcd | Error (ppm) | i-FIT (norm) | Compound | Fragments | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0.85 | 184.0646 | — | — | C5H13NO4S | 184.0644 | 1.1 | 0.4 | Choline-O-sulfate | — | 59 and 60 |
| 2 | 2.50 | 306.0558 | — | — | C21H7NO2 | 306.0555 | 1.0 | 3.2 | Unknown | — | — |
| 3 | 3.96 | — | — | 481.2625 | C20H42O11 | 481.2625 | 0.0 | 1.6 | Unknown | — | — |
| 4 | 4.20 | — | — | 191.0324 | C8H8O4 | 191.0320 | 2.1 | 0.6 | Xanthofusin | 126 | 19 |
| 5 | 4.28 | — | 217.0479 | C10H10O4 | 189.0528 | −0.9 | 1.0 | Pestaloficiol W | 189 | 21 | |
| 6 | 5.08 | 243.0867 | — | — | C11H14O6 | 243.0869 | −0.8 | 1.0 | Multicolic acid | 189 | 19 |
| 7 | 5.45 | 191.0671 | — | — | C9H12O3Na | 191.0684 | −6.3 | 0.2 | Pestalotiopyrone C | 176, 151 | 22 |
| 8 | 5.19 | — | 207.0253 | — | C5H7N2O7 | 207.0253 | −8.7 | 0.3 | Unknown | — | — |
| 9 | 6.67 | 273.0748 | — | — | C15H12O5 | 273.0763 | −5.5 | 0.0 | Djalonensone | — | 59 |
| 10 | 8.13 | — | 313.0352 | — | C16H10O7 | 313.0348 | 1.3 | 1.4 | 8-(Methoxycarbonyl)-1-hydroxy-9-oxo-9H-xanthene-3-carboxylic acid | 248 | 25 and 60 |
| 11 | 9.60 | — | — | 433.1272 | C23H22O7 | 433.1263 | 2.1 | 0.1 | Rotenolone (2) | 410, 433, 393, 365 | 29 |
| 12 | 9.76 | — | — | 433.1273 | C23H22O7 | 433.1263 | 2.3 | 0.8 | Tephrosin (3) | 410, 433, 393, 365 | 28 |
| 13 | 10.84 | — | — | 608.3793 | C32H51N5O5 | 608.3788 | 0.8 | 0.8 | Cyclo-Phe-Leu-Val-Leu-Leu | 120, 261, 360, 473, 558, 586, 608 | 61 |
| 14 | 11.28 | 600.4107 | — | 622.3952 | C33H53N5O5 | 600.4125 | −3.0 | 0.4 | Cyclo-l-Phe-d-Leu-l-Leu-l-Leu-l-lle (1) | 120, 261, 374, 487, 572, 600 | — |
| 15 | 12.32 | — | — | 602.4296 | C36H57N3O3 | 602.4291 | −0.3 | 4.2 | Unknown | — | — |
| 16 | 13.32 | 425.2102 | — | — | C29H28O3 | 425.2117 | −3.1 | 4.1 | Unknown | — | — |
Fig. 2. Structures of compounds 1–3.
Choline-O-sulfate is a common metabolite in fungi, accounting for up to 0.6% in Aspergillus and Penicillium species. This compound is a reservoir of C, N and S that prevents the cell starvation during elemental depletion events, besides its role seems to be related to osmoprotection.18
Xanthofusin is a tetronic acid isolated from Fusiccoccum sp. that exhibits strong antifungal activity against the phytopathogen Phytophthora infestans. Multicolic acid is found in Penicillium sclerotiorum and considered a quorum sensing molecule, namely, a compound able to induce intra and interspecific microbial responses. Xanthofusin and multicolic acid are butyrolactones formed in vivo via oxidative cleavage of polyketide-derived aromatic intermediates.19,20
Pestaloficiol W and pestalotiopyrone C are polyketides reported previously for Pestalotiopsis. This genus consists of endophytic fungi commonly isolated from higher tropical plants and its BGC's encode chiefly PKS.21,22
Djalonensone (also named alternariol 9-methyl ether) is a dibenzo-α-pyrone biosynthesized via polyketide synthase and isolated previously from many endophytic fungal strains of Alternaria and Colletotrichum, besides the plant Anthocleista djalonesis.23 The cytotoxicity of djalonensone was demonstrated against SW1116 human colon adenocarcinoma cells (IC50 = 14.0 μg mL−1).24
The xanthone 8-(methoxycarbonyl)-1-hydroxy-9-oxo-9H-xanthene-3-carboxylic acid was isolated from Penicillium sp. and tested against KB and KBv200 cells earlier, however it did not exhibit any cytotoxic effect.25 Xanthones are also biosynthesized through polyketide synthase pathway.
Afterwards, sephadex fractionation of the broth extract yielded three fractions, whose cytotoxicity was evaluated at the concentrations of 5 and 50 μg mL−1: fraction A had a cell growth inhibition of 2% and 8.5%, B suppressed 17% and 45%, while fraction C inhibited 66% and 94%, respectively. Since the fractions B and C were the two most active ones, we refractionated them by SPE cartridges and HPLC, resulting in the isolation of compounds 1–3. Their structures were determined by interpretation of its 1D and 2D NMR spectra (ESI Fig. S1–S21†), HRESIMS experiments and comparison with published data.
Compound 1 (Fig. 2) was isolated as a white amorphous solid (mp 276.1 °C; [α]20 −13.0) and its HRESIMS exhibited a molecular ion peak at m/z 600.4107 [M + H]+ indicating the molecular formula C33H54N5O5 (calcd for m/z 600.4125, 3.0 ppm), hence an unsaturation degree of nine. The FTIR spectrum displayed bands compatible with absorptions from amide (3320 and 1712 cm−1, axial deformations of N–H and C O) and aromatic groups (1648 and 1537 cm−1). Moreover, the NMR data were consistent with a peptide structure. In fact, the 1H NMR spectrum (ESI Fig. S1†) showed signals to five amide hydrogens at δH 8.51 (H-8, Phe), 8.62 (H-15, Leu1), 8.51 (H-22, Leu2), 7.50 (H-29, Leu3) and 8.32 (H-36, lle) indicating five amino acid moieties, supported by the set of signals at δH 4.65 (t, J = 7.8 Hz, H-2), 4.42 (t, J = 15.0 Hz, H-24), 4.25 (dd, J = 10.8 and 4.8 Hz) and 4.20 (t, J = 7.8 Hz, H-10) corresponding to five azomethine hydrogens. In addition, a multiplet at δH 7.19–7.25 (5H) along with signals at δH 0.71–2.38 were assigned for a monosubstituted aromatic ring and alkyl groups, respectively. 13C NMR and HMBC-NMR spectra revealed signals to five amide carbonyls at δC 172.40 (C-1-Phe), 173.1 (C-9-Leu1), 173.2 (C-16-Leu2), 172.6 (C-23-Leu3) and 172.5 (C-30-lle) which showed cross-peaks with the amide hydrogens: δH 8.62 (H-15-Leu1) → δC 172 40 (C-1-Phe), δH 8.51 (H-22-Leu2) → δC 173.1 (C-9-Leu1), δH 7.50 (H-29-Leu3) → δC 173.2 (C-16-Leu2). The HMBC spectrum was also crucial to define the complete assignments of each amino acid residue. Additionally, the sequence of aminoacid residues was ratified by MS fragments observed in the UPLC-ESI-QTOF spectrum: m/z 147 (Phe), m/z 113 (Leu) and m/z 113 (lle). Finally, acid hydrolysis followed by chiral GC-MS analysis enabled to determine the configuration of the aminoacids residues of 1 as l-Phe-d-Leu1-l-Leu2-l-Leu3-l-lle. In our cytotoxicity assay, compound 1 was unable to inhibit the growth of HCT-116 cells. However, in a previous study, its epimer (cycle l-Phe-l-Leu-l-Leu-l-Leu-l-lle) isolated from an unidentified endophytic fungus showed inhibitory activity of 67% against Bel-7402 cell at a concentration of 15 μg mL−1.26 Biosynthetically, the cyclopeptides l-Phe-d-Leu1-l-Leu2-l-Leu3-l-lle (1) together with cyclo-Phe-Leu-Val-Leu-Leu are derived from NRPS pathway.
Compounds 2 (yellow powder, mp 139.6–141.0 °C; [α]21D −24.84) and 3 (yellow resin, [α]21D −7.06) were characterized as isomers according to LC-MS analysis, exhibiting the molecular formula C23H22O7Na (m/z 433.1272 for 2 and m/z 433.1273 for 3, calcd for m/z 433.1263). Their 1H and 13C NMR resonances suggested to be from prenylated flavonoids. HSQC and HMBC NMR correlations revealed that 2 and 3 were the rotenoids rotenolone and tephrosin, respectively, in agreement with literature data.27,28 Also, their stereochemistry was based on NMR data and optical rotation.27–29
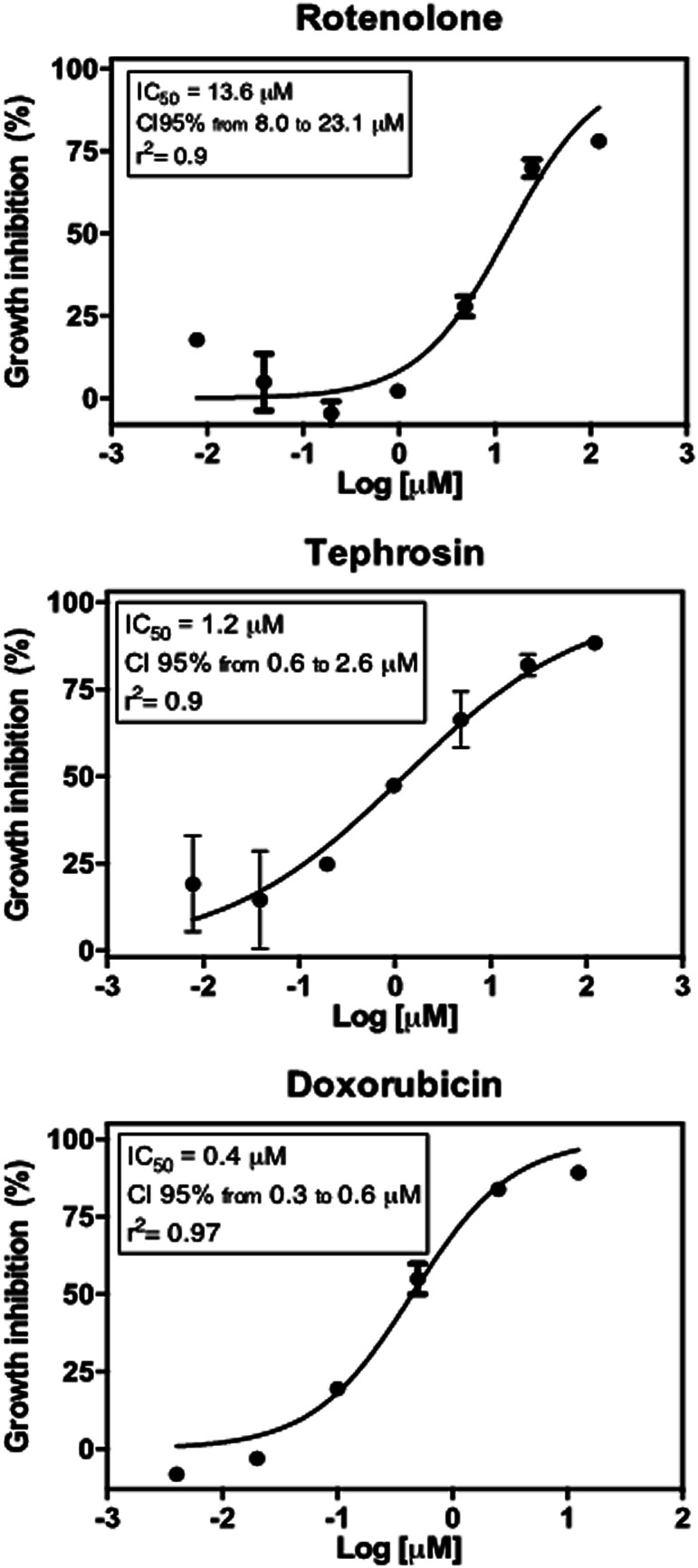
Tephrosin (3) displayed significant cytotoxic effect against the HCT 116 cancer cells with IC50 value of 1.2 μM, while compound rotenolone (2) was moderately active against this cell line (IC50 = 13.6 μM) (Fig. 3). The cytotoxicity of rotenoids has been well-documented in the literature. For instance, rotenolone (2) had moderate antiproliferative activity on many cancer cell lines such as A2780 cells (ovarian), BT-549 (breast), DU 145 NSCLC NCI-H460 (prostate) and HCC-2998 (colon).29 In contrast, Mittraphab et al. (2018) reported the potent growth inhibition of rotenolone (2) and tephrosin (3) against tumoral (HCT-116) and normal colon cells (CCD841).30 Furthermore, in spite of high cytotoxicity against colon cells, both rotenolone and tephrosin inhibited weakly hepatocarcinoma (HepG2) and cervical carcinoma (CaSki) cells.30 Nevertheless, the anti-angiogenic effect on HUVEC cells and apoptotic effects of these compounds were further observed against the cancer cells HepG2, C26, LL2 and B16.31 Earlier, Choi et al. (2010) demonstrated that tephrosin (3) exerts its antitumor effect on the human colon cancer cells (HT-29) by inducing internalization and degradation of inactivated epidermal growth factor receptors.32
Fig. 3. Antiproliferative effect of rotenolone and tephrosin against human colon cancer cells (HCT 116) after 72 h incubation by the MTT assay. Doxorubicin was used as positive control. Graphs are presented as concentration–effect curves. Insets show inhibition concentration mean (IC50) values along with their respective confidence intervals of 95% (CI95%) and r square (r2) values.
The rotenoids rotenolone (2) and tephrosin (3) are likely biosynthesized from a polyketide synthase. So far, rotenoids have been found only in plants, mainly species from the genera Derris and Tephrosia (Leguminosae).33,34 Therefore, this is the first report of a fungus producing of rotenoids. On the other hand, there is no report of rotenoids in M. urundeuva, the host plant of the fungus studied. Furthermore, when an ethanol extract obtained from the same plant was analyzed by LC-MS, we did not detect the rotenoids 2 and 3 (data not shown).
In order to evaluate the encoded secondary metabolome, we performed Illumina DNA sequencing of P. stromaticum genomic DNA. A total of approximately 1.5 gigabase pairs (Gbp) of sequence data which was assembled in 2571 contigs by Spades.35 The total contigs size is about 42 Mbp and the coverage was 8.48×, with a G + C% of 57.03. Assembly quality was evaluated using QUAST,36 leading to L50 and N50 values of 5775 and 2369, respectively. Genome completeness evaluation of the genome assemblage was then performed by searching of a set of fungal Benchmarking Universal Single-Copy Orthologs from OrthoDB.37 BUSCO analysis revealed that 92.4% of BUSCOs could be found in the P. stromaticum draft genome, being 32.4% of them fragmented. This data highlights the quality of the sequencing and assembly process.
We automatically annotated the P. stromaticum genome using an unsupervised training in Genemark-ES.38 A total of 14 421 genes could be detected. The number of exons in the genes varies from 1 to 27, with an average exon number of 2.83. From the complete predicted proteome, 91.41% displayed at least one conserved domain according to InterProScan analysis (ESI Table 1†).
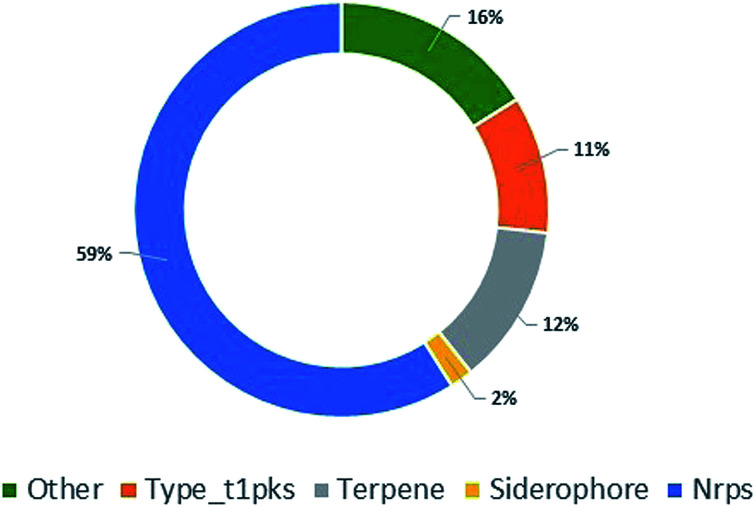
To further explore P. stromaticum secondary metabolome, the annotated draft genome was submitted to the Antibiotics & Secondary Metabolite Analysis SHell (antiSMASH) server for Biosynthetic Gene Cluster (BGC) prediction. Fifty-six contigs were shown to codify for non-ribosomal peptide synthetases (NRPS), type I polyketide synthase, terpene synthase, among other, representing segments of these classes of BGCs (Fig. 4). NRPS and type I PKS, representing respectively 59 and 11% of the contigs from P. stromaticum secondary metabolome. These are classes of large multifunctional and modular enzymes that build the core structures of complex peptides and polyketides, respectively from simple amino acid and malonyl building blocks. In NPRS and type I PKS co-evolved modularity, three basal core domains perform each step of compound elongation: adenylation (A)/acyltransferase (AT) domain recognize the specific amino acid/acetyl-CoA substrate of specificity and activates it through a thioester linkage to the peptide/acetyl – carrier protein domain (PCP/ACP), and the condensation (C)/ketosynthase (KS) domain catalyze the peptide/carbon–carbon bond for elongating the compound with one more building block. Scaffolds can also undergo further modifications as heterocyclization and, epimerization in NRPS and reduction and/or methylation of ketone and carbonyl groups in PKS.1,10
Fig. 4. Pie chart showing the classes of biosynthetic gene clusters (BGCs) for specialized metabolites found in P. stromaticum genome.
Core biosynthetic genes from fifty-two of the found contigs (92%) presented hit (∼41% amino acid sequence identity and 93% protein coverage) against referential proteins cataloged on the Minimum Information about a Biosynthetic Gene cluster (MIBiG) repository (ESI Table 2†). No compound-BGC matching was expected since none of the chemically detected compounds has their respective BGCs deposited on MIBiG repository or, for the best of our knowledge, has been yet characterized. However, seventeen core biosynthetic genes coding to NRPS enzymes (51%) presented hit to multi-modular NRPS enzymes involved in the biosynthesis of cyclic peptides, including: the cyclic tetrapeptides cyclosporin (SimA), fungisporin (HcpA) and apicidin (APS1) as well as the cyclohexadepsipeptide destruxins (DtxS1) (ESI Table 2†).39–42
Other 10 NRPS core genes (30%) presented homology to hybrid polyketide synthase/nonribosomal peptide synthetase LcsA (8 related core genes) and EasA (2 related core genes), respectively related to production of leucinostatin A and emericellamide A.43,44 Finally, 5 NRPS genes (15%) presented similarity to the trimodular NRPS Afu6g12080, which is associated to biosynthesis of fumiquinazoline (FQ) peptidyl alkaloids (ESI Table 2†).45 Furthermore, 3 of the 23 adenylation (A) domains detected at P. stromaticum NRPS modules, had their substrate of specificity predicted by antiSMASH as Leu amino acids residues, one of which is related with the biosynthesis of cyclopeptides. Therefore, this gene is potentially involved in biosynthesis of the cyclo-l-Phe-d-Leu-l-Leu-l-Leu-l-lle (1), and so, good target for specific gene mutation/deletion experiments followed by new chemical characterization, in order to confirm gene product–compound correlation and further characterize the entire BGC.
When considering the non-redundant protein sequences database (nr, NCBI), approximately 44.6% of the proteins encoded on contigs of P. stromaticum secondary metabolome presented higher similarities (BLASTp) to NRPS, PKS and terpene BGCs presented at the genome of Neofusicoccum parvum UCRNP2 and Diplodia corticola, both plant pathogens endophytic fungi also grouped at the family Botryosphaeriaceae (ESI Table 2†). Indeed, 16 (∼29%) of those contigs could actually be mapped to these fungi BGCs (with ∼75% pairwise identity) (ESI Table 2†). Interestingly, six of the NRPS contigs presenting similarity to the leucinostatin A biosynthetic NRPS, LcsA, were mapped exactly on same order to the genes UCRNP2_4860 and MPH_12472l, coding to highly similar (E value = 0, 78% identities and 85% positives) 9 modules NRPS respectively signed as hypothetical protein and a putative aminoadipate-semialdehyde dehydrogenase of N. parvum and D. corticola (ESI Table 2,† for N. parvum gene). When analyzed at antiSMASH, the contig containing UCRNP2_4860 showed similarity (∼20%) to leucinostatin A BGCs. Therefore, our results suggest that, as plant pathogenic Botryosphaeriaceae N. parvum and D. corticola, P. stromaticum genome encodes a BGCs related with leucinostatin A BGC. No lipopeptide could be detected on our chemical characterization of this fungus, although such result can be related with a multitude of factors, including BGC silence under the used growth conditions.
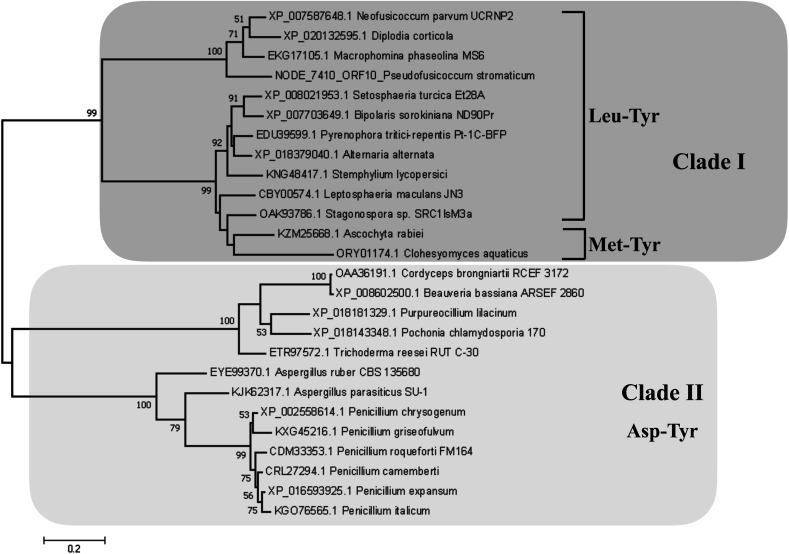
Detection of rotenoids on P. stromaticum chemical extracts was intriguing. In plants, rotenoids are formed from the biosynthesis of flavonoids, a multi-step pathway that has among its key enzymes chalcone synthase (CHS) and chalcone isomerase (CHI).46 Recently, overexpression of CHS and CHI was correlated with enhancement of rotenoids production in transgenic culture lines of the Tibetan herbal plant Mirabilis himalaica.47 Based on such information, we performed local BLAST searches using plant-derived CHS and CHI proteins as query against P. stromaticum draft genome to retrieve a CHI-like protein of 318 amino acid residues (Fig. 5), while no CHS homolog could be found. PSI-BLAST using P. stromaticum CHI-like protein as query recruited highly similar (query cover ≥ 98%, protein identity ≥ 76%) putative CHI-like proteins from N. parvum, D. seriata and Macrophomina phaseolina, besides other distantly related fungi CHI-like proteins (∼30% identity). Alignment of 26 selected fungi CHI-like proteins with Medicago sativa chalcone–flavonone isomerase (GenBank accession P28012) showed that fungi and plant enzymes share several blocks of sequence conservation, including key amino acid residues of secondary structure forming CHI substrate-binding cleft,48 keeping the hydrophobicity of the cleft highly conserved (Fig. 5). Maximum likelihood phylogenetic reconstitution grouped fungi CHI-like enzymes into two main clades, one including a subclade formed by proteins from Botryosphaeriaceae species and with P. stromaticum Chi-like homolog as outgroup (Fig. 6). Interestingly, the two substrate-specificity determining amino acid residues in plant CHI: Thr 190 and Met 191 for chalcone, or Ser–Ile respectively for 6′-deoxychalcone, are consistently replaced by Tyr–Phe in clade I or Asp–Phe in clade II, indicating that: (i) fungi CHI-like enzymes recognize chalcanoids other than (2S)-naringenin and (ii) have more than one substrate specificities (Fig. 6). These results corroborate previous genome mining analysis showing that fungi and bacteria orthologues of plant CHI retain the enzyme family fold and key catalytic amino acid residues preserved, and so, are assumed to be enzymatically active proteins.49 Additionally, detection of CHI but not of CHS in P. stromaticum genome is also sustained, since these genes are found in exclusion one to the other in fungi genomes, a perspective that also corroborates that fungal CHI-like enzymes substrate is not chalcone.49 Interestingly, M. urundeuva, produces chalcones (urundeuvines),17 and such chalcone or other plant-host derived precursor could be used by P. stromaticum rotenoids biosynthesis.
Fig. 5. Alignment of fungi CHI-like and referential plant CHI amino acids sequences. Legend: 1 – Medicago sativa, P28012, 2 – Pseudofusicoccum stromaticum; 3 – Diplodia corticola, XP_020132595.1; 4 – Neofusicoccum parvum UCRNP2, XP_007587648.1; 5 – Macrophomina phaseolina MS6, EKG17105.1; 6 – Alternaria alternata, XP_018379040.1; 7 – Pyrenophora tritici-repentis Pt-1C-BFP XP_001930494.1; 8 – Bipolaris sorokiniana ND90Pr, XP_007703649.1; 9 – Setosphaeria turcica Et28A, XP_008021953.1; 10 – Stemphylium lycopersici, KNG48417.1; 11 – Leptosphaeria maculans JN3, XP_003844053; 12 – Stagonospora sp. SRC1lsM3a, OAK93786.1; 13 – Ascochyta rabiei, KZM25668.1; 14 – Clohesyomyces aquaticus ORY01174.1; 15 – Trichoderma reesei RUT C-30, ETR97572.1; 16 – Purpureocillium lilacinum, XP_018181329.1; 17 – Pochonia chlamydosporia 170, XP_018143348.1; 18 – Cordyceps brongniartii RCEF 3172, OAA36191.1; 19 – Beauveria bassiana ARSEF 2860, XP_008602500.1; 20 – Penicillium chrysogenum XP_002558614.1; 21 – Penicillium griseofulvum, KXG45216.1; 22 – Penicillium expansum, XP_016593925.1; 23 – Penicillium italicum, KGO76565.1; 24 – Penicillium camemberti, CRL27294.1; 25 – Penicillium roqueforti FM164, CDM33353.1; 26 – Aspergillus ruber CBS 135680, EYE99370.1; 27 – Aspergillus parasiticus SU-1, KJK62317.1. Secondary structure of CHI from Medicago sativa (alfalfa; P28012) is shown on top of the figure, with α-helices and β-strands shown in gold and blue rectangles respectively. α-helices and β-strands forming the substrate-binding clef are highlighted in bold.
Fig. 6. Maximum Likelihood reconstituted tree of fungi chalcone isomerase-like (CHI-like) proteins. The tree with the highest log likelihood (−5855.99) is shown. Percentages (>0.5) of trees in which the associated taxa clustered together is shown next to the branches. All positions containing gaps and missing data were eliminated. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. All positions containing gaps and missing data were eliminated. There was a total of 275 positions in the final dataset. Main clades (clade I and II) are highlighted in dark and light shade boxes. Substrate-specificity determining amino acid residues, according to plant CHI crystallography are shown for each fungi CHI-like clade or subclade.
There are several examples of endophytic fungi capable to biosynthesize secondary metabolites from their host plants. For instance, the biosynthesis of the antineoplastic compound camptothecin by the fungus Fusarium solani involves key biosynthetic plant enzyme.3 Also, the anticancer natural drugs paclitaxel and vincristine, isolated originally from plants Taxus brevifolia and Catharanthus roseus, were also shown to be produced by their respective endophytes Pestalotiopsis microspora and Fusarium oxysporum, as well as in fungi colonizing plants not producing these metabolites.2,3
Experimental
General experimental procedures
The 1D and 2D NMR spectra were accomplished on an Agilent DD2 spectrometer equipped with a 5 mm One Probe, operating at 600 MHz for 1H NMR and 150 MHz for 13C NMR. The chemical shift values (δ) were expressed in parts per million (ppm) and coupling constants in Hz. The NMR spectra of compound 1 were recorded in 600 μL of CD3OD (Cambridge Isotope Laboratories-CIL, 99.8%), whereas compound 2 and 3 were solubilized in CDCl3 (CIL, 99.8%). Optical rotations were measured on a JASCO P-2000 polarimeter at 20 °C using methanol or chloroform as solvent according to solubility. LC-HRMS analysis were performed in an Acquity Xevo UPLC-ESI-QTOF system (Waters) fitted to a Waters BEH C-18 column (1.7 μm, 2.1 × 150 mm) at 40 °C, injecting 5 μL of sample. The mobile phase was a combination of A (0.1% formic acid in water) and B (0.1% formic acid in acetonitrile), ranging from 5 to 95% of B (v/v) for 15 min at a flow rate of 0.4 mL min−1. Mass spectra were recorded in positive and negative modes in a mass range of 50–1180 Da. In prior to run, all samples and solvents were filtered through 0.22 μm PTFE membranes (Simplepure). LC-MS grade acetonitrile was purchased from Sigma-Aldrich. The putative metabolite annotation of the broth extract was achieved according to the molecular formulas provided by MassLynx 4.1 software, taking into account their accurate masses (error > 5 ppm) and isotopic patterns (i-fit), as well as their MS fragmentation patterns when compared with the in silico MS/MS database CSI:FingerID.9 Furthermore, the annotation of the compounds was supported by literature survey (Scifinder database) on the previous occurrence in Botryosphaeriaceae fungi, mainly in those with higher identity of BGC's according to the genome mining. Additionally, a blank sample was injected in order to discard possible contaminants.
The compounds 1–3 were isolated on a Waters 2489 HPLC chromatograph coupled to a Waters 2555 UV detector set at 254 nm. The chromatographic separations were achieved on Waters Sunfire preparative column C-18 OBS (19 mm × 100 mm, 5 μm), using an isocratic mobile phase of H2O–MeOH (30 : 70) at a flow rate of 16 mL min−1 and injecting 100 μL of sample per run. All samples and solvents were filtered through 0.45 μm PTFE membranes (Simplepure) before the run. HPLC grade methanol was from Tedia. SPE cartridges were Chromabond C-18 (Macherey-Nagel), containing 500 mg and 1 g of adsorbant. The FTIR analysis was performed on a Varian/Agilent Model 660-IR instrument in ATR mode (4000–800 cm−1), with a resolution of 4 cm−1 by collecting 32 scans. GC-MS analysis was performed on a CG-7890B/MSD-5977A Agilent instrument fitted to a CP-Chirasil-l-Val column (Varian 25 mm × 0.25 mm, 0.12 μm), using helium carrier gas at a flow rate of 1.00 mL min−1, injector temperature of 200 °C, detector temperature of 150 °C, transfer line temperature of 280 °C. Chromatographic oven was set to 100 °C for 1 min, next it was raised to 180 °C at 5 °C min−1 and kept constant at 180 °C for 5 min.
Isolation, identification and growth of the fungus
The fungus was isolated from Myracrodruon urundeuva Fr. All. (Anacardiaceae) branches collected in Tauá County (Ceará State, Brazil-S 05°53′05.7′′ W 40°28′03.8′′, at 304 m above sea level) and identified as Pseudofusicoccum stromaticum by DNA sequencing of the regions ITS1/ITS4.50 The isolate has been deposited in the collection of microorganisms from Embrapa Agroindústria Tropical. The isolate was grown on plates containing 2% peptone, malt dextrose agar (MDPA, Becton, Dickson and Company) and 2 mg mL−1 of chloramphenicol (Sigma), agar (Oxoid) at 25 ± 2 °C with periods of 12 h of light and 12 h of dark during six days. The colonies formed on MDPA were transferred to plates containing agar (AA, Oxoid) and incubated at 26 ± 1 °C under ultraviolet light for 15 days to induce sporulation. Afterwards, P. stromaticum strain was grown in six erlenmeyers of 2 L, each containing 250 mL of potato dextrose culture medium (BD), 200 g and 20 g potato dextrose for 21 days at 25 ± 2 °C.
Obtaining of P. stromaticum extracts
P. stromaticum extract was obtained by liquid–liquid partition with ethyl acetate (AcOEt) of the broth. The broth was separated from the mycelium by vacuum filtration and next partitioned with ethyl acetate (AcOEt, 3 × 400 mL). The AcOEt phases were pooled, treated with anhydrous NaSO4, filtered and distilled on an IKA RV10 rotaevaporator at 40 °C, yielding a dark solid (770 mg).
Isolation of the compounds 1–3
P. stromaticum AcOEt extract (750 mg) was chromatographed on Sephadex LH-20 glass column (Sigma-Aldrich), using MeOH as eluent. The 15 fractions obtained were pooled into three fractions: A (37.9 mg), B (403.1 mg) and C (84.8 mg). Fraction B was fractionated on a SPE-C18 (1 g) cartridge and eluted with a gradient of H2O : MeOH (100 : 0; 75 : 25; 50 : 50; 25 : 75; 0 : 100), yielding five subfractions respectively: B1 (208.7 mg); B2 (8.5 mg); B3 (9.2 mg); B4 (24.1 mg) and B5 (34.9 mg). The fraction B5 was purified on a SPE-C18 cartridge (500 mg), providing a white solid named compound 1. Fraction C (IC50 2.91 μg mL−1), which was the most bioactive fraction, was fractionated on SPE-C18 cartridge and eluted with a gradient H2O : MeOH (100 : 0; 90 : 10; 80 : 20; 60 : 40; 50 : 50; 40 : 60; 30 : 70; 20 : 80; 10 : 90; 0 : 100) to give 10 subfractions (C1–C10). Subfraction C7 fraction was subjected to separation by HPLC (MeOH : H2O; 70 : 30 isocratic), to afford compounds 2 and 3.
Absolute configuration cyclopeptide
The absolute configuration of the amino acids residues was determined according to method described earlier.51 Briefly, Compound 1 (100 μg) was hydrolyzed with HCl 6 (500 μL) at 110 °C for 24 h, treated with 10% HCl/MeOH (500 μL) at 100 °C for 30 min and dried with nitrogen gas. Next, the mixture of amino acids was reacted with acetic anhydride/CH2Cl2 (1 : 1, 500 μL) at 100 °C for 5 min. Finally, it was dried in nitrogen gas, dissolved in dichloromethane and filtered through a 0.45 nm nylon filter. Then, 1 μL of sample was analyzed by GC-MS. This same procedure was performed for the l-amino acids standards, except for hydrolysis with HCl. The absolute configuration was based on their retention times.
Spectrometric data of the metabolites
Cyclo l-Phe-d-Leu1-l-Leu2-l-Leu3-l-lle (1)
Solid amorphous. mp 276.1 °C. [α]20D = −13.06 (c. 0.16; MeOH). HRESI MS (+) m/z: 622.3963 ([M + Na]+ calculated for C33H53N5O5Na 622.3944). IR νmax: 3320, 3270, 2930, 2865, 1712, 1648, 1537, 1448, 1377, 1255, 1077 cm−1, 1H NMR (600 MHz, CD3OD): Phe: 4.65 (1H, t, J = 7.8 Hz, H-2), 3.00 (1H, dd, J = 7.2 Hz, 13.2 Hz; H-3a), 2.87 (1H, dd, J = 7.2 Hz; 13.8 Hz, H-3b), 7.22 (2H, m, H-5), 7.25 (2H, m, H-6), 7.19 (1H, m, H-7), 8.51 (NH, H-8), Leu1: 4.20 (1H, t, J = 7.8 Hz, H-10), 1.47 (1H, m, H-11a), 1.41 (1H, m, H-11b), 1.32 (1H, m, H-12), 0.83 (3H, m, H-13), 0.89 (3H, m, H-14), 8.62 (NH; H-15), Leu2: 4.25 (1H, dd, J = 4.8 Hz; 10.8 Hz, H-17), 1.62 (2H, m, H-18), 1.67 (1H, m, H-19), 0.94 (3H, d, J = 5.4 Hz, H-20), 0.88 (3H, d, J = 6 Hz, H-21), 8.51 (NH; H-22), Leu3: 4.42 (1H, t, J = 15 Hz, H-24), 1.68 (1H, m, H-25a), 1.54 (1H, m, H-25b), 1.55 (1H, m, H-26), 0.96 (3H, d, J = 6.6 Hz, H-27), 0.98 (3H, d, J = 6 Hz, H-28), 7.50 (NH; H-29), lle: 3.30 (1H, d, J = 6.6 Hz, H-31), 2.38 (1H, m, H-32), 1.52 (1H, m, H-33a), 1.08 (1H, m, H-33b), 0.86 (3H, d, J = 6.6 Hz, H-34), 0.71 (3H, d, J = 6.0 Hz, H-35), 8.32 (NH; H-36). 13C NMR (150 MHz, CD3OD): Phe: 172.4 (C O; C-1), 53.9 (CH–N; C-2), 38.2 (CH2; C-3), 136.7 (C; C-4), 129.0 (CH; C-5), 128.0 (CH; C-6), 126.3 (CH; C-7), Leu1: 173.1 (C O; C-9), 52.4 (CH–N; C-10), 38.6 (CH2; C-11), 24.3 (CH; C-12), 21.4 (CH3, C-13), 21.4 (CH3; C-14), Leu2: 173.2 (C O, C-16), 52.9 (CH–N, C-17), 40.0 (CH2, C-18), 24.7 (CH, C-19), 21.6 (CH3, C-20), 19.9 (CH3, C-21), Leu3: 172.6 (C O, C-23), 52.3 (CH–N, C-24); 40.1 (CH2, C-25), 24.8 (CH, C-26), 22.0 (CH3, C-27), 21.2 (CH3, C-28), lle: 172.5 (C O, C-30), 65.47 (CH–N, C-31), 33.3 (CH, C-32), 25.3 (CH2, C-33), 8.9 (CH3, C-34), 14.3 (CH3, C-35).
Rotenolone (2)
Yellow amorphous powder. mp 139.6–141 °C. [α]20D = −24.84 (c. 0.07; MeOH). HRESI MS (+): 433.1272 ([M + Na]+ calcd for C23H22O7Na 433.1263). 1H NMR (600 MHz, CDCl3): 6.53 (1H, s, H-6′), 6.46 (1H, s, H-3′), 2.90 (2H, dd, J = 7.8 Hz; 15.6 Hz, H-4′′), 5.23 (1H, t, J = 8.4 Hz, H-5′′), 4.59 (1H, dd, J = 3 Hz, 12.6 Hz, H-2′′), 4.47 (1H, d, J = 10.8 Hz, H-2′′); 4.56 (1H, d, J = 3 Hz, H-2), 5.04 (2H, d, J = 78 Hz, H-8′′), 1.74 (3H, s, H-7′′), 6.52 (1H, d, J = 8.4 Hz, H-6); 7.81 (1H, d, J = 8.4 Hz, H-5), 3.79 (3H, s, H-5′a), 3.70 (3H, s, H-4′a). 13C NMR (150 MHz, CDCl3): 109.4 (CH, C-6′), 157.5 (C-5′), 162.8 (C-4′), 101.0 (CH, C-3′), 144.3 (C-2′), 30.9 (CH2; C-4′′), 87.9 (CH, C-5′′), 63.8 (CH2, C-2′), 76.1 (CH, C-2), 142.4 (C-6′′), 148.4 (C-8a), 111.9 (CH2, C-8′′), 113.2 (C-8), 17.0 (CH3, C-7′′), 151.0 (C-7), 104.9 (CH, C-6), 130.0 (CH, C-5), 111.8 (C-4a), 191.0 (C O, C-4), 67.3 (C-3), 108.6 (C-1′), 55.8 (OCH3, C-5′a), 56.5 (OCH3, 4′a).
Tephrosin (3)
Yellow resin. [α]20D = −7.06 (c. 0.15; CHCl3). HRESI-MS (+): 433.1273 ([M + Na]+ calcd for C23H22O7Na 433.1263). 1H NMR (600 MHz, CDCl3): 1.37 (3H, s, H-8′′), 1.43 (3H, s, H-7′′), 3.71 (3H, s, H-4′a), 3.79 (3H, s, H-5′a), 4.62 (1H, d, H-2′′); 4.48 (1H, dd, J = 11.4 Hz, H-2′′); 4.55 (1H, d, J = 1.8 Hz, H-2); 5.54 (1H, d, J = 10.2 Hz, H-5′′); 6.46 (1H, d; J = 9 Hz, H-6); 6.47 (1H, s, H-3′); 6.54 (1H, s, H-6′); 6.59 (1H, d, J = 10.8 Hz, H-4′′); 7.72 (1H, d, J = 9 Hz, H-5). 13C NMR (150 MHz, CDCl3): 26.82 (CH3, C-8′′); 27.21 (CH3, C-7′′), 55.7 (OCH3, C-4′a), 54.9 (OCH3, C-5′a), 63.4 (CH2, C-2′); 76.5 (CH, C-2), 127.8 (CH, C-5′′), 111.2 (CH, C-6), 100.9 (CH, C-3′), 109.1 (CH, C-6′), 114.7 (CH, C-4′′), 128.9 (CH, C-5), 67.2 (C-3), 78.0 (C-6′′), 108.0 (C-1′), 108.1 (C-8), 112.0 (C-4a), 143.5 (C-5′), 148.0 (C-2′), 151.9 (C-4′), 156.2 (C-8a), 161.5 (C-7), 191.5 (C-4).
Cytotoxicity assay
The antiproliferative effect was evaluated against a human colon cancer cell line HCT-116 (purchased from Banco de Células do Rio de Janeiro, Brazil). The cells were grown in RPMI 1640 medium (GIBCO) supplemented with 10% fetal bovine serum (v/v), maintained in an incubator at 37 °C and atmosphere containing 5% CO2. The compounds (1–3) were tested at concentrations ranging from 0.32 ng mL−1 to 50 μg mL−1 during 72 h and the effect on cell proliferation was evaluated in vitro using the MTT [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide] assay.52 Doxorubicin and DMSO (0.5% v/v) were used as positive control and negative control, respectively. Inhibition concentration mean values (IC50) were calculated, along with their respective confidence intervals of 95% (CI95%), by non-linear regression using GraphPad Prism v6.0 software.
gDNA sequencing, annotation and BGC mining
P. stromaticum gDNA was purified as previously reported for environmental samples.53 Briefly, the mycelium of P. stromaticum was grown as described earlier and pulverized with a sterile mortar and pestle after liquid nitrogen freezing. Approximately 150 mg of the resulting powdered culture was suspended in 1 mL of a CTAB lyses buffer for freezing/thawing lyses cycles (5 min at −80 °C and 5 min at 65 °C). After two phenol/chloroform extractions, DNA was precipitated with sodium acetate and isopropanol, washed with 70% ethanol and eluted with 10 mM Tris buffer (4 °C, overnight). Resulting double-stranded DNA integrity, purity and concentration were then respectively measured by agarose gel (1% agarose, 0.5X TBE buffer) running, spectrophotometry at NanoDrop (Thermo Fisher Scientific) and fluorometric quantification with Qubit (Thermo Fisher Scientific). Approximately 4 ng of high weight and pure gDNA was used to generate a genomic library preparation with Nextera XT DNA Library Preparation Kit (Illumina), following manufacturer's instructions. Sequencing was performed at MiSeq sequencer (Illumina) with the MiSeq Reagent Kits v3 (600 cycles) chemistry, at CeGenBio facility, in Universidade Federal do Ceará (htpps://www.cegenbio.ufc.br). Raw sequence data was then assembled into contigs with SPAdes 3.9.0 using default parameters for Illumina paired-end reads.35 Assembled genomic contigs statistics were retrieved using QUAST 4.5 and completeness of genome assembly calculated using BUSCO v. 3.0.2.36,37 The gene content of the draft genome sequence was evaluated using Genemark-ES v. 4.30 with default parameters.54 Genome mining for natural products BGCs was performed with ANTISMASH 4.0.0rc1 (FungiSMASH) software using P. stromaticum GenBank genomic annotations as input, with default parameters.
Chalcone isomerase-like alignment and phylogenetic analysis
P. stromaticum putative CHI protein was retrieved by local tbasltn analysis against a BLAST database created from P. stromaticum genome assemblage and using Lotus japonicus CHI (CAD69022.1) as query. PSI-BLAST analysis under default settings and using P. stromaticum CHI-like protein as query recruited closely (%7E76% identity) and distant (%7E30% identity) related fungi CHI-like proteins after 5 iterations.55 Fungi CHI-like and referential plant CHI (GenBank accession P28012) proteins were aligned using MAFFT encapsulated at Geneious software version 10.2 (https://www.geneious.com).56 Evolutionary analyses were then conducted in MEGA7.57 Phylogenetic reconstruction was performed using the Maximum Likelihood method based on the Le_Gascuel model with Gamma-distributed rates among sites,58 as indicated as best fitting model according the Bayesian Information Criterion (BIC).
Conclusions
Here we implemented bioguided-fractionation and high-resolution mass spectrometry in combination with genome mining to uncover the metabolic versatility of Myracrodruon urundeuva's endophytic fungus, P. stromaticum. Of particular relevance was the isolation of tephrosin as the compound responsible for the antiproliferative activity of P. stromaticum against a colorectal cancer cell line (HCT-116). Recovering of a chalcone isomerase-like protein with preserved catalytic fold also pave an exciting path for clarifying the biosynthesis of plant-related rotenoids in fungi. Inversely, possible correlations between genetic coded NRPS BGCs with the production of the purified cyclopeptide 1 and a leucinostatin A-related lipopeptide conserved in P. stromaticum and plant pathogens of Botryosphaeriaceae family, are also fertile subjects for further exploration in future studies.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
We are grateful to the financial support of Banco do Nordeste do Brasil (Convênio 7385) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq # 400764/2014-8), as well as to the Center for Genomics and Bioinformatics (CeGenBio) of Drug Research and Development Center at Universidade Federal do Ceará for technical support.
Electronic supplementary information (ESI) available: HRESIMS, 1D- and 2D-NMR spectra of compounds 1–3, along with BGCs found for P. stromaticum. See DOI: 10.1039/c8ra06824k
Notes and references
- Brakhage A. A. Nat. Rev. Microbiol. 2013;11:21–30. doi: 10.1038/nrmicro2916. [DOI] [PubMed] [Google Scholar]
- Kharwar R. N. Mishra A. Gond S. K. Stierle A. Stierle D. Nat. Prod. Rep. 2011;28:1208–1228. doi: 10.1039/C1NP00008J. [DOI] [PubMed] [Google Scholar]
- Kusari S. Pandey S. P. Spiteller M. Phytochemistry. 2013;91:81–87. doi: 10.1016/j.phytochem.2012.07.021. [DOI] [PubMed] [Google Scholar]
- Jia M. Chen L. Xin H. L. Zheng C. J. Rahman K. Han T. Qin L. P. Front. Microbiol. 2016;7:906. doi: 10.3389/fmicb.2016.00906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber T. Kim H. U. Synth. Syst. Biotechnol. 2016;1:69–79. doi: 10.1016/j.synbio.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scossa F. Benina M. Alseekh S. Zhang Y. Fernie A. R. Planta Med. 2018;84:855–873. doi: 10.1055/a-0630-1899. [DOI] [PubMed] [Google Scholar]
- Nielsen K. F. Månsson M. Frisvad J. C. Larsen T. O. J. Nat. Prod. 2011;74:2338–2348. doi: 10.1021/np200254t. [DOI] [PubMed] [Google Scholar]
- Bouslimani A. Sanchez L. M. A. Garg N. Dorrestein P. C. Nat. Prod. Rep. 2014;31:718. doi: 10.1039/C4NP00044G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dührkop K. Shen H. Meusel M. Rousu J. Böcker S. Proc. Natl. Acad. Sci. U. S. A. 2015;112:12580–12585. doi: 10.1073/pnas.1509788112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M. Carver J. J. Phelan V. V. Sanchez L. M. Garg N. Peng Y. Nguyen D. D. Watrous J. Kapono C. A. Luzzatto-Knaan T. Porto C. Bouslimani A. Melnik A. V. Meehan M. J. Liu W. T. Crüsemann M. Boudreau P. D. Esquenazi E. Sandoval-Calderón M. Kersten R. D. Pace L. A. Quinn R. A. Duncan K. R. Hsu C. C. Floros D. J. Gavilan R. G. Kleigrewe K. Northen T. Dutton R. J. Parrot D. Carlson E. E. Aigle B. Michelsen C. F. Jelsbak L. Sohlenkamp C. Pevzner P. Edlund A. McLean J. Piel J. Murphy B. T. Gerwick L. Liaw C. C. Yang Y. L. Humpf H. U. Maansson M. Keyzers R. A. Sims A. C. Johnson A. R. Sidebottom A. M. Sedio B. E. Klitgaard A. Larson C. B. CAB P. Torres-Mendoza D. Gonzalez D. J. Silva D. B. Marques L. M. Demarque D. P. Pociute E. O'Neill E. C. Briand E. Helfrich E. J. N. Granatosky E. A. Glukhov E. Ryffel F. Houson H. Mohimani H. Kharbush J. J. Zeng Y. Vorholt J. A. Kurita K. L. Charusanti P. McPhail K. L. Nielsen K. F. Vuong L. Elfeki M. Traxler M. F. Engene N. Koyama N. Vining O. B. Baric R. Silva R. R. Mascuch S. J. Tomasi S. Jenkins S. Macherla V. Hoffman T. Agarwal V. Williams P. G. Dai J. Neupane R. Gurr J. C Rodríguez A. M. Lamsa A. Zhang C. Dorrestein K. Duggan B. M. Almaliti J. Allard P. M. Phapale P. Nothias L. F. Alexandrov T. Litaudon M. Wolfender J. L. Kyle J. E. Metz T. O. Peryea T. Nguyen D. T. VanLeer D. Shinn P. Jadhav A. Müller R. Waters K. M. Shi W. Liu X. Zhang L. Knight R. Jensen P. R. Palsson B. O. Pogliano K. Linington R. G. Gutiérrez M. Lopes N. P. Gerwick W. H. Moore B. S. Dorrestein P. C. Bandeira N. Nat. Biotechnol. 2016;34:828–837. doi: 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber T. Blin K. Duddlela S. Krug D. Kim H. U. Bruccoleri R. Lee S. Y. Fischbach M. A. Muller R. Wolfgang W. Breitling R. Takano E. Medema M. H. Nucleic Acids Res. 2015;43:237–243. doi: 10.1093/nar/gkv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema M. H. Kottmann R. Yilmaz P. Cummings M. Biggins J. B. Blin K. de Bruijn I. Chooi Y. H. Claesen J. Coates R. C. Cruz-Morales P. Duddela S. Düsterhus S. Edwards D. J. Fewer D. P. Garg N. Geiger C. Gomez-Escribano J. P. Greule A. Hadjithomas M. Haines A. S. Helfrich E. J. Hillwig M. L. Ishida K. Jones A. C. Jones C. S. Jungmann K. Kegler C. Kim H. U. Kötter P. Krug D. Masschelein J. Melnik A. V. Mantovani S. M. Monroe E. A. Moore M. Moss N. Nützmann H. W. Pan G. Pati A. Petras D. Reen F. J. Rosconi F. Rui Z. Tian Z. Tobias N. J. Tsunematsu Y. Wiemann P. Wyckoff E. Yan X. Yim G. Yu F. Xie Y. Aigle B. Apel A. K. Balibar C. J. Balskus E. P. Barona-Gómez F. Bechthold A. Bode H. B. Borriss R. Brady S. F. Brakhage A. A. Caffrey P. Cheng Y. Q. Clardy J. Cox R. J. De Mot R. Donadio S. Donia M. S. van der Donk W. A. Dorrestein P. C. Doyle S. Driessen A. J. Ehling-Schulz M. Entian K. D. Fischbach M. A. Gerwick L. Gerwick W. H. Gross H. Gust B. Hertweck C. Höfte M. Jensen S. E. Ju J. Katz L. Kaysser L. Klassen J. L. Keller N. P. Kormanec J. Kuipers O. P. Kuzuyama T. Kyrpides N. C. Kwon H. J. Lautru S. Lavigne R. Lee C. Y. Linquan B. Liu X. Liu W. Luzhetskyy A. Mahmud T. Mast Y. Méndez C. Metsä-Ketelä M. Micklefield J. Mitchell D. A. Moore B. S. Moreira L. M. Müller R. Neilan B. A. Nett M. Nielsen J. O'Gara F. Oikawa H. Osbourn A. Osburne M. S. Ostash B. Payne S. M. Pernodet J. L. Petricek M. Piel J. Ploux O. Raaijmakers J. M. Salas J. A. Schmitt E. K. Scott B. Seipke R. F. Shen B. Sherman D. H. Sivonen K. Smanski M. J. Sosio M. Stegmann E. Süssmuth R. D. Tahlan K. Thomas C. M. Tang Y. Truman A. W. Viaud M. Walton J. D. Walsh C. T. Weber T. Van Wezel G. P. Wilkinson B. Willey J. M. Wohlleben W. Wright G. D. Ziemert N. Zhang C. Zotchev S. B. Breitling R. Takano E. Glöckner F. O. Nat. Chem. Biol. 2015;11:625–631. doi: 10.1038/nchembio.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema M. H. Fischbach M. A. Nat. Chem. Biol. 2015;11:639–648. doi: 10.1038/nchembio.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber T. Kim H. U. Synth. Syst. Biotechnol. 2016;1:69–79. doi: 10.1016/j.synbio.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darsih C. Prachyawarakorn V. Wiyakrutta S. Mahidol C. Ruchirawat S. Kittakoop P. RSC Adv. 2015;5:70595–70603. doi: 10.1039/C5RA13735G. [DOI] [Google Scholar]
- Wibowo M. Prachyawarakorn V. Aree T. Mahidol C. Ruchirawat S. Kittakoop P. Phytochemistry. 2016;122:126–138. doi: 10.1016/j.phytochem.2015.11.016. [DOI] [PubMed] [Google Scholar]
- Viana G. S. B. Bandeira M. A. M. Matos F. J. A. Phytomedicine. 2003;10:189–195. doi: 10.1078/094471103321659924. [DOI] [PubMed] [Google Scholar]
- Cregut M. Durand M.-J. Thouand G. Microb. Ecol. 2014;67:350–357. doi: 10.1007/s00248-013-0328-7. [DOI] [PubMed] [Google Scholar]
- Breinholt J. Damtoft S. Frederiksen L. B. Hansen L. B. Jensen S. R. Phytochemistry. 1995;39:1359–1362. doi: 10.1016/0031-9422(95)00175-7. [DOI] [Google Scholar]
- Raina S. Odell M. Keshavarz T. J. Biotechnol. 2010;148:91–98. doi: 10.1016/j.jbiotec.2010.04.009. [DOI] [PubMed] [Google Scholar]
- Wu G. Zhou H. Zhang P. Wang X. Li W. Zhang W. Liu H. W. Keller N. P. An Z. Yin W. B. Org. Lett. 2016;18:1832–1835. doi: 10.1021/acs.orglett.6b00562. [DOI] [PubMed] [Google Scholar]
- Xu J. Aly A. H. Wray V. Proksch P. Tetrahedron Lett. 2011;52:21–25. doi: 10.1016/j.tetlet.2010.10.131. [DOI] [Google Scholar]
- Mao Z. Sun W. Fu L. Luo H. Lai D. Zhou L. Molecules. 2014;19:5088–5108. doi: 10.3390/molecules19045088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H. W. Huang W. Y. Song Y. C. Chen J. R. Tan R. X. Helv. Chim. Acta. 2005;88:2861–2864. doi: 10.1002/hlca.200590228. [DOI] [Google Scholar]
- Shao C. Wang C. Wei M. Gu Y. Xia X. She Z. Lin Y. Magn. Reson. Chem. 2008;46:1066–1069. doi: 10.1002/mrc.2293. [DOI] [PubMed] [Google Scholar]
- Li H. J. Lin Y. C. Yao J. H. Vrijmoed L. L. P. Jones E. B. G. J. Asian Nat. Prod. Res. 2004;3:185–191. doi: 10.1080/102860201653237. [DOI] [PubMed] [Google Scholar]
- Abidi S. L. J. Heterocycl. Chem. 1983;20:1687–1692. doi: 10.1002/jhet.5570200649. [DOI] [Google Scholar]
- Luyengi L. Lee I. S. Mar W. Fong H. H. S. Pezzuto J. M. Kinghorn A. D. Phytochemistry. 1994;36:1523–1526. doi: 10.1016/S0031-9422(00)89755-1. [DOI] [Google Scholar]
- Harinantenaina L. Brodie P. J. Slebodnick C. Callmander M. W. Rakotobe E. Randrianasolo S. Randrianaivo R. Rasamison V. E. TenDyke K. Shen Y. Suh E. M. Kingston D. G. I. J. Nat. Prod. 2010;73:1559–1562. doi: 10.1021/np100430r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittraphab Y. Ngamrojanavanich N. Shimizu K. Matsubara K. Pudhom K. Planta Med. 2018;84:779–785. doi: 10.1055/s-0044-100797. [DOI] [PubMed] [Google Scholar]
- Ye H. Fu A. Wu W. Li Y. Wang G. Tang M. Li S. He S. Zhong S. Lai H. Yang J. Xiang M. Peng A. Chen L. Fitoterapia. 2012;83:1402–1408. doi: 10.1016/j.fitote.2012.08.001. [DOI] [PubMed] [Google Scholar]
- Choi S. Choi Y. Dat N. T. Hwangbo C. Lee J. J. Lee J. H. Cancer Lett. 2010;293:23–30. doi: 10.1016/j.canlet.2009.12.012. [DOI] [PubMed] [Google Scholar]
- Takashima J. Chiba N. Yoneda K. Ohsaki A. J. Nat. Prod. 2002;65:611–613. doi: 10.1021/np010126p. [DOI] [PubMed] [Google Scholar]
- Vasconcelos J. N. Santiago G. M. P. Lima J. Q. Mafezoli J. Lemos T. L. G. Silva F. R. L. Lima M. A. S. Pimenta A. T. A. Braz-Filho R. Arriaga A. M. C. Cesarin-Sobrinho D. Quim. Nova. 2012;35:1097–1100. doi: 10.1590/S0100-40422012000600005. [DOI] [Google Scholar]
- Bankevich A. Nurk S. Antipov D. Gurevich A. A. Dvorkin M. Kulikov A. S. Lesin V. M. Nikolenko S. I. Pham S. Prjibelski A. D. Pyshkin A. V. Sirotkin A. V. Vyahhi N. Tesler G. Alekseyev M. A. Pevzner P. A. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich A. Saveliev V. Vyahhi N. Tesler G. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão F. A. Waterhouse R. M. Ioannidis P. Kriventseva E. V. Zdobnov E. M. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- Ter-Hovhannisyan V. Lomsadze A. Chernoff Y. O. Borodovsky M. Genome Res. 2008;18:1979–1990. doi: 10.1101/gr.081612.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushley K. E. Raja R. Jaiswal P. Cumbie J. S. Nonogaki M. Boyd A. E. Owensby C. A. Knaus B. J. Elser J. Miller D. Di Y. McPail K. L. Spatafora J. W. PLoS Genet. 2013;9:e1003496. doi: 10.1371/journal.pgen.1003496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali H. Ries M. I. Lankhorst P. P. Van der Hoeven R. A. M. Schouten O. L. Noga M. Hankemeier T. Van Peij N. N. M. E. Bovenberg R. A. L. Vreeken R. J. Driessen A. J. M. PLoS One. 2014;9:98212. doi: 10.1371/journal.pone.0098212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J. M. Lee S. Lee J. Baek S. R. Kim J. C. Yun S. H. Park S. Y. Kang S. Lee Y. W. Mol. Microbiol. 2010;76:456–466. doi: 10.1111/j.1365-2958.2010.07109.x. [DOI] [PubMed] [Google Scholar]
- Wang B. Kang Q. Lu Y. Bai L. Wang C. Proc. Natl. Acad. Sci. U. S. A. 2012;109:1287–1292. doi: 10.1073/pnas.1115983109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G. Liu Z. Lin R. Li E. Mao Z. Ling J. Yang Y. Yin W. B. Xie B. PLoS Pathog. 2016;12:e1005685. doi: 10.1371/journal.ppat.1005685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang Y. M. Szewczyk E. Nayak T. Davidson A. D. Sanchez J. F. Lo H. C. Ho W. Y. Simityan H. Kuo E. Praseuth A. Watanabe K. Oakley B. R. Wang C. C. Chem. Biol. 2008;15:527–532. doi: 10.1016/j.chembiol.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames B. D. Haynes S. W. Gao X. Evans B. S. Kelleher N. L. Tang Y. Walsh C. T. Biochemistry. 2011;50:8756–8769. doi: 10.1021/bi201302w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crombie L. Whiting D. A. Phytochemistry. 1998;49:1479–1507. doi: 10.1016/S0031-9422(98)00178-2. [DOI] [PubMed] [Google Scholar]
- Lan X. Quan H. Xia X. Yin W. Zheng W. Biotechnol. Appl. Biochem. 2016;63:419–426. doi: 10.1002/bab.1376. [DOI] [PubMed] [Google Scholar]
- Jez J. M. Bowman M. E. Dixon R. A. Noel J. P. Nat. Struct. Biol. 2000;7:786–791. doi: 10.1038/79025. [DOI] [PubMed] [Google Scholar]
- Gensheimer M. Mushegian A. Protein Sci. 2004;13:540–544. doi: 10.1110/ps.03395404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves F. J. T. Freire F. C. O. Lima J. S. Melo J. G. M. Câmara M. P. S. Summa Phytopathol. 2016;42:43–52. doi: 10.1590/0100-5405/2099. [DOI] [Google Scholar]
- He R. Wang B. Toshiyuki W. Manyuan W. Lianca Z. Ikuro A. J. Braz. Chem. Soc. 2013;24:1926–1932. [Google Scholar]
- Mosmann T. J. Immunol. Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- Garcia G. D. Gregoracci G. B. Santos-Ede O. Meirelles P. M. Silva G. G. Edwards R. S. T. Gotoh K. Nakamura S. Iida T. de Moura R. L. Thompson F. L. Microbiol. Ecol. 2013;65:1076–1086. doi: 10.1007/s00248-012-0161-4. [DOI] [PubMed] [Google Scholar]
- Lomsadze A. Burns P. D. Borodovsky M. Nucleic Acids Res. 2014;42:e119. doi: 10.1093/nar/gku557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F. Madden T. L. Schäffer A. A. Zhang J. Zhang Z. Miller W. Lipman D. J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K. Misawa K. Kuma K. Miyata T. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S. Stecher G. Tamura K. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le S. Q. Gascuel O. Mol. Biol. Evol. 2008;25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- Xiao J. Zhang Q. Gao Y. Q. Tang J. J. Zhang A. L. Gao J. M. J. Agric. Food Chem. 2014;62:3584–3590. doi: 10.1021/jf500054f. [DOI] [PubMed] [Google Scholar]
- Klitgaard A. Iversen A. Andersen M. R. Larsen T. O. Frisvad J. C. Nielsen K. F. Anal. Bioanal. Chem. 2014;406:1933–1943. doi: 10.1007/s00216-013-7582-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimming O. Fleischhacker F. Nollmann F. I. Bode H. B. ChemBioChem. 2014;15:1290–1294. doi: 10.1002/cbic.201402065. [DOI] [PubMed] [Google Scholar]