Abstract
Aim
To provide a detailed genomic-epidemiological description of a complex multi-ward SARS-CoV-2 outbreak, which originated in the crowded emergency department (ED) in our hospital during the third wave of the COVID-19 pandemic, and was elucidated promptly by local whole-genome sequencing (WGS).
Methods
SARS-CoV-2 was detected by reverse transcriptase real-time polymerase chain reaction on viral RNA extracted from nasopharyngeal swabs. WGS was performed using an Oxford MinION Mk1C instrument following the ARTIC v3 sequencing protocol. High-quality consensus genomes were assembled with the artic-ncov2019 bioinformatics pipeline and viral phylogenetic trees were built, inferred by maximum-likelihood. Clusters were defined using a threshold of 0–1 single nucleotide polymorphisms (SNPs) between epidemiologically linked sequences.
Results
In April 2021, outbreaks of COVID-19 were declared on two wards at University Hospital Limerick after 4 healthcare-associated SARS-CoV-2 infections were detected by post-admission surveillance testing. Contact tracing identified 12 further connected cases; all with direct or indirect links to the ED ‘COVID Zone’. All sequences were assigned to the Pangolin B.1.1.7 lineage by WGS, and SNP-level analysis revealed two distinct but simultaneous clusters of infections. Repeated transmission in the ED was demonstrated, involving patients accommodated on trolleys in crowded areas, resulting in multiple generations of infections across three inpatient hospital wards and subsequently to the local community. These findings informed mitigation efforts to prevent cross-transmission in the ED.
Conclusion
Cross-transmission of SARS-CoV-2 occurred repeatedly in an overcrowded emergency department. Viral WGS elucidated complex viral transmission networks in our hospital and informed infection, prevention and control practice.
Keywords: SARS-CoV-2, Whole-genome sequencing, COVID-19, Outbreak investigation, Genomic epidemiology, Hospital overcrowding
Introduction
COVID-19, caused by SARS-CoV-2, is a global public health concern that has reached pandemic status [1] and caused significant morbidity and mortality worldwide. In Ireland, the ‘third-wave’ of the pandemic was characterized by a dramatic rise in community infections in early 2021, following phased easing of public health non-pharmaceutical interventions and the widespread transmission of the B.1.1.7 Alpha Variant of Concern (VOC) [2,3]. During this time, a surge in COVID-19 hospitalizations threatened to overwhelm the Irish healthcare system, and despite robust infection prevention and control (IPC) efforts, healthcare-acquired COVID-19 infections and nosocomial outbreaks were frequently observed [4]. As such, the prevention of SARS-CoV-2 transmission was recognized as critical to protecting patients and healthcare workers in the acute hospital setting [4].
Whole-genome sequencing (WGS) has emerged as an important tool for characteriszing the epidemiology and patterns of transmission of a wide variety of microbial pathogens [[5], [6], [7]], and has been shown to offer higher resolution for comparative analyses during outbreak investigations than traditional typing techniques [8]. In recent years, the development of affordable, high-throughput, massively parallel next-generation sequencing (NGS) platforms has meant that sequencing is now feasible for diagnostic purposes in routine clinical microbiology laboratories [5,8], and can deliver actionable results to IPC teams in hospitals within clinically meaningful timeframes [9,10]. Throughout the COVID-19 pandemic, the unprecedented scale of genomic data generated worldwide has provided important insights into the evolution and pathogenesis of the SARS-CoV-2 virus. Integrated genomic surveillance networks have informed public health decision-making in many jurisdictions [11,12] while, on a local scale, high-resolution single nucleotide polymorphism (SNP)-level analysis has helped to resolve nosocomial [9,[13], [14], [15]] and community [16] COVID-19 outbreaks. Despite this, limitations to WGS exist, and in real-world settings phylogenetic reconstruction of outbreaks can only approximate true chains of transmission as unsampled cases will invariably exist; therefore, genetic data must be combined with epidemiological information and knowledge of background genomic diversity for accurate interpretation of results.
To improve our understanding of SARS-CoV-2 transmission within our hospital and inform IPC practice, WGS was introduced in early 2021. When a complex nosocomial outbreak was declared in April of that year, genomic data were combined with an enhanced epidemiological investigation to elucidate mechanisms of viral transmission.
Methods
Setting
University Hospital Limerick (UHL) is a tertiary-referral hospital in Ireland, with an inpatient capacity of 531 beds. The hospital's new emergency department (ED), which serves the entire mid-west region of the country and a population of almost 500,000 people, was opened in 2017 and is built to modern specifications, with 49 single treatment rooms and mechanical ventilation supplied to all care areas. In accordance with national guidelines [17], patients presenting to the ED undergo an initial risk assessment and are allocated to specific COVID-19 or non-COVID-19 pathways. Those with suspected or confirmed COVID-19 receive care in one of three dedicated zones within the department, designated Zone B, Zone C and Zone D. At the time of this outbreak, surgical face masks were worn universally by healthcare workers (HCWs) and offered to patients; within dedicated COVID zones, filtered face-piece respirators (FFP-2) were used by those providing direct patient care. During periods when inpatient bed-space capacity is exceeded by the number of acute presentations, patients are accommodated on trolleys situated along corridors in the ED and also on hospital wards, and priority for isolation is given to confirmed COVID-19 cases. When available, patients with suspected or confirmed COVID-19 requiring admission to hospital are placed in single rooms on dedicated COVID-19 wards supplied with mechanical ventilation. However, outside of dedicated COVID-19 units, many of the general inpatient wards have older infrastructure, and are comprised of multi-bedded, open-plan, ‘Nightingale-style’ bays accommodating up to 14 inpatients, relying exclusively on natural ventilation. During the study period, post-admission nasal swab surveillance testing of inpatients for SARS-CoV-2 was conducted 72-hourly. Routine surveillance testing of asymptomatic HCWs was not performed either by polymerase chain reaction (PCR) or antigen testing, in line with contemporaneous national IPC guidance. However, in the context of outbreak investigations and on the advice of the Outbreak Control Team (OCT), PCR testing of both asymptomatic staff and patients was conducted in specified areas at day 0 (first discussion at outbreak meeting) and day 7.
Case definitions
SARS-CoV-2 infections were diagnosed in individuals who returned an ‘RNA detected’ result by real-time, quantitative reverse-transcriptase PCR (RT-qPCR) on nasopharyngeal swabs, performed on either the Alinity M (Abbott, Abbott Park, IL, USA), Respibio (Serosep, Limerick, Ireland) or GeneXpert (Cepheid, Sunnyvale, CA, USA) diagnostic SARS-CoV-2 platforms. Samples were collected in the context of suspected COVID-19 infection, as part of routine post-admission surveillance testing, or as part of contact tracing efforts during outbreak investigations. Healthcare-associated infections (HCAIs) and community-acquired infections (CAIs) were categorized in accordance with national surveillance definitions [18]. ‘Probable’ outbreak cases were defined as those in which a combination of both epidemiological and genomic data provided support for viral transmission, while ‘possible’ outbreak cases were those linked by epidemiological or genomic information alone.
WGS and comparative genome analysis
All HCAI cases were reviewed by the OCT, and PCR-positive samples with cycle threshold (CT) values of <30 were selected for WGS in suspected outbreak cases. Nucleic acid re-extraction was performed using a Liferiver EX3600 automated nucleic acid extractor system (Liferiver Bio-tech, San Diego, CA, USA) and SARS-CoV-2 whole-genome libraries were prepared using the ARTIC v3 tiled amplicon multiplex PCR protocol [19]. Final libraries were sequenced on a MinION Mk1C instrument (Oxford Nanopore Technologies, Oxford, UK). Using the artic-ncov2019 SARS-CoV-2 bioinformatics pipeline [20], high-quality consensus genomes were assembled with a minimum of 94% genome completion and Pangolin v2.0 [21] was utilized for viral lineage assignment. Analysis of run quality was with the ‘Ncov-tools’ QC pipeline [22]. Multi-way sequence alignments were performed using MAFFT v7.407 [23], and viral phylogenetic trees inferred by maximum-likelihood were generated with IQ-TREE v1.6.12 [24] and visualized with iTOL v.6 [25]. USHeR [26] was utilized for rapid placement of outbreak sequences on a global phylogeny and for interrogation of mutations. In keeping with published thresholds for differentiating clusters of nosocomial SARS-CoV-2 infections [13], 0–1 pairwise SNPs supported viral transmission between epidemiologically linked cases. Plots of individual SNPs relative to a reference sequence were generated with ‘snipit’ [27] and all outbreak sequences were deposited in the Global Initiative on Sharing All Influenza Data (GISAID) repository [28] (accession numbers are provided in the Supplementary data.)
Ethical approval
Ethical approval for this study was granted by the hospital Research and Ethics Committee (Reference 141/2021).
Results
A COVID-19 outbreak was declared at UHL on 13th April 2021 after post-admission surveillance testing of four inpatients across two inpatient medical wards yielded positive results. All four sentinel cases were asymptomatic and in hospital for more than seven days prior to testing positive; meeting the definition for HCAI. Genomic and epidemiological investigation identified seven further linked hospitalized cases, along with five cases sampled in the community or at local healthcare facilities. Temporospatial links to patients in the ED COVID Zone C at UHL between 30th March and 1st April 2021 were demonstrated and, while an initial IPC assessment declared a single outbreak, WGS revealed two simultaneous but distinct clusters.
Case characteristics and outcomes of hospitalized patients
Case characteristics, including outcomes at 30 days from diagnosis, for hospitalized cases are summarized in Table I .
Table I.
Case characteristics of hospitalized cases
| Case | Age | Gender | Indication for hospitalization | Ward diagnosed | Date diagnosed | Outcome (30 days) | Distinguishing SNP patterna | |
|---|---|---|---|---|---|---|---|---|
| Cluster 1 | 1 | 90 | Male | Falls | ED Zone C | 19/03/21 | In hospital | H, I |
| 2 | 81 | Male | Acute cholecystitis | ED Zone C | 31/03/21 | Recovered; discharged | H, I, J | |
| 3 | 71 | Female | Chest pain | ED Zone C | 11/04/21 | ICU admission; recovered | H, I, J | |
| 4 | 87 | Female | Vasculitis | Ward B | 04/04/21 | In hospital | H, I, J, K | |
| 5 | 77 | Female | Falls | Community | 12/04/21 | Recovered | H, I, J, K | |
| 6 | 83 | Female | UTI | Ward B | 10/04/21 | In hospital | H, I, J, K, L | |
| 7 | 88 | Female | Falls | Ward B | 09/04/21 | Recovered; discharged | X | |
| 8 | 82 | Female | UTI | Community | 13/04/21 | Recovered | X | |
| Cluster 2 | 9 | 96 | Female | TIA | ED Zone C | 30/03/21 | Recovered; discharged | M |
| 10 | 76 | Male | Aspiration pneumonia | Ward C | 04/04/21 | Recovered; discharged | M | |
| 11 | 87 | Male | B-Cell lymphoma | Ward C | 06/04/21 | Died | M |
ED, emergency department; ICU, intensive care unit; SNP, single nucleotide polymorphism; TIA, transient ischaemic attack; UTI, urinary tract infection.
H = C15732T, I= C29546T, J = C17474T; K = C25611A; L = T23752C; M = G4288A; X = Not sequenced. Patient 7: cycle threshold (CT) value >30: did not meet criteria for whole-genome sequencing (WGS). Patient 8: tested in the community: sample could not be retrieved for WGS.
Description of cluster 1
The suspected index case, designated Patient 1, was transferred to UHL on 19th March 2021, testing positive for the presence of SARS-CoV-2 RNA on admission. He had been identified as a close-contact of a COVID-19 infection at another hospital 10 days earlier. He was initially placed on a dedicated COVID-19 ward but, two days post-admission, he required transfer to a two-bedded room on a non-COVID ward (Ward A) due to a requirement for cardiac telemetry, which was only available in certain areas of the hospital. During this time, transmission-based precautions were applied and no other patients were accommodated in this room. Shortly after being admitted to Ward A, Patient 1 experienced a seizure necessitating the administration of high-flow oxygen in his room and was transferred to a dedicated COVID-19 ward for higher acuity care.
Two hours after Patient 1 left his room, Patient 2 was admitted to the adjacent bed-space from a different area on ward A, where he spent a total of 14 h before transfer to another ward. He was discharged on 30th March, but re-presented to the ED Zone C on 31st March after a clinical deterioration. Before the result of his admission PCR test was available, which confirmed the presence of SARS-CoV-2 RNA, he spent 11 h on a trolley in the ED. During this time, he was in close proximity (approximately 4 m) to Patient 3, a female patient also located on a trolley. After a negative COVID-19 test, Patient 3 was discharged on 31st March, but re-presented with COVID-19 pneumonia requiring direct admission to the intensive care unit (ICU) on 11th April. Patient 4, one of the sentinel cases, also spent time in the ED Zone C on 30th March after transfer from another healthcare facility, overlapping with Patients 2 and 3. In the week prior to transfer to UHL, she had returned a total of four negative COVID-19 PCR tests. Following a period of 24 h in ED Zone C, she was moved to a general medical inpatient ward (Ward B) where, five days later, she tested positive for SARS-CoV-2 on routine surveillance testing. Although she was asymptomatic at the time of diagnosis, she deteriorated shortly thereafter and was transferred to a dedicated COVID-19 ward. However, during her time on Ward B, Patient 4 overlapped with four patients who were subsequently designated as HCAI outbreak cases: Patients 5, 6, 7 and 8.
Patient 5 was a woman treated on Ward B between 27th March and 8th April 2021, and subsequently diagnosed with COVID-19 infection in the community on 12th April: despite multiple negative COVID swabs during her inpatient stay, she became symptomatic shortly after leaving hospital, and was identified as a close contact of Patient 4. Another sentinel case, Patient 6, attended ED Zone C on 1st April, testing negative for COVID-19 on presentation. She was admitted to a multi-bedded bay on Ward B and SARS-CoV-2 RNA was subsequently detected through surveillance testing on 10th April. Patients 7 and 8, who were both present on the same multi-bedded bay on Ward B as Patient 6 for five days, were both deemed ‘possible’ outbreak cases as their samples were unavailable for WGS: Patient 7 was also asymptomatic and diagnosed on a surveillance test performed on 9th April with a CT value >30, while Patient 8 became symptomatic shortly after leaving hospital and was diagnosed in the community. Temporal and geographic links between cases within Clusters 1 and 2 are illustrated in Figure 1 .
Figure 1.
Gannt chart illustrating temporal and geographic links between cases. Within cluster 1, the putative chain of transmission from Patient 1 to Patient 8 is supported, beginning in Ward A, through the emergency department on 30th–31st March, and subsequently on to Ward B. Within Cluster 2, Patient 9 is the proposed source of introduction, overlapping with Patient 10 in the emergency department, who in turn shared time with Patient 11 on Ward C. HDU, high-dependency unit; ICU, intensive care unit.
Viral WGS and pairwise SNP analysis was performed on available samples from Patients 1 to 6, with preliminary results available within seven days of outbreak declaration, supporting the hypothesis that Patient 1 was the source of introduction into UHL. The viral genomes recovered from Patients 1 and 2 differed by a single SNP (C17474T); given that their time in the same room on Ward A was separated by a 2-hour window, and no HCWs sampled on the ward returned positive results, viral transmission from Patient 1 to Patient 2 via an airborne route or via an environmental surface in the room was felt to be most plausible. Sampling of frequent touch surfaces in this room was performed in the weeks following the putative transmission event from Patient 1 to Patient 2, though SARS-CoV-2 RNA was not detected from these samples. A stepwise accumulation of two further SNPs over sequential generations of infection over a 21-day period supported a direction of transmission from Patient 1 to Patient 6, which is illustrated in Figure 2 .
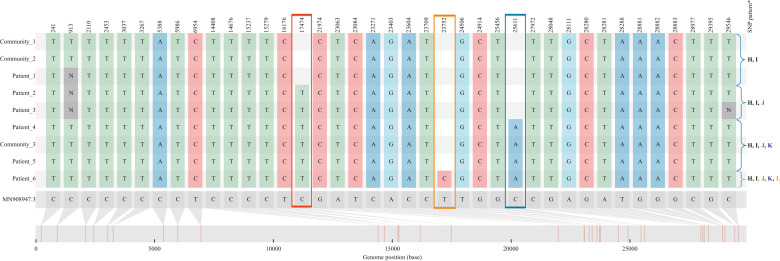
Figure 2.
Single nucleotide polymorphism (SNP) analysis of Cluster 1 using snipit, displaying outbreak sequences relative to reference MN9089047.3 (in grey) and genome position of each SNP. From the top row down, a stepwise accumulation of three mutations is seen over sequential generations of infection, from Patient 1 to Patient 6. The viral sequences recovered from Patient 2 onwards differed from those obtained from Patient 1 (and Community Cases 1 and 2) by the accumulation of single non-synonymous SNP, C17474T (highlighted within red box), leading to a missense mutation causing an amino acid change of Threonine to Isoleucine at codon 5737 of ORF1ab. Patient 2 and Patient 3 shared indistinguishable viral genomes, which offered support for transmission while both were accommodated in close proximity on trolleys in Emergency Department Zone C. The sequences obtained from Patient 4, Patient 5 and Community sequence 3 revealed one further SNP difference, C25611A (blue box), while in the sequence from Patient 6 diverged further by another single SNP; T23752C (orange box). In keeping with the annotation used in Table I, the SNP pattern distinguishing viral genomes in Cluster 1 from other sequences in the local region are highlighted on the right-hand side. ∗ H = C15732T, I= C29546T, J = C17474T; K = C25611A; L = T23752C.
Description of cluster 2
Patient 9 was transferred to UHL from a nursing home, and also received care in the ED Zone C on 30th March 2021, testing positive for the presence of SARS-CoV-2 RNA on presentation. She spent a total of 10 h in Zone C, overlapping temporally and geographically with patients subsequently assigned to Cluster 1 by WGS. Her time in ED also overlapped with Patient 10, a patient who was also diagnosed with HCAI on 4th April through post-admission surveillance testing on Ward C where, in turn, she shared time with Patient 11, an immunosuppressed man who tested positive on 6th April. Illustrated by Figure 3 , WGS performed on samples received from Patients 9, 10 and 11 demonstrated indistinguishable viral genomes with a shared unique G2488A mutation, which supported a direction of transmission from Patient 9 to 10 in the ED zone C, and subsequently from Patient 10 to Patient 11 on Ward C.
Figure 3.
(a) Maximum-likelihood viral phylogenetic tree rooted in Wuhan MN908947.3, including all B.1.1.7 viral genomes submitted to Global Initiative on Sharing All Influenza Data (GISAID) from the Mid-West region of Ireland (including counties Limerick, Clare and Tipperary, N = 422) between 1st March and 30th April 2021. Cluster 1 is highlighted in red and Cluster 2 in green. (b) Singler nucleotide polymorphism (SNP) annotated viral phylogenetic tree rooted on MN908947.3 and pruned to highlight outbreak sequences: both clusters were preserved with 100% ultrafast bootstrap support (nodes with support values ≥97% are annotated). SNPs unique to each clade are highlighted in blue: Cluster 1 was defined by the presence of two nucleotide mutations C15732T and C29546T, while sequences in Cluster 2 all shared a unique G4288A mutation.
Links to community sequences
As illustrated by Figure 3, outbreak sequences were placed on a viral phylogenetic tree which included over 400 Irish B.1.1.7 lineage genomes from the local region submitted to GISAID in the months preceding and following the outbreak. Five additional identical sequences were uncovered by this analysis (Community Cases 1 to 5), and were identified as ‘possible’ outbreak cases. These were sequenced at the UCD National Virus Reference Laboratory (NVRL), Dublin, Ireland, and assembled viral genomes were retrieved from GISAID for inclusion in our phylogenetic analysis. Plausible links were established between each of these community sequences and cases within Clusters 1 and 2, which supported a continuous chain of transmission between individuals rather than the introduction of unrelated but identical sequences into UHL by chance: Community Cases 1 and 2 were indistinguishable from the sequence recovered from Patient 1, and were both collected from patients with COVID-19 at the local hospital where he received care prior to transfer to UHL, while Community Case 3 was a household contact of Patient 4. Community Cases 4 and 5 were collected from individuals from the same town as Patient 9, the suspected primary case in Cluster 2, and shared identical viral genomes.
Infection prevention and control response
Following the outbreak declaration on Wards A, B and C, PCR testing of all patients and HCWs took place, with no further cases identified. Cross-transmission demonstrated by WGS facilitated focused IPC interventions; in the ED, point-of-care testing using the Abbott ID NOW™ instrument (Abbott, Abbott Park, IL, USA) for SARS-CoV-2 was introduced to rapidly detect asymptomatic infections and provide overnight testing when the virology laboratory was closed (8pm to 8am), while on Wards B and C, portable HEPA filtration units have been installed. Furthermore, decisions on the postponement of scheduled care across the hospital to assist with efforts to alleviate ED overcrowding were also informed by WGS.
Discussion
This study describes the experience of using WGS, performed and analysed locally in near real-time, to characterize a nosocomial COVID-19 outbreak linked to an overcrowded ED in a tertiary hospital in Ireland, during a period of high community prevalence of SARS-CoV-2. Findings supported viral spread between patients accommodated on trolleys and transmission across multiple inpatient hospital wards, resulting in significant morbidity.
Crowded conditions in hospitals, and particularly emergency departments, are known to pose considerable risks to timely, safe and efficient patient care [29] and such conditions have been associated with both transmission of SARS-CoV-2 [30] and increased patient mortality during the COVID-19 pandemic [31]. At our institution, overcrowding has been identified as an ongoing challenge for many years, attributed to insufficient inpatient bed-space capacity in the hospital and wider region, while the practice of boarding extra patients on trolleys in the ED and other areas of the hospital during busy periods has been highlighted as a particular concern [32]. This outbreak highlights how such practices and conditions acted as barriers to the implementation of optimal IPC standards, including the maintenance of adequate distance between patients awaiting SARS-CoV-2 test results in dedicated COVID-19 care zones. However, it is important to recognize that crowding was inevitable within the context of available bed capacity (both total and single room) and the unprecedented numbers of acute presentations to our ED during the third pandemic wave. Despite the temporary curtailment of elective care at our hospital in early 2021 to facilitate the provision of unscheduled care, repeated surges in COVID-19 admissions unmasked longstanding deficits in healthcare capacity, manifesting as overcrowding, virus cross-transmission and, ultimately, worse patient outcomes. Identifying and addressing such capacity deficits is crucial to avoid nosocomial transmission during future surges of infectious diseases including COVID-19.
Interestingly, we note parallels between this outbreak and the original SARS-CoV-1 epidemic 20 years ago, whereby a large nosocomial outbreak occurred following the introduction and spread of SARS-Coronavirus Type 1 into the ED of a hospital in Toronto. Similar to our experience, onward transmission was observed from an individual with an occult healthcare-acquired infection, who was initially discharged from hospital after spending time in close proximity to an index case, and was not isolated upon re-presentation to ED shortly thereafter [33]. However, in contrast to the tools available to investigators in 2003, access to viral WGS in our diagnostic laboratory enabled prompt reconstruction of this outbreak.
Throughout the COVID-19 pandemic, genomic data have informed IPC responses to nosocomial COVID-19 outbreaks in many countries [9,[13], [14], [15]] and shed light on the dynamics of viral transmission. In this study, pairwise-SNP analysis of viral genomes from inpatients and community cases quickly elucidated the source of introduction into our hospital and, in combination with a detailed epidemiological investigation, resolved the chain of transmission between patients. This facilitated targeted IPC interventions in areas of the hospital where cross-transmission was demonstrated, including the introduction of point-of-care testing in our ED and the installation of portable HEPA filters on inpatient wards.
Of note, comparative SNP analysis provided support for an airborne or environmentally mediated transmission event between Patient 1 and Patient 2, who were temporally separated by a 2-h window on Ward A, and whose viral sequences differed by a single mutation. Although the relative contribution of airborne viral particles and fomites to onward transmission of SARS-CoV-2 has been subject to debate, laboratory studies have demonstrated potentially infectious virus is detectable for up to 3 h in aerosols and for up to 72 h on plastic and stainless steel [34], while experiments conducted on hospital wards have recovered viable SARS-CoV-2 RNA from patients' rooms even in the absence of aerosol-generating procedures [35]. While PCR testing of environmental surfaces in the room on Ward A did not detect viral RNA, it was performed 2 weeks after the event, with terminal cleaning of all surfaces in the room (using a chlorine-dioxide-based solution with potent activity against SARS-CoV-2 [36]) taking place after the room was vacated by Patient 1. Air sampling was not performed which, combined with the above limitations, meant the precise route of transmission could not be established. However, given that enhanced contact tracing which included PCR testing of all asymptomatic patients and HCWs did not identify other cases, IPC measures in place for HCWs (which included universal use of surgical face-masks and FFP-2 masks in COVID zones) were shown to be effective, and indirect transmission via aerosols or an environmental surface in the room was judged to be the most plausible mechanism.
In addition to providing support for directionality of transmission between patients, individual genomic mutations may also offer insights into patient outcomes and the predicted pathogenicity of SARS-CoV-2; there is evidence to suggest that the C25611A mutation detected in Patients 4, 5 and 6, which causes a synonymous mutation in the Orf3A region may be associated with increased disease severity relating to changes in RNA secondary structure [37]. However, no clear correlation with outcomes of the patients harbouring this mutation were apparent in this study.
Despite the power of WGS to resolve outbreaks in healthcare settings and delineate chains of transmission, its limitations must also be considered when making inferences based on similar genetic sequences alone. The moderate substitution rate in SARS-CoV-2 as a result of viral replication with proofreading activity [38] and the frequent occurrence of convergent mutations [39] means that genetically indistinguishable viruses may be recovered from unrelated individuals, and can lead to false assumptions about links between cases. This limitation is highlighted by Abbas et al. [40] who found that, due to low genetic diversity between cases sequenced during a nosocomial SARS-CoV-2 outbreak in a rehabilitation centre, genomic data offered little to the outbreak investigation. The relative sensitivity and specificity of SNP thresholds for defining linkage between and within clusters can vary according to the viral genomic diversity and prevalence at a given time-point in the pandemic; this is highlighted by Lumley et al. [13] who, in the context of a large analysis of nosocomial infections during the winter of 2020, demonstrated that a threshold of 0–1 SNPs combined with epidemiological data captured the majority of cases genuinely linked to a given cluster whilst minimizing over-calling of linkage in non-nosocomial cases. Conversely, we show that in prolonged outbreaks the index case may differ from subsequent connected cases by two or more SNPs after multiple generations of transmission; however, links between these cases could only be demonstrated by intensive contact tracing and rigorous sampling and in many circumstances (including this study whereby WGS could not be performed on samples from Patients 7 and 8) complete phylogenetic reconstruction is limited by the existence of unsampled or unidentified intermediates.
This study also underlines the importance of integrated pathways to combine community and hospital patient metadata with genomic information, as previously unrecognized relationships between hospital samples and those collected in the local community were uncovered only through collaboration with our regional public health team and by access to consensus genomes sequenced elsewhere through the public genome repository GISAID. Internationally, coordinated national WGS networks have been crucial to pandemic responses by informing public health policy [12] and by enabling rapid sequencing which is prioritized at the point of need [41]. In the UK, the CLIMB-COVID digital infrastructure [42], which incorporates bioinformatics tools such as CIVET [43] has enabled integration of sample-associated metadata with genomic sequencing data and has facilitated prompt analysis of outbreaks in the context of background genomic diversity. In April 2021, when this outbreak took place, such a resource did not exist in Ireland and sequencing of SARS-CoV-2 had been implemented predominantly for genomic surveillance purposes on a national scale [11]. With some exceptions [14], limited infrastructure or capacity existed for the analysis of local hospital outbreaks in real time. To address this need, the Irish National SARS-CoV-2 Surveillance and Whole Genome Sequencing Programme was established in January 2021 [44], which has overseen plans to develop a distributed network of sequencing laboratories in six hospitals (including UHL) across the country, linking with Public Health and a central ‘Hub’ at UCD NVRL. With support from a European Centre for Disease Control (ECDC) Health Emergency preparedness and Response Authority (HERA) Incubator grant [45], this Hub-and-Spoke model will be introduced in early 2022, and aims to facilitate both prompt reactive analysis in the context of local outbreaks and enhanced national genomic surveillance.
In conclusion, cross-transmission of SARS-CoV-2 occurred repeatedly in our overcrowded ED involving patients accommodated in close proximity on trolleys, resulting in onward spread throughout the hospital. WGS, combined with a detailed epidemiological investigation and collaboration with regional Public Health, characterized the outbreak promptly and facilitated focused IPC interventions in high-risk areas of the hospital. The experience of developing and implementing WGS has informed local IPC strategies and has led to our site being designated a regional spoke in a national sequencing network.
Acknowledgements
The authors thank the staff within the clinical molecular laboratory and the Infection Control department at UHL, the whole-genome sequencing team at the UCD NVRL and Dr Margaret Morris-Downes, Surveillance Scientist, Department of Public Health, HSE Mid-West.
Conflict of interest statement
None declared.
Funding sources
None.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jhin.2022.04.015.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Cucinotta D., Vanelli M. WHO Declares COVID-19 a Pandemic. Acta Biomed. 2020 Mar 19;91(1):157–160. doi: 10.23750/abm.v91i1.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Epidemiological report of COVID-19 cases in Ireland during the third pandemic wave; November 22nd to February 21st2021. Epi Insight. March 2021;22(2) https://www.hpsc.ie/epi-insight/volume222021/ Available at: [last accessed January 2022] [Google Scholar]
- 3.Lima, V. The Pandemic One Year on: Trends and Statistics Between Three Waves of the COVID-19 Pandemic in Ireland. Available at: https://publicpolicy.ie/papers/the-pandemic-one-year-on-trends-and-statistics-between-three-waves-of-the-covid-19-pandemic-in-ireland/[last accessed January 2022].
- 4.Brief Update on COVID -19 Outbreaks and Hospital acquired cases in Acute Hospitals. Acute Operations HSE and AMRIC Lead. 27th January 2021 https://assets.gov.ie/126556/996860e1-2b03-4690-b69b-fedef14aadfe.pdf Available at: [last accessed January 2022] [Google Scholar]
- 5.Eyre D.W. Infection prevention and control insights from a decade of pathogen whole-genome sequencing. J Hosp Infect. 2022;122:180–186. doi: 10.1016/j.jhin.2022.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eyre D.W., Cule M.L., Wilson D.J., Griffiths D., Vaughan A., O’Connor L., et al. Diverse sources of C. difficile infection identified on whole-genome sequencing. N Engl J Med. 2013 Sep 26;369(13):1195–1205. doi: 10.1056/NEJMoa1216064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Price J.R., Cole K., Bexley A., Kostiou V., Eyre D.W., Golubchik T., et al. Modernising Medical Microbiology informatics group. Transmission of Staphylococcus aureus between health-care workers, the environment, and patients in an intensive care unit: a longitudinal cohort study based on whole-genome sequencing. Lancet Infect Dis. 2017 Feb;17(2):207–214. doi: 10.1016/S1473-3099(16)30413-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mitchell S.L., Simner P.J. Next-generation sequencing in clinical microbiology: are we there yet? Clin Lab Med. 2019 Sep;39(3):405–418. doi: 10.1016/j.cll.2019.05.003. [DOI] [PubMed] [Google Scholar]
- 9.Francis R.V., Billam H., Clarke M., Yates C., Tsoleridis T., Berry L., et al. COVID-19 Genomics UK (COG-UK) consortium. The Impact of Real-Time Whole-Genome Sequencing in Controlling Healthcare-Associated SARS-CoV-2 Outbreaks. J Infect Dis. 2022;225:10–18. doi: 10.1093/infdis/jiab483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Siqueira G.M.V., Pereira-Dos-Santos F.M., Silva-Rocha R., Guazzaroni M.E. Nanopore sequencing provides rapid and reliable insight into microbial profiles of intensive care units. Front Public Health. 2021;9:710985. doi: 10.3389/fpubh.2021.710985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mallon P.W.G., Crispie F., Gonzalez G., Tinago W., Garcia Leon A.A., McCabe M., et al. Whole-genome sequencing of SARS-CoV-2 in the Republic of Ireland during waves 1 and 2 of the pandemic. medRxiv. 2021 02.09.21251402. [Google Scholar]
- 12.Oude Munnink B.B., Nieuwenhuijse D.F., Stein M., O'Toole Á., Haverkate M., Mollers M., et al. Dutch-Covid-19 response team. Author Correction: Rapid SARS-CoV-2 whole-genome sequencing and analysis for informed public health decision-making in the Netherlands. Nat Med. 2020;26(9):1405–1410. doi: 10.1038/s41591-020-0997-y. 26(11):1802. Erratum for Nat Med. 2020. [DOI] [PubMed] [Google Scholar]
- 13.Lumley S.F., Constantinides B., Sanderson N., Rodger G., Street T.L., Swann J., et al. Epidemiological data and genome sequencing reveals that nosocomial transmission of SARS-CoV-2 is underestimated and mostly mediated by a small number of highly infectious individuals. J Infect. 2021;83(4):473–482. doi: 10.1016/j.jinf.2021.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lucey M., Macori G., Mullane N., Sutton-Fitzpatrick U., Gonzalez G., Coughlan S., et al. Whole-genome sequencing to track severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) transmission in nosocomial outbreaks. Clin Infect Dis. 2021;72:e727–e735. doi: 10.1093/cid/ciaa1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Løvestad A.H., Jørgensen S.B., Handal N., Ambur O.H., Aamot H.V. Investigation of intra-hospital SARS-CoV-2 transmission using nanopore whole-genome sequencing. J Hosp Infect. 2021;111:107–116. doi: 10.1016/j.jhin.2021.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hare D., Gonzalez G., Dean J., McDonnell K., Carr M.J., De Gascun C.F. Genomic epidemiological analysis of SARS-CoV-2 household transmission. Access Microbiol. 2021:3. doi: 10.1099/acmi.0.000252. 000252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Health Service Executive . Health Service Executive; Dublin: 2020. Interim guidance for adult unscheduled care pathway in the COVID-19 era; the acute floor.https://hse.drsteevenslibrary.ie/ld.php?content_id=33212096 Available at: [last accessed January 2022] [Google Scholar]
- 18.COVID-19 hospital acquired case definition. Health Protection Surveillance Centre. Available at: https://www.hpsc.ie/a-z/respiratory/coronavirus/novelcoronavirus/casedefinitions/covid-19hospitalacquiredcasedefinitionforireland/ [last accessed January 2022].
- 19.Quick J. ARTIC v3 sequencing protocol. Available at: https://www.protocols.io/view/ncov-2019-sequencing-protocol-v3-locost-bh42j8ye [last accessed January 2022].
- 20.ARTIC SARS-CoV-2 bioinformatics protocol. Available at: https://artic.network/ncov-2019/ncov2019-bioinformatics-sop.html [last accessed April 2021].
- 21.Rambaut A., Holmes E.C., O'Toole A., Hill V., McCrone J.T., Ruis C., et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nature Microbiol. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simpson J. Ncov-tools v.1.8.0 July 7 2021. Available at: https://github.com/jts/ncov-tools [last accessed August 2021].
- 23.Katoh K., Misawa K., Kuma K., Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Minh B.Q., Schmidt H.A., Chernomor O., Schrempf D., Woodhams M.D., von Haeseler A., et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 2020;37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Letunic I., Bork P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49:W293–W296. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turakhia Y., Thornlow B., Hinrichs A.S., De Maio N., Gozashti L., Lanfear R., et al. Ultrafast sample placement on existing tRees (UshER) enables real-time phylogenetics for the SARS-CoV-2 pandemic. Nat Genet. 2021;53:809–816. doi: 10.1038/s41588-021-00862-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’ Toole, A. snipit. Available at: https://github.com/aineniamh/snipit [last accessed January 2022].
- 28.Global Initiative for Sharing All Influenza Data (GISAID). Available at: https://www.gisaid.org/[last accessed August 2021].
- 29.Richardson D.B., Mountain D. Myths versus facts in emergency department overcrowding and hospital access block. Med J Aust. 2009;190:369–374. doi: 10.5694/j.1326-5377.2009.tb02451.x. [DOI] [PubMed] [Google Scholar]
- 30.Rana K., Sharma B., Lakshmi P.V.M., Kaur M., Singh M.P., Singh R., et al. Nosocomial outbreak of SARS-CoV-2 in a non-COVID zone of a tertiary care hospital of North India: need to upgrade infection control practices. J Prim Care Community Health. 2021;12 doi: 10.1177/21501327211050753. 21501327211050753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iacobucci G. Overcrowding and long delays in A&E caused over 4000 deaths last year in England, analysis shows. BMJ. 2021;375 doi: 10.1136/bmj.n2835. n2835. [DOI] [PubMed] [Google Scholar]
- 32.Health Information and Quality Authority. Report of the unannounced inspection of University Hospital Limerick. Available at: https://www.hiqa.ie/system/files?file=inspectionreports/university-hospital-limerick-29-october-2020.pdf [last accessed January 2022].
- 33.Varia M., Wilson S., Sarwal S., McGeer A., Gournis E., Galanis E., et al. Hospital Outbreak Investigation Team. Investigation of a nosocomial outbreak of severe acute respiratory syndrome (SARS) in Toronto, Canada. CMAJ. 2003;169(4):285–292. [PMC free article] [PubMed] [Google Scholar]
- 34.van Doremalen N., Bushmaker T., Morris D.H., Holbrook M.G., Gamble A., Williamson B.N., et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N Engl J Med. 2020;382:1564–1567. doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lednicky J.A., Lauzard M., Fan Z.H., Jutla A., Tilly T.B., Gangwar M., et al. Viable SARS-CoV-2 in the air of a hospital room with COVID-19 patients. Int J Infect Dis. 2020;100:476–482. doi: 10.1016/j.ijid.2020.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hatanaka N., Xu B., Yasugi M., Morino H., Tagishi H., Miura T., et al. Chlorine dioxide is a more potent antiviral agent against SARS-CoV-2 than sodium hypochlorite. J Hosp Infect. 2021;118:20–26. doi: 10.1016/j.jhin.2021.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mehta P., Alle S., Chaturvedi A., Swaminathan A., Saifi S., Maurya R., et al. Clinico-genomic analysis reveals mutations associated with COVID-19 disease severity: possible modulation by RNA structure. Pathogens. 2021;10:1109. doi: 10.3390/pathogens10091109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robson F., Khan K.S., Le T.K., Paris C., Demirbag S., Barfuss P., et al. Coronavirus RNA proofreading: molecular basis and therapeutic targeting. Mol Cell. 2020;79:710–727. doi: 10.1016/j.molcel.2020.07.027. Erratum in: Mol Cell. 2020;80:1136-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin D.P., Weaver S., Tegally H., San J.E., Shank S.D., Wilkinson E., et al. The emergence and ongoing convergent evolution of the SARS-CoV-2 N501Y lineages. Cell. 2021;184:5189–5200.e7. doi: 10.1016/j.cell.2021.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abbas M., Robalo Nunes T., Cori A., Cordey S., Laubscher F., Baggio S., et al. Explosive nosocomial outbreak of SARS-CoV-2 in a rehabilitation clinic: the limits of genomics for outbreak reconstruction. J Hosp Infect. 2021;117:124–134. doi: 10.1016/j.jhin.2021.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.COVID-19 Genomics UK (COG-UK) consortiumcontact@cogconsortium.uk An integrated national scale SARS-CoV-2 genomic surveillance network. Lancet Microbe. 2020;1:e99–100. doi: 10.1016/S2666-5247(20)30054-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nicholls S.M., Poplawski R., Bull M.J., Underwood A., Chapman M., Abu-Dahab K., et al. COVID-19 Genomics UK (COG-UK) Consortium. CLIMB-COVID: continuous integration supporting decentralised sequencing for SARS-CoV-2 genomic surveillance. Genome Biol. 2021;22:196. doi: 10.1186/s13059-021-02395-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O’Toole Á., Hill V., Jackson B., Dewar R., Sahadeo N., Colquhoun R., et al. Genomics-informed outbreak investigations of SARS-CoV-2 using civet. medRxiv. 2021 doi: 10.1371/journal.pgph.0000704. 12.13.21267267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.SARS-CoV-2 Surveillance & Whole Genome Sequencing Working Group: Proposal for a National SARS-CoV-2 Surveillance & Whole Genome Sequencing Programme. Available at: https://assets.gov.ie/200008/47f6d0d6-ff1b-4180-a930-1f8655913f92.pdf [last accessed March 2022].
- 45.HERA incubator grants. Available at: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX:52021DC0078 [last accessed February 2022].
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.