Dear Editor,
The Omicron variant has recently become a dominant variant globally with four lineages such as BA.1, BA.2, BA.3, and B.1.1.529. But little is known about which parts of the world the BA.2 lineage dominates. Here we find out that BA.2 has its central cluster of infection installed in Denmark. It should be noted that 79.4% of the total BA.2 sequences belong to Denmark (as of January 18, 2022). The BA.2 lineage phylogenetically consists of five groups: Sweden/Denmark, Philippines, Hong Kong, India, and China. The ORF3a protein of BA.2, which is spreading in Denmark, contains a specific mutation called H78Y, and this mutation is not widely found in other countries. The Philippines, Hong Kong, India, and China groups of BA.2 also have their own unique mutations. A future experimental study is warranted to understand the significance of these mutations to reveal why this lineage is widely spreading in Denmark, which we hope will help design antiviral and antibodies to control the spread of BA.2 lineage in Denmark and other countries.
The Omicron variant, first detected in Africa in November 2021, has spread to six continents in just 2 months, affecting more than 150 countries/territories. 1 Recently, this Omicron variant has been divided into four lineages: BA.1, BA.2, BA.3, and B.1.1.529. 1 , 2 , 3 Of the Omicron sequences submitted to GISAID, the BA.1 lineage is reported to be approximate >98%, the BA.2 to approximately 1% sequence, and the BA.3 around 0.1% sequence. 2 , 4 All this suggests that the Omicron variant has been pushed to the point where it represents the BA.1 lineage. In this case, the Omicron variant's second most widespread lineage of BA.2 is spreading and dominating in which part of the world and the genetic variation within it is not fully known. Identifying the BA.2 lineage spreading pattern and its genetic diversity can be expected to support the formulation of administrative policies governing and controlling this lineage spread and the formulation of antiviral or antibody therapies. In the present study, we have systematically analyzed the genetic diversity of the BA.2 lineage and how this lineage spreads.
In this study, first, we did the phylogenetic analysis to find the genetic diversity in the Omicron variant, in which the Omicron variant was divided into four lineages: B.1.1.529, BA.1, BA.2, and BA.3 (Figure 1A). Of these four lineages, B.1.1.529 appears to be the parental lineage of the Omicron variant, and then the BA.1 lineage seems to be the closest to this B.1.1.529 lineage (Figure 1A). BA.2 has significant diversity from the B.1.1.529 and BA.1 lineages, while BA.3 has the intermediate lineage to BA.1 and BA.2 (Figure 1A). Consistent with previous reports of mutations in the spike protein of BA.3 is a combination of mutations in the spike protein of the BA.1 and BA.2 lineages. 4 Then, as of January 19, 2022, BA.1 was 97.69% (415 222/425 055), BA.2 was 1.62% (6888/425 055), BA.3 was 0.016% (66/425 055), and B.1.1.529 was 0.68% (2879/425 055) of the sequences submitted to GISAID worldwide. Of the total 6888 BA.2 sequences, 5469 (79.40%) sequences are found in Denmark alone (Figure 1B). Meanwhile, the BA.1 sequences from Denmark were 20 639 (4.97%) (Figure 1C), and the B.1.1.529 was 15 (0.52%) (Figure 1D). It is noteworthy that the BA.3 lineage was not detected in Denmark until January 19, 2022. As of January 19, 2022, the global ratio of the BA.1 and BA.2 sequences is 60.28 (BA.1): 1 (BA.2), while in Denmark, it is 3.77 (BA.1): 1 (BA.2). This indicates that one BA.2 sequence is detected worldwide for every 60.28 BA.1 sequences, but one BA.2 sequence is detected for every 3.77 BA.1 sequences in Denmark. All of this makes it clear that the BA.2 lineage dominates Denmark as a cluster. Also, mutation A22786T in BA.2 is said to have changed to A22786C (https://github.com/cov-lineages/pango-designation/issues/410), so we started to show preference in these mutations. Our analysis revealed that A22786C appeared 45 375 times and A22786T 13 times in the BA.2 sequences submitted worldwide from November 8, 2021 to February 08, 2022 (https://cov-spectrum.org/). It is noteworthy that A22786C appeared 28 443 times and A22786T 12 times in the BA.2 sequences submitted from Denmark during the same period (https://cov-spectrum.org/). From these analyzes, it is clear that A22786C dominates globally and in Denmark. Moreover, both of these nucleotide mutations (A22786T and A22786C) form the same R408S mutation in the spike protein (https://github.com/cov-lineages/pango-designation/issues/410). Therefore, it can be believed that these two nucleotide mutations do not cause any change in the functional state of the protein. However, these nucleotide mutations can be expected to cause some effect when designing PCR primers/probes in this specific region.
Figure 1.
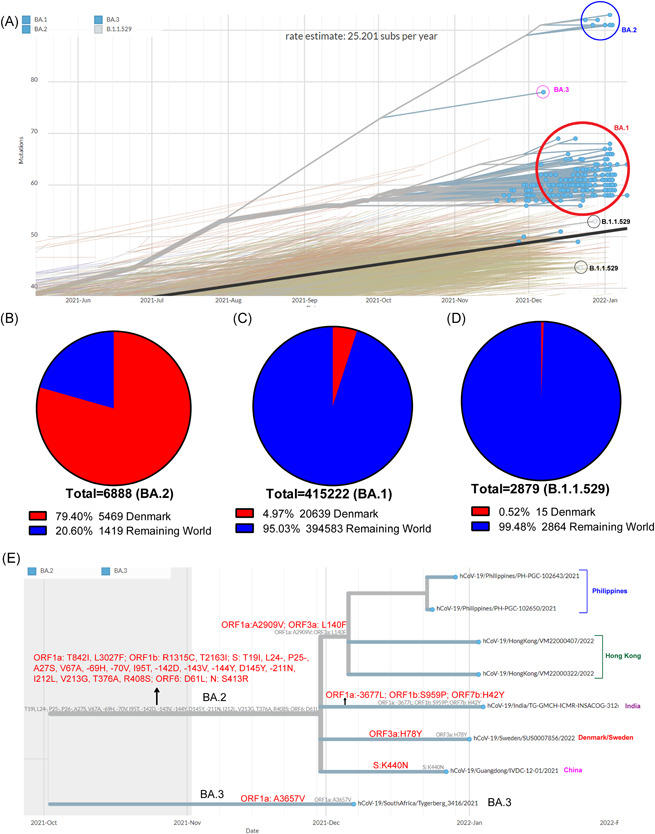
Genetic diversity in the Omicron variant. (A) Phylogenetic tree depicting the relationship and substitution rate of different lineages of Omicron variant. This Phylogenetic tree was created using the Nextstrain online tool, with options such as nCoV, GISAID sequences, global sequences, PANGO Lineage, and CLOCK tree in this online tool for the Omicron variant (https://www.gisaid.org/phylodynamics/global/nextstrain/, January 14, 2022). (B and C) Depicting the part of Omicron variant's different lineages sequences detected in Denmark against Worldwide (as of January 18, 2022, sequences submitted in GISAID); (B) represent BA.2 lineage; (C) represent BA.1 lineage, and (D) represent B.1.1.529 lineage. (E) Phylogenetic tree displaying the presence of five different subgroups in the BA.2 lineage and their specific mutations. This Phylogenetic tree was created using the Nextstrain online tool, with options such as nCoV, GISAID sequences, global sequences, PANGO Lineage, and Rectangular tree in this online tool for the Omicron variant (https://www.gisn/aid.org/phylodynamics/global/nextstrai, January 14, 2022)
Next, we explore whether any subgroups within the BA.2 lineage extend specifically to geographical locations. In this analysis, we considered that there are five subgroups within the BA.2 lineage, and they are subdivided into geographical areas, namely Sweden/Denmark, Philippines, Hong Kong, India, and China (Figure 1E). Of these, we note that the Philippines and Hong Kong subgroups differ significantly from other sub‐groups (Figure 1E). Also, these Philippines and Hong Kong subgroups have unique mutations found in ORF1a: A2909V and ORF3a: L140F (Figure 1E). Until January 18, 2022, mutations ORF1a: A2909V and ORF3a: L140F in the BA.2 lineage were primarily found in the Philippines, Hong Kong, and Japan (only 10–20 sequences), with this mutation occurring in just two or three sequences in Europe (Figure 2A,B). ORF1a: A2909V is a mutation in nonstructural proteins 4 (Nsp4); although no experimental studies have been performed on the functional significance of this specific mutation, it is well‐known that Nsp4 is a protein required for virus replication. 5 , 6 Similarly, no experimental studies have been performed on the functional significance of ORF3a: L140F mutation, although it is essential to note that ORF3 is a protein that contributes to the viral release of viral ion channels (viroporins). 7 Then we noticed that in India, subgroup (BA.2) has unique mutations of ORF1a: ‐3677L, ORF1b: S959P, and ORF7b: H42Y (Figure 1E). As of January 18, 2022, ORF1a: ‐3677L mutations in the BA.2 lineage are primarily found in India (114 sequences) (Figure 2C), ORF1b: S959P mutations are mostly found in India (73 sequences), and Singapore (25 sequences) (Figure 2D), and ORF7b: H42Y mutations are found only in tiny sequences. ORF1a: ‐3677L and ORF1b: S959P are mutations in Nsp6 and Nsp13, respectively, although no experimental studies have been performed on the functional significance of these specific mutations; it is well‐known that Nsp6 is the protein required for virus replication 5 , 6 and the Nsp13 helicase protein. 8 , 9
Figure 2.
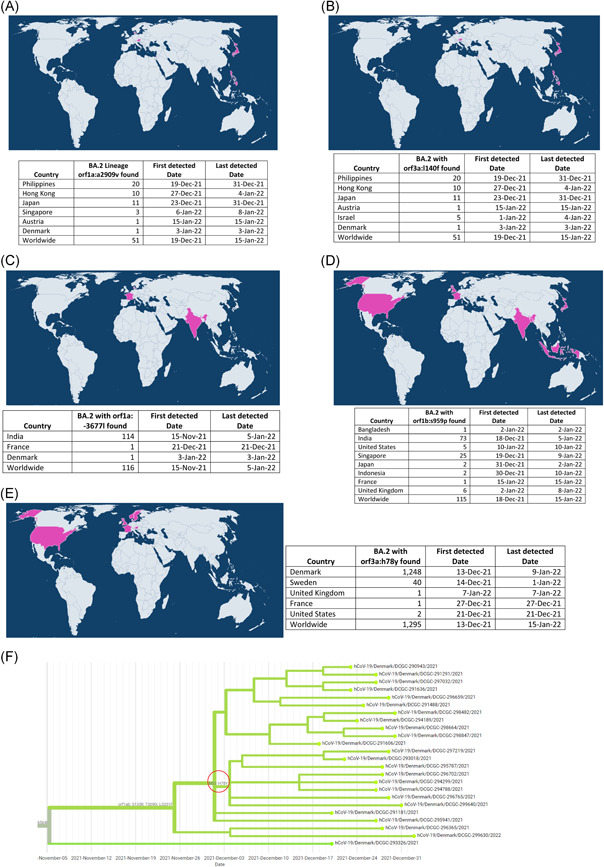
The five subgroups with the BA.2 lineage express different and distinct mutations in different geographical regions. (A and B) Depicting the expression of a specific mutation in the ORF1a: A2909V and ORF3a: L140F in the Philippines and Hong Kong subgroups and its global distribution, as well as numbers sequences, has this mutation in the BA.2 lineage; (A) represent ORF1a: A2909V mutation, and (B) represent ORF3a: L140F mutation. This information is derived from the online tool “SARS‐CoV‐2 (hCoV‐19) Mutation Reports” (https://outbreak.info/compare-lineages, January 18, 2022). (C and D) Depicting the expression of a specific mutation in the ORF1a: −3677L and ORF1b: S959P in the Indian subgroup and its global distribution and numbers sequences have this mutation in the BA.2 lineage. (C) represent ORF1a: −3677L mutation, and (D) represent ORF1b: S959P mutation. This information is derived from the online tool “SARS‐CoV‐2 (hCoV‐19) Mutation Reports” (E) Depicting the expression of a specific mutation in the ORF3a: H78Y in the Sweden/Denmark subgroup and its global distribution and numbers sequences have this mutation in the BA.2 lineage. This information is derived from the online tool “SARS‐CoV‐2 (hCoV‐19) Mutation Reports” (https://outbreak.info/compare-lineages, January 18, 2022). (F) Phylogenetic tree depicting the presence of ORF3a: H78Y mutation in the BA.2 sequences submitted from Denmark. This Phylogenetic tree was created using the Nextstrain online tool, with options such as nCoV, GISAID sequences, Denmark sequences, PANGO Lineage, and Rectangular tree in this online tool for the Omicron variant (https://www.gisaid.org/phylodynamics/denmark/, January 14, 2022)
Finally, we note that there is a unique mutation in the Sweden/Denmark sub‐group called ORF3a: H78Y (Figure 1E). Until January 18, 2022, mutations ORF3a: H78Y in the BA.2 lineage mainly were detected in Denmark (1,248 sequences) and Sweden (40 sequences) (Figure 2E). Furthermore, phylogenetic analysis confirmed the presence of this ORF3a: H78Y mutation in most BA.2 lineages circulating in Denmark (Figure 2F). Much of the experimental studies on the role of this ORF3a: H78Y mutation in the activity of the virus has not yet been carried out, but ORF3 is the protein needed to create viral ion channels (viroporins) and thereby facilitate virus release. 7 Is this ORF3a: H78Y mutation the reason why the BA.2 lineage is so widespread in Denmark; or if genetic makeup of Denmark people, predisposing causes like dietary habits, climate; or large‐scale research into whether any animals in Denmark are infected with this lineage and continue to transmit BA.2 from those animals to humans can be expected to help control the spread of BA.2 in Denmark and other countries. Furthermore, it is hoped that this BA.2 lineage may be able to control and treat the spread of BA.2 lineage by designing antiviral and antibody therapies against specific mutations and identification of other predisposing causes.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
All the authors contributed significantly to this manuscript. Perumal A. Desingu analyzed and wrote the first draft, K. Nagarajan reviewed the manuscript. All the authors reviewed and approved the final submission.
ACKNOWLEDGMENTS
Perumal A. Desingu is a DST‐INSPIRE faculty is supported by research funding from the Department of Science and Technology (DST/INSPIRE/04/2016/001067), Government of India, and Core grant from the Science and Engineering Research Board (SERB) (CRG/2018/002192), Department of Science and Technology (DST), Government of India.
DATA AVAILABILITY STATEMENT
Not applicable.
REFERENCES
- 1. Desingu PA, Nagarajan K, Dhama K. Emergence of Omicron third lineage BA.3 and its importance. J Med Virol. 2022. 10.1002/jmv.27601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Enhancing response to Omicron SARS‐CoV‐2 variant: Technical brief and priority actions for Member States. World Health Organization.
- 3. Majumdar S, Sarkar R. Mutational and phylogenetic analyses of the two lineages of the Omicron variant. J Med Virol. 2021. 10.1002/jmv.27558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Desingu PA, Nagarajan K, Dhama K. Emergence of Omicron third lineage BA.3 and its importance. J Med Virol. 2022. 10.1002/jmv.27601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mariano G, Farthing RJ, Lale‐Farjat SLM, Bergeron JRC. Structural characterization of SARS‐CoV‐2: where we are, and where we need to be. Front Mol Biosci. 2020;7:605236. 10.3389/fmolb.2020.605236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Thomas S. Mapping the nonstructural transmembrane proteins of severe acute respiratory syndrome coronavirus 2. J Comput Biol. 2021;28:909‐921. 10.1089/cmb.2020.0627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tan Y, Schneider T, Shukla PK, Chandrasekharan MB, Aravind L, Zhang D. Unification and extensive diversification of M/Orf3‐related ion channel proteins in coronaviruses and other nidoviruses. Virus Evol. 2021;7:veab014. 10.1093/ve/veab014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. White MA, Lin W, Cheng X. Discovery of COVID‐19 inhibitors targeting the SARS‐CoV‐2 Nsp13 Helicase. J Phys Chem Lett. 2020;11:9144‐9151. 10.1021/acs.jpclett.0c02421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Newman JA, Douangamath A, Yadzani S, et al. Structure, mechanism and crystallographic fragment screening of the SARS‐CoV‐2 NSP13 helicase. Nat Commun. 2021;12:4848. 10.1038/s41467-021-25166-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.


