Abstract
Although individuals with coronavirus disease 2019 (COVID‐19) are known to be at increased risk for other conditions resulting from pathogenic changes (including metaplastic or anaplastic) in the lungs and other organs and organ systems, it is still unknown whether COVID‐19 affects childhood intelligence. The present two‐sample Mendelian randomization study aims to identify the genetic causal link between COVID‐19 and childhood intelligence. Four COVID‐19 genetic instrumental variants (IVs) were chosen from the largest genome‐wide association studies (GWAS) for COVID‐19 (hospitalized vs. population) (6406 cases and 902 088 controls of European ancestry). The largest childhood intelligence GWAS (n = 12 441 individuals of European ancestry) was used to evaluate the effect of the identified COVID‐19‐associated genetic IVs on childhood intelligence. We found that as the genetic susceptibility to COVID‐19 increased, childhood intelligence followed a decreasing trend, according to mr_egger (β = −0.156; p = 0.601; odds ratio [OR] = 0.856; 95% confidence interval [CI]: 0.522–1.405), simple mode (β = −0.126; p = 0.240; OR = 0.882; 95% CI: 0.745–1.044), and weighted mode (β = −0.121; p = 0.226; OR = 0.886; 95% CI: 0.758–1.036) analyses. This trend was further demonstrated by the weighted median (β = −0.134; p = 0.031; OR = 0.875; 95% CI: 0.774–0.988) and the inverse variance weighted (β = −0.152; p = 0.004; OR = 0.859; 95% CI: 0.776–0.952). Our analysis suggests a causal link between genetically increased COVID‐19 and decreased childhood intelligence. Thus, COVID‐19 may be a risk factor for declines in childhood intelligence.
Keywords: childhood intelligence, COVID‐19 pandemic, genetic variants, genome‐wide association study, Mendelian randomization
1. INTRODUCTION
In late 2019, severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2)‐induced respiratory distress syndrome named coronavirus disease 2019 (COVID‐19) was identified in humans. 1 The ongoing COVID‐19 pandemic has affected millions worldwide and continues to present a serious public health threat. 2 Researchers have estimated that 30% of COVID‐19 survivors may be affected by the long‐term consequences of the illness, known as post‐COVID‐19 condition after they recover from acute illness. 3
COVID‐19 and cancer demonstrate some clinical and molecular similarities in the four major signaling pathways: immune checkpoint signaling, type I interferon, cytokine, and androgen receptor. 4 Histopathological analyses revealed diffuse alveolar damage including viral cytopathic changes, metaplastic epithelial changes, intra‐alveolar hemorrhaging, and pulmonary edema. These studies suggest that COVID‐19 infection may result in an increased risk of other diseases such as lung cancer.
COVID‐19 can infect people of ages, including children. 5 Children comprise a small percentage of the total number of COVID‐19 cases and usually present with milder symptoms than adults. 5 Only 6% of pediatric cases have been classified as severe and critical. 6 The symptoms of severe and critical COVID‐19 are acute respiratory distress syndrome, hypoxia (defined as blood oxygen saturation of less than 92%), shock, and the failure of organs such as the heart and kidneys. 5 , 6 Children with both mild and severe COVID‐19 have experienced long‐term symptoms including tiredness, fatigue, headache, trouble sleeping, trouble concentrating, muscle and joint pain, and cough. In addition, the levels of cardiac troponin, a marker of myocardial (heart muscle) injury, are elevated in an unexpected number of patients hospitalized with COVID‐19. 7 One underexplored area of long COVID is how COVID‐19 may affect children's intelligence.
Using genetic variants independent of many factors that bias observational studies, Mendelian randomization (MR) studies have many advantages in assessing the causal association between an exposure and an outcome. 8 , 9 , 10 , 11 , 12 , 13 Thus, we used a two‐sample MR study to identify the causal genetic link between COVID‐19 and childhood intelligence.
2. METHODS
2.1. COVID‐19 genetic instrumental variants (IVs)
The largest GWAS for COVID‐19 (hospitalized vs. population) RELEASE 4 was described by the COVID‐19 Host Genetics Initiative in 2020. 14 This GWAS data set is based on 6406 cases and 902 088 controls with European ancestry. The summary data set is available in https://gwas.mrcieu.ac.uk/datasets/ebi-a-GCST010779. Four independent COVID‐19 genetic IVs were obtained based on the following three criteria: (1) genome‐wide significance threshold p < 5 × 10−8; (2) r 2 < 0.001, indicating no linkage disequilibrium between single‐nucleotide polymorphisms (SNPs); (3) no effects on other potential risk factors including body mass index, smoking, and blood pressure. Detailed information about these IVs is shown in Table 1.
Table 1.
COVID‐19 genetic IVs.
| SNP | EA | NEA | EAF | β | SE | p Value |
|---|---|---|---|---|---|---|
| rs2166172 | C | A | 0.410 | 0.124 | 0.022 | 2.74E−08 |
| rs143334143 | A | G | 0.088 | 0.229 | 0.036 | 1.28E−10 |
| rs2269899 | T | C | 0.685 | 0.124 | 0.022 | 3.24E−08 |
| rs13050728 | C | T | 0.649 | −0.153 | 0.023 | 1.91E−11 |
Abbreviations: β, the regression coefficient based on the COVID‐19 effect allele; COVID‐19, coronavirus disease 2019; EA, effect allele; EAF, effect allele frequency; IVs, instrumental variants; NEA, non‐effect allele; SE, standard error; SNP, single‐nucleotide polymorphism.
2.2. Childhood intelligence GWAS
Benyamin et al. 15 described the largest GWAS for childhood. This GWAS consists of 12 441 individuals of European ancestry. The profile of this GWAS is provided in Table 2. The summary data set is available at https://gwas.mrcieu.ac.uk/datasets/ebi-a-GCST001837.
Table 2.
GWAS for childhood intelligence.
| GWAS ID | Year | Trait | Sample size | nsnp | Population | PMID |
|---|---|---|---|---|---|---|
| ebi‐a‐GCST001837 | 2013 | Intelligence (childhood) | 12 441 | 1 374 543 | European | 23358156 |
Abbreviations: GWAS, genome‐wide association study; GWAS ID, GWAS identity; nsnp, the number of single‐nucleotide polymorphism; PMID, PubMed unique identifier.
2.3. Association of COVID‐19 genetic IVs with childhood intelligence GWAS
Potential proxy SNPs were identified by the LD proxy tool (r 2 > 0.8) when COVID‐19 genetic IVs could not be found in childhood intelligence summary statistics. However, we were able to successfully extract four independent COVID‐19 genetic IVs from the childhood intelligence GWAS summary data set. The association of four independent COVID‐19 genetic IVs with childhood intelligence GWAS is shown in Table 3.
Table 3.
Association of COVID‐19 genetic IVs with childhood intelligence GWAS.
| SNP | Exposure (COVID‐19) GWAS | Outcome (childhood intelligence) GWAS | ||||
|---|---|---|---|---|---|---|
| β | SE | p Value | β | SE | p Value | |
| rs13050728 | −0.153 | 0.023 | 1.91E−11 | 0.017 | 0.014 | 0.227 |
| rs143334143 | 0.229 | 0.036 | 1.28E−10 | −0.041 | 0.026 | 0.113 |
| rs2166172 | 0.124 | 0.022 | 2.74E−08 | −0.015 | 0.013 | 0.262 |
| rs2269899 | 0.124 | 0.022 | 3.24E−08 | −0.027 | 0.014 | 0.048 |
Abbreviations: β, the regression coefficient based on COVID‐19 raising effect allele; COVID‐19, coronavirus disease 2019; GWAS, genome‐wide association study; IVs, instrumental variants; SE, standard error; SNP, single‐nucleotide polymorphism.
2.4. Pleiotropy test
Both mr_egger_intercept and PRESSO methods 16 , 17 were used to test the pleiotropy of four independent COVID‐19 genetic IVs in the childhood intelligence GWAS data set. The results of the pleiotropy test are shown in Table 4. A p value > 0.05 represents no significant pleiotropy of the four independent COVID‐19 genetic IVs in childhood intelligence GWAS.
Table 4.
Pleiotropy and heterogeneity test of COVID‐19 genetic IVs in childhood intelligence GWAS.
| Pleiotropy test | Heterogeneity test | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| mr_egger | PRESSO | mr_egger | IVW | ||||||
| Intercept | SE | p Value | p Value | Q | Q_df | p Value | Q | Q_df | p Value |
| 0.001 | 0.037 | 0.989 | 0.896 | 0.676 | 2 | 0.713 | 0.677 | 3 | 0.879 |
Note: p value > 0.05 represent no significant pleiotropy. Q_p value > 0.05 represents no significant heterogeneity.
Abbreviations: COVID‐19, coronavirus disease 2019; GWAS, genome‐wide association study; IVs, instrumental variants; IVW, inverse variance weighted; MR, Mendelian randomization; SE, standard error.
2.5. Heterogeneity test
Both mr_egger and inverse variance weighted (IVW) in Cochran's Q statistic 18 , 19 were used to test the heterogeneity of four independent COVID‐19 genetic IVs in the childhood intelligence GWAS data set. The results of the heterogeneity test are shown in Table 4. A p value > 0.05 represents no significant heterogeneity in the four independent COVID‐19 genetic IVs in the childhood intelligence GWAS.
2.6. MR analysis
mr_egger, weighted median, IVW, simple mode, and weighted mode methods 16 , 20 , 21 were used to analyze the causal association of COVID‐19 with childhood intelligence. The results of the MR analysis are shown in Table 5. A p value < 0.05 represents a causal association of COVID‐19 with childhood intelligence.
Table 5.
The causal association of COVID‐19 with childhood intelligence.
| Method | nsnp | β | SE | p Value | OR | OR_lci95 | OR_uci95 |
|---|---|---|---|---|---|---|---|
| mr_egger | 4 | −0.156 | 0.253 | 0.601 | 0.856 | 0.522 | 1.405 |
| Weighted median | 4 | −0.134 | 0.062 | 0.031 | 0.875 | 0.774 | 0.988 |
| IVW | 4 | −0.152 | 0.052 | 0.004 | 0.859 | 0.776 | 0.952 |
| Simple mode | 4 | −0.126 | 0.086 | 0.240 | 0.882 | 0.745 | 1.044 |
| Weighted mode | 4 | −0.121 | 0.080 | 0.226 | 0.886 | 0.758 | 1.036 |
Note: p < 0.05 represents the causal association of the increased COVID‐19 with childhood intelligence.
Abbreviations: β, the regression coefficient based on COVID‐19 raising effect allele; COVID‐19, coronavirus disease 2019; IVW, inverse variance weighted; MR, Mendelian randomization; nsnp, number of single‐nucleotide polymorphism; OR, odds ratio; OR_lci95, lower limit of 95% confidence interval for OR; OR_uci95, upper limit of 95% confidence interval for OR; SE, standard error.
2.7. Single SNP effect analysis
“mr” and “mr_scatter_plot” were used to test the individual putative causal effects of COVID‐19 on childhood intelligence (Figure 1). “mr_singlesnp” and “mr_forest_plot” were used to determine single SNP effect size for COVID‐19 on childhood intelligence (Figure 2). “mr_singlesnp” and “mr_leaveoneout_plot” were used to analyze the effect of leave‐one‐out of the four independent COVID‐19 genetic IVs of childhood intelligence (Figure 3).
Figure 1.
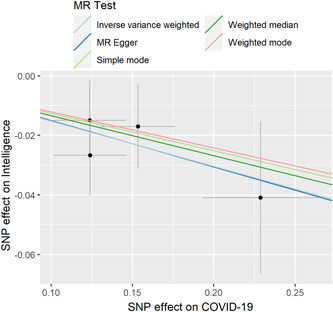
Individual estimates about the putative causal effect of COVID‐19 on childhood intelligence. The x‐axis shows the SNP effect and SE on each of COVID‐19 IVs. The y‐axis shows the SNP effect and SE on childhood intelligence. The regression line for mr_egger, weighted median, IVW, simple mode, and weighted mode is shown. COVID‐19, coronavirus disease 2019; IV, instrumental variant; IVW, inverse variance weighted; MR, Mendelian randomization; SE, standard error; SNP, single‐nucleotide polymorphism.
Figure 2.
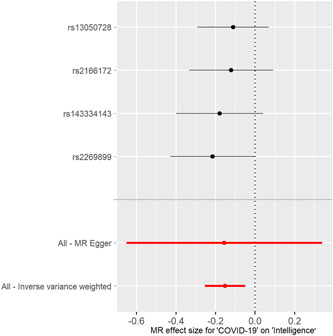
Forest plot of COVID‐19 associated with childhood intelligence. The x‐axis shows MR effect size for COVID‐19 on childhood intelligence. The y‐axis shows the analysis for each of SNPs. COVID‐19, coronavirus disease 2019; MR, Mendelian randomization; SNP, single‐nucleotide polymorphism.
Figure 3.
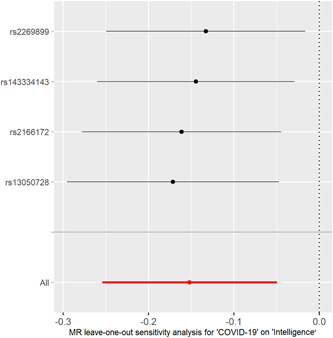
MR leave‐one‐out sensitivity analysis for the effect of COVID‐19 SNPs on childhood intelligence. The x‐axis shows MR leave‐one‐out sensitivity analysis for COVID‐19 on childhood intelligence. The y‐axis shows the analysis for the effect of leave‐one‐out of SNPs on childhood intelligence. COVID‐19, coronavirus disease 2019; MR, Mendelian randomization; SNP, single‐nucleotide polymorphism.
3. RESULTS
3.1. No significant pleiotropy or heterogeneity was observed among the four COVID‐19 genetic IVs
Four COVID‐19 genetic IVs (Table 1) were successfully extracted from the childhood intelligence GWAS data set (Table 2). The association of the four COVID‐19 genetic IVs with childhood intelligence GWAS is shown (Table 3).
Both mr_egger_intercept and PRESSO methods suggested no significant pleiotropy in the four independent COVID‐19 genetic IVs in the childhood intelligence GWAS (Table 4). Both mr_egger and IVW in Cochran's Q statistic showed no significant heterogeneity in the four independent COVID‐19 genetic IVs in the childhood intelligence GWAS (Table 4). Therefore, all selected COVID‐19‐associated genetic variants can be taken as the effective IVs in this MR study.
3.2. COVID‐19 genetically reduces childhood intelligence
We found that as COVID‐19 genetically increased, childhood intelligence had an decreased trend using mr_egger (β = −0.156; p = 0.601; odds ratio [OR] = 0.856; 95% confidene interval [CI]: 0.522–1.405), simple mode (β = −0.126; p = 0.240; OR = 0.882; 95% CI: 0.745–1.044), and weighted mode (β =−0.121; p = 0.226; OR = 0.886; 95% CI: 0.758–1.036) (Table 5). This trend was further demonstrated by weighted median (β =−0.134; p = 0.031; OR = 0.875; 95% CI: 0.774–0.988) and IVW (β = −0.152; p = 0.004; OR = 0.859; 95% CI: 0.776–0.952) (Table 5). Altogether, our analysis suggests a causal link between genetically increased COVID‐19 and reduced childhood intelligence.
3.3. Single COVID‐19 SNP effect is robust without obvious bias
The individual MR estimates (mr_egger, weighted median, IVW, simple mode, and weighted mode) demonstrate that as the effect of a single SNP on COVID‐19 increased, the suppressive effect of a single SNP on childhood intelligence increased (Figure 1). All effect size analyses suggest that each effect of COVID‐19 SNPs on childhood intelligence was robust (Figure 2). MR leave‐one‐out sensitivity analysis suggested that removing a specific SNP of the four COVID‐19 SNPs did not change the results (Figure 3). Altogether, these results indicate that our data were robust without obvious bias.
4. DISCUSSION
The present MR study showed that genetic predisposition to a higher COVID‐19 was genetically associated with reduced childhood intelligence. Thus, to protect childhood intelligence, COVID‐19 prevention in children is essential.
Although COVID‐19 primarily attacks the lungs, it can also affect many other organs such as the brain, heart, and kidneys. One previous study provided objective neuroimaging evidence for the coexistence of recoverable and long‐term unrecovered changes in the brain at 10‐month after acute COVID‐19. 22 Moreover, COVID‐19 has been found to lead to psychiatric illness. 23 Our study suggests that genetic predisposition to a higher COVID‐19 reduced childhood intelligence.
The human brain develops over a prolonged period. 24 , 25 , 26 The infant's brain is relatively immature with simultaneous traits of competence and vulnerability. 26 It possesses an immense capacity to learn, remodel, and adapt, but is vulnerable and sensitive to environmental exposures. 26 , 27 , 28 , 29 , 30 Neurodevelopmental processes such as myelination and synaptogenesis and external cues such as maternal interaction and physical skin‐to‐skin “kangaroo” care play important roles in optimal brain development. 26 , 31 , 32 , 33 , 34 The brain's adaptive plasticity is promoted by positive and enriching environments 26 , 35 , 36 , 37 , 38 , 39 and impaired by neglect, insecurity, stress, and lack of stimulation. 26 , 40 , 41 , 42 Our results suggest that COVID‐19 genetically reduces childhood intelligence. Thus, the brain development of infants and young children may be impaired by COVID‐19 infection.
MR studies use genetic variants independent of many factors that bias observational studies in assessing the causal association of an exposure with an outcome. 8 , 9 , 10 , 11 , 12 Thus, the genetical effect of COVID‐19 on childhood intelligence, identified by our present MR study, is not related to the lack of a stimulating environment during the pandemic but to COVID‐19 infection. There is clear evidence that SARS‐CoV‐2 could infect neurons in the brain organoids, killing some and reducing the formation of synapses. 43 , 44 Animal studies have proposed that SARS‐CoV‐2 can infect cerebrovascular endothelium and brain parenchyma via the angiotensin‐converting enzyme 2 receptor, resulting in COVID‐19‐related neuronal damage via apoptosis and necrosis. 45 In addition, SARS‐CoV‐2‐induced immune‐mediated demyelinating disease, cerebrovascular damage, neurodegeneration, and depression are among the neurological complications described. 46 Thus, COVID‐19 infection may reduce childhood intelligence by SARS‐CoV‐2‐induced neurological complications via direct damage and indirect immune responses.
One strength of our study is that because both COVID‐19 genetic IVs and childhood intelligence GWAS were from populations with European ancestries, it removed the influence of population stratification. Second, four different analytical methods demonstrated no significant pleiotropy or heterogeneity in severe COVID‐19 genetic IVs. Third, all five MR analyses (mr_egger, weighted median, IVW, simple mode, and weighted mode) showed the causal link between genetically increased COVID‐19 and decreased childhood intelligence.
This study has several limitations. First, because four independent COVID‐19 genetic IVs and childhood intelligence GWAS are from European ancestry, our conclusions cannot necessarily be extrapolated to populations of other ancestries. Second, randomized controlled trials are necessary to clarify whether COVID‐19 can reduce childhood intelligence.
In conclusion, although our analysis suggests a causal link between genetically increased COVID‐19 and reduced childhood intelligence, it is necessary for future studies to examine the mechanism underlying this link.
AUTHOR CONTRIBUTIONS
Renxi Wang conceived, initiated, and supervised the project. Gaizhi Zhu, Shan Zhou, Yaqi Xu, Ran Gao, and Huan Li collected and analyzed the data. Wenting Su and Gencheng Han contributed to the interpretation of the results. Renxi Wang wrote a draft of the manuscript. All authors critically reviewed and revised the manuscript, and agreed to the published version of the manuscript.
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
ETHICS STATEMENT
This study was approved by the Ethics Committee of Beijing Institute of Brain Disorders in Capital Medical University and the ethical code number is AEEI‐2021‐077.
ACKNOWLEDGMENTS
This study was supported by the National Natural Science Foundation of China (82071758 and 31770956). The authors would like to thank ieu open gwas project (https://gwas.mrcieu.ac.uk/datasets/) for providing summary results data for these analyses.
Zhu G, Zhou S, Xu Y, et al. Mendelian randomization study on the causal effects of COVID‐19 on childhood intelligence. J Med Virol. 2022;94:3233‐3239. 10.1002/jmv.27736
Gaizhi Zhu and Shan Zhou contributed equally to this study as the first authors.
Contributor Information
Wenting Su, Email: wtsu@ccmu.edu.cn.
Gencheng Han, Email: genchenghan@163.com.
Renxi Wang, Email: renxi_wang@ccmu.edu.cn.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are openly available in ieu open gwas project at https://gwas.mrcieu.ac.uk/datasets/ebi-a-GCST001837 for childhood intelligence GWAS and https://gwas.mrcieu.ac.uk/datasets/ebi-a-GCST010779 for COVID‐19 GWAS. The MR analysis code can be found at https://mrcieu.github.io/TwoSampleMR/articles/index.html.
REFERENCES
- 1. Rai P, Kumar BK, Deekshit VK, Karunasagar I, Karunasagar I. Detection technologies and recent developments in the diagnosis of COVID‐19 infection. Appl Microbiol Biotechnol. 2021;105(2):441‐455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Majumder J, Minko T. Recent developments on therapeutic and diagnostic approaches for COVID‐19. AAPS J. 2021;23(1):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lechner‐Scott J, Levy M, Hawkes C, Yeh A, Giovannoni G. Long COVID or post COVID‐19 syndrome. Mult Scler Relat Disord. 2021;55:103268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zong Z, Wei Y, Ren J, Zhang L, Zhou F. The intersection of COVID‐19 and cancer: signaling pathways and treatment implications. Mol Cancer. 2021;20(1):76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Galindo R, Chow H, Rongkavilit C. COVID‐19 in children: clinical manifestations and pharmacologic interventions including vaccine trials. Pediatr Clin North Am. 2021;68(5):961‐976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dong Y, Mo X, Hu Y, et al. Epidemiology of COVID‐19 among children in China. Pediatrics. 2020;145(6):e20200702. [DOI] [PubMed] [Google Scholar]
- 7. Abbasi J. Researchers investigate what COVID‐19 does to the heart. JAMA. 2021;325(9):808‐811. [DOI] [PubMed] [Google Scholar]
- 8. Gao R, Xu Y, Zhu G, et al. Genetic variation associated with COVID‐19 is also associated with endometrial cancer. J Infect. 2022. 10.1016/j.jinf.2022.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang R. Genetic variation of interleukin‐1 receptor type 1 is associated with severity of COVID‐19 disease. J Infect. 2021;84(2):e19‐e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang R. Mendelian randomization study updates the effect of 25‐hydroxyvitamin D levels on the risk of multiple sclerosis. J Transl Med. 2022;20(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xu Y, Gao R, Zhu G, et al. Genetic variation of allergic disease is associated with the susceptibility to COVID‐19. J Infect. 2022. 10.1016/j.jinf.2022.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhu G, Zhou S, Xu Y, et al. Mendelian randomization study on the causal effects of omega‐3 fatty acids on rheumatoid arthritis. Clin Rheumatol. 2022. 10.1007/s10067-022-06052-y [DOI] [PubMed] [Google Scholar]
- 13. Zhou S, Zhu G, Xu Y, et al. Mendelian randomization study on the putative causal effects of omega‐3 fatty acids on low back pain. Front Nutr. 2022;9:819635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. COVID‐19 Host Genetics Initiative . The COVID‐19 host genetics initiative, a global initiative to elucidate the role of host genetic factors in susceptibility and severity of the SARS‐CoV‐2 virus pandemic. Eur J Hum Genet. 2020;28(6):715‐718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Benyamin B, Pourcain B, Davis OS, et al. Childhood intelligence is heritable, highly polygenic and associated with FNBP1L. Mol Psychiatry. 2014;19(2):253‐258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50(5):693‐698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu H, Zhang Y, Zhang H, et al. Effect of plasma vitamin C levels on Parkinson's disease and age at onset: a Mendelian randomization study. J Transl Med. 2021;19(1):221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Greco MF, Minelli C, Sheehan NA, Thompson JR. Detecting pleiotropy in Mendelian randomisation studies with summary data and a continuous outcome. Stat Med. 2015;34(21):2926‐2940. [DOI] [PubMed] [Google Scholar]
- 19. Liu G, Zhang S, Cai Z, et al. PICALM gene rs3851179 polymorphism contributes to Alzheimer's disease in an Asian population. Neuromolecular Med. 2013;15(2):384‐388. [DOI] [PubMed] [Google Scholar]
- 20. Liu G, Zhao Y, Jin S, et al. Circulating vitamin E levels and Alzheimer's disease: a Mendelian randomization study. Neurobiol Aging. 2018;72(189):e1‐e9. [DOI] [PubMed] [Google Scholar]
- 21. Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR‐Egger method. Eur J Epidemiol. 2017;32(5):377‐389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tian Tian JW, Chen Tao, Li Jia, et al. Long‐term follow‐up of dynamic brain changes in patients recovered from COVID‐19 without neurological manifestations. JCI Insight. 2022;7(4):e155827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xia J, Chen S, Li Y, et al. Immune response is key to genetic mechanisms of SARS‐CoV‐2 infection with psychiatric disorders based on differential gene expression pattern analysis. Front Immunol. 2022;13:798538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Casey BJ, Giedd JN, Thomas KM. Structural and functional brain development and its relation to cognitive development. Biol Psychol. 2000;54(1‐3):241‐257. [DOI] [PubMed] [Google Scholar]
- 25. Johnson MH. Functional brain development in humans. Nat Rev Neurosci. 2001;2(7):475‐483. [DOI] [PubMed] [Google Scholar]
- 26. Deoni SC, Beauchemin J, Volpe A, Da Sa V, RESONANCE Consortium . Impact of the COVID‐19 pandemic on early child cognitive development: initial findings in a longitudinal observational study of child health. medRxiv. 2021. 10.1101/2021.08.10.21261846 [DOI] [Google Scholar]
- 27. Luby J, Belden A, Botteron K, et al. The effects of poverty on childhood brain development: the mediating effect of caregiving and stressful life events. JAMA Pediatr. 2013;167(12):1135‐1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Monk C, Georgieff MK, Osterholm EA. Research review: maternal prenatal distress and poor nutrition—mutually influencing risk factors affecting infant neurocognitive development. J Child Psychol Psychiatry. 2013;54(2):115‐130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Blair C, Raver CC. Poverty, stress, and brain development: new directions for prevention and intervention. Acad Pediatr. 2016;16(3, suppl):S30‐S36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pulli EP, Kumpulainen V, Kasurinen JH, et al. Prenatal exposures and infant brain: review of magnetic resonance imaging studies and a population description analysis. Hum Brain Mapp. 2019;40(6):1987‐2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kolb B. Brain and behavioural plasticity in the developing brain: neuroscience and public policy. Paediatr Child Health. 2009;14(10):651‐652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. He S, Ma J, Liu N, Yu X. Early enriched environment promotes neonatal GABAergic neurotransmission and accelerates synapse maturation. J Neurosci. 2010;30(23):7910‐7916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fields RD. A new mechanism of nervous system plasticity: activity‐dependent myelination. Nat Rev Neurosci. 2015;16(12):756‐767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ismail FY, Fatemi A, Johnston MV. Cerebral plasticity: Windows of opportunity in the developing brain. Eur J Paediatr Neurol. 2017;21(1):23‐48. [DOI] [PubMed] [Google Scholar]
- 35. Dobbing J. The influence of early nutrition on the development and myelination of the brain. Proc R Soc Lond Ser B. 1964;159:503‐509. [DOI] [PubMed] [Google Scholar]
- 36. Pollitt E. Poverty and child development: relevance of research in developing countries to the United States. Child Dev. 1994;65(Spec No. 2):283‐295. [PubMed] [Google Scholar]
- 37. Bradley RH, Corwyn RF. Socioeconomic status and child development. Annu Rev Psychol. 2002;53:371‐399. [DOI] [PubMed] [Google Scholar]
- 38. Swain JE, Lorberbaum JP, Kose S, Strathearn L. Brain basis of early parent–infant interactions: psychology, physiology, and in vivo functional neuroimaging studies. J Child Psychol Psychiatry. 2007;48(3‐4):262‐287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Poulain T, Vogel M, Kiess W. Review on the role of socioeconomic status in child health and development. Curr Opin Pediatr. 2020;32(2):308‐314. [DOI] [PubMed] [Google Scholar]
- 40. Rees S, Inder T. Fetal and neonatal origins of altered brain development. Early Hum Dev. 2005;81(9):753‐761. [DOI] [PubMed] [Google Scholar]
- 41. Bonnier C. Evaluation of early stimulation programs for enhancing brain development. Acta Paediatr. 2008;97(7):853‐858. [DOI] [PubMed] [Google Scholar]
- 42. de Oliveira KHD, de Almeida GM, Gubert MB, et al. Household food insecurity and early childhood development: systematic review and meta‐analysis. Matern Child Nutr. 2020;16(3):e12967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Marshall M. How COVID‐19 can damage the brain. Nature. 2020;585(7825):342‐343. [DOI] [PubMed] [Google Scholar]
- 44. Mukerji SS, Solomon IH. What can we learn from brain autopsies in COVID‐19? Neurosci Lett. 2021;742:135528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Aghagoli G, Gallo Marin B, Katchur NJ, Chaves‐Sell F, Asaad WF, Murphy SA. Neurological involvement in COVID‐19 and potential mechanisms: a review. Neurocrit Care. 2021;34(3):1062‐1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mahalakshmi AM, Ray B, Tuladhar S, et al. Does COVID‐19 contribute to development of neurological disease? Immun Inflamm Dis. 2021;9(1):48‐58. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are openly available in ieu open gwas project at https://gwas.mrcieu.ac.uk/datasets/ebi-a-GCST001837 for childhood intelligence GWAS and https://gwas.mrcieu.ac.uk/datasets/ebi-a-GCST010779 for COVID‐19 GWAS. The MR analysis code can be found at https://mrcieu.github.io/TwoSampleMR/articles/index.html.


