Abstract
Pooled data from 2352 hospitalized coronavirus disease 2019 (COVID‐19) patients with viral RNA in feces across 46 studies were analyzed and the pooled prevalence of fecal RNA was 46.8% (95% confidence interval [CI]: 0.383–0.554). The pooled analysis showed that the occurrence of total gastrointestinal (GI) symptoms was 28.5% (95% CI: 0.125–0.44) in COVID‐19 patients with fecal RNA, that of both respiratory and GI symptoms was 21.9% (95% CI: 0.09–0.346), that of only GI symptoms was 19.8% (95% CI: 0.107–0.288), and that of only respiratory symptoms was 50.5%(95% CI: 0.267–0.744). The pooled data showed no significant difference in positive fecal RNA between severe and nonsevere cases (odds ratio = 2.009, p = 0.079, 95% CI: 0.922–4.378). During hospital admission, after samples from the respiratory system tested negative for viral RNA, 55.4% (95% CI: 0.418–0.669) of the patients with positive fecal RNA had persistent shedding of fecal RNA and pooled results from the other 4 studies including 848 discharged patients with nucleic acid‐negative stool samples indicated that the occurrence of repositive stool swabs was 18.1% (95% CI: 0.028–0.335), that of repositive respiratory swabs was 22.8% (95% CI: 0.003–0.452), that of both repositive stool and respiratory swabs was 19.1% (95% CI: 0.019–0.363), and that of only repositive stool swabs was 9.6% (95% CI: 0.010–0.203). The digestive tract may be an important organ involved in COVID‐19 infection and in the excretion of the virus. Because of the potential risk of fecal–oral transmission, giving emphasis on stool swab tests can help increase the detection rate of asymptomatic carriers and reduce missed diagnoses.
Keywords: COVID‐19, fecal, meta‐analysis, systematic review
1. INTRODUCTION
The emergence of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) is still an ongoing global health crisis due to the coronavirus disease 2019 (COVID‐19), with severe threats to public health because of a very high transmissibility rate. Apart from respiratory symptoms, gastrointestinal (GI) manifestations are common in patients with COVID‐19 and, in some cases, GI symptoms may precede the respiratory symptoms. 1 , 2 , 3 The positive detection of RNA from SARS‐CoV‐2 in feces suggests that the virus can replicate and exist in the digestive tract. 4 , 5 It was subsequently found that SARS‐CoV‐2 binds to the angiotensin‐converting enzyme 2 (ACE2) expressed in the upper esophagus and stratified epithelial cells, and absorptive enterocytes from the ileum and colon, which is the entry point for the virus to the epithelial cells. 6 , 7 The presence of new mutations may enable an increase in the viral tropism of the digestive tract. 8 At present, the GI symptoms in different studies are quite different in patients with COVID‐19 with nucleic acid‐positive stool samples, 9 , 10 , 11 which poses an important diagnostic challenge to clinicians on initial presentation. The presence of SARS‐CoV‐2 in stool samples and the potential of fecal–oral transmission is critical for our understanding of COVID‐19; therefore, more attention should be given to these patients.
In addition, the recurrence of SARS‐CoV‐2 viral RNA in patients makes the pandemic more complex and some countries are facing a resurgence of the disease. This increases healthcare costs and the financial burden to families and societies. As viral loads in stool and perianal swabs appear to decline slower than in throat swabs, 12 the concern is the infectivity of SARS‐CoV‐2 in feces in the late stages of infection and recurrent viral RNA positivity in recovered COVID‐19 patients. It is unclear whether patients with COVID‐19 with positive long‐term fecal nucleic acid tests have the risk of infection. Thus, we performed a systematic review and meta‐analysis of studies reporting the disease course in patients with COVID‐19 with nucleic acid‐positive stool samples and recurrence of SARS‐CoV‐2 viral RNA in stool samples. This might help inform public health protocols for contact tracing and quarantine.
2. METHODS
2.1. Information sources and literature search
Three databases including PubMed, EMBASE, and the Cochrane Library were systematically searched from the inception of the databases to May 15, 2021. A principal electronic search strategy was developed for PubMed and then applied to the other databases. This search was done in two parts and the following search terms alone or matched with the Boolean operators “AND” or “OR” were used: “diarrhea,” “gastrointestinal,” “digestive,” “feces,” “fecal,” “stool,” “rectal swab,” “anal swab,” “COVID‐19,” “severe acute respiratory syndrome coronavirus 2,” “SARS‐CoV‐2,” “novel coronavirus,” “2019‐nCoV,” “recurrence,” “discharge,” and “recovery.” No language or geographic restrictions were imposed. We focused on full‐text articles, but abstracts were considered if relevant. In addition, relevant review articles and references were examined for thorough assessment for existing literature. All articles were managed with Endnote X9.2 (Thompson and Reuters)/EndNote(version X9.2) and duplicates were removed.
2.2. Election criteria
Two reviewers (ZJQ and LGX) independently screened the titles and abstracts according to these eligibility criteria. A third reviewer (HXL) subsequently reviewed the full‐text articles and identified articles for inclusion. Disagreement was discussed and subsequently resolved via consensus. The inclusion criteria included the following: (1) study population: COVID‐19 patients (including adult or pediatric patients and pregnant women) provided data on stool/anal/perianal viral RNA; (2) study design: case series, prospective/retrospective cohort study, case–control study, and randomized controlled trials. There was no language restriction. The exclusion criteria were small studies (N < 5), review articles, meta‐analyses, editorials, and other forms (e.g., commentary).
2.3. Data extraction
A data extraction sheet was created and the study characteristics, source of data, patient characteristics, and outcome of interest were collected. Two of the authors (ZJQ and LGX) independently extracted data and potential discrepancies were resolved by the third author (GHT).
Disease severity was performed according to World Health Organization interim guidance, 13 mainly on the basis of the symptoms present at diagnosis; patients with pulse oxygen saturation (SpO2) < 90% or need of intensive care unit care, or with acute respiratory distress syndrome were classified as having severe disease.
The discharge criteria according to the discharge recommendations of the European Centre for Disease Prevention and Control: 14 (a) no fever lasting longer than 3 days, (b) resolved respiratory symptoms, (c) substantially improved acute exudative lesions on chest computed tomography (CT) images, (d) at least two consecutive negative reverse transcriptase PCR (RT‐PCR) test results in respiratory samples (with samples separated by at least 1 day), and (e) appearance of specific IgG when a serological test is available.
2.4. Data analysis
Our analysis includes cumulative descriptive statistics expressed as counts (n) and percentages (%) with a comparative analysis for the selected studies. The quantitative variables with normal distribution are presented as the mean ± SD and those with skewed distribution as median or range. We computed the odds ratios (ORs) as our effect estimate using the Mantel–Haenszel method with random effects, with a study confidence interval (CI) of 95%. Depending on the heterogeneity between studies, a fixed‐ or random‐effects model was used to estimate the average effect and its precision. We used the I 2 statistic and Cochran's Q test to assess statistical heterogeneity. The publication bias was evaluated by the visual inspection of funnel plot and Begger's regression tests. The publication bias was done to assess the effect of each study on the pooled effect size. A p < 0.05 was considered statistically significant. All statistical analyses were performed using the STATA software (version 15.0, Stata Corp. LP).
Trial sequential analysis (TSA) was performed for the nucleic acid‐positive stool of patients with diarrhea compared with those without diarrhea, using the TSA software 0.9.5.10 Beta (Copenhagen Trial Unit; Figure 3). The thresholds for the Z values using O'Brien‐Fleming α‐spending function were adjusted to control the risk of type 1 error. The cumulative Z curve represents the trial data. The risk of type 2 error was controlled using the β‐spending function and futility boundaries. Random‐effects modeling were applied. A two‐sided CI with 95% confidence level was used to indicate statistical significance. We estimated the information size for the analyses based on the achievement of 80% power and 10% relative risk reduction between the two groups.
Figure 3.
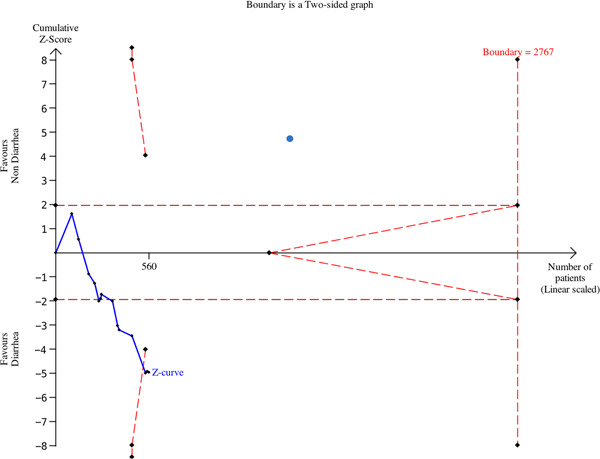
Trial sequential analysis for the nucleic acid‐positive stool of patients with diarrhea compared with those without diarrhea
3. RESULT
Our literature search identified 1809 citations from PubMed, EMBASE, and Cochrane Library database (Figure 1), of which 834 studies were removed after initial screening for duplicates. Further studies were excluded using the title and abstract review in 975 studies. A total of 205 articles were assessed for eligibility, and after excluding 116 studies, which did not provide data on stool viral RNA, 25 small studies (N < 5), and 4 studies that lack available dates, 50 studies were included in the final analysis (46 studies with data on stool viral load of patients with COVID‐19 at the first hospitalization and 4 studies with data on stool viral RNA positivity in recovered COVID‐19 patients).
Figure 1.
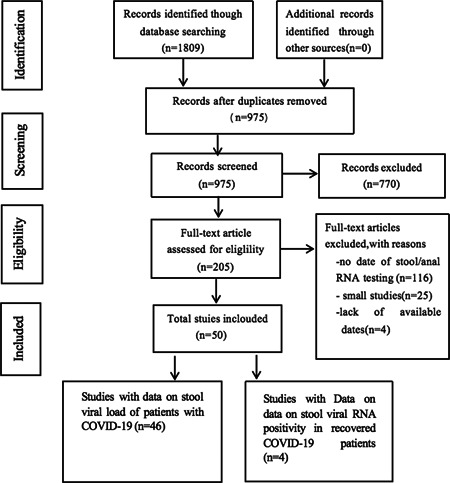
The workflow of the selection process in coronavirus disease 2019 (COVID‐19) patients with the date of virus RNA in stool
3.1. Characteristics of fecal SARS‐CoV‐2 RNA
The characteristics of the included studies with data of viral RNA in stool samples at the first hospitalization are shown in Table 1, including sites of patient recruitment, sample size, age, sex, SARS‐CoV‐2 RNA‐positive result in stool sample, duration of virus shedding in stool, disease severity, and GI symptoms on presentation. There were 46 studies that reported the prevalence of fecal SARS‐CoV‐2 RNA in patients with COVID‐19 infection confirmed by respiratory samples: 38 (82.6%) studies were from China (3 in Hong Kong) and 8 (17.4%) were from other countries (Singapore, United States, France, Germany, Italy, Korea, and India). Of the 2352 patients with COVID‐19, who tested for viral RNA in stool samples from the 46 studies, 735 were reported to have positive stool specimens 4 , 5 , 11 , 12 , 13 , 14 , 15 , 16 , 17 , 18 , 19 , 20 , 21 , 22 , 23 , 24 , 25 , 26 , 27 , 28 , 29 , 30 , 31 , 32 , 33 , 34 , 35 , 36 , 37 , 38 , 39 , 40 , 41 , 42 , 43 , 44 , 45 , 46 , 47 , 48 , 49 , 50 , 51 , 54 (46.8% CI: 0.383–0.554; I 2 = 96.8%; Figure 2 and Table 2). The median age of patients with positive fecal RNA was 41.6 ± 4.24 years and 55.4% were male.
Table 1.
Characteristics of studies included in COVID‐19 patients
| Study | Design | Country | Study period | COVID‐19 patients (n) | Positive fecal RNA of COVID‐19 patients (n, %) | Age of positive fecal RNA patients (years) | Man in positive fecal RNA patients (n, %) | Positive fecal RNA of severe cases (n, %) vs. positive fecal RNA of nonsevere cases (n, %) | Positive fecal RNA patients with severe disease (n, %) vs. fecal virus negative patients with severe disease (n, %) | Positive fecal RNA patients: RNA positive in stool and negative in respiratory samples: (n, %) | Clinical symptoms of positive fecal RNA patients |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Zhang et al. 15 | Cohort study | China | n.a. | 39 | 4 (10.3) | n.a. | n.a. | 0 (0.0) vs. 4 (8.7) | 0 (0.0) vs. 3 (10.0) | n.a. | n.a. |
| Wang et al. 5 | Case series | China | Jan 1–Feb 17, 2020 | 153 | 44 (28.7) | n.a. | n.a. | n.a. | n.a. | 1/6 (16.7) | n.a. |
| Young et al. 16 | Case series | Singapore | Jan 23–Feb 3, 2020 | 8 | 4 (50.0) | n.a. | n.a. | n.a. | n.a. | 1/4 (25.0) | n.a. |
| Jiehao et al. 17 | Case series | China | Jan 19–Feb 3, 2020 | 6 | 5 (83.3) | Median: 7 (0.6–9) | 2 (33.3) | n.a. | n.a. | 5/5 (100.0) | 5 Patients with respiratory symptoms |
| Ling et al. 18 | Retrospective study | China | Jan 20–Feb 10, 2020 | 66 | 54 (81.8) | Median: 44.0 (34.0–62.0) | 38 (56.7) | n.a. | n.a. | 43/54 (78.2) | n.a. |
| Han et al. 19 | Retrospective study | China | Feb 13–Feb 29, 2020 | 22 | 12 (54.5) | Mean: 43.3 (±13.8) | 5 (24.0) | n.a. | n.a. | n.a. | 9 Patients with respiratory symptoms, 9 patients with GI symptoms |
| Lei et al. 20 | Cohort study | China | Jan 22–Feb 12, 2020 | 7 | 4 (57.1) | n.a. | n.a. | n.a. | n.a. | 2/4 (50.0) | n.a. |
| Lo et al. 21 | Case series | China | Jan 21–Feb 16, 2020 | 10 | 10 (100.0) | Median: 54 (27–64) | 3 (30.0) | n.a. | n.a. | 5/10 (50.0) | n.a. |
| Yin et al. 22 | Retrospective study | China | Jan 19–Feb 7, 2020 | 33 | 8 (24.2) | n.a. | 5 (62.5) | n.a. | n.a. | n.a. | 5 Patients with respiratory symptoms, 5 patients with GI symptoms |
| The COVID‐19 Investigation Team 23 | Retrospective study | US | Jan–Feb, 2020 | 10 | 7 (70.0) | Range: 30–69 | 3 (60.0) | n.a. | n.a. | 1/7 (14.3) | 4 Patients with respiratory symptoms, 1 patient with GI symptoms |
| Lescure et al. 24 | Case series | French | Jan 24–Jan 29, 2020 | 5 | 2 (40.0) | 30, 46 | 0 (0.0) | n.a. | n.a. | 1/2 (50.0) | 2 Patients with respiratory symptoms |
| Xie et al. 25 | Retrospective study | China | Feb 27, 2020 | 9 | 8 (88.9) | Median: 43 (26–59) | 4 (50.0) | n.a. | n.a. | n.a. | 5 Patients with respiratory symptoms, 1 patient with GI symptoms |
| Chen et al. 26 | Retrospective study | China | Feb 26, 2020 | 28 | 11 (39.3) | n.a. | n.a. | 8 (66.7) vs. 3 (18.7) | 8 (72.7) vs. 4 (23.5) | n.a. | n.a. |
| Wu et al. 27 | Retrospective study | China | Jan 16–Mar 15, 2020 | 74 | 41 (55.4) | n.a. | n.a. | n.a. | n.a. | 31/41 (75.6) | n.a. |
| Peng et al. 28 | Retrospective study | China | Jan 22–Feb 29, 2020 | 9 | 2 (22.2) | 41, 49 | 2 (100.0) | n.a. | n.a. | n.a. | 2 Patients with respiratory symptoms, 1 patient with GI symptoms |
| Zheng et al. 29 | Cohort study | China | Jan 19–Feb 15, 2020 | 96 | 55 (57.3) | n.a. | n.a. | 42 (56.8) vs. 13 (59.1) | 42 (76.4) vs. 32 (78.1) | n.a. | n.a. |
| Zhang et al. 30 | Retrospective study | China | Jan 29–Feb 10, 2020 | 14 | 5 (35.7) | n.a. | n.a. | n.a. | n.a. | 3/5 (60.0) | n.a. |
| Xiao et al. 31 | Case series | China | Jan, 2020 | 28 | 12 (42.9) | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. |
| Xiao et al. 4 | Retrospective study | China | Feb 1–Feb 14, 2020 | 97 | 39 (53.42) | Median: 49 (0.83–78) | n.a. | n.a. | n.a. | 17/39 (43.6) | n.a. |
| Yongchen et al. 32 | Observational study | China | n.a. | 15 | 5/15 (33.3) | n.a. | n.a. | n.a. | n.a. | 3/5 (60.0) | n.a. |
| Wei et al. 33 | Retrospective study | China | Jan 19–Feb 7, 2020 | 84 | 28 (33.3) | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. |
| Tan et al. 34 | Retrospective study | China | Jan 27–Mar 10, 2020 | 10 | 3 (33.3) | Median: 8.8 (3.6–9.4) | n.a. | n.a. | n.a. | n.a. | 1 Patients with GI symptoms |
| Chen et al. 35 | Retrospective study | China | Jan 26–Feb 6, 2002 | 22 | 12 (54.5) | Median: 35 (29–48) | 8 (66.7) | 0 (0.0) vs. 12 (60.0) | 0 (0.0) vs. 2 (20.0) | 9/12 (75.0) | n.a. |
| Tan et al. 36 | Retrospective study | China | Jan 17–Feb 29, 2020 | 13 | 1 (7.7) | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. |
| Wölfel et al. 37 | Case series | Germany | n.a. | 9 | 8 (88.9) | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. |
| Xu et al. 11 | Case series | China | Jan 22–Feb 20, 2020 | 10 | 8 (80.0) | n.a. | 5 (62.5) | n.a. | n.a. | 7/8 (87.5) | 7 Patients with respiratory symptoms, 3 patients with GI symptoms |
| Yuan et al. 38 | Case series | China | n.a. | 6 | 6 (100.0) | Median: 60 (range: 37–71) | 3 (60.0) | n.a. | n.a. | 1/6 (16.7) | 4 Patients with respiratory symptoms, 1 patient with GI symptoms |
| Chen et al. 39 | Retrospective study | China | Jan 20–Feb 9, 2020 | 42 | 28 (66.7) | Median: 51.5 (43–62) | 12 (42.86) | 9 (81.8) vs 19 (61.2) | 9 (32.14 vs. 2 (14.29) | 11/28 (39.3) | n.a. |
| Lin et al. 12 | Retrospective study | China | Jan 17–Feb 25, 2020 | 65 | 31 (47.7) | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. |
| Wu et al. 67 | Observation study | China | Jan 31–Feb 29, 2020 | 244 | 24 (10.0) | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. |
| Hua et al. 41 | Retrospective study study | China | To Feb 29, 2020 | 35 | 32 (91.4) | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. |
| Chen et al. 42 | Observation study | China | Jan 17–Mar 2, 2020 | 97 | 52 (53.6) | Mean: 45.27 (±20.47) | 31 (59.6) | 15 (57.7) vs. 37 (57.7) | 15 (59.6) vs. 11 (73.3) | n.a. | n.a. |
| Cheung et al. 43 | Observation study | China:Hong Kong | Jan 20–Jan 29, 2020 | 59 | 9 (15.3) | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. |
| Tong et al. 44 | Observation study | China | Feb 1–Feb 28, 2020 | 262 | 32 (12.21) | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. |
| Park et al. 45 | Retrospective cohort study | Korea | Apr 4–Apr 24, 2020 | 46 | 2 (4.3) | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. |
| Lin et al. 46 | Retrospective study | China | Jan 20–Feb 20, 2020 | 217 | 46 (21.2) | 53 (41–62) | 6 (37.5) vs. 40 (19.9) | 6 (13.0) vs. 10 (5.8) | n.a. | n.a. | |
| Wang et al. 47 | Observation study | China | Feb 6– Feb 22, 2020 | 69 | 20 (28.99) | Median: 43 (I31.25–51.0) | 13 (35.0) | n.a. | n.a. | 11/20 (55.0) | n.a. |
| Liu et al. 48 | Observational study | China | Jan 31–Mar 16, 2020 | 47 | 2/47 (4.3) | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. |
| Li et al. 49 | Retrospective study | China | Between 9 and 28 February 2020 | 13 | 5 (38.0) | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. |
| Effenberger et al. 50 | Observation study | Austria | n.a. | 40 | 12 (25.0) | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. |
| De Ioris et al. 51 | Observation study | Italy | March 16, 2020–April 8, 2020 | 22 | 15 (68) | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. |
| Huang et al. 68 | Retrospective study | China | Jan 26–Feb 25, 2020 | 16 | 11 (68.8) | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. |
| To et al. 52 | Observation study | China: Hong Kong | Jan 22–Feb 12, 2020 | 23 | 4 (17.4) | n.a. | n.a | 3 (75.0) vs. 1 (5.2) | 3 (33.3) vs. 1 (7.7) | n.a. | n.a. |
| Xu et al. 53 | Observation study | China | Jan 13–Feb 27, 2020 | 51 | 1 (1.9) | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. |
| Zuo et al. 54 | Observation study | China:Hong Kong | Feb 5–Mar 17, 2020 | 15 | 11 (73.3) | n.a. | n.a. | n.a. | n.a. | n.a. | 11 Patients with respiratory symptoms, 1 patient with GI symptoms |
Abbreviations: COVID‐19, coronavirus disease 2019; n.a., not applicable.
Figure 2.
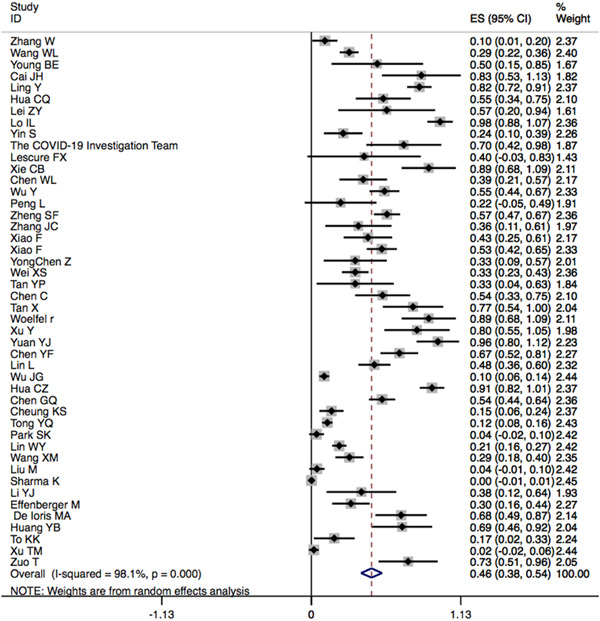
Pooled prevalence of detectable severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) RNA in fecal samples of patients with confirmed coronavirus disease 2019 (COVID‐19) infection
Table 2.
Summarizing results of pooled estimates of stool virus RNA tests in COVID‐19 patients
| Sample size | Test of association | Effect size | Heterogeneity | Publication bias | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Characteristics | Number studies | Event | Total | Model | Estimate | 95% CI | p | I 2 | p | p (Begger's) |
| Positive fecal RNA of COVID‐19 patients | 46 | 735 | 2352 | Random | 0.468 | 0.383 −0.554 | <0.001 | 96.8% | <0.001 | 0.338 |
| Nucleic acid was positive in stool swabs and negative in respiratory swabs of positive fecal RNA COVID‐19 patients | 17 | 152 | 282 | Random | 0.554 | 0.418–0.669 | 0.0481 | 79.0% | <0.001 | 0.484 |
| Positive fecal RNA of COVID‐19 patients with GI symptoms | 14 | 73 | 143 | Random | 0.659 | 0.453–0.865 | <0.001 | 88.8% | <0.001 | 0.4243 |
| Positive fecal RNA of COVID‐19 patients without GI symptoms | 14 | 138 | 446 | Random | 0.331 | 0.207–0.455 | <0.001 | 90.10% | <0.001 | 0.055 |
| Positive fecal RNA patients with total GI symptoms | 11 | 23 | 70 | Random | 0.285 | 0.125–0.44 | <0.001 | 68.0% | 0.001 | 0.184 |
| Positive fecal RNA patients: with both GI and respiratory symptoms | 11 | 19 | 70 | Random | 0.219 | 0.09–0.346 | 0.001 | 56.9% | <0.001 | 0.102 |
| Positive fecal RNA patients: only with GI symptoms | 11 | 4 | 70 | Fixed | 0.198 | 0.107–0.288 | 0.001 | 0.0% | 0.901 | 0.119 |
| Positive fecal RNA patients: only with respiratory symptoms | 11 | 34 | 70 | Random | 0.505 | 0.267–0.744 | <0.001 | 86.4% | <0.001 | 0.435 |
| Test of association | Effect size | Heterogeneity | ||||||||
| Characteristics 1 | Event 1/Total 1 (n) | Characteristics 2 | Event 2/Total 2 (n) | Model | OR | 95% CI | p | I 2 | p | P (Egger's) |
| Positive fecal RNA of COVID‐19 patients with diarrhea | 88/179 | Positive fecal RNA of COVID‐19 patients without diarrhea | 115/381 | Random | 2.961 | 1.355–6.473 | 0.007 | 54.6% | 0.007 | 0.783 |
| Positive fecal RNA of severe cases | 83/139 | Positive fecal RNA of nonsevere cases | 129/416 | Fixed | 2.009 | 0.922–4.378 | 0.079 | 49.1% | 0.056 | 0.902 |
| Severe disease of positive fecal RNA patients | 83/227 | Severe disease of negative fecal RNA patients | 65/364 | Fixed | 1.533 | 0.949–2.475 | 0.081 | 0.3% | 0.862 | 0.711 |
Abbreviations: CI, confidence interval; COVID‐19, coronavirus disease 2019; GI, gastrointestinal; OR, odds ratio.
Fourteen studies 11 , 19 , 22 , 23 , 24 , 25 , 28 , 30 , 33 , 34 , 38 , 39 , 43 including 609 patients reported the prevalence of fecal SARS‐CoV‐2 RNA in patients with different clinical symptoms. Pooled results indicated that 65.9% (95% CI: 0.453–0.865, I 2 = 88.8%) of the 143 COVID‐19 patients with GI symptoms tested positive for RNA in stool samples, whereas 33.1% (95% CI: 0.207–0.451, I 2 = 90.1%) of the 469 COVID‐19 patients without GI symptoms were positive for RNA in stool samples. Pooled results from 14 studies 11 , 12 , 19 , 21 , 23 , 24 , 25 , 28 , 33 , 39 , 42 , 43 , 47 including 560 patients indicated that the proportion of patients with COVID‐19 with nucleic acid‐positive stool samples was markedly increased in patients with diarrhea compared with those without diarrhea (OR = 2.961, 95% CI: 1.355–6.473, p = 0.007, I 2 = 54.6%).
In 8 studies 15 , 26 , 29 , 35 , 39 , 42 , 46 , 52 including 632 COVID‐19 patients, 139 patients were categorized as severe cases. Eighty‐three of 139 severe cases and 129 of 416 nonsevere cases tested positive for fecal RNA. The pooled data showed no significant difference between the two groups for positive fecal RNA (OR = 2.009, p = 0.079, 95% CI: 0.922–4.378, I 2 = 49.1%). There were 83 severe cases in the nucleic acid‐positive stool group (227 patients) and 65 severe cases in the nucleic acid‐negative stool group (365 patients). The pooled data showed no significant difference in the severity of illness between the two groups (OR = 1.533, p = 0.081, 95% CI: 0.949–2.47, I 2 = 0.3%).
Eleven studies 11 , 17 , 19 , 22 , 25 , 28 , 34 , 38 including 69 COVID‐19 patients with positive stool RNA had available data on respiratory symptoms and GI symptoms. The pooled prevalence of the total GI symptoms was 28.5% (95% CI: 0.125–0.44, I 2 = 68.0%), both respiratory symptoms and GI symptoms was 21.9% (95% CI: 0.09–0.346, I 2 = 56.9%), only respiratory symptoms (without GI symptoms) was 50.5% (95% CI: 0.267–0.744, I 2 = 86.4%), and only GI symptoms (without respiratory symptoms) was 19.8% (95% CI: 0.107–0.288, I 2 = 0.0%).
The shedding of SARS‐CoV‐2 RNA in feces or respiratory samples was assessed in 17 studies, 5 , 11 , 16 , 17 , 18 , 20 , 21 , 23 , 24 , 27 , 30 , 31 , 32 , 35 , 38 , 47 152 of 282 patients with positive fecal RNA (pooled prevalence: 55.4%, 95% CI: 0.418–0.669, I 2 = 79.0%) still presented with nucleic acid‐positive stool samples after the virus was negative in their respiratory samples.
3.2. Repositive of SARS‐CoV‐2 RNA
In addition, 4 studies, 55 , 56 , 57 , 58 including 848 discharged COVID‐19 patients with negative RT‐PCR tests in respiratory and stool samples, had available data on repositive tests for SARS‐CoV‐2 RNA. The proportion of repositive tests in stool samples was 18.1% (95% CI: 0.028–0.335, I 2 = 88.9%) and persisted from 2 days to 21 days after discharge. The proportion of repositive tests in respiratory samples was 22.8% (95% CI: 0.003–0.452, I 2 = 98.9%) and persisted from 2 to 19 days after discharge. By combining 3 of these studies 55 , 57 , 58 (including 229 discharged patients), the proportion of repositive tests in both stool samples and respiratory samples was 19.1% (95% CI: 0.019–0.363,I 2 = 94.4%), and the proportion of repositive tests only in stool samples was 9.6%(95% CI: 0.010–0.203, I 2 = 76.2%). Most of the patients with repositive fecal RNA presented as asymptomatic or with mild‐to‐moderate symptoms and no severe cases were reported; there were no self‐infection reports and no close contacts were found to be infected in these patients.
3.3. Publication bias and sensitivity analysis
The funnel plot of clinical parameters is shown in Figures S1−S8 and Begger's tests are shown in Table 2. There was no publication bias in this study. In sensitivity analysis, it revealed that the study performed by Wu et al. 67 and Xu et al. 53 contributed in the significant heterogeneity observed in SARS‐CoV‐2 RNA‐positive result in stool samples; the study by Zuo et al. 54 contributed in the significant heterogeneity observed in only respiratory symptoms of patients with positive fecal RNA; study by Lin et al. 46 contributed in the significant heterogeneity observed in patients with COVID‐19 with GI symptoms tested positive for fecal RNA; study by Chen et al. 35 contributed in the significant heterogeneity observed in COVID‐19 patients without GI symptoms tested positive for fecal RNA; and the study by Yuan et al. 55 contributed in the significant heterogeneity observed in repositive tests for SARS‐CoV‐2 RNA in both respiratory samples and in stool samples, and in repositive tests for SARS‐CoV‐2 RNA only in stool samples (Table 3).
Table 3.
Characteristics of studies in discharged COVID‐19 pateints with repositive SARS‐CoV‐2 RNA
| Study | Design | Country | Study period | COVID‐19 patients (n) | Repositive of SARS‐CoV‐2 viral RNA in stool samples (%) | Repositive of SARS‐CoV‐2 viral RNA in respiratory samples (%) | Repositive of SARS‐CoV‐2 viral RNA in respiratory and stool samples (%) | Repositive of SARS‐CoV‐2 viral RNA only in stool samples (%) |
|---|---|---|---|---|---|---|---|---|
| Yuan et al. 55 | Cohort study | China | Apr, 2020 | 182 | 7 (3.8) | 12 (6.6) | 0 (3.1) | 8 (29.6) |
| Lu et al. 56 | Retrospective | China | Jan 23–Feb 19, 2020 | 619 | 19/68 (27.9)a | 87 (14.0) | n.a. | n.a. |
| Ma et al. 57 | Retrospective | China | Jan 30–Mar, 2020 | 27 | 8 (29.2) | 0 (0.0) | 0 (3.1) | 7 (3.8) |
| Zheng et al. 58 | Observation study | China | Jan 25–Feb 26, 2020 | 20 | 3 (15.0) | 2 (10.0) | 2 (66.7) | 1 (5.0) |
Abbreviation: COVID‐19, coronavirus disease 2019.
A total of 68 patients tested for RNA in stool samples of which 19 were positive.
3.4. TSA
The nucleic acid‐positive stool of patients with diarrhea compared with those without diarrhea: the cumulative Z‐value curve crossed the traditional boundary value and crossed the TSA threshold line (Figure 3), which meant a positive conclusion had been reached before the expected amount of information had been reached. Patients with COVID‐19 with nucleic acid‐positive stool samples was increased in patients with diarrhea compared with those without diarrhea.
4. DISCUSSION
In the current SARS‐CoV‐2 pandemic, the tropism of the virus to the GI tract and its positive detection in stool are attracting increasing attention. In this meta‐analysis, we noted that 46.8% of patients had detectable stool viral RNA during the course of illness and the proportion of patients with COVID‐19 with nucleic acid‐positive stool was markedly increased in patients with diarrhea symptoms compared with those without diarrhea, indicating that patients with COVID‐19 with positive RNA in feces samples are more likely to experience GI symptoms such as diarrhea, possibly because GI epithelial cells express ACE2 and SARS‐CoV‐2 binds to the ACE2 before cleavage by the host transmembrane serine protease 2. 4 , 7 , 59 Virus‐specific RNA and proteins can then be synthesized in the cytoplasm to assemble new virions, which can be released into the GI tract. 7 , 59 , 60 This indicates that the digestive system might be vulnerable to COVID‐19 infection and fecal–oral transmission may be another route for SARS‐CoV‐2 spread. 7 , 57
Despite the high positive rate of viral RNA in stool samples, only 28.5% of patients with nucleic acid‐positive stool had GI symptoms. For most COVID‐19 patients, respiratory symptoms were the main complaints at admission instead of GI symptoms. In our meta‐analysis, 50.5% of patients with nucleic acid‐positive stool samples had only respiratory symptoms but no GI symptoms. Therefore, fecal nucleic acid examinations may be missed in two‐thirds of patients without GI symptoms. Notably, it has been found that SARS‐CoV‐2 detection was positive in the fecal swabs but negative in respiratory swabs of patients during the visit. Li et al. 61 presented a case on mild SARS‐CoV‐2 infection in a baby with PCR‐negative oropharyngeal/nasopharyngeal (OP/NP) swabs and normal chest CT, but her anal swabs remained positive for 8 days. Therefore, patients who only have positive RT‐PCR tests in stool samples may be clinically ignored. Pauci‐symptomatic and asymptomatic individuals represent a major concern for diagnosis and viral transmission. Furthermore, false‐negative results of OP/NP swabs ranged from 1% to 30% in previous studies. 62 , 63 To reduce the rate of missed diagnosis, it is proposed to perform SARS‐CoV‐2 RT‐PCR testing on fecal samples as part of routine analyses for the detection of SARS‐CoV‐2. 64
SARS‐CoV‐2 RNA can be detected not only in fecal samples from severe cases but also in fecal samples from nonsevere cases. The pooled data showed no significant difference in positive fecal RNA between the two groups; therefore, fecal SARS‐CoV‐2 RNA tests are also important for patients with mild disease.
The elimination of SARS‐CoV‐2 from the digestive system may be much later and harder than that from the respiratory system, as ACE2 is abundantly expressed in gastric, duodenal, and rectal epithelia in patients with COVID‐19, which may lead to virus internalization and accumulation in these organs. 4 , 7 In our meta‐analysis, 55.4% of patients with positive stool RNA still had persistent positive viral RNA in the feces after the pharyngeal swabs turned negative. The potential recurrence of the disease in discharged patients with two sequential negative OP swab tests collected 24 h apart from the clearance of viral RNA in patient stool samples is delayed. 65 , 66 In our meta‐analysis, we noted that some patients with COVID‐19 (18.1%) tested positive for SARS‐CoV‐2 RNA in fecal samples after discharge. To reduce the number of false negatives, it is important to consider a combined assessment of both fecal and respiratory specimens for patients, especially at the time of discharge and during convalescence. 39 , 65
Furthermore, even if viral nucleic acid examinations in stool were negative at discharge, there is still a possibility of repositive tests for SARS‐CoV‐2 RNA. It is still uncertain whether the recurrence of SARS‐CoV‐2 RNA among discharged COVID‐19 patients could be contagious. 66 In our analysis, 9.6% of discharged patients tested positive again for SARS‐CoV‐2 RNA in stool samples but negative in respiratory samples; the possibility cannot be excluded that the virus may be transmitted through the digestive tract. Therefore, to prevent the spread of the pandemic, it is important to monitor patients, and respiratory and fecal samples should be tested regularly after discharge. 66 Patients need to pay close attention to hand hygiene and try to avoid sharing toilets with family members after discharge. Attention should be paid to standard and transmission‐based precautions for patients until the negative conversion of SARS‐CoV‐2 RNA in feces. 39
In conclusion, the detection of fecal SARS‐CoV‐2 RNA in patients with COVID‐19 is common, and the repositive tests of viral RNA are not unusual in discharged patients. As the respiratory RNA test results may not be consistent with those from stool samples, giving emphasis on stool swab tests can help increase the detection rate of asymptomatic carriers and reduce the number of false negatives. In addition, the possibility of fecal–oral transmission is unclear and the virus may be transmitted through the digestive tract; therefore, quarantine and other such policies should be maintained during convalescence even after discharge.
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
AUTHORS CONTRIBUTIONS
Jin‐Qiu Zhou, Gong‐Xiang Liu, Xiao‐Li Huang, and Hua‐Tian Gan wrote the manuscript and participated in the literature review. Jin‐Qiu Zhou analyzed the data, wrote the manuscript, and participated in literature review. Gong‐Xiang Liu participated in the literature review. Xiao‐Li Huang and Hua‐Tian Gan supervised, designed, and checked the quality of the study. All authors read and approved the final manuscript.
Supporting information
Supporting information.
ACKNOWLEDGMENTS
The present work was supported by Science Foundation from the Science Technology Department of Sichuan Province, PR China (No. 2019YFS0262) and 1.3.5 Project for Disciplines of Excellence, West China Hospital, Sichuan University.
Zhou J‐Q, Liu G‐X, Huang X‐L, Gan H‐T. The importance of fecal nucleic acid detection in patients with coronavirus disease (COVID‐19): a systematic review and meta‐analysis. J Med Virol. 2022;94:2317‐2330. 10.1002/jmv.27652
Contributor Information
Xiao‐Li Huang, Email: huangxiaoli@scu.edu.cn.
Hua‐Tian Gan, Email: ganhuatian123@163.com.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Mao R, Qiu Y, He JS, et al. Manifestations and prognosis of gastrointestinal and liver involvement in patients with COVID‐19: a systematic review and meta‐analysis. Lancet Gastroenterol Hepatol. 2020;5(7):667‐678. Gastrointestinal symptoms & stool viral RNA positivity rate in COVID‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kumar VCS, Mukherjee S, Harne PS, et al. Novelty in the gut: a systematic review and meta‐analysis of the gastrointestinal manifestations of COVID‐19. BMJ Open Gastroenterol. 2020;7(1):e000417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hayashi Y, Wagatsuma K, Nojima M, et al. The characteristics of gastrointestinal symptoms in patients with severe COVID‐19: a systematic review and meta‐analysis. J Gastroenterol. 2021;56(5):409‐420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Xiao F, Tang M, Zheng X, Liu Y, Li X, Shan H. Evidence for gastrointestinal infection of SARS‐CoV‐2. Gastroenterology. 2020;158(6):1831‐1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang WL, Xu Y, Gao, R , et al. Detection of SARS‐CoV‐2 in different types of clinical specimens. JAMA. 2020;323(18):1843‐1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wan Y, Shang, Graham J. R, et al. Receptor recognition by novel coronavirus from Wuhan: an analysis based on decade‐long structural studies of SARS. J Virol. 2020;94:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gu J, Han B, Wang J. COVID‐19: gastrointestinal manifestations and potential fecal‐oral transmission. Gastroenterology. 2020;158(6):1518‐1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jimenez DG, Rodríguez‐Belvís MV, Gonzalez F, et al. COVID‐19 gastrointestinal manifestations are independent predictors of PICU admission in hospitalized pediatric patients. Pediatr Infect Dis J. 2020;39(12):e459‐e462. [DOI] [PubMed] [Google Scholar]
- 9. Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus‐infected pneumonia in Wuhan, China. JAMA. 2020;323(11):1061‐1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jiang X, Luo M, Zou Z, Wang X, Chen C, Qiu J. Asymptomatic SARS‐CoV‐2 infected case with viral detection positive in stool but negative in nasopharyngeal samples lasts for 42 days. J Med Virol. 2020;92(10):1807‐1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xu Y, Li X, Zhu B, et al. Characteristics of pediatric SARS‐CoV‐2 infection and potential evidence for persistent fecal viral shedding. Nat Med. 2020;26:502‐505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lin L, Jiang X, Zhang Z, et al. Gastrointestinal symptoms of 95 cases with SARS‐CoV‐2 infection. Gut. 2020;69(6):997‐1001. [DOI] [PubMed] [Google Scholar]
- 13. WHO . Global surveillance for human infection with coronavirus disease (COVID‐19). Accessed at https://www.who.int/publications/i/item/10665-332299
- 14. Centre for Disease Prevention and Control . Guidance for discharge and ending isolation in the context of widespread community transmission of COVID‐19‐first update. Available at: https://www.ecdc.europa.eu/sites/default/files/documents/covid-19-guidance-discharge-and-ending-isolation-firstupdate.pdf
- 15. Zhang W, Du RH, Li B, et al. Molecular and serological investigation of 2019‐nCoV infected patients: implication of multiple shedding routes. Emerg Microbes Infect. 2020;9(1):386‐389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Young BE, Ong S, Kalimuddin S, et al. Epidemiologic features and clinical course of patients infected with SARS‐CoV‐2 in Singapore. JAMA. 2020;323(15):1488‐1494. Erratum in: JAMA. 2020 Apr 21;323(15):1510. 10.1001/jama.2020.3204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jiehao C, Jin X, Daojiong L, et al. A case series of children with 2019 novel coronavirus infection: clinical and epidemiological features. Clin Infect Dis. 2020;71(6):1547‐1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ling Y, Xu SB, Lin YX, et al. Persistence and clearance of viral RNA in 2019 novel coronavirus disease rehabilitation patients. Chin Med J. 2020;133(9):1039‐1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Han C, Duan C, Zhang S, et al. Digestive symptoms in COVID‐19 patients with mild disease severity: clinical presentation, stool viral RNA testing, and outcomes. Am J Gastroenterol. 2020;115(6):916‐923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lei Z, Cao H, Jie Y, et al. A cross‐sectional comparison of epidemiological and clinical features of patients with coronavirus disease (COVID‐19) in Wuhan and outside Wuhan, China. Travel Med Infect Dis. 2020;35:101664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lo IL, Lio CF, Cheong HH, et al. Evaluation of SARS‐CoV‐2 RNA shedding in clinical specimens and clinical characteristics of 10 patients with COVID‐19 in Macau. Int J Biol Sci. 2020;16(10):1698‐1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yin S, Peng Y, Ren Y, et al. The implications of preliminary screening and diagnosis: clinical characteristics of 33 mild patients with SARS‐CoV‐2 infection in Hunan, China. J Clin Virol. 2020;128:104397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. The COVID‐19 Investigation Team . Clinical and virologic characteristics of the first 12 patients with coronavirus disease 2019 (COVID‐19) in the United States. Nat Med. 2020;26(6):861‐868. [DOI] [PubMed] [Google Scholar]
- 24. Lescure FX, Bouadma L, Nguyen D, et al. Clinical and virological data of the first cases of COVID‐19 in Europe: a case series. Lancet Infect Dis. 2020;20(6):697‐706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xie C, Jiang L, Huang G, et al. Comparison of different samples for 2019 novel coronavirus detection by nucleic acid amplification tests. Int J Infect Dis. 2020;93:264‐267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen W, Lan Y, Yuan X, et al. Detectable 2019‐nCoV viral RNA in blood is a strong indicator for the further clinical severity. Emerg Microbes Infect. 2020;9(1):469‐473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wu Y, Guo C, Tang L, et al. Prolonged presence of SARS‐CoV‐2 viral RNA in faecal samples. Lancet Gastroenterol Hepatol. 2020;5(5):434‐435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Peng L, Liu J, Xu W. SARS‐CoV‐2 can be detected in urine, blood, anal swabs, and oropharyngeal swabs specimens. J Med Virol. 2020;92(9):1676‐1680. 10.1002/jmv.25936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zheng S, Fan J, Yu F, et al. Viral load dynamics and disease severity in patients infected with SARS‐CoV‐2 in Zhejiang province, China, January‐March 2020: retrospective cohort study. BMJ. 2020;369:m1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang J, Wang S, Xue Y. Fecal specimen diagnosis 2019 novel coronavirus‐infected pneumonia. J Med Virol. 2020;92(6):680‐682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xiao F, Sun J, Xu Y, et al. Infectious SARS‐CoV‐2 in feces of patient with severe COVID‐19. Emerg Infect Dis. 2020;26(8):1920‐1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yongchen Z, Shen H, Wang X, et al. Different longitudinal patterns of nucleic acid and serology testing results based on disease severity of COVID‐19 patients. Emerg Microbes Infect. 2020;9(1):833‐836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wei XS, Wang X, Niu YR, et al. Diarrhea is associated with prolonged symptoms and viral carriage in corona virus disease 2019. Clin Gastroenterol Hepatol. 2020;18(8):1753‐1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tan Y, Tan B, Pan J, Wu J, Zeng S, Wei H. Epidemiologic and clinical characteristics of 10 children with coronavirus disease 2019 in Changsha, China. J Clin Virol. 2020;127:104353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen C, Gao G, Xu Y, et al. SARS‐CoV‐2‐positive sputum and feces after conversion of pharyngeal samples in patients with COVID‐19. Ann Intern Med. 2020;172(12):832‐834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tan X, Huang J, Zhao F, Zhou Y, Li JQ, Wang XY. Clinical features of children with SARS‐CoV‐2 infection: an analysis of 13 cases from Changsha, China]. Zhongguo Dang Dai Er Ke Za Zhi. 2020;22(4):294‐298. (Chinese). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wölfel R, Corman VM, Guggemos W, et al. Virological assessment of hospitalized patients with COVID‐2019. Nature. 2020;581(7809):465‐469. [DOI] [PubMed] [Google Scholar]
- 38. Yuan Y, Wang N, Ou X. Caution should be exercised for the detection of SARS‐CoV‐2, especially in the elderly. J Med Virol. 2020;92(9):1641‐1648. [DOI] [PubMed] [Google Scholar]
- 39. Chen Y, Chen L, Deng Q, et al. The presence of SARS‐CoV‐2 RNA in the feces of COVID‐19 patients. J Med Virol. 2020;92(7):833‐840. [DOI] [PubMed] [Google Scholar]
- 40. Chaimayo C, Kaewnaphan B, Tanlieng N, et al. Rapid SARS‐CoV‐2 antigen detection assay in comparison with real‐time RT‐PCR assay for laboratory diagnosis of COVID‐19 in Thailand. Virol J. 2020;17(1):177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hua CZ, Miao ZP, Zheng JS, et al. Epidemiological features and viral shedding in children with SARS‐CoV‐2 infection. J Med Virol. 2020;92(11):2804‐2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen GQ, Luo WT, Zhao CH, et al. Comparison of clinical characteristics between fecal/perianal swab nucleic acid‐positive and ‐negative patients with COVID‐19. J Infect Dev Ctries. 2020;14(8):847‐852. [DOI] [PubMed] [Google Scholar]
- 43. Cheung KS, Hung IFN, Chan PPY, et al. Gastrointestinal manifestations of SARS‐CoV‐2 infection and virus load in fecal samples from a Hong Kong cohort: systematic review and meta‐analysis. Gastroenterology. 2020;159(1):81‐95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tong Y, Bao A, Chen H, et al. Necessity for detection of SARS‐CoV‐2 RNA in multiple types of specimens for the discharge of the patients with COVID‐19. J Transl Med. 2020;18(1):411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Park S, Lee CW, Park DI, et al. Detection of SARS‐CoV‐2 in fecal samples from patients with asymptomatic and mild COVID‐19 in Korea. Clin Gastroenterol Hepatol. 2021;19(7):1387‐1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lin W, Xie Z, Li Y, et al. Association between detectable SARS‐COV‐2 RNA in anal swabs and disease severity in patients with coronavirus disease 2019. J Med Virol. 2021;93(2):794‐802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang X, Zheng J, Guo L, et al. Fecal viral shedding in COVID‐19 patients: clinical significance, viral load dynamics and survival analysis. Virus Res. 2020;289:198147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liu M, Li Q, Zhou J, et al. Value of swab types and collection time on SARS‐COV‐2 detection using RT‐PCR assay. J Virol Methods. 2020;286:113974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li Y, Hu Y, Yu Y, et al. Positive result of Sars‐Cov‐2 in faeces and sputum from discharged patients with COVID‐19 in Yiwu, China. J Med Virol. 2020;92(10):1938‐1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Effenberger M, Grabherr F, Mayr L, et al. Faecal calprotectin indicates intestinal inflammation in COVID‐19. Gut. 2020;69(8):1543‐1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. De Ioris MA, Scarselli A, Atti MLCD, et al. Dynamic viral severe acute respiratory syndrome coronavirus 2 RNA shedding in children: preliminary data and clinical consideration from a Italian Regional Center. J Pediatric Infect Dis Soc. 2020;9(3):366‐369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. To KK, Tsang OT, Leung WS, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS‐CoV‐2: an observational cohort study. Lancet Infect Dis. 2020;20(5):565‐574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Xu T, Chen C, Zhu Z, et al. Clinical features and dynamics of viral load in imported and non‐imported patients with COVID‐19. Int J Infect Dis. 2020;94:68‐71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zuo T, Zhang F, Lui GCY, et al. Alterations in gut microbiota of patients with COVID‐19 during time of hospitalization. Gastroenterology. 2020;159(3):944‐955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yuan B, Liu HQ, Yang ZR, et al. Recurrence of positive SARS‐CoV‐2 viral RNA in recovered COVID‐19 patients during medical isolation observation. Sci Rep. 2020;10(1):11887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lu J, Peng J, Xiong Q, et al. Clinical, immunological and virological characterization of COVID‐19 patients that test re‐positive for SARS‐CoV‐2 by RT‐PCR. EBioMedicine. 2020;59:102960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ma X, Su L, Zhang Y, Zhang X, Gai Z, Zhang Z. Do children need a longer time to shed SARS‐CoV‐2 in stool than adults? J Microbiol Immunol Infect. 2020;53(3):373‐376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zheng KI, Wang XB, Jin XH, et al. A case series of recurrent viral RNA positivity in recovered COVID‐19 Chinese patients. J Gen Intern Med. 2020;35(7):2205‐2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Letko M, Marzi A, Munster V. Functional assessment of cell entry and receptor usage for SARS‐CoV‐2 and other lineage B betacoronaviruses. J Nature Microbiol. 2020;5(4):562‐569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhang H, Kang ZJ, Gong HY, et al. (January 30, 2020). The digestive system is a potential route of 2019‐nCov infection: a bioinformatics analysis based on single‐cell transcriptomes. Preprint at bioRxiv. 10.1101/2020.01.30.927806 [DOI]
- 61. Li J, Feng J, Liu TH, Xu FC, Song GQ. An infant with a mild SARS‐CoV‐2 infection detected only by anal swabs: a case report. Braz J Infect Dis. 2020;24(3):247‐249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Long DR, Gombar S, Hogan CA. Occurrence and timing of subsequent SARS‐CoV‐2 RT‐PCR positivity among initially negative patients. Clin Infect Dis. 2021;72(2):323‐326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Arevalo‐Rodriguez I, Buitrago‐Garcia D, Simancas‐Racines D, et al. False‐negative results of initial RT‐PCR assays for COVID‐19: a systematic review. PLoS One. 2020;15(12):e0242958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Brogna B, Brogna C, Petrillo M, et al. SARS‐CoV‐2 detection in fecal sample from a patient with typical findings of COVID‐19 pneumonia on CT but negative to multiple SARS‐CoV‐2 RT‐PCR tests on oropharyngeal and nasopharyngeal swab samples. Medicina. 2021;57(3):290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wang Y, Chen X, Wang F, et al. Value of anal swabs for SARS‐COV‐2 detection: a literature review. Int J Med Sci. 2021;18(11):2389‐2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Dao TL, Hoang VT, Gautret P. Recurrence of SARS‐CoV‐2 viral RNA in recovered COVID‐19 patients: a narrative review. Eur J Clin Microbiol Infect Dis. 2021;40(1):13‐25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wu J, Liu J, Li S, et al. Detection and analysis of nucleic acid in various biological samples of COVID‐19 patients. Travel Med Infect Dis. 2020;37:101673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Huang Y, Chen S, Yang Z, et al. SARS‐CoV‐2 viral load in clinical samples from critically Ill patients. Am J Respir Crit Care Med. 2020;201(11):1435‐1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


