Abstract
Numerous variants of the severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) pandemic have evolved. Viral variants may evolve with harmful susceptibility to the immunity established with the existing COVID‐19 vaccination. These variants are more transmissible, induce relatively extreme illness, have evasive immunological features, decrease neutralization using antibodies from vaccinated persons, and are more susceptible to re‐infection. The Centers for Disease Control and Prevention (CDC) has categorized SARS‐CoV‐2 mutations as variants of interest (VOI), variants of concern (VOC), and variants of high consequence (VOHC). At the moment, four VOC and many variants of interest have been defined and require constant observation. This review article summarizes various variants of SARS‐CoV‐2 surfaced with special emphasis on VOCs that are spreading across the world, as well as several viral mutational impacts and how these modifications alter the properties of the virus.
Keywords: Delta plus variant, Delta variant, mutation, Omicron variant, SARS‐CoV‐2, vaccination, viral variant
1. INTRODUCTION
The severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) infection has resulted in a great rise in morbidity and mortality all across the globe. 1 , 2 Over 4000 SARS‐CoV‐2 mutations have been detected as the worldwide SARS‐CoV‐2 outbreak proceeds. Attempts are being made to detect viral mutations and different viral strains. The ultimate goal of the current research toward COVID‐19 is to discover new viral mutations fast and determine their potential consequences. 3 A mutation occurs when the pattern of the gene is altered. The order of these nucleotides in RNA or DNA determines the amino‐acid sequence. Proteins are constructed from amino acids and are species‐specific. 4
“A mutation in a viral genome can alter the encoded amino acid sequences, which can cause the virus to replicate. Mutations are classified into two types: deletion and substitution.” 5 Substitution can originally be referred to as a proofreading process, but deletions cannot. The genome present in SARS‐CoV‐2 consists of 14 open reading frames (ORFs) of which two‐thirds are responsible for encoding 16 nonstructural proteins (NSP 1−16) necessary to make up the replicase complex. 6 The remaining one‐third ORFs are involved in the encoding of four proteins that are S (Spike), N (Nucleocapsid), M (Membrane), E (Envelope), and nine accessory protein‐ORFs. 7 S (Spike) proteins are required for entry of COVID‐19 virus into host cells. 8 NSP 14 performs proofreading for the SARS‐CoV‐2 virus. Mutations are always to be anticipated, despite the fact that not all mutations are purposeful or advantageous to the virus. 9 Viral variants are the outcome of mutations that occur throughout viral replication. A mutation is an alteration in a genome of a virus pattern that differs from the typical pattern, including a replacement, removal, or inclusion. 10 The SARS‐CoV‐2 virus is no exception, and multiple variants of the same have been reported all over the globe since its inception. Considering a growing frequency of instances identified viral variants with mutation sites in the viral spike protein's receptor‐binding domain (RBD) region have garnered widespread interest, the RBD is the primary focus of neutralizing antibodies generated after infection of SARS‐CoV‐2. 8 , 11 , 12 , 13 , 14 , 15 Some abnormalities in the S protein, like those reported in the N‐terminal domain (NTD), may also affect neutralizing antibody capacity. 16 A reconstituted SARS‐CoV‐2 (virus or genetic mutant) may have additional mutations that separate it from the basic pattern or common viral variants widely circulated in humans. 17
The proliferation of mutations poses a significant barrier for vaccination‐based protection and management of the SARS‐CoV‐2 outbreak. Existing SARS‐CoV‐2 vaccines have been approved for immediate application. Those vaccines that are in clinical trials have also demonstrated substantial benefits in terms of offering effective coverage toward novel viral variants. 18 This review encompasses the impact of identified variants on neutralizing antibodies and the preventive effect of various vaccines. We have also proposed ways for using present vaccines toward variants as well as generating upcoming vaccines.
2. VARIANTS OF THE SARS‐COV‐2
SARS‐CoV‐2 variants can have a variety of features. Testing results may be affected if a patient sample contains SARS‐CoV‐2 viral mutations. 19 Multiple factors, including the variant sequence, examination system, and the incidence of change in the population, are used to analyze the influence of mutations on test performance. 20 Typically, transcription or translation error in the viral genome is the main reason for mutation. 21 It has been shown that RNA viruses undergo mutation at higher rates than DNA viruses with mutation rates from 10−6 to 10−4 substitutions per nucleotide, per round of copying. 22 The high rate of mutation is correlated with an increase in evolvability and enhanced virulence, which is a beneficial survival trait for viruses. Mutation in viruses causes both geno‐ and phenotypic changes as seen in an influenza A virus. The main reason for mutation in the influenza A virus is the re‐assortment of viral genomes from different strains. 23 The mutation causes the change in patterns of influenza A subtype H3N2 which is responsible for antigenic evolution in humans. 24 Influenza viruses are ever‐shifting in two ways, antigenic drift and antigenic shift. The first is responsible for causing small mutation in genes of the virus which leads to changes in HA (hemagglutinin) and NA (neuraminidase), which are surface proteins and later creates a substantial shift in influenza A viral surface protein produces new HA and/or NA surface proteins, that are relevant to human infection. 25 In conjunction with the SARS‐CoV‐2 interagency committee, the CDC defined three categories of SARS‐CoV‐2 variants: variants of interest (VOI), variants of concern VOC, and variants of high consequence (VOHC). These variants are continually evolving as a result of the number of additional alterations (Figure 1). 26
Figure 1.
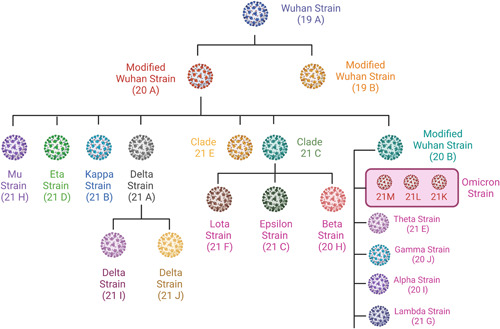
Variants of SARS‐CoV‐2 and their clade. (SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2).
2.1. VOI
It is a variation associated with altered receptor binding, reduced neutralization by antibodies generated in response to past infection or immunization, reduced therapeutic efficacy, possible diagnostic effect, or an anticipated rise in infectivity or growth of the disease. This variation has a nonidentical sequence of receptor binding. 27 Antibodies that are created against additional infection or vaccination can reduce the neutralization. The efficacy of treatment gets reduces which leads to the possible impact of diagnosis or an anticipated increase in infectiousness, or intensity of the disease. 28 Table 1 summarizes VOI that are being monitored to date.
Table 1.
VOI of SARS‐CoV‐2 as per the World Health Organization (WHO)
| Variant name | WHO label | First detected in |
|---|---|---|
| C.37 | Lambda 29 | Peru, December 2020 |
| B.1.621 | Mu 30 | Colombia, January 2021 |
| B.1.526 | Iota 31 | In New York, the United States in November 2020 |
| P.2 | Zeta 31 | In Brazil in April 2020 |
Abbreviations: SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2; VOI, variants of interest.
A mutation, known as D614G, is responsible for one specific viral activity. It is reflected in the reality that viruses with this alteration have a higher transmission rate than viruses without this variation. 32 In the starting phase of the pandemic, this mutation was one of the first documented mutation in the United States, after having initially circulated in Europe. 33 “The phylogenetic assignment of named global outbreak lineage (PANGOLIN),” also known as Pango lineage terminology, was used to designate SARS‐CoV‐2 variants. 34 The phylogeny of SARS‐CoV‐2 is divided into two primary lineages, A and B, as per nomenclature. The most typical lineage of variations is debated. 35
2.2. VOC
A variation characterized by increased amplitude, a substantial decrease in treatment potency or vaccine performance due to neutralization with antibodies produced following previous sickness or inoculation, or diagnostic identification errors (Figure 2). 36 The transmission rate is excessive in this variant type of SARS‐CoV‐2 (Tables 2 and 3). A high rate of transmission leads to more acute disease. 42 There is a possible decrease in neutralization by antibodies produced from earlier illness or immunization. 43 The efficacy of medicines or vaccinations is decreased, or diagnostic recognition fails. 44 Table 4 summarizes all the potential mutations of SARS‐CoV‐2 variants and their impact.
Figure 2.
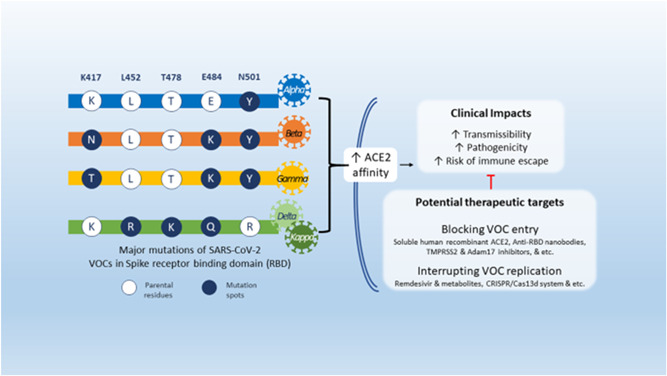
Mutations of SARS‐CoV‐2 VOCs, their clinical implications, and potential therapeutic targets (adopted under Creative Commons Attribution 4.0 International License from Khateeb and Zhang 17 ). SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2; VOC, variants of concern.
Table 2.
| Lineage | The most common countries | Description |
|---|---|---|
| A (A.1, A.2, A.2.2, A.2.3, A.2.4, A.2.5, A.2.5.1, A.2.5.2, A.2.5.3, A.3, etc.) | The USA, Arab countries, Japan, China, Germany, etc. | Lineage A is the reason for the pandemic. China is featured in this genealogy with a wide variety of industries including the vast majority of foreign trade partners like Japan, Australia, the United States, South Korea, and Europe. |
| B (B.1, B.1.1, B.1.1.1, B.1.1.3, B.1.1.4, B.1.1.5, B.1.1.7, Q.1, Q.2, B.58, B.59, B.60, B.61, etc.) | The United Kingdom, The USA, Germany, Spain, Japan, Belgium, Peru, etc. | This is the second most prevalent haplotype. |
| A broad European lineage whose origins generally overlap to the Northern Italian pandemic in early 2020. | ||
| C (C.1, C.1.1, C,2, C.2.1, C.3, C.2, etc.) | South Africa, Zambia, The USA, Mozambique, etc. | B.1.1.1.1's alias |
| D (D.2, D.3, D.4, D.5) | Australia, UK, Denmark, Ireland, Sweden, Bangladesh | Alias of B.1.1.25.2, B.1.1.25.3, B.1.1.25.5 |
| G.1 | United Kingdom | B.1.258.2.1 is an abbreviation for the UK lineage B.1.258.2.1 |
Abbreviations: PANGO, phylogenetic assignment of named global outbreak; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
Table 3.
VOC identified by the CDC and the WHO for SARS‐CoV‐2
| Variant name | WHO label | Spike protein substitutions | Transmissibility | Immune evasiveness | Vaccine effectiveness | First detected In |
|---|---|---|---|---|---|---|
| B.1.1.7 | Alpha 31 , 39 | 69del,70del,144del, (E484K*),N501Y,A570D,D614G,P681H,T7161,S982A, D1118H (K1191N*) | + + + | _ _ | Yes | In the United Kingdom, September 2020 |
| B.1.351 | Beta 39 | D80A, D215G, 241del, 243del, K417N, E484K, N501Y, D614G, A701V | + | + + + + | Yes | in South Africa, May 2020 |
| B.1.617.2 | Delta 40 | T19R, (G142D*), 156del,R158G,L452R,T478K,D614,E484K,N501Y,D614G,A701V | + + + + | + + | Yes | In India, April 2021 |
| P.1 | Gamma 9 , 41 | L18F, T20N,P26S,D138Y,R190S,K4171T,E484K,N501Y,D614G,H655Y,T1027I | ++ | + + | Yes | In Japan/Brazil, November 2020 |
| B.1.429 | Epsilon 35 | S13I,W152C,L452R,L452R,D614 | + | + | Yes | In California, USA, March 2020 |
| B.1.427 | Epsilon 41 | L452R, D614 | + | + | Yes | In California, USA, March 2020 |
Abbreviations: CDC, Centers for Disease Control and Prevention; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2; VOC, variants of concern; WHO, World Health Organization.
Table 4.
SARS‐CoV‐2 mutations of different variants of concern
| Virus structure protein | SARS‐CoV‐2 genome site | Role | Mutation | Variants of concern | |||||
|---|---|---|---|---|---|---|---|---|---|
| Alpha variant | Beta variant | Gamma variant | Epsilon variant | Delta variant | Omicron variant | ||||
| Spike protein | ORF1ab | Binding protein regulation | PLpro: T183I | Yes | No | No | No | No | No |
| PLpro: A890D | |||||||||
| PLpro: I14127 | |||||||||
| Nsp6: | |||||||||
| S106K | |||||||||
| RdRp: | |||||||||
| P323L | |||||||||
| nsp2: | No | Yes | No | No | No | No | |||
| T85I | |||||||||
| PLpro: | |||||||||
| K837N | |||||||||
| 3CL: | |||||||||
| K90R | |||||||||
| nsp6: | |||||||||
| S106K | |||||||||
| RdRP: | |||||||||
| P323L | |||||||||
| Lpro: K38R | No | No | No | No | No | Yes | |||
| PLpro: S1265I | |||||||||
| PLpro: Δ1266 | |||||||||
| PLpro: A1892T | |||||||||
| nsp4: T492I | |||||||||
| 3CL: P132H | |||||||||
| nsp6:L105F | |||||||||
| nsp6: Δ106‐108 | |||||||||
| nsp6: I189V | |||||||||
| RdRP: P323L | |||||||||
| nsp14: I42V | |||||||||
| PLpro:S370L | No | No | Yes | No | No | No | |||
| PLpro:K977Q | |||||||||
| nsp6:S106K | |||||||||
| nsp6:Δ107‐109 | |||||||||
| RdRP:P323L | |||||||||
| nsp13:E341D | |||||||||
| nsp4:V167L | No | No | No | No | Yes | No | |||
| RdRP:P323L | |||||||||
| RdRP:G671S | |||||||||
| nsp13:P77L | |||||||||
| RBD | Increase the binding affinity of the virus | K417N | No | Yes | Yes | No | No | No | |
| G339D | No | No | No | No | No | Yes | |||
| S371L | |||||||||
| S373P | |||||||||
| S375F | |||||||||
| K417N | |||||||||
| RBM | Increase transmissibility and replication | N501Y | Yes | Yes | Yes | No | No | No | |
| E484K | No | Yes | Yes | No | No | No | |||
| L452R | No | No | No | No | Yes | No | |||
| T478K | |||||||||
| N440K | No | No | No | No | No | Yes | |||
| G446S | |||||||||
| S477N | |||||||||
| T478K | |||||||||
| E484A | |||||||||
| Q493R | |||||||||
| G496S | |||||||||
| Q498R | |||||||||
| N501Y | |||||||||
| SD1 | A570D | Yes | No | No | No | No | No | ||
| Y505H | No | No | No | No | No | Yes | |||
| SD2 | D614G | Yes | Yes | No | No | No | No | ||
| H655Y | No | No | Yes | No | No | No | |||
| D614G | No | No | No | No | Yes | No | |||
| T547K | No | No | No | No | No | Yes | |||
| D614G | |||||||||
| H655Y | |||||||||
| S1/S2 | P681H | Yes | No | No | No | No | No | ||
| T7161 | |||||||||
| A701V | No | Yes | No | No | No | No | |||
| P681R | No | No | No | No | Yes | No | |||
| D950N | |||||||||
| N679K | No | No | No | No | No | Yes | |||
| P681H | |||||||||
| N764K | |||||||||
| D796Y | |||||||||
| N856K | |||||||||
| Q954H | |||||||||
| N969K | |||||||||
| L981F7t5 | |||||||||
| N | ORF8: Q27* | Yes | No | No | No | No | No | ||
| ORF8: R521 | |||||||||
| ORF8: Y73C | |||||||||
| N: D3L | |||||||||
| N: R203K | |||||||||
| N: G204R | |||||||||
| N: 5235F | |||||||||
| ORF3a: Q57H | No | Yes | No | No | No | No | |||
| ORF3a: S171L | |||||||||
| E: P71L | |||||||||
| N: T205I | |||||||||
| ORF3a: S26L | No | No | No | No | Yes | No | |||
| M:I82T | |||||||||
| ORF7a:V82A | |||||||||
| ORF7a:T120I | |||||||||
| ORF8:D119I | |||||||||
| ORF8:Δ120‐121 | |||||||||
| N:D63G | |||||||||
| N:R203M | |||||||||
| N:D377Y | |||||||||
| E: T9I | No | No | No | No | No | Yes | |||
| M: D3G | |||||||||
| M: Q19E | |||||||||
| M: A63T | |||||||||
| N: P13L | |||||||||
| N:Δ31‐33 | |||||||||
| N: R203K | |||||||||
| N: G204R | |||||||||
| Outside of spike protein | Enhanced transmissibility | Nsp6: Δ107‐109 | Yes | No | No | No | No | No | |
| NTD | Evasion of antibody neutralization | Δ69‐70 | Yes | No | No | No | No | No | |
| Δ144‐145 | |||||||||
| A67V | No | No | No | No | No | Yes | |||
| Δ69‐70 | |||||||||
| T95I | |||||||||
| G142D | |||||||||
| Δ143‐145 | |||||||||
| N211I | |||||||||
| Δ212 | |||||||||
| 215EPEins | |||||||||
| L18F | No | Yes | No | No | No | No | |||
| T20N | |||||||||
| P26S | |||||||||
| D138Y | |||||||||
| R190S | |||||||||
| ORF3a: S253P | No | No | Yes | No | No | No | |||
| ORF8:E92K | |||||||||
| N:P80R | |||||||||
| N:R203K | |||||||||
| N:G204R | |||||||||
| T19R | No | No | No | No | Yes | No | |||
| G142D | |||||||||
| E156G | |||||||||
| Δ157‐158 | |||||||||
Abbreviation: SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
2.2.1. Alpha variant
WHO has reported that instances of Alpha variants have been diagnosed in around 170 countries and in various territories across the globe. 36 The United Kingdom, Japan, Alaska, the United States of America, and Turkey have faced the severe effect of this alpha variant. More than 10 000 cases of Alpha variant are reported in these countries. 45 Other than these countries, Canada, Mexico, Brazil have also reported more than 8000 confirmed cases and in India, Peru, Russia, China there were around a thousand cases reported of Alpha variants. 46
This is the first VOC reported in the WHO study on variant classification. This strain clade 201/501Y.V1, Pango lineage B.1.1.7, and GISAID clade are all recognized Alpha variant by various scientific names. 47 In the United Kingdom, the very first case of the SARS‐Cov‐2 Alpha variant was detected in September 2020.
Furthermore, the Alpha variant is related to a greater death rate in patients in comparison with other variants. 48 The alteration in the S protein of the virus is the main reason behind its mutation. This new variant also carries NTD and RBD mutations, which play a vital part in the binding of a virus with host cells via the angiotensin‐converting enzyme 2 (ACE2) receptor. S1 subunit of viral spike protein, which is made up of the NTD and the RBD, is essential for defining tissue tropism and host ranges. 49
The N501Y mutation in the RBD of the spike protein, and a few additional mutations, identify the Alpha variant. 50 Among these, there are two deletion mutations in the NTD of the S protein, HV69‐70del and Y144del (also known as Y145del due to the presence of tyrosine at both positions). 51 SARS‐CoV‐2 variants with membrane (M) protein alterations, such as I82T and V70L, have recently been identified as a potential cause of concern. The Alpha variant was revealed to be the result of the consecutive acquisitions of mutations in M Protein: V70L in November 2020 and the unique S Protein: D178H mutation in early February 2021. 52 Pfizer‐BioNTech, Moderna, AstraZeneca‐Oxford, Johnson and Johnson, and Novavax have all proven that their vaccines, based on various designs, can all be effective against this variation. 52 In Phase 3 clinical trial done in the United Kingdom for the Novavax vaccine, for example, showed an efficacy of 89.3% against an Alpha variant (NCT04611802).
SARS‐CoV‐2 mutations are frequent; the COVID‐19 Genomics UK (COG‐UK) Consortium reports that around 4000 mutations have been identified in its spike protein alone. There are 23 mutations in VOC‐202012/01: 14 nonsynonymous mutations, 3 deletions, and 6 identical mutations. 53 Furthermore, the Alpha variant is related to a greater death rate in patients in comparison with other variants. The alteration in the S protein of the virus is the main reason behind its mutation. 53 , 54 Two vaccinations with either BNT162b2 or ChAdOx1 nCoV‐19 demonstrated good protection against Alpha variant and reduces the viral transmission. 50 Another study estimated the efficacy of the Pfizer vaccine of roughly 90%.
2.2.2. Beta variant
Numerous official designations are assigned to the beta variant, including strain clade 20H/501.V2 and Pango lineage B.1.351. 55 The first incidence of the SARS‐Cov‐2 beta strain was identified in the United Kingdom in May 2020, and it was mostly discovered in South Africa. 56 The mutation caused an increase in transmissibility and also the neutralizing capacity of the virus. 26 There are three mutations of significant importance in the spike area of the lineage, B.1.351 genome, K417N, E484K, N501Y, and a further five spike mutations, L18F, D80A, D215G, R246I, A701V, that have so far raised little concern. Aside from the spike area, it also has K1655N, a deletion of SGF 3675‐3677, P71L, and T205I. 57
According to the weekly update released by WHO on June 22, 2021, the cases of beta variants are reported in almost 119 countries. 58 Due to the mutation, there is a reduction in the susceptibility of a virus toward the combination of some monoclonal antibody treatment, like a combination of bamlanivimab and estesevimab. 47 In the RBD of spike protein, notable mutations include N501Y, K417N, and E484K, which can increase the protein's affinity for the human ACE2 receptor. 59 The E484K mutation may allow an individual to evade the immune system's response. 60 According to an in vitro test, all existing vaccines generate antibodies with decreased neutralizing activity against beta variants. 58 , 61 Overall vaccine efficacy for COVID‐19 of any severity was 33.5% up to 14 days after the first vaccine dose. 56 Safety and immunogenicity study of a SARS‐CoV‐2 variant vaccine (mRNA‐1273.351) is currently ongoing sponsored by The National Institute of Allergy and Infectious Diseases (NIAID) (NCT04785144).
2.2.3. Gamma variant
Lineage P.1, frequently referred to as the gamma variant, is a cause of COVID‐19. This variant contains 17 amino acid substitutions and among them, 10 are in its spike protein. 60 This variant was found in the Institute of Infectious Diseases (NIID), Japan. It was later transmitted in Brazil. 62 This variant comprises two subvariants 28‐AM‐1 and 28‐AM‐2 that both carry mutation K417T, E484K, and N501Y. Gamma variant is particularly found from the other Brazilian zeta variant (Lineage P.2). 63 The immunological escape mutation (E484K) is present in this variant. 64 SARS‐CoV‐2 variants gamma contains 10 defining mutations in its spike protein, including N501Y and E484K, in addition to eight other mutations (four of which are synonymous genetic variants) in its ORFs (ORF1a and ORF1b), one of which is a set of deletions. It also possesses two mutations in its ORF8 gene, one of which is an insertion, and one in its N gene. 65 After vaccination with Moderna or Pfizer, the gamma variant has been demonstrated to be relatively resistant to neutralization by convalescent plasma and vaccine sera. 66 The severity of the disease toward death was minor (3.8−4.8‐fold). 67 CoronaVac, an inactivated vaccine has been demonstrated to be 50% effective in preventing sickness 14 days after the first dose in a two‐dose regimen. 68 Over 1000 cases of this variant are diagnosed in Brazil and the United States of America, and less than 100 instances are detected in India, Canada, Australia, and Mexico. 60 In February 2021, more COVID‐19 individuals with no comorbidities were admitted to the ICU. Gamma was discovered to be prevalent in adolescent ICU patients in February 2021. 64 Reinfection by gamma is widespread and may play a large role in epidemics where gamma is ubiquitous, emphasizing the ongoing hazard variations of concern pose even in situations where big epidemics have occurred. 69 Although the clinical significance and transmissibility of reinfections were not investigated, the projected reinfection rates imply that the gamma variation may cause a greater infection risk than earlier non‐gamma versions. As the majority of blood donors had asymptomatic or oligosymptomatic illnesses, the found protection against reinfection does not generalize to cohorts of exclusively hospitalized or symptomatic people. 64 , 69 , 70
2.2.4. Epsilon variant
These resembling variants, B.1.427 and B.1.429 (epsilon variant), were initially identified in California (USA). In the beginning, they were designated as CA VUI1 but afterward WHO classified them and labeled them “epsilon” on May 31, 2021. These variants have a 20% higher efficiency than the original virus and can rapidly transmit from one individual to another. 71 The researchers studied the neutralizing incidence of the epsilon variants on antibodies present in the specimens using plasma from a COVID‐19 recovered individual, along with an entirely immunized person and concluded that their potency was reduced. 72 An estimated rise in the transmission rate is high and it is found in multiple other states of the United States. 73 , 74 It has L452R mutation in S protein which was discovered in the RBD. It increases infectivity because of the interaction between spike protein and ACE2 receptors. 75 W152C has been shown to diminish sensitivity to numerous NTD‐binding monoclonal antibodies, implying yet another involvement in immune evasion. 76 Currently, it is considered a VOI. This variant demonstrated lower susceptibility to neutralization by convalescent (4–6.7‐fold) and postvaccination sera (2–2.9‐fold). 58
2.2.5. Delta variant
B.1.617.2 (Delta variant) is a variant of lineage B.1.617 of SARS‐CoV‐2, which is the reason for India's second wave in this pandemic of COVID‐19. On May 31, 2021 WHO named this variant as “delta variant.” 77 Initially, the Delta variant was detected in India on May 7, 2021. Public health England (PHE) put the Delta variant in the category VOC from a variant under investigation (VUI). The spike protein mutations 19R, (G142D), 156del, 157del, R158G, L452R, T478K, D614G, P681R, and D950N identify and distinguish this variant. Several of these mutations, as well as the loss of a portion of the NTD, may affect immune responses aimed at the critical antigenic areas of RBD, that is, 452, 478, 156, and 157. The P681R mutation alters an amino acid right adjacent to the furin cleavage site, a crucial step, allowing the virus to penetrate human cells and thereby increasing viral infectivity. 78 The spike protein of the new coronavirus is 1273 amino acids long. The RBD of the spike protein is perhaps the most important portion since it is crucial for connecting the SARS‐CoV‐2 virus toward the human ACE2 proteins on certain cells, allowing the virus to invade those cells. 79 This variant is made up of a mutation in the gene−gene that expresses the SARS‐CoV‐2 spike protein, which leads to substitutions in T478K, P681R, and L452R, that are designated to influence the infectiousness of the virus including its ability to be neutralized using antibodies against the formerly propagating form of COVID‐19 virus. 80 Fragments 319−541 define the RBD. The receptor‐binding motif, which connects the spike protein to the human ACE2 receptor, is a critical governance motif in RBD. Any mutation that arises around amino acid residues 319−541—particularly between 438 and 506—may have a major influence on the infectivity of the virus, modes of transmission, intensity, and/or immunity‐evading capability. 81 According to the PHE report, attacks of Delta variant were diagnosed more than 51%−67% than an Alpha variant. 82 Another critical mutation in the RBD, L452R, increases cell transmission efficiency, allowing the variation to spread fast from one individual to another. This mutation is expected to permit 18%–24% increased transmissibility and a 20‐fold reduction in neutralizing titers from the vaccinated individual, as well as resistance to neutralization by particular antibodies. 83 The B.1.617.1 strain is 6.8‐fold more resistant to neutralization by sera from COVID‐19 convalescent and Moderna and Pfizer vaccinated patients, according to a live virus experiment. 84 Despite this, the B.1.617.1 variant was neutralized by the majority of sera from convalescent patients and all sera from vaccinated persons. 85 The mRNA vaccines evaluated here are likely to protect against the B.1.617.1 mutation. 86 Clinical data from vaccinated people should be used to better investigate this. In the UK experiment, the two‐dose Pfizer vaccination was shown to be 87.9% effective against this variant (93.4% effective against B.1.1.7); the two‐dose AstraZeneca vaccine was found to be 59.8% effective against this variant and 66.1% effective against B.1.1.7 (NCT04516746). Two weeks following the second treatment, Pfizer‐BioNTech and Oxford‐AstraZeneca were 88% and 60% effective against the SARS‐CoV‐2 Delta strain, respectively. However, 3 weeks after the initial dose, both of these vaccinations are only 33% effective against the Delta form. The neutralization of the Pfizer‐BioNTech and Moderna vaccines was reduced (Figure 3). 87
Figure 3.
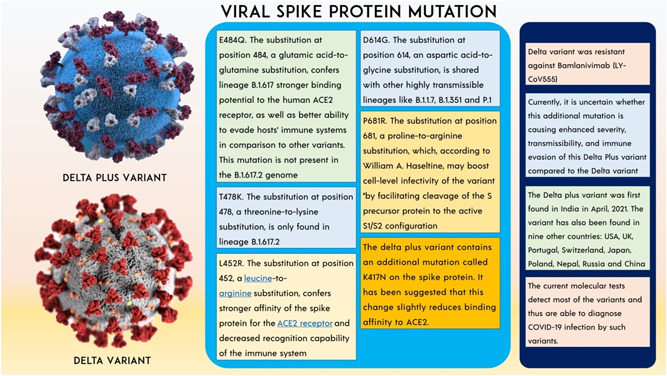
The consequences of Delta variant and spike protein mutation.
Delta plus variant (delta‐AY.1)
B.1.617.2 is somewhat more infectious than B.1.1.7. The main unanswered question is how much more viral it will become. Also, how long will it take to come out of lockdown? If HIV becomes significantly more contagious, individuals will be unable to control the spread, and some form of social distance limitations will very probably be required in the future. Even if the infection rate continues to rise exponentially, we are still likely to see severe illness, more hospitalizations, and tremendous pressure on the health service, including among people who have been immunized. The result of Delta variant mutation, obtaining spike protein mutation K417N to form AY.1 which is Delta plus variant. 88 The P871R mutation is one of the most important in Delta Plus, as it occurs in the furin binding site and improves the efficiency of entry into the cell via the furin cleavage site. It causes syncytia, which allows the virus to infect many cells via the cell‐to‐cell transfer mechanism without leaving the cell. Even monoclonal antibodies are ineffective in this case and are expected to lose some efficiency in the Delta form. 40 , 89
Delta plus variant (delta‐AY.4.2)
As per records published to GISAID, the AY.4.2 lineage of COVID‐19 is a subvariant of the Delta strain, has surfaced in six states of India, showing 17 instances documented yet. 90 However, a team of specialists is still investigating this novel strain, which is considered to be responsible for the latest transmission surge in the United Kingdom. British officials have speculated that AY.4.2 might be significantly more communicable than Delta, albeit there is presently no proof that it triggered more extreme infections or rendered immunizations worthless. 91 Two potential instances of the AY.4.2 strain were detected in India, and the items were transferred to a laboratory for genomic decoding. The alteration A1711V, which alters the virus's NSP3 protein, and serves a variety of functions in viral replication, is the characterizing modification in AY.4.2. Nevertheless, the consequences of these changes are unclear. 90 , 91
2.2.6. Omicron (C.1.2) variant
In November, Omicron was detected in Botswana. Many nations, particularly South Africa, have discovered a novel strain of COVID‐19 known as C.1.2. On November 26, 2021, it was recognized as a VOC. 1 Furthermore, instances of the novel variety have been recorded in Mauritius, England, Switzerland, New Zealand, Portugal, and the Democratic Republic of the Congo (DRC). 92 According to some scientists, this new variant is more certain to be transferrable and can, to a certain degree, avoid the immunity established by vaccinations. 92 Researchers discovered that this new C.1.2 variant is evolving and mutating at a faster pace inside its genome than other VOC or VOI along with the Delta variant. Six of South Africa's nine regions (along with the East and West Capes) had reported instances of C.1.2 strain as of August 13, 2021. 93 Concerns are raised by a large number of spike mutations (at least 32 mutations). “The variant is related to the lambda and beta variants, which are linked to innate immunity. K417N, N440K, G446S, S477N, T478K, E484A, Q493K, G496S, Q498R, N501Y, Y505H, and P681H are the most common spike protein mutations found in omicron variant.” 94 According to preliminary laboratory findings, three doses of the Pfizer‐BioNTech COVID‐19 vaccine neutralize the Omicron variation (B.1.1.529 lineage), but two doses had much lower neutralization titers. 95 Omicron has a mutation known as N501Y, which allows the virus to attach to human cells more firmly. This mutation was found in the Alpha variant as well, and it was connected to its infectivity. According to computational modeling, the variant may also be immune to cell‐mediated immune function. 96 With the upsurge of Omicron VOC, countries must now definitely contemplate reinstating WHO‐recommended fundamental healthcare and social disease standard precautions such as wearing well‐fitting masks, hand hygiene, physical distance, improving indoor ventilation, and avoiding crowded areas if unimmunized. Nations must also speed up COVID‐19 vaccination campaigns. 95 According to the data, “the third dose of BNT162b2 increases neutralizing antibody titers by 25‐fold when compared to two doses against the Omicron variant; titers after the booster dose are comparable to titers seen after two doses against the wild‐type virus, which are linked to high levels of protection. As the mutations in the Omicron form do not alter 80 percent of epitopes in the spike protein identified by CD8+ T cells, two doses may still protect against severe illness.” 97 As per WHO, 94 “on 26 November, the WHO's Technical Advisory Group on SARS‐CoV‐2 virus evolution declared PANGO lineage B.1.1.529 a VOC and designated it with the greek letter omicron. The heavily mutated Omicron coronavirus variant is likely to spread internationally and poses a very high risk of infection surges that could have severe consequences in some part of the globe (28 countries and Territories).” The viral spike protein has reported 32 mutations, 15 of which are in the RBD and influence viral disease transmission, immunological evasion, and vaccine tolerance. 96 , 98 SARS‐CoV‐2 contains many mutations, and each mutation affects the virus's protein binding site differently. The table below contains a thorough explanation of VOC mutation. 31 , 35 , 79 , 99 , 101
As per WHO, 94 “On 26 November, the WHO's Technical Advisory Group on SARS‐CoV‐2 Virus Evolution declared PANGO lineage B.1.1.529 a variant of concern and designated it with the Greek letter omicron. The heavily mutated Omicron coronavirus variant is likely to spread internationally and poses a very high risk of infection surges that could have severe consequences in some part of the globe (28 countries and Territories).” Omicron has a great amount of formerly known mutations in other VOCs, involving at least 32 alterations in the spike protein alone compared with 16 mutations in the existing extremely contagious delta form, and several other viral replication proteins including in NSP12 and NSP14. 95 The likely evolving pattern of the Omicron variation includes the possibility of circulation among chronically infected people. The emergence of the novel variation during the winter wave in various South African nations was undetected owing to poorer genome sequencing in some countries. 94 Spike mutations may have improved Spike's capacity to bind to the ACE2 receptor on host cells. Due to the huge number of mutations observed in the Omicron form, a secret animal reservoir might be responsible. The poor vaccination rate in Africa may have aided in the spread of the Omicron form. 94 Omicron has a mutation known as N501Y, which allows the virus to attach to human cells more firmly. This mutation was found in the Alpha variant as well, and it was connected to its infectivity. According to computational modeling, the variant may also be immune to cell‐mediated immune function. 96 , 102 With the upsurge of Omicron VOC, countries must now definitely contemplate reinstating WHO‐recommended fundamental healthcare and social disease standard precautions such as wearing well‐fitting masks, hand hygiene, physical distance, improving indoor ventilation, and avoiding crowded areas if unimmunized. Nations must also speed up COVID‐19 vaccination campaigns. 95 , 103
Convalescent sera from standard COVID‐19 cohorts have performed poorly in neutralizing omicron. 104 First, unlike Delta and other variations, Omicron prefers a cathepsin‐dependent (E64d‐sensitive) entrance path over a TMPRSS‐like proteases‐dependent (Camostat‐sensitive) entrance route. Such results may reflect the shift in viral tropism in host cells having varying levels of TMPRSS‐like protease, and they point to a mixture of TMPRSS‐like and cathepsin inhibitors as a safe therapy for all SARS‐CoV‐2 strains. Second, despite the P681H mutation, the fusogenicity of Mu and Omicron is much lower than that of other variations. Third, in accordance with fusogenicity, the proinflammatory action of Omicron S protein is mitigated. Fourth, the substantial mutations confer on Mu and Omicron variants the greatest capacity to evade immune protection from vaccination and mNAbs. 105 Altogether, S protein mutations in Lambda, Mu, and Omicron variations change pathogenicity, fusogenicity, and immune function, posing a serious danger to current therapeutic and prophylactic techniques and emphasizing the significance of enforcing strong epidemic prevention measures. A research study conducted by Li et al. demonstrated that “molnupiravir and nirmatrelvir potently inhibited the infection of SARS‐CoV‐2 Omicron variant. The combination of molnupiravir and nirmatrelvir exerted synergistic antiviral activity.” 106 Table 4 summarizes SARS‐CoV‐2 mutations of different VOCs.
2.3. VOHC
It is proven that precautionary measures or medical countermeasures (MCMs) have remarkably lowered their effectivity in the case of VOHCs as compared with that of the abovementioned previously circulating variants. 26 A piece of information to WHO under the nternational health regulations (IHR) is essential in case of these VOHCs, which is further, reported to CDC, which is an announcement to establish certain approaches to avert the transmission and guidance to update to solve this health crisis. Recently, SARS‐CoV‐2 variants showed infection to the degree of severity. 26
Due to COVID‐19, the single most essential action required to manage the continuing SARS‐CoV‐2 epidemic is adequate vaccine administration. Even though numerous vaccines are being given under emergency use authorization, global immunization coverage will only be attained when vaccine supply surpasses vaccine demand. 28 Governments and international private companies have invested billions of dollars in developing viable COVID‐19 vaccines. More than 20 vaccines, including those from Pfizer and Moderna, BioNTech, and Sinopharm, have already been disseminated, with around half of the world's population having been properly immunized. Vaccines are subjected to extensive testing for safety and efficacy before they are licensed for use in the general population. 107 Several prestigious institutes, universities, and major pharmaceutical corporations throughout the world have successfully generated COVID‐19 vaccine candidates that have advanced to clinical trials. However, newly discovered variations may have an impact on their protective effects. 108 Several reaction tactics have been proposed, including speeding major rollouts of existing vaccinations, enhancing vaccine immunogenicity through increased immunization doses, and accelerating next‐generation vaccines against variations. 28 , 109 In this crucial time, the world is preparing the most wide‐reaching and most challenging immunization campaign and leveraging the vaccine's pharmaceutical production capabilities of delivering supplies of vaccines. Vaccination producers are now researching booster doses, which are additional doses of the same vaccine, as well as reformulated vaccinations to target particular variations. 110 SARS‐CoV‐2 is constantly developing and mutating, giving birth to a variety of variations with varying degrees of infectivity and mortality. 111 The virus, which first arose in China, mutated multiple times before causing havoc and taking countless lives globally as part of the continuing COVID‐19 epidemic. 112 Following the Alpha, Beta, Gamma, and Delta variants, the most recently emerged VOC is the Omicron (B.1.1.529), which has evolved as a result of the accumulation of high numbers of mutations, particularly in the spike protein, raising concerns about its potential to dodge pre‐existing immunity obtained through vaccination or infection, and also outperforming antibodies‐based therapies. 113 The Omicron is extremely transmissible and spreads quicker than any prior version; however, it may cause milder symptoms than earlier forms. The Omicron can evade immune system defenses, and coronavirus disease 2019 vaccinations are less effective against the Omicron version. 114
As of January 31, 2022, there have been more than 9.70 billion vaccine doses have been delivered globally, and over 46.7% global population is fully vaccinated. Despite differences in immunization efforts among countries, every effort is being taken to treat and prevent this virus. 115
3. VARIANTS AND VACCINE EFFICACY
Since March 2020, we are facing a global pandemic because of COVID‐19 and its different variants' mutation and this pandemic is having profound social and economic consequences globally. To tackle this hazardous condition, a vaccination strategy is found to be beneficial. In the manufacturing of COVID‐19 vaccines, the focus was on its forms of molecular, particular, and cell‐based types. All the vaccines fundamentally target to produce an antibody‐mediated immune response. 116 Efforts toward developing safe vaccines are taking place all across the world. Currently, approximately 149 vaccine approaches toward SARS‐CoV‐2 are being developed. 109 At the time of writing, there are 168 vaccine candidates and 536 vaccine trials ongoing in more than 62 countries. There are 40 vaccines in phase I clinical trials, 58 in phase II trials, 62 in phase III trials, 33 approved vaccines, around 10 vaccine candidates are in phase IV post‐licensure surveillance, and 8 vaccines that are not further progressing. 117 , 118 The number of SARS‐CoV‐2 variations has increased as the virus has spread over the world. 119 The implementation of long‐term lockdowns to restrict the transmission of SARS‐CoV‐2 is not practicable owing to significant economic and social damage. As a result, worldwide public health measures, along with mass immunization, are the most viable way to contain the SARS‐CoV‐2 outbreak. 3 A COVID‐19 vaccine that is successful will very certainly involve both neutralizing antibodies and a Th1‐driven cellular element. In this section, we analyze the influence of variant of concern on the immune responses generated with the four most commonly used vaccines, as well as their effectiveness. 120 Pfizer, Moderna, BioNTech developed the m‐RNA based vaccine while Covishield is an adenovirus vaccine. 121 Russia invented the recombinant adenovirus vaccine Sputnik V and there were many other vaccines are also developed. 122
Furthermore, if some of the VOC have a higher risk of transmission or pathogenicity, the significance of effective public health interventions and immunization programs will grow. 72 , 123 The international reaction must be both prompt and scientific. It is not hard to adapt vaccines to target mutations. Concerns have been raised concerning the ability to exist vaccinations to defend against new virus strains. 79 , 124 S‐glycoprotein mutations may influence transmission kinetics and the possibility of immunological escape. 125 Vaccination decreases the incidence of delta variant infection and speeds up viral clearance, according to several studies. 119 Despite this, fully vaccinated persons with breakthrough infections have peak viral loads comparable with unprotected patients and may easily spread illness in home settings, including completely vaccinated contacts. 126 When an immunized group has concluded the main vaccination dose, booster doses are given when immunity in that community has dropped below a rate judged adequate over time. The goal of a booster dosage is to re‐establish insufficient vaccination efficacy. Booster vaccination doses lowered both symptomatic and asymptomatic infection incidence similarly. 1 , 2 , 12 , 46 , 127
Over 74% of global civilization has gotten one dose of the COVID‐19 vaccine, and 54% is completely immunized against the disease. It has been provided in more than 6.70 billion doses across the world and 36.67 million doses are administered each day. Even in low‐income nations, just 1.2% of the population has got at least one dosage of the drug. 15 , 128 Table 5 provides brief information about the vaccine efficacy on the different variants of SARS‐CoV‐2.
Table 5.
Variants of SARS‐CoV‐2 and vaccine efficiency
| Vaccine platform | EUA vaccine candidate | Company name | % Efficacy of vaccine during Phase 3 trial | Effectiveness against variants | ||||
|---|---|---|---|---|---|---|---|---|
| Alpha variant | Beta variant | Gamma variant | Delta variant | Omicron variant | ||||
| mRNA (Nucleic acid vaccine) | Comirnaty (BNT162b2) 129 , 130 | Pfizer, and BioNTech | 95% | Yes | Yes | Yes | Yes | Yes |
| Moderna COVID‐19 vaccine (m‐RNA‐1273) 14 | Moderna, BARDA, and NIAID | 94% | No | Yes | Yes | No | Yes | |
| Moderna spikevax 14 | Moderna | 90% | Yes | No | Yes | Yes | Yes | |
| DNA (Nucleic acid vaccine) | ZyCoV‐D 131 | Zydus Cadila | 90% | Yes | Yes | Yes | Yes | Yes |
| Nonreplicating viral vector vaccine | COVID‐19 vaccine AstraZeneca (AZD1222); also known as Vaxzevria and Covishield 7 | BARDA, OWS | 76% | Yes | Yes | No | Yes | Yes |
| Sputnik V 132 | Gamaleya Research Institute, Acellena Contract Drug Research, and Development | 91% | Yes | Yes | No | Yes | Yes | |
| Sputnik light 7 | Gamaleya Research Institute, Acellena Contract Drug Research, and Development | 79.4% | Yes | No | Yes | No | Yes | |
| JNJ‐78436735 133 , 134 | Janssen vaccines (Johnson & Johnson) | 85% | Yes | Yes | Yes | No | Yes | |
| Convidicea 135 , 136 (PakVac, Ad5‐nCoV) | CanSino Biologics | 65.7% | Yes | Yes | No | Yes | No | |
| Inactivated vaccine Inactivated vaccine | CoronaVac 137 | Sinovac | 51% | Yes | No | Yes | No | No |
| BBIBP‐CorV | Beijing Institute of Biological Products; China National Pharmaceutical Group (Sinopharm) | 78.1% | No | Yes | No | Yes | No | |
| Covaxin 138 | Bharat Biotech | 77.8% | Yes | No | Yes | No | Yes | |
| KoviVac | Chumakov Center | 58% | No | Yes | No | Yes | No | |
| Turkovac | Health Institutes of Turkey | 60% | No | Yes | Yes | Yes | Yes | |
| KCONVAC | Minhai Biotechnology Co. | – | No | Yes | No | No | No | |
| FAKHRAVAC (MIVAC) | Organization of Defensive Innovation and Research | – | Yes | No | No | Yes | Yes | |
| QazVac | Research Institute for Biological Safety Problems (RIBSP) | 96% | — | – | Yes | Yes | — | |
| Inactivated (Vero Cells) | Sinopharm (Wuhan) | 79% | — | Yes | No | Yes | No | |
| COVIran Barekat | Shifa Pharmed Industrial Co. | 93.5% | — | No | Yes | Yes | No | |
| Covilo | Sinopharm (Beijing) | 79% | — | No | No | Yes | No | |
| Protein subunit vaccine | EpiVacCorona | Federal Budgetary Research Institution, State Research Center of Virology and Biotechnology | 79% | No | No | Yes | No | No |
| SpikoGen | Vaxine/CinnaGen Co. | 60% | Yes | Yes | No | Yes | No | |
| Aurora‐CoV | Vector State Research Center of Virology and Biotechnology | 90% | — | — | No | — | No | |
| COVOVAX (Novavax formulation) | Serum Institute of India | 96.4% | Yes | — | No | — | No | |
| Razi Cov Pars | Razi Vaccine and Serum Research Institute | 90% | — | Yes | No | Yes | No | |
| Recombinant SARS‐CoV‐2 Vaccine (CHO Cell) | National Vaccine and Serum Institute | – | Yes | Yes | No | Yes | No | |
| Nuvaxovid | Novavax | 92.6% | Yes | Yes | No | Yes | No | |
| MVC‐COV1901 | Medigen | – | Yes | — | No | Yes | No | |
| Soberana Plus | Instituto Finlay de Vacunas Cuba | 91.2% | — | — | No | Yes | No | |
| Soberana 02 | Instituto Finlay de Vacunas Cuba | 92.4% | — | — | No | Yes | No | |
| Zifivax | Anhui Zhifei Longcom | 82% | — | — | No | Yes | No | |
| Corbevax | Biological E Limited | 90% | No | Yes | No | No | No | |
| Abdala | Center for Genetic Engineering and Biotechnology (CIGB) | 92% | No | Yes | No | Yes | No | |
Abbreviations: EUA, Emergency Use Authorization; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
Some of these vaccines are currently in clinical testing, and their efficacy and effectiveness against various emerging viral variants are still being studied. 139 And over 7.9% of COVID‐19 vaccination programs have been delivered globally over a year of lockdowns and social isolation, and around 19.4% of people have been completely immunized. 140 We, humans, are racing against time to develop immunity to this elusive virus, whose ability to mutate and evolve seems to be outpacing our ability to achieve herd immunity. Due to the new variants, it may be a sprint to the finish line. 141
We, humans, are racing against time to develop immunity to this elusive virus, whose ability to mutate and evolve seems to be outpacing our ability to achieve herd immunity. Due to the new variants, it may be a sprint to the finish line. 142
These variants are concerning for several reasons. First, the SARS‐CoV‐2 VOC spread at least 20%−50% more quickly from person to person. This encourages them to infect more people and grow faster and farther, gradually becoming the dominant paradigm. Second, SARS‐CoV‐2 VOC can create more acute illness, as well as an uptick in hospitalizations and deaths. In other words, they may be more virulent. 143 According to Richard Lessells, “If a virus is going through an evolutionary process inside the host, then it is quite likely that it would be adapting to be better at entering the cells and evading the immune response; this could lead to a variant with enhanced transmissibility and enhanced immune evasion.”
Herbal remedies, 144 drug repurposing, and nanotechnology‐based formulations are also proved to be efficient in disease management. 140 , 141 , 142 , 143 , 144 , 145 , 146 , 147 We can see the consequences: tragic deaths, worldwide epidemic outbreaks, and lockdowns. 148 Research on vaccine efficacy, particular groundbreaking illnesses, and the capacity of postvaccination serum to destroy emerging variant viruses are major elements of assessing vaccination's efficiency in managing COVID‐19 in an arena of developing viral variants. Finally, a concentrated and well‐coordinated public health effort, as well as quick and broad adoption of effective vaccinations, is required to stay ahead of the inevitable emergence of variations that might severely expedite the pandemic's progression.
4. CONCLUSION
Generally, viruses mutate to adapt and sustain themselves in the environment. The critical thing here would be remembering this fact about COVID‐19 as and when this situation is resolved. The need for instruments that enable quick identification and close monitoring of SARS‐CoV‐2 VOCs is higher than ever because these variants are more communicable and hence put more strain on health services. Non‐Spike variants should be targeted for research into their involvement in escaping innate immunity and enhancing SARS‐CoV‐2 proliferation, as well as their relevance to viral viability more broadly. As viral variants have the ability to evade naturally acquired and vaccine‐induced immunity, the invention of next‐generation vaccines that trigger widely neutralizing action against present and possible future SARS‐CoV‐2 variants is the main objective. Control of transcription and replication by both public health interventions and fair vaccination dissemination is crucial in lowering the danger of novel variant creation. A validated immunization technique that is effective against the majority of VOCs is urgently needed. Scientists should consider nasal vaccination as well, as it delivers localized immunity. Furthermore, we should maintain extreme monitoring in following all preventative measures to limit the transmission of SARS‐Co‐2.
AUTHOR CONTRIBUTIONS
Conceptualization: Vivek P. Chavda. Writing—original draft preparation: Aayushi B. Patel, Vivek P. Chavda, and Darsh D. Vaghasiya. Writing—review and editing: Vivek P. Chavda. All authors have read and agreed to the published version of the manuscript. Vivek P. Chavda, Darsh D. Vaghasiya, and Aayushi B. Patel dedicate this article to L.M. College of Pharmacy on the 75th Year celebration. Figure 1 is created with Biorender.com. For viral variant‐related information, we have also referred to https://nextstrain.org/ and GISAID.
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
ACKNOWLEDGMENT
VP Chavda wants to dedicate this work to LM College of pharmacy as a part of the 75th year celebration of the college.
Chavda VP, Patel AB, Vaghasiya DD. SARS‐CoV‐2 variants and vulnerability at the global level. J Med Virol. 2022;94:2986‐3005. 10.1002/jmv.27717
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.
REFERENCES
- 1. Chavda VP, Apostolopoulos V. Omicron variant (B.1.1.529) of SARS‐CoV‐2: threat for the elderly? Maturitas. 2022;158:78‐81. 10.1016/j.maturitas.2022.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Basu D, Chavda VP, Mehta AA. Therapeutics for COVID‐19 and post COVID‐19 complications: an update. Curr Res Pharmacol Drug Discov. 2022;3:100086. 10.1016/j.crphar.2022.100086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bian L, Gao F, Zhang J, et al. Effects of SARS‐CoV‐2 variants on vaccine efficacy and response strategies. Expert Rev Vaccines. 2021;20(4):365‐373. 10.1080/14760584.2021.1903879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kames J, Holcomb DD, Kimchi O, et al. Sequence analysis of SARS‐CoV‐2 genome reveals features important for vaccine design. Sci Rep. 2020;10(1):15643. 10.1038/s41598-020-72533-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sanjuán R, Domingo‐Calap P. Mechanisms of viral mutation. Cell Mol Life Sci. 2016;73(23):4433‐4448. 10.1007/s00018-016-2299-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Snijder EJ, Decroly E, Ziebuhr J. The nonstructural proteins directing coronavirus RNA synthesis and processing. Adv Virus Res. 2016;96:59‐126. 10.1016/bs.aivir.2016.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chavda VP, Kapadia C, Soni S, et al. A global picture: therapeutic perspectives for COVID‐19. Immunotherapy. 2022;14:351‐371. 10.2217/imt-2021-0168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chavda VP, Gajjar N, Shah N, Dave DJ. Darunavir ethanolate: repurposing an anti‐HIV drug in COVID‐19 treatment. Eur J Med Chem Reports. 2021;3:100013. 10.1016/j.ejmcr.2021.100013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yadav PD, Sapkal GN, Abraham P, et al. Neutralization of variant under investigation B.1.617 with sera of BBV152 vaccinees. Clin Infect Dis. 2021;74:366‐368. 10.1093/cid/ciab411 [DOI] [PubMed] [Google Scholar]
- 10. Pokhrel S, Kraemer BR, Lee L, Samardzic K, Mochly‐Rosen D. Increased elastase sensitivity and decreased intramolecular interactions in the more transmissible 501Y.V1 and 501Y.V2 SARS‐CoV‐2 variants' spike protein—an in silico analysis. PLoS ONE. 2021;16(5):e0251426. 10.1371/journal.pone.0251426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chavda VP, Pandya R, Apostolopoulos V. DNA vaccines for SARS‐CoV‐2: towards third generation vaccination era. Expert Rev Vaccines. 2021;20:1549‐1560. 10.1080/14760584.2021.1987223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chavda VP, Apostolopoulos V. Mucormycosis—an opportunistic infection in the aged immunocompromised individual: a reason for concern in COVID‐19. Maturitas. 2021;58:58‐61. 10.1016/j.maturitas.2021.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chavda VP, Vora LK, Pandya AK, Patravale VB. Intranasal vaccines for SARS‐CoV‐2: from challenges to potential in COVID‐19 management. Drug Discov Today. 2021;26(11):2619‐2636. 10.1016/j.drudis.2021.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chavda VP, Hossain MK, Beladiya J, Apostolopoulos V. Nucleic acid vaccines for COVID‐19: a paradigm shift in the vaccine development arena. Biologics. 2021;1(3):337‐356. 10.3390/biologics1030020 [DOI] [Google Scholar]
- 15. Chavda VP, Vora LK, Vihol DR. COVAX‐19® vaccine: completely blocks virus transmission to non‐immune individuals. Clin Complement Med Pharmacol. 2021;1(1):100004. 10.1016/j.ccmp.2021.100004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ramesh S, Govindarajulu M, Parise RS, et al. Emerging SARS‐CoV‐2 variants: a review of its mutations, its implications and vaccine efficacy. Vaccines. 2021;9(10):1‐35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Khateeb J, Li Y, Zhang H. Emerging SARS‐CoV‐2 variants of concern and potential intervention approaches. Crit Care. 2021;25(1):244. 10.1186/s13054-021-03662-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hodgson SH, Mansatta K, Mallett G, Harris V, Emary KRW, Pollard AJ. What defines an efficacious COVID‐19 vaccine? A review of the challenges assessing the clinical efficacy of vaccines against SARS‐CoV‐2. Lancet Infect Dis. 2021;21(2):e26‐e35. 10.1016/S1473-3099(20)30773-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tahan S, Parikh BA, Droit L, Wallace MA, Burnham CD, Wang D. SARS‐CoV‐2 E gene variant alters analytical sensitivity characteristics of viral detection using a commercial reverse transcription‐PCR assay. J Clin Microbiol. 2022;59(7):e00075‐21. 10.1128/JCM.00075-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tang YE. Laboratory diagnosis of COVID‐19: current issues and challenges. J Clin Microbiol. 2022;58(6):e00512‐20. 10.1128/JCM.00512-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brégeon D, Doetsch PW. Transcriptional mutagenesis: causes and involvement in tumour development. Nat Rev Cancer. 2011;11(3):218‐227. 10.1038/nrc3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Arvin AM, Greenberg HB. New viral vaccines. Virology. 2006;344(1):240‐249. 10.1016/j.virol.2005.09.057 [DOI] [PubMed] [Google Scholar]
- 23. Donati Zeppa S, Agostini D, Piccoli G, Stocchi V, Sestili P. Gut microbiota status in COVID‐19: an unrecognized player? Front Cell Infect Microbiol. 2020;10:576551. 10.3389/fcimb.2020.576551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Koelle K, Rasmussen DA. The effects of a deleterious mutation load on patterns of influenza A/H3N2' s antigenic evolution in humans. eLife. 2015;4(2007):1‐31. 10.7554/eLife.07361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. CDC . How the Flu Virus Can Change: “Drift” and “Shift”. Centers for Disease Control and Prevention, National Center for Immunization and Respiratory Diseases (NCIRD); 2019. Accessed February 22, 2022. https://www.cdc.gov/flu/about/viruses/change.htm [Google Scholar]
- 26. CDC . What You Need to Know About Variants. National Center for Immunization and Respiratory Diseases (NCIRD), Division of Viral Diseases; 2021. Accessed February 21, 2022. https://www.cdc.gov/coronavirus/2019-ncov/variants/about-variants.html#:%7E:text=Newvariantsofthevirus,virusthatcausesCOVID-19 [Google Scholar]
- 27. Konings F, Perkins MD, Kuhn JH, et al. SARS‐CoV‐2 variants of interest and concern naming scheme conducive for global discourse. Nat Microbiol. 2021;6(7):821‐823. 10.1038/s41564-021-00932-w [DOI] [PubMed] [Google Scholar]
- 28. Koch T, Fathi A, Addo MM. The COVID‐19 vaccine landscape. Adv Exp Med Biol. 2021;1318:549‐573. 10.1007/978-3-030-63761-3_31 [DOI] [PubMed] [Google Scholar]
- 29. Romero PE, Dávila‐Barclay A, Salvatierra G. The emergence of Sars‐CoV‐2 variant Lambda (C.37) in South America. Microbiol Spectr. 2022;9(2):e00789-21. 10.1128/Spectrum.00789-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Menni C, Klaser K, May A, et al. Vaccine side‐effects and SARS‐CoV‐2 infection after vaccination in users of the COVID Symptom Study app in the UK: a prospective observational study. Lancet Infect Dis. 2021;21(7):939‐949. 10.1016/S1473-3099(21)00224-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Annavajhala MK, Mohri H, Wang P, et al. A novel SARS‐CoV‐2 variant of concern, B.1.526, identified in New York. medRxiv Prepr Serv Heal Sci. Published online February 2021. 10.1101/2021.02.23.21252259 [DOI] [Google Scholar]
- 32. Wu K, Werner AP, Moliva JI, et al. mRNA‐1273 vaccine induces neutralizing antibodies against spike mutants from global SARS‐CoV‐2 variants. bioRxiv Prepr Serv Biol. Published online January 2021. 10.1101/2021.01.25.427948 [DOI] [Google Scholar]
- 33. Meng B, Kemp SA, Papa G, et al. Recurrent emergence of SARS‐CoV‐2 spike deletion H69/V70 and its role in the Alpha variant B.1.1.7. Cell Rep. 2021;35(13):109292. 10.1016/j.celrep.2021.109292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. O'Toole Á, Pybus OG, Abram ME, Kelly EJ, Rambaut A. Pango lineage designation and assignment using SARS‐CoV‐2 spike gene nucleotide sequences. BMC Genomics. 2022;23(1):121. 10.1186/s12864-022-08358-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Deng X, Garcia‐Knight MA, Khalid MM, et al. Transmission, infectivity, and antibody neutralization of an emerging SARS‐CoV‐2 variant in California carrying a L452R spike protein mutation. medRxiv Prepr Serv Heal Sci. Published online March 2021. 10.1101/2021.03.07.21252647 [DOI] [Google Scholar]
- 36. Choi JY, Smith DM. SARS‐CoV‐2 variants of concern. Yonsei Med J. 2021;62(11):961‐968. 10.3349/ymj.2021.62.11.961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rambaut A, Holmes EC, O'toole Á, et al. A dynamic nomenclature proposal for SARS‐CoV‐2 lineages to assist genomic epidemiology. Nat Microbiol. 2020;5(11):1403‐1407. 10.1038/s41564-020-0770-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Saxena SK, Kumar S, Ansari S, et al. Transmission dynamics and mutational prevalence of the novel severe acute respiratory syndrome coronavirus‐2 Omicron Variant of Concern. J Med Virol. 2022;94:2160‐2166. 10.1002/jmv.27611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jangra S, Ye C, Rathnasinghe R, et al. SARS‐CoV‐2 spike E484K mutation reduces antibody neutralisation. The Lancet Microbe. 2021;2:283. 10.1016/S2666-5247(21)00068-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chavda VP, Apostolopoulos V. Global impact of delta plus variant and vaccination. Expert Rev Vaccines. 2022;28:1‐4. 10.1080/14760584.2022.2044800 [DOI] [PubMed] [Google Scholar]
- 41. Greaney AJ, Loes AN, Crawford KHD, et al. Comprehensive mapping of mutations in the SARS‐CoV‐2 receptor‐binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe. 2021;29(3):463‐476. 10.1016/j.chom.2021.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Aleem A, Akbar Samad AB, Slenker AK. Emerging variants of SARS‐CoV‐2 and novel therapeutics against coronavirus (COVID‐19). StatPearls [Internet]. StatPearls Publishing; 2022. [PubMed] [Google Scholar]
- 43. Burki T. The origin of SARS‐CoV‐2 variants of concern. Lancet Infect Dis. 2022;22(2):174‐175. 10.1016/S1473-3099(22)00015-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Division of Viral Diseases . SARS‐CoV‐2 Variant Classifications and Definitions. CDC Science, National Center for Immunization and Respiratory Diseases (NCIRD), Division of Viral Diseases; 2021. Accessed May 23, 2021. https://www.cdc.gov/coronavirus/2019-ncov/cases-updates/variant-surveillance/variant-info.html [Google Scholar]
- 45. Domingo P, de Benito N. Alpha variant SARS‐CoV‐2 infection: how it all starts. EBioMedicine. 2021;74:74. 10.1016/j.ebiom.2021.103703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Singanayagam A, Hakki S, Dunning J, et al. Community transmission and viral load kinetics of the SARS‐CoV‐2 delta (B.1.617.2) variant in vaccinated and unvaccinated individuals in the UK: a prospective, longitudinal, cohort study. Lancet Infect Dis. 2021;21:363. 10.1016/S1473-3099(21)00648-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Roquebert B, Trombert‐paolantoni S, Haim‐boukobza S, et al. The SARS‐CoV‐2 B. 1. 351 lineage (VOC β) is outgrowing the B 1. 1. 7 lineage (VOC α) in some French regions in April 2021. Euro Surveill. 2021;26(23):2100447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Davies NG, Jarvis CI, Group CC‐W, et al. Increased mortality in community‐tested cases of SARS‐CoV‐2 lineage B.1.1.7. Nature. 2021;593(7858):270‐274. 10.1038/s41586-021-03426-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hart WS, Miller E, Andrews NJ, et al. Generation time of the alpha and delta SARS‐CoV‐2 variants: an epidemiological analysis. Lancet Infect Dis. 2022. 10.1016/S1473-3099(22)00001-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Eyre DW, Taylor D, Purver M, et al. Effect of Covid‐19 vaccination on transmission of alpha and delta variants. N Engl J Med. 2022;386(8):744‐756. 10.1056/NEJMoa2116597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Duong D. Alpha, Beta, Delta, Gamma: what's important to know about SARS‐CoV‐2 variants of concern? CMAJ. 2021;193(27):E1059‐E1060. 10.1503/cmaj.1095949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shen L, Bard JD, Triche TJ, Judkins AR, Biegel JA, Gai X. Rapidly emerging SARS‐CoV‐2 B.1.1.7 sub‐lineage in the United States of America with spike protein D178H and membrane protein V70L mutations. Emerge Microbes Infect. 2021;10(1):1293‐1299. 10.1080/22221751.2021.1943540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Walker AS, Vihta K‐D, Gethings O, et al. Tracking the emergence of SARS‐CoV‐2 alpha variant in the United Kingdom. N Engl J Med. 2021;385(27):2582‐2585. 10.1056/NEJMc2103227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lessells RJ. SARS‐CoV‐2 variants of concern: the knowns and unknowns. Anaesthesia, Crit Care Pain Med. 2021;40(3):100868. 10.1016/j.accpm.2021.100868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Joshi G, Borah P, Thakur S, Sharma P, Mayank Poduri R. Exploring the COVID‐19 vaccine candidates against SARS‐CoV‐2 and its variants: where do we stand and where do we go? Hum Vaccin Immunother. 2021;17:1‐27. 10.1080/21645515.2021.1995283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhou D, Dejnirattisai W, Supasa P, et al. Evidence of escape of SARS‐CoV‐2 variant B.1.351 from natural and vaccine‐induced sera. Cell. 2021;184(9):2348‐2361. 10.1016/j.cell.2021.02.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Moores LK, Tritschler T, Brosnahan S, et al. Prevention, diagnosis, and treatment of VTE in patients with coronavirus disease 2019: CHEST Guideline and Expert Panel Report. Chest. 2020;158(3):1143‐1163. 10.1016/j.chest.2020.05.559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Garcia‐Beltran WF, Lam EC, St Denis K, et al. Multiple SARS‐CoV‐2 variants escape neutralization by vaccine‐induced humoral immunity. Cell. 2021;184(9):2372‐2383. 10.1016/j.cell.2021.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yadav PD, Sarkale P, Razdan A, et al. Isolation and characterization of SARS‐CoV‐2 Beta variant from UAE travelers. J Infect Public Health. 2022;15(2):182‐186. 10.1016/j.jiph.2021.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Dejnirattisai W, Zhou D, Supasa P, et al. Antibody evasion by the P.1 strain of SARS‐CoV‐2. Cell. 2021;184(11):2939‐2954. 10.1016/j.cell.2021.03.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. World Health Organization . COVID‐19 weekly epidemiological update 22. World Heal Organ. 2021;(December):1‐3. [Google Scholar]
- 62. Bhuiyan MSA, Amin Z. Bakar AMSA, et al. Factor influences for diagnosis and vaccination of avian infectious bronchitis virus (Gammacoronavirus) in chickens. Vet Sci. 2021;8(3):47. 10.3390/vetsci8030047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Pal M, Berhanu G, Desalegn C, Kandi V. Severe acute respiratory syndrome coronavirus‐2 (SARS‐CoV‐2): an update. Cureus. 2020;12(3):e7423. 10.7759/cureus.7423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Nonaka CKV, Gräf T, Barcia C, et al. SARS‐CoV‐2 variant of concern P.1 (Gamma) infection in young and middle‐aged patients admitted to the intensive care units of a single hospital in Salvador, Northeast Brazil, February 2021. Int J Infect Dis. 2021;111:47‐54. 10.1016/j.ijid.2021.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Brant AC, Tian W, Majerciak V, Yang W, Zheng Z‐M. SARS‐CoV‐2: from its discovery to genome structure, transcription, and replication. Cell Biosci. 2021;11(1):136. 10.1186/s13578-021-00643-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Collier DA, Ferreira IATM, Kotagiri P. Age‐related immune response heterogeneity to SARS‐CoV‐2 vaccine BNT162b2. Nature. 2021;596(7872):417‐422. 10.1038/s41586-021-03739-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Toovey OTR, Harvey KN, Bird PW, Tang JWW. Introduction of Brazilian SARS‐CoV‐2 484K.V2 related variants into the UK. J Infect. 2021;82(5):e23‐e24. 10.1016/j.jinf.2021.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wang Z, Schmidt F, Weisblum Y, et al. mRNA vaccine‐elicited antibodies to SARS‐CoV‐2 and circulating variants. Nature. 2021;592(January):616‐622. 10.1038/s41586-021-03324-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Prete CA Jr, Buss LF, Buccheri R, et al. Reinfection by the SARS‐CoV‐2 Gamma variant in blood donors in Manaus, Brazil. BMC Infect Dis. 2022;22(1):127. 10.1186/s12879-022-07094-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Vargas‐Herrera N, Araujo‐Castillo RV, Mestanza O, Galarza M, Rojas‐Serrano N, Solari‐Zerpa L. SARS‐CoV‐2 Lambda and Gamma variants competition in Peru, a country with high seroprevalence. Lancet Reg Heal—Am. 2022;6:6. 10.1016/j.lana.2021.100112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Martin Webb L, Matzinger S, Grano C, et al. Identification of and surveillance for the SARS‐CoV‐2 variants B.1.427 and B.1.429—Colorado, January–March 2021. MMWR Morb Mortal Wkly Rep. 2021;70(19):717‐718. 10.15585/mmwr.mm7019e2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Otto SP, Day T, Arino J, et al. The origins and potential future of SARS‐CoV‐2 variants of concern in the evolving COVID‐19 pandemic. Curr Biol. 2021;31(14):R918‐R929. 10.1016/j.cub.2021.06.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Khandia R, Singhal S, Alqahtani T, et al. Emergence of SARS‐CoV‐2 Omicron (B.1.1.529) variant, salient features, high global health concerns and strategies to counter it amid ongoing COVID‐19 pandemic. Environ Res. 2022;209:112816. 10.1016/j.envres.2022.112816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Gonzalez‐Parra G. Analysis of delayed vaccination regimens: a mathematical modeling approach. Epidemiologia. 2021;2(3):271‐293. 10.3390/epidemiologia2030021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Thiagarajan K. Covid‐19: India is at centre of global vaccine manufacturing, but opacity threatens public trust. BMJ. 2021;372:n196. 10.1136/bmj.n196 [DOI] [PubMed] [Google Scholar]
- 76. McCallum M, Bassi J, De Marco A, et al. SARS‐CoV‐2 immune evasion by the B.1.427/B.1.429 variant of concern. Science. 2021;373(6555):648‐654. 10.1126/science.abi7994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Lauring AS, Malani PN. Variants of SARS‐CoV‐2. JAMA. 2021;326(9):880. 10.1001/jama.2021.14181 [DOI] [PubMed] [Google Scholar]
- 78. Alkhatib M, Svicher V, Salpini R, et al. SARS‐CoV‐2 variants and their relevant mutational profiles: update summer 2021. Microbiol Spectr. 2022;9(3):e01096‐21. 10.1128/Spectrum.01096-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Harvey WT, Carabelli AM, Jackson B, et al. SARS‐CoV‐2 variants, spike mutations and immune escape. Nat Rev Microbiol. 2021;19:614‐424. 10.1038/s41579-021-00573-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Lauring AS, Hodcroft EB. Genetic variants of SARS‐CoV‐2—what do they mean? JAMA. 2021;325(6):529‐531. 10.1001/jama.2020.27124 [DOI] [PubMed] [Google Scholar]
- 81. Cherian S, Potdar V, Jadhav S, et al Convergent evolution of SARS‐CoV‐2 spike mutations, L452R, E484Q and P681R, in the second wave of COVID‐19 in Maharashtra, India. bioRxiv. 2021. 2021.04.22.440932 10.1101/2021.04.22.440932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Twohig KA, Nyberg T, Zaidi A, et al. Hospital admission and emergency care attendance risk for SARS‐CoV‐2 delta (B.1.617.2) compared with alpha (B.1.1.7) variants of concern: a cohort study. Lancet Infect Dis. 2022;22(1):35‐42. 10.1016/S1473-3099(21)00475-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Chakraborty C, Bhattacharya M, Sharma AR. Present variants of concern and variants of interest of severe acute respiratory syndrome coronavirus 2: their significant mutations in S‐glycoprotein, infectivity, re‐infectivity, immune escape and vaccines activity. Rev Med Virol. 2021;32(2):e2270. 10.1002/rmv.2270 [DOI] [Google Scholar]
- 84. Lopez Bernal J, Andrews N, Gower C, et al. Effectiveness of COVID‐19 vaccines against the B. 1. 617. 2 variant. N Engl J Med. 2021;385(7):585‐594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Edara V‐V, Lai L, Sahoo MK, et al. Infection and vaccine‐induced neutralizing antibody responses to the SARS‐CoV‐2 B.1.617.1 variant. bioRxiv Prepr Serv Biol. Published online May 2021. 10.1101/2021.05.09.443299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Liu J, Liu Y, Xia H, et al. BNT162b2‐elicited neutralization of B.1.617 and other SARS‐CoV‐2 variants. Nature. 2021;596(7871):273‐275. 10.1038/s41586-021-03693-y [DOI] [PubMed] [Google Scholar]
- 87. Iacobucci G. Covid‐19: single vaccine dose is 33% effective against variant from India, data show. BMJ. 2021;373:n1346. 10.1136/bmj.n1346 [DOI] [PubMed] [Google Scholar]
- 88. Das A, Ahmed R, Akhtar S, Begum K, Banu S. An overview of basic molecular biology of SARS‐CoV‐2 and current COVID‐19 prevention strategies. Gene Rep. 2021;23:101122. 10.1016/j.genrep.2021.101122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Thakur V, Bhola S, Thakur P, et al. Waves and variants of SARS‐CoV‐2: understanding the causes and effect of the COVID‐19 catastrophe. Infection. 2021;16:1‐16. 10.1007/s15010-021-01734-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Kannan SR, Spratt AN, Cohen AR, et al. Evolutionary analysis of the Delta and Delta Plus variants of the SARS‐CoV‐2 viruses. J Autoimmun. 2021;124:102715. 10.1016/j.jaut.2021.102715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Planas D, Veyer D, Baidaliuk A, et al. Reduced sensitivity of SARS‐CoV‐2 variant Delta to antibody neutralization. Nature. 2021;596:276‐280. [DOI] [PubMed] [Google Scholar]
- 92. Gaspar‐Marques J, van Zeller M, Carreiro‐Martins P, Chaves Loureiro C. Severe asthma in the era of COVID‐19: a narrative review. Pulmonology. 2021;28:34‐43. 10.1016/j.pulmoe.2021.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Lennon A. COVID‐19: what do we know about the C.1.2 variant. Medical News Today. September 10, 2021.
- 94. WHO . Classification of Omicron (B.1.1.529): SARS‐CoV‐2 variant of concern. Accessed November 30, 2021. https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern
- 95. Petersen E, Ntoumi F, Hui DS, et al. Emergence of new SARS‐CoV‐2 Variant of Concern Omicron (B.1.1.529)—highlights Africa's research capabilities, but exposes major knowledge gaps, inequities of vaccine distribution, inadequacies in global COVID‐19 response and control efforts. Int J Infect Dis. 2022;114:268‐272. 10.1016/j.ijid.2021.11.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Callaway E. Heavily mutated Omicron variant puts scientists on alert. Nature News. 2021. Accessed November 30, 2021. https://www.nature.com/articles/d41586-021-03552-w [DOI] [PubMed]
- 97. Moss P. The T cell immune response against SARS‐CoV‐2. Nat Immunol. 2022;23(2):186‐193. 10.1038/s41590-021-01122-w [DOI] [PubMed] [Google Scholar]
- 98. Variant Technical Group (UK Health Security Agency). SARS‐CoV‐2 Variants of Concern and Variants under Investigation in England; 2021. Acessed December 12, 2021. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/1036501/Technical_Briefing_29_published_26_November_2021.pdf
- 99. Drożdżal S, Rosik J, Lechowicz K, et al. An update on drugs with therapeutic potential for SARS‐CoV‐2 (COVID‐19) treatment. Drug Resist Updat. 2021;59(2):100794. 10.1016/j.drup.2021.100794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Winger A, Caspari T. The spike of concern — the novel variants of SARS‐CoV‐2. Viruses. 2021;13(6):1‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Standford University . Coronavirus Antiviral & Resistance Database; 2021. Accessed February 22, 2022. https://covdb.stanford.edu/
- 102. Kandimalla R, Chakraborty P, Vallamkondu J, et al. Counting on COVID‐19 vaccine: insights into the current strategies, progress and future challenges. Biomedicines. 2021;9(11):1740. 10.3390/biomedicines9111740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Klemeš JJ, Jiang P, Fan YVan, Bokhari A, Wang X‐C. COVID‐19 pandemics Stage II—energy and environmental impacts of vaccination. Renew Sustain Energy Rev. 2021;150:111400. 10.1016/j.rser.2021.111400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Lu Y, Zhu Y, Cui M, Cheng Z, Hong P. Post‐recovery enhancement of anti‐variant neutralisation after severe COVID‐19. The Lancet Microbe. 2022. 10.1016/S2666-5247(22)00032-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Du X, Tang H, Gao L, et al. Omicron adopts a different strategy from Delta and other variants to adapt to host. Signal Transduct Target Ther. 2022;7(1):45. 10.1038/s41392-022-00903-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Li P, Wang Y, Lavrijsen M, et al. SARS‐CoV‐2 Omicron variant is highly sensitive to molnupiravir, nirmatrelvir, and the combination. Cell Res. 2022;32(3):322‐324. 10.1038/s41422-022-00618-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Yadav PD, Kumar S. Comment global emergence of SARS‐CoV‐2 variants: new foresight needed for improved vaccine efficacy. Lancet Infect Dis. 2021;3099(21):2‐3. 10.1016/S1473-3099(21)00687-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Keegan LT, Truelove S, Lessler J. Analysis of vaccine effectiveness against COVID‐19 and the emergence of delta and other variants of concern in Utah. JAMA Netw Open. 2021;4(12):e2140906. 10.1001/jamanetworkopen.2021.40906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Le TT, Cramer JP, Chen R, Mayhew S. The COVID‐19 vaccine development landscape. Nat Rev Drug Discov. 2020;19:305‐306. 10.1038/d41573-020-0 [DOI] [PubMed] [Google Scholar]
- 110. Mallah SI, Ghorab OK, Al‐Salmi S, et al. COVID‐19: breaking down a global health crisis. Ann Clin Microbiol Antimicrob. 2021;20(1):35. 10.1186/s12941-021-00438-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. To KK, Sridhar S, Chiu KH, et al. Lessons learned 1 year after SARS‐CoV‐2 emergence leading to COVID‐19 pandemic. Emerg Microbes Infect. 2021;10(1):507‐535. 10.1080/22221751.2021.1898291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Logette E, Lorin C, Favreau C, et al. A machine‐generated view of the role of blood glucose levels in the severity of COVID‐19. Front Public Heal. 2021;9:695139. 10.3389/fpubh.2021.695139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Bui N‐N, Lin Y‐T, Huang S‐H, Lin C‐W. Haplotype distribution of SARS‐CoV‐2 variants in low and high vaccination rate countries during ongoing global COVID‐19 pandemic in early 2021. Infect Genet Evol. 2022;97:105164. 10.1016/j.meegid.2021.105164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Safiabadi Tali SH, LeBlanc JJ, Sadiq Z, et al. Tools and techniques for severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2)/COVID‐19 detection. Clin Microbiol Rev. 2021;34(3):1‐63. 10.1128/CMR.00228-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Vaccine G, Plan A. Global vaccine action plan. Vaccine. 2013;31:B5‐B31. 10.1016/j.vaccine.2013.02.015 [DOI] [PubMed] [Google Scholar]
- 116. Singh R, Kang A, Luo X, et al. COVID‐19: current knowledge in clinical features, immunological responses, and vaccine development. FASEB J. 2021;35(3):1‐23. 10.1096/fj.202002662R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Kim JH, Marks F, Clemens JD. Looking beyond COVID‐19 vaccine phase 3 trials. Nat Med. 2021;27(2):205‐211. 10.1038/s41591-021-01230-y [DOI] [PubMed] [Google Scholar]
- 118. WHO . COVID‐19 Vaccine Tracker and Landscape, 2022. Accessed March 3, 2022. https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines [Google Scholar]
- 119. Rees H, Ropero AM, Sc B, et al. Special Report SARS‐CoV‐2 variants and vaccines, 2021:1‐8.
- 120. Flanagan KL, Best E, Crawford NW, et al. Progress and pitfalls in the quest for effective SARS‐CoV‐2 (COVID‐19) vaccines. Front Immunol. 2020;11(October):1‐24. 10.3389/fimmu.2020.579250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Moore JP, Offit PA. SARS‐CoV‐2 Vaccines and the growing threat of viral variants. JAMA. 2021;325(9):821‐822. 10.1001/jama.2021.1114 [DOI] [PubMed] [Google Scholar]
- 122. Loo K‐Y, Letchumanan V, Ser H‐L, et al. COVID‐19: insights into potential vaccines. Microorganisms. 2021;9(3):605. 10.3390/microorganisms9030605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Ren S‐Y, Wang W‐B, Gao R‐D, Zhou A‐M. Omicron variant (B.1.1.529) of SARS‐CoV‐2: mutation, infectivity, transmission, and vaccine resistance. World J Clin Cases. 2022;10(1):1‐11. 10.12998/wjcc.v10.i1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Ciotti M, Ciccozzi M, Pieri M, Bernardini S. The COVID‐19 pandemic: viral variants and vaccine efficacy. Crit Rev Clin Lab Sci. 2022;59(1):66‐75. 10.1080/10408363.2021.1979462 [DOI] [PubMed] [Google Scholar]
- 125. Wald A. Booster vaccination to reduce SARS‐CoV‐2 transmission and infection. JAMA. 2022;327:327‐328. 10.1001/jama.2021.23726 [DOI] [PubMed] [Google Scholar]
- 126. Rubin R. Sorting out whether vitamin D deficiency raises COVID‐19 risk. JAMA. 2021;325(4):329‐330. 10.1001/jama.2020.24127 [DOI] [PubMed] [Google Scholar]
- 127. Chavda VP, Apostolopoulos V. Is booster dose strategy sufficient for Omicron variant of SARS‐CoV‐2? Vaccines. 2022;10(3):367. 10.3390/vaccines10030367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Sallam M, Dababseh D, Eid H, et al. Low COVID‐19 vaccine acceptance is correlated with conspiracy beliefs among university students in Jordan. Int J Environ Res Public Health. 2021;18(5):2407. 10.3390/ijerph18052407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Lustig Y, Zuckerman N, Nemet I, et al. Neutralising capacity against Delta (B.1.617.2) and other variants of concern following Comirnaty (BNT162b2, BioNTech/Pfizer) vaccination in health care workers, Israel. Euro Surveill. 2021;26(26):2100557. 10.2807/1560-7917.ES.2021.26.26.2100557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Collie S, Champion J, Moultrie H, Bekker L‐G, Gray G. Effectiveness of BNT162b2 vaccine against Omicron variant in South Africa. N Engl J Med. 2021;386(5):494‐496. 10.1056/NEJMc2119270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Chavda VP, Pandya R, Apostolopoulos V. DNA vaccines for SARS‐CoV‐2: toward third‐generation vaccination era. Expert Rev Vaccines. 2021;20(12):1549‐1560. 10.1080/14760584.2021.1987223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Ikegame S, Siddiquey MNA, Hung C‐T, et al. Neutralizing activity of Sputnik V vaccine sera against SARS‐CoV‐2 variants. Nat Commun. 2021;12(1):4598. 10.1038/s41467-021-24909-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Sadoff J, Le Gars M, Shukarev G, et al. Interim results of a phase 1‐2a trial of Ad26.COV2.S Covid‐19 vaccine. N Engl J Med. Published online January2021;384:1824‐1835. 10.1056/NEJMoa2034201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Duerr R, Dimartino D, Marier C, et al. Dominance of Alpha and Iota variants in SARS‐CoV‐2 vaccine breakthrough infections in New York City. J Clin Invest. 2021;131(18):e152702. 10.1172/JCI152702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Doroftei B, Ciobica A, Ilie O‐D, Maftei R, Ilea C. Mini‐review discussing the reliability and efficiency of COVID‐19 vaccines. Diagnostics. 2021;11(4):579. 10.3390/diagnostics11040579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Rao GSNK, Gowthami B, Naveen NR, Samudrala PK. An updated review on potential therapeutic drug candidates, vaccines and an insight on patents filed for COVID‐19. Curr Res Pharmacol Drug Discov. 2021;2:100063. 10.1016/j.crphar.2021.100063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Vacharathit V, Aiewsakun P, Manopwisedjaroen S, et al. CoronaVac induces lower neutralising activity against variants of concern than natural infection. Lancet Infect Dis. 2021;21(10):1352‐1354. 10.1016/S1473-3099(21)00568-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Bharat Biotech . COVAXIN®—India' s First Indigenous COVID‐19 Vaccine. Vol 154, 2021:1‐6. [Google Scholar]
- 139. Shetty R, Ghosh A, Honavar SG, Khamar P, Sethu S. Therapeutic opportunities to manage COVID‐19/SARS‐CoV‐2 infection: present and future. Indian J Ophthalmol. 2020;68(5):693‐702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Chakraborty C, Sharma AR, Bhattacharya M, Agoramoorthy G, Lee S‐S. Asian‐origin approved COVID‐19 vaccines and current status of COVID‐19 vaccination program in Asia: a critical analysis. Vaccines. 2021;9(6):600. 10.3390/vaccines9060600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Peacocke EF, Heupink LF, Frønsdal K, Dahl EH, Chola L. Global access to COVID‐19 vaccines: a scoping review of factors that may influence equitable access for low and middle‐income countries. BMJ Open. 2021;11(9):e049505‐e049505. 10.1136/bmjopen-2021-049505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Veranda P. How worried should you be about coronavirus variants? A virologist explains his concerns. DownToEarth. April 9, 2021. Accessed March 2, 2022. https://www.downtoearth.org.in/blog/health/how-worried-should-you-be-about-coronavirus-variants-a-virologist-explains-his-concerns-76384
- 143. Eyawo O, Viens AM. Rethinking the central role of equity in the global governance of pandemic response. J Bioeth Inq. 2020;17(4):549‐553. 10.1007/s11673-020-10001-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Chavda VP, Patel AB, Vihol D, et al. Herbal remedies, nutraceuticals, and dietary supplements for COVID‐19 management: an update. Clin Complement Med Pharmacol. 2022:100021. 10.1016/j.ccmp.2022.100021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Chavda V, Sheta S, Changani D, Chavda D. New bioinformatics platform‐based approach for drug design. In: Balamurugan S, Krishnan A, Goyal D, Chandrasekaran B, Pandi B, eds. Comput Bioinform. Scrivener Publishing; 2021:101‐120. 10.1002/9781119654803.ch6 [DOI] [Google Scholar]
- 146. Chavda VP, Chen R, Patel AB, Chen Z‐S. Phytochemical delivery through Transferosome (Phytosome): an advanced transdermal drug delivery for complementary medicine. Front Pharmacol. 2022;13:850862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Chavda VP. Nanotherapeutics and nanobiotechnology. In: Shyam M, Shivendu R, Nandita D, Raghvendra M, Sabu T, eds. Applications of Targeted Nano Drugs and Delivery Systems. Elsevier; 2019:1‐13. [Google Scholar]
- 148. Nie J‐B. In the shadow of biological warfare: conspiracy theories on the origins of COVID‐19 and enhancing global governance of biosafety as a matter of urgency. J Bioeth Inq. 2020;17(4):567‐574. 10.1007/s11673-020-10025-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. US FDA . Emergency Use Authorization (EUA) for etesevimab. fda.gov. 2021. Accessed February 22, 2022. https://www.fda.gov/media/145802/download
- 150. US FDA . Casirivimab with imdevimab—USFDA Factsheet. fda.gov. 2021. Accessed February 22, 2022. https://www.fda.gov/media/145612/download
- 151. Chavda V, Thalkari Y, Marwadi S. New strategies in drug discovery. Comput Bioinform. 2021;28:25‐48. 10.1002/9781119654803.ch2 [DOI] [Google Scholar]
- 152. Chavda VP, Ertas YN, Walhekar V, et al. Advanced computational methodologies used in the discovery of new natural anticancer compounds. Front Pharmacol. 2021;12:702611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Chavda VP. Chapter 4—nanobased nano drug delivery: a comprehensive review. In: Mohapatra SS, Ranjan S, Dasgupta N, Mishra RK, Thomas SBT‐A of TND and DS, eds. Micro and Nano Technologies. Elsevier; 2019:69‐92. [Google Scholar]
- 154. Shah Dhaval, Chavda Vivek, Nasal HT. Medication conveyance framework: an approach for brain delivery from essential to cutting edge. Res Rev J Med. 2016;6(1):14‐27. [Google Scholar]
- 155. Chen R‐P, Chavda VP, Patel AB, Chen Z‐S. Phytochemical delivery through transferosome (phytosome): an advanced transdermal drug delivery for complementary medicines. Front Pharmacol. 2022;13:850862. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.


