Abstract
We developed a nonculture method to predict the susceptibility of Neisseria meningitidis to penicillin G. The penA gene was amplified and submitted to restriction fragment length polymorphism analysis. This approach was first validated with a collection of 75 meningococcal strains of known phenotypes. It was next successfully applied to 29 clinical samples.
Treatment of meningococcal infections is a medical emergency and requires a rapid diagnosis by the isolation of bacteria from cerebrospinal fluid (CSF), blood, or other body fluids (synovial, pericardial, etc.). However, the isolation of Neisseria meningitidis by culture is frequently hindered by early antibiotic treatment, which is highly recommended whenever meningococcal infection is suspected (6, 21). Several methods for nonculture diagnosis of N. meningitidis have been recently reported. These methods rapidly (1 to 2 h) identify N. meningitidis by amplifying one target gene, such as that encoding 16S rRNA (12, 14), IS1106 (17), porB (29), dhpS (13), ctrA (9), or crgA (25). The serogroup, which reflects the capsule immunospecificity, is subsequently predicted by PCR amplification of serogroup-specific allele of the siaD gene for sialic acid-containing serogroups (B, C, Y, and W135) and by PCR amplification of the mynB gene for the mannoseamine-containing serogroup A (3, 4, 8, 19, 24, 25). These nonculture methods provide essential data for assessing the etiological diagnosis of the disease, but they do not provide information about antibiotic susceptibility. Penicillin G is still effective for the treatment of meningococcal infections. However, meningococcal strains with reduced susceptibility to penicillin (Peni), for which MICs range from 0.125 to 1 μg/ml (11), have been increasingly reported in several countries (18). Penicillin-resistant strains (Penr) for which MICs are >1 μg/ml are now emerging (Table 1). This phenotype is thought to be due to a reduction in the affinity of penicillin-binding protein 2 (PBP2), encoded by an altered penA gene, for penicillin (16, 22). Mosaic structures in penA result from horizontal DNA exchange by transformation between commensal Neisseria species and N. meningitidis (5, 22, 23). We have previously used restriction fragment length polymorphism (RFLP) and DNA sequencing to show that penicillin-susceptible strains (Pens) of different geographic origins, antigenic formulas, and genetic lineages have the same penA allele, whereas Peni strains have a variety of different penA alleles (1). Moreover, penA sequences from Peni strains are highly divergent, particularly in the transpeptidase-encoding region (nucleotides 718 to 1743). This approach was shown to be suitable for the analysis of genetic relatedness between different penA alleles. The aim of the present study was to develop a nonculture method to predict the susceptibility of N. meningitidis to penicillin G.
TABLE 1.
Distribution of the 75 N. meningitidis strains tested in this study according to PCR-RFLP and susceptibility to penicillin (MICs)
| Penicillin MIC (μg/ml) for N. meningitidis | No. of strains in PCR-RFLP
|
|||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | |
| Pens(n = 39): MIC < 0.125 | 39 | |||||
| Peni(n = 36): 0.125 ≤ MIC ≤ 1 | ||||||
| 0.125 | 5a | 1 | 3 | 2 | 1 | |
| 0.19 | 1 | 2 | 1 | |||
| 0.25 | 4 | 1 | 1 | |||
| 0.38 | 3 | 3 | ||||
| 0.5 | 1 | 1 | 1 | |||
| 0.75 | 1 | |||||
| 1b | 2b | |||||
| 1.5 | 1b | |||||
| >32c | 1 | |||||
Strains susceptible to amoxicillin (MIC = 0.125 or 0.19 μg/ml).
Strains isolated in 1999 and 2000.
β-Lactamase-positive strain.
DNA sequence alignment revealed conserved regions in the transpeptidase-encoding region of the penA gene of both Pens and Peni strains (1). Two oligonucleotides were designed on the basis of these conserved sequences: AA-1 (5′-ATCGAACAGGCGACGATGTC-3′; nucleotides 1237 to 1256) and 99–2 (5′-GATTAAGACGGTGTTTTGACGG-3′; nucleotides 1728 to 1748). They amplify a 500-bp fragment of the 3′ end of the penA gene. These oligonucleotides were first tested on a collection of 75 meningococcal strains for which there were different MICs of penicillin G (39 Pens and 36 Peni strains).
Penicillin G susceptibility was tested by the agar dilution method on G medium with G supplement (Sanofi Diagnostic Pasteur, Marnes La Coquette, France). The strains were tested with inocula of 108 CFU/ml on plates containing penicillin G at the following concentrations: 0.06, 0.125, 0.25, 0.5, and 0.75 μg/ml. The MIC was defined as the lowest concentration of penicillin G that inhibited visible growth after 18 h of incubation in 5% CO2 at 37°C. Penicillin G susceptibility was also tested by the diffusion method on G medium (Etest; AB-Biodisk, Solna, Sweden). β-Lactamase activity was detected with a Cefinaseä disk (BioMérieux, Marcy l'Etoile, France).
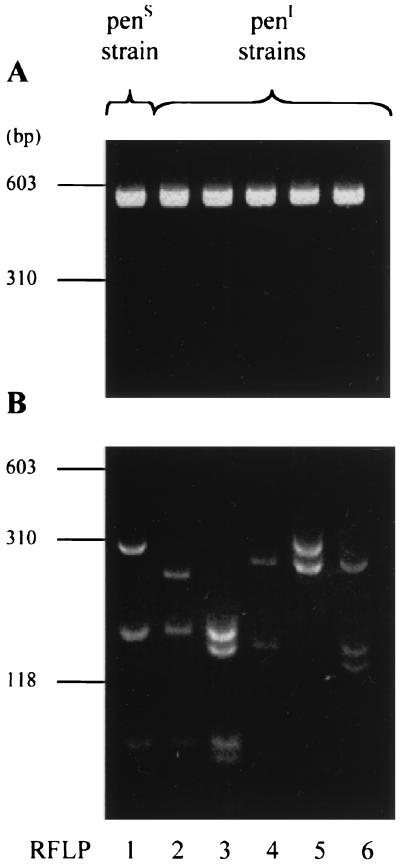
PCR was performed as previously described (1, 25). Amplification products of the expected size (500 bp) were obtained for all of the strains (Fig. 1A). After digestion with TaqI and separation on a 4% agarose gel, six different profiles were obtained (Fig. 1B). An arbitrary number was assigned to each profile (patterns 1 to 6). The results of PCR-RFLP from 75 Pens and Peni N. meningitidis strains are shown in the Table 1. On the basis of the RFLPs, two classes of strains were identified. (i) All Pens strains for which the MIC was <0.125 μg/ml had the same pattern (RFLP1). (ii) Peni strains for which the MIC was 0.125 μg/ml had altered patterns (RFLP2 to -6) compared to Pens strains. However, for five strains for which the MIC was 0.125 μg/ml, we found RFLP1 as for Pens strains. MICs for these strains were controlled by Etest and the agar dilution method. These strains could have been misclassified as Peni strains, as we have previously suggested for such strains for which the MIC of penicillin G is at the breakpoint level (1). The fact that these strains are susceptible to amoxicillin supports this interpretation (Table 1). This observation underlines the technical difficulties in determining meningococcal susceptibility to penicillin by MIC tests. Indeed, 14 European Reference Laboratories using identical methods, media, and strains reported differences in MIC determinations. Only molecular approaches showed complete agreement in detection of Peni meningococcal strains (2). The PCR-RFLP results with the penA gene were highly consistent with susceptibility to penicillin. No correlation was observed between a particular RFLP pattern and MICs in Peni strains. Indeed, the MICs for strains with the same RFLP could be different (Table 1). Molecular approaches are therefore able to overcome technical problems of MIC determination and to detect meningococcal strains with reduced susceptibility to penicillin G, regardless of the MIC for them. Treatment could be immediately adapted.
FIG. 1.
Agarose gel electrophoresis of PCR-amplified DNA fragment of penA gene (A) and corresponding restriction fragment length patterns after digestion by TaqI (B) of Pens and Peni N. meningitidis strains (one representative strain by RFLP).
Our PCR-RFLP assay was subsequently used to test 29 clinical samples from various biological fluids obtained from different patients with suspected meningococcal infection (Table 2). Samples were treated as previously described (25), and PCR was performed with 15 μl of each sample. PCR was first used to amplify the crgA gene as recently described (25) and all of the samples were found to contain meningococcal DNA. Culture methods also found that 10 of the samples were positive. This allowed the MICs of penicillin G for them to be determined (Table 2). We next amplified the penA gene. Amplification products of the expected size (500 bp) were obtained for all 29 samples. Analysis of restriction patterns after digestion with TaqI showed that 20 samples had the same RFLP as the Pens strains (RFLP1), whereas 9 samples had different RFLPs, as observed for Peni strains. A complete correlation was observed between the PCR-RFLP results for biological samples, the PCR-RFLP results from bacterial DNA extracts, and the MIC of penicillin G for the corresponding strains (Table 2).
TABLE 2.
Clinical samples tested in this study and PCR-RFLP results, penicillin MICs, and PCR-RFLP results of the corresponding strains
| Clinical sample | Sample sourcea | No. (type) of PCR-RFLP samples | Penicillin MIC (μg/ml) | No. (type) of PCR-RFLP strains |
|---|---|---|---|---|
| L209 | CSF | 4 (Peni) | 0.125 | 4 (Peni) |
| L239 | CSF | 1 (Pens) | NAb | |
| L257 | CSF | 1 (Pens) | NA | |
| L263 | CSF | 5 (Peni) | NA | |
| S197 | Serum | 5 (Peni) | NA | |
| S209 | Serum | 5 (Peni) | NA | |
| L302 | CSF | 1 (Pens) | 0.064 | 1 (Pens) |
| S234 | Serum | 4 (Peni) | 0.125 | 4 (Peni) |
| S235 | Serum | 1 (Pens) | 0.064 | 1 (Pens) |
| S238 | Serum | 1 (Pens) | 0.125 | 1 (Pens) |
| L358 | CSF | 1 (Pens) | 0.047 | 1 (Pens) |
| L363 | CSF | 1 (Pens) | 0.047 | 1 (Pens) |
| S247 | Serum | 1 (Pens) | NA | |
| L374 | CSF | 1 (Pens) | NA | |
| S248 | Serum | 2 (Peni) | NA | |
| S250c | PCF | 1 (Pens) | 0.094 | 1 (Pens) |
| L380 | CSF | 1 (Pens) | NA | |
| L387 | CSF | 1 (Pens) | 0.064 | 1 (Pens) |
| L393 | CSF | 1 (Pens) | NA | |
| L395 | CSF | 6 (Peni) | NA | |
| L396c | PF | 1 (Pens) | NA | |
| L398 | CSF | 2 (Peni) | NA | |
| S263 | Serum | 1 (Pens) | NA | |
| S270 | Serum | 1 (Pens) | 0.032 | 1 (Pens) |
| L399 | CSF | 1 (Pens) | NA | |
| L400 | CSF | 3 (Peni) | NA | |
| L405 | CSF | 1 (Pens) | NA | |
| L412 | CSF | 1 (Pens) | NA | |
| L415 | CSF | 1 (Pens) | 0.032 | 1 (Pens) |
Samples tested were CSF, serum, pericardial fluid (PCF), and pleural fluid (PF).
NA, not available (no positive culture for these samples).
For these strains, another sample (CSF or serum) was also available and gave the same result.
Molecular detection of genes associated with bacterial resistance to antibiotics has been developed for several species. Methicillin resistance in Staphylococcus aureus and coagulase-negative staphylococci results from the synthesis of a novel PBP encoded by the mecA gene. The PCR-based detection of the mecA gene is becoming the standard method for the detection of methicillin resistance (27, 28). Resistance to vancomycin encoded by the van genes (vanA, vanB, vanC, vanD, vanE, and vanG) is now widespread in enterococci. Therefore, amplification assays for the detection of the van genes have been developed (7, 20). β-Lactamase-producing Haemophilus influenzae can be detected by PCR with specific primers for the blaTEM and blaROB β-lactamase genes (26).
Early antibiotic treatment is the major element in the immediate management of meningococcal infections. The use of rapid and specific methods of nonculture diagnosis and the development of a molecular approach for the prediction of meningococcal susceptibility to penicillin are needed because of the increasing number of culture-negative cases. The use of PCR to detect reduced susceptibility to penicillin was recently reported with seven different PCRs with the penA gene (15). A strain was considered to be Peni if it failed to produce at least one PCR product. However, this approach was not tested in clinical samples. Our approach only uses one PCR product that is produced in all Pens and Peni strains and is easily and directly applicable for clinical samples. Our data clearly suggest that the detection of an altered penA allele is always correlated with reduced susceptibility to penicillin. Moreover, the combination of this molecular approach with the phenotypic antibiotic susceptibility tests to penicillin may enable us to clearly classify strains as Pens or Peni and to overcome the difficulties encountered for strains for which the MIC of penicillin G is close to the breakpoint level of 0.125 μg/ml (1).
The acquisition of β-lactamases seems to be rare in N. meningitidis, and resistance to penicillin G is now evolving by alteration of PBP2. Penr strains (MIC >1 μg/ml) may be expected in the near future, analogous to Streptococcus pneumoniae and as suggested by the recent characterization in our laboratory of meningococcal strains for which the MICs were 1 and 1.5 μg/ml (Table 1). The threshold of 1 μg/ml corresponds to the therapeutic concentration in the CSF obtained during treatment with penicillin G (10). The emergence of meningococcal strains for which the MIC was >1 μg/ml may provoke treatment failure. The development of a reliable molecular approach to surveillance is clinically relevant and is expected to anticipate and enhance detection of the resistant strains in order to administer an adequate antibiotic treatment. Moreover, this nonculture detection of meningococcal strains with altered susceptibility to penicillin G provides information that otherwise is inaccessible in culture-negative cases of meningococcal infections. It also complements our approach combining nonculture diagnosis and serogroup prediction of meningococcal infections (25).
Acknowledgments
We thank Magaly Ducos for help with MIC determination.
This work was supported by the Institut Pasteur. A. Antignac was supported by a fellowship from the Caisse Nationale d'Assurance Maladie.
REFERENCES
- 1.Antignac A, Kriz P, Tzanakaki G, Alonso J-M, Taha M-K. Polymorphism of Neisseria meningitidis penA gene associated with reduced susceptibility to penicillin. J Antimicrob Chemother. 2001;47:285–296. doi: 10.1093/jac/47.3.285. [DOI] [PubMed] [Google Scholar]
- 2.Arreaza L, Vazquez J A. Abstracts of the 6th European Monitoring Group on Meningococci. 2001. Proposal for standardization of media and methods to be used on the determination of the susceptibility level of resistance to antibiotics in Neisseria meningitidis, pp. 40–43. Orebro, Sweden. [Google Scholar]
- 3.Borrow R, Claus H, Guiver M, Smart L, Jones D M, Kaczmarski E B, Frosch M, Fox A J. Non-culture diagnosis and serogroup determination of meningococcal B and C infection by a sialyltransferase (siaD) PCR ELISA. Epidemiol Infect. 1997;118:111–117. doi: 10.1017/s0950268896007261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borrow R, Claus H, Chaudhry U, Guiver M, Kaczmarski E B, Frosch M, Fox A J. siaD PCR ELISA for confirmation and identification of serogroup Y and W135 meningococcal infections. FEMS Microbiol Lett. 1998;159:209–214. doi: 10.1111/j.1574-6968.1998.tb12862.x. [DOI] [PubMed] [Google Scholar]
- 5.Bowler L D, Zhang Q-Y, Riou J-Y, Spratt B G. Interspecies recombination between the penA genes of Neisseria meningitidis and commensal Neisseria species during the emergence of penicillin resistance in N. meningitidis: natural events and laboratory simulation. J Bacteriol. 1994;176:333–337. doi: 10.1128/jb.176.2.333-337.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cartwright K, Reilly S, White D, Stuart J. Early treatment with parenteral penicillin in meningococcal disease. Br Med J. 1992;305:143–147. doi: 10.1136/bmj.305.6846.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dutka-Malen S, Evers S, Courvalin P. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J Clin Microbiol. 1995;33:24–27. doi: 10.1128/jcm.33.1.24-27.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frosch M, Edwards U, Bousset K, Krausse B, Weisgerber C. Evidence for a common molecular origin of the capsule gene loci in gram-negative bacteria expressing group II capsular polysaccharides. Mol Microbiol. 1991;5:1251–1263. doi: 10.1111/j.1365-2958.1991.tb01899.x. [DOI] [PubMed] [Google Scholar]
- 9.Guiver M, Borrow R, Marsh J, Gray S J, Kaczmarski E B, Howells D, Boseley P, Fox A J. Evaluation of the Applied Biosystems automated Taqman polymerase chain reaction system for the detection of meningococcal DNA. FEMS Immunol Med Microbiol. 2000;28:173–179. doi: 10.1111/j.1574-695X.2000.tb01473.x. [DOI] [PubMed] [Google Scholar]
- 10.Hibber J P, Nelson J D. A pharmacologic evaluation of penicillin in children with purulent meningitis. N Engl J Med. 1977;297:410–413. doi: 10.1056/NEJM197708252970802. [DOI] [PubMed] [Google Scholar]
- 11.Hughes J H, Biedenbach D J, Erwin M E, Jones R N. E test as susceptibility test and epidemiologic tool for evaluation of Neisseria meningitidis isolates. J Clin Microbiol. 1993;31:3255–3259. doi: 10.1128/jcm.31.12.3255-3259.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kotilainen P, Jalava J, Meurman O, Lehtonen O-P, Rintala E, Seppala O-P, Eeorala E, Nikkari S. Diagnosis of meningococcal meningitis by broad-range bacterial PCR with cerebrospinal fluid. J Clin Microbiol. 1998;36:2205–2209. doi: 10.1128/jcm.36.8.2205-2209.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kristiansen B-E, Ask E, Jenkins A, Fermer C, Radstrom P, Skold O. Rapid diagnosis of meningococcal meningitidis by polymerase chain reaction. Lancet. 1991;337:1568–1569. doi: 10.1016/0140-6736(91)93262-8. [DOI] [PubMed] [Google Scholar]
- 14.Ley B E, Linton C J, Bennett D M C, Jalal H, Foot A B M, Millar M R. Detection of bacteraemia in patients with fever and neutropenia using 16S rRNA gene amplification by polymerase chain reaction. Eur J Clin Microbiol Infect Dis. 1998;17:247–253. doi: 10.1007/BF01699981. [DOI] [PubMed] [Google Scholar]
- 15.Maggs A F, Logan J M, Carter P E, Pennington T H. The detection of penicillin insensitivity in Neisseria meningitidis by polymerase chain reaction. J Antimicrob Chemother. 1998;42:303–307. doi: 10.1093/jac/42.3.303. [DOI] [PubMed] [Google Scholar]
- 16.Mendelman P M, Campos J, Chaffin D O, Serfass D A, Smith A L, Sáez-Nieto J A. Relative penicillin G resistance in Neisseria meningitidis and reduced affinity of penicillin-binding protein 3. Antimicrob Agents Chemother. 1987;32:706–709. doi: 10.1128/aac.32.5.706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Newcombe J, Cartwright K, Palmer W H, McFadden J. PCR of peripheral blood for diagnosis of meningococcal disease. J Clin Microbiol. 1996;34:1637–1640. doi: 10.1128/jcm.34.7.1637-1640.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oppenheim B. Antibiotic resistance in Neisseria meningitidis. Clin Infect Dis. 1997;24:S98–S101. doi: 10.1093/clinids/24.supplement_1.s98. [DOI] [PubMed] [Google Scholar]
- 19.Orvelid P, Backman A, Olcen P. PCR identification of the group A Neisseria meningitidis gene in cerebrospinal fluid. Scand J Infect Dis. 1999;31:481–483. doi: 10.1080/00365549950164003. [DOI] [PubMed] [Google Scholar]
- 20.Patel R, Uhl J R, Kohner P, Hopkins M K, Cockerill F R., III Multiplex PCR detection of vanA, vanB, vanC-1, and vanC-2/3 genes in enterococci. J Clin Microbiol. 1997;35:703–707. doi: 10.1128/jcm.35.3.703-707.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quagliarello V J, Scheld W M. Treatment of bacterial meningitis. N Engl J Med. 1996;336:708–716. doi: 10.1056/NEJM199703063361007. [DOI] [PubMed] [Google Scholar]
- 22.Spratt B G, Zhang Q-Y, Jones D M, Hutchison A, Brannigan J A, Dowson C G. Recruitment of a penicillin-binding protein gene from Neisseria flavescens during the emergence of penicillin resistance in Neisseria meningitidis. Proc Natl Acad Sci USA. 1989;86:8988–8992. doi: 10.1073/pnas.86.22.8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spratt B G, Bowler L D, Zhang Q-Y, Zhou J, Smith J M. Role of interspecies transfer of chromosomal genes in the evolution of penicillin resistance in pathogenic and commensal Neisseria species. J Mol Evol. 1992;34:115–125. doi: 10.1007/BF00182388. [DOI] [PubMed] [Google Scholar]
- 24.Swartley J S, Liu L-J, Miller Y K, Martin L E, Edupuganti S, Stephens D S. Characterization of the gene cassette required for the biosynthesis of the (-16)-linked N-acetyl-d-mannosamine-1-phosphate capsule of serogroup A Neisseria meningitidis. J Bacteriol. 1998;180:1533–1539. doi: 10.1128/jb.180.6.1533-1539.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taha M-K. Simultaneous approach for nonculture PCR-based identification and serogroup prediction of Neisseria meningitidis. J Clin Microbiol. 2000;38:855–857. doi: 10.1128/jcm.38.2.855-857.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tenover F C, Huang M B, Rasheed J K, Persing D H. Development of PCR assays to detect ampicillin resistance genes in cerebrospinal fluid samples containing Haemophilus influenzae. J Clin Microbiol. 1994;32:2729–2737. doi: 10.1128/jcm.32.11.2729-2737.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tokue Y, Shoji S, Satoh K, Watanabe A, Motomiya M. Comparison of a polymerase chain reaction assay and a conventional microbiologic method for detection of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1992;36:6–9. doi: 10.1128/aac.36.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ubukata K, Nakagami S, Nitta A, Yamane A, Kawakami S, Sugiura M, Konno M. Rapid detection of the mecA gene in methicillin-resistant staphylococci by enzymatic detection of polymerase chain reaction products. J Clin Microbiol. 1992;30:1728–1733. doi: 10.1128/jcm.30.7.1728-1733.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Urwin R, Kaczmarski E B, Guiver M, Fox A J, Maiden M C. Amplification of the meningococcal porB gene for non-culture serotype characterization. Epidemiol Infect. 1998;120:257–262. doi: 10.1017/s0950268898008656. [DOI] [PMC free article] [PubMed] [Google Scholar]