Abstract
The coronavirus disease‐2019 (COVID‐19) pandemic is still challenging public health systems worldwide, particularly with the emergence of novel SARS‐CoV‐2 variants with mutations that increase their transmissibility and immune escape. This is the case of the variant of concern Omicron that rapidly spread globally. Here, using epidemiological and genomic data we compared the situations in South Africa as the epicenter of emergence, United Kingdom, and with particular interest New York City. This rapid global dispersal from the place of first report reemphasizes the high transmissibility of Omicron, which needed only two weeks to become dominant in the United Kingdom and New York City. Our analyses suggest that as SARS‐CoV‐2 continues to evolve, global authorities must prioritize equity in vaccine access and continued genomic surveillance. Future studies are still needed to fully unveil the biological properties of Omicron, but what is certain is that vaccination, large‐scale testing, and infection prevention efforts are the greatest arsenal against the COVID‐19 pandemic.
Keywords: epidemiology, genetic variability evolution, SARS coronavirus classification
The coronavirus disease‐2019 (COVID‐19) pandemic continues to burden health systems worldwide, in part due to the emergence of novel severe acute respiratory syndrome coronavirus‐2 (SARS‐CoV‐2) variants that contain mutations associated with increased transmission and immune evasion. A dramatic example is the recent emergence of the lineage B.1.1.529, named by the World Health Organization as the Omicron variant of concern (VOC) on November 24, 2021, less than 2 weeks after its first detection in specimens collected on November 11, 2021 in Botswana and November 14, 2021 in South Africa. 1 , 2 Within weeks, the Omicron VOC was already reported in more than 67 countries around the globe, driven by a set of mutations across its genome proposed to be associated with increased transmissibility and immune escape. 1 , 3 In some regions of the world, particularly in international travel hubs during the winter holiday season, Omicron became the dominant circulating variant within a very short time frame. In the context of the recent arrival and rapid dominance of the Omicron VOC in New York City (NYC), we compared the reported data for COVID‐19 transmission, dynamics, and genomic diversity of Omicron for NYC, compared with data from the United Kingdom (UK) due to its impressive sequencing coverage, and South Africa as an early epicenter of Omicron emergence (SA).
Omicron has attracted the attention of global authorities due to recent reports of incomplete immune escape (41 times reduction in neutralizing titers) from healthy, uninfected individuals who received (2 or 3) vaccine doses (Pfizer‐BioNtech). In addition, neutralizing activity was decreased in sera from patients vaccinated with two doses of Pfizer‐BioNtech or Moderna or heterologous AstraZeneca/BioNTech among others. 4 , 5 These findings may correlate with the observation that Omicron appears to be more contagious, but not more lethal. Recent evidence from South Africa suggests a reduced risk of hospitalization and a reduced risk of severe disease when compared to patient populations infected with the earlier Delta VOC. Given these features, we studied the number of cases, deaths, the effective reproduction number (R t is the average number of new infections caused by a single infected individual at time t in the partially susceptible population) and phylogenomic characteristics of Omicron viruses in NYC compared to UK and SA using publicly available epidemiological and genomic data until January 30, 2022. 6 , 7 , 8 , 9 , 10
Following identification of the first Omicron case in NYC at the Icahn School of Medicine at Mount Sinai on December 2, 2021 from a specimen collected on November 27, there has been a dramatic increase in new detections, reaching approximately 42 000 cases per day for NYC alone (Figure 1A). 6 Notably, the number of deaths has not increased at the rate seen during earlier waves, consistent with data from SA and the UK (Figure 1B,C). Although R t values increased in NYC, UK, and SA with the arrival of Omicron (Figure 1, right), but not the number of deaths and hospitalizations (Figure 1, left). 7 , 8 , 9 It is interesting to note that the magnitude of the peak caused by Omicron did not reflect the R t changes caused by other variants (i.e., Alpha or Delta), these other VOCs caused 5000 and 1500 cases per day, respectively, versus Omicron reaching over 40 000 daily cases, corroborating the high transmissibility of Omicron. In NYC, the high vaccination rate (74.2% fully vaccinated) may have helped prevent severe disease and hospitalizations in those infected (Figure 1). In contrast, in NYC, hospitalizations in children under 12‐year old have increased, highlighting the vulnerability of this high‐risk unvaccinated population to serious illness due to COVID‐19, as well the potential to jeopardize transmission dynamics. 11 , 12
Figure 1.
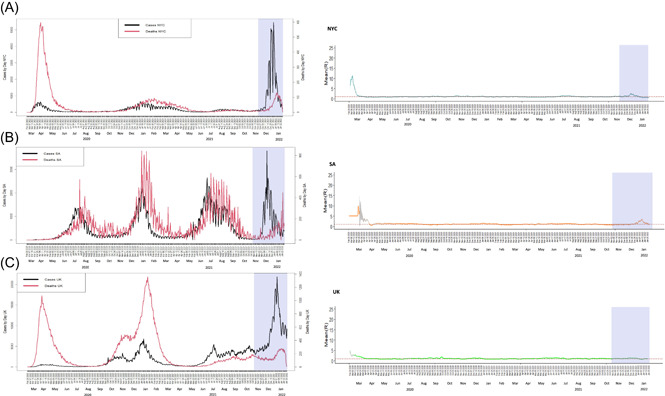
Number of cases (black line) and deaths (red line) reported over time from the start of the pandemic (first case reported in each geographical region) to January 28, in (A) New York, (B) South Africa, and (C) United Kingdom. For each case, the shaded region represents the time from the first case of the Omicron variant reported in GISAID to the cut‐off date (January 28). For the same period, we performed an evaluation to quantify transmissibility throughout an epidemic from the analysis of time series of incidence, as described in Cori et al. (2013), using Epiestim Package (WHO model). Results of modeling of R t were plotted to each case, (A‐NYC) New York, (B‐SA) South Africa, and (C‐UK) UK. For each case, the shaded region represents the time period from the first case of the Omicron variant reported in GISAID to the cut‐off date (January 28)
It is likely that high transmissibility and large numbers of infected persons has enabled the Omicron VOC to diversify rapidly into three sublineages: BA.1 reported in 122 countries, BA.2 reported in 54 countries, and BA.3 reported in 12 countries. We analyzed 18 680 publicly available genomes in GISAID from November 23 (putative arrival of Omicron in NYC) to January 28, 2022 and compared them with 306 genomes from UK, 1394 from SA and 2365 from representative lineages around the globe. All Omicron sequences clustered in a larger, well‐defined group from the NextClade reference genomes (representative of all SARS‐CoV‐2 lineages) (Figure 2A) and its corresponding sublineages. Most NYC genomes clustered with genomes from UK and SA, suggesting a putative introduction to NYC. When stratified by chronologic weeks, most genomes from November 23–27 (Week 1) are from SA, the geographic origin of Omicron. The number of reported genomes in the UK and NYC increase during the following weeks, and in the last week (January 28, 2022), no genomes from SA are reported (Figure 2B). This period subdivision highlights the sequencing effort of SA as the epicenter of Omicron emergence and the further spread of Omicron to UK and NYC.
Figure 2.
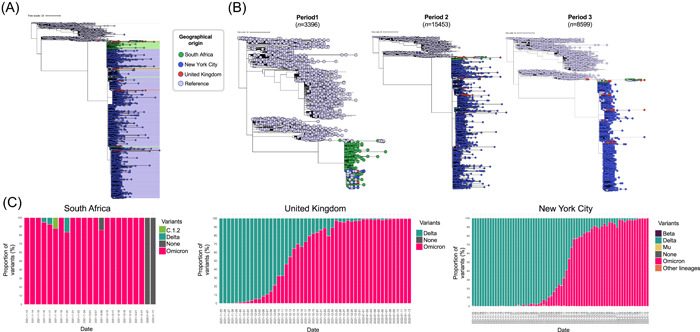
Phylogenomic relationships and temporal distribution of variants sequences in South Africa, United Kingdom and New York City. (A) Phylogenetic analysis of the Omicron variant sequences from South Africa (SA), United Kingdom (UK), and New York City (NY). A dataset with 22 745 Omicron genomes (from November 5 until January 28), consisting in 306 from UK (red dots), 1394 from SA (green dots), 18 680 from NY (blue dots) and 2365 reference genomes of other SARS‐CoV‐2 lineages (light blue) were used. (B) Temporal phylogenetic analysis of the Omicron variant sequences from SA, UK, and NY. Temporal comparison of Omicron genomes in three periods, Period 1 (n = 1031, November 5–December 3), Period 2 (n = 13 088, December 4–31), and Period 3 (n = 6234, January 3–28). (C) Proportion of SARS‐CoV‐2 variants in SA, UK, and NY
This rapid global dispersal from the place of first report reemphasizes the high transmissibility of Omicron, which needed only 2 weeks to become dominant in the UK and NYC (Figure 2C). In these areas, putative introductions of Omicron through travel from other countries were followed by rapid, adaptive community transmission, and/or by divergent transmissibility profiles. Now, robust phylogenomic studies are needed to rule out the putative cryptic introduction of Omicron in NYC.
In conclusion, current data indicate that Omicron is highly transmissible, with a high capability for immune evasion, but with relatively lower morbidity and mortality in comparison with earlier VOCs, particularly among vaccinated populations. Also, current data shows that it took only a couple of weeks to note a decrease in the number of cases. As the pandemic continues and SARS‐CoV‐2 continues to evolve, global authorities must prioritize equity in vaccine access and continued genomic surveillance. Future studies are still needed to fully unveil the biological properties of Omicron, but what is certain is that vaccination, large‐scale testing, and infection prevention efforts are the greatest arsenal against the COVID‐19 pandemic.
PSP (MOUNT SINAI PATHOGEN SURVEILLANCE) STUDY GROUP
H. Alshamarry, B. Alburquerque, A. Amoako, S. Aslam, C. Cognigni, M. Espinoza‐Moraga, K. Farrugia, A. S. Gonzalez‐Reiche, Z. Khalil, M. Laporte, I. Mena, J. Polanco, A. Rooker, L. Sominsky, and A. van de Guchte.
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
AUTHOR CONTRIBUTIONS
Juan D. Ramírez, Emilia M. Sordillo, and Matthew Hernández conceived the idea and wrote the manuscript. Juan D. Ramírez, Sergio Castañeda, Nathalia Ballesteros, Marina Muñoz, Radhika Banu, Paras Shrestha, Feng Chen, Huanzhi Shi, and PSP study group analyzed the data. Harm van Bakel, Viviana Simon, and Carlos Cordon‐Cardo provided information and valuable comments. All authors revised the final version of the manuscript.
Supporting information
Supporting information.
ACKNOWLEDGMENTS
The authors thank and acknowledge all authors and laboratories around the globe contributing their genomes on GISAID. All used genomes in these analyses are listed on Table S1. This publication is based on work supported by The Pershing Square Foundation for the Mount Sinai Health System COVID‐19 Saliva Testing Project.
Ramírez JD, Castañeda S, Ballesteros N, et al. Hotspots for SARS‐CoV‐2 Omicron variant spread: Lessons from New York City. J Med Virol. 2022;94:2911‐2914. 10.1002/jmv.27691
REFERENCES
- 1.GISAID ‐ Initiative. Accessed December 25, 2021. https://www.gisaid.org/
- 2. Shu Y, McCauley J. GISAID: Global Initiative on Sharing All Influenza Data—from vision to reality. Euro Surveill. 2017;22:30494. 10.2807/1560-7917.ES.2017.22.13.30494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. VanBlargan LA, Errico JM, Halfmann PJ, et al. An infectious SARS‐CoV‐2 B.1.1.529 Omicron virus escapes neutralization by therapeutic monoclonal antibodies. Nat Med. 2022. 10.1038/s41591-021-01678-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Garcia‐Beltran WF, St Denis KJ, Hoelzemer A, et al. mRNA‐based COVID‐19 vaccine boosters induce neutralizing immunity against SARS‐CoV‐2 Omicron variant. Cell. 2022;185:457‐466. 10.1016/J.CELL.2021.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lippi G, Mattiuzzi C, Henry BM. Neutralizing potency of COVID‐19 vaccines against the SARS‐CoV‐2 Omicron (B.1.1.529) variant. J Med Virol. 2022. 10.1002/JMV.27575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.COVID‐19 Daily Counts of Cases, Hospitalizations, and Deaths | NYC Open Data. Accessed December 25, 2021. https://data.cityofnewyork.us/Health/COVID-19-Daily-Counts-of-Cases-Hospitalizations-an/rc75-m7u3
- 7.Coronavirus (COVID‐19) Cases ‐ Our World in Data. Accessed December 25, 2021. https://ourworldindata.org/covid-cases
- 8.COVID‐19 South African Coronavirus News And Information. Accessed December 25, 2021. https://sacoronavirus.co.za/
- 9.Cases in the UK | Coronavirus in the UK. Accessed December 25, 2021. https://coronavirus.data.gov.uk/details/cases
- 10.COVID‐19 Data in New York | Department of Health. Accessed December 25, 2021. https://coronavirus.health.ny.gov/covid-19-data-new-york
- 11.COVID‐19: Data on Vaccines ‐ NYC Health. Accessed Accessed December 25, 2021. https://www1.nyc.gov/site/doh/covid/covid-19-data-vaccines.page#doses
- 12.Coronavirus (COVID‐19) Vaccinations ‐ Humanitarian Data Exchange. Accessed December, 25 2021. https://data.humdata.org/dataset/covid-19-vaccinations
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.


