Abstract
Coronavirus disease of 2019 (COVID‐19) is a pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2). Mutations of mitochondrial DNA (mtDNA) are becoming increasingly common in various diseases. This study aims to investigate mutations in the cytochrome‐b (CYB) and adenosine triphosphatase‐6 (ATPase‐6) genes of mtDNA in COVID‐19 patients. The association between mtDNA mutations and clinical outcomes is investigated here. In the present study, mutations of the mtDNA genes CYB and ATPase‐6 were investigated in COVID‐19 (+) (n = 65) and COVID‐19 (−) patients (n = 65). First, we isolated DNA from the blood samples. After the PCR analyses, the mutations were defined using Sanger DNA sequencing. The age, creatinine, ferritin, and CRP levels of the COVID 19 (+) patients were higher than those of the COVID‐19 (−) patients (p = 0.0036, p = 0.0383, p = 0.0305, p < 0.0001, respectively). We also found 16 different mutations in the CYB gene and 14 different mutations in the ATPase‐6 gene. The incidences of CYB gene mutations A15326G, T15454C, and C15452A were higher in COVID‐19 (+) patients than COVID‐19 (−) patients; p < 0.0001: OR (95% CI): 4.966 (2.215−10.89), p = 0.0226, and p = 0.0226, respectively. In contrast, the incidences of A8860G and G9055A ATPase‐6 gene mutations were higher in COVID‐19 (+) patients than COVID‐19 (−) patients; p < 0.0001: OR (95%CI): 5.333 (2.359−12.16) and p = 0.0121 respectively. Yet, no significant relationship was found between mtDNA mutations and patients' age and biochemical parameters (p > 0.05). The results showed that the frequency of mtDNA mutations in COVID‐19 patients is quite high and it is important to investigate the association of these mutations with other genetic mechanisms in larger patient populations.
Keywords: ATPase‐6, COVID‐19, CYB, mtDNA, PCR, Sanger sequencing
Abbreviations
- ALT
alanine aminotransferase
- AST
aspartate aminotransferase
- BUN
blood urea nitrogen
- COVID‐19
2019 coronavirus
- CRP
C‐reactive protein
- CYB
cytochrome‐b
- HCT
hematocrit
- HGB
hemoglobin
- LDH
lactate dehydrogenase
- LYM
lymphocytes
- MCV
mean corpuscular volume
- mtDNA
mitochondrial DNA
- NEU
neutrophils
- PCR
polymerase chain reaction
- PLT
platelets
- WBCs
white blood cells
1. INTRODUCTION
The coronavirus disease of 2019 (COVID‐19) pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), has become a serious public health threat globally, endangering millions of people in a growing number of countries. 1 COVID‐19 began in the Chinese city of Wuhan and has since extended to almost all countries of the world. 2 Many recent studies have described the epidemiological and clinical characteristics of symptomatic patients infected with SARS‐CoV‐2 remains largely unknown. 2 SARS‐CoV‐2 causes numerous cellular and systemic events that significantly impact the intracellular and extracellular mitochondrial activities and can lead to disease progression and severity. 3 Mitochondria play an essential role in the host's response to viral infection and immunity, which is the key to antiviral signaling and exacerbating inflammatory processes. Mitochondria have been identified as potential targets in SARS‐CoV‐2 infection. 4 The relationship between mitochondrial DNA (mtDNA) damage and COVID‐19 infection is based on oxidative damage. mtDNA is vulnerable and exposed to oxidative stress as a result of metabolic function. In COVID‐19 infection, increased reactive oxygen species (ROS) production can affect cell organelles including mitochondria. 5
Mitochondria are crucial for cellular energy production. 6 mtDNA is a closed circular molecule that encodes 13 polypeptides which form oxidative phosphorylation complexes in humans. 7 mtDNA mutations and deletions are associated with oxidative stress, mitochondrial malfunction, and cell death. 7 mtDNA mutations can be occasional, genetic, or Mendelian in nature. Moreover, they can include mtDNA rearrangements such as deletions, inversions, or duplications, as well as point mutations. 8 mtDNA damage plays a key role in human aging, cancer, and neurological disorders. Point mutations of single bases or deletions of the 16.5‐kb mitochondrial genome are the leading footprints of mtDNA damage. 9 Because the human mitochondrial genome is so tiny compared with the nuclear genome, mitochondrial genetics poses unique clinical and research questions. 10 Mitochondrial ATPase 6 (mt‐ATP6) is a component of ATP synthase, a large enzyme that catalyzes the last stage of oxidative phosphorylation and is encoded by the mitochondrial genome. 11 , 12 mtDNA encodes three subunits of these complexes (I, III, and IV). The mitochondrial cytochrome‐b (mt‐CytB) gene encodes the mt‐CytB protein, which is the only component of the respiratory complex III encoded by the mitochondrial genome that plays a key role in the electron transport system. 13
Thus, the present study was performed to evaluate mitochondrial cytochrome‐b (CYB) and ATPase‐6 gene mutations in COVID‐19‐positive and ‐negative patients. We also investigated the association between these gene mutations and the clinical biochemical demographic features in COVID‐19 patients.
2. MATERIALS AND METHODS
2.1. Collection of COVID‐19‐positive and ‐negative blood samples
COVID‐19‐positive (n = 65) and negative (control group) blood samples (n = 65) were included in this study. All blood samples were collected from individuals admitted to the Emergency Department of Ordu University Research Hospital with suspected COVID‐19. Individuals with suspected COVID‐19 and showing symptoms (e.g., cough, sore throat, shortness of breath, myalgia, fever, loss of taste, and chest pain) and who had undertaken polymerase chain reaction (PCR) tests were enrolled in the study. Blood samples were picked up randomly. The patients were separated into two groups according to whether the PCR test was positive or negative. Sampling was not conducted according to the severity of the disease. At the time of sample collection, vaccinations had not yet started in our country. Therefore, all individuals are unvaccinated. The necessary permissions for sample collection were obtained from the Turkish Republic's Ministry of Health and from Ordu University's Clinical Ethics Committee (number: 2021/26). All blood samples were collected in a hemogram tube (EDTA).
2.2. Genomic DNA extraction from blood samples
The Eco→Tech DNA isolation kit (Cat no: EcoBGD‐50x) was used to isolate genomic DNA from blood. For DNA isolation, 200 µl of blood was used for each sample and the isolation was successfully performed by following the protocol recommended by the manufacturer of the corresponding kit.
First, 200 µl of EcoSpin Lysis Buffer was added to each 200 µl whole‐blood sample and mixed well. Then, 20 µl RNase A was added to the mixture from Step 1 and mixed well, which was incubated for 3 min at room temperature. Next, 20 µl Proteinase K was added to the mixture and mixed well, which was incubated for 10 min at 55°C. Then, 400 µl EcoSpin Binding Buffer was added and mixed well. After the washing and elution steps, the isolation was successfully completed.
The NanoDrop instrument Take3 Plate (BioTek) was used to measure the concentration of the DNA samples. The absorption ratios at 260 and 280 nm were used to evaluate the DNA purity. A ratio of approximately 1.8 is universally considered “pure” for DNA. All of the DNA samples were stored at −20°C before PCR.
2.3. PCR and Sanger sequence analysis of CYB and ATPase‐6 genes
The SensoQuest Labcycler device (thermalcycler) was used for the PCR stage. For each PCR reaction, 25 µl of EcoTaq 2× PCR Master Mix, 2 µl of forward primer (10 µM), 2 µl of reverse primer (10 µM), 10 pg−500 µg template DNA, and ddH2O were used. Preoptimized primers were preferred 14 (Table 1). PCR conditions were set as follows: 5 min at 95°C, 2 min at 94°C, 1 min at 61°C, 2 min at 72°C, 10 min at 72°C, and pause at 4°C for CYB and ATPase‐6 genes. After PCR analysis, PCR products were run with 3 µl of ethidium bromide on a 2% gel. A 50 bp marker was used. Bands of 675 and 1064 bp amplified for CYB and ATPase‐6 genes, respectively, were visualized in UV light.
Table 1.
Primer lists
| CYB | Forward, 5'‑TATCCGCCATCCCATACATT‑3' |
| Reverse, 5'‑GGTGATTCCTAGGGGGTTGT‑3 | |
| ATPase‐6 | Forward, 5'‑AACGAAAATCTGTTCGCTTCAT‑3' |
| Reverse, 5'‑ATGTGTTGTCGTGCAGGTAGAG‑3' |
The ABI3500 (Applied Biosciences) instrument was used for DNA sequencing. Before performing sequence analysis, the PCR products were cleared with exoSAP. After Sanger sequencing, analysis was performed using MITOMAP and Chromas Lite 2.1 (Technelysium) software.
2.4. Identifying the variants of CYB and ATPase‐6 genes by seven in silico programs
We used seven bioinformatics tools, Polymorphism Phenotyping v2 (PolyPhen‐2), Protein Analysis Trough Evolutionary Relationship (PANTHER), Sorting Intolerant From Tolerant (SIFT), Protein Variation Effect Analyzer (PROVEAN), Mutation Assessor, Single Nucleotide Polymorphism Annotation Platform (SNAP) and Combined Annotation Dependent Depletion (CADD) to predict the functional effects of the variants of CYB and ATPase‐6 gene.
2.5. Hematological and biochemical tests
Routine test data of individuals admitted with suspected COVID‐19 to the Emergency Department of Ordu University Research Hospital were used as hematological and biochemical parameters. In this study, parameters such as creatinine, white blood cells (WBCs), hemoglobin (HGB), platelets (PLT), neutrophils (NEU), lymphocytes (LYM), aspartate aminotransferase (AST), alanine aminotransferase (ALT), lactate dehydrogenase (LDH), C‐reactive protein (CRP), ferritin, blood urea nitrogen (BUN), hematocrit (HCT), and mean corpuscular volume (MCV) were evaluated. The means and standard deviations of test results obtained for the positive and negative samples were determined. We then performed statistical analyses by comparing the clinical data of the positive and negative samples.
2.6. Statistical analysis
The program GraphPad Prism 7.04 was used for all of the statistical analyses. The normal distribution of the data was demonstrated by the Shapiro−Wilk normality test. We used mean ± SD to describe the normally distributed variables. The association between COVID‐19 (+) and COVID‐19 (−) patients and clinical parameters was examined using the Mann−Whitney U test. A χ 2 test was conducted to calculate the association between the mutation types. The association between mutations and clinical parameters was also examined using the Mann−Whitney U test. Values for p < 0.05 were accepted as statistically significant.
3. RESULTS
3.1. COVID‐19‐positive and ‐negative individuals' clinical and demographic data distribution
The ages of our COVID‐19 (+) patients ranged from 26 to 87 years, while those of COVID‐19 (−) patients ranged in age from 18 to 92 years. In addition to COVID‐19 disease, 52.31% (34/65) of our patients were also suffering from other diseases such as chronic obstructive pulmonary disease (COPD) (21.54%), cardiovascular disease (34.84%), hypertension (49.23%), diabetes (12.31%), chronic kidney disease (6.15%), neurological disease (12.31%), and hepatitis B (33.84%). Moreover, 18.47% (12/65) of our patients had other diseases, such as COPD (12.31%), cardiovascular disease (6.15%), hypertension (15.38%), diabetes (1.54%), chronic kidney disease (1.54%), neurological disease (4.62%), and hepatitis B (9.23%) in addition to COVID‐19 (−) individuals.
The age, creatinine, ferritin, and CRP levels of COVID‐19 (+) patients were higher than those of COVID‐19 (−) patients (p = 0.0036, p = 0.0383, p = 0.0305, and p < 0.0001, respectively). However, there were higher levels of AST, ALT, LDH, WBC, PLT, NEU, LYM, BUN, HCT, and MCV in COVID‐19 (+) patients than those in COVID‐19 (−) patients (p > 0.05). Only HGB values were higher in COVID‐19 (−) patients compared with COVID‐19 (+) patients (p > 0.05). The age, hematological, and biochemical parameters of COVID‐19 (+) and COVID‐19 (−) patients are listed in Table 2.
Table 2.
Characteristics of COVID‐19‐positive patients and negative samples
|
Parameters (Reference range) |
COVID‐19 (+) patients (n = 65) (mean ± SD) |
COVID‐19 (−) samples (n = 65) (mean ± SD) |
p |
|---|---|---|---|
| Age (years) | 62.35 ± 19.71 | 51.58 ± 21.21 | **0.0036 |
|
Creatinine (0.50−0.90 mg/dl) |
1.11 ± 1.07 | 0.90 ± 0.68 | * 0.0383 |
|
AST (0−32 U/L) |
23.94 ± 10.78 | 22.97 ± 14.62 | 0.3839 |
|
ALT (0−33 U/L) |
23.8 ± 21.72 | 23.23 ± 13.73 | 0.6020 |
|
LDH (135−214 U/L) |
238.06 ± 79.48 | 226.77 ± 86.47 | 0.3523 |
|
Ferritin (30−400 µg/L) |
254.18 ± 281.08 | 130.50 ± 107.44 | * 0.0305 |
|
WBCs (4.49−12.6 × 103/μl) |
8.41 ± 5.79 | 7.94 ± 3.67 | 0.7620 |
|
HGB (12.3‐15.3 g/dl) |
12.58 ± 2.05 | 12.91 ± 2.05 | 0.2543 |
|
PLT (150−450 × 103/μl) |
240.88 ± 79.67 | 230.31 ± 89.65 | 0.6855 |
|
NEU (1.8−6.98 × 103/μl) |
5.80 ± 3.35 | 5.37 ± 3.34 | 0.3322 |
|
LYM (1.26−3.35 × 103/μl) |
1.96 ± 3.36 | 1.87 ± 1.37 | 0.1794 |
|
CRP (0−0.5 mg/dl) |
5.18 ± 5.88 | 2.81 ± 6.01 | **** 0.0001 |
|
BUN (6−18 mg/dl) |
18.82 ± 14.78 | 16.06 ± 7.12 | 0.7882 |
|
HCT (%) (35.7−43.8) |
38.47 ± 5.18 | 37.93 ± 5.834 | 0.8828 |
|
MCV (82.9−98 fl) |
86.72 ± 6.59 | 86.23 ± 7.93 | 0.9434 |
Note: Mann−Whitney U test was used to calculate the association between the variables.
Abbreviations: ALT, alanine aminotransferase; AST, aspartate aminotransferase; BUN, blood urea nitrogen; COVID‐19, Coronavirus disease of 2019; CRP, C‐reactive protein; HCT, hematocrit; HGB, hemoglobin, LDH, lactate dehydrogenase; LYM, lymphocyte; MCV, mean corpuscular volume; NEU, neutrophils; PLT, platelets, WBCs, white blood cells.
p < 0.05 is statistically significant.
3.2. CYB and ATPase‐6 gene PCR results
In all COVID‐19 (+) and COVID‐19 (−) patients included in the study, the mtDNA CYB and ATPase‐6 genes were amplified by PCR. The 1064 bp (CYB) and 675 bp (ATPase‐6) PCR products were analyzed in a 2% agarose gel (Figure 1A, B).
Figure 1.
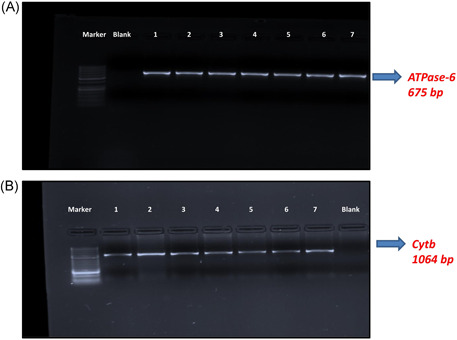
ATPase‐6 and CYB gel electrophoresis image
3.3. CYB and ATP Sanger DNA sequence analysis results
The human mitochondrial genome sequence used to identify mutations was “Cambridge Reference Series” (http://www.mitomap.org) and analyzed using Chromas Lite software. The sequence analysis of the detected mutations is shown in Table 3. Sixteen different mutations were detected in the CYB gene and 14 different mutations were detected in the ATPase‐6 gene in COVID‐19 (+) patients. Seven different mutations (G15431A, T15747C, A15758G, C15452A, T15674C, A15326G, and T15693C) were missense type (nonsynonymous substitution), which causes amino acid alteration in the CYB gene. Six different mutations (G9055A, A8836G, T9070G, A8860G, A8701G, and G8950A) were missense type in the ATPase‐6 gene. Eight mutations were synonymous substitution (not alter amino acids) type in both CYB (C15574T, T15310C, A15607G, G15301A, C15338T, T15454C, T15622C, and A15562G) and ATPase‐6 gene (G8856A, A8901G, G8994A, G9123A, G8697A, C8943T, T8772C, and G8865A). In total, 90 missense mutations were determined in the patients.
Table 3.
Distribution of CYB and ATPase‐6 mtDNA mutations in COVID‐19‐positive patients and negative samples
| Gene | Nucleoid position | Nucleotide exchange | Amino acid change | Mutation type |
Mutation rate (number of mutations/total number of patients) |
||
|---|---|---|---|---|---|---|---|
| Patient group | Control group | * p | |||||
| CYB | 15431 | G → A | A229T | Missense | 1/65 | 0/65 | 0.3154 |
| CYB | 15574 | C → T | F276F | Transition | 1/65 | 0/65 | 0.3154 |
| CYB | 15310 | T → C | I188I | Transition | 2/65 | 0/65 | 0.1541 |
| CYB | 15747 | T →C | I334T | Missense | 1/65 | 0/65 | 0.3154 |
| CYB | 15758 | A → G | I338V | Missense | 1/65 | 0/65 | 0.3154 |
| CYB | 15607 | A → G | K287K | Transition | 3/65 | 0/65 | 0.0797 |
| CYB | 15301 | G → A | L185L | Transition | 0/65 | 1/65 | 0.3154 |
| CYB | 15338 | C → T | L198L | Transition | 1/65 | 0/65 | 0.3154 |
| CYB | 15452 | C → A | L236I | Missense | 5/65 | 0/65 | * 0.0226 |
| CYB | 15454 | T → C | L236L | Transition | 5/65 | 0/65 | * 0.0226 |
| CYB | 15622 | T → C | L292L | Transition | 1/65 | 0/65 | 0.3154 |
| CYB | 15674 | T → C | S310P | Missense | 1/65 | 0/65 | 0.3154 |
| CYB | 15326 | A → G | T194A | Missense | 36/65 | 13/65 | ****0.0001 |
| CYB | 15562 | A → G | W272W | Transition | 1/65 | 0/65 | 0.3154 |
| CYB | 15804 | T: | frmshft | Deletion | 1/65 | 0/65 | 0.3154 |
| CYB | 15693 | T → C | M316T | Missense | 1/65 | 0/65 | 0.3154 |
| ATPase‐6 | 8856 | G → A | A110A | Transition | 1/65 | 0/65 | 0.3154 |
| ATPase‐6 | 9055 | G → A | A177T | Missense | 6/65 | 0/65 | * 0.0121 |
| ATPase‐6 | 8901 | A → G | L125L | Transition | 1/65 | 0/65 | 0.3154 |
| ATPase‐6 | 8994 | G → A | L156L | Transition | 1/65 | 0/65 | 0.3154 |
| ATPase‐6 | 9123 | G → A | L199L | Transition | 1/65 | 0/65 | 0.3154 |
| ATPase‐6 | 8836 | A → G | M104V | Missense | 3/65 | 0/65 | 0.0797 |
| ATPase‐6 | 8697 | G → A | M57M | Transition | 2/65 | 0/65 | 0.1541 |
| ATPase‐6 | 8943 | C → T | P139P | Transition | 1/65 | 0/65 | 0.3154 |
| ATPase‐6 | 9070 | T → G | S182A | Missense | 1/65 | 0/65 | 0.3154 |
| ATPase‐6 | 8860 | A → G | T112A | Missense | 32/65 | 10/65 | ****0.0001 |
| ATPase‐6 | 8701 | A → G | T59A | Missense | 1/65 | 0/65 | 0.3154 |
| ATPase‐6 | 8772 | T → C | T82T | Transition | 1/65 | 0/65 | 0.3154 |
| ATPase‐6 | 8865 | G → A | V113V | Transition | 1/65 | 0/65 | 0.3154 |
| ATPase‐6 | 8950 | G → A | V142I | Missense | 1/65 | 0/65 | 0.3154 |
Note: χ 2 test was used to calculate the association between the variables.
p < 0.05 is statistically significant.
3.4. Distribution of CYB and ATPase‐6 gene mutations in COVID‐19 (+) patient and control groups
The mutations, nucleotide, and amino acid changes, mutation rates, and p values detected in the CYB and ATPase‐6 genes are shown in Table 3. In addition, the rates of CYB and ATPase‐6 mutations in the COVID‐19 (+) and COVID‐19 (−) patients are shown.
G15431A (n = 1), C15574T (n = 1), T15310C (n = 2), T15747C (n = 1), A15758G (n = 1), A15607G (n = 3), C15338T (n = 1), T15454C (n = 5), C15452A (n = 5), T15622C (n = 1), T15674C (n = 1), A15326G (n = 36), A15562G (n = 1), T15693C (n = 1), and T15804: frame shift (n = 1) mtDNA mutations were detected in the CYB gene in COVID‐19 patients. In COVID‐19 (−) patients, G15301A (n = 1) and A15326G (n = 13) mtDNA mutations were found in the CYB gene.
G8856A (n = 1), G9055A (n = 6), A8901G (n = 1), G8994A (n = 1), G9123A (n = 1), A8836G (n = 3), G8697A (n = 2), C8943T (n = 1), T9070G (n = 1), A8860G (n = 32), A8701G (n = 1), T8772C (n = 1), G8865A (n = 1), and G8950A (n = 1) mtDNA mutations were detected in the ATPase‐6 gene in COVID‐19 (+) patients. In the COVID‐19 (−) patients, A8860G (n = 10) mtDNA mutations were found in the ATPase‐6 gene.
COVID‐19 (+) patients had significantly more A15326G, T15454C, and C15452A mutations in the CYB gene than COVID‐19 (−) patients; p < 0.0001, OR (95% CI): 4.966 (2.215−10.89); p = 0.0226 and p = 0.0226, respectively. In contrast, A8860G and G9055A mutations of the ATPase‐6 gene were more frequent in COVID‐19 (+) patients than in COVID‐19 (−) patients; p < 0.0001, OR (95%CI): 5.333 (2.359−12.16) and p = 0.0121; respectively) (Table 3).
A15326G and A8860G mutations were the most frequent mutation types in both the COVID‐19 patient and control groups.
3.5. Results of in silico analysis predicting the effects of human CYB and ATPase‐6 gene variants
We used seven different in silico variants prediction tools, PolyPhen‐2, PANTHER, SIFT, PROVEAN, Mutation Assessor, SNAP, and CADD, to predict the functional effects of the variants of CYB and ATPase‐6 genes. As a result of the in silico analysis, we found that 13 missense types (nonsynonymous substitution) were found to have various effects on diseases (Table 4). G15431A and T15674C were predicted to be deleterious variants in the CYB gene by in silico programs. Moreover, G9055A, A8836G, and A8860G were predicted to be deleterious variants in the ATPase‐6 gene.
Table 4.
Results of in silico analysis predicting the effects of human CYB and ATPase‐6 gene variants
| Gene | Nucleoid position | Amino acid change | Polyphen2 | PANTHER | SIFT | PROVEAN | Mutation assessor | SNAP | CADD |
|---|---|---|---|---|---|---|---|---|---|
| CYB | 15431 | A229T | Benign | Neutral | Neutral | Neutral | Low impact | Neutral | Deleterious |
| CYB | 15747 | I334T | Benign | Neutral | Neutral | Neutral | Low impact | Neutral | Neutral |
| CYB | 15758 | I338V | Benign | Neutral | Neutral | Neutral | Medium impact | Neutral | Neutral |
| CYB | 15452 | L236I | Benign | Neutral | Neutral | Neutral | Neutral impact | Neutral | Neutral |
| CYB | 15674 | S310P | Possible damaging | Disease | Neutral | Deleterious | High impact | Neutral | Deleterious |
| CYB | 15326 | T194A | Benign | Neutral | Neutral | Neutral | Neutral impact | Neutral | Neutral |
| CYB | 15693 | M316T | Benign | Neutral | Neutral | Neutral | Neutral impact | Disease | Neutral |
| ATPase‐6 | 9055 | A177T | Possible damaging | Disease | Neutral | Deleterious | Low impact | Neutral | Deleterious |
| ATPase‐6 | 8836 | M104V | Possible damaging | Disease | Neutral | Neutral | Low impact | Disease | Deleterious |
| ATPase‐6 | 9070 | S182A | Benign | Neutral | Neutral | Neutral | Neutral impact | Neutral | Neutral |
| ATPase‐6 | 8860 | T112A | Benign | Neutral | Neutral | Deleterious | Medium impact | Disease | Neutral |
| ATPase‐6 | 8701 | T59A | Benign | N/A | Neutral | Neutral | Neutral impact | Neutral | Neutral |
| ATPase‐6 | 8950 | V142I | Benign | Neutral | Neutral | Neutral | Neutral impact | Neutral | Neutral |
3.6. Clinical parameters in COVID‐19 patients according to CYB and ATPase‐6 gene mutational distribution
The age, hematological, and biochemical test results of the samples with and without (wild‐type) CYB and ATP mutations are shown in Table 5. No significant relationship was found between the mtDNA mutations and patients' age, hematological, and biochemical parameters (p > 0.05). The mean age of patients with mutations (CYB and ATP) was lower than that of patients without mutations. The mean ALT, LDH, and MCV levels of patients with mutations (CYB and ATP) were higher than those of patients without mutations. The mean creatinine, ferritin, HGB, and PLT levels of patients with mutations (CYB and ATP) were lower than those of patients without mutations. Among the patients carrying the CYB gene mutation, 69.44% carried additional disease and among the patients carrying the ATPase‐6 gene mutation, 65.63% carried additional disease.
Table 5.
Characteristics of COVID‐19‐positive patients with CYB and ATPase‐6 gene mutation or wild type (n = 65)
| CYB (mean ± SD) | ATPase‐6 (mean ± SD) | |||||
|---|---|---|---|---|---|---|
| Parameters(Reference range) | Mutant | Wild type | * p | Mutant | Wild type | * p |
| Age (years) | 61.44 ± 22.09 | 63.48 ± 16.60 | 0.6819 | 60.16 ± 23.17 | 64.48 ± 15.72 | 0.7272 |
|
Creatinine (0.50−0.90 mg/dl) |
0.98 ± 0.39 | 1.27 ± 1.54 | 0.7406 | 0.94 ± 0.29 | 1.27 ± 1.46 | 0.8680 |
|
AST (0−32 U/L) |
24.61 ± 11.29 | 23.10 ± 10.25 | 0.6817 | 23.5 ± 8.97 | 24.36 ± 12.42 | 0.8576 |
|
ALT (0−33 U/L) |
27.31 ± 27.37 | 19.45 ± 10.33 | 0.4403 | 24.44 ± 19.91 | 23.18 ± 23.64 | 0.7866 |
|
LDH (135−214 U/L) |
241.39 ± 86.27 | 233.93 ± 71.43 | 0.7308 | 247.94 ± 88.26 | 228.49 ± 69.96 | 0.2975 |
|
Ferritin (30−400 µg/L) |
237.92 ± 253.14 | 274.36 ± 315.80 | 0.9660 | 218.70 ± 189.05 | 288.60 ± 347.72 | 0.9196 |
|
WBCs (4.49−12.6 × 103/μl) |
8.91 ± 7.38 | 7.78 ± 2.84 | 0.9087 | 7.98 ± 3.12 | 8.83 ± 7.58 | 0.9351 |
|
HGB (12.3‐15.3 g/dl) |
12.51 ± 2.17 | 12.67 ± 1.93 | 0.8211 | 12.48 ± 2.16 | 12.68 ± 1.97 | 0.8170 |
|
PLT (150−450 × 103/μl) |
230.83 ± 64.61 | 253.35 ± 94.87 | 0.3845 | 239.97 ± 74.29 | 241.76 ± 85.71 | 0.9403 |
|
NEU (1.8−6.98 × 103/μl) |
6.00 ± 3.66 | 5.55 ± 2.96 | 0.8725 | 5.63 ± 2.90 | 5.96 ± 3.77 | 0.8121 |
|
LYM (1.26−3.35 × 103/μl) |
2.24 ± 4.45 | 1.60 ± 0.88 | 0.9765 | 1.63 ± 0.78 | 2.27 ± 4.66 | 0.6458 |
|
CRP (0−0.5 mg/dl) |
5.52 ± 6.61 | 4.75 ± 4.92 | 0.7681 | 4.85 ± 5.77 | 5.49 ± 6.07 | 0.3783 |
|
BUN (6−18 mg/dl) |
20.44 ± 16.70 | 20.04 ± 15.68 | 0.9347 | 19.25 ± 16.017 | 21.24 ± 16.42 | 0.4791 |
|
HCT (%) (35.7−43.8) |
38.64 ± 5.72 | 38.51 ± 5.29 | 0.8467 | 38.36 ± 5.80 | 38.80 ± 5.25 | 0.9558 |
|
MCV (82.9−98 fl) |
87.48 ± 6.23 | 85.37 ± 6.29 | 0.2078 | 87.44 ± 6.52 | 85.66 ± 6.04 | 0.3522 |
Note: Mann−Whitney U test was used to calculate the association between the variables.
Abbreviations: ALT, alanine aminotransferase; AST, aspartate aminotransferase; BUN, blood urea nitrogen; COVID‐19, Coronavirus disease of 2019; CRP, C‐reactive protein; HCT, hematocrit; HGB, hemoglobin, LDH, lactate dehydrogenase; LYM, lymphocyte; MCV, mean corpuscular volume; NEU, neutrophils; PLT, platelets, WBCs, white blood cells.
p < 0.05 is statistically significant.
4. DISCUSSION
Mutations in the human mitochondrial genome are linked to several diseases, most of which are inherited from the mother and all of which are related to abnormalities in oxidative energy metabolism. 15 These diseases are currently incurable and virtually untreatable and have a wide range of penetrance, symptoms, and prognosis. 16 mtDNA mutations have been detected in many body fluids, including urine and saliva 17 and serum. 18 Mutations in OXPHOS mtDNA genes do not always result in alterations in the encoded protein. 19
Viruses affect mitochondrial function, metabolism, and innate immune signaling. 20 Metabolism, calcium regulation, airway contractility in the lung, gene and protein balance, oxidative stress, and apoptosis are all affected by mitochondrial dysfunction. Mitochondrial dysfunction affects homeostatic cellular processes such as aging and senescence. 21 Mitochondria are proving to be significant in COVID‐19 pathogenesis due to their function in innate antiviral immunity and inflammation. 22
To the best of our knowledge, no previous research has investigated the association between CYB and ATPase‐6 gene mutations in Turkish COVID‐19 patients. In this study, the significance of CYB and ATPase‐6 gene mutations in COVID‐19 patients was investigated.
Clinical and laboratory findings were used to determine disease severity. The laboratory findings were not specific to COVID‐19 infection; however, they were used to estimate patients' prognosis. Higher WBCs and NEU count, lymphopenia, thrombocytopenia, CRP, LDH, creatine kinase (CK), troponin, increased liver enzymes, impairment of coagulation mechanisms, and rinsed cytokines are related to the severity of COVID‐19. 23 , 24 Some biomarkers, including C‐reactive protein (CRP) and ferritin, have been reported to be useful in the literature. 23 , 25 Zhang et al. 25 reported that CRP level increased in COVID‐19 patients. Moreover, with the increase in the CRP level in COVID‐19, the development of ARDS and death can be observed. 26 During viral infections, the concentration of circulating ferritin rises and can indicate viral replication. 27 Ferritin levels have been reported to increase in tables where a cytokine storm is observed, such as COVID‐19. 28 The laboratory findings of our patients showed that COVID‐19 (+) patients had higher age, creatinine, ferritin, and CRP levels than COVID‐19 (−) patients. Moreover, COVID‐19 (+) patients had lower HGB values than COVID‐19 (−) patients. In accordance with the literature, the laboratory findings vary according to disease severity. 29 Although the heterogeneity of the patients does not reflect the results confirmed by the literature, we can say that the laboratory findings were related to the severity of disease in our patients.
More mutations occur in mtDNA than in nuclear DNA, and a correlation has been found between ROS increase and the age‐related increase in mutant mtDNA. 30 Considering that SARS‐CoV‐2 indirectly produces the production of ROS, the cells of elderly people might be exposed to more ROS than those of healthy younger people when infected with this virus. 5 In our literature search, we did not find any articles addressing CYB and ATPase‐6 variations in COVID‐19 patients. However, mtDNA variations have been studied in many other diseases. According to the results of our Sanger analysis, 16 different mutations were found in the CYB gene and 14 different mutations were found in the ATPase‐6 gene. Moreover, missense and synonymous substitutions were detected in CYB and ATPase‐6 genes. The COVID‐19 (+) patient group and the negative control group had the more common mt15326 A → G (in CYB) and mt8860 A → G (in ATPase‐6) missense mutations. The COVID‐19 (+) patient had significantly more A15326G, T15454C, and C15452A mutations in the CYB gene than the COVID‐19 (−) patients. In addition, the COVID‐19 (+) patients had higher frequency of A8860G and G9055A missense mutations of the ATPase‐6 gene than in the COVID‐19 (−) patients. However, no significant relationship was detected between CYB and ATPase‐6 variants and the age and biochemical parameters of the patients. These results demonstrated that mtDNA is a part of cells that might be affected by COVID‐19 infection. mtDNA mutations and clinical parameters can reflect disease severity more effectively if they are studied with an extended patient population.
Mitochondrial genetic changes have been proven to affect metabolic parameters and can play a role in bioenergetic pathways, metabolic rates, and energy consumption depending on the ethnic background. 31 , 32 Given the results of studies on other diseases associated with mtDNA variations, according to Mao et al., 33 variations in the MT‐ATP6 and MT‐CYB genes might play a role in the unexpected fertilization disorder. The A8860G mutation in the ATPase‐6 gene was detected in 79%−91.66% in breast tumors, 75%−100% in other types of cancers 32 , 33 and 92.85%−100% in neurodegenerative diseases. 34 , 35 In the present study, 32 COVID‐19 (+) patients had A8860G missense mutation in the ATPase‐6 gene. In another study, the frequencies of transversions in the ATP6 and CytB genes were found to be 96% and 97%, respectively, while the frequency of transversions in the ATP6 gene was found to be 4% and 3% in the CytB gene. 13 Pirola et al. 36 reported that NASH is associated with hereditary alterations in the cellular respirasome of the liver, including high cytochrome‐b diversity and mtDNA damage, which can lead to widespread cellular effects. Li et al. 37 discovered three additional significant mitochondrial variants, mutations A4769G, A8860G, and A15326G, in samples taken from all 15 individuals in the family. They also assumed that these variants do not have much effect on mitochondrial work. In the current study, 36 COVID‐19 patients had A15326G missense mutation in the CYB gene. G15431A and T15674C were predicted to be deleterious variants of the CYB gene by in silico programs. Moreover, G9055A, A8836G, and A8860G were predicted to be deleterious variants in the ATPase‐6 gene by in silico programs. Our study indicates that these variants may change the secondary structure of the CYB and ATPase‐6 proteins and subsequently can reduce their enzyme activity. The alteration of the enzyme activity can affect ROS and disease severity. In addition, these data suggest that more studies will need to be conducted to reveal the effects of these A8860G and A15326G mtDNA mutations in COVID‐19 disease.
The current study has some limitations that should be mentioned. We examined CYB and ATPase‐6 mutations and clinical parameters of patients with COVID‐19. First, we were unable to analyze other mtDNA genes (ND1 and D310). Second, we did not have the opportunity to work with many patients. Therefore, additional research and study with larger patient populations are required to substantiate our findings and demonstrate their relevance.
To the best of our knowledge, there is no publication in the relevant literature that focuses on the association between CYB and ATPase‐6 mutations and COVID‐19 patients. As this is a pilot study, more data on mtDNA variants need to be collected to highlight the association between known mitochondrial variants and COVID‐19 patients. The high prevalence of mtDNA mutations in COVID‐19 patients suggests that they play a key role in the disease and alter patients' energy metabolism. We suggest that clinicians should consider the genetic background of patients when evaluating them.
AUTHOR CONTRIBUTIONS
Conceptualization, methodology, software, validation, formal analysis: Ebubekir Dirican, Gonca Gülbay, İsmail Erkan Aydın, Şeyda Tuba Savrun, Ülkü Karaman. Investigation, resources, data curation: Ebubekir Dirican, Gonca Gülbay, İsmail Erkan Aydın, Şeyda Tuba Savrun, Ülkü Karaman. Writing—original draft, Writing—review and editing: Ebubekir Dirican, Gonca Gülbay, İsmail Erkan Aydın, Şeyda Tuba Savrun, Ülkü Karaman. Visualization, supervision, project administration: Ebubekir Dirican, Gonca Gülbay, İsmail Erkan Aydın, Şeyda Tuba Savrun, Ülkü Karaman.
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
Dirican E, Savrun ŞT, Aydın İE, Gülbay G, Karaman Ü. Analysis of mitochondrial DNA cytochrome‐b (CYB) and ATPase‐6 gene mutations in COVID‐19 patients. J Med Virol. 2022;94:3138‐3146. 10.1002/jmv.27704
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Majumder J, Minko T. Recent developments on therapeutic and diagnostic approaches for COVID‐19. AAPS J. 2021;23(1):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rothan HA, Byrareddy SN. The epidemiology and pathogenesis of coronavirus disease (COVID‐19) outbreak. J Autoimmun. 2020;109:102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Saleh J, Peyssonnaux C, Singh KK, Edeas M. Mitochondria and microbiota dysfunction in COVID‐19 pathogenesis. Mitochondrion. 2020;54:1‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Costa TJ, Potje SR, Fraga‐Silva TFC, et al. Mitochondrial DNA and TLR9 activation contribute to SARS‐CoV‐2‐induced endothelial cell damage. Vascul Pharmacol. 2022;142:106946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ganji R, Reddy PH. Impact of COVID‐19 on mitochondrial‐based immunity in aging and age‐related diseases. Front Aging Neurosci. 2021;12:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Taanman J‐W. The mitochondrial genome: structure, transcription, translation and replication. Biochim Biophys Acta—Bioenerg. 1999;1410(2):103‐123. [DOI] [PubMed] [Google Scholar]
- 7. Roubicek DA, Souza‐Pinto NCde. Mitochondria and mitochondrial DNA as relevant targets for environmental contaminants. Toxicology. 2017;391:100‐108. [DOI] [PubMed] [Google Scholar]
- 8. Craigen WJ, Mitochondrial DNA. Mitochondrial DNA mutations: an overview of clinical and molecular aspects. Mutations. 2012;837:3‐15. [DOI] [PubMed] [Google Scholar]
- 9. Birch‐Machin MA. The role of mitochondria in ageing and carcinogenesis. Clin Exp Dermatol. 2006;31(4):548‐552. [DOI] [PubMed] [Google Scholar]
- 10. Taylor RW, Turnbull DM. Mitochondrial DNA mutations in human disease. Nat Rev Genet. 2005;6(5):389‐402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Anderson S, Bankier AT, Barrell BG, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290(5806):457‐465. [DOI] [PubMed] [Google Scholar]
- 12. Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23(2):147. [DOI] [PubMed] [Google Scholar]
- 13. Demir D, Türkkahraman D, Aktaş Samur A, Lüleci G, Akçurin S, Alper ÖM. Mitochondrial ATPase subunit 6 and cytochrome B gene variations in obese Turkish children. J Clin Res Pediatr Endocrinol. 2014;6(4):209‐215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Avcilar T, Kirac D, Ergec D, et al. Investigation of the association between mitochondrial DNA and p53 gene mutations in transitional cell carcinoma of the bladder. Oncol Lett. 2016;12(4):2872‐2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schon EA, DiMauro S, Hirano M. Human mitochondrial DNA: roles of inherited and somatic mutations. Nat Rev Genet. 2012;13(12):878‐890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gammage PA, Viscomi C, Simard M‐L, et al. Genome editing in mitochondria corrects a pathogenic mtDNA mutation in vivo. Nat Med. 2018;24(11):1691‐1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fliss MS. Facile detection of mitochondrial DNA mutations in tumors and bodily fluids. Science. 2000;287(5460):2017‐2019. [DOI] [PubMed] [Google Scholar]
- 18. Okochi O, Hibi K, Uemura T, et al. Detection of mitochondrial DNA alterations in the serum of hepatocellular carcinoma patients. Clin Cancer Res. 2002;8(9):2875‐2878. [PubMed] [Google Scholar]
- 19. Grzybowska‐Szatkowska L, Ślaska B, Rzymowska J, Brzozowska A, Floriańczyk B. Novel mitochondrial mutations in the ATP6 and ATP8 genes in patients with breast cancer. Mol Med Rep. 2014;10(4):1772‐1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Elesela S, Lukacs NW. Role of mitochondria in viral infections. Life. 2021;11(3):232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kauppila TES, Kauppila JHK, Larsson N‐G. Mammalian mitochondria and aging: an update. Cell Metab. 2017;25(1):57‐71. [DOI] [PubMed] [Google Scholar]
- 22. Prasun P. COVID‐19: a mitochondrial perspective. DNA Cell Biol. 2021;40(6):713‐719. [DOI] [PubMed] [Google Scholar]
- 23. Du R‐H, Liang L‐R, Yang C‐Q, et al. Predictors of mortality for patients with COVID‐19 pneumonia caused by SARS‐CoV‐2: a prospective cohort study. Eur Respir J. 2020;55(5):2000524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang G, Zhang J, Wang B, Zhu X, Wang Q, Qiu S. Analysis of clinical characteristics and laboratory findings of 95 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a retrospective analysis. Respir Res. 2020;21:565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang J, Dong X, Cao Y, et al. Clinical characteristics of 140 patients infected with SARS‐CoV‐2 in Wuhan, China. Allergy. 2020;75(7):1730‐1741. [DOI] [PubMed] [Google Scholar]
- 26. Terpos E, Ntanasis‐Stathopoulos I Elalamy I, et al. Hematological findings and complications of COVID‐19. Am J Hematol. 2020;95(7):834‐847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li Y, Hu Y, Yu J, Ma T. Retrospective analysis of laboratory testing in 54 patients with severe‐ or critical‐type 2019 novel coronavirus pneumonia. Lab Investig. 2020;100(6):794‐800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497‐506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Henry BM, de Oliveira MHS, Benoit S, Plebani M, Lippi G. Hematologic, biochemical and immune biomarker abnormalities associated with severe illness and mortality in coronavirus disease 2019 (COVID‐19): a meta‐analysis. Clin Chem Lab Med. 2020;58(7):1021‐1028. [DOI] [PubMed] [Google Scholar]
- 30. Reddy PH, Beal MF. Amyloid beta, mitochondrial dysfunction and synaptic damage: implications for cognitive decline in aging and Alzheimer's disease. Trends Mol Med. 2008;14(2):45‐53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fuku N, Oshida Y, Takeyasu T, et al. Mitochondrial ATPase subunit 6 and cytochrome b gene polymorphisms in young obese adults. Biochem Biophys Res Commun. 2002;290(4):1199‐1205. [DOI] [PubMed] [Google Scholar]
- 32. Guo L‐J, Oshida Y, Fuku N, et al. Mitochondrial genome polymorphisms associated with type‐2 diabetes or obesity. Mitochondrion. 2005;5(1):15‐33. [DOI] [PubMed] [Google Scholar]
- 33. Mao G‐H, Wang Y‐N, Xu M, Wang W‐L, Tan L, Tao S‐B. Polymorphisms in the MT‐ATP6 and MT‐CYB genes in in vitro fertilization failure. Mitochondrial DNA. 2015;26(1):20‐24. [DOI] [PubMed] [Google Scholar]
- 34. Tan D‐J, Bai R‐K, Wong L‐JC. Comprehensive scanning of somatic mitochondrial DNA mutations in breast cancer. Cancer Res. 2002;62(4):972‐976. [PubMed] [Google Scholar]
- 35. Czarnecka AM, Klemba A, Krawczyk T, et al. Mitochondrial NADH‐dehydrogenase polymorphisms as sporadic breast cancer risk factor. Oncol Rep. 2010;23(2):531‐535. [PubMed] [Google Scholar]
- 36. Pirola CJ, Garaycoechea M, Flichman D, Castaño GO, Sookoian S. Liver mitochondrial DNA damage and genetic variability of Cytochrome b—a key component of the respirasome—drive the severity of fatty liver disease. J Intern Med. 2021;289(1):84‐96. [DOI] [PubMed] [Google Scholar]
- 37. Li W, Zhang W, Li F, Wang C. Mitochondrial genetic analysis in a Chinese family suffering from both mitochondrial encephalomyopathy with lactic acidosis and stroke‐like episodes and diabetes. Int J Clin Exp Pathol. 2015;8(6):7022‐7027. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


