Abstract
Gene therapy is a promising method for the treatment of vascular disease; however, successful strategies depend on the development of safe and effective delivery technologies with specific targeting to a diseased point of vasculature. Reactive oxygen species (ROS) are overproduced by vascular smooth muscle cells (VSMCs) at critical stages of atherosclerosis progression. Therefore, ROS were exploited as a stimulus for vascular targeted gene delivery in this study. A combination of bio-conjugation methods and controlled reverse addition-fragmentation chain-trasfer (RAFT) polymerization was utilized to synthesize a new ROS-cleavable, pH-responsive mPEG113-b-CP5K-b-PDMAEMA42-b-P(DMAEMA22-co-BMA40-co-PAA24) (PPDDBP) polymer as a nanocarrier for plasmid DNA (pDNA) delivery. The ros degradability of PPDDBP polymers was confirmed by SIN-1-mediated cleavage of CP5K peptide linkers through a shift in GPC chromatogram with an appearance of mPEG shoulder peak and an increase in zeta potential (ζ). The polyplex nanocarrier also demonstrated effective PDNA loading, serum stability, and hemocompatibility, indicating its excellent performance under physiological conditions. The polyplexes demonstrated ideal pH responsiveness for endosomal escape and effective ROS responsiveness for improved targeting in an in vitro model of pathogenic VSMCs in terms of both uptake and expression of reporter gene. These data suggest this novel nanocarrier polyplex system is a promising gene delivery tool for preventing or treating areas of high ROS, such as atherosclerotic lesions.
1. Introduction
Atherosclerosis is accompanied by progressive thickening of a vessel wall resulting from abnormal behavior of vascular cells in response to disturbed blood flow with inflammatory signals. Currently, to treat this disease it is common practice to reopen the blocked vessel with a stent; however, rapid re-occlusion (restenosis) occurs because the underlying problem of cell health is not addressed. Advances in vascular gene delivery over the past decade have demonstrated potential in addressing clinical issues that arise from restenosis by directly controlling the behaviors of the problematic cell types.1 To date, viral vectors are the most effective gene delivery platform due to their high transfection efficiency; however, safety issues such as immunogenicity and pathogenicity inhibit the broader clinical implementation of these systems.2,3 To address these issues, cationic polymers and lipids have been explored as alternative delivery platforms due to their facile synthesis, tunable properties and demonstrated biosafety.4-6 However, low transfection efficiency in vascular cells (due to inefficient uptake) is considered to be a significant hurdle for clinical translation.7 In particular, none of the current gene delivery platforms demonstrated successful transfection into pathological vascular smooth muscle cells (VSMCs).
It is well known that overproduction of reactive oxygen species (ROS) is indicative of abnormal VSMC behavior,8,9 including the process of restenosis.10 However, the development of ROS-responsive polymeric nanocarriers that can target and deliver therapeutics into ROS-overproducing VSMCs is still quite limited.11 Recently, Xia et al. demonstrated an intracellular ROS-degradable cationic polymers containing thioketal units that effectively delivered plasmid DNA (pDNA) into cancer cells, suggesting that specific delivery to cells with high ROS is a feasible approach.12 Several previous studies demonstrated that polyplexes formed from pDNA and polycations have high transfection efficiency;13 however, low affinity of these polycations to pDNA can result in premature dissociation of polyplexes under physiological conditions. This problem can be overcome by an increased ratio of polycation to pDNA (N/P ratio), but at the expense of increased cellular toxicity. Therefore, the successful clinical application of cell-based gene therapy requires a stable, non-viral polyplex system that addresses both the extra- and intracellular environmental barriers without compromising biocompatibility.
Here, we report the successful utilization of PEGylated, oligoproline peptide-derived nanocarriers to improve pDNA transfection into VSMCs that are overproducing ROS due to inflammatory activation.14-16 Our nanocarrier is dual-responsive as it is programmed to react first to extracellular ROS, and then to intracellular pH, imparting a specific functionality for application only in high ROS environments.12,17-19 Each copolymer block was designed to incorporate one or more desired properties, such as effective condensation with pDNA, serum stability, ROS degradability, pH responsiveness, hemocompability, cellular biocompatibility or improved transfection efficiency. This work represents a significant improvement over the current state-of-the-art delivery methods. The extracellular ROS-cleavable, pH-responsive polymer, mPEG113-b-CP5K-b-PDMAEMA42-b-P(DMAEMA22-co-BMA40-co-PAA24) (PPDDBP) was synthesized by combined approach of bio-conjugation methods and controlled reversible addition–fragmentation chain-transfer (RAFT) polymerization. The first block contains methoxy polyethylene glycol (mPEG) as a corona-forming hydrophilic block to shield the cationic poly(2-dimethylamino)ethyl methacrylate (PDMAEMA) block from nonspecific interactions with blood and tissue components until the polyplexes encounter a high ROS environment.20 Though PEG shielding of polyplexes is known to reduce the efficiency of cellular uptake21,22 and endosomal escape,23 pathophysiological stimuli, (including glutathione and MMPs) have been successfully employed for targeted dePEGylation of cationic polyplex to rescue drug and gene delivery capabilities.20,21,24
To induce dePEGylation under high ROS environments, a proline oligomer, cysteine-(proline)5-lysine (CP5K) peptide was chosen as an ROS degradable second block, based on our previous work.25-27 The third block, hydrophilic PDMAEMA contains positively charged tertiary amines, which facilitate effective condensation of nucleic acids through interaction with negatively-charged phosphate groups.28 The fourth block, a core-stabilizing hydrophobic terpolymer P(DMAEMA-co-BMA-co-PAA) is composed of N,N-dimethylaminoethyl methacrylate (DMAEMA), N-butyl methacrylate (BMA), and 2-propylacrylic acid (PAA). This terpolymer block is ampholytic in nature under physiological pH with approximately equimolar ratios of positively charged DMAEMA and negatively charged PAA, together with hydrophobic butyl methacrylate (BMA).29,30 The low pH within the endosome triggers protonation of the negatively-charged PAA with a simultaneous increase in the positive charge of the DMAEMA. This increased positive charge destabilizes the polyplex core and causes membrane disruption, thereby enabling endosomal escape of the nanocarrier.21,31,32 As a control, we used a non-ROS-cleavable polymer without the CP5K peptide to demonstrate enhanced uptake of these polyplexes under ROS rich environments.
The polyplex nanocarrier was evaluated for its size, charge, morphology, ROS degradability, pDNA condensation, serum stability and blood compatibility. To demonstrate the feasibility of ROS-responsive gene delivery into pathogenic VSMCs, we additionally investigated cellular uptake, cytotoxicity, and in vitro transfection efficiency with human coronary artery smooth muscle cells (HCASMCs).
2. Materials and methods
2.1. Materials
All chemicals and reagents were purchased either from Sigma-Aldrich or Fisher Scientific, USA and used without purification unless otherwise noted. 2-Propyl acrylic acid (PAA),33 N-hydroxyl succinimide functional 4-cyano-4-(ethylsulfanylthiocarbonyl) sulfanylpentanoic acid (NHS-ECT)21 and 4-cyano-4-(ethylsulfanyl-thiocarbonyl) sulfanylpentanoic acid (ECT)34 were synthesized following previously reported protocols. The monomers, N,N-dimethylaminoethyl methacrylate (DMAEMA), and N-butyl methacrylate (BMA), were filtered twice through alumina columns just before polymerization. Maleimide end-functionalized methoxypolyethylene glycol (mPEG113-MAL) was purchased from JenKem Technology USA (Allen, TX, USA). Rink amide-MBHA resin and Fmoc-protected l-amino acids were purchased from EMD Biosciences (Gibbstown, NJ, USA). Luciferase reporter plasmid (pPK-CMV-R3) with 5500 base pairs was purchased from Promokine (Heidelberg, Germany). Penicillin-streptomycin, fetal bovine serum (FBS) and lipofectamine2000 (LF2K) transfection reagent were purchased from Life Technologies (Carlsbad, CA, USA). 3-Morpholinosydnonimine (SIN-1) was purchased from Life Technologies as a package of 1 mg plastic vials. Label IT® Fluorescein Plasmid Delivery Control was purchased from Mirus Bio (Madison, WI, USA). A bifunctional HS-CP5K-NH2 peptide (CPPPPPK) was synthesized by the standard FMOC-based solid phase method on a Rink amide-MBHA resin using PS3 synthesizer (Protein Technologies, Tucson, AZ, USA) and characterized by liquid chromatography-mass spectrometry (LC-MS).25
2.2. Synthesis of ROS cleavable mPEG113-b-CP5K-b-PDMAEMA42-b-P(DMAEMA22-co-BMA40-co-PAA24) (PPDDBP)
This polymer was synthesized using the following steps.
2.2.1. Synthesis of mPEG113-b-CP5K-NH2.
A degassed solution of triethyl amine (Et3N) (0.176 mM, 0.024 mL) in a methanol/acetonitrile mixture (1 : 1 ratio, 25 mL) was added to a previously dried flask containing mPEG113-MAL (0.08 mM, 0.4 g) and CP5K peptide (0.12 mM, 93 mg), followed by stirring at room temperature (RT) for 24 h. The reaction mixture was purified by dialysis against methanol using 2k MW cutoff tubing for 24 h to yield purified mPEG113-b-CP5K-NH2.
2.2.2. Synthesis of mPEG113-b-CP5K-ECT.
A previously degassed solution of triethyl amine (0.08 mM, 0.012 mL) in anhydrous methanol (10 mL) and DMF (10 mL) was added to a dried, degassed flask containing mPEG113-b-CP5K-NH2 (0.04 mM, 0.231 g) and NHS-ECT (0.1 mM, 0.036 g) with stirring for 48 h. The reaction mixture was purified by precipitating in cold diethyl ether. Dried polymers were used for UV based absorption measurement at 310 nm to determine the percentage ECT conjugation to mPEG113-b-CP5K-NH2.
2.2.3. Synthesis of mPEG113-b-CP5K-b-pDMAEMA42-ECT.
mPEG113-b-CP5K-ECT (0.072 g, 0.012 mM), DMAEMA (0.27 mL, 1.17 mM) and AIBN (0.21 mg, 0.0013 mM) were dissolved in dioxane (5 mL) in a 15 mL glass ampoule and degassed by purging with nitrogen gas for 30 min. The degassed solution was submerged in a preheated oil bath at 70 °C for 8 h. The crude polymerization mixture was purified by dialysis (2k MW cutoff tubing) against methanol and vacuum dried.
2.2.4. Synthesis of mPEG113-b-CP5K-b-PDMAEMA42-b-P(DMAEMA22-co-BMA40-co-PAA24) (PPDDBP).
mPEG113-b-CP5K-b-PDMAEMA42-ECT (0.062 g, 0.005 mM), DMAEMA (0.399 mM, 0.067 mL), BMA (0.74 mM, 0.117 mL), PAA (0.285 mM, 0.032 g), AIBN (0.08 mg, 0.0005 mM), dioxane (4.5 mL) and DMF (1.5 mL) were placed in a dry glass ampoule, and the solution was degassed by nitrogen bubbling for 30 min. Polymerization was conducted at 70 °C for 24 h. The crude polymer mixture was purified by dialysis (2k MW cutoff tubing) first against methanol and then against water, and finally lyophilized.
2.3. Synthesis of control polymer without ROS cleavable CP5K peptide, mPEG113-b-PDMAEMA50-b-P(DMAEMA16-co-BMA42-co-PAA17) (PDDBP)
This block copolymer was synthesized through following steps.
2.3.1. Synthesis of mPEG113-ECT.
mPEG based RAFT macro chain trasfer agent (CTA) was synthesized following a previously published protocol.34 In a 100 mL flask, mPEG113-OH (2 mmol, 10 g, Mn = 5 kDa,), ECT (4 mmol, 1.045 g) and DMAP (10 mg) were dissolved in anhydrous dichloromethane (100 mL) by stirring at RT. Dicyclohexylcarbodimide (DCC, 4 mmol, 0.82 g) was then added to the solution and stirred for 48 h under nitrogen atmosphere. The white precipitate of dicyclohexyl urea was filtered. The filtrate was concentrated under vacuum and further precipitated three times into cold diethyl ether. The precipitated polymer was dried under vacuum. 1H NMR spectra revealed 81% conjugation of ECT to mPEG. The molecular weight (Mn) and polydispersity (PDI) of polymer were 5300 and 1.05, respecively, as determined by gel permeation chromatography (GPC).
2.3.2. Synthesis of mPEG113-b-PDMAEMA50-ECT.
In a glass test tube, mPEG113-ECT (0.064 g, 0.012 mmol, Mn 5300), DMAEMA (0.27 mL, 1.17 mmol), and AIBN (0.21 mg, 0.0012 mmol) were dissolved in dioxane (5 mL) and degassed by purging with nitrogen for 30 minutes. The degassed solution was submerged in a preheated oil bath at 70 °C for 8 h. The final polymerization mixture was quenched by exposure to air at RT. The polymer was purified by precipitation into tenfold excess cold diethyl ether, and then dried under vacuum. The polymer Mn and PDI were 14 800 and 1.55, respectively, as determined by GPC.
2.3.3. Synthesis of mPEG113-b-PDMAEMA50-b-P(DMAEMA16-co-BMA42-co-PAA17) (PDDBP).
A chain extension of mPEG113-b-pDMAEMA50-ECT was carried out to prepare mPEG113-b-PDMAEMA50-b-P(DMAEMA16-co-BMA42-co-PAA17) by RAFT polymerization of DMAEMA, BMA and PAA. Briefly, mPEG113-b-PDMAEMA50-ECT (0.075 g, 0.005 mmol, Mn 15 000), DMAEMA (0.067 mL, 0.399 mmol), BMA (0.74 mmol, 0.117 mL), PAA (0.285 mM, 0.032 g), AIBN (0.08 mg, 0.0005 mmol), dioxane (4.5 mL) and DMF (1.5 mL) were placed in a dry ampoule, and the solution was degassed by bubbling nitrogen for 30 minutes. The polymerization was performed at 70 °C for 24 h. The crude polymerization mixture was purified by precipitation twice in cold diethylether. The polymer Mn and PDI were 25 300 and 1.22, respectively, as determined by GPC.
2.4. Polymer characterization
1H NMR spectra of organic compounds and polymers were collected in CDCl3 with a Brüker 400 MHz spectrometer. GPC (Agilent Technologies, Santa Clara, CA, USA) was used to determine the Mn and PDI of polymers in dimethylformamide (DMF) +0.1 M LiBr mobile phase at 60 °C through three serial Tosoh Biosciences TSKGel Alpha columns (Tokyo, Japan). A Wyatt miniDAWN TREOS light scattering (LS) detector (Wyatt Technology Corp., Santa Barbara, CA, USA) and Agilent refractive index (RI) detector were used to calculate absolute Mn based on dn/dc values experimentally determined through off-line injection into the RI detector.
2.5. Preparation and characterization of polymer micelles
To prepare polymer micelles, either lyophilized PPDDBP or non-cleavable PDDBP (10 mg) was dissolved in 200 μL of methanol, followed by addition of pH 4.0 buffer (200 μL). Micelle formation was induced by drop wise addition of pH 8.0 buffer (10 mL) to the magnetically stirred polymer suspension using a syringe pump. Polymer micelles were filtered through a 0.2 micron syringe filter just before size and zeta potential measurements by dynamic light scattering (DLS, Malvern Zetasizer Nano ZS, Malvern, UK). To measure critical micelle concentration (CMC) of PPDDBP micelles, a range of polymer concentration from 0.01 to 1 mg ml−1 in DPBS was prepared, and stability of micelles was measured by DLS.35 ROS mediated cleavage of peptide and subsequent changes in zeta potential of micelles in PBS were determined in the presence of 4 mM SIN-1 (a generator of peroxynitrite that decomposes into nitric oxide and superoxide in an aqueous solution).
2.6. Assessment of ROS-triggered detachment of PEG from PPDDBP by GPC
To confirm ROS-mediated dePEGylation of PPDDBP polymer through cleavage of CP5K peptide, lyophilized PPDDBP (10 mg mL−1) was incubated with 1 mM SIN-1 in DPBS at RT for 16 h. The polymer solution was then lyophilized and dissolved into DMF to run in GPC. The cleavage of CP5K peptide was confirmed by comparing GPC chromatograms of PPDDBP before and after SIN-1 treatment.
2.7. Preparation and characterization of polyplexes
Lyophilized PPDDBP or noncleavable PDDBP polymer at various N/P ratios were mixed with pDNA (1.5 μg mL−1) in a 100 mM citric acid/sodium citrate buffer solution (pH 4.0). After 30 min incubation at RT, polyplex formation was induced by addition of pH 8.0 buffer to raise the pH up to 7.4. The size and zeta potential (ζ) of the polyplexes (N/P 10) were determined using DLS (Malvern Zetasizer Nano ZS). To see ROS mediated dePEGylation of polyplexes, changes in zeta potential of polyplexes (N/P 10) were measured in the presence of SIN-1 (4 mM) over 24 h. Transmission electron microscopy (TEM) samples were prepared on carbon film-backed copper grid (Electron Microscopy Sciences, Hatfield, PA, USA) by addition of a polyplex solution (5 μL, N/P 10) for 60 seconds, followed by dry blotting. Next, the samples were counter stained with filtered 3% uranyl acetate (5 μL) as a negative stain for 20 seconds and again blotted dry. The samples were then dried under vacuum overnight and imaged using a FEI Tecnai Osiris TEM at 200 kV.
2.8. Assessment of pDNA loading by agarose gel electrophoresis
The ability of PPDDBP to condense pDNA was assessed by agarose gel electrophoresis. The polyplexes at various N/P ratios (1, 2, 5, 10, 20, and 30) were loaded with 10 μg ml−1 pDNA, and the samples were run into 0.5% agarose gel containing 0.05 μg ml−1 ethidium bromide (EtBr). Naked pDNA was used as a positive control. The gel electrophoresis was performed in TAE (1×) buffer at 100 V for 30 min. The bands were visualized and imaged with a UV trans-illuminator.
2.9. Stability of polyplexes in the presence of serum and SDS decomplexation
PPDDBP polyplexes (N/P 10) were incubated in a high concentration (50% w/v) of fetal bovine serum (FBS) at 37 °C for 0.25, 0.5, 1, 2, 3, and 6 h. As a harsh control condition to prove the DNase protection capability of the polyplexes, a solution of sodium dodecyl sulfate (SDS, 1% w/v) was added to cause release of pDNA. The samples were analyzed by gel electrophoresis as described above.
2.10. Hemolysis assay
The blood cytocompatibility of polymer alone and pDNA/PPDDBP polyplexes was measured by a pH dependent hemolysis assay.23 Briefly, red blood cells (RBCs) were isolated from anonymous, consenting human donors according to well established protocols.23 10 μL solutions of PPDDBP or polymer/pDNA polyplexes (at polymer concentrations of 20–800 μg ml−1) were incubated with RBCs (190 μL) in DPBS in four different pH conditions (7.4. 6.8, 6.2 and 5.8) that mimic extracellular and endosomal trafficking. The RBCs treated with DPBS and 1% (v/v) Triton X-100 were used as negative and positive controls, respectively. After 1 h of incubation at 37 °C, RBCs were centrifuged at 500 g for 5 min, and supernatant was measured using the M100 Pro plate reader by recording the absorbance at 541 nm to determine the percent hemolysis of samples compared to the positive control treated with Triton-X-100.
2.11. Cell viability
Cell viability of HCASMCs was measured by a resazurin assay.36 HCASMCs were seeded in 24-well plates at a density of 4 × 104 cells per well with 500 μL of SMC medium (Cell Applications Inc, San Diego, CA) and allowed to adhere for 24 h. Afterwards, the culture media was replaced with a fresh solution of media containing polyplexes (1.5 μg mL−1) with various N/P ratio (1, 2, 4, 8, 10, and 20) or lipofectamine 2000 (LF2K) (N/P10). After 24 h of culture, cells were incubated with fresh media containing resazurin sodium salt (5 μM) (Sigma-Aldrich) for 4 h at 37 °C in the dark. The incubated culture media was then transferred to a new 96-well plate (100 μL per well), and the fluorescence intensity was measured by a plate reader at 530–560 nm excitation wavelength and 590 nm emission wavelength. The percent cell viability for each group was calculated based on a standard curve obtained from cell culture on tissue culture polystyrene (TCPS) (n = 3).
2.12. Measurement of intracellular ROS levels in HCASMCs by flow cytometry
Intracellular ROS levels were measured by CM-H2DCFDA staining with flow cytometric analysis.37 HCASMCs (4.0 × 104 cells per well in a 24-well plate) were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco Cell Culture, Carlsbad, CA, USA) containing 2% FBS and 1% streptomycin/penicillin for 24 h (n = 3). A freshly prepared, sterile CM-H2DCFDA solution (10 mM) in dimethylsulfoxide (DMSO) (8.65 μL) was added to the media (8.65 mL) to obtain 10 μM concentration, and this media was incubated with cells in the dark for 20 min at 37 °C. The cells were washed twice with DPBS (500 μL) to remove any free residual dye and detached using 0.05% trypsin (200 μL) for 5 min. Then fresh media (500 μL) was added to the cells, and the fluorescence intensity of CM-H2-DCFDA was analyzed by running 10 000 cells per sample in flow cytometry (FACSCalibur flow cytometer, BD Biosciences, Franklin Lakes, NJ, USA) at excitation at 488 nm with a 530/30 filter.
2.13. Cellular uptake of polyplexes by flow cytometry
To evaluate cellular uptake of polyplexes, HCASMCs (1.0 × 105 cells per well in a 12-well plate) were cultured in SMC medium (Cell Applications Inc.) for 24 h. Cells were then treated with lipopolysaccharide (LPS, 1 μg mL−1) and interferon gamma (IFN)-γ (100 units per mL) in fresh culture media for 6 h to induce ROS production. Polyplexes were prepared from either ROS cleavable PPDDBP or non-cleavable PDDBP (both with N/P 10) by loading fluorescein labeled pDNA (Mirus Bio) at a final concentration of 1.5 μg mL−1. Cells with or without LPS/IFN-γ were then incubated with these polyplexes (n = 3) for 6 h, washed twice with DPBS, and detached with 0.05% trypsin. Cellular uptake of particles was measured by FACSCalibur flow cytometry (BD Biosciences) and analyzed with the BD Cell Quest ProTM software.
2.14. Imaging intracellular distribution of polyplexes by confocal laser scanning microscopy
To track intracellular localization of polyplexes, HCASMCs (1 × 104 cells per well) were cultured in a Falcon 8-well culture slide (BD Biosciences) for 24 h. HCASMCs were then treated with LPS (1 μg mL−1) and IFN-γ (100 units per mL) in fresh culture media for 6 h to induce ROS production. Polyplexes (N/P 10) were prepared by loading fluorescein labelled pDNA (Mirus Bio) at the final concentration of 1.5 μg mL−1 to ROS cleavable PPDDBP, and control PDDBP. HCASMCs with or without LPS/IFN-γ were incubated with the polyplexes with (n = 3) for 6 h, and then treated with Lysotracker (Life Technologies, NY, USA) for 1h and imaged by confocal laser scanning microscopy (Nikon, TE2000). The confocal images were analyzed using ImageJ (NIH, Maryland, USA). For measuring in vitro plasmid uptake by HCASMCs, the average intensity by green fluorescent plasmids within the total individual cell area was measured for N = 10–15 cells per each condition using ImageJ (NIH, US), and averaged for each condition.
2.15. In vitro transfection efficiency
In vitro transfection efficiency of polyplexes in HCASMCs was assessed by a luciferase gene reporter assay. HCASMCs were cultured in 96-well plates (12 000 cells per well for ~70–80% confluent) in SMC medium for 24 h. Cells were then treated with LPS (1 μg mL−1) and IFN-γ (100 units per mL) for 6 h to induce ROS production, followed by treatment with PPDDBP and LF2K polyplexes at N/P 10 ratio for 24 h at 37 °C. Luciferin (150 μg mL−1) was added to the samples, and bioluminescence from cells was measured using a Xenogen IVIS Imaging system 200 series (Perkin Elmer, Waltham, MA, USA). Cells were then washed twice with DPBS and lysed with 200 μL of lysis buffer (Promega, Madison, WI, USA). The protein content was using a microBCA protein assay reagent kit (Pierce) and reading absorbance at 562 nm using the M1000 Pro platen reader. The luciferase expression was reported in relative light units (RLU) normalized to mg protein (RLU per mg).
2.16. Statistical analysis
The experimental results are reported as means ± standard error mean (SEM). Data from each experiment were initially analyzed using single factor analysis of variance, and comparisons between individual samples were then performed using an unpaired Student’s t-test. For all statistics p < 0.05 was considered significantly different.
3. Results and discussion
3.1. Polymer synthesis and characterization
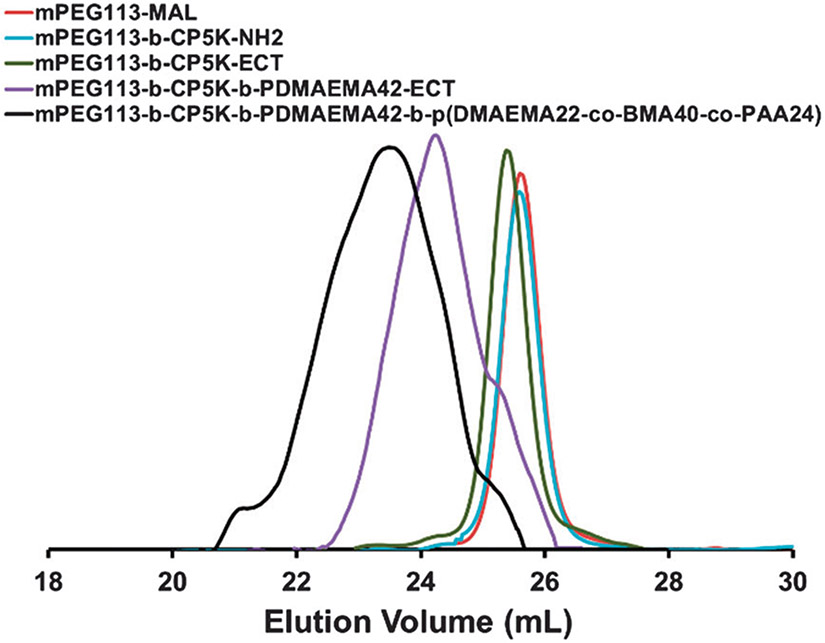
The ROS-cleavable oligo-proline peptide (CP5K) conjugated PEGylated copolymer, PPDDBP was synthesized as described in Scheme 1. First, the oligo-proline peptide (SH-CP5K-NH2) containing five proline units (P5) between a thiol (−SH) group of cysteine at one end and a primary amine (−NH2) of lysine at the other terminus was synthesized by fluorenylmethyloxycarbonyl-chloride (FMOC)-based peptide synthesis.25 The formation of the CP5K peptide was confirmed by the presence of single characteristic mass peak at m/z 776 in LC-MS spectrum (Fig. S1, ESI†). The peptide, CP5K was conjugated with 5 kDa maleimide-functionalized methoxypolyethylene glycol (mPEG113-MAL) through the thiol group of cysteine end by maleimide–thiol coupling chemistry. The conjugation of mPEG113-b-CP5K-NH2 was confirmed by a left-shift in the GPC chromatogram (Fig. 1). Second, in order to build the other polymer blocks on this mPEG-peptide conjugate, the amine group of mPEG113-b-CP5K-NH2 was coupled with NHS-ECT, yielding mPEG113-b-CP5K-ECT, a macro initiator for RAFT polymerization.
Scheme 1.
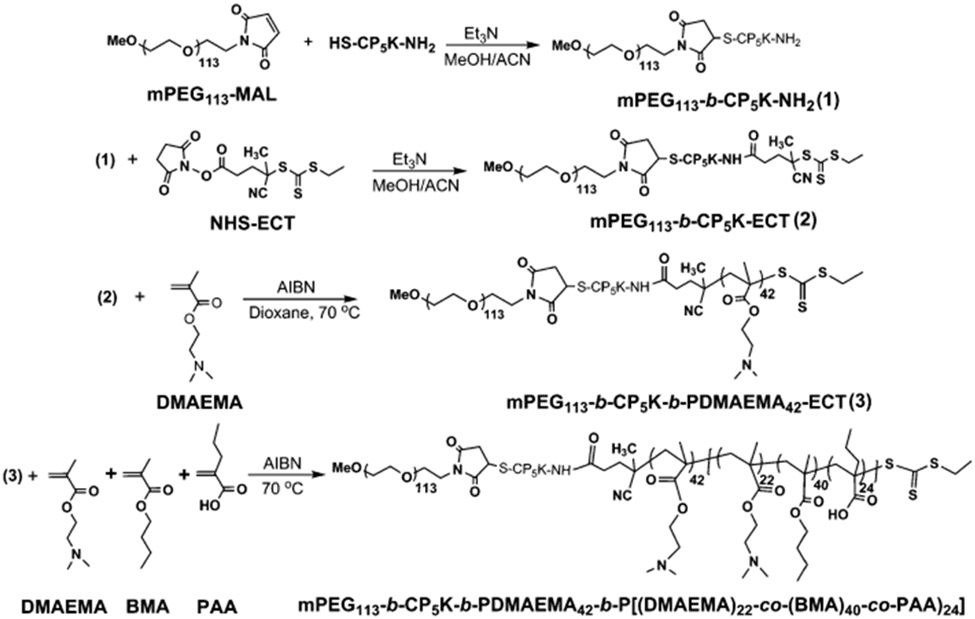
Synthesis of mPEG113-b-CP5K-b-PDMAEMA42-b-P(DMAEMA22-co-BMA40-co-PAA24) (PPDDBP) via bio-conjugation methods and RAFT polymerization.
Fig. 1.
GPC refractive index detector traces of mPEG113-MAL, mPEG113-b-CP5K-NH2, mPEG113-b-CP5K-ECT, mPEG113-b-CP5K-b-PDMAEMA42-ECT, and mPEG113-b-CP5K-b-PDMAEMA42-b-P(DMAEMA22-co-BMA40-co-PAA24) (PPDDBP).
The formation of RAFT macro chain transfer agent (CTA) was confirmed by a shift in GPC chromatogram and by UV absorption of the trithiocarbonate group of CTA at 310 nm. Based on the CTA extinction coefficient, the conjugation efficiency of 4-cyano-4-(ethylsulfanylthiocarbonyl) sulfanylpentanoic acid (ECT) to mPEG113-b-CP5K-NH2 was 98%. This macro-CTA was then used for RAFT polymerization of DMAEMA using azobisisobutyronitrile (AIBN) in dioxane, yielding mPEG113-b-CP5K-b-PDMAEMA42-ECT. The number of repeating DMAEMA units in the PDMAEMA block of copolymer was calculated from 1H NMR spectra by comparing the integration of CH2 protons of DMAEMA at 4.15 ppm to the −OCH2 peak of mPEG block at 3.65 ppm. Finally, a chain extension of mPEG113-b-CP5K-b-pDMAEMA42-ECT was performed to prepare mPEG113-b-CP5K-b-PDMAEMA42-b-P(DMAEMA22-co-BMA40-co-PAA24) (PPDDBP) by RAFT polymerization of DMAEMA, BMA and PAA in dioxane/DMF mixture (3 : 1) using AIBN at 70 °C for 24 h (Fig. 1).
The numbers of repeating DMAEMA, BMA, and PAA units in the random terpolymer block of the final copolymer were calculated by relative integration of the mPEG block peak at 3.65 ppm with CH2 protons from DMAEMA, BMA and PAA at 4.15, 4.0 and 1.2 ppm, respectively (Table 1 and Fig. S2, ESI†).21
Table 1.
Molecular weight of polymers prepared by bio-conjugation methods and controlled RAFT polymerization
| S. No. | Polymer | M n,NMR a | M n,GPC b | PDIc |
|---|---|---|---|---|
| 1 | mPEG113-MAL | 5000 | 5000 | 1.05 |
| 2 | mPEG113-b-CP5K-NH2 | 5776 | 5700 | 1.05 |
| 3 | mPEG113-b-CP5K-ECT | 6135 | 6000 | 1.09 |
| 4 | mPEG113-b-CP5K-b-PDMAEMA42-ECT | 12 737 | 13 700 | 1.15 |
| 5 | mPEG113-b-CP5K-b-PDMAEMA42-b-P(DMAEMA22-co-BMA40-co-PAA24)(PPDDBP) | 24 610 | 31 500 | 1.22 |
Molecular weight (Mn,NMR) of polymers calculate by 1H NMR.
Number average molecular weight of polymer by GPC in DMF solvent.
Polydispersity index (PDI) of polymers.
To prepare non-cleavable control polymer mPEG113-b-PDMAEMA50-b-P(DMAEMA16-co-BMA42-co-PAA17) (PDDBP) without the CP5K peptide, the first terminal hydroxyl group of 5k mPEG was conjugated with the carboxylic group of ECT by a DCC/DMAP method, producing mPEG113-ECT as a RAFT macro CTA. This CTA was used to further build the second block as PDMAEMA. Subsequently, mPEG113-b-PDMAEMA50-ECT was utilized for the third block as P(DMAEMA-co-BMA-co-PAA) random copolymer under similar polymerization conditions as described for PPDDBP (Scheme S1, Table S1, Fig. S3 and S4, ESI†).
3.2. Preparation and characterization of polymer micelles
In aqueous media, ROS-responsive PPDDBP and uncleavable PDDBP assembled into stable micelles through hydrophilic PEG and PDMAEMA as corona and hydrophobic P(DMAEMA-co-BMA-co-PAA) as core forming block with an average diameters of 37 and 46 nm, respectively (Fig. S5A, ESI†).
Their ζ-potential values were 1.54 mV and 0.9 mV (Fig. S5B, ESI†), respectively, indicating mPEG-mediated shielding of positively-charged PDMAEMA. The PPDDBP micelles were stable down to 0.1 mg ml−1 concentration but were destabilized at 0.01 mg ml−1 concentration. This observation indicates that the CMC of these micelles is between 0.1–0.01 mg mL−1 (Fig. S6, ESI†).
3.3. Confirmation of ROS-triggered dePEGylation of PPDDBP
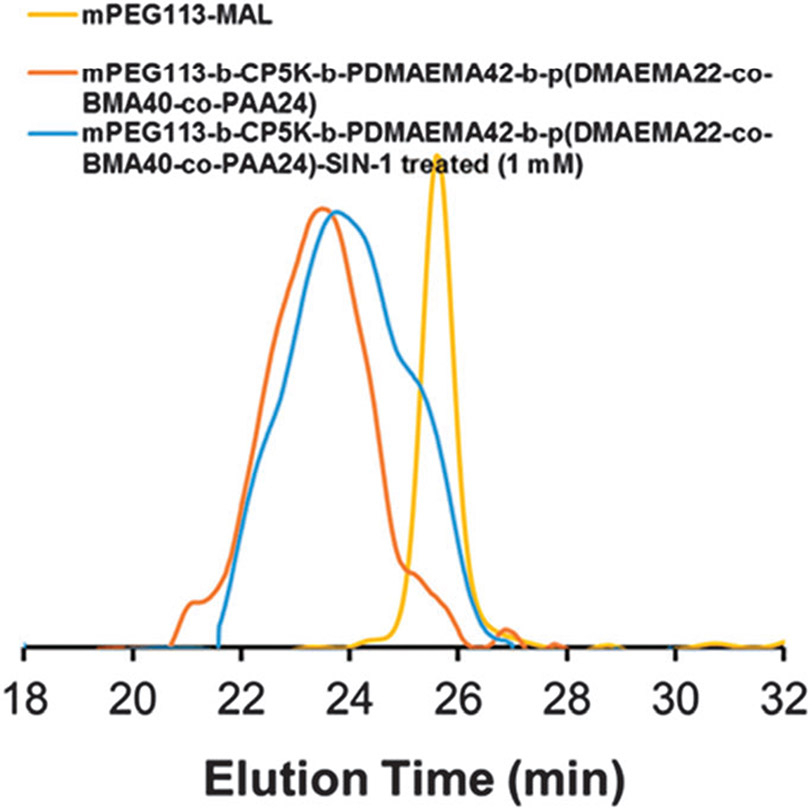
To confirm ROS-mediated dePEGylation of the nanocarrier, PPDDBP copolymer micelles were incubated overnight with a 1 mM dose of SIN-1, a producer of peroxynitrite,38,39 in DPBS at pH 7.4. Fig. 2 shows GPC chromatograms of mPEG113-MAL and PPDDBP polymer before and after SIN-1 treatment. In the presence of SIN-1, a clear decrease in the polymer molecular weight with an appearance of a mPEG shoulder peak indicates ROS-mediated dePEGylation of polymer micelles through cleavage of CP5K peptide. DePEGylation also increased the ζ-potential from 1.54 mV to 8.45 mV for PPDDBP micelles (Fig. S5B, ESI†) over a 20 hour incubation with 4 mM concentration of SIN-1. In contrast, the zeta potential of the control PDDBP micelles remained unchanged (Fig. S5B, ESI†). The increase in the zeta potential of the PPDDBP micelles compared to control (PDDBP) after SIN-1 treatment suggests that the cleavage of mPEG block from PPDDBP results in an exposure of the positively-charged amine groups of the PDMAEMA block in aqueous solution. To investigate the effect of SIN-1 mediated dePEGylation on size and stability of micelles, PPDDBP polymer micelles (1 mg mL−1) in DPBS (pH 7.4) were incubated with SIN-1 (4 mM) for 24 h. DLS measurement indicated that the size of polymer micelles (47 nm) remained unchanged after SIN-1 treatment, indicating the micelle stability was maintained by hydrophilic PDMAEMA corona and hydrophobic P(DMAEMA-co-BMA-co-PAA) core. (Fig. S7, ESI†).
Fig. 2.
GPC chromatograms of PPDDBP show ROS-mediated detachment of PEG block from PPDDBP polymer through cleavage of CP5K peptide linkers after overnight treatment of SIN-1 (1 mM), evidenced by the appearance of shoulder peak of PPDDBP (blue) overlapping the PEG peak (yellow).
These results suggest that in ROS-rich microenvironments, the PPDDBP micelles would improve cellular uptake by triggering the interaction of positively-charged polymer micelles with the negatively-charged plasma.
3.4. Preparation and characterization of PPDDBP/pDNA polyplexes
The size and ζ-potential of polyplexes were measured to determine their compatibility for intravenous delivery.40 To determine the stability of the polyplexes (N/P 10), we used DLS to characterize the size distribution, and observed a unimodal peak with an average diameter of 120 nm (Fig. 3A). The size distribution of polyplexes was further examined by TEM, which confirmed monodisperse particles with spherical morphology (Fig. 3B). The smaller size of the polyplexes in the TEM image compared to that measured by DLS can be explained by dehydration of the hydrophilic segments during TEM sample preparation.41
Fig. 3.
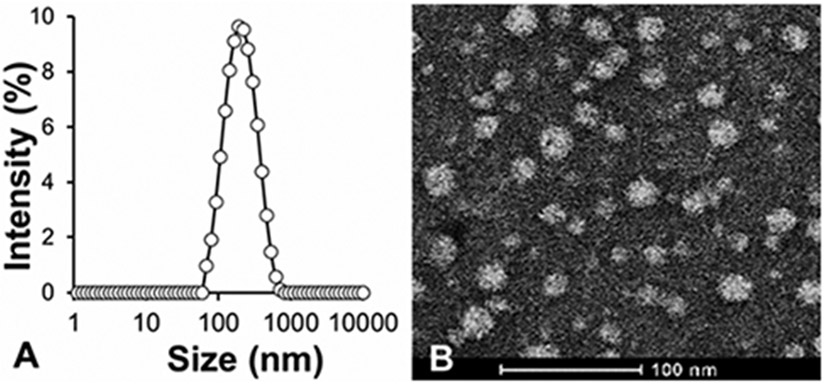
Size and morphology of PPDDBP/pDNA polyplexes (A) DLS-based size measurement PPDDBP/pDNA polyplexes in DPBS (pH 7.4) at N/P 10 shows formation of stable polyplexes with an average diameter of 120 nm (PDI = 0.27). (B) TEM images of PPDDBP/pDNA polyplexes confirm the formation of stable polyplexes with spherical morphology.
To further verify the ROS-mediated dePEGylation from polyplexes, PPDDBP/pDNA polyplexes (N/P 10) were incubated overnight with SIN-1 under similar condition as described for PPDDBP micelles in Fig. 4. The polyplex showed an increase in ζ-potential from 1.55 mV to 8.75 mV after SIN-1 treatment. This observation indicates the polyplexes can undergo dePEGylation in response ROS overproduction, thereby improving cellular uptake through enabling positively charged PDMAEMA to interact with the negatively charged plasma membrane of target cells.20,42
Fig. 4.
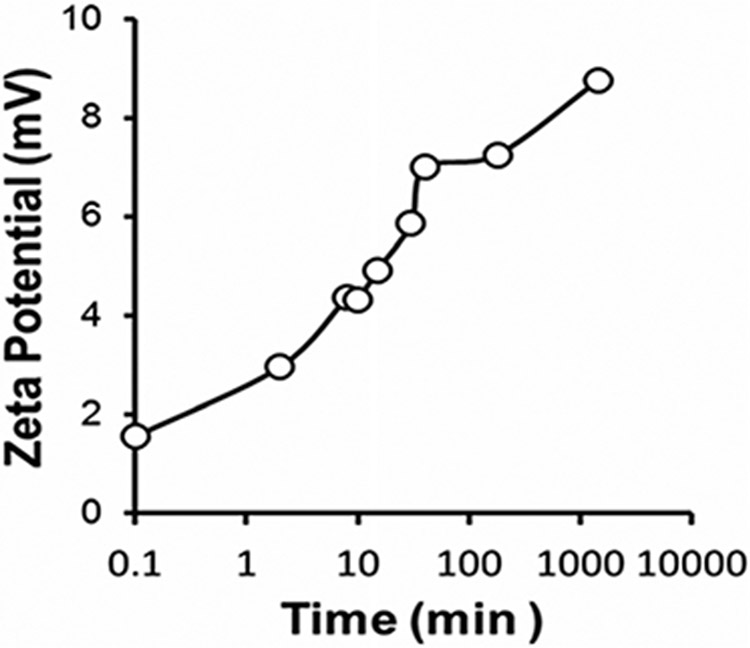
ROS responsiveness of polyplexes by DLS based ζ-potential. An increase in the ζ-potential of polyplexes from 1.55 to 8.75 mV after SIN-1 (4 mM) treatment for 24h further confirms the detachment of PEG block via a ROS-mediated cleavage of CP5K peptide linkers.
The condensation of PPDDBP with pDNA was examined by gel electrophoresis at various N/P ratios (1, 2, 5, 10, 20, and 30) and compared to free pDNA to determine the minimum amount of PPDDBP polymer required for effective polyplex formation with pDNA (Fig. S8, ESI†). Regardless of the N/P ratios, all the polyplex groups showed no free pDNA, which indicates effective condensation of pDNA.43 To investigate the stability of polyplexes and protection of pDNA in serum, PPDDBP/pDNA polyplexes at N/P 10 ratios were incubated with 50% serum for 0.25, 0.5, 1, 2, 3, and 6 h. As shown in Fig. S9, ESI,† a complete retention of pDNA within polyplexes was observed whereas free pDNA degraded in the presence of serum. In another gel experiment, protection of pDNA from serum-derived DNAses by PPDDBP was accessed by addition of 1% SDS to release pDNA from polyplexes in incubation with 50% serum. Notably, a solution of SDS (1% w/v) solution in PBS was added to each sample to release the pDNA from polyplexes. The appearance of released pDNA bands with persistent intensity relative to free pDNA in PBS indicates PPDDBP completely protected pDNA from FBS-derived DNAses at 37 °C (Fig. S9, ESI†).
A major barrier to intracellular gene delivery is entrapment and degradation of polyplexes in the endo-lysosomal pathway.44 To test if our nanocarrier can overcome this hurdle, hemolysis of the nanocarrier before and after polyplex formation was evaluated with three different polymer concentrations (1, 5, and 40 μg mL−1) at four different pH conditions (7.4. 6.8, 6.2 and 5.8) that mimic extracellular and endo-lysosomal trafficking.23 As shown in Fig. S10, ESI,† minimal hemolysis of RBCs was observed for all test concentrations at pH 7.4 compared to Triton-X-100 (positive control) indicating excellent hemocompatibility of PPDDBP under physiological conditions. Both polymers alone (Fig. S10A, ESI†) and polyplexes (Fig. S10B, ESI†) increased hemolysis of RBCs as pH decreased from 7.4 to 5.8. This result indicates that polyplexes can rupture RBC membranes at endosomal pH, thereby enabling successful escape from endo-lysosomal degradation.
3.5. Cytotoxicity assay
Cytotoxicity is a major limitation of liposomes, the most widely accepted delivery platform. Therefore, cytocompatibility of polyplexes prepared at different N/P ratios (1, 2, 4, 8, 10 and 20) with HCASMCs was evaluated by resazurin assay (Fig. 5).36 Cells showed more than 85% cell viability with PPDDBP polyplexes (N/P ratio at and below 10) while lipofectamine 2000 (LF2K) exhibited significantly lower cell viability. This result suggests better cytocompatibility of PPDDBP/pDNA polyplexes compared to commercial LF2K polyplexes under similar environments.
Fig. 5.
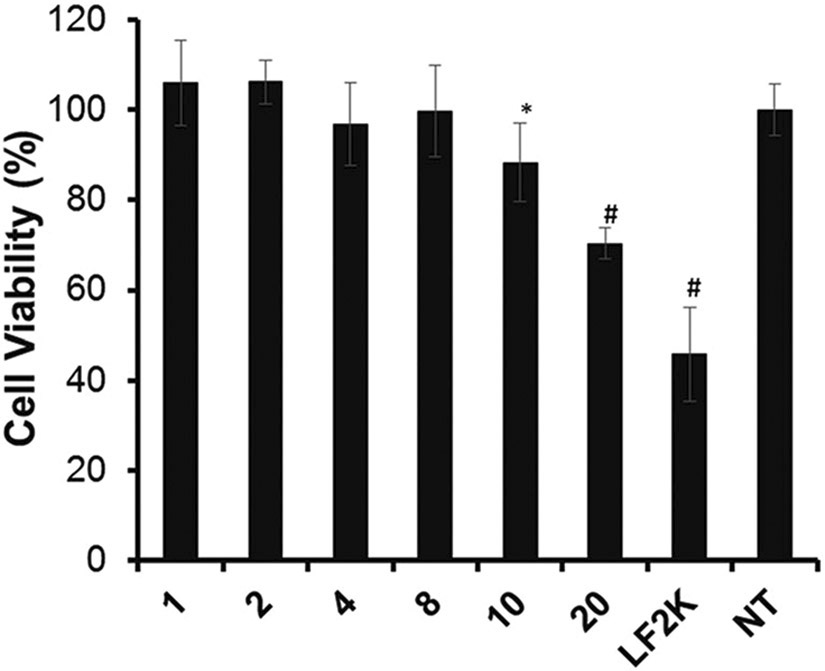
In vitro cytotoxicity of PPDDBP/pDNA polyplexes at different N/P ratio (1, 2, 4, 8, 10, and 20) was assessed by resazurin assay. Human coronary artery smooth muscle cells (HCASMCs) were incubated with PPDDBP/pDNA polyplexes or lipofectamine 2000 (LF2K) for 24 h before the endpoint assay (n = 3); *p < 0.05 vs. no treatment (NT); #p < 0.001 vs. NT.
3.6. Measurement of intracellular ROS production in HCASMCs
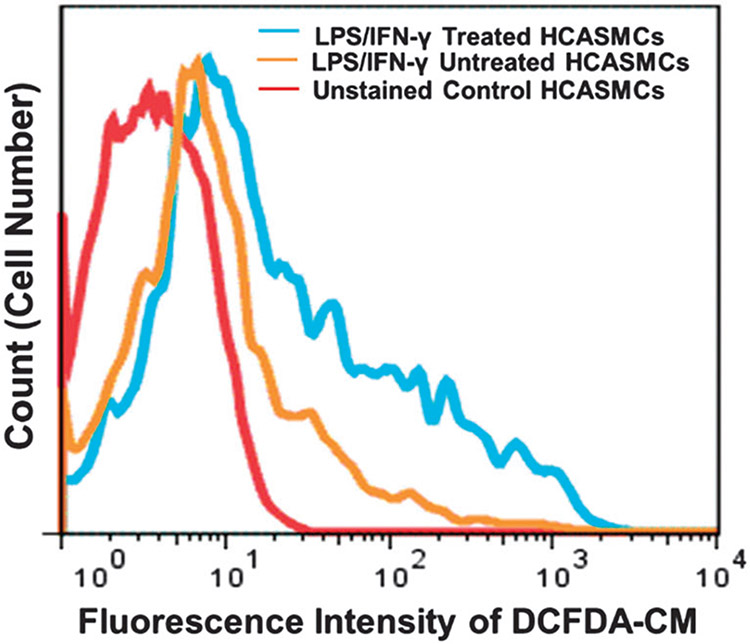
ROS are overproduced when VSMCs become dysfunctional under pathological vascular environments. Thus, ROS overproduction is likely an effective pathological stimulus for triggering therapeutic release from delivery systems.9 To generate an in vitro model of oxidative stress, ROS production of HCASMCs was induced by treatment of LPS (1 μg mL−1) and IFN-γ (100 units per mL) for 6 h.25 Intracellular ROS production was then measured by CM-H2DCFDA staining with flow cytometric analysis.37 LPS/IFN-γ-treated HCASMCs showed a significant shift in the mean fluorescence intensity of intracellular CM-H2DCFDA when compared with untreated HCASMCs (Fig. 6). These conditions were used to test ROS-responsive pDNA transfection of polyplexes.
Fig. 6.
Flow cytometric analysis of intracellular ROS levels in HCASMCs stained with CM-H2DCFDA (10 μM) after treatment with or without LPS (1 μg mL−1)/IFN-γ (100 units per mL) for 8 h. The LPS/IFN-γ-treated HCASMCs showed more ROS production compared to untreated cells (n = 3).
3.7. Cellular uptake of polyplexes
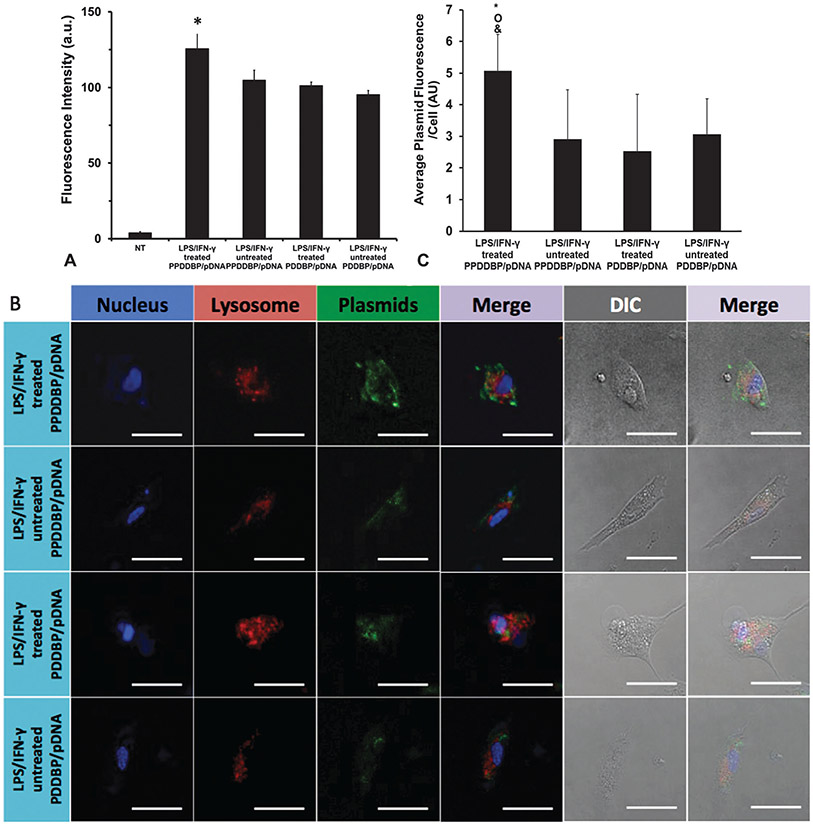
To test if CP5K cleavage in an ROS-rich environment would improve nanoparticle uptake and subsequent transfection, cellular uptake of polyplexes prepared by condensation of fluorescein-labeled pDNA either with ROS-cleavable PPDDBP or control PDDBP was quantitatively determined by flow cytometry (Fig. 7A) LPS/IFN-γ-treated HCASMCs showed improved uptake of PPDDBP polyplexes when compared to untreated HCASMCs or PDDBP conditions. This result suggests that ROS overproduction of HCASMCs in response to LPS/IFN-γ treatment induced dePEGylation of PPDDBP through CP5K peptide and thereby resulted in exposure of cationic PDMAEMA. These processes improved cellular uptake. Confocal microscopy images (Fig. 7B) and quantitative image analysis (Fig. 7C) showed that pDNA (green) was sequestered to endosomes (red) for untreated HCASMCs with PPDDBP, or with either control PDDBP condition, as evidenced by colocalization of the two colors. In stark contrast, treated HCASMCs with PPDDBP exhibited free pDNA within the cell, indicating that the pH-responsive nature of the underlying polyplex core was active once dePEGylation occurred, thereby enabling lysosomal escape of polyplexes.
Fig. 7.
Cellular uptake of polyplexes prepared using either PPDDBP or non-cleavable PDDBP with fluorescein-labeled pDNA was assessed in HCASMCs after 6 h incubation in the presence or the absence of LPS/IFN-γ. (A) Flow cytometry analysis indicates enhanced internalization of LPS /IFN-γ treated PPDDBP/pDNA polyplexes compared to other treatment groups. NT represents the non-treated group. *p < 0.05, where * is in comparison to the other test groups. (B) Confocal images of internalized polyplexes demonstrated distribution of plasmid (green), endosomes (red), and nuclei (blue). The cells were stained with DAPI and endosomal dye Lysotracker to visualize the nucleus and endosomes, respectively. Scale bar: 50 μm. (C) Confocal image analysis of polyplexes supports that LPS/IFN-γ treated PPDDBP/pDNA condition enhance uptake of plasmids significantly compared to all three other conditions. *, o, & indicates p < 0.05, where * is in comparison to LPS/IFN-γ untreated PPDDBP/pDNA, o to LPS/IFN-γ treated PDDBP/pDNA, and & to LPS/IFN-γ treated PPDDBP/pDNA, with N = 10–15 cells in each condition.
3.8. In vitro gene transfection
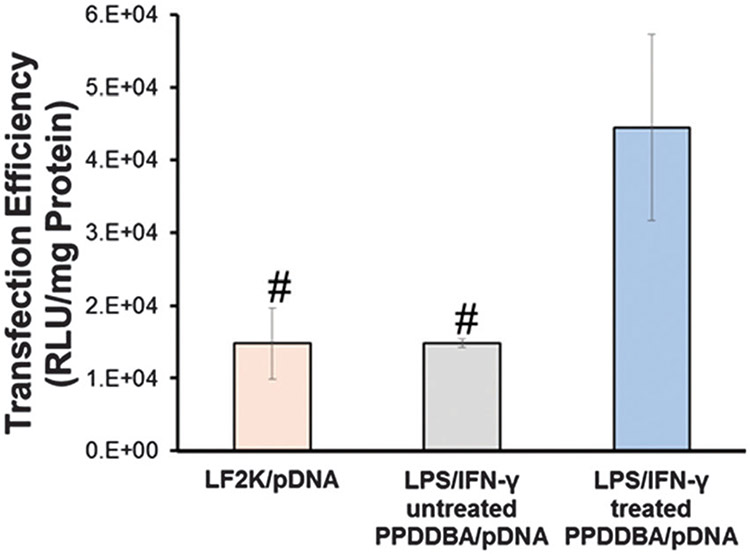
Transfection efficiency of PPDDBP/pDNA polyplexes with and without LPS/IFN-γ treatment was further evaluated in HCASMCs using a luciferase reporter (Fig. 8). PPDDBP polyplexes (N/P 10) were compared with commercially available lipofectamine (LF2K) as a positive control. The transfection efficiency of PPDDBP polyplexes was about 2.5 fold higher in HCASMCS pretreated with LPS/IFN-γ than that of untreated HCASMCS or commercially-available LF2K. The excellent cytocompatibility and higher cellular uptake of polyplexes under ROS-rich environments contributed to high transfection efficiency in HCASMCs when treated with LPS/IFN-γ.
Fig. 8.
Transfection efficiency of polyplexes was assessed in HCASMCs using PPDDBP or LF2K containing luciferase pDNA. Luminescence produced by cells post transfection was normalized to total protein. Data are expressed as mean ± SD (n = 3), #p < 0.05 compared with LPS/IFN-γ untreated PPDDBP/pDNA polyplexes.
4. Conclusions
In summary, the present study demonstrates the use of a new ROS-responsive polyplex nanocarrier as a non-viral gene delivery platform for targeting pathological vascular micro-environments that exhibit high levels of oxidative stress. A dual ROS and pH-responsive polyplex nanocarrier was synthesized by a combination of site-specific orthogonal bio-conjugation and RAFT polymerization methods, and was characterized for use in gene delivery applications. ROS-mediated dePEGylation of the polyplex nanocarrier was confirmed by the increase in ζ-potential in DLS and the appearance of a PEG shoulder in the GPC chromatogram. The polyplexes were shown to maintain hemolytic capability at endosomal pH and to protect pDNA from DNAses at a high serum concentration. When ROS was overproduced in HCASMCs in response to LPS/IFN-γ treatment, higher cellular uptake of polyplexes and enhanced transfection of pDNA were observed, compared with the controls. The collective data suggests the functional significance of this polyplex nanocarrier as a new class of dual stimuli-responsive polyplex platform for intravascular gene delivery.
Supplementary Material
Acknowledgements
This research work was funded and supported by NSF CAREER CBET 1056046 and NIH UH2 TR000491. DLS and TEM measurements were conducted through the use of the core facilities of the Vanderbilt Institute of Nanoscale Sciences and Engineering (VINSE), which was supported by NSF EPS 1004083.
Footnotes
Electronic supplementary information (ESI) available: Additional supporting figures. See DOI: 10.1039/c5tb00988j
References
- 1.Theoharis S, Manunta M and Tan PH, Expert Opin. Biol. Ther, 2007, 7, 627–643. [DOI] [PubMed] [Google Scholar]
- 2.Kay MA, Glorioso JC and Naldini L, Nat. Med, 2001, 7, 33–40. [DOI] [PubMed] [Google Scholar]
- 3.Thomas CE, Ehrhardt A and Kay MA, Nat. Rev. Genet, 2003, 4, 346–358. [DOI] [PubMed] [Google Scholar]
- 4.Liu Z, Zhang Z, Zhou C and Jiao Y, Prog. Polym. Sci, 2010, 35, 1144–1162. [Google Scholar]
- 5.Yin H, Kanasty RL, Eltoukhy AA, Vegas AJ, Dorkin JR and Anderson DG, Nat. Rev. Genet, 2014, 15, 541–555. [DOI] [PubMed] [Google Scholar]
- 6.Brito L, Little S, Langer R and Amiji M, Biomacromolecules, 2008, 9, 1179–1187. [DOI] [PubMed] [Google Scholar]
- 7.Golda A, Pelisek J, Klocke R, Engelmann MG, Rolland PH, Mekkaoui C and Nikol S, J. Vasc. Res, 2007, 44, 273–282. [DOI] [PubMed] [Google Scholar]
- 8.de Gracia Lux C, Joshi-Barr S, Nguyen T, Mahmoud E, Schopf E, Fomina N and Almutairi A, J. Am. Chem. Soc, 2012, 134, 15758–15764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clempus RE and Griendling KK, Cardiovasc. Res, 2006, 71, 216–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Madamanchi NR, Vendrov A and Runge MS, Arterioscler., Thromb., Vasc. Biol, 2005, 25, 29–38. [DOI] [PubMed] [Google Scholar]
- 11.Cai H and Harrison DG, Circ. Res, 2000, 87, 840–844. [DOI] [PubMed] [Google Scholar]
- 12.Shim MS and Xia Y, Angew. Chem., Int. Ed, 2013, 52, 6926–6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones CH, Chen C-K, Ravikrishnan A, Rane S and Pfeifer BA, Mol. Pharmaceutics, 2013, 10, 4082–4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griendling KK, Sorescu D and Ushio-Fukai M, Circ. Res, 2000, 86, 494–501. [DOI] [PubMed] [Google Scholar]
- 15.Tardif J-C, Grégoire J and L’Allier P, Am. J. Cardiovasc. Drugs, 2002, 2, 323–334. [DOI] [PubMed] [Google Scholar]
- 16.Martin JR, Gupta MK, Page JM, Yu F, Davidson JM, Guelcher SA and Duvall CL, Biomaterials, 2014, 35, 3766–3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee SH, Gupta MK, Bang JB, Bae H and Sung H-J, Adv. Healthcare Mater, 2013, 2, 908–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joshi-Barr S, de Gracia Lux C, Mahmoud E and Almutairi A, Antioxid. Redox Signaling, 2014, 21, 730–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song C-C, Du F-S and Li Z-C, J. Mater. Chem. B, 2014, 2, 3413–3426. [DOI] [PubMed] [Google Scholar]
- 20.Li J, Ge Z and Liu S, Chem. Commun, 2013, 49, 6974–6976. [DOI] [PubMed] [Google Scholar]
- 21.Li H, Yu SS, Miteva M, Nelson CE, Werfel T, Giorgio TD and Duvall CL, Adv. Funct. Mater, 2013, 23, 3040–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nie S, Nanomedicine, 2010, 5, 523–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Evans BC, Nelson CE, Yu SS, Beavers KR, Kim AJ, Li H, Nelson HM, Giorgio TD and Duvall CL, J. Visualized Exp, 2013, 73, e50166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cai X, Dong C, Dong H, Wang G, Pauletti GM, Pan X, Wen H, Mehl I, Li Y and Shi D, Biomacromolecules, 2012, 13, 1024–1034. [DOI] [PubMed] [Google Scholar]
- 25.Yu SS, Koblin RL, Zachman AL, Perrien DS, Hofmeister LH, Giorgio TD and Sung H-J, Biomacromolecules, 2011, 12, 4357–4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stadtman ER and Levine RL, Amino Acids, 2003, 25, 207–218. [DOI] [PubMed] [Google Scholar]
- 27.Lee SH, Boire TC, Lee JB, Gupta MK, Zachman AL, Rath R and Sung H-J, J. Mater. Chem. B, 2014, 2, 7109–7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Agarwal S, Zhang Y, Maji S and Greiner A, Mater. Today, 2012, 15, 388–393. [Google Scholar]
- 29.Convertine AJ, Benoit DSW, Duvall CL, Hoffman AS and Stayton PS, J. Controlled Release, 2009, 133, 221–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goloub T, de Keizer A and Cohen Stuart MA, Macromolecules, 1999, 32, 8441–8446. [Google Scholar]
- 31.Nelson CE, Gupta MK, Adolph EJ, Shannon JM, Guelcher SA and Duvall CL, Biomaterials, 2012, 33, 1154–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nelson CE, Kim AJ, Adolph EJ, Gupta MK, Yu F, Hocking KM, Davidson JM, Guelcher SA and Duvall CL, Adv. Mater, 2014, 26, 607–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tirrel DA and Ferrito M, Macromol. Synth, 1992, 11, 59–62. [Google Scholar]
- 34.Nelson CE, Kintzing JR, Hanna A, Shannon JM, Gupta MK and Duvall CL, ACS Nano, 2013, 7, 8870–8880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gupta MK, Meyer TA, Nelson CE and Duvall CL, J. Controlled Release, 2012, 162, 591–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O’Brien J, Wilson I, Orton T and Pognan F, Eur. J. Biochem, 2000, 267, 5421–5426. [DOI] [PubMed] [Google Scholar]
- 37.Eruslanov E and Kusmartsev S, in Advanced Protocols in Oxidative Stress II, ed. Armstrong D, Humana Press, 2010, ch. 4, vol. 594, pp. 57–72. [Google Scholar]
- 38.Kuzkaya N, Weissmann N, Harrison DG and Dikalov S, Biochem. Pharmacol, 2005, 70, 343–354. [DOI] [PubMed] [Google Scholar]
- 39.Hogg N, Darley-Usmar VM, Wilson MT and Moncada S, Biochem. J, 1992, 281, 419–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Merdan T, Kopeček J and Kissel T, Adv. Drug Delivery Rev, 2002, 54, 715–758. [DOI] [PubMed] [Google Scholar]
- 41.Cabral H, Matsumoto Y, Mizuno K, Chen Q, Murakami M, Kimura M, Terada Y, Kano MR, Miyazono K, Uesaka M, Nishiyama N and Kataoka K, Nat. Nanotechnol, 2011, 6, 815–823. [DOI] [PubMed] [Google Scholar]
- 42.Takae S, Miyata K, Oba M, Ishii T, Nishiyama N, Itaka K, Yamasaki Y, Koyama H and Kataoka K, J. Am. Chem. Soc, 2008, 130, 6001–6009. [DOI] [PubMed] [Google Scholar]
- 43.Putnam D, Nat. Mater, 2006, 5, 439–451. [DOI] [PubMed] [Google Scholar]
- 44.Layek B and Singh J, Biomacromolecules, 2013, 14, 485–494. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.