Abstract
The COVID‐19 disease caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) primarily affects the lung, particularly the proximal airway and distal alveolar cells. NKX2.1+ primordial lung progenitors of the foregut (anterior) endoderm are the developmental precursors to all adult lung epithelial lineages and are postulated to play an important role in viral tropism. Here, we show that SARS‐CoV‐2 readily infected and replicated in human‐induced pluripotent stem cell‐derived proximal airway cells, distal alveolar cells, and lung progenitors. In addition to the upregulation of antiviral defense and immune responses, transcriptomics data uncovered a robust epithelial cell‐specific response, including perturbation of metabolic processes and disruption in the alveolar maturation program. We also identified spatiotemporal dysregulation of mitochondrial heme oxygenase 1 (HMOX1), which is associated with defense against antioxidant‐induced lung injury. Cytokines, such as TNF‐α, INF‐γ, IL‐6, and IL‐13, were upregulated in infected cells sparking mitochondrial ROS production and change in electron transport chain complexes. Increased mitochondrial ROS then activated additional proinflammatory cytokines leading to an aberrant cell cycle resulting in apoptosis. Notably, we are the first to report a chemosensory response resulting from SARS‐CoV‐2 infection similar to that seen in COVID‐19 patients. Some of our key findings were validated using COVID‐19‐affected postmortem lung tissue sections. These results suggest that our in vitro system could serve as a suitable model to investigate the pathogenetic mechanisms of SARS‐CoV‐2 infection and to discover and test therapeutic drugs against COVID‐19 or its consequences.
Keywords: chemosensory response, induced pluripotent stem cells, inflammatory response, lung epithelial cells, mitochondrial damage, SARS‐CoV‐2
In this study, we showed that human‐induced pluripotent stem cell‐derived airway and alveolar epithelial cells were permissive to live severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) infection that triggered a massive inflammatory response, sets off chemosensory changes, and impaired proximal‐distal lung patterning. These alterations were associated with mitochondrial dysfunction eventually leading to a pulmonary fibrosis‐like situation similar to that seen in COVID‐19 patients. Our results highlight the potential applications of this in vitro platform in studying SARS‐CoV‐2 infection and anti‐viral screening of compounds.

1. INTRODUCTION
The global pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) has led to over 6.0 million deaths worldwide as of April 2022. It is primarily transmitted via respiratory droplets and the lungs are the most important organ involved (Pei et al., 2021). It causes fever, shortness of breath, fatigue, diarrhea, and loss of smell and taste. The infection is initiated in the proximal airways and the alveolar type 2 (AT2) cells of the distal lung (Mulay et al., 2021). The damage to the alveolar cells is associated with inflammation, which triggers focal capillary micro‐thrombus formation causing pulmonary parenchymal fibrosis and the eventual death of the patient (Kommoss et al., 2020).
Researchers are still trying to understand how COVID‐19 infection leads to acute respiratory distress syndrome (ARDS), severe pneumonia, and multiple organ failure all of which contribute to high patient mortality. In addition, the consequences of “lung Covid syndrome” have created a substantial societal burden. Even though vaccines such as an inactivated virus or a modified version of the virus have been developed, potential therapeutic drugs in the form of antivirals etc. are still lacking. One major obstacle in developing effective treatment is the lack of understanding of viral pathogenesis due to the unavailability of a proper model system. Using patient cells is advantageous, but there is an acute shortage of patient tissues, particularly from the early stages of the disease (Huang et al., 2020). Further, the inability to grow and expand these primary cells for extended periods of time, compared to immortalized cell lines, has made them difficult to use. Animal models like mice, rats, hamsters etc., help achieve a better understanding of in vivo tissue‐specific and systemic host−pathogen interactions. However, the genetic and tissue‐level variations among species are a major drawback in extrapolating such results to the human system. Nevertheless, cell lines can be used for in vitro studies as a viable model system to investigate the cellular and molecular effects of viral infection and also for evaluating a potential therapeutic drug (Pei et al., 2021). Although various SARS‐CoV‐2 infection studies have used multiple cell lines, such as Caco‐2, Calu‐3, HEK 293T, and Huh7 etc., there are limitations such as low viral titers, lack of appropriate immune responses, cytogenetic instability, insufficient physiological relevance, and low and inconsistent expression of ACE2 and TMPRSS2 (Abo et al., 2020).
The foundation of our model lies in a differentiation protocol that generates biopotential lung progenitor cells eventually giving rise to both airway and alveolar cells. Employing this 2D model, we sought to understand the underlying mechanism involved in SARS‐CoV‐2 infection and the pathogenesis of the severe disease. First, we tested the permissiveness of this model system by infecting the cultures at various multiplicities of infection (MOI) and progressive time points. Following infection, among other parameters, we observed the presence of viral spike protein inside the cell establishing its permissibility. We then performed bulk RNA sequencing to study the molecular changes and compared the observed cellular responses with published reports using primary cells and patient samples. We noticed massive inflammation, fibrosis, and cell death sequentially, reminiscent of COVID‐19 pathology. A deeper analysis of the transcriptomics data helped identify the possible role of mitochondria in COVID‐19. Modulation of electron transport chain (ETC) complexes postinfection marked by heme oxygenase 1 (HMOX1) dysregulation and other indicators pointed toward mitochondrial damage as a key factor in the pathogenesis of severe COVID‐19 infection. Our model suggests that the cells manifest changes that may be relevant to alterations in sensory functions (taste change) in COVID‐19 patients, thus highlighting its physiological relevance when compared to other in vitro models in use. Lastly, we validated some of our key findings in lung tissue samples from COVID‐19 patients.
2. MATERIALS AND METHODS
2.1. Differentiation of hiPSC into lung epithelial cells
All stem cell‐related experiments were approved by the Institutional Committee for Stem Cell Research (ICSCR), Indian Council of Medical Research (ICMR), New Delhi. Lung differentiation was carried out using a previously established protocol (Banerjee et al., 2018; Surendran et al., 2019). In principle, undifferentiated iPSCs were converted to desired lung lineages via the formation of definitive endoderm and anterior foregut endoderm (AFE) thus simulating the in vivo events of lung development. De novo‐generated cells were cryopreserved at the progenitor stage (Day 14) and proximal airway and distal alveolar cell stages (Day 21) after adapting them to air−liquid interface (ALI) cultures for 7−14 days. Live cells were washed twice with 1X DPBS and incubated for 3 min at 37°C with prewarmed Accutase (Gibco). Cells were flushed from the wells using 1 ml pipette tips and centrifuged at 1000 rpm for 2 min at room temperature (RT). The supernatant was discarded and the cell pellet was resuspended in CryoStor CS10 (Stem Cell Technologies) freezing medium at 1 million cells per vial. It was then frozen and stored in liquid nitrogen (LN2) until further use. Each batch of cells was characterized using stage‐specific markers to test their identity, purity, and potency.
2.2. SARS‐CoV‐2 infection
Cells were partially thawed in a 37°C water bath and immediately transferred to a 15 ml falcon tube containing a sterile medium, allowing the cells to completely thaw in the presence of a prewarmed medium. The cells were pelleted at 1000 rpm for 2 min at RT and the pellet was resuspended in an appropriate culture medium. Cells were then seeded onto 48‐well tissue culture plates at a density of 200,000 cells per well and media was replenished every other day for 48−72 h before infection. The cells were transported to a BSL3 high containment facility for SARS‐CoV2 infection. Cultures were infected with a plaque‐purified B.6 lineage of SARS‐CoV‐2 (Anantharaj et al., 2021) MOI of 5.0 PFU/cell. Growth media was aspirated, and then the cell monolayer was washed once with plain DMEM. The stock virus was diluted in DMEM supplemented with 2% FBS and 100 µl of this solution was added to the cells. Cultures were incubated at 37°C in a CO2 incubator for 1 h. After 1 h of virus adsorption, cells were washed twice with plain DMEM, and finally, 300 µl of the specific growth media was added per well of the 48‐well plate and incubated at 37°C, 5% CO2 in the CO2 incubator. At indicated time points, supernatants were collected and stored at −80° for plaque assay. Cells were either homogenized in Trizol (Gibco) to isolate RNA or fixed in chilled methanol and stored at −20°C for immunostaining.
2.3. RNA extraction and RT‐PCR
Cells were lysed in Trizol and total RNA was extracted using the conventional phenol‐chloroform method. Viral RNA PCR was carried out using the 2019‐nCoV CDC Probe and Primer kit for SARS‐CoV‐2 (Biosearch Technologies) to detect the N gene using real‐time RT‐PCR following the manufacturer's instruction. TaqMan™ RNase P assay kit (Thermo Fisher Scientific) was used as an endogenous control for normalization. Fold change was calculated using method with uninfected control and RNAseP. The RNA was converted to cDNA using the RevertAid reverse transcription kit (Thermo Fisher Scientific) as per the manufacturer's protocol, and quantitative PCR was performed for human genes using the QuantStudio5 Real Time PCR system (Applied Biosystems). Fold change estimations were based on double normalization with β‐actin and undifferentiated iPSC. The human‐specific primers used for PCR are listed in Table S1.
2.4. Plaque assay
SARS‐CoV‐2 titer was determined by plaque assay using a 10‐fold serial dilution method and was performed on the Vero/E6 monolayer. Briefly, 150 K Vero/E6 cells were seeded in DMEM (Himedia) supplemented with 10% FBS, 1X penicillin‐streptomycin antibiotic solution (Himedia). 24 h after seeding of cells, the monolayer was washed with PBS. One hundred and twenty‐five microlitres of 10‐fold serially diluted virus inoculum was added and incubated for 1 h at 37°C in a CO2 incubator. The plate was moved slightly every 10 min so that monolayer was covered by the inoculum and did not dry. After 1 h of incubation, the inoculum was removed and CMC was overlaid. The plate was left undisturbed in the CO2 incubator for 48 h. Plates were fixed using 3.7% formaldehyde solution for 30 min inside BSL3 after aspirating the overlay medium followed by staining with crystal violet solution. Plates were washed using tap water and dried at RT. The plaque number was counted manually.
2.5. Immunocytochemistry
Infected cultures fixed in chilled methanol were placed on ice for 10 min. Cells were washed twice with PBS, once with ice‐cold PBS, and then once with PBS at RT followed by blocking with 0.2% BSA‐PBS for 10 min at RT. Cells were incubated with the (Genscript) antibody at 1:1000 for 1 h at RT and counterstained with secondary antibody‐ goat anti‐mouse AF488 (Life Technologies) at 1:500 dilution for 30 min in the dark at RT. Cells were washed with PBS three times and stained with 4ʹ,6‐diamidino‐2‐phenylindole (DAPI) (molecular probes) at 1:10,000 dilution for 10 min. Cells were washed with PBS and images were captured using an Olympus DP80 microscope. Differentiated cells for immunocytochemistry were fixed with 4% paraformaldehyde and stained using a previously described protocol (Surendran et al., 2019). Images were captured using Olympus fluorescent microscope CKX53. Image analysis was performed using ImageJ (NIH) and graphics editing software (Photoshop, Adobe, https://www.adobe.com). The antibodies used for staining are listed in Table S2.
2.6. RNA sequencing and data interpretation
Total RNA from control and 5.0 MOI infected lung epithelial cell types cultured for 24 and 72 h underwent quality checks and was subjected to bidirectional RNAseq library construction on the Illumina HiSeq platform (Illumina, http://www.illumina.com). For the RNAseq data analysis, the unwanted sequences were removed using Bowtie2 and paired‐end reads were aligned using the HISAT2 program, which was further used to estimate the expression of the transcripts using the cufflinks program and reported as FPKM (Fragment per kilo per million) units. Principle component analysis (PCA) and hierarchical clustering were performed with the normalized data to find the correlation between the samples. The differentially expressed genes (DEG) were identified using DESeq.2 analysis and logarithmic fold change calculated in comparison with the control. Further, Gene ontology (GO) enrichment analysis was carried out using Amigo2 for DEGs. Data are represented as heat maps using Microsoft excel and a protein network was made using the string protein interaction database. The DEGs were further validated by qPCR using SYBR green reaction dye using Quantstudio5 (Applied Biosystems). Fold change was calculated by method using uninfected cells as control after normalization with β‐actin.
2.7. Statistical analysis and data availability
Results are represented as mean ± standard error (SE) with technical triplicates. Statistical significance was calculated using the Student's t‐test—95% confidence interval and p ≤ 0.05 was considered statistically significant. The RNAseq data have been deposited in Gene Expression Omnibus and are accessible through accession number GSE190193.
2.8. Hematoxylin and eosin (H&E) staining and immunohistochemistry of lung tissue sections
Postmortem specimens of lung tissue from five COVID‐19 positive cases were received in 10% neutral buffered formalin after obtaining Institutional Ethics Committee (IEC) approval. Representative blocks were taken for further processing based on clinical descriptions. The H&E stained slides were examined and a report was generated in each of the cases. Representative blocks were selected from the five cases and 3−4 µm‐thick sections were cut on charged slides. The primary antibodies tested were CC10, SPC, IL‐6, and IL‐13. The immunohistochemistry was done by the peroxidase‐conjugated polymer method, including antigen retrieval using the automated method—Ventana Benchmark GX machine. Universal DAB was the detection kit with hematoxylin counterstain. Before running the IHC on the COVID‐19 positive lung blocks, the markers were standardized on normal lung tissue sections.
3. RESULTS
3.1. SARS‐CoV‐2 readily infects and replicates inside human iPSC‐derived lung lineages
We generated biopotential lung progenitor cells from iPSC, which after adapting to ALI were further differentiated to distal alveolar and proximal airway lung cell types (Figure 1a). Human iPSCs were directed toward anteriorized foregut endoderm cells positive for NKX2.1, which serves as a common precursor for upper airway and alveolus (Figure 1b). Alveolar epithelial cells expressed SOX9, which is present in the distal tips of the branching epithelium, balancing proliferation and differentiation (Figure 1b). Distal lung bud marker FOXP2 and surfactant protein C (SP‐C) were also present in these differentiated cells (Figure 1b). The proximal airway comprises various cell types and we were able to identify most of them in our differentiated cultures. Because we have not used any sorting technique, it was not surprising to see a heterogeneous mixture of cell types specific to lung lineage. Proximal epithelium‐specific transcription factor SOX2, goblet cell marker FOXJ1, ciliated cell marker ARL13B, tight junction protein ZO‐1, basal cell protein P63, and Clara cell secretory protein CC10 were expressed in the differentiated proximal airway cultures (Figure 1c).
Figure 1.
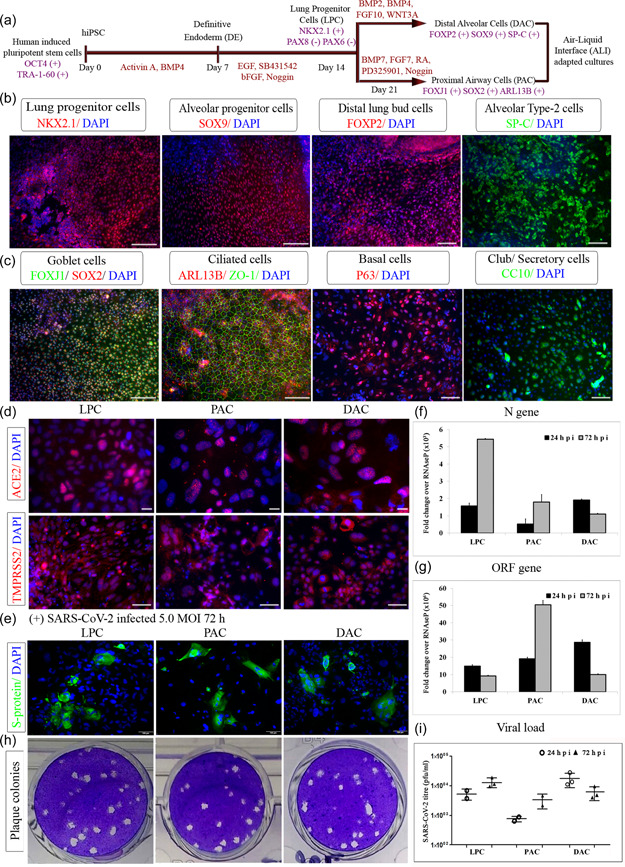
Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) infection and replication inside induced pluripotent stem cells (iPSC)‐derived lung epithelial lineages. (a) Differentiation snapshot capturing various stages involved in the generation of lung cells from iPSC along with distinct markers expressed along different lung cell types (b) Lung progenitor cells—transcription factor NKX2.1, alveolar progenitor—SOX9, developing lung bud marker‐ FOXP2, and distal alveolar type II cells expressing SP‐C (c) Proximal airway cells expressing goblet cell—FOXJ1 costained with SOX2, ciliated cell—ARL13B costained with tight junction protein ‐ZO1, basal cell‐ P63, Clara cell secretory protein—CC10 (d) Lung cell types showing ACE2 and TMPRSS2 at protein levels (e) Immunofluorescence analysis of SARS‐CoV‐2 spike protein (green) in uninfected lung cells. Nuclei were counterstained with DAPI (blue) (f, g) Real‐time PCR levels of viral N gene and ORF gene in the cell after 24 and 72 h of SARS‐CoV‐2 infection (h) Representative plaque assay plate picture of SARS‐CoV‐2 infection (i) Plaque assay plates quantified and levels of viral infection virus in culture supernatant represented as titer values. Scale bars represent 100 µm
We then examined the presence of receptor proteins, which are responsible for the entry and replication of the SARS‐CoV‐2 virus inside the host. We observed strong and consistent expression of ACE2 and TMPRSS2 across different stages of lung differentiation (Figure 1d). To determine an optimum MOI and incubation period for these cells, a range of MOIs of SARS‐CoV‐2 from 0.1 to 10.0 and incubation periods from 24 to 120 h were tested. While 5.0 MOI caused a small cytopathic effect, cytotoxicity was not seen (data not shown). Hence, for the following experiments, cells were infected with 5.0 MOI SARS‐CoV‐2 for 1 h and then cultured with a lung cell type‐specific culture medium until harvest. SARS‐CoV‐2 readily infected the lung epithelial cells and the presence of the virus was confirmed using an antibody against spike protein (Figure 1e). Viral replication and or gene expression were measured by qPCR with SARS‐CoV‐2 nucleoprotein gene (N gene) and ORF gene in control versus infected samples. Detection of N gene expression, when compared to RNaseP, indicated the presence of viral mRNA. N gene expression increased at 24 hpi (hpi) and 72 hpi in progenitor and proximal cell types (Figure 1f). The ORF gene was significantly upregulated across all the infection time points (Figure 1g). A quantitative viral plaque assay using culture supernatants ascertained this data, showing a similar trend for viral replication. The difference in viral load in the culture supernatant between 24 and 72 hpi was statistically significant (Figure 1h,i). Transcripts of viral genes were undetected in uninfected sets across all lung lineages. Although most publications report the airway cells as the primary entry for the virus, we found significantly high levels of infection in lung progenitors and alveolar cells as well. Thus, we established an in vitro platform that comprises all the relevant types of pulmonary cells and encompasses the complete infection and spread route for SARS‐CoV‐2 within the lungs.
3.2. Acute inflammatory and antiviral immune responses after SARS‐CoV2 infection
To understand the host defense response to SARS‐CoV‐2 infection, we performed RNA sequencing of the infected lung cells. Transcriptomic analysis was carried out with lung progenitors, proximal airway, and distal alveolar cell types all infected with 5.0 MOI for 24 and 72 h. PCA revealed no clustering among the samples indicating differences in their genotypic profiles (data not shown). However, we observed significant differences in transcriptomic profiles between SARS‐CoV‐2‐infected cells and their respective uninfected controls. The total number(s) of significantly up or downregulated genes is represented in tabular form (Figure 2a). GO analysis revealed that most of the upregulated genes were associated with viral clearance processes as a primary cellular defense response across all infected cell types (Figure 2b). As reported in several publications, we also observed upregulation of a series of inflammatory cytokines and interferon signaling pathways represented as fold enrichment (Figure 2c).
Figure 2.
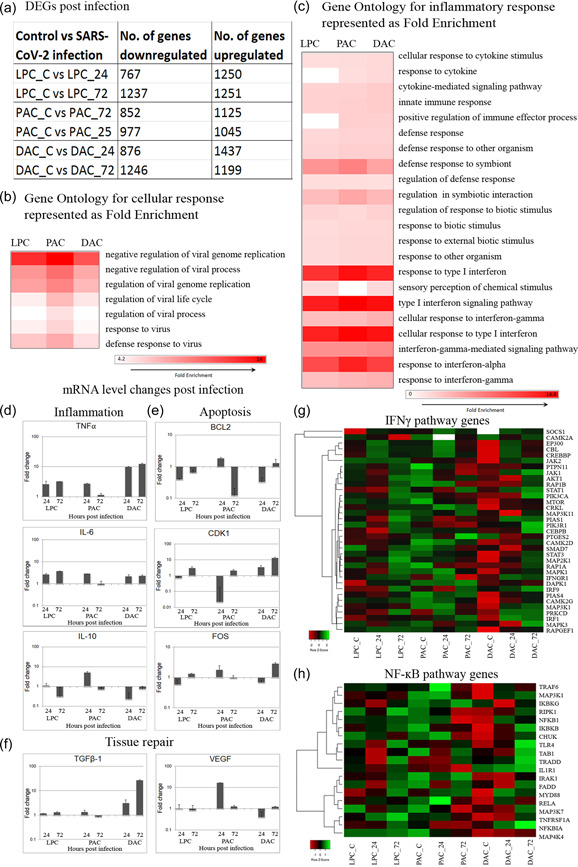
Severe acute respiratory syndrome coronavirus 2 infection results in a massive inflammatory response. (a) Number of significantly up/downregulated genes identified by comparing with respective uninfected control represented in a table. (b, c) Gene ontology fold enrichment for infection and inflammatory response. Heat map color, white to red. (d−f) Real‐time PCR validation of selected genes (d) Inflammation—TNF‐α, IL‐6, IL‐13 (e) Apoptosis—BCL2, CDK1, FOS (f) Tissue repair—TGFβ1, VEGF (g) Heat map of genes from key signaling pathway involved in inflammation—IFNγ, NF‐κΒ; heat map color, red to green through black. NF‐kB, nuclear factor kappa B; TGF, tumor growth factor; NF, tumor necrosis factor; VEGF, vascular endothelial growth factor
We validated this finding using qPCR and detected significant upregulation in proinflammatory cytokines TNF‐α and IL‐6 and concomitant downregulation of anti‐inflammatory cytokine IL‐10 in infected cells, thereby complementing the GO studies (Figure 2d). Furthermore, upregulation of apoptosis‐related genes suggests that viral infection triggered apoptosis. FOS gene, which induces p53‐dependent apoptosis was upregulated and antiapoptotic gene BCL2 was downregulated (Figure 2e). Additionally, we observed dysregulation of CDK2, which regulates G1 arrest and prevents cellular apoptosis (Figure 2e). Angiogenesis promoting VEGF and TGF‐β1 at 72 hpi was higher in alveolar cells, an observation that suggests an attempt to repair an injured alveolar‐capillary membrane (Figure 2f). Differential gene expression for crucial genes in the IFNγ and NF‐κappaΒ pathways is represented as a heat map (Figure 2g,h), indicating sharp upregulation at 72 hpi across all cell types further supporting the physiological relevance of our in vitro model.
3.3. SARS‐CoV‐2 exposure prompted alterations in structural gene expression and deregulation of the airway and alveolar programs
RNA sequencing data analysis identified a set of lung programming genes that are drastically dysregulated upon infection (based on log2 fold change >2 and <−2 in comparison to respective uninfected controls). Proximal epithelium‐specific transcription factor SOX2, which regulates the basal cells of the airway was significantly downregulated across all infected samples. Contrastingly, distal epithelium‐specific transcription factor, SOX9, which controls proliferation and differentiation of alveolar cells was increased in infected samples (Figure 3a,b). Similarly, we observed a decrease in other airway‐specific genes TP63, FOXJ1, and surfactant protein SFTPC. Likewise, Tenascin C (TNC), which is associated with tissue repair, and HOPX, which is involved in alveolar homeostasis, were downregulated in proximal cells but slightly upregulated in distal cells (Figure 3a,b). Muc5ac was downregulated (greater than fourfold) indicating a disruption in cellular lubrication and chemical barrier formation (Figure 3c). At the same time, aquaporin 5 (Aqp5) showed significant dysregulation (threefold) across all cell types, indicating a loss of integrity in the epithelial barrier and impaired lung physiology (Figure 3c).
Figure 3.
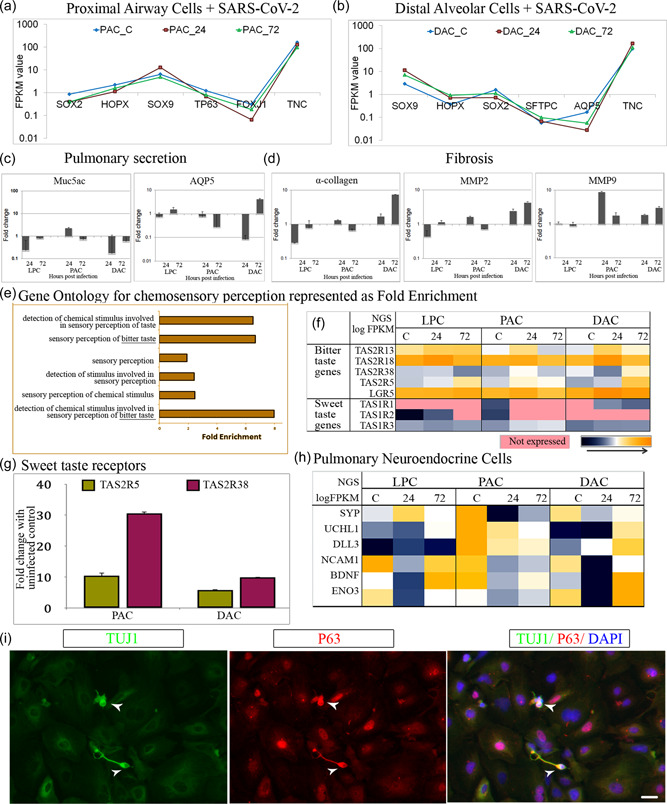
Severe acute respiratory syndrome coronavirus 2 infection negatively impacts alveolar maturation, induces fibrosis, and triggers chemosensory changes. (a, b) messanger RNA (mRNA) level changes in lung developmental program represented as FPKM values for (a) proximal airway genes and (b) distal alveolar genes (c, d) Real‐time polymerase chain reaction (PCR) validation of selected genes (c) Pulmonary secretion—Muc5ac, AQP5 (d) Fibrosis—α‐collagen, MMP2, MMP9 (e) Gene ontology showing changes in sensory perception functional modules (f) Genes responsible for bitter taste upregulated in infected samples and sweet taste downregulated (g) qPCR validation of bitter taste gene—TAS2R5, TAS2R38 (h) Heat map showing expression of PNEC genes. (i) Proximal airway cells expressing PNEC marker TUJ1 costained with basal cell marker P63. Heat map color, blue to yellow through white. Scale bars represent 100 µm
Increased levels of TGF‐β1 in alveolar cells can induce epithelial to mesenchymal transition (EMT), which promotes transdifferentiation of AT1/AT2 to fibroblast/myofibroblast. Lending support to this theory, we noticed upregulation (twofold) of matrix metalloproteinases (MMPs) and increased (fivefold) collagen deposition leading to fibro‐proliferation of alveolar cells (Figure 3d). SARS‐CoV‐2 activates macrophages to produce inflammatory cytokines like TGF‐β1 and IL‐6. It could similarly trigger TGF‐β1 expression in alveolar cells thus promoting pulmonary fibrosis. Based on these results, we infer that the proximal airway cells were critically affected by SARS‐CoV‐2 infection in a fashion reminiscent of lung injury and fibrosis. Furthermore, pulmonary fibrosis‐like features observed in our in vitro lung model upon SARS‐CoV‐2 infection are in alignment with clinical evidence of post‐COVID‐19 fibrosis in affected patients (Rai et al., 2021).
3.4. Host cells exhibit chemosensory changes after SARS‐CoV‐2 infection
Our novel observation concurs with the emerging central role of support cells in COVID pathophysiology, especially how it alters chemical perception (Cooper et al., 2020). Interestingly, GO analysis of the DEGs revealed alterations in sensory perception and a change in taste toward bitterness (Figure 3e). A closer look at the transcriptomics data highlighted gross chemosensory changes, including upregulation of genes related to bitterness in infected samples. Additionally, the genes related to sweet taste were completely downregulated or not expressed in infected samples (Figure 3f) compared to uninfected controls. Bitter taste receptors TAS2R5 and TAS2R38, which are responsible for the ability to taste both 6‐n‐propylthiouracil and phenylthiocarbamide, were significantly upregulated in the airway and alveolar cells postinfection (Figure 3g). Although this observation was relevant to COVID‐19‐induced ageusia in patients, we were curious to know how this biological process was stimulated in a model made up of pulmonary cells.
Airway epithelial progenitors and basal stem cells could give rise to pulmonary neuroendocrine cells (PNEC) during embryogenesis (Gu et al., 2014; Montoro et al., 2018). Therefore, we speculated that the observed chemosensory response could be due to the presence of a small population of PNEC in our human airway cultures. In support of this, we identified a candidate set of PNEC‐specific genes expressed in uninfected lung cells that were considerably upregulated upon infection at 72 hpi (Figure 3h). Furthermore, coexpression of TUJ1 and basal cell marker P63 (Figure 3i) helped us distinguish these rare TUBB3 positive airway basal stem cells that give rise to distinct subpopulations of PNECs (Mou et al., 2021).
3.5. Impaired mitochondrial metabolism leads to oxidative stress
Existing literature suggests that oxidative stress is an acute response to COVID‐19 infection because the pathophysiology is very similar to that seen in ALI. ACE2 also affects mitochondrial functions and low levels of ACE2 are associated with decreased ATP production and altered activation of NADPH oxidase 4 (NOX4) in the mitochondria (K. K. Singh et al., 2020). Interestingly, we detected decreased levels of ACE2 immediately after infection across the cell types, but we are not sure of the underlying mechanism. We then tested the levels of NOX4 in the infected cells and found it decreasing in progenitor and airway cell types but increasing in alveolar cells (Figure 4a). This could have been driven by NOX‐induced TGF‐β1‐mediated conversion of fibroblasts into myofibroblasts, leading to fibrosis in alveolar cells (Amara et al., 2010). This was clearly demonstrated in our model marked by high TGF‐β1 and collagen deposition.
Figure 4.
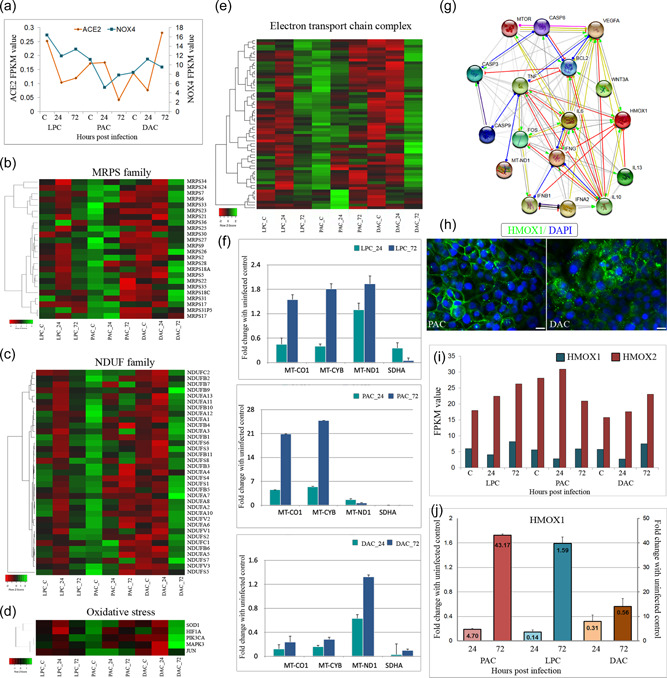
Mitochondrial damage is associated with inflammation and apoptosis. (a) mRNA level changes in ACE2 and NOX4 are represented as FPKM values (b, c) MRPS and NDUF family genes are represented as heat maps. (d) Genes related to reactive oxygen species are represented as heat maps (e) Key mitochondrial genes involved in the electron transport chain are represented as heat maps (f) Real‐time qPCR validation of mitochondrial electron transport chain complex genes—MT‐CO1, MT‐CYB, MT‐ND1, and SDHA across all cell types upon infection (g) Protein network analysis showing a direct correlation between mitochondrial HMOX with inflammatory and apoptotic genes. (h) Airway and alveolar lung cells expressing heme oxygenase 1 (HMOX1) protein (i) messanger RNA level changes in HMOX1 and HMOX2 represented as FPKM value (j) Real‐time qPCR analysis showing the dysregulation of HMOX1 during the course of infection. Heat map color, red to green through black. Scale bars represent 100 µm
A massive inflammatory response caused by infection could increase mitochondrial ROS production and perturb the ETC complexes (Ganji & Reddy, 2021). We investigated the levels of mitochondrial ribosomal proteins encoded by nuclear genes (MRPS gene family), which aid in the mitochondrion protein synthesis within the cell. We found significant dysregulation across infected samples, indicating a pivotal role of mitochondria in SARS‐CoV‐2 infection (Figure 4b). Further, we tested the members of the NADH ubiquinone oxidoreductase (NDUF) gene family, which is crucial for initiating the ETC for ATP production inside the cell. A similar pattern of dysregulation was also seen in MRPS genes (Figure 4c). In addition, we found that some of the reactive oxygen species (ROS) related genes and most of the ETC complex molecules were severely dysregulated upon infection (Figure 4d,e). This could lead to an imbalance in the oxidative phosphorylation cycle, resulting in an increase in ROS. We validated the mRNA levels of some of the molecules by qPCR and established that MT‐CO1, MT‐CYB, MT‐ND1, and SDHA; representative genes from ETC complexes I−IV showed a similar expression trend to sequencing data (Figure 4f).
3.6. Upregulation of HMOX1 as a response to mitochondrial damage
Heme oxygenase has been identified to play a vital role as a defense molecule activated in injured lung tissue against injury to prevent anti‐inflammatory and antiapoptotic effects. HMOX1 is the most inducible isoform involved in preventing vascular inflammation and is used as a therapeutic target for acute lung injury in multiple pulmonary disease models (Fredenburgh et al., 2007). To understand the cytoprotective effects of HMOX1, we created a protein network using the STRING database with network edges, indicating the predicted mode of action between two proteins. We included HMOX1 and key inflammatory and apoptotic genes to illustrate their interaction in the normal state (Figure 4g). Some of the direct interactions include HMOX1 and anti‐inflammatory molecule IL‐10, HMOX1, and VEGF mutually activating each other, and proinflammatory cytokine IL‐13 inhibits HMOX1. These findings also support the known role of HMOX1 in preventing inflammation and promoting angiogenesis upon lung damage.
We further checked the presence of HMOX1 in our in vitro differentiated cells and fluorescent microscopy showed considerable levels of protein expression (Figure 4h). NGS data showed that HMOX1 was slightly downregulated at 24 hpi, which could be due to a sudden spike in the inflammatory cascade after infection (Figure 4i). At 72 hpi, HMOX1 was highly upregulated across all lung cell types, something that has been demonstrated previously in ARDS patients' lung tissue (Fredenburgh et al., 2007). The levels of HMOX2, which is a constitutive form, were observed to be consistent irrespective of infection and HMOX3 was not present (Figure 4i). Lastly, we verified the mRNA levels of HMOX1 by qPCR and found them to be matching the FPKM values (Figure 4j). Upregulation of HMOX1 in the face of inflammatory response and apoptosis concurs with its established role as a defense molecule activated for damage control.
3.7. Postmortem lung samples from COVID‐19 patients validate key in vitro findings
We further investigated whether the SARS‐CoV‐2 induced responses and phenotypic remodeling observed in our vitro model are analogous to that seen in clinical settings. We obtained autopsy specimens of deceased individuals diagnosed with COVID‐19. Patients included in the study were identified to be positive either by RT‐PCR or Rapid Antigen Test. Patient lung sections stained with H&E showed edema leading to thickening of the alveolar septa and lack of air spaces (Figure 5a). Histopathological features also revealed hemorrhage and inflammatory cell infiltrations in the interalveolar spaces with pneumonic consolidation and features of diffuse alveolar damage (Figure 5b).
Figure 5.
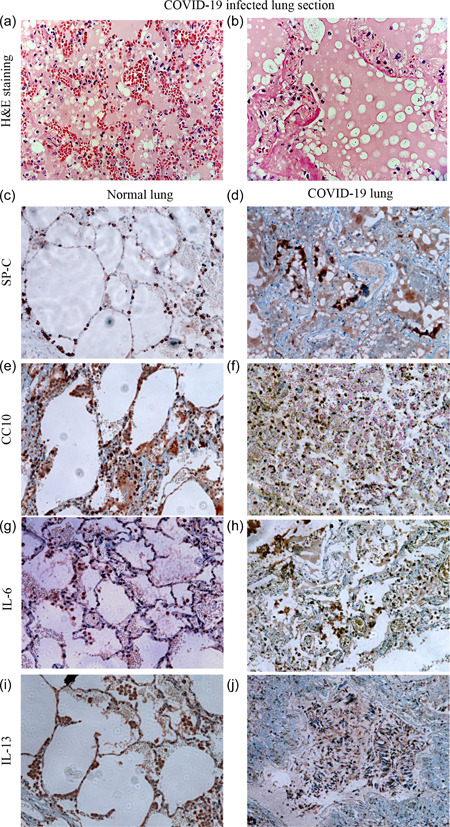
Disruption of alveolar spaces and inflammation in COVID‐19 infected lung tissues support in vitro findings. (a, b) Hematoxylin and eosin‐stained COVID lung section showing (a) ARDS like pathology with edema and hyaline membrane (b) features of pneumonia (c−f) Lung surfactant—SPC, secretory protein CC10 highlights the alveolar lining cells in normal lung and disrupted staining along the alveolar walls with no proper air spaces (g−j) Inflammatory cytokines—IL‐6, IL‐13 stained macrophages in normal lung and massive cellular infiltration in COVID lung. Scale bars represent 100 µm
We studied pathology further by immunohistochemistry for crucial alveolar markers. Surfactants (SP‐C) found on the alveolar lining of the lung play an essential role in the formation of lamellar bodies and in reducing surface tension (Figure 5c). Club or Clara cell marker CC10 was detected in the distal airways, contributing to the maintenance of airway integrity and repair (Figure 5e). In COVID‐19 positive sections, both SP‐C and CC10 showed disruption and discontinuous staining when compared to normal lungs due to edema and cellular infiltration (Figure 5d,f). Furthermore, we examined the levels of inflammatory cytokines secreted by alveolar macrophages, including IL‐6 and IL‐13. In the control lung, IL‐6 and IL‐13 were lightly expressed in macrophages, seen as faint staining near the alveolar lining (Figure 5g,i). In contrast, COVID‐19‐positive sections exhibited an increased staining of IL‐6 and IL‐13 positive alveolar macrophages that spilled into the alveolar air spaces with scattered neutrophils and lymphocytes (Figure 5h,j). Notably, our in vitro model shows similar pathophysiological features with respect to the alveolar damage and inflammatory response seen in COVID‐19 positive lung tissues.
4. DISCUSSION
While some of the available vaccines have proven effective against COVID‐19, several other vaccine candidates are still under development. Moreover, the effectiveness varies according to the viral variant, some even escaping the protective effect. Further, none of the vaccines prevent infection. The recent disaster sparked by the emergence of a new SARS‐CoV‐2 variant, Omicron, (B.1.1.529) clearly indicates that this pandemic is far from over. In fact, it is predicted that more variants may emerge to trigger additional pandemic waves in the future. Understanding the cellular processes and molecular interactions impacted in the host cells responding to a virus infection will be necessary to understand the virus pathogenesis and to develop better intervention strategies.
With respect to viral cell tropism, besides corroborating the previously reported ciliated and AT2 cells as target cells (Djidrovski et al., 2021; Huang et al., 2020), lung progenitor cells from the AFE were also found to be permissive to SARS‐CoV‐2 infection. This is a key finding since progenitor cells found in the adult lung are involved in lung branching morphogenesis, cell growth, maturation, injury, and repair (Kotton & Morrisey, 2014). Furthermore, it was reported that lung stem or progenitor cells could be infected by SARS‐CoV‐2, which may lead to defects in regeneration capacity, partially accounting for the severity of SARS‐CoV‐2 infection (Valyaeva et al., 2020).
Once inside the cell, SARS‐CoV‐2 prompts a massive inflammatory response, which is similar to a cell‐autonomous or ‘‘epithelial only” host response to pathogens (Huang et al., 2020). Our model responded to SARS‐CoV‐2 infection within 24 h and dysregulation in NF‐κΒ signaling is prominent across the airway, alveolar, and lung progenitor cells. The aberrant gene expression observed in relation to inflammation, host defense, lung development, surfactant production, apoptosis, fibrosis, and tissue repair is clinically relevant. Like others (Huang et al., 2020; Katsura et al., 2020) we witnessed perturbation in IFN‐γ signaling presenting significant differential expression across three cell types after SARS‐CoV‐2 infection at 24 and 72 h compared to noninfected cells (Figure 2g). Our finding shows that the type‐II IFN pathway is activated and contradicts the more prevalent activation of the IFN‐I and IFN‐III pathways in cells due to viral infection. However, our data is consistent with earlier studies, showing delayed host innate immune responses after SARS‐CoV (2003) infection (Menachery et al., 2014). At the same time, it underscores the need for dynamic analyses of host responses using multiple MOIs and at different times after infection. After innate immunity is triggered upon viral infection, cytokines, such as TNF‐α, INF‐γ, IL‐6, and IL‐13. are activated in infected cells, causing an increase in mitochondrial ROS production through gene expression upregulation and ETC modulation (Saleh et al., 2020). Mitochondrial ROS then stimulates additional proinflammatory cytokine production as the virus persists leading to a “cytokine storm.” This immune response could also prompt the mitochondria to deviate from ATP production toward ROS production, which can harm the mitochondria, leading to apoptosis (Saleh et al., 2020).
We have demonstrated that ACE2 and TMPRSS2 are robust but heterogeneously expressed, which is similar to what was observed in adult human AT2s in vivo (Abo et al., 2020). Entry of SARS‐CoV‐2 inside the cell using the ACE2 receptor results in a failure of ACE2 conversion of angiotensin II to angiotensin. This excess angiotensin II stimulates NADPH oxidase thus generating high levels of ROS. As a result, cortisol stimulates the release of ATP via activation of mineralocorticoid receptor (MR), which then acts on purinergic receptors leading to an increase in intracellular calcium (Edwards et al., 2021). Purinergic receptors are known to play an important role in both taste and smell (Eddy et al., 2009; Hegg et al., 2003) and ATP signaling has been shown to be crucial for communication from taste buds to gustatory nerves (Finger et al., 2005). In our model, ACE2 decreased significantly from 24 to 72 h and calcium signaling genes like CAMKK, RAC1, and TG2 (data not shown) increased considerably after SARS‐CoV‐2 infection. Therefore, it is likely that the SARS‐CoV‐2 virus might well activate ATP‐mediated odor suppression as a novel protective mechanism.
Mitochondria play a key role in the host's response to viral infection and immunity (Takumi Koshiba, 2013). Using machine learning models, Wu et al. determined that the SARS‐CoV‐2 RNA genome and all subgenomic RNAs were enriched in the host mitochondria and nucleolus (Wu et al., 2020), and investigations into the SARS‐CoV‐2 hijacking of host mitochondria may lead to novel approaches to prevent and treat COVID‐19 (K. K. Singh et al., 2020). We observed that across cell types, SARS‐CoV‐2 reduced nuclear‐encoded mitochondrial (NEM) gene host response related to cellular respiration and Complex I. Complex I is one of the main contributors to ROS production and downregulation of many NDUF and MRPS family of genes after SARS‐CoV‐2 infection could be behind lower ROS production, thus facilitating SARS‐CoV‐2 propagation. SARS‐CoV‐2 ORF9c has been previously reported to interact with mitochondrial NDUFAF1, NDUFB9, MRPS2, MRPS5, MRPS25, and MRPS27 (Gordon et al., 2020). This could explain the direct interaction between SARS‐CoV‐2 and these Complex I and mitochondrial ribosome proteins observed in our transcriptomic data after SARS‐CoV‐2 infection.
HMOX1 is a cytoprotective enzyme that plays a crucial role in the defense against oxidant and inflammation‐induced lung injury during ARDS and idiopathic pulmonary fibrosis (Fredenburgh et al., 2007). Among the three isoforms, HO‐1 is the inducible isoform and is thought to be an oxidative stress‐responsive protein (Morse & Choi, 2005). Likewise, the dysregulation of HMOX1 and not HMOX2 noticed in proximal airway cells after SARS‐CoV‐2 infection could result from oxidant−antioxidant imbalance, thus contributing to the pathogenesis of lung fibrosis in COVID‐19. This is a significant finding as modulation of HO‐1 has been hypothesized as a potential therapeutic target for COVID‐19 via suppression of viral replication by increasing IFNs (K. K. Singh et al., 2020).
There are many other interesting and novel observations from this study that provide critical insights for understanding the pathophysiology of SARS‐CoV‐2 infection. Airway secretory mucin gene (muc5ac) was downregulated, which could lead to neutrophil trafficking into the lungs (Koeppen et al., 2013). Aquaporins regulate the osmotic pressure during normal lung functioning (Wittekindt & Dietl, 2019) and the water channel protein AQP5, which maintains the alveolar epithelial barrier was significantly downregulated upon SARS‐CoV‐2 infection. Tissue repair gene TNC was upregulated, which has been shown to induce lung injury due to activation of multiple inflammatory cytokines, such as pathogens and damage‐associated molecular patterns (PAMP and DAMP) (Midwood & Orend, 2009). Further, downregulation of BCL2 inhibits the intrinsic pathway and PAMP‐induced apoptosis (Leibowitz & Yu, 2010). EMT features transdifferentiation of AT1/AT2 to fibroblast/myofibroblast, resulting in increased MMPs and collagen deposition. Likewise, increased expression of MMPs and collagen was observed in distal alveolar cells following SARS‐CoV‐2 infection. Since SARS‐CoV‐2 infection eventually leads to pulmonary fibrosis, HOPX upregulation could be a potential indicator of progression to pulmonary fibrosis in our model (Ota et al., 2018). Persistent alveolar activation of TGF‐β, the release of PDGF and IL‐6 from alveolar epithelial cells, immune cells, and myofibroblasts leads to the proliferation of myofibroblasts and the development of fibrosis (John et al., 2021). Imbalance in the expression of SOX2/SOX9 and PITX2/CTNNB1 indicates perturbed proximal‐distal lung patterning (Banerjee et al., 2018).
Most COVID‐19 patients suffer from sensory dysfunction (anosmia), which starts with their sense of smell, but because the smell is necessary to taste the flavor, the symptoms are often connected (Hannum et al., 2021). Since ACE2 receptors are not found in the olfactory nerves and taste buds, the high incidence of anosmia and ageusia caused by SARS‐CoV‐2 is without directly infecting these cell types. Further, it is well known that taste receptors are expressed far beyond the tongue, from the airway and gastrointestinal epithelia to the pancreas and brain (Yamamoto & Ishimaru, 2013). Bitter taste receptors (TAS2Rs) are G‐protein coupled receptors divided into 36 subunits, depending on the species (over 25 subunits in humans). With respect to the lung, the presence of TAS2Rs has been shown in airway ciliated cells (Shah et al., 2009), sinonasal epithelial cells (Lee et al., 2012), solitary chemosensory cells (Gulbransen et al., 2008), and bronchial smooth muscle cells (Robinett et al., 2014). Interestingly, GO analysis identified the expression of a subset of TAS2R bitter receptor genes but not TAS1R sweet receptor genes, which was further confirmed by qPCR and immunostaining. However, we failed to detect many other TAS2Rs, possibly because their expression level is below the detection limit or because these receptors are only weakly expressed in the iPSC‐derived lung epithelial lineages. Olfactory receptors in the airway were first identified in PNECs, a rare yet multifunctional epithelial cells and suggested that they act as intrapulmonary chemosensors (Gu et al., 2014). Hor et al. (2020) successfully differentiated human iPSCs to iPNECs with a gene expression profile like that of primary fetal PNECs. Interestingly, they adapted this iPNEC differentiation protocol from their previously established airway epithelial differentiation protocol (Firth et al., 2014), which was quite similar to our lung differentiation protocol (Banerjee et al., 2018; Surendran et al., 2019). Based on this information, we established the expression of major PNEC marker genes, including SYP, UCHL1, DLL3, NCAM1, BDNF, and ENO2, consistent with the phenotype of primary human PNECs (Branchfield et al., 2016). We also demonstrated the presence of rare TUJ1+/P63+ positive solitary PNEC in our airway cultures, which is in line with a recent study showing the ability of airway basal stem cells to generate TUBB3+ PNEC (Mou et al., 2021). Within the lung milieu, activation of proneural transcription factor ASCL1 is required for cells to form the pulmonary neuroendocrine lineage (Linnoila, 2006), whereas the Notch‐HES1/HEY1 pathway regulates the non‐neuroendocrine fate of lung endoderm by repressing proneural genes like ASCL1 (Henke et al., 2009). Although we recently adapted our monolayer cultures of lung epithelial cells to ALI (Surendran et al., 2020), we have not used G‐secretase/Notch inhibitor. This clearly explains the presence of HES1 and the absence of ASCL1 in our lung lineage cells (data not shown). Nonetheless, with <1% of the cellular composition of the adult lung being PNECs and given the heterogeneity in iPSC‐based differentiation it is not ideal to make head to head comparisons with respect to quantification based on in vitro results.
5. CONCLUSION
In the current study, we focused on the transcriptional response elicited by the host (lung) cells to SARS‐CoV‐2 infection to understand viral cell tropism and early cell response to viral infection. SARS‐CoV‐2 virus anchored, replicated, and demonstrated its hallmark features, such as inflammatory response, apoptosis, oxidative stress, and skewed proximal‐distal lung patterning in our hiPSC‐derived model (Figure 6). A deeper dive indicated upregulation in HMOX1 and its role in inflammation and mitochondrial ETC (Figure 6). Interestingly, and for the first time, we identified alterations in chemosensory perception and taste changes using an in vitro model. Therefore, hiPSCs‐derived pulmonary cells could offer a robust and biologically relevant model to investigate the underlying mechanism of SARS‐CoV‐2 infection and to discover and test therapeutic drugs for COVID‐19.
Figure 6.
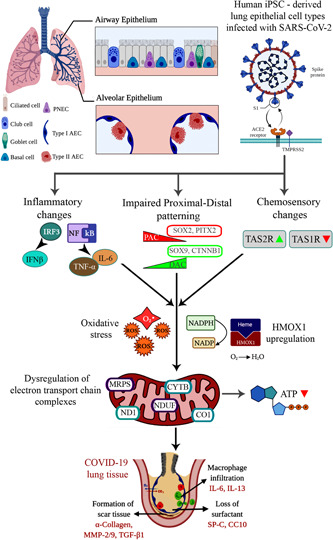
A schematic representing the working model of our study, starting from entry of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) inside the lung epithelial cell types, triggering an inflammatory response, chemosensory changes, and impaired proximal‐distal lung patterning. These alterations eventually resulted in mitochondrial dysfunction and pulmonary fibrosis via specific cellular and molecular events. HMOX1, heme oxygenase 1; iPSC, induced pluripotent stem cells.
AUTHOR CONTRIBUTIONS
Design of experiments, collection and assembly of data, data analysis and interpretation, and manuscript writing: Harshini Surendran. Design of experiments, collection and assembly of data, data analysis, and interpretation: Saurabh Kumar. Growing of cell lines and their characterization, and data collection: Swathi Narasimhaiah. Collection and assembly of data, data interpretation, and presentation: Anuradha Ananthamurthy. Provision of the human lung tissue samples for the study, and verification of the results: PS Varghese. Provision of suitable laboratory facility, design, and conception: George A. D'Souza and Guruprasad Medigeshi. Conception and design, data analysis and interpretation, manuscript writing, and financial support: Rajarshi Pal. The authors read and approved the final manuscript.
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
Supporting information
Supporting information.
ACKNOWLEDGMENTS
The authors are thankful to Dr. Jyotsna Dhawan, CSIR‐Center for Cellular and Molecular Biology, Hyderabad, and Prof. Ramaswamy Subramaniam, Purdue University, Indiana for help in conducting pilot experiments with the SARS‐CoV‐2 virus at the beginning of this project. The authors would like to thank Dr. Ruchi Sharma, NEI/NIH, Bethesda, MD for proofreading the manuscript and MedGenome Labs for the RNA sequencing work. This study was funded in part by a grant from the Biotechnology Industry Research Assistance Council (BIRAC), a Govt. of India Enterprise (#BT/COVID0046/01/20). Centre for Cellular and Molecular Platforms (C‐CAMP), Bengaluru is acknowledged for support in form of incubation and core facilities.
Surendran, H. , Kumar, S. , Narasimhaiah, S. , Ananthamurthy, A. , Varghese, P. , D'Souza, G. A. , Medigeshi, G. , & Pal, R. (2022). SARS‐CoV‐2 infection of human induced pluripotent stem cells‐derived lung lineage cells evokes inflammatory and chemosensory responses by targeting mitochondrial pathways. Journal of Cellular Physiology, 237, 2913–2928. 10.1002/jcp.30755
REFERENCES
- Abo, K. M. , Ma, L. , Matte, T. , Huang, J. , Alysandratos, K. D. , Werder, R. B. , Mithal, A. , Beermann, M. L. , Lindstrom‐Vautrin, J. , Mostoslavsky, G. , Ikonomou, L. , Kotton, D. N. , Hawkins, F. , Wilson, A. , & Villacorta‐Martin, C. (2020). Human iPSC‐derived alveolar and airway epithelial cells can be cultured at air‐liquid interface and express SARS‐CoV‐2 host factors. bioRxiv: The Preprint Server for Biology , 132639. 10.1101/2020.06.03.132639 [DOI]
- Amara, N. , Goven, D. , Prost, F. , Muloway, R. , Crestani, B. , & Boczkowski, J. (2010). NOX4/NADPH oxidase expression is increased in pulmonary fibroblasts from patients with idiopathic pulmonary fibrosis and mediates TGF‐B1‐induced fibroblast differentiation into myofibroblasts. Thorax, 65(8), 733–738. 10.1136/thx.2009.113456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaj, A. , Gujjar, S. , Verma, N. , Khan, N. A. , Shaman, H. , Sharanabasava, P. , Das, A. , Pandey, R. , Pandey, A. K. , & Medigeshi, G. R. (2021). Resolution of viral load in mild COVID‐19 patients is associated with both innate and adaptive immune responses. Journal of Clinical Virology, 21(146), 105060. 10.1016/j.jcv.2021.105060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee, P. , Surendran, H. , Bharti, K. , Morishita, K. , Varshney, A. , & Pal, R. (2018). Long noncoding RNA RP11‐380D23.2 drives distal‐proximal patterning of the lung by regulating PITX2 expression. Stem Cells, 36(2), 218–229. 10.1002/stem.2740 [DOI] [PubMed] [Google Scholar]
- Branchfield, K. , Nantie, L. , Verheyden, J. M. , Sui, P. , Wienhold, M. D. , & Sun, X. (2016). Pulmonary neuroendocrine cells function as airway sensors to control lung immune response. Science, 351(6274), 707–710. 10.1126/science.aad7969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, K. W. , Brann, D. H. , Farruggia, M. C. , Bhutani, S. , Pellegrino, R. , Tsukahara, T. , Weinreb, C. , Joseph, P. V. , Larson, E. D. , Parma, V. , Albers, M. W. , Barlow, L. A. , Datta, S. R. , & Di Pizio, A. (2020). COVID‐19 and the chemical senses: Supporting players take center stage. Neuron, 22, 107(2), 219–233. 10.1016/j.neuron.2020.06.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djidrovski, I. , Georgiou, M. , Hughes, G. L. , Patterson, E. I. , Casas‐Sanchez, A. , Pennington, S. , Biagini, G. A. , Moya‐Molina, M. , van den Bor, J. , Smit, M. J. , Chung, G. , Lako, M. , & Armstrong, L. (2021). SARS‐CoV‐2 infects an upper airway model derived from induced pluripotent stem cells. Stem Cells, 39(10), 1310–1321. 10.1002/stem.3422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy, M. C. , Eschle, B. K. , Barrows, J. , Hallock, R. M. , Finger, T. E. , & Delay, E. R. (2009). Double P2X2/P2X3 purinergic receptor knockout mice do not taste NaCl or the artificial sweetener SC45647. Chemical Senses, 34(9), 789–797. 10.1093/chemse/bjp068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, C. , Klekot, O. , Halugan, C. , & Korchev, Y. (2021). Follow your nose: A key clue to understanding and treating COVID‐19. Frontiers in Endocrinology (Lausanne), 12, 747744. 10.3389/fendo.2021.747744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finger, T. E. , Danilova, V. , Barrows, J. , Bartel, D. L. , Vigers, A. J. , Stone, L. , Hellekant, G. , & Kinnamon, S. C. (2005). ATP signaling is crucial for communication from taste buds to gustatory nerves. Science, 310(5753), 1495–1499. 10.1126/science.1118435 [DOI] [PubMed] [Google Scholar]
- Firth, A. L. , Dargitz, C. T. , Qualls, S. J. , Menon, T. , Wright, R. , Singer, O. , Gage, F. H. , Khanna, A. , & Verma, I. M. (2014). Generation of multiciliated cells in functional airway epithelia from human induced pluripotent stem cells. Proceedings of National Academy of Sciences (USA), 111(17), E1723–E1730. 10.1073/pnas.1403470111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredenburgh, L. E. , Perrella, M. A. , & Mitsialis, S. A. (2007). The role of heme oxygenase‐1 in pulmonary disease. American Journal of Respiratory Cellular and Molecular Biology, 36(2), 158–165. 10.1165/rcmb.2006-0331TR [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganji, R. , & Reddy, P. H. (2021). Impact of COVID‐19 on mitochondrial‐based immunity in aging and age‐related diseases. Frontiers in Aging Neurosciences, 12, 614650. 10.3389/fnagi.2020.614650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, D. E. , Jang, G. M. , Bouhaddou, M. , Xu, J. , Obernier, K. , White, K. M. , O'Meara, M. J. , Rezelj, V. V. , Guo, J. Z. , Swaney, D. L. , Tummino, T. A. , Hüttenhain, R. , Kaake, R. M. , Richards, A. L. , Tutuncuoglu, B. , Foussard, H. , Batra, J. , Batra, K. , Modak, M. , … Krogan, N. J. (2020). A SARS‐CoV‐2 protein interaction map reveals targets for drug repurposing. Nature, 583(7816), 459–468. 10.1038/s41586-020-2286-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, X. , Karp, P. H. , Brody, S. L. , Pierce, R. A. , Welsh, M. J. , Holtzman, M. J. , & Ben‐Shahar, Y. (2014). Chemosensory functions for pulmonary neuroendocrine cells. American Journal of Respiratory Cellular and Molecular Biology, 50(3), 637–646. 10.1165/rcmb.2013-0199OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulbransen, B. D. , Clapp, T. R. , Finger, T. E. , & Kinnamon, S. C. (2008). Nasal solitary chemoreceptor cell responses to bitter and trigeminal stimulants in vitro. Journal of Neurophysiology, 99(6), 2929–2937. 10.1152/jn.00066.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannum, M. E. , Koch, R. J. , Ramirez, V. A. , Marks, S. S. , Toskala, A. K. , Herriman, R. D. , Lin, C. , Joseph, P. V. , & Reed, D. R. (2021). Taste loss as a distinct symptom of COVID‐19: A systematic review and meta‐analysis. medRxiv: The Preprint Server for Health Sciences , 21264771. 10.1101/2021.10.09.21264771 [DOI]
- Hegg, C. C. , Greenwood, D. , Hua, W. , Han, P. , & Lucero, M. T. (2003). Activation of purinergic receptor subtypes modulates odor sensitivity. Journal of Neurosciences, 23(23), 8291–8301. 10.1523/jneurosci.23-23-08291.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henke, R. M. , Meredith, D. M. , Borromeo, M. D. , Savage, T. K. , & Johnson, J. E. (2009). Ascl1 and Neurog2 form novel complexes and regulate Delta‐like3 (Dll3) expression in the neural tube. Developmental Biology, 328(2), 529–540. 10.1016/j.ydbio.2009.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hor, P. , Punj, V. , Calvert, B. A. , Castaldi, A. , Miller, A. J. , Carraro, G. , Stripp, B. R. , Brody, S. L. , Spence, J. R. , Ichida, J. K. , Ryan Firth, A. L. , & Borok, Z. (2020). Efficient generation and transcriptomic profiling of human iPSC‐derived pulmonary neuroendocrine cells. iScience, 23(5):101083. 10.1016/j.isci.2020.101083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, J. , Hume, A. J. , Abo, K. M. , Werder, R. B. , Villacorta‐Martin, C. , Alysandratos, K. D. , Beermann, M. L. , Simone‐Roach, C. , Lindstrom‐Vautrin, J. , Olejnik, J. , Suder, E. L. , Bullitt, E. , Hinds, A. , Sharma, A. , Bosmann, M. , Wang, R. , Hawkins, F. , Burks, E. J. , Saeed, M. , … Kotton, D. N. (2020). SARS‐CoV‐2 infection of pluripotent stem cell‐derived human lung alveolar type 2 cells elicits a rapid epithelial‐intrinsic inflammatory response. Cell Stem Cell, 27(6), 962–973.e7. 10.1016/j.stem.2020.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John, A. E. , Joseph, C. , Jenkins, G. , & Tatler, A. L. (2021). COVID‐19 and pulmonary fibrosis: A potential role for lung epithelial cells and fibroblasts. Immunological Reviews, 302(1), 228–240. 10.1111/imr.12977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsura, H. , Sontake, V. , Tata, A. , Kobayashi, Y. , Edwards, C. E. , Heaton, B. E. , Konkimalla, A. , Asakura, T. , Mikami, Y. , Fritch, E. J. , Lee, P. J. , Heaton, N. S. , Boucher, R. C. , Randell, S. H. , Baric, R. S. , & Tata, P. R. (2020). Human lung stem cell‐based alveolospheres provide insights into SARS‐CoV‐2‐mediated interferon responses and pneumocyte dysfunction. Cell Stem Cell, 27(6), 890–904. 10.1016/j.stem.2020.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeppen, M. , McNamee, E. N. , Brodsky, K. S. , Aherne, C. M. , Faigle, M. , Downey, G. P. , Colgan, S. P. , Evans, C. M. , Schwartz, D. A. , & Eltzschig, H. K. (2013). Detrimental role of the airway mucin Muc5ac during ventilator‐induced lung injury. Mucosal Immunology, 6(4), 762–775. 10.1038/mi.2012.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kommoss, F. , Schwab, C. , Tavernar, L. , Schreck, J. , Wagner, W. L. , Merle, U. , Jonigk, D. , Schirmacher, P. , & Longerich, T. (2020). The pathology of severe COVID‐19‐related lung damage. Deutsches Arzteblatt International, 117(29‐30), 500–506. 10.3238/arztebl.2020.0500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshiba, T. (2013). Mitochondrial‐mediated antiviral immunity. Biochimica et Biophysica Acta, 1833(1), 225–232. 10.1016/j.bbamcr.2012.03.005 [DOI] [PubMed] [Google Scholar]
- Kotton, D. N. , & Morrisey, E. E. (2014). Lung regeneration: Mechanisms, applications and emerging stem cell populations. Nature Medicine, 20(8), 822–832. 10.1038/nm.3642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, R. J. , Xiong, G. , Kofonow, J. M. , Chen, B. , Lysenko, A. , Jiang, P. , Abraham , V. , Doghramji, L. , Adappa, N. D. , Palmer, P. N. , Kennedy, D. W. , Beauchamp, G. K. , Doulias, P. T. , Ischiropoulos, H. , Kreindler, J. L. , Reed, D. R. , & Cohen, N. A. (2012). T2R38 taste receptor polymorphisms underlie susceptibility to upper respiratory infection. Journal of Clinical Investigation, 122(11), 4145–4159. 10.1172/JCI64240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibowitz, B. , & Yu, J. (2010). Mitochondrial signaling in cell death via the Bcl‐2 family. Cancer Biology and Therapy, 9(6), 417–422. 10.4161/cbt.9.6.11392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnoila, R. I. (2006). Functional facets of the pulmonary neuroendocrine system. Laboratory Investigation, 86(5), 425–444. 10.1038/labinvest.3700412 [DOI] [PubMed] [Google Scholar]
- Menachery, V. D. , Eisfeld, A. J. , Schäfer, A. , Josset, L. , Sims, A. C. , Proll, S. , Fan, S. , Li, C. , Neumann, G. , Tilton, S. C. , Chang, J. , Gralinski, L. E. , Long, C. , Green, R. , Williams, C. M. , Weiss, J. , Matzke, M. M. , Webb‐Robertson, B. J. , Schepmoes, A. A. , … Baric, R. S. (2014). Pathogenic influenza viruses and coronaviruses utilize similar and contrasting approaches to control interferon‐stimulated gene responses. mBio, 5(3), e01174–14. 10.1128/mBio.01174-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midwood, K. S. , & Orend, G. (2009). The role of tenascin‐C in tissue injury and tumorigenesis. Journal of Cellular Communication and Signaling, 3(3−4), 287–310. 10.1007/s12079-009-0075-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoro, D. T. , Haber, A. L. , Biton, M. , Vinarsky, V. , Lin, B. , Birket, S. E. , Yuan, F. , Chen, S. , Leung, H. M. , Villoria, J. , Rogel, N. , Burgin, G. , Tsankov, A. M. , Waghray, A. , Slyper, M. , Waldman, J. , Nguyen, L. , Dionne, D. , Rozenblatt‐Rosen, O. , … Rajagopal, J (2018). A revised airway epithelial hierarchy includes CFTR‐expressing ionocytes. Nature, 560, 319–324. 10.1038/s41586-018-0393-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse, D. , & Choi, A. M. (2005). Heme oxygenase‐1: From bench to bedside. American Journal of Respiratory Critical Care and Medicine, 172(6), 660–670. 10.1164/rccm.200404-465SO [DOI] [PubMed] [Google Scholar]
- Mou, H. , Yang, Y. , Riehs, M. A. , Barrios, J. , Shivaraju, M. , Haber, A. L. , Montoro, D. T. , Gilmore, K. , Haas, E. A. , Paunovic, B. , Rajagopal, J. , Vargas, S. O. , Haynes, R. L. , Fine, A. , Cardoso, W. V. , & Ai, X. (2021). Airway basal stem cells generate distinct subpopulations of PNECs. Cell Reports, 35(3):109011. 10.1016/j.celrep.2021.109011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulay, A. , Konda, B. , Garcia, G., Jr. , Yao, C. , Beil, S. , Villalba, J. M. , Koziol, C. , Sen, C. , Purkayastha, A. , Kolls, J. K. , Pociask, D. A. , Pessina, P. , de Aja, J. S. , Garcia‐de‐Alba, C. , Kim, C. F. , Gompert, B. , Arumugaswami, V. , & Stripp, B. R. (2021). SARS‐CoV‐2 infection of primary human lung epithelium for COVID‐19 modeling and drug discovery. Cell Reports, 35(5):109055. 10.1016/j.celrep.2021.109055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota, C. , Ng‐Blichfeldt, J. P. , Korfei, M. , Alsafadi, H. N. , Lehmann, M. , Skronska‐Wasek, W. , M De Santis, M. , Guenther, A. , Wagner, D. E. , & Königshoff, M. (2018). Dynamic expression of HOPX in alveolar epithelial cells reflects injury and repair during the progression of pulmonary fibrosis. Scientific Reports, 8(1), 12983. 10.1038/s41598-018-31214-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei, R. , Feng, J. , Zhang, Y. , Sun, H. , Li, L. , Yang, X. , He, J. , Xiao, S. , Xiong, J. , Lin, Y. , Wen, K. , Zhou, H. , Chen, J. , Rong, Z. , & Chen, X. (2021). Host metabolism dysregulation and cell tropism identification in human airway and alveolar organoids upon SARS‐CoV‐2 infection. Protein and Cell, 12(9), 717–733. 10.1007/s13238-020-00811-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai, D. K. , Sharma, P. , & Kumar, R. (2021). Post Covid 19 pulmonary fibrosis. Is it real threat? Indian Journal of Tuberculosis, 68(3), 330–333. 10.1016/j.ijtb.2020.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinett, K. S. , Koziol‐White, C. J. , Akoluk, A. , An, S. S. , Panettieri, R. A., Jr. , & Liggett, S. B. (2014). Bitter taste receptor function in asthmatic and nonasthmatic human airway smooth muscle cells. American Journal of Respiratory Cell and Molecular Biology, 50(4), 678–683. 10.1165/rcmb.2013-0439RC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh, J. , Peyssonnaux, C. , Singh, K. K. , & Edeas, M. (2020). Mitochondria and microbiota dysfunction in COVID‐19 pathogenesis. Mitochondrion, 54, 1–7. 10.1016/j.mito.2020.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, A. S. , Ben‐Shahar, Y. , Moninger, T. O. , Kline, J. N. , & Welsh, M. J. (2009). Motile cilia of human airway epithelia are chemosensory. Science, 325(5944), 1131–1134. 10.1126/science.1173869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, K. K. , Chaubey, G. , Chen, J. Y. , & Suravajhala, P. (2020). Decoding SARS‐CoV‐2 hijacking of host mitochondria in COVID‐19 pathogenesis. American Journal of Physiology Cell Physiology, 319(2), C258–C267. 10.1152/ajpcell.00224.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surendran, H. , Nandakumar, S. , & Pal, R. (2020). Human induced pluripotent stem cell‐derived lung epithelial system for SARS‐CoV‐2 infection modeling and its potential in drug repurposing. Stem Cells and Development, 29(21), 1365–1369. 10.1089/scd.2020.0152 [DOI] [PubMed] [Google Scholar]
- Surendran, H. , Rajamoorthy, M. , & Pal, R. (2019). Differentiating human induced pluripotent stem cells (iPSCs) into lung epithelial cells. Current Protocols in Stem Cell Biology, 49(1), e86. 10.1002/cpsc.86 [DOI] [PubMed] [Google Scholar]
- Valyaeva, A. A. , Zharikova, A. A. , Kasianov, A. S. , Vassetzky, Y. S. , & Sheval, E. V. (2020). Expression of SARS‐CoV‐2 entry factors in lung epithelial stem cells and its potential implications for COVID‐19. Scientific Reports, 10(1), 17772. 10.1038/s41598-020-74598-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittekindt, O. H. , & Dietl, P. (2019). Aquaporins in the lung. Pflugers Archiv: European Journal of Physiology, 471(4), 519–532. 10.1007/s00424-018-2232-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, K. E. , Fazal, F. M. , Parker, K. R. , Zou, J. , & Chang, H. Y. (2020). RNA‐GPS predicts SARS‐CoV‐2 RNA residency to host mitochondria and nucleolus. Cell Systems, 11(1), 102–108.e3. 10.1016/j.cels.2020.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, K. , & Ishimaru, Y. (2013). Oral and extra‐oral taste perception. Seminars in Cell and Developmental Biology, 24(3), 240–246. 10.1016/j.semcdb.2012.08.005 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.


