ABSTRACT
Erwinia amylovora is a plant-pathogenic bacterium that causes fire blight disease in many economically important plants, including apples and pears. This bacterium produces three exopolysaccharides (EPSs), amylovoran, levan, and cellulose, and forms biofilms in host plant vascular tissues, which are crucial for pathogenesis. Here, we demonstrate that ProQ, a conserved bacterial RNA chaperone, was required for the virulence of E. amylovora in apple shoots and for biofilm formation in planta. In vitro experiments revealed that the deletion of proQ increased the production of amylovoran and cellulose. Prc is a putative periplasmic protease, and the prc gene is located adjacent to proQ. We found that Prc and the associated lipoprotein NlpI negatively affected amylovoran production, whereas Spr, a peptidoglycan hydrolase degraded by Prc, positively regulated amylovoran. Since the prc promoter is likely located within proQ, our data showed that proQ deletion significantly reduced the prc mRNA levels. We used a genome-wide transposon mutagenesis experiment to uncover the involvement of the bacterial second messenger c-di-GMP in ProQ-mediated cellulose production. The deletion of proQ resulted in elevated intracellular c-di-GMP levels and cellulose production, which were restored to wild-type levels by deleting genes encoding c-di-GMP biosynthesis enzymes. Moreover, ProQ positively affected the mRNA levels of genes encoding c-di-GMP-degrading phosphodiesterase enzymes via a mechanism independent of mRNA decay. In summary, our study revealed a detailed function of E. amylovora ProQ in coordinating cellulose biosynthesis and, for the first time, linked ProQ with c-di-GMP metabolism and also uncovered a role of Prc in the regulation of amylovoran production.
IMPORTANCE Fire blight, caused by the bacterium Erwinia amylovora, is an important disease affecting many rosaceous plants, including apple and pear, that can lead to devastating economic losses worldwide. Similar to many xylem-invading pathogens, E. amylovora forms biofilms that rely on the production of exopolysaccharides (EPSs). In this paper, we identified the RNA-binding protein ProQ as an important virulence regulator. ProQ played a central role in controlling the production of EPSs and participated in the regulation of several conserved bacterial signal transduction pathways, including the second messenger c-di-GMP and the periplasmic protease Prc-mediated systems. Since ProQ has recently been recognized as a global posttranscriptional regulator in many bacteria, these findings provide new insights into multitiered regulatory mechanisms for the precise control of virulence factor production in bacterial pathogens.
KEYWORDS: ProQ, Prc, NlpI, cellulose, fire blight, exopolysaccharide, c-di-GMP, cyclic di-GMP
INTRODUCTION
Erwinia amylovora is a Gram-negative enterobacterium and the causal agent of fire blight, one of the most devastating diseases of rosaceous plants such as apples and pears (1, 2). This pathogen initiates infection predominantly through flowers or the shoot tips of branches of susceptible host plants, and infection occurs through natural openings in flower nectaries or leaves (3). In the apoplast, E. amylovora transits to the endophytic infection stage. Bacterial cells move through cortical parenchyma cell layers and can also form biofilms in the plant vascular tissue xylem, resulting in the restriction of water transport and, eventually, the death of the host plant (1, 4–7).
A wide range of pathogenicity and virulence factors are required for endophytic infection by E. amylovora (4, 8). The hypersensitive response and pathogenicity (hrp) type III secretion system (T3SS) and the exopolysaccharide (EPS) amylovoran are essential pathogenicity factors as T3SS- or amylovoran-deficient mutants of E. amylovora are nonpathogenic (9–11). The T3SS is a well-studied protein secretion/translocation system that is known to deliver effector proteins from bacterial cells directly into the plant cell cytoplasm where these proteins then manipulate host responses (12, 13). The mechanism of amylovoran as a pathogenicity factor of E. amylovora continues to be enigmatic; however, several studies have demonstrated that amylovoran is the main EPS produced by E. amylovora and a critical component of biofilms formed in vitro (6, 14, 15). Other EPSs produced by E. amylovora are levan and cellulose. Levan is a homopolymer of fructose synthesized extracellularly from sucrose by levansucrase (16), and cellulose, a homopolymer of glucose (17), is synthesized by enzymes encoded within the bcs operon (18). Both levan and cellulose are virulence factors of E. amylovora because mutants unable to produce either of these EPSs are defective in in vitro and in planta biofilm formation and are reduced in virulence in host plants (6, 18).
To coordinately modulate the expression of virulence-related genes, E. amylovora utilizes a sophisticated regulatory network, including two-component signal transduction systems, nucleotide signaling, transcriptional and posttranscriptional regulators, and small regulatory RNAs (sRNAs) (4). Of these, the ubiquitous bacterial second messenger bis-(3′–5′)-cyclic dimeric GMP (c-di-GMP) is one of the most critical nodes. c-di-GMP is a global regulator in many bacteria that is best known for its role in promoting motile-to-sessile lifestyle transitions (19–21). In E. amylovora, c-di-GMP represses flagellum-based motility mechanisms and the expression of T3SS regulon genes while promoting biofilm formation and the production of the EPSs amylovoran and cellulose (22, 23). c-di-GMP activates cellulose biosynthesis via allosteric binding to the PilZ domain of the cellulose catalytic subunit BcsA in E. amylovora (18), which has also been reported in other bacteria (24–26). Intracellular levels of c-di-GMP are controlled by two kinds of enzymes exerting opposing functions. The GGDEF domain-containing diguanylate cyclases (DGCs) synthesize c-di-GMP from guanosine-5′-triphosphate, and the EAL or the HD-GYP domain-containing phosphodiesterases (PDEs) degrade c-di-GMP into 5′-phosphoguanylyl-(3′–5′)-guanosine or GMP, respectively (19, 27–29). At least eight proteins containing EAL, GGDEF, or both domains have been identified in E. amylovora (22, 23); however, little is known about the environmental stimuli that trigger the enzymatic activities of these proteins or the regulators that control the expression of their protein-encoding genes.
The regulation of virulence in E. amylovora is also known to be affected at the posttranscriptional level by sRNAs such as ArcZ that are dependent on the sRNA chaperone Hfq (30, 31). To date, this sRNA-mediated regulation in E. amylovora has been shown to be controlled by effects on the translation of mRNAs of other regulatory genes such as flhDC or effects on the stability of the mRNA of the leucine regulatory protein Lrp (32, 33). While the role of Hfq in controlling a global posttranscriptional network that is of particular importance in virulence regulation has been known for some time (34), the RNA-binding protein ProQ has been more recently discovered (35–37). ProQ was shown to associate with a class of highly structured sRNAs that is distinct from Hfq-dependent sRNAs, and the expression of approximately 16% of the Salmonella enterica genome was altered in a proQ deletion mutant (35). ProQ has subsequently been shown to be an important regulator of virulence in S. enterica serovar Typhimurium (38), and the RNA interactome of ProQ overlaps Hfq in Escherichia coli (39).
We hypothesized that ProQ would function in virulence regulation in E. amylovora by impacting EPS biosynthesis and biofilm formation. In this work, we demonstrated that proQ is essential for E. amylovora virulence in an apple shoot assay and is required for in planta biofilm formation. We also determined that the production of amylovoran and cellulose was significantly affected upon the deletion of proQ and identified distinct mechanisms underlying the regulation of these EPSs. Furthermore, ProQ was found to repress intracellular c-di-GMP levels, and the regulatory roles of ProQ on the c-di-GMP metabolic enzymes were elucidated.
RESULTS
Deletion of proQ inhibits E. amylovora virulence in an apple shoot assay.
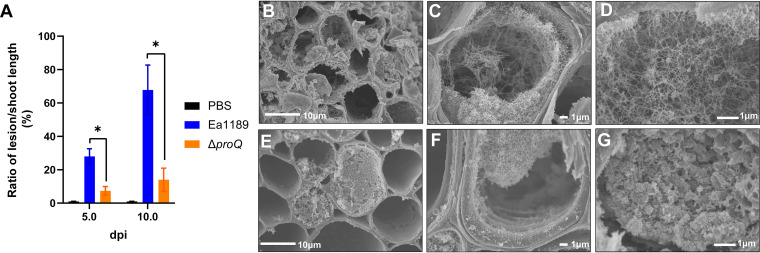
To determine whether ProQ is involved in E. amylovora virulence, we generated a proQ deletion mutant in E. amylovora strain Ea1189 and assessed its ability to cause disease in the host plant apple. Cells of wild-type (WT) Ea1189 or the Ea1189 ΔproQ mutant were inoculated into actively growing shoots of apple through wounding. At 5 days postinoculation (dpi), we observed a significant reduction in disease progression in shoots inoculated with Ea1189 ΔproQ, with an average ratio of lesion/shoot length of approximately 7%, relative to a 28% ratio in shoots inoculated with WT Ea1189 (Fig. 1A). At 10 dpi, Ea1189 ΔproQ-inoculated shoots continued to exhibit minor necrosis, whereas those inoculated with WT bacteria further extended their lesion lengths to >60% of the total shoot length (Fig. 1A).
FIG 1.
Impact of ProQ on Erwinia amylovora virulence and in planta biofilm formation. (A) The virulence of wild-type (WT) E. amylovora strain Ea1189 and Ea1189 ΔproQ was examined in apple shoots at 5 and 10 days postinoculation (dpi), respectively. The ratio of lesion length to shoot length for disease severity was calculated. Shoots treated with phosphate-buffered saline (PBS) were used as negative controls. All results are from one representative experiment. Three independent experiments were conducted, and three replicates were used for each experiment. Error bars indicate standard errors of the means. Asterisks indicate statistically significant differences in the means (P < 0.01 by Student’s t test). (B to G) Micrographs of xylem vessels infected by WT Ea1189 (B to D) or Ea1189 ΔproQ (E to G). Images were taken using a scanning electron microscope at 7 dpi.
Since the formation of biofilms in the host plant vascular system plays an important role for E. amylovora pathogenesis in apple shoots (40), we investigated the impact of the deletion of proQ on biofilm formation in planta by visualizing cross sections of the midrib of apple leaves inoculated with WT Ea1189 or Ea1189 ΔproQ at 7 dpi. Micrographs captured using scanning electron microscopy (SEM) showed that biofilms formed by Ea1189 were present in almost all xylem vessels (Fig. 1B). The production of a fibrillar material was found, likely contributing to the colonization of bacterial cells on the xylem wall (Fig. 1C), bacterial biofilm development, as well as the further expansion of the biofilm inward, spanning xylem vessels (Fig. 1D). Interestingly, xylem vessels of leaf midribs infected by Ea1189 ΔproQ were mostly empty (Fig. 1E). The Ea1189 ΔproQ mutant bacteria could form biofilms, as shown in Fig. 1F, but only on one side of the xylem wall and failed to occupy the entire xylem vessels, unlike WT Ea1189 (Fig. 1C and D). In a small number of Ea1189 ΔproQ-infected xylem vessels (approximately 5% of the total xylem vessels examined), we observed a unique cellular matrix blocking the xylem vessels (Fig. 1G), which appeared to be structurally different from the WT Ea1189-formed biofilms (Fig. 1D). Further investigation is needed as these could be bacterial biofilms formed by Ea1189 ΔproQ or could be plant secretions that act as a defense mechanism. No visible biofilm structures were observed in xylem vessels from apple plants treated with phosphate-buffered saline (PBS) buffer as a negative control (data not shown). Taken together, our data suggest that a functional proQ gene is required for biofilm formation and successful infection by E. amylovora in a host plant.
The deletion of proQ enhances the production of two EPSs, amylovoran and cellulose.
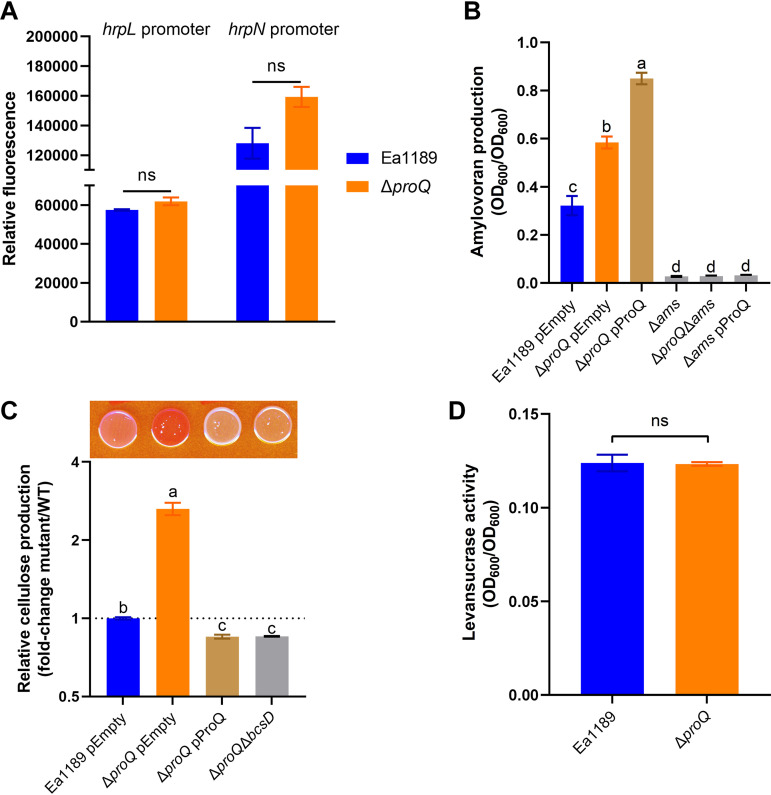
To investigate whether and which pathogenicity or virulence factors were affected by the deletion of proQ in E. amylovora, we examined the expression of T3SS regulon genes and the production of three EPSs, amylovoran, levan, and cellulose, in Ea1189 ΔproQ, and compared them with those of WT Ea1189. The promoter activities of two T3SS regulon genes, hrpL (encoding the master regulator) and hrpN (encoding the harpin protein), were not significantly different in Ea1189 ΔproQ compared to WT Ea1189 (Fig. 2A), suggesting that ProQ is likely not involved in the transcriptional regulation of the T3SS. In contrast, the production of amylovoran was significantly increased when proQ was deleted (Fig. 2B). We failed to complement this phenotype in Ea1189 ΔproQ because the in trans expression of proQ from the low-copy-number plasmid pCL1920 further elevated the production of amylovoran by 1.4-fold relative to that of Ea1189 ΔproQ harboring the empty vector (Fig. 2B). Meanwhile, the deletion of the ams operon responsible for amylovoran biosynthesis led to the complete abolishment of amylovoran production in either the proQ deletion or overexpression strain (Fig. 2B).
FIG 2.
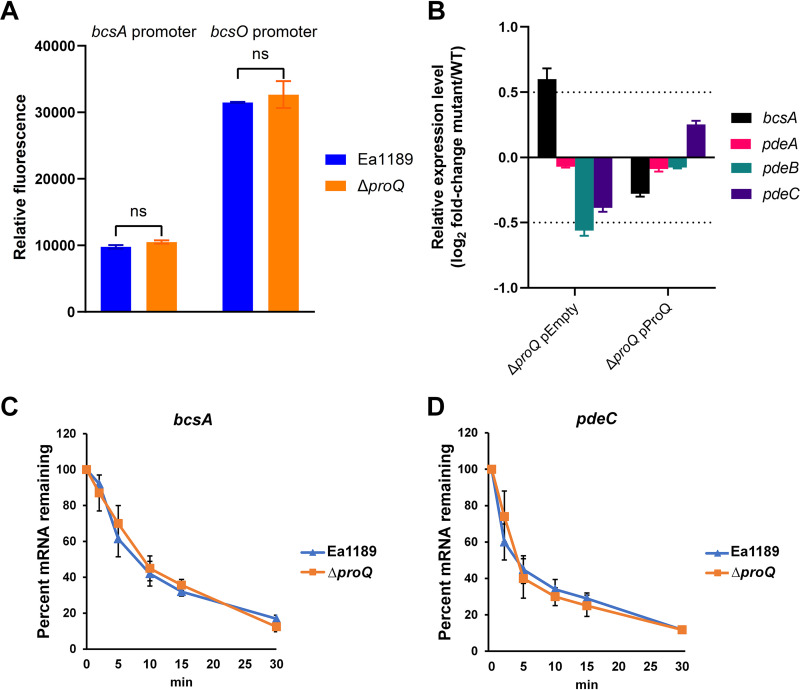
Impact of the deletion of proQ on various virulence determinants of Erwinia amylovora. (A) Promoter activities of hrpL and hrpN were determined in wild-type (WT) Ea1189 and Ea1189 ΔproQ, respectively. All results are from one representative experiment. Three independent experiments were conducted, and three replicates were used for each experiment. Error bars indicate standard errors of the means. ns (not significant), P > 0.05 by Student’s t test. (B) Amylovoran production was determined in WT Ea1189 harboring the empty vector pCL1920, Ea1189 ΔproQ harboring pCL1920, Ea1189 ΔproQ harboring pCL1920-proQ, Ea1189 Δams, Ea1189 ΔproQ Δams, and Ea1189 Δams harboring pCL1920-proQ. (C) Cellulose production was determined in WT Ea1189 harboring pCL1920, Ea1189 ΔproQ harboring pCL1920, Ea1189 ΔproQ harboring pCL1920-proQ, and Ea1189 ΔproQ ΔbcsD. The image was captured at 24 h postinoculation. Relative cellulose production was calculated as described in Materials and Methods. Values are representative of results from three independent experiments. Three replicates were used in each experiment. Error bars indicate standard errors of the means. Different lowercase letters above the bars indicate statistically significant differences between treatments (P < 0.05 by Fisher’s least significant difference test). (D) Levansucrase activity was determined in WT Ea1189 and Ea1189 ΔproQ. One representative experiment was chosen, and three independent experiments were performed. Error bars indicate standard errors of the means. ns, P > 0.05 by Student’s t test.
The production of cellulose was assessed by qualitatively evaluating the color of E. amylovora cells grown on plates containing a red dye, Congo red, that binds to cellulose (18). As shown in Fig. 2C, WT Ea1189 exhibited a pink colony, whereas Ea1189 ΔproQ developed a red colony with a calculated redness >2.5-fold higher than that of WT Ea1189, suggesting that ProQ represses cellulose production in E. amylovora. We confirmed this regulation via the in trans expression of proQ (pCL1920-proQ) and the deletion of the essential cellulose biosynthesis gene bcsD in the proQ mutant background, respectively, and found white colonies for both strains (Fig. 2C). Finally, the activities of levansucrase responsible for levan production (16) were comparable between WT Ea1189 and Ea1189 ΔproQ (Fig. 2D). Collectively, our data suggest that the production of two EPSs, amylovoran and cellulose, was significantly affected when proQ was deleted in E. amylovora.
Prc inhibits amylovoran production via the Prc-NlpI-Spr regulatory pathway.
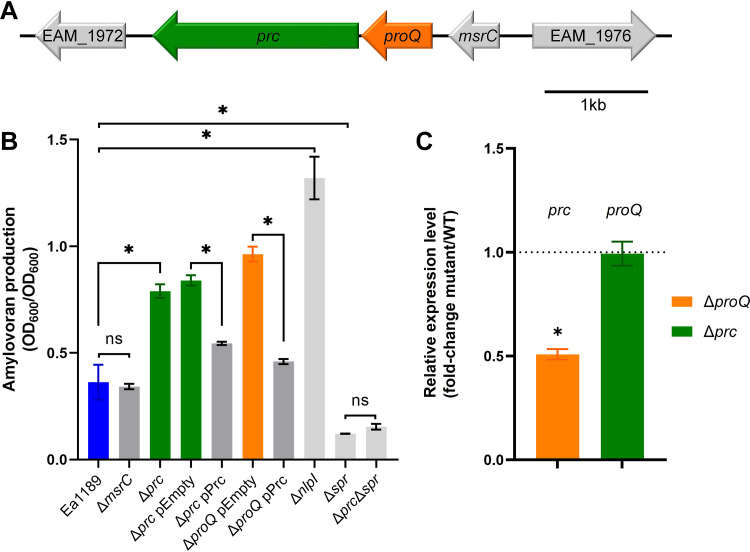
Genomic analysis showed that proQ is located between msrC (also known as yebR, a methionine sulfoxide reductase-encoding gene) and prc (a periplasmic protease-encoding gene) in E. amylovora (Fig. 3A); this gene synteny is conserved in other enterobacteria such as Dickeya dadantii and Escherichia coli (41, 42). Furthermore, Kerr et al. (42) reported that a putative prc promoter was located within the coding DNA sequence (CDS) of proQ in Escherichia coli. This led us to hypothesize that the phenotypic changes that we observed in the E. amylovora proQ mutant (Fig. 2) were due to a putative reduction in the expression of prc due to the deletion of the prc promoter. To validate this hypothesis, two independent deletion mutants were constructed, Ea1189 Δprc and Ea1189 ΔmsrC, and the ability of each mutant to produce amylovoran was examined. Indeed, the deletion of msrC had no impact on amylovoran production, whereas the deletion of prc resulted in a significant increase in amylovoran production by approximately 2.5-fold relative to Ea1189 (Fig. 3B). More importantly, unlike the above-described results showing that complementation of proQ failed to restore the phenotype in Ea1189 ΔproQ (Fig. 2B), complementation of prc lowered amylovoran production in both Ea1189 Δprc and Ea1189 ΔproQ to nearly WT levels (Fig. 3B), suggesting that Prc negatively controls the production of amylovoran in E. amylovora. In addition, our data showed that the mRNA levels of prc were reduced by 2-fold in Ea1189 ΔproQ compared with those in Ea1189, while the mRNA levels of proQ were comparable with or without the presence of gene prc (Fig. 3C).
FIG 3.
Amylovoran production is controlled by the Prc-NlpI-Spr pathway in Erwinia amylovora. (A) Schematic drawing of the genomic content, including genes EAM_1972, prc, proQ, msrC, and EAM_1976. (B) Amylovoran production was measured in wild-type (WT) Ea1189, Ea1189 ΔmsrC, Ea1189 Δprc, Ea1189 Δprc harboring the empty vector pBBR1-MCS5, Ea1189 Δprc harboring pBBR1-MCS5-prc, Ea1189 ΔproQ harboring pBBR1-MCS5, Ea1189 ΔproQ harboring pBBR1-MCS5-prc, Ea1189 ΔnlpI, Ea1189 Δspr, and Ea1189 Δprc Δspr. (C) RNA levels of prc were measured in WT Ea1189 and Ea1189 ΔproQ, while RNA levels of proQ were measured in WT Ea1189 and Ea1189 Δprc. Three independent experiments were performed with three replicates in each experiment. Values are from one representative experiment. Error bars indicate standard errors of the means. Asterisks indicate statistically significant differences in the means (P < 0.05 by Student’s t test). ns, not significant.
In Escherichia coli, the Prc homolog has been well studied in the synthesis of the bacterial cell wall, as Prc interacts with the outer membrane lipoprotein NlpI, and these proteins together degrade the peptidoglycan (PG) hydrolase Spr (also known as MepS) (43–45). We found that the deletion of nlpI in E. amylovora Ea1189 greatly enhanced the production of amylovoran (Fig. 3B). In contrast, the deletion of spr abolished amylovoran production in both the WT and Ea1189 Δprc backgrounds (Fig. 3B). Together, these data imply that the Prc-NlpI-Spr regulatory pathway is conserved in E. amylovora and is involved in the regulation of amylovoran production.
Genome-wide transposon mutagenesis reveals that c-di-GMP signaling is involved in the ProQ-mediated regulation of cellulose production.
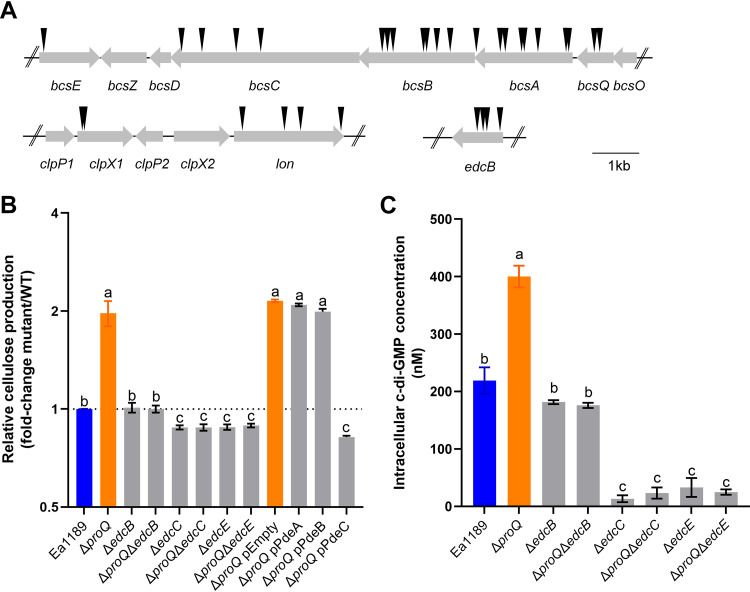
We excluded the possibility that Prc contributes to ProQ-mediated cellulose production because complementation of prc in Ea1189 ΔproQ did not influence cellulose production (see Fig. S1 in the supplemental material). We also showed that ProP, a homolog of the Escherichia coli osmoregulatory transporter known to be controlled by ProQ (46, 47), is not required for the regulation of cellulose since the deletion of proP had a negligible impact on cellulose production in Ea1189 (Fig. S1). Therefore, to further understand the molecular mechanism of the regulation of cellulose by ProQ, we conducted a transposon mutagenesis screen in the Ea1189 ΔproQ mutant background using the mariner transposon miniHimar RB1 (48). We aimed to identify the regulatory components involved in ProQ-mediated cellulose production by comparing the colony morphology of individual transposon insertion mutants on Congo red plates with that of Ea1189 ΔproQ in a high-throughput manner. In total, 7,000 transposon insertion mutants (an average of 2 transposon insertions per gene) were generated, and 50 mutants were found to form colonies with reduced red color relative to Ea1189 ΔproQ (Table S1). Among them, 22 transposon mutants were shown to contain the transposon insertion within one of several genes of the bcs operon, including bcsA, bcsB, bcsC, bcsE, and bcsQ (Fig. 4A), validating our transposon mutagenesis and screening method. Four mutants had transposons inserted in lon (Fig. 4A), encoding a protease (49), and two mutants had transposons inserted upstream of lon in clpX1 (Fig. 4A), which encodes an ATPase associated with the protease ClpP and plays a pleiotropic role in E. amylovora (50). In addition, four mutants had a transposon insertion in edcB (Fig. 4A), which encodes a DGC necessary for c-di-GMP biosynthesis in E. amylovora (23). To validate the cellulose phenotype observed in the transposon mutants, we deleted edcB in Ea1189 ΔproQ and confirmed that the double mutant Ea1189 ΔproQ ΔedcB exhibited WT levels of cellulose production (Fig. 4B).
FIG 4.
c-di-GMP plays a key role in ProQ-mediated cellulose production. (A) Schematic drawing of the selected transposon insertions. Black arrows indicate the transposon insertion sites. (B) Cellulose production was determined in wild-type (WT) Erwinia amylovora strain Ea1189, Ea1189 ΔproQ, Ea1189 ΔedcB, Ea1189 ΔproQ ΔedcB, Ea1189 ΔedcC, Ea1189 ΔproQ ΔedcC, Ea1189 ΔedcE, Ea1189 ΔproQ ΔedcE, Ea1189 ΔproQ harboring the empty vector pBBR1-MCS5, Ea1189 ΔproQ harboring pBBR1-MCS5-pdeA, Ea1189 ΔproQ harboring pBBR1-MCS5-pdeB, and Ea1189 ΔproQ harboring pBBR1-MCS5-pdeC. (C) Intracellular c-di-GMP levels were measured in WT Ea1189, Ea1189 ΔproQ, Ea1189 ΔedcB, Ea1189 ΔproQ ΔedcB, Ea1189 ΔedcC, Ea1189 ΔproQ ΔedcC, Ea1189 ΔedcE, and Ea1189 ΔproQ ΔedcE. One representative experiment was chosen, and three independent experiments with three replicates were performed. Error bars indicate standard errors of the means. Different lowercase letters above the bars indicate statistically significant differences between treatments (P < 0.05 by Fisher’s least significant difference test).
ProQ represses intracellular c-di-GMP levels in E. amylovora.
Previous studies demonstrated that at least four DGC-encoding genes, including edcB, are actively involved in c-di-GMP signaling in E. amylovora (18, 23). To address the question of whether EdcB functions as the sole DGC in ProQ-mediated cellulose production, the regulation of the cellulose biosynthesis of two additional DGCs was evaluated in Ea1189 ΔproQ. As shown in Fig. 4B, cellulose production was significantly reduced in the double mutants Ea1189 ΔproQ ΔedcC and Ea1189 ΔproQ ΔedcE, highlighting the importance of c-di-GMP signaling in the regulation of cellulose by ProQ. We also measured the intracellular levels of c-di-GMP in WT Ea1189, Ea1189 ΔproQ, and several DGC-derived single- and double-deletion mutants using ultraperformance liquid chromatography coupled with tandem mass spectrometry (UPLC-MS/MS). In agreement with the data from cellulose production assays (Fig. 4B), the deletion of proQ increased c-di-GMP levels relative to WT Ea1189 (Fig. 4C), which could be complemented by pCL1920-proQ (Fig. S2). Also, the deletion of either edcB, edcC, or edcE reduced the elevated c-di-GMP levels observed in Ea1189 ΔproQ (Fig. 4C); however, we observed drastic reductions in c-di-GMP levels in single and double mutants of edcC or edcE but not edcB compared with those in WT Ea1189 (Fig. 4C). Among the three PDEs responsible for the degradation of c-di-GMP, only pdeC overexpression significantly lowered cellulose production in Ea1189 ΔproQ, whereas pdeA or pdeB overexpression did not (Fig. 4B). Taken together, these data suggest that ProQ negatively modulates intracellular c-di-GMP levels to control cellulose biosynthesis in E. amylovora.
Impact of ProQ on transcriptional and posttranscriptional regulation of cellulose biosynthesis and c-di-GMP metabolism genes.
A recent study demonstrated that a ProQ homolog represses the level of bcsA transcripts in the soft rot phytopathogen D. dadantii (41). To better understand how ProQ controls cellulose biosynthesis in E. amylovora, promoter activities and RNA levels of cellulose biosynthesis genes were compared between WT Ea1189 and Ea1189 ΔproQ. Our data showed that the deletion of proQ did not affect the promoter activities of bcsA or bcsO (Fig. 5A), indicating that ProQ is likely not controlling the transcription of cellulose biosynthesis genes. However, the levels of bcsA mRNA were elevated by approximately 50% when proQ was deleted, which was restored by the in trans complementation of proQ (Fig. 5B). Given that ProQ was found to predominantly bind the 3′ untranslated regions (UTRs) of target RNAs (38, 39, 51) and that our data showed that ProQ reduced bcsA transcript levels without affecting transcription, we examined the effect of the deletion of proQ in Ea1189 on the stability of the bcsA transcript. Interestingly, no significant differences in bcsA transcript stability were found between WT Ea1189 and Ea1189 ΔproQ (Fig. 5C), suggesting that ProQ affects bcsA via a mechanism that is not through destabilizing the bcsA transcript.
FIG 5.
Impacts of ProQ on cellulose- and c-di-GMP-related genes in Erwinia amylovora. (A) Promoter activities of bcsA and bcsO were determined in wild-type (WT) Ea1189 and Ea1189 ΔproQ. (B) RNA levels of bcsA, pdeA, pdeB, and pdeC were measured in WT Ea1189 harboring the empty vector pCL1920, Ea1189 ΔproQ harboring pCL1920, and Ea1189 ΔproQ harboring pCL1920-proQ. (C and D) The transcript stabilities of bcsA (C) and pdeC (D) were measured in WT Ea1189 and Ea1189 ΔproQ following the addition of rifampin at time zero. All experiments were conducted at least three times with three replicates in each experiment. Values are from one representative experiment. Error bars indicate standard errors of the means. ns (not significant), P > 0.05 by Student’s t test.
We also assessed the consequences of proQ deletion on the mRNA levels of three DGC-encoding genes, edcB, edcC, and edcE, and found no impact (data not shown). On the other hand, slight reductions in the mRNA levels of two PDE-encoding genes, pdeB and pdeC, in the absence of proQ were observed (Fig. 5B), which supports the cellulose production and intracellular c-di-GMP level data (Fig. 4). A complementation experiment restored the mutant phenotype (Fig. 5B). In an attempt to evaluate the impact of ProQ on the stability of RNAs of PDE-encoding genes, we tested pdeC as our data showed that the overexpression of pdeC alone could complement the cellulose production in Ea1189 ΔproQ (Fig. 4B). However, no significant difference in pdeC transcript stabilities was observed in the presence or absence of proQ (Fig. 5D).
DISCUSSION
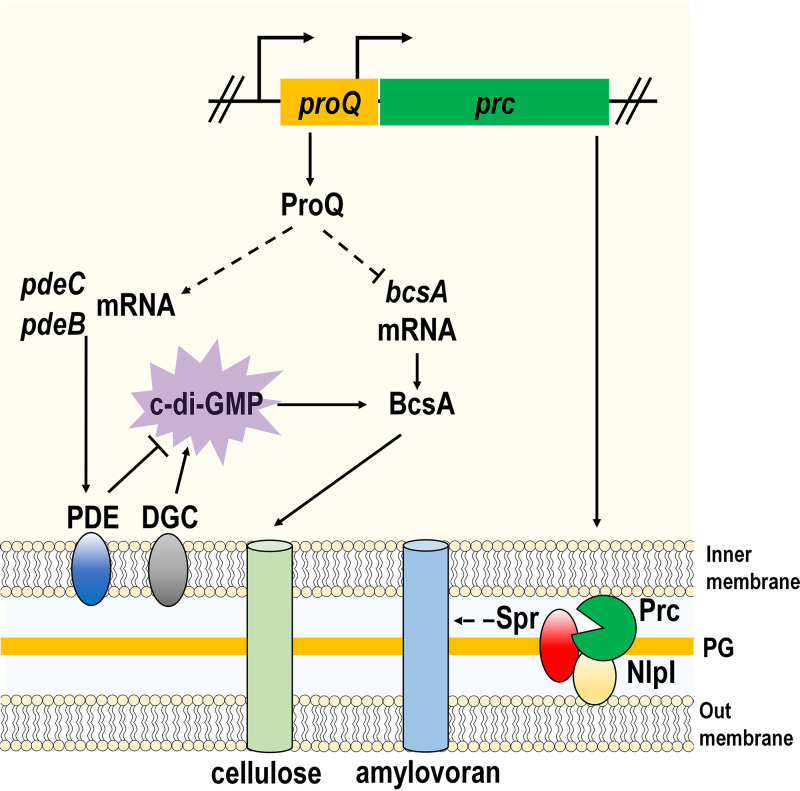
In this study, we characterized the biological and molecular functions of the global RNA-binding protein ProQ in E. amylovora. Our findings revealed a novel regulatory cascade where ProQ acts as a hub that controls cellulose biosynthesis via c-di-GMP signaling. In addition, we uncovered a previously uncharacterized role of Prc, a periplasmic protease whose promoter is located within proQ, in the negative regulation of amylovoran production (Fig. 6).
FIG 6.
Working model for ProQ/Prc-mediated regulation of amylovoran production and cellulose biosynthesis in Erwinia amylovora. ProQ controls cellulose biosynthesis in a c-di-GMP-dependent manner. In brief, ProQ positively contributes to the expression of two PDE-encoding genes, pdeB and pdeC. The function of PDEs is to degrade c-di-GMP that is synthesized by DGCs. c-di-GMP binds to BcsA and activates its catalytic activity to produce cellulose. ProQ also inhibits the mRNA levels of bcsA, but the underlying mechanism remains unknown. The regulation of amylovoran is dependent on Prc. Prc, whose transcriptional initiation requires proQ, is a periplasmic protease. Prc interacts with a lipoprotein, NlpI, and degrades a peptidoglycan hydrolase, Spr. Spr positively regulates amylovoran production, and the mechanism needs further investigation. ⊥ represents the negative control; → represents the positive control. The dotted lines indicate regulatory mechanisms identified in this study.
Global posttranscriptional regulators that participate in the modulation of c-di-GMP have been discovered in several bacterial species. For example, Bellows et al. (52) reported that Hfq negatively regulates intracellular c-di-GMP levels in the plague pathogen Yersinia pestis, which has also been reported in the plant pathogen D. dadantii (53). Moreover, in Escherichia coli, CsrA, a well-studied carbon storage regulator and an RNA-binding protein (54), directly binds to mRNAs of DGC-encoding genes, and the deletion of csrA leads to modestly increased intracellular levels of c-di-GMP (55). Similarly, we found that ProQ negatively regulates c-di-GMP levels likely by maintaining the expression of PDE-encoding genes in E. amylovora (Fig. 4 and 5). This result emphasizes the influence of RNA-binding proteins on c-di-GMP metabolism and implies cross talk between different posttranscriptional regulatory processes in bacteria. In support of this, Silva-Rohwer et al. (56) recently proposed a multitiered regulation in controlling c-di-GMP metabolism in Y. pestis that requires both Hfq and CsrA. Unlike Hfq and CsrA, which bind target RNAs via defined sequence motifs, ProQ predominantly recognizes highly structured RNAs and, in most cases, stabilizes them (37, 39, 57). However, our data indicated that this might not be true for pdeC because ProQ did not interfere with the stability of the pdeC mRNA (Fig. 5D). Thus, further investigation is needed to better understand the mechanism of ProQ-mediated c-di-GMP signaling in E. amylovora.
The multiplicity and redundancy of the c-di-GMP metabolic enzymes raise the question of their respective functions in various aspects of c-di-GMP signaling. In E. amylovora, high intracellular levels of c-di-GMP achieved by the overexpression of DGC-encoding genes led to hyper-biofilm-forming phenotypes and the enhanced production of both amylovoran and cellulose (18, 23). In line with this, our results showed that the deletion of either edcC or edcE abrogated cellulose production and significantly reduced c-di-GMP levels in E. amylovora in a ProQ-independent manner (Fig. 4). These results are particularly intriguing and indicate that several functionally redundant DGCs cooperatively contribute to cellulose production in E. amylovora. Since c-di-GMP promotes cellulose via allosteric binding to BcsA (18), it is likely that both EdcC and EdcE are required to synthesize c-di-GMP to maintain its binding affinity for BcsA, and EdcB, whose overexpression was previously found to increase cellulose but not amylovoran production (18), functions as a cellulose-specific DGC allowing further inductions of this EPS when needed. Why and how we identified only EdcB but no other DGCs in our transposon mutagenesis study remain unknown, which could be due to insertion bias of the transposon (58). Finally, a recent study reported that all three PDEs, PdeA, PdeB, and PdeC, are active in E. amylovora (22). However, our data demonstrated that only pdeC overexpression restored the cellulose phenotype in Ea1189 ΔproQ (Fig. 4B). As an increasing number of studies have demonstrated the temporal and spatial regulation of c-di-GMP signaling in the cell (19, 26, 59), the expression patterns and localization of different DGCs and PDEs for the regulation of diverse cellular behaviors of E. amylovora are currently being investigated as a follow-up to this study.
Prc is a soluble periplasmic protease that belongs to the large family of C-terminal processing proteases (60). In Escherichia coli, a group of studies reported that Prc and its lipoprotein adaptor NlpI participate in PG biosynthesis by degrading Spr, a PG hydrolase responsible for maintaining overall PG turnover (43, 45, 61). PG, a highly cross-linked sacculus comprising alternating N-acetylglucosamine and N-acetylmuramic acid, is an important structural element in the bacterial cell wall, and the main function of PG is to preserve cell integrity (62). We found that the inactivation of Prc or NlpI significantly enhanced the production of amylovoran in an Spr-dependent manner (Fig. 3), and similar observations have also been reported for Escherichia coli as two penicillin-binding proteins involved in hydrolyzing PG are also required for EPS production and biofilm formation (63). These results, collectively, lead to an interesting question: How does PG affect EPS? A recent study conducted in Pseudomonas aeruginosa provided a possible explanation: Gheorghita et al. (64) discovered that AlgL, a periplasmic lyase (65), functions as a homeostasis enzyme for the production of the EPS alginate by clearing the periplasmic space of accumulated polymers. Therefore, it is likely that active PG metabolism is required to modulate the overall production of amylovoran in E. amylovora.
Another explanation for Prc-NlpI-Spr-mediated amylovoran production is associated with the Rcs phosphorelay system, a conserved two-component signal transduction system among enterobacteria (66). In line with our observations (Fig. 3), Huang et al. (67) identified Prc as a major virulence regulator in extraintestinal pathogenic Escherichia coli and further demonstrated that the accumulation of Spr due to prc deletion could be sensed as a signal to trigger the Rcs system. Given that the Rcs system transcriptionally activates the expression of amylovoran biosynthesis genes (68), Prc could repress amylovoran production by inactivating the Rcs system in E. amylovora.
Besides c-di-GMP, our transposon mutagenesis study identified several known E. amylovora virulence regulators such as Lon and ClpXP (see Table S1 in the supplemental material). Interestingly, these proteins are highly conserved cytosolic ATP-dependent proteases and are known for their ability to degrade damaged or misfolded proteins (69, 70). Lee and Zhao (50) reported that ClpXP, consisting of an AAA+ ATPase, ClpX, and a proteolytic chamber, ClpP, is intimately involved in the regulation of amylovoran production, T3SS gene expression, and motilities, and Lee et al. later characterized Lon in controlling the same virulence determinants of E. amylovora (49). However, direct evidence demonstrating that ClpXP or Lon regulates E. amylovora cellulose production is lacking. In addition, Lon as a c-di-GMP effector protein has been reported in several bacterial species but not yet in E. amylovora (71, 72). In the facultative human pathogen Vibrio cholerae, for example, Joshi and colleagues showed that under conditions of high c-di-GMP levels, c-di-GMP directly binds to Lon to inhibit its protease activity, resulting in a derepression of the Lon-mediated repression of motility and type VI secretion system-dependent killing (72). Our findings indicated that Lon is required for elevated cellulose production in Ea1189 ΔproQ. Since c-di-GMP positively contributes to the biosynthesis of cellulose (18), it is unlikely that the putative c-di-GMP-dependent repression of the protease activity of Lon plays a dominant role in regulating ProQ-mediated cellulose production.
In summary, this work provides novel insights into the roles of ProQ, a newly defined RNA-binding protein, in E. amylovora. We showed that ProQ was a virulence factor of E. amylovora in the host plant apple and that the deletion of proQ positively affected the production of amylovoran and cellulose under in vitro conditions. Using transposon mutagenesis, we established a link between c-di-GMP signaling and ProQ-mediated cellulose production. Further experiments demonstrated that ProQ had a mild negative regulatory effect on BcsA and repressed intracellular c-di-GMP levels in E. amylovora. Taken together, the data in this study highlight the importance of posttranscriptional and posttranslational regulators and nucleotide signaling in the regulation of key bacterial virulence factors, and to our knowledge, this is the first report implicating ProQ in manipulating the c-di-GMP signaling network required for the biosynthesis of cellulose.
MATERIALS AND METHODS
Bacterial strains, plasmids, primers, and media.
The bacterial strains and plasmids used in this study are listed in Table 1. The E. amylovora or Escherichia coli strains were grown in Luria-Bertani (LB) broth medium at 28°C and 37°C, respectively. Modified basal medium A (MBMA) [KH2PO4 at 3 g/L, K2HPO4 at 7 g/L, (NH4)2SO4 at 1 g/L, citric acid at 0.5 g/L, MgSO4 at 0.03 g/L (23)] containing 1% (wt/vol) sorbitol was used to determine amylovoran production. The expression of T3SS genes of E. amylovora was measured in Hrp-inducing minimal medium (73). Antibiotics were added as needed to media at the following concentrations: ampicillin (Ap) at 100 μg/mL, chloramphenicol (Cm) at 10 μg/mL, gentamicin (Gm) at 15 μg/mL, kanamycin (Km) at 30 μg/mL, and spectinomycin (Sp) at 100 μg/mL. The E. amylovora strain Ea1189 chromosome and plasmid sequences were retrieved from the National Center for Biotechnology Information database under the accession numbers CP055227 and CP055228 (74). Oligonucleotide primers used for cloning are listed in Table 2.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Reference or source |
|---|---|---|
| Strains | ||
| Erwinia amylovora | ||
| Ea1189 | Wild type | 87 |
| ΔproQ | ΔproQ::Cm; Cmr; EAM_1974 deletion mutant in Ea1189 | This study |
| Δprc | Δprc::Cm; Cmr; EAM_1973 deletion mutant in Ea1189 | This study |
| ΔmsrC | ΔmsrC::Cm; Cmr; EAM_1975 deletion mutant in Ea1189 | This study |
| Δams | Δams; clean mutant; deletion of the 12-gene ams operon in Ea1189 | 87 |
| ΔproP | ΔproP::Cm; Cmr; EAM_3312 deletion mutant in Ea1189 | This study |
| ΔnlpI | ΔnlpI::Cm; Cmr; EAM_3066 deletion mutant in Ea1189 | This study |
| Δams ΔproQ | Δams ΔproQ::Cm; Cmr; deletion mutant of the 12-gene ams operon and EAM_1974 in Ea1189 | This study |
| ΔproQ ΔbcsD | ΔbcsD ΔproQ::Cm; Cmr; EAM_3384 and EAM_1974 deletion mutant in Ea1189 | This study |
| Δspr | Δspr::Km; Kmr; EAM_2227 deletion mutant in Ea1189 | This study |
| Δprc Δspr | Δprc::Cm Δspr::Km; Kmr and Cmr; EAM_1973 and EAM_2227 deletion mutant in Ea1189 | This study |
| ΔedcB | ΔedcB::Cm; Cmr; EAM_0564 deletion mutant in Ea1189 | This study |
| ΔproQ ΔedcB | ΔproQ ΔedcB::Cm; Cmr; EAM_1974 and EAM_0564 deletion mutant in Ea1189 | This study |
| ΔedcC | ΔedcC; clean mutant; EAM_1504 deletion mutant in Ea1189 | 23 |
| ΔedcE | ΔedcE; clean mutant; EAM_2435 deletion mutant in Ea1189 | 23 |
| ΔproQ ΔedcC | ΔedcC ΔproQ::Cm; Cmr; EAM_1504 and EAM_1974 deletion mutant in Ea1189 | This study |
| ΔproQ ΔedcE | ΔedcE ΔproQ::Cm; Cmr; EAM_2435 and EAM_1974 deletion mutant in Ea1189 | This study |
| Escherichia coli | ||
| DH5α | supE44 ΔlacU169 (ϕ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Lab stock |
| S17-1 λpir | λ(pir) hsdR pro thi; chromosomally integrated RP4-2 Tc::Mu Km::Tn7 | Lab stock |
| Plasmids | ||
| pKD4 | Template plasmid for kanamycin cassette; Kmr | 75 |
| pKD3 | Template plasmid for chloramphenicol cassette; Cmr | 75 |
| pKD46 | Arabinose-inducible lambda red recombinase; Apr | 75 |
| pBBR1-MCS5 | Broad-host-range plasmid; Gmr | 88 |
| pCL1920 | Low-copy-no. plasmid; lac promoter; Spr | 89 |
| p1920-proQ | proQ cloned into pCL1920; Spr | This study |
| pBBR1-prc | prc cloned into pBBR1-MCS5; Gmr | This study |
| pBBR1-pdeA | pdeA cloned into pBBR1-MCS5; Gmr | 22 |
| pBBR1-pdeB | pdeB cloned into pBBR1-MCS5; Gmr | 22 |
| pBBR1-pdeC | pdeC cloned into pBBR1-MCS5; Gmr | 22 |
| pPROBE-NT | Promoter-probe vector; promoterless gfp; Apr | 77 |
| pNT-hrpN | pPROBE-NT containing a putative hrpN promoter-gfp transcriptional fusion; Apr | 15 |
| pNT-hrpL | pPROBE-NT containing a putative hrpL promoter-gfp transcriptional fusion; Apr | 15 |
| pNT-bcsO | pPROBE-NT containing a putative bcsO promoter-gfp transcriptional fusion; Apr | This study |
| pNT-bcsA | pPROBE-NT containing a putative bcsA promoter-gfp transcriptional fusion; Apr | This study |
Apr, ampicillin resistance; Cmr, chloramphenicol resistance; Kmr, kanamycin resistance; Gmr, gentamicin resistance; Spr, spectinomycin resistance.
TABLE 2.
Oligonucleotide primers used in this studya
| Primer | Sequence (5′–3′) | Use |
|---|---|---|
| proQ-mut1 | GTAATCAGGAAATTTCATGGAAAATCAACCTAAGTTGAATAGCACTAAAGgtgtaggctggagctgcttc | proQ deletion |
| proQ-mut2 | CTGTTCATGCCCTGATTAGCCTCCGTATCAGAACTGCAAGTGTTCTGCGCcatatgaatatcctcctta | |
| msrC-mut1 | CAACTATGGTTAAACAATGACTAAAGCAGAATTTTATACTGAACTCAATCgtgtaggctggagctgcttc | msrC deletion |
| msrC-mut2 | CAGGCGACATTATAATGACCCCTTCAGCAATTGCTACGTGATCAAACACTcatatgaatatcctcctta | |
| prc-mut1 | CCGTTCTCATTACCGTCAGTTCGATCTCAATAAAGAATTCTCCGCTAAAAgtgtaggctggagctgcttc | prc deletion |
| prc-mut2 | GCGGATGCTGTGCCTGTTGGGCTGACTAAGCTATTGAGCGGGGTTGAAAAcatatgaatatcctcctta | |
| nlpI-mut1 | CAAGCGGGTAACAGGATGTTATTCCCAATGTTTGTTTTCGGGAGTGGTACgtgtaggctggagctgcttc | nlpI deletion |
| nlpI-mut2 | CTTTTCGAAATTCGGGCAATAAAAGTTCGTCAGCTATTGCTGGTCAGATTcatatgaatatcctcctta | |
| spr-mut1 | CCATTTTCTAACGACTTCGTCGTTAAGGACTTCAAGGGATCACACATATAgtgtaggctggagctgcttc | spr deletion |
| spr-mut2 | CACAGGCAGCGTTTTTTTTTCTCGATTTCGCAGAGTTCAAGGGATTGAAAcatatgaatatcctcctta | |
| edcB-mut1 | CTTAACGGTGCTGGCACGTTGATATTTCAGGGCAGTCACGAGTAAATATAgtgtaggctggagctgcttc | edcB deletion |
| edcB-mut2 | CCCCGGCTTTTAGCCAGATACAATGCCCGGTCAGCATGGGCCAGCGCCTTcatatgaatatcctcctta | |
| proP-mut1 | CAGATAATGACAGGGTATTCTATATGAAATTACGTAGGAAGCGTGTTAAGgtgtaggctggagctgcttc | proP deletion |
| proP-mut2 | AGATGTGAGCGTTTGGCTTCAAGCTCAGCAAGCTGCTGGTTAATATCTGCcatatgaatatcctcctta | |
| proQ-F-pCL1920 | AAATTCTAGATTATATGAACGGCTTGAAGG | proQ expression from pCL1920 |
| proQ-Rc-pCL1920 | TTTTAAGCTTAGACTGTTCATGCCCTGATT | |
| prc-F-MCS5 | AAAATCTAGACACTTGCAGTTCTGATACGGAGGC | prc expression from pBBR1-MCS5 |
| prc-Rc-MCS5 | TTTTGGATCCTGCCTGTTGGGCTGACTAAGCTAT | |
| bcsA-p1 | cgactctagaggatccccATTAAAGGCAATGACGGCGCTTGCTGATTTG | bcsA promoter in pPROBE-NT |
| bcsA-p2 | aattcgagctcggtacccGACTTTATTCATGAGTCATCCTGGAAAGCATA | |
| bcsO-p1 | cgactctagaggatccccCAGGACGAAGGTCGGTTACCCTGAATGCTGCC | bcsO promoter in pPROBE-NT |
| bcsO-p2 | aattcgagctcggtacccCATCATAACTTTTCATCAGTATGATCCCCAAG | |
| bcsA-q1 | AACCACGCCATGCAAATCAC | bcsA qRT-PCR |
| bcsA-q2 | AGTAGTGCGGCGTCTGTAAC | |
| pdeA-q1 | CCAACAGCGCTCAACCTTTC | pdeA qRT-PCR |
| pdeA-q2 | AATCAGGTTCTGCCCTTCGG | |
| pdeB-q1 | CCGCAAATTTTCACAGGGCA | pdeB qRT-PCR |
| pdeB-q2 | CAACACAGCCAGCTTATGCG | |
| pdeC-q1 | CACCAGGGCAAGAACCAGAT | pdeC qRT-PCR |
| pdeC-q2 | CTCTGCCAGCCCCTGTAAAA | |
| proQ-q1 | TCAAAGTCACCGCGGGTAAA | proQ qRT-PCR |
| proQ-q2 | TATCATTGCCAGGCCGGAAG | |
| prc-q1 | AGGCCTGTTTATTCCCGGTG | prc qRT-PCR |
| prc-q2 | CCAGCGGGCCTTTGTAGTAA | |
| LAD1-1 | ACGATGGACTCCAGAGCGGCCGCVNVNNNGGAA | Transposon mutagenesis |
| LAD1-2 | ACGATGGACTCCAGAGCGGCCGCBNBNNNGGTT | |
| LAD1-3 | ACGATGGACTCCAGAGCGGCCGCVVNVNNNCCAA | |
| LAD1-4 | ACGATGGACTCCAGAGCGGCCGCBDNBNNNCGGT | |
| AC-1 | ACGATGGACTCCAGAG | |
| SP-1 | GACCGAGATAGGGTTGAGTGTTGTTCC | |
| SP-2 | ATCTGGGAATCATTTGAAGGTTGGTAC | |
| Himar1 | CATTTAATACTAGCGACGCCATCT | |
Underlining sequences are restriction sites recognized by restriction enzyme XbaI, HindIII, or BamHI; lowercase sequences are flanking sequences of the Cm in pKD3, the Km in pKD4, or the multiple cloning site in pPROBE-NT.
Mutant construction and complementation assays.
The proQ, prc, msrC, nlpI, spr, edcB, and proP genes were deleted from the genome of E. amylovora Ea1189 using the red recombinase method (75). In brief, specific primers (Table 2) were designed to include 50-nucleotide homology sequences of flanking regions upstream and downstream of each target gene and sequences of flanking regions of the short flippase recognition target (FRT) sites of the Cm or Km resistance cassette from the plasmid pKD3 or pKD4, respectively. PCR was performed to amplify recombination fragments, which were then purified using the QIAquick PCR purification kit (Qiagen, Hilden, Germany). Purified PCR fragments were then electroporated into E. amylovora containing the helper plasmid pKD46, and the resulting recombinants were plated onto LB plates supplemented with Cm or Km for the selection of strains with chromosomal deletions. Mutations validated via PCR confirmation using outside primers were further confirmed by sequencing.
To generate complementation strains, the putative promoter and open reading frame (ORF) regions of target genes were amplified and cloned into the plasmids pBBR1-MCS5 and pCL1920, respectively (Table 1). The resulting plasmids were then confirmed by sequencing and electroporated into E. amylovora cells.
Virulence assay in apple shoots.
For apple shoot assays, bacterial cells were first cultured in LB medium and then resuspended in 0.5× PBS buffer (Thermo Fisher Scientific, Waltham, MA, USA) at a density of 2 × 108 CFU/mL. Actively growing shoots from 1-year-old potted apple trees (Malus × domestica cv. Gala on M9 rootstock) were inoculated with E. amylovora by cutting with scissors dipped in a bacterial suspension (6). The necrotic lesion length was measured with a ruler from the point of inoculation regularly for up to 2 weeks. Two independent experiments were conducted, with a minimum of three shoots per tree and three trees per treatment in each experiment.
In planta biofilm formation assay.
Biofilms formed by E. amylovora WT strain Ea1189 or Ea1189 ΔproQ in planta were visualized using SEM. Briefly, bacterial cells were inoculated into actively growing shoots of 1-year-old potted cv. Gala apple trees using the scissor dip method. At 7 dpi, the midrib of infected apple leaves was sectioned into 1-cm sections. Samples were then fixed in paraformaldehyde/glutaraldehyde, followed by tissue dehydration, critical point drying, sectioning, mounting, and osmium coating. SEM analyses were performed as described previously (6, 76).
Green fluorescent protein-based transcriptional activity assay.
To determine the transcriptional activities of hrpL, hrpN, bcsO, and bcsA, putative promoter regions of each target gene were amplified and cloned into the pPROBE-NT vector. pPROBE-NT is a promoter-probe vector that contains a promoterless green fluorescent protein (GFP) gene, whose transcription is fully reliant on the integrated promoters (77, 78). The resulting plasmids, including pNT-hrpL, pNT-hrpN, pNT-bcsO, and pNT-bcsA, were transferred into E. amylovora by electroporation. Bacterial cells containing the reporter plasmid pNT-hrpL or pNT-hrpN were first cultured in LB medium overnight and then inoculated at 1:100 into Hrp-inducing minimal medium at 28°C for 16 h. Cells containing pNT-bcsO or pNT-bcsA were first grown in LB medium and then inoculated at 1:100 into fresh LB medium at 28°C for 16 h. To measure the GFP intensity, bacterial cells washed with 0.5× PBS buffer were assayed in a Tecan (Männedorf, Switzerland) Spark plate reader with excitation at 488 nm and emission detection at 435 nm. GFP fluorescence was normalized to the optical density at 600 nm (OD600) values of bacterial cultures.
Amylovoran production and levansucrase activity assays.
An amylovoran production assay was conducted via a cetylpyridinium chloride (CPC)-based turbidity assay (79). Cells of E. amylovora from LB cultures grown overnight were inoculated 1:100 into MBMA supplemented with 1% sorbitol at 28°C for 48 h. The supernatants of bacterial cultures were then mixed with 0.1 volumes of CPC (50 mg/mL) and incubated at 28°C for 10 min. To determine amylovoran production, turbidity values of mixtures at the OD600 were measured and normalized to the OD600 values of the cultures grown in MBMA.
Levansucrase activity assays were performed as previously described (80). The supernatants of bacterial cultures in LB medium were mixed 1:1 with 2 M sucrose in PBS buffer. Mixtures were then incubated with shaking (220 rpm) at 28°C for 24 h, and the resulting turbidity, due to the catalyzation of sucrose by levansucrase, was measured at the OD600. Levansucrase activity was calculated by normalizing the OD600 values of turbidity to the OD600 of the bacterial culture.
Cellulose production assay.
To determine the production of cellulose in E. amylovora, a total of 5 μL (approximately 2.5 × 107 cells) of bacterial cultures grown overnight in LB medium was spotted onto LB agar plates lacking sodium chloride and amended with Congo red (40 μg/mL) and Coomassie brilliant blue (20 μg/mL). Plates were then incubated at 28°C and photographed at 24 h postinoculation. To measure the redness of colonies representing the production of cellulose, images were split into 3 RGB (red, green, and blue) channels using ImageJ (81), and the resulting green images were chosen to quantify the redness. Color densities of the entire bacterial colonies were measured using the default settings of ImageJ, with lower values representing higher redness.
Transposon mutagenesis assay.
Transposon mutagenesis was performed by conjugating the Ea1189 ΔproQ::Cm mutant strain with Escherichia coli S17-1 λpir containing the transposon miniHimar RB1 (48). Conjugates were plated onto LB agar plates containing Km and Cm and incubated at 28°C for 2 days. Mutants grown on plates were then picked and inoculated into LB broth for cellulose production assays. Those that exhibited reduced redness compared to Ea1189 ΔproQ::Cm were preserved. To identify the transposon insertion sites, a previously described thermal asymmetric interlaced PCR method was used, with a few modifications (82). In brief, genomic DNA of transposon mutants was purified using the phenol-chloroform method (83) and used as the template for PCR. Four individual PCRs were conducted per mutant using a transposon-specific primer, SP-1, and one of four random primers, LAD1-1, LAD1-2, LAD1-3, and LAD1-4 (Table 2), followed by the following program: 1 cycle of 93°C for 2 min and 95°C for 1 min; 11 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 1 min 20 s; 1 cycle of 94°C for 30 s, 25°C for 2 min, and 72°C for 3 min; 26 cycles of 94°C for 20 s, 58°C for 30 s, and 72°C for 1 min 20 s; and 1 cycle of 72°C for 5 min. Four individual secondary PCRs were conducted per mutant using the primary PCR products as the template and primers AC-1 and SP-2 (Table 2), followed by the following program: 2 cycles of 94°C for 2 min, 65°C for 30 s, and 72°C for 40 s; 14 cycles of 94°C for 1 min, 68°C for 30 s, 72°C for 40 s, 94°C for 1 min, 68°C for 30 s, 72°C for 40 s, 94°C for 1 min, 50°C for 30 s, and 72°C for 40 s; and 1 cycle of 72°C for 5 min. The resulting PCR products were validated via gel electrophoresis and sequenced using transposon-specific primer Himar1 (Table 2).
Isolation of RNA and quantitative real-time PCR.
Total RNAs of E. amylovora cultured in LB broth were extracted using the RNeasy minikit method (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Genomic DNA was removed using Turbo DNA-free DNase (Ambion, Austin, TX, USA), and cDNA was synthesized using TaqMan reverse transcription (RT) reagents (Applied Biosystems, Foster City, CA, USA). To evaluate the mRNA levels of proQ, prc, thiO, and thiG, quantitative real-time PCR (qRT-PCR) was conducted using cDNA as the template. Levels of cDNAs from different samples were quantified using SYBR green PCR master mix (Applied Biosystems, Foster City, CA, USA), and the relative expression levels of each target gene were calculated using the 2–ΔΔCT method (84), with the recA gene as the internal control (85). Primers used for qRT-PCR are listed in Table 2.
RNA stability assay.
For RNA stability assays, bacterial cultures grown overnight were inoculated into fresh medium at a ratio of 1:100 at 28°C for 16 h, followed by rifampin treatment at a final concentration of 500 μg/mL. Total RNAs were isolated from samples immediately after rifampin treatment and at subsequent time points thereafter, including 2, 5, 10, 15, and 30 min. cDNAs were synthesized using total RNAs as the template, and qRT-PCR was performed to determine RNA stability. The gene-specific primers bcsA-q1 and bcsA-q2 (Table 2) were used to amplify the targeted region, bp 595 to 736, within the bcsA coding sequence, and primers pdeC-q1 and pdeC-q2 (Table 2) were used to amplify the targeted region, bp 1171 to 1272, within the pdeC coding sequence. Cycle threshold values of the sample taken immediately upon the addition of rifampin were used to set 100% mRNA remaining.
Quantification of intracellular c-di-GMP concentrations.
Intracellular levels of c-di-GMP were quantified using UPLC-MS/MS as described previously (86). Bacterial cultures grown overnight were inoculated 1:100 into fresh LB medium and incubated at 28°C until the OD600 reached approximately 0.8, corresponding to the mid- to late exponential growth phase. Bacterial cells from 1-mL cultures were then collected by centrifugation at 1,500 × g for 30 min, resuspended in 100 μL of extraction buffer (40% acetonitrile and 40% methanol in 0.1 N formic acid), and incubated for 15 min at −20°C. After centrifugation for 5 min at 21,000 × g to pellet insoluble debris, 10 μL of the supernatant containing c-di-GMP was analyzed by UPLC-MS/MS on a Quattro Premier XE instrument. Intracellular levels of c-di-GMP were quantified by comparison against a standard curve generated by using chemically synthesized c-di-GMP (Axxora Life Science Inc., San Diego, CA).
Statistical analysis.
Means and standard deviations of experimental results were calculated using Excel, and mean comparisons were performed using two-tailed Student’s t test (Microsoft, Redmond, WA).
ACKNOWLEDGMENTS
This project was supported by Michigan State University AgBioResearch.
We thank Jingsheng Xu from the Chinese Academy of Agricultural Sciences for sharing valuable information and helpful discussion on the thermal asymmetric interlaced PCR technique, and we thank Jared Zaporski for construction of the E. amylovora Ea1189 ΔedcB mutant.
Footnotes
Supplemental material is available online only.
Contributor Information
George W. Sundin, Email: sundin@msu.edu.
Gladys Alexandre, University of Tennessee at Knoxville.
REFERENCES
- 1.Malnoy M, Martens S, Norelli JL, Barny M-A, Sundin GW, Smits TH, Duffy B. 2012. Fire blight: applied genomic insights of the pathogen and host. Annu Rev Phytopathol 50:475–494. 10.1146/annurev-phyto-081211-172931. [DOI] [PubMed] [Google Scholar]
- 2.Griffith CS, Sutton TB, Peterson PD. 2003. Fire blight: the foundation of phytobacteriology. APS Press, St Paul, MN. [Google Scholar]
- 3.Thomson SV. 2000. Epidemiology of fire blight, p 9–37. In Vanneste JL (ed), Fire blight: the disease and its causative agent, Erwinia amylovora. CABI Publishing, Wallingford, United Kingdom. [Google Scholar]
- 4.Kharadi RR, Schachterle JK, Yuan X, Castiblanco LF, Peng J, Slack SM, Zeng Q, Sundin GW. 2021. Genetic dissection of the Erwinia amylovora disease cycle. Annu Rev Phytopathol 59:191–212. 10.1146/annurev-phyto-020620-095540. [DOI] [PubMed] [Google Scholar]
- 5.van der Zwet T, Orolaza-Halbrendt N, Zeller W. 2012. Fire blight: history, biology, and management. American Phytopathological Society, St Paul, MN. [Google Scholar]
- 6.Koczan JM, McGrath MJ, Zhao Y, Sundin GW. 2009. Contribution of Erwinia amylovora exopolysaccharides amylovoran and levan to biofilm formation: implications in pathogenicity. Phytopathology 99:1237–1244. 10.1094/PHYTO-99-11-1237. [DOI] [PubMed] [Google Scholar]
- 7.Zeng Q, Puławska J, Schachterle J. 2021. Early events in fire blight infection and pathogenesis of Erwinia amylovora. J Plant Pathol 103:S13–S24. 10.1007/s42161-020-00675-3. [DOI] [Google Scholar]
- 8.Piqué N, Miñana-Galbis D, Merino S, Tomás J. 2015. Virulence factors of Erwinia amylovora: a review. Int J Mol Sci 16:12836–12854. 10.3390/ijms160612836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuan X, Hulin MT, Sundin GW. 2021. Effectors, chaperones, and harpins of the type III secretion system in the fire blight pathogen Erwinia amylovora: a review. J Plant Pathol 103:S25–S39. 10.1007/s42161-020-00623-1. [DOI] [Google Scholar]
- 10.Oh C-S, Kim JF, Beer SV. 2005. The Hrp pathogenicity island of Erwinia amylovora and identification of three novel genes required for systemic infection. Mol Plant Pathol 6:125–138. 10.1111/j.1364-3703.2005.00269.x. [DOI] [PubMed] [Google Scholar]
- 11.Bellemann P, Geider K. 1992. Localization of transposon insertions in pathogenicity mutants of Erwinia amylovora and their biochemical characterization. J Gen Microbiol 138:931–940. 10.1099/00221287-138-5-931. [DOI] [PubMed] [Google Scholar]
- 12.Büttner D, He SY. 2009. Type III protein secretion in plant pathogenic bacteria. Plant Physiol 150:1656–1664. 10.1104/pp.109.139089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coburn B, Sekirov I, Finlay BB. 2007. Type III secretion systems and disease. Clin Microbiol Rev 20:535–549. 10.1128/CMR.00013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geider K. 2000. Exopolysaccharides of Erwinia amylovora: structure, biosynthesis, regulation, role in pathogenicity of amylovoran and levan, p 117–140. In Vanneste JL (ed), Fire blight: the disease and its causative agent, Erwinia amylovora. CABI Publishing, Wallingford, United Kingdom. [Google Scholar]
- 15.Yuan X, McGhee G, Slack S, Sundin GW. 2021. A novel signaling pathway that connects thiamine biosynthesis, bacterial respiration, and production of the exopolysaccharide amylovoran in Erwinia amylovora. Mol Plant Microbe Interact 34:1193–1208. 10.1094/MPMI-04-21-0095-R. [DOI] [PubMed] [Google Scholar]
- 16.Seemüller E, Beer S. 1977. Isolation and partial characterization of two neutral proteases of Erwinia amylovora. J Phytopathol 90:12–21. 10.1111/j.1439-0434.1977.tb02880.x. [DOI] [Google Scholar]
- 17.Ross P, Mayer R, Benziman M. 1991. Cellulose biosynthesis and function in bacteria. Microbiol Rev 55:35–58. 10.1128/mr.55.1.35-58.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Castiblanco LF, Sundin GW. 2018. Cellulose production, activated by cyclic di-GMP through BcsA and BcsZ, is a virulence factor and an essential determinant of the three-dimensional architectures of biofilms formed by Erwinia amylovora Ea1189. Mol Plant Pathol 19:90–103. 10.1111/mpp.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hengge R. 2009. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol 7:263–273. 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 20.Jenal U, Reinders A, Lori C. 2017. Cyclic di-GMP: second messenger extraordinaire. Nat Rev Microbiol 15:271–284. 10.1038/nrmicro.2016.190. [DOI] [PubMed] [Google Scholar]
- 21.Römling U, Galperin MY, Gomelsky M. 2013. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 77:1–52. 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kharadi RR, Castiblanco LF, Waters CM, Sundin GW. 2019. Phosphodiesterase genes regulate amylovoran production, biofilm formation, and virulence in Erwinia amylovora. Appl Environ Microbiol 85:e02233-18. 10.1128/AEM.02233-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edmunds AC, Castiblanco LF, Sundin GW, Waters CM. 2013. Cyclic di-GMP modulates the disease progression of Erwinia amylovora. J Bacteriol 195:2155–2165. 10.1128/JB.02068-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuan X, Khokhani D, Wu X, Yang F, Biener G, Koestler BJ, Raicu V, He C, Waters CM, Sundin GW, Fang T, Yang C-H. 2015. Cross-talk between a regulatory small RNA, cyclic-di-GMP signalling and flagellar regulator FlhDC for virulence and bacterial behaviours. Environ Microbiol 17:4745–4763. 10.1111/1462-2920.13029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morgan JL, McNamara JT, Zimmer J. 2014. Mechanism of activation of bacterial cellulose synthase by cyclic di-GMP. Nat Struct Mol Biol 21:489–496. 10.1038/nsmb.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richter AM, Possling A, Malysheva N, Yousef KP, Herbst S, von Kleist M, Hengge R. 2020. Local c-di-GMP signaling in the control of synthesis of the E. coli biofilm exopolysaccharide pEtN-cellulose. J Mol Biol 432:4576–4595. 10.1016/j.jmb.2020.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paul R, Weiser S, Amiot NC, Chan C, Schirmer T, Giese B, Jenal U. 2004. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev 18:715–727. 10.1101/gad.289504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whiteley CG, Lee D-J. 2015. Bacterial diguanylate cyclases: structure, function and mechanism in exopolysaccharide biofilm development. Biotechnol Adv 33:124–141. 10.1016/j.biotechadv.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 29.Schmidt AJ, Ryjenkov DA, Gomelsky M. 2005. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J Bacteriol 187:4774–4781. 10.1128/JB.187.14.4774-4781.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeng Q, Sundin GW. 2014. Genome-wide identification of Hfq-regulated small RNAs in the fire blight pathogen Erwinia amylovora discovered small RNAs with virulence regulatory function. BMC Genomics 15:414. 10.1186/1471-2164-15-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zeng Q, McNally RR, Sundin GW. 2013. Global small RNA chaperone Hfq and regulatory small RNAs are important virulence regulators in Erwinia amylovora. J Bacteriol 195:1706–1717. 10.1128/JB.02056-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schachterle JK, Sundin GW. 2019. The leucine-responsive regulatory protein Lrp participates in virulence regulation downstream of small RNA ArcZ in Erwinia amylovora. mBio 10:e00757-19. 10.1128/mBio.00757-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schachterle JK, Zeng Q, Sundin GW. 2019. Three Hfq-dependent small RNAs regulate flagellar motility in the fire blight pathogen Erwinia amylovora. Mol Microbiol 111:1476–1492. 10.1111/mmi.14232. [DOI] [PubMed] [Google Scholar]
- 34.Vogel J, Luisi BF. 2011. Hfq and its constellation of RNA. Nat Rev Microbiol 9:578–589. 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smirnov A, Förstner KU, Holmqvist E, Otto A, Günster R, Becher D, Reinhardt R, Vogel J. 2016. Grad-seq guides the discovery of ProQ as a major small RNA-binding protein. Proc Natl Acad Sci USA 113:11591–11596. 10.1073/pnas.1609981113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olejniczak M, Storz G. 2017. ProQ/FinO‐domain proteins: another ubiquitous family of RNA matchmakers? Mol Microbiol 104:905–915. 10.1111/mmi.13679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holmqvist E, Vogel J. 2018. RNA-binding proteins in bacteria. Nat Rev Microbiol 16:601–615. 10.1038/s41579-018-0049-5. [DOI] [PubMed] [Google Scholar]
- 38.Westermann AJ, Venturini E, Sellin ME, Förstner KU, Hardt W-D, Vogel J. 2019. The major RNA-binding protein ProQ impacts virulence gene expression in Salmonella enterica serovar Typhimurium. mBio 10:e02504-18. 10.1128/mBio.02504-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Melamed S, Adams PP, Zhang A, Zhang H, Storz G. 2020. RNA-RNA interactomes of ProQ and Hfq reveal overlapping and competing roles. Mol Cell 77:411–425. 10.1016/j.molcel.2019.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koczan JM, Lenneman BR, McGrath MJ, Sundin GW. 2011. Cell surface attachment structures contribute to biofilm formation and xylem colonization by Erwinia amylovora. Appl Environ Microbiol 77:7031–7039. 10.1128/AEM.05138-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leonard S, Villard C, Nasser W, Reverchon S, Hommais F. 2021. RNA chaperones Hfq and ProQ play a key role in the virulence of the plant pathogenic bacterium Dickeya dadantii. Front Microbiol 12:687484. 10.3389/fmicb.2021.687484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kerr CH, Culham DE, Marom D, Wood JM. 2014. Salinity-dependent impacts of ProQ, Prc, and Spr deficiencies on Escherichia coli cell structure. J Bacteriol 196:1286–1296. 10.1128/JB.00827-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singh SK, Parveen S, SaiSree L, Reddy M. 2015. Regulated proteolysis of a cross-link-specific peptidoglycan hydrolase contributes to bacterial morphogenesis. Proc Natl Acad Sci USA 112:10956–10961. 10.1073/pnas.1507760112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tadokoro A, Hayashi H, Kishimoto T, Makino Y, Fujisaki S, Nishimura Y. 2004. Interaction of the Escherichia coli lipoprotein NlpI with periplasmic Prc (Tsp) protease. J Biochem 135:185–191. 10.1093/jb/mvh022. [DOI] [PubMed] [Google Scholar]
- 45.Su M-Y, Som N, Wu C-Y, Su S-C, Kuo Y-T, Ke L-C, Ho M-R, Tzeng S-R, Teng C-H, Mengin-Lecreulx D, Reddy M, Chang C-I. 2017. Structural basis of adaptor-mediated protein degradation by the tail-specific PDZ-protease Prc. Nat Commun 8:1516. 10.1038/s41467-017-01697-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kunte H, Jr, Crane RA, Culham DE, Richmond D, Wood JM. 1999. Protein ProQ influences osmotic activation of compatible solute transporter ProP in Escherichia coli K-12. J Bacteriol 181:1537–1543. 10.1128/JB.181.5.1537-1543.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Milner JL, Wood JM. 1989. Insertion proQ220::Tn5 alters regulation of proline porter II, a transporter of proline and glycine betaine in Escherichia coli. J Bacteriol 171:947–951. 10.1128/jb.171.2.947-951.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bouhenni R, Gehrke A, Saffarini D. 2005. Identification of genes involved in cytochrome c biogenesis in Shewanella oneidensis, using a modified mariner transposon. Appl Environ Microbiol 71:4935–4937. 10.1128/AEM.71.8.4935-4937.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee JH, Ancona V, Zhao Y. 2018. Lon protease modulates virulence traits in Erwinia amylovora by direct monitoring of major regulators and indirectly through the Rcs and Gac‐Csr regulatory systems. Mol Plant Pathol 19:827–840. 10.1111/mpp.12566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee JH, Zhao Y. 2017. ClpXP-dependent RpoS degradation enables full activation of type III secretion system, amylovoran production, and motility in Erwinia amylovora. Phytopathology 107:1346–1352. 10.1094/PHYTO-06-17-0198-R. [DOI] [PubMed] [Google Scholar]
- 51.Smirnov A, Wang C, Drewry LL, Vogel J. 2017. Molecular mechanism of mRNA repression in trans by a ProQ-dependent small RNA. EMBO J 36:1029–1045. 10.15252/embj.201696127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bellows LE, Koestler BJ, Karaba SM, Waters CM, Lathem WW. 2012. Hfq-dependent, co-ordinate control of cyclic diguanylate synthesis and catabolism in the plague pathogen Yersinia pestis. Mol Microbiol 86:661–674. 10.1111/mmi.12011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yuan X, Zeng Q, Khokhani D, Tian F, Severin GB, Waters CM, Xu J, Zhou X, Sundin GW, Ibekwe AM, Liu F, Yang CH. 2019. A feed-forward signaling circuit controls bacterial virulence through linking cyclic di-GMP and two mechanistically distinct sRNAs; ArcZ and RsmB. Environ Microbiol 21:2755–2771. 10.1111/1462-2920.14603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Romeo T, Babitzke P. 2018. Global regulation by CsrA and its RNA antagonists. Microbiol Spectr 6:RWR-0009-2017. 10.1128/microbiolspec.RWR-0009-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jonas K, Edwards AN, Simm R, Romeo T, Römling U, Melefors Ö. 2008. The RNA binding protein CsrA controls cyclic di‐GMP metabolism by directly regulating the expression of GGDEF proteins. Mol Microbiol 70:236–257. 10.1111/j.1365-2958.2008.06411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Silva-Rohwer AR, Held K, Sagawa J, Fernandez NL, Waters CM, Vadyvaloo V. 2021. CsrA enhances cyclic-di-GMP biosynthesis and Yersinia pestis biofilm blockage of the flea foregut by alleviating Hfq-dependent repression of the hmsT mRNA. mBio 12:e01358-21. 10.1128/mBio.01358-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bauriedl S, Gerovac M, Heidrich N, Bischler T, Barquist L, Vogel J, Schoen C. 2020. The minimal meningococcal ProQ protein has an intrinsic capacity for structure-based global RNA recognition. Nat Commun 11:2823. 10.1038/s41467-020-16650-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cain AK, Barquist L, Goodman AL, Paulsen IT, Parkhill J, van Opijnen T. 2020. A decade of advances in transposon-insertion sequencing. Nat Rev Genet 21:526–540. 10.1038/s41576-020-0244-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hengge R. 2021. High-specificity local and global c-di-GMP signaling. Trends Microbiol 29:993–1003. 10.1016/j.tim.2021.02.003. [DOI] [PubMed] [Google Scholar]
- 60.Rawlings ND, O’Brien E, Barrett AJ. 2002. MEROPS: the protease database. Nucleic Acids Res 30:343–346. 10.1093/nar/30.1.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vollmer W, Joris B, Charlier P, Foster S. 2008. Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol Rev 32:259–286. 10.1111/j.1574-6976.2007.00099.x. [DOI] [PubMed] [Google Scholar]
- 62.Vollmer W, Blanot D, De Pedro MA. 2008. Peptidoglycan structure and architecture. FEMS Microbiol Rev 32:149–167. 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- 63.Mallick S, Kiran S, Maiti TK, Ghosh AS. 2021. PBP4 and PBP5 are involved in regulating exopolysaccharide synthesis during Escherichia coli biofilm formation. Microbiology (Reading) 167:e001031. 10.1099/mic.0.001031. [DOI] [PubMed] [Google Scholar]
- 64.Gheorghita AA, Wolfram F, Whitfield GB, Jacobs HM, Pfoh R, Wong SSY, Guitor AK, Goodyear MC, Berezuk AM, Khursigara CM, Parsek MR, Howell PL. 2022. The Pseudomonas aeruginosa homeostasis enzyme AlgL clears the periplasmic space of accumulated alginate during polymer biosynthesis. J Biol Chem 298:101560. 10.1016/j.jbc.2021.101560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Farrell EK, Tipton PA. 2012. Functional characterization of AlgL, an alginate lyase from Pseudomonas aeruginosa. Biochemistry 51:10259–10266. 10.1021/bi301425r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Majdalani N, Gottesman S. 2005. The Rcs phosphorelay: a complex signal transduction system. Annu Rev Microbiol 59:379–405. 10.1146/annurev.micro.59.050405.101230. [DOI] [PubMed] [Google Scholar]
- 67.Huang W-C, Lin C-Y, Hashimoto M, Wu J-J, Wang M-C, Lin W-H, Chen C-S, Teng C-H. 2020. The role of the bacterial protease Prc in the uropathogenesis of extraintestinal pathogenic Escherichia coli. J Biomed Sci 27:14. 10.1186/s12929-019-0605-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang D, Korban SS, Zhao Y. 2009. The Rcs phosphorelay system is essential for pathogenicity in Erwinia amylovora. Mol Plant Pathol 10:277–290. 10.1111/j.1364-3703.2008.00531.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chung CH, Goldberg AL. 1981. The product of the lon (capR) gene in Escherichia coli is the ATP-dependent protease, protease La. Proc Natl Acad Sci USA 78:4931–4935. 10.1073/pnas.78.8.4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Baker TA, Sauer RT. 2012. ClpXP, an ATP-powered unfolding and protein-degradation machine. Biochim Biophys Acta 1823:15–28. 10.1016/j.bbamcr.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Osbourne DO, Soo VW, Konieczny I, Wood TK. 2014. Polyphosphate, cyclic AMP, guanosine tetraphosphate, and c-di-GMP reduce in vitro Lon activity. Bioengineered 5:264–268. 10.4161/bioe.29261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Joshi A, Mahmoud SA, Kim S-K, Ogdahl JL, Lee VT, Chien P, Yildiz FH. 2020. c-di-GMP inhibits LonA-dependent proteolysis of TfoY in Vibrio cholerae. PLoS Genet 16:e1008897. 10.1371/journal.pgen.1008897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Huynh TV, Dahlbeck D, Staskawicz BJ. 1989. Bacterial blight of soybean: regulation of a pathogen gene determining host cultivar specificity. Science 245:1374–1377. 10.1126/science.2781284. [DOI] [PubMed] [Google Scholar]
- 74.Yu M, Singh J, Khan A, Sundin GW, Zhao YF. 2020. Complete genome sequence of the fire blight pathogen Erwinia amylovora strain Ea1189. Mol Plant Microbe Interact 33:1277–1279. 10.1094/MPMI-06-20-0158-A. [DOI] [PubMed] [Google Scholar]
- 75.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97:6640–6645. 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Slack SM, Zeng Q, Outwater CA, Sundin GW. 2017. Microbiological examination of Erwinia amylovora exopolysaccharide ooze. Phytopathology 107:403–411. 10.1094/PHYTO-09-16-0352-R. [DOI] [PubMed] [Google Scholar]
- 77.Miller WG, Leveau JH, Lindow SE. 2000. Improved gfp and inaZ broad-host-range promoter-probe vectors. Mol Plant Microbe Interact 13:1243–1250. 10.1094/MPMI.2000.13.11.1243. [DOI] [PubMed] [Google Scholar]
- 78.Leveau JH, Lindow SE. 2001. Predictive and interpretive simulation of green fluorescent protein expression in reporter bacteria. J Bacteriol 183:6752–6762. 10.1128/JB.183.23.6752-6762.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bellemann P, Bereswill S, Berger S, Geider K. 1994. Visualization of capsule formation by Erwinia amylovora and assays to determine amylovoran synthesis. Int J Biol Macromol 16:290–296. 10.1016/0141-8130(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 80.Hildebrand M, Aldridge P, Geider K. 2006. Characterization of hns genes from Erwinia amylovora. Mol Genet Genomics 275:310–319. 10.1007/s00438-005-0085-5. [DOI] [PubMed] [Google Scholar]
- 81.Abràmoff MD, Magalhães PJ, Ram SJ. 2004. Image processing with ImageJ. Biophotonics Int 11:36–42. [Google Scholar]
- 82.Liu Y-G, Chen Y. 2007. High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences. Biotechniques 43:649–656. 10.2144/000112601. [DOI] [PubMed] [Google Scholar]
- 83.Sambrook J, Russell DW. 2006. Purification of nucleic acids by extraction with phenol:chloroform. CSH Protoc 2006:pdb.prot4455. 10.1101/pdb.prot4455. [DOI] [PubMed] [Google Scholar]
- 84.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 85.Takle GW, Toth IK, Brurberg MB. 2007. Evaluation of reference genes for real-time RT-PCR expression studies in the plant pathogen Pectobacterium atrosepticum. BMC Plant Biol 7:50. 10.1186/1471-2229-7-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Massie JP, Reynolds EL, Koestler BJ, Cong J-P, Agostoni M, Waters CM. 2012. Quantification of high-specificity cyclic diguanylate signaling. Proc Natl Acad Sci USA 109:12746–12751. 10.1073/pnas.1115663109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhao Y, Sundin GW, Wang D. 2009. Construction and analysis of pathogenicity island deletion mutants of Erwinia amylovora. Can J Microbiol 55:457–464. 10.1139/w08-147. [DOI] [PubMed] [Google Scholar]
- 88.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176. 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 89.Lerner CG, Inouye M. 1990. Low copy number plasmids for regulated low-level expression of cloned genes in Escherichia coli with blue/white insert screening capability. Nucleic Acids Res 18:4631. 10.1093/nar/18.15.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 and S2 and Table S1. Download aem.00239-22-s0001.pdf, PDF file, 0.4 MB (364.9KB, pdf)