Abstract
Although vaccines are currently used to control the coronavirus disease 2019 (COVID‐19) pandemic, treatment options are urgently needed for those who cannot be vaccinated and for future outbreaks involving new severe acute respiratory syndrome coronavirus virus 2 (SARS‐CoV‐2) strains or coronaviruses not covered by current vaccines. Thus far, few existing antivirals are known to be effective against SARS‐CoV‐2 and clinically successful against COVID‐19. As part of an immediate response to the COVID‐19 pandemic, a high‐throughput, high content imaging–based SARS‐CoV‐2 infection assay was developed in VeroE6 African green monkey kidney epithelial cells expressing a stable enhanced green fluorescent protein (VeroE6‐eGFP cells) and was used to screen a library of 5676 compounds that passed Phase 1 clinical trials. Eight drugs (nelfinavir, RG‐12915, itraconazole, chloroquine, hydroxychloroquine, sematilide, remdesivir, and doxorubicin) were identified as inhibitors of in vitro anti–SARS‐CoV‐2 activity in VeroE6‐eGFP and/or Caco‐2 cell lines. However, apart from remdesivir, toxicity and pharmacokinetic data did not support further clinical development of these compounds for COVID‐19 treatment.
Keywords: antiviral agents, coronavirus, SARS coronavirus
1. INTRODUCTION
Coronaviruses (CoVs) are enveloped, positive‐sense single‐stranded RNA viruses; of the seven members of the CoV family known to infect humans, most cause mild respiratory disease. 1 In the last two decades, beta CoVs have caused outbreaks of severe respiratory disease, including severe acute respiratory syndrome (SARS) in 2002 and 2003, caused by SARS‐CoV‐1, 2 followed by Middle Eastern respiratory syndrome (MERS) in 2012, caused by MERS‐CoV. 3 In late 2019, an outbreak of a novel respiratory syndrome, coronavirus disease 2019 (COVID‐19), was reported in Wuhan, China. 4 Common presenting symptoms of COVID‐19 caused by Alpha variants include cough, fever, loss of taste or smell, and fatigue. 5 However, the more recent Delta variant seems to present slightly different symptoms, such as headache, runny nose, throat ache, fever, and coughing. 5 In cases that progress to severe disease, patients commonly experience dyspnea and hypoxemia followed by respiratory failure. 6 COVID‐19 and its etiologic agent, SARS coronavirus 2 (SARS‐CoV‐2), have spread globally since the initial outbreak, infecting >400 million people and leading to ≥6 million deaths.
Even before the World Health Organization declared COVID‐19 a public health emergency of international concern on January 30, 2020, repurposing of existing drugs and drug candidates was explored to accelerate the traditional research and development timelines to provide a rapid response to this unmet medical need. 7 Drug repurposing had been applied previously for SARS‐CoV‐1, MERS‐CoV, and other viruses. 7 , 8 Early in the pandemic, the development of cell‐based systems was critical for the rapid evaluation of drugs with antiviral activity against SARS‐CoV‐2. Similar to SARS‐CoV‐1, Vero cells and Caco‐2 cells were found to be susceptible to infection with SARS‐CoV‐2. 9 , 10 Therefore, a previously published high‐throughput screening (HTS) assay for SARS‐CoV‐1 11 using authentic infection of VeroE6 African green monkey kidney epithelial cells expressing a stable enhanced green fluorescent protein (VeroE6‐eGFP) was adapted, further developed, and miniaturized for screening of antiviral drugs against SARS‐CoV‐2, isolated from a Belgian patient. The resulting assay is an HTS 384‐well cell‐based SARS‐CoV‐2 infection assay with a high content imaging (HCI) readout of fluorescence that provides a measure for cytopathic effect (CPE). In parallel, a cellular toxicity assay was developed using ATPlite™; any toxicity of the compound to the cell line is then evaluated by luminescence.
To identify potential candidates for rapid clinical development, 5676 chemical structures that had passed Phase 1 clinical studies with applications in a variety of therapeutic fields, including oncology, neuroscience, and infectious diseases, were screened. The compounds were evaluated in a seven‐point dose–response curve, starting at 20 µM, for their ability to inhibit SARS‐CoV‐2–induced CPE without causing general cellular toxicity. Selected hits were further evaluated in a second cellular model using Caco‐2 cells that are susceptible to SARS‐CoV‐2 infection. 12 , 13
2. MATERIALS AND METHODS
2.1. Assembly of the library
Janssen Pharmaceutica maintains a regularly updated database of compounds that have been approved or tested in a successfully completed clinical Phase 1 study. One of the many uses of this Phase One Passed Structures (POPS) database is to serve as a starting point to identify high‐priority compounds for possible repurposing. The goal of the POPS database is to be highly enriched in druggable, well‐documented, and diverse compounds from all disease areas, including oncology, neuroscience, and infectious diseases. The database is updated on a regular basis, both from a (virtual) annotation standpoint and with a physically available set of compounds that can be screened. Due to previous acquisition and synthesis efforts and active purchasing in the first months of 2020, approximately 5500 compounds that had sufficient availability and passed quality control for purity were identified and plated.
2.2. Plate production
Plates were freshly prepared to ensure high‐quality assays and were submitted for screening. CELLSTAR® 384‐well plates (Greiner Bio‐One) were prespotted with a 300 nl compound in dimethyl sulfoxide (DMSO; 300 nl 100% DMSO for control wells), and each plate was spotted in triplicate. An Echo® 555 Liquid Handler (Labcyte Inc.) was used to spot the compounds at a final concentration of 20 µM. Spotted plates were frozen and transported to the Rega Institute of the KU Leuven, Belgium.
2.3. Cell cultures
VeroE6‐eGFP cells were cloned and validated in‐house as previously described. 11 Cells were cultured and maintained in Dulbecco's modified Eagle medium (DMEM; Gibco) supplemented with 10% (vol/vol) heat‐inactivated fetal bovine serum (FBS; Biowest), 0.75% sodium bicarbonate (Gibco), and 50 U/ml penicillin‐streptomycin (Gibco). Caco‐2 cells (human colon carcinoma cell line; obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen) were cultured in a minimal essential medium supplemented with 10% FBS with penicillin (100 IU/ml) and streptomycin (100 μg/ml). Cells were maintained at 37°C in 5% CO2.
2.4. SARS‐CoV‐2 preparation
SARS‐CoV‐2‐Belgium (strain BetaCov/Belgium/GHB‐03021/2020) was recovered from a nasopharyngeal swab taken from an asymptomatic patient returning from Wuhan, China. Virus stocks were inoculated and passaged first in HuH‐7 cells and then five times in VeroE6‐eGFP cells before storage at –80°C. Passage 6 was used for all VeroE6‐eGFP experiments (viral titer: 3.0 × 106 TCID50/ml). All manipulations were performed in a licensed and certified biosafety level 3 (BSL‐3) facility at the Laboratory of Virology and Chemotherapy at the Rega Institute of the KU Leuven, Belgium.
SARS‐CoV‐2/FFM1 (strain hCoV‐19/Germany/FrankfurtFFM1/2020) was isolated from a German patient sample and passaged twice in Caco‐2 cells before storage at –80°C. All passages were sequenced using a MinION platform (Oxford Nanopore). Viral titers were determined for the last three passages by end‐point dilution assays. All manipulations were performed in a licensed and certified BSL‐3 facility at the Laboratory Medizinische Mikrobiologie at the Universitats Klinikum Frankfurt (Goethe University) in Germany.
2.5. Antiviral and toxicity assays in VeroE6‐eGFP cells
Each compound from the library was evaluated for antiviral activity using a cell‐based VeroE6‐eGFP assay. Two assays were developed simultaneously: the first one used whole well fluorescence measured with a multimode plate reader (PR; Tecan, Infinite M1000 Pro), and the other used HCI (Thermofisher, Arrayscan XTI). Spotted 384‐well plates were seeded with 30 µl with 2000 (PR assay) or 8000 (HCI assay) VeroE6‐eGFP cells in DMEM 2% FBS in each well. An additional 30 µl medium was added to the cell controls (CCs). The plates used in the HCI assay were incubated overnight or approximately 20 h before infection in a humidified incubator at 37°C with 5% CO2. After cell seeding (PR assay) or overnight incubation (HCI assay), the plates were transferred to the BSL‐3+ Caps‐It robotics system where 30 µl SARS‐CoV‐2 was added using a noncontact liquid handling system (Tecan, EVO 100) to achieve a multiplicity of infection (MOI) of 0.001. After infection, the plates were automatically transferred to the system's integrated incubators (37°C with 5% CO2) for 5 (PR assay) or 4 (HCI assay) days before performing the readout (Figure 1).
Figure 1.
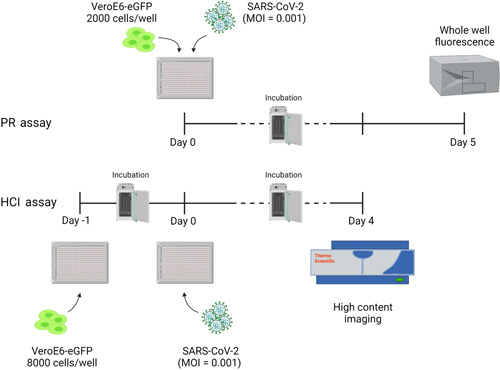
Schematic overview of PR and HCI assays using prespotted 384‐well plates with compounds in a dose–response curve format at a final starting concentration ranging from 100 to 25 µM. PR assay (top): 2000 cells/well were seeded on Day 0 and infected on the same day, after which a PR readout was performed on Day 5. HCI assay (bottom): 8000 cells/well were seeded on Day –1 and infection with SARS‐CoV‐2 was performed on Day 0, after which an HCI readout was performed on Day 4. Plates were incubated at 37°C and 5% CO2 between days. This figure was made in BioRender. eGFP, enhanced green fluorescent protein; HCI, high content imaging; MOI, the multiplicity of infection; PR, plate reader; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2; VeroE6‐eGFP, VeroE6 African green monkey kidney epithelial cells expressing a stable enhanced green fluorescent protein
Whole well fluorescence settings on the PR were set on 488 nm excitation and 507 nm emission bandwidths with 20 µs integration time and 4 reads per well. PR data were obtained in .csv format. Image acquisition on the high‐content imagers was set to 485/20 nm excitation wavelength with an exposure time of 23 ms. The emission signal was captured with a multiband BGRFRN filter set and a dichroic mirror using widefield microscopy technology. One field per well was imaged using a 5× objective with 2 × 2 binning and 1104 × 1104 pixel resolution. Image analysis was performed on the acquired images using the HCS studio software. A custom image analysis protocol was developed based on the Spot Detector BioApplication. A background correction was performed to remove the nonspecific signal from the raw image files. With the entire well selected as the region of interest, a fixed threshold for pixel intensity was set to measure the eGFP signal. SARS‐CoV‐2 induced CPE in VeroE6‐eGFP cells, leading to a marked reduction of the eGFP signal. In contrast, a strong eGFP signal was observed under conditions without the virus. A quality control assessment and data analysis were performed using several output features: SpotTotalAreaCh2, SpotTotalIntensityCh2, and ValidObject Count. Following assay completion, plates were automatically decontaminated by the Caps‐It system.
Compounds were screened in dose–response, and dose–response curves plotting normalized values versus compound concentrations were generated in Genedata Screener® (version 17.0.6—Standard). A smart fit strategy with an automatic model selection without automatic masking was used for plotting the graphs and calculating EC50 (concentration of the compound that inhibited 50% of the virus‐induced CPE) values. A low threshold for hit selection was determined to minimize the risks of having false negatives. Only the whole well fluorescence data and the HCI output feature “SpotTotalAreaCh2” were used for calculation. When one of three replicate data points deviated, it was masked and not taken into consideration for calculations or fitting.
The PR assay was developed first, and data analysis could be performed on the extracted data using the already established informatics tools of Genedata Screener®. The HCI assay was developed during the initial screening using the PR assay. Since the extracted HCI data were more robust and accurate, the HCI assay was used instead of the PR assay during this screening campaign. Due to the urgency of the screen, data points of >900 approved (based on the robust Z′ factor [RZ′]) assay plates were already collected using the PR assay.
Toxicity was assessed using the ATPlite™ kit by measuring luminescence after a reaction of ATP with added d‐luciferin and luciferase. VeroE6‐eGFP cells (40 μl) were seeded at 2000 cells/well in prespotted 384‐well plates. The plates were then placed for 5 days in a humidified incubator with 5% CO2. After incubation, steps were followed according to the ATPlite™ manufacturer's instructions. The luminescence read‐out was performed on the ViewLux™ PR (PerkinElmer). Data were analyzed by nonlinear curve fitting (four‐parameter fit) from a dose–response curve using GraphPad Prism to calculate CC50 (cytotoxic concentration of the compound that reduced cell viability to 50%).
2.6. Antiviral and toxicity assay in Caco‐2 cells
After compounds with potential antiviral activity in VeroE6‐eGFP cells were identified, confirmation of their inhibition of virus‐induced CPE was performed in Caco‐2 cells. Caco‐2 cells were cultured for 72 h on 96‐well plates (50 000 cells/well) and infected with SARS‐CoV‐2/FFM1 at an MOI of 0.01. After a 48‐h incubation of the virus, cells, and compound in MEM supplemented with 1% FBS, the CPE was visually scored by two independent laboratory technicians. Optical densities were measured at 560/620 nm in a Multiskan Reader. Cell viability in Caco‐2 cells in the absence of virus was assessed using the 3‐(4,5‐dimethylthiazol‐2‐yl)–2,5‐diphenyltetrazolium bromide (MTT) assay, as previously described. 14 Data were analyzed by four‐parameter curve fitting from a dose–response curve using GraphPad Prism to calculate the EC50 and CC50.
2.7. Development and optimization of a HTS pipeline for both assays
Assay quality performance (inter‐ and intraplate data comparison) and optimization (cell density, viral input, incubation time, Z′, signal/noise) were performed on 384‐well plates spotted with reference compounds. Raw data were normalized using the following formula:
where z i is the normalized value of the dataset, xi is the raw value of the dataset, min(x) is the minimum raw value of the dataset, and max(x) is the maximum raw value of the dataset. The Z′ value, which encompasses the dynamic range of the assay and well‐to‐well variability, was calculated for each plate as a measure of assay quality. 15 , 16 All compounds were screened in a dose–response (seven dilutions) starting from 20 µM with half dilution steps and tested in triplicate on separate plates.
3. RESULTS
3.1. High‐throughput assay development
At the start of the pandemic, an immediate public health response was needed but tools to screen for antiviral agents were not available at that time. Due to the high sequence homology between SARS‐CoV‐1 and SARS‐CoV‐2, the primary focus was on VeroE6 cells, which were previously shown to be readily susceptible to infection with SARS‐CoV‐2. 9 , 10 Indeed, SARS‐CoV‐2 infected the cells and caused pronounced CPE, especially once the virus had been adapted to cell culture (three to five passages on VeroE6 cells). Next, a SARS‐CoV‐1 antiviral assay in VeroE6‐eGFP cells 11 was adapted to be used for screening with SARS‐CoV‐2 (Figure 1). In this system, SARS‐CoV‐2 infection causes cellular CPE and loss of fluorescent signal. Compound‐mediated antiviral activity reduces CPE, leading to increased fluorescence compared with controls. To monitor CPE and inhibition thereof by antiviral molecules, the assay was developed in parallel tracks whereby readout was performed either by fluorescence (using a GFP PR) or HCI at low resolution to visualize and quantify CPE (Figure 1).
In uninfected (CC) conditions, the mean normalized signal for the PR readout was 79.8 ± 10.9 (% CV = 13.7); for the HCI readout, the mean normalized signal was 94.8 ± 2.41 (% CV = 2.55). Under infected (virus control [VC]) conditions, a high % CV was calculated for both readouts with a large variation between the data points; because the mean value was low (PR < 5 and HCI < 1), the % CV was highly sensitive to small changes. In this type of biological assay, unavoidable “eGFP cell debris,” found in residual organic waste after virus‐induced cell death, contributes to variation in eGFP. For HTS quality assessment, the RZ′ and signal to noise ratio (S/N) were calculated for each method (PR readout: mean RZ′ 0.51 and S/N 27.3; HCI readout: mean RZ′ 0.91 and S/N 362) (Figure 2).
Figure 2.
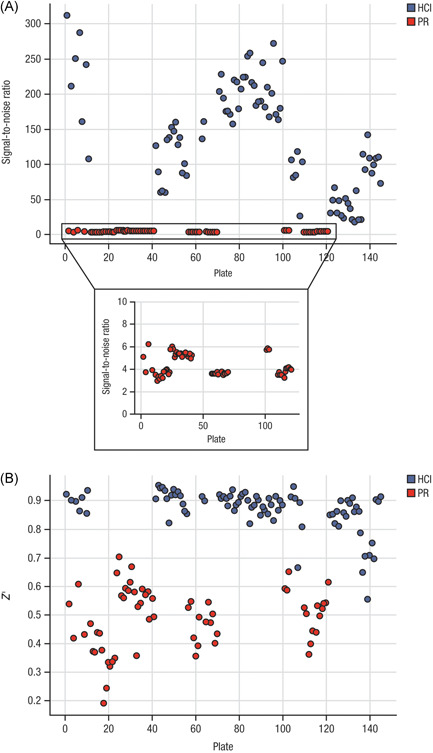
Signal‐to‐noise (A) and Z′ (B) comparison between PR (red) and HCI (blue) assays. The signal‐to‐noise ratio (raw data CC/raw data VC) for the PR assay ranged from 1.84 to 6.22 with Z′ values ranging from –0.26 to 0.72. The HCI assay yielded much higher values for both parameters; the signal‐to‐noise ratio ranged from 17.95 to 312.3 while Z′ values ranged from 0.57 to 0.95. CC, uninfected conditions; HCI, high content imaging; PR, plate reader; VC, infected conditions.
Reference compounds were used to verify the inhibition of SARS‐CoV‐2 infected in VeroE6‐eGFP cells. Twelve compounds were selected based on what was known at the time regarding activity against SARS‐CoV‐1, MERS, and SARS‐CoV‐2: chloroquine, 8 , 17 , 18 , 19 cinchocaine, 20 colchicine, 21 hydroxychloroquine, 22 ponatinib, 23 indomethacin, 23 loperamide, 8 lopinavir, 8 nelfinavir, 24 posaconazole, 25 remdesivir, 18 , 19 and saperconazole. 25 These compounds were spotted 200 times more concentrated in quadruplicate at 300 nl/well, with a final concentration per well ranging from 10 to 100 µM depending on the compound, followed by a seven‐point dilution with a dilution factor of two. The EC50 value was calculated based on the obtained values from the HCI and PR readouts for each compound using the GeneData screener as described in Section 2.
Intra‐ and interplate comparisons were evaluated under uninfected conditions (CC) and SARS‐CoV‐2 infected conditions (VC) for both PR and HCI readout methods. In this step, plates containing reference compounds were seeded with VeroE6‐eGFP cells as previously described for the PR and HCI antiviral assays to obtain CPE‐induced eGFP signal reduction after SARS‐CoV‐2 infection. Intraplate variation was evaluated by calculating the mean eGFP output signal and standard deviation (SD); these values were calculated from one plate under 32 uninfected conditions and 16 infected conditions. Interplate variation values were calculated using the data from all four individual experiments (four plates; CC, 128 data points; VC, 64 data points). To assess reproducibility, measurements were obtained in four independent experiments. Additionally, the RZ′ and S/N were calculated for each experiment to determine HTS quality. The assay performance of both the PR and HCI antiviral assays is shown in Table 1.
Table 1.
Assay performance of PR and HCI methods
| Intraplate repeatability | Interplate reproducibility | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Experiment 1 | Experiment 2 | Experiment 3 | Experiment 4 | |||||||
| PR | HCI | PR | HCI | PR | HCI | PR | HCI | PR | HCI | |
| N | 32 | 32 | 32 | 32 | 32 | 32 | 32 | 32 | 128 | 128 |
| Uninfected cells (CC) | ||||||||||
| Mean | 79.2 | 93.3 | 81.1 | 93.6 | 82.4 | 96.6 | 76.3 | 95.6 | 79.8 | 94.8 |
| SD | 11.5 | 2.46 | 9.56 | 2.79 | 9.49 | 1.71 | 13.0 | 2.69 | 11.1 | 2.79 |
| N | 16 | 16 | 16 | 16 | 16 | 16 | 16 | 16 | 64 | 64 |
| SARS‐CoV‐2–infected cells (VC) | ||||||||||
| Mean | 2.86 | 0.16 | 2.24 | 0.26 | 4.30 | 0.37 | 2.95 | 0.40 | 3.09 | 0.30 |
| SD | 1.88 | 0.11 | 1.08 | 0.26 | 3.22 | 0.16 | 1.58 | 0.18 | 2.18 | 0.20 |
| Plate metrics | ||||||||||
| RZ′ | 0.50 | 0.92 | 0.59 | 0.90 | 0.54 | 0.93 | 0.42 | 0.90 | 0.49 | 0.90 |
| S/N | 5.05 | 584 | 7.84 | 364 | 5.08 | 258 | 3.69 | 240 | 5.42 | 319 |
Note: Metrics were performed on normalized data, representing the percentage of green fluorescent signal.
Abbreviations: CC, cell control; HCI, high content imaging; PR, plate reader; RZ′, robust Z′ factor; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2; SD, standard deviation; S/N, signal to background; VC, virus control.
Among the 12 compounds selected for assay development, six resulted in a dose–response relation on both readouts: chloroquine, cinchocaine, hydroxychloroquine, lopinavir, nelfinavir, and remdesivir. The mean and SD of the computed EC50 values from 16 replicates (four intraplate replicates and four plate replicates) and the toxicity data (CC50) computed from five replicates are shown in Table 2. Cinchocaine (PR: 59.6 µM vs. HCI: 63.2 µM), hydroxychloroquine (PR: 13.5 µM vs. HCI: 17.7 µM), nelfinavir (PR: 3.5 µM vs. HCI: 4.2 µM), and remdesivir (PR: 3.6 µM vs. HCI: 2.6 µM) showed comparable EC50 values with no significant differences between both readouts (p >0.05). Chloroquine and lopinavir showed higher discrepancies between PR and HCI readouts, which can likely be attributed to physical differences (whole well fluorescence vs. image analysis) and experimental differences (PR: 2000 cells/well, seeding and infection on Day 0, reading 5 dpi; HCI: 8000 cells/well, seeding on Day –1 and infection on Day 0, reading 4 dpi). Remdesivir exhibited in vitro inhibition of SARS‐CoV‐2 with minimal effect on cell viability and was further used as a positive control.
Table 2.
Mean antiviral activity (EC50) and cytotoxicity (CC50) values of reference compounds used for assay development
| Compound | HCI EC50 (µM) | PR EC50 (µM) | PR CC50 (µM) | |||
|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | |
| Chloroquine | 19.5 | 6.47 | 6.15 | 1.59 | 38.1 | 9.88 |
| Cinchocaine | 63.2 | 40.7 | 59.6 | 38.8 | 38.3 | 4.94 |
| Hydroxychloroquine | 17.7 | 0.32 | 13.5 | 9.04 | >50 | N/A |
| Loperamide | 8.91a | N/A | 11.4 | 0.81 | 25.0 | 5.54 |
| Lopinavir | 7.77 | 0.34 | 43.2 | 17.2 | 31.7 | 4.30 |
| Nelfinavir | 4.21 | 1.44 | 3.50 | 0.17 | 13.5 | 3.16 |
| Remdesivir | 2.60 | 0.80 | 3.62 | 1.10 | >100 | N/A |
Note: EC50 values computed from 16 replicates (four intraplate replicates and four plate replicates). CC50 values computed from five replicates.
Abbreviations: CC50, concentration of the compound that reduced 50% of the cell viability; CPE, cytopathic effect; EC50, concentration of the compound that inhibited 50% of the virus‐induced CPE; HCI, high content imaging; N/A, not available; PR, plate reader; SD, standard deviation.
Could not be reproduced in later experiments.
3.2. Drug repurposing screen
Slightly cytotoxic compounds may result in false negatives in this assay (i.e., activity caused by interference with cell proliferation). Therefore, all compounds were evaluated using dose–response curves, in which bell‐shaped curves indicate upcoming cytotoxicity. In addition, every compound was tested in a parallel toxicity assay.
The drug repurposing screening hit rate was approximately 4.0%. All compounds were screened at the highest concentration available in 100% DMSO stock, which was either 100, 50, or 25 µM final concentration. Of the 5676 compounds screened, 228 compounds were found to be active (EC50 value <20 µM in either PR or HCI readout) (Figure 3 and Table S1). All compounds were redissolved from neat and were tested again in seven‐point dilution with ½ dilution steps dose–response (starting at the highest possible concentration: 100, 50, or 25 µM, final concentration) in triplicate for confirmation. In the confirmation run, 52 compounds remained active (maximum percent inhibition ≥30%). A thorough evaluation of these hits was made based on the quality of the curve, the chemistry, potential/known mode of action, literature, and toxicity to select only those with potential clinical relevance.
Figure 3.
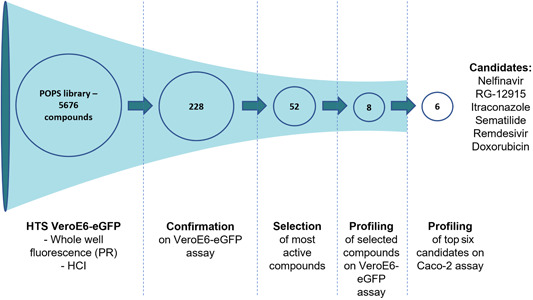
Schematic overview of the screening campaign used on the Janssen POPS library containing 5676 compounds. The high‐throughput screen was performed on 384‐well plates with the described PR or HCI VeroE6‐eGFP assay. After the initial screening, 228 compounds showed activity against SARS‐CoV‐2 with EC50 values <20 µM in either assay. These compounds were retested in triplicate in the same assay for confirmation, which yielded 52 active compounds. Finally, the eight most promising compounds were selected for further profiling, and the top six candidates were selected for additional testing using the Caco‐2 assay. EC50, concentration of the compound that inhibited 50% of the infection; HCI, high content imaging; POPS, Phase One Passed Structures; PR, plate reader; VeroE6‐eGFP, VeroE6 African green monkey kidney epithelial cells expressing a stable enhanced green fluorescent protein.
Finally, eight compounds with antiviral activity against SARS‐CoV‐2 in the VeroE6‐eGFP cells were selected for further investigation: chloroquine, doxorubicin, hydroxychloroquine, itraconazole, nelfinavir, remdesivir, RG‐12915, and sematilide (Table 3).
Table 3.
Observed antiviral activity (EC50) against SARS‐CoV‐2 and cytotoxicity (CC50) of the eight compounds identified using the VeroE6‐eGFP cell assay a
| Compound | Class | EC50, µM (min, max) | Repeats | CC50, µM (min, max) | Repeats |
|---|---|---|---|---|---|
| Sematilide | Antiarrhythmic | 3.9 (N/A) | 1 | >20 (N/A) | 1 |
| Remdesivir | Nucleotide analogue | 5.4 (1.15, 19.7) | 18 | >100 (>100, >100) | 10 |
| Nelfinavir | Protease inhibitor | 5.7 (3.29, >50) | 18 | 12.9 (10.3, 13.7) | 10 |
| Hydroxychloroquine | Antimalarial | 6.7 (N/A) | 1 | >50 (N/A) | 1 |
| RG‐12915 | 5‐HT3 receptor antagonist | 6.8 (N/A) | 1 | 19.7 (N/A) | 1 |
| Chloroquine | Antimalarial | 7.3 (3.13, >50) | 12 | 35.1 (29.9, 50.3) | 10 |
| Doxorubicin | Anthracycline | 8.4 (5.93, 10.9) | 4 | >20 (N/A) | 1 |
| Itraconazole | Antifungal | >100 (5.13, >100) | 8 | >100 (>100, >100) | 8 |
Abbreviations: 5‐HT3, 5‐hydroxytryptamine; CC50, cytotoxic concentration of the compound that reduced cell viability to 50%; CPE, cytopathic effect; EC50, concentration of the compound that inhibited 50% of the virus‐induced CPE; HCI, high content imaging; N/A, not available; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
Activity readout of the assay was based on HCI of fluorescent cells; cell toxicity was measured using ATPlite in uninfected cells.
3.3. Confirmation of the top six hits in a Caco‐2 infection assay
To exclude cell line–specific modes of antiviral activity, six compounds were further evaluated using a Caco‐2 infection assay and a viral strain isolated from a different source (SARS‐CoV‐2/FFM1). Hydroxychloroquine and chloroquine were excluded from this evaluation because they had previously been found inactive in SARS‐CoV‐2–infected Caco‐2 cells. 26 This system has been used previously for SARS‐CoV‐1 11 and was recently deployed for another drug repurposing screen. 26 All compounds were evaluated with a dose–response curve to obtain an EC50 value based on visual inspection of CPE. In parallel, a CC50 value was obtained using a similar assay but without viral infection. Remdesivir and itraconazole exhibited a higher potency in Caco‐2 cells compared to Vero cells; other compounds showed similar EC50 values in both cell lines. Sematilide was not active in Caco‐2 cells. Dose–response curves of CPE and cytotoxicity readouts are shown in Figure 4; EC50 and CC50 values are summarized in Table 4.
Figure 4.
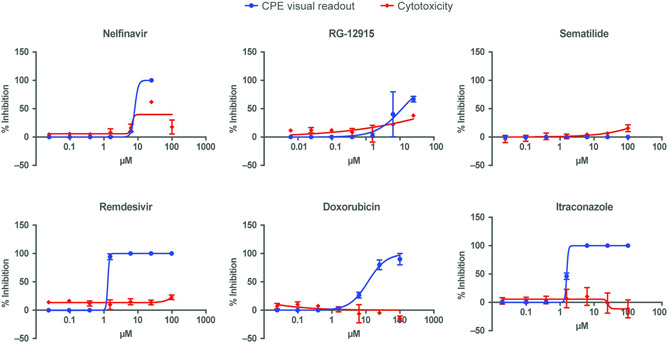
Effect on CPE assays and viability of Caco‐2 cells of identified compounds. CPE, cytopathic effect
Table 4.
Observed antiviral activity (EC50) against SARS‐CoV‐2 and cytotoxicity (CC50) of the six compounds evaluated using the Caco‐2 cell assay
| Compounda | EC50 visual CPE (µM) | CC50 (µM) |
|---|---|---|
| Remdesivir (positive control) | 1.3 | >100 |
| Itraconazole | 1.6 | >100 |
| Nelfinavir | 8.1 | 6.7 |
| Doxorubicin | 12 | >100 |
| RG‐12915 | 12 | >25 |
| Sematilide | >100 | >100 |
Abbreviations: CC50, cytotoxic concentration of the compound that reduced cell viability to 50%; CPE, cytopathic effect; EC50, concentration of the compound that inhibited 50% of the virus‐induced CPE; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
Hydroxychloroquine and chloroquine were not tested in this cell line.
4. DISCUSSION
Early in the pandemic, there was an urgent need for the treatment of COVID‐19, and the most rapid solution involved repurposing drugs already validated in the clinic. To accelerate drug repurposing, libraries have been screened in computational models to target viral proteins, including the receptor‐binding domain of the spike protein, ACE2, viral Plpro, 3Clpro, and Mpro. 19 , 27 In parallel, cell‐based screening using a variety of cell lines, including VeroE6, Huh7, Calu‐3, and Caco‐2 cells infected with SARS‐CoV‐2, were developed to identify potential candidates from repurposing libraries. 10 , 28 , 29 , 30 , 31
The drug repurposing library screen described in this study used a VeroE6‐eGFP cell‐based SARS‐CoV‐2 infection assay to identify therapeutic drug candidates. This cell line is known to be susceptible to SARS‐CoV‐1 and SARS‐CoV‐2 infection and shows a reduced eGFP signal upon cell death. 11 The HTS screening assay developed in this study was robust (Table 1), automated, and represents a scalable and efficient system for identifying antiviral compounds. Although some previously developed assays have used HCI, 10 , 28 , 29 few have compared HCI and PR readout, as described in this study. In developing HTS assays, factors such as data storage, data acquisition speed, and logistics should be considered, especially when speed is of utmost importance, such as during a pandemic. In addition to the advantages of speed and storage afforded by PR, all parameters for HTS quality (RZ′, S/N), intra‐ and interplate variation were overall substantially better using HCI compared with PR readout (Tables 1 and 2, Figure 2). This made HCI the preferred readout for screening.
A library of 5676 compounds that passed Phase 1 was screened. The primary hit rate was 4.0%, which is relatively high among experimental repurposing screening studies that usually report hit rates of less than 2%. 31 , 32 , 33 This rather high hit rate is due to the low threshold that was set for primary hit selection and because toxicity was not taken into account for primary hit selection. In total, 52 hits were confirmed. After elimination of toxic compounds, screening artifacts (e.g., vitamin B2 and orantinib cause a false positive signal due to fluorescence of the compound), and unwanted modes of action such as influence on lysosomal function or phospholipidosis, 34 eight compounds remained (Table 3). Five of these compounds also exhibited antiviral activity in Caco‐2 cells, a validated model for SARS‐CoV research, 35 showing that these compounds were active across different cell types.
Of the eight selected compounds, only remdesivir, an adenosine nucleotide prodrug that inhibits the viral RNA‐dependent RNA polymerase, 19 , 36 has been approved by the US Food and Drug Administration (FDA). In the current study, remdesivir consistently produced EC50 values of single‐digit micromolar magnitude in VeroE6‐eGFP cells and in Caco‐2 cells; these values were comparable to those reported in previous studies (range, 0.77–7.28 µM). 9 , 10 , 26 , 28 , 30 Remdesivir was the first drug approved by the FDA for the treatment of patients with COVID‐19 requiring hospitalization in the United States and is conditionally recommended by Infectious Diseases Society of America guidelines for use in hospitalized patients with severe COVID‐19. 37
Although rare examples of successful drug repurposing exist (e.g., sildenafil citrate 38 and thalidomide 39 ) the outcome in the case of SARS‐CoV‐2 and COVID‐19 has been disappointing but not unexpected. Although thorough off‐target screening was conducted to limit side effects, some candidate compounds progressed to clinical development yet ultimately failed endpoint criteria. 40 Nevertheless, this unprecedented effort was of critical importance during the sudden onset of a pandemic. Many lessons were learned regarding rapid assay design and deployment in a high biosafety environment. In drug discovery campaigns, assay design should incorporate mechanisms designed to reduce the risk of false positives, such as counter screens to validate the physiology of results. 41 This screen and many others 42 used the VeroE6 cell line. Although this line has proven valuable for rapid screening and clear CPE readouts, the occurrence of false positives shows that additional data in other models are invaluable in filtering out the most promising hits. In addition to these technical learnings, there were valuable lessons learned concerning the contextualization of data and the importance of scientific communication.
Pharmacological parameters of compounds (such as pharmacokinetics, pharmacodynamics, tissue distribution, and tolerability), as well as cytotoxicity/cytostatic assays, should be carefully considered when evaluating the feasibility of clinical applications. 40 For example, chloroquine and its analog hydroxychloroquine inhibit SARS‐CoV‐2 entry and replication in VeroE6 cells, 44 yet failed in clinical trials. 37 , 45 Nonetheless, screening this repurposing library contributed to our understanding of fundamental features of SARS‐CoV‐2 and the rapid design/deployment needed for high‐throughput assays, which may be instrumental during future outbreaks.
5. CONCLUSIONS
The findings of this study were comparable with other large screening studies that identified potential anti–SARS‐CoV‐2 agents and compounds, although in vivo efficacy, toxicity, and pharmacokinetic investigation of the selected hits in this report did not support novel applications against SARS‐CoV‐2. 10 , 12 , 26 , 28 , 30 , 46 , 47 , 48 , 49 , 50 Together, these studies demonstrate that drug repurposing technology can identify potential agents for rapidly spreading novel pathogens, as in the case of remdesivir, which has been used to treat patients with severe COVID‐19. Although preclinical drug repurposing studies may provide a means of identifying potential agents and avoiding some of the limitations of traditional clinical development, the findings of this study suggest that results from in silico or in vitro studies should be interpreted with caution. Evaluations of compounds in vivo and in clinical trials are necessary to support the use of identified compounds in clinical settings.
CONFLICTS OF INTEREST
The authors declare the following financial interests/personal relationships that may be considered as potential competing interests: S. C. received research funding from Janssen for this study. W. C., D. J., S. D. J., P. L., and J. N. are employees from KU Leuven and received funding from Janssen for this study. L. V., C. V. d. E., C. B., S. D. M., M. V. L., and E. V. D. are employees of Janssen and may be stock owners of Johnson & Johnson. D. B. and J. C. have nothing to declare.
AUTHOR CONTRIBUTIONS
All authors contributed to the data interpretation and development of the manuscript, and all authors approved the manuscript for submission. In addition, Winston Chiu, Lore Verschueren, and Christophe Buyck contributed to study conceptualization and data curation and analysis. Christel Van den Eynde contributed to data curation and data analysis. Sandra De Meyer contributed to study conceptualization. Dirk Jochmans contributed to study conceptualization and methodology and had supervisory duties. Denisa Bojkova, Sandra Ciesek, and Jindrich Cinatl contributed to the methodology. Steven De Jonghe contributed to study conceptualization and methodology. Pieter Leyssen contributed to data interpretation and had supervisory duties. Johan Neyts and Marnix Van Loock contributed to study conceptualization and had supervisory duties. Ellen Van Damme contributed to study conceptualization, data curation and data analysis, methodology, project administration, and had supervisory duties.
Supporting information
Supporting information.
ACKNOWLEDGMENTS
We thank Joost Schepers and Kayvan Abbasi for their excellent technical assistance. The authors also wish to thank Dr. Ruxandra Draghia‐Akli for the interesting discussion and guidance. Medical writing support for the development of this manuscript was provided by Kurt Kunz, MD, MPH, and Catherine DeBrosse, PhD, of Cello Health Communications/MedErgy, and was funded by Janssen Pharmaceutica. This study has been executed as part of the Corona Accelerated R&D in Europe (CARE) project under grant agreement number 101005077 with the Innovative Medicines Initiative 2 Joint Undertaking (JU). The JU receives support from the European Union's Horizon 2020 research and innovation program EFPIA, Bill & Melinda Gates Foundation, Global Health Drug Discovery Institute, and the University of Dundee. The content of this publication only reflects the authors' views and the JU is not responsible for any use that may be made of the information it contains. This project has received funding from the European Union's Horizon 2020 research and innovation program under grant agreement number 101003627. Part of this study was performed using the “Caps‐It” research infrastructure (project ZW13‐02) that was financially supported by the Hercules Foundation (FWO) and Rega Foundation, KU Leuven. This study has been funded in part with Federal funds from the Office of the Assistant Secretary for Preparedness and Response, Biomedical Advanced Research and Development Authority (BARDA), under OTA number HHSO100201800012C. Employees of the sponsor, Janssen Pharmaceutica NV, contributed to the study design, data analysis and interpretation, the writing of the report, and the decision to submit the manuscript for publication.
Chiu W, Verschueren L, Van den Eynde C, et al. Development and optimization of a high‐throughput screening assay for in vitro anti‐SARS‐CoV‐2 activity: evaluation of 5676 Phase 1 Passed Structures. J Med Virol. 2022;94:3101‐3111. 10.1002/jmv.27683
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions. Data are available upon reasonable request.
REFERENCES
- 1. Dhama K, Khan S, Tiwari R, et al. Coronavirus disease 2019‐COVID‐19. Clin Microbiol Rev. 2020;33(4):e0028‐0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Peiris JS, Lai ST, Poon LL, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361(9366):1319‐1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367(19):1814‐1820. [DOI] [PubMed] [Google Scholar]
- 4. Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727‐733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. ZOE Study Group . ZOE COVID Symptom Study. Accessed November 30, 2021. https://covid.joinzoe.com/us-2
- 6. Berlin DA, Gulick RM, Martinez FJ. Severe covid‐19. N Engl J Med. 2020;383(25):2451‐2460. [DOI] [PubMed] [Google Scholar]
- 7. Mercorelli B, Palu G, Loregian A. Drug repurposing for viral infectious diseases: how far are we? Trends Microbiol. 2018;26(10):865‐876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. de Wilde AH, Jochmans D, Posthuma CC, et al. Screening of an FDA‐approved compound library identifies four small‐molecule inhibitors of Middle East respiratory syndrome coronavirus replication in cell culture. Antimicrob Agents Chemother. 2014;58(8):4875‐4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mellott DM, Tseng CT, Drelich A, et al. A clinical‐stage cysteine protease inhibitor blocks SARS‐CoV‐2 infection of human and monkey cells. ACS Chem Biol. 2021;16(4):642‐650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mirabelli C, Wotring JW, Zhang CJ, et al. Morphological cell profiling of SARS‐CoV‐2 infection identifies drug repurposing candidates for COVID‐19. Proc Natl Acad Sci USA. 2021;118(36):e2105815118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ivens T, Van den Eynde C, Van Acker, et al. Development of a homogeneous screening assay for automated detection of antiviral agents active against severe acute respiratory syndrome‐associated coronavirus. J Virol Methods. 2005;129(1):56‐63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Van Damme E, De Meyer S, Bojkova D, et al. In vitro activity of itraconazole against SARS‐CoV‐2. J Med Virol. 2021;93(7):4454‐4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. De Meyer S, Bojkova D, Cinatl J, et al. Lack of antiviral activity of darunavir against SARS‐CoV‐2. Int J Infect Dis. 2020;97:7‐10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bojkova D, Bechtel M, McLaughlin KM, et al. Aprotinin inhibits SARS‐CoV‐2 replication. Cells. 2020;9(11):2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McClure R, Redha R, Vinson P, Pham W. A robust and scalable high‐throughput compatible assay for screening amyloid‐beta‐binding compounds. J Alzheimers Dis. 2019;70(1):187‐197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 1999;4(2):67‐73. [DOI] [PubMed] [Google Scholar]
- 17. Vincent MJ, Bergeron E, Benjannet S, et al. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol J. 2005;2:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang M, Cao R, Zhang L, et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019‐nCoV) in vitro. Cell Res. 2020;30(3):269‐271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wu C, Liu Y, Yang Y, et al. Analysis of therapeutic targets for SARS‐CoV‐2 and discovery of potential drugs by computational methods. Acta Pharm Sin B. 2020;10(5):766‐788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pant S, Singh M, Ravichandiran V, Murty USN, Srivastava HK. Peptide‐like and small‐molecule inhibitors against Covid‐19. J Biomol Struct Dyn. 2021;39(8):2904‐2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tardif JC, Bouabdallaoui N, L'Allier PL, et al. Colchicine for community‐treated patients with COVID‐19 (COLCORONA): a phase 3, randomised, double‐blinded, adaptive, placebo‐controlled, multicentre trial. Lancet Respir Med. 2021;9(8):924‐932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu J, Cao R, Xu M, et al. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS‐CoV‐2 infection in vitro. Cell Discov. 2020;6:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gordon DE, Jang GM, Bouhaddou M, et al. A SARS‐CoV‐2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583(7816):459‐468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yamamoto N, Yang R, Yoshinaka Y, et al. HIV protease inhibitor nelfinavir inhibits replication of SARS‐associated coronavirus. Biochem Biophys Res Commun. 2004;318(3):719‐725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Turlington M, Chun A, Tomar S, et al. Discovery of N‐(benzo[1,2,3]triazol‐1‐yl)‐N‐(benzyl)acetamido)phenyl) carboxamides as severe acute respiratory syndrome coronavirus (SARS‐CoV) 3CLpro inhibitors: identification of ML300 and noncovalent nanomolar inhibitors with an induced‐fit binding. Bioorg Med Chem Lett. 2013;23(22):6172‐6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ellinger B, Bojkova D, Zaliani A, et al. A SARS‐CoV‐2 cytopathicity dataset generated by high‐content screening of a large drug repurposing collection. Sci Data. 2021;8(1):70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mohamed K, Yazdanpanah N, Saghazadeh A, Rezaei N. Computational drug discovery and repurposing for the treatment of COVID‐19: a systematic review. Bioorg Chem. 2021;106:104490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen CZ, Xu M, Pradhan M, et al. Identifying SARS‐CoV‐2 entry inhibitors through drug repurposing screens of SARS‐S and MERS‐S pseudotyped particles. ACS Pharmacol Transl Sci. 2020;3(6):1165‐1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Riva L, Yuan S, Yin X, et al. Discovery of SARS‐CoV‐2 antiviral drugs through large‐scale compound repurposing. Nature. 2020;586(7827):113‐119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Touret F, Gilles M, Barral K, et al. In vitro screening of a FDA approved chemical library reveals potential inhibitors of SARS‐CoV‐2 replication. Sci Rep. 2020;10(1):13093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Smith E, Davis‐Gardner ME, Garcia‐Ordonez RD, et al. High‐throughput screening for drugs that inhibit papain‐like protease in SARS‐CoV‐2. SLAS Discov. 2020;25(10):1152‐1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhu T, Cao S, Su PC, et al. Hit identification and optimization in virtual screening: practical recommendations based on a critical literature analysis. J Med Chem. 2013;56(17):6560‐6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhu W, Xu M, Chen CZ, et al. Identification of SARS‐CoV‐2 3CL protease inhibitors by a quantitative high‐throughput screening. ACS Pharmacol Transl Sci. 2020;3(5):1008‐1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tummino TA, Rezelj VV, Fischer B, et al. Phospholipidosis is a shared mechanism underlying the in vitro antiviral activity of many repurposed drugs against SARS‐CoV‐2. bioRxiv. 2021. 10.1101/2021.03.23.436648 [DOI] [Google Scholar]
- 35. Bojkova D, Klann K, Koch B, et al. Proteomics of SARS‐CoV‐2‐infected host cells reveals therapy targets. Nature. 2020;583(7816):469‐472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gordon CJ, Tchesnokov EP, Woolner E, et al. Remdesivir is a direct‐acting antiviral that inhibits RNA‐dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J Biol Chem. 2020;295(20):6785‐6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bhimraj A, Morgan RL, Shumaker AH, et al. Infectious Diseases Society of America guidelines on the treatment and management of patients with COVID‐19. Accessed June 8, 2021. https://www.idsociety.org/practice-guideline/covid-19-guideline-treatment-and-management/ [DOI] [PMC free article] [PubMed]
- 38. Tzoumas N, Farrah TE, Dhaun N, Webb DJ. Established and emerging therapeutic uses of PDE type 5 inhibitors in cardiovascular disease. Br J Pharmacol. 2020;177(24):5467‐5488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stephens TD, Brynner R. Dark remedy: the impact of thalidomide and its revival as a vital medicine. Reading, MA. Oxford: Perseus; 2002. [Google Scholar]
- 40. Sourimant J, Aggarwal M, Plemper RK. Progress and pitfalls of a year of drug repurposing screens against COVID‐19. Curr Opin Virol. 2021;49:183‐193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Han Y, Duan X, Yang L, et al. Identification of SARS‐CoV‐2 inhibitors using lung and colonic organoids. Nature. 2021;589(7841):270‐275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kanimozhi G, Pradhapsingh B, Singh Pawar C, Khan HA, Alrokayan SH, Prasad NR. SARS‐CoV‐2: pathogenesis, molecular targets and experimental models. Front Pharmacol. 2021;12:638334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hoffmann M, Mösbauer K, Hofmann‐Winkler H, et al. Chloroquine does not inhibit infection of human lung cells with SARS‐CoV‐2. Nature. 2020;585(7826):588‐590. [DOI] [PubMed] [Google Scholar]
- 44. Yao X, Ye F, Zhang M, et al. In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2). Clin Infect Dis. 2020;71(15):732‐739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kashour Z, Riaz M, Garbati MA, et al. Efficacy of chloroquine or hydroxychloroquine in COVID‐19 patients: a systematic review and meta‐analysis. J Antimicrob Chemother. 2021;76(1):30‐42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jeon S, Ko M, Lee J, et al. Identification of antiviral drug candidates against SARS‐CoV‐2 from FDA‐approved drugs. Antimicrob Agents Chemother. 2020;64(7):e00819‐e00820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zandi K, Amblard F, Musall K, et al. Repurposing nucleoside analogs for human coronaviruses. Antimicrob Agents Chemother. 2020;65(1):e01652‐01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tripathi PK, Upadhyay S, Singh M, et al. Screening and evaluation of approved drugs as inhibitors of main protease of SARS‐CoV‐2. Int J Biol Macromol. 2020;164:2622‐2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Weston S, Coleman CM, Haupt R, et al. Broad anti‐coronavirus activity of Food and Drug Administration‐approved drugs against SARS‐CoV‐2 in vitro and SARS‐CoV in vivo. J Virol. 2020;94(21):e01218‐01220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jan JT, Cheng TR, Juang YP, et al. Identification of existing pharmaceuticals and herbal medicines as inhibitors of SARS‐CoV‐2 infection. Proc Natl Acad Sci USA. 2021;118(5):e2021579118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions. Data are available upon reasonable request.


