Abstract
Purpose
We tested polyplexes of a diblock polymer containing a pH-responsive, endosomolytic core (dimethylaminoethyl methacrylate and butyl methacrylate; DB) and a zwitterionic Poly (methacryloyloxyethyl phosphorylcholine) (PMPC) corona for the delivery of plasmid DNA (pDNA) to glioblastoma cells.
Methods
We studied the physicochemical characteristics of the DNA polyplexes such as particle hydrodynamic diameter and surface potential. Cytocompatibility of free PMPC-DB polymer and pDNA polyplexes with U-87MG and U-138MG glioma cell lines were evaluated using the ATP assay. The transfection activity of luciferase pDNA polyplexes was measured using a standard luciferase assay. Anti-proliferative, apoptotic, and cell migration inhibitory activities of PMPC-DB/Interferon-beta (IFN-β1) pDNA polyplexes were examined using ATP assay, flow cytometry, and wound closure assay, respectively.
Results
PMPC-DB copolymer condensed pDNA into nanosized polyplexes. DNA polyplexes showed particle diameters ranging from ca. 100–150 nm with narrow polydispersity indices and near electroneutral zeta potential values. PMPC-DB/Luciferase pDNA polyplexes were safe and showed an 18-fold increase in luciferase expression compared to the gold standard PEI polyplexes in U-87MG cells. PMPC-DB/IFN-β1 polyplexes induced apoptosis, demonstrated anti-proliferative effects, and retarded cell migration in glioblastoma cells.
Conclusion
The results described herein should guide the future optimization of PMPC-DB/DNA delivery systems for in vivo studies.
Keywords: DNA polyplexes, interferon-beta1, glioblastoma, transfection, zwitterionic phosphorylcholine polymer
INTRODUCTION
DNA therapeutics offer great potential for the treatment of intractable diseases including genetic disorders, neurodegenerative diseases, infections, and malignancies by modulating gene and/or protein epression [1]. The clinical approval of Gendicine and Advexine for head and neck squamous cell carcinoma led to over 1000 clinical trials accelerating the development of plasmid DNA-based gene therapeutics for a wide range of human disorders [2, 3]. However, the delivery of naked plasmid DNA (pDNA) is associated with challenges such as nuclease degradation, short half-life, unpredictable pharmacokinetic profile, and poor cellular uptake [2]. Therefore, safe and efficient delivery systems are required for the successful clinical application of pDNA that not only improve its pharmacokinetic profile but also increase delivery to the target tissue. Synthetic non-viral vectors including polyelectrolyte polyplexes of lipids (lipoplexes) and polymers (polyplexes) of DNA are promising alternatives to the immunogenic viral vectors. These are attractive due to the flexibility in chemistry allowing them to rationally design the carriers and make them less immunogenic compared to the viral carriers. For efficient pDNA transfection, DNA polyplexes require a wide range of functionalities such as polyplex stability in the blood and extracellular fluids, cellular targeting and internalization, endosomal escape, and the nuclear import of the DNA payload [4]. Modulation of structure and surface chemistry have allowed polyplexes to be protected from physiological degradation, conferred stealth protection, facilitated target cell interaction, and conferred endosomolytic activity to allow DNA release into the cytoplasm [4, 5].
2-Methacryloxyethyl phosphorylcholine (MPC)-based copolymers are clinically utilized due to their biocompatibility and the physicochemical aspects and transfection efficiency of their nucleic acid polyplexes have been studied earlier [6]. Optimized dimethylaminoethyl methacrylate (DMAEMA)-MPC copolymers demonstrated stable DNA condensation and showed very little non-specific cellular association [6]. Our coworkers previously developed and tested a zwitterionic nanocarrier based on a pH-responsive, endosomolytic core-forming block composed of DMAEMA and butyl methacrylate (BMA) and a zwitterionic polymeric MPC (PMPC) corona for siRNA delivery [5, 7]. The cationic DMAEMA moieties can electrostatically complex the pDNA whereas the hydrophobic BMA chains further stabilize the polyplex core [8]. In general, the balance between the cationic and hydrophobic block components are known to modulate the membrane binding, cellular uptake, endosomal escape, and decrease the cytotoxicity of DNA polyplexes [8, 9]. The DMAEMA-BMA diblock copolymer (abbreviated henceforth as DB copolymer) exhibits colloidal stability, blood serum stability, and pH-responsive membrane lytic activity in the endolysosomal range [9, 10] allowing efficient escape of the polyplexes into the cytoplasm. Zwitterionic PMPC, used in FDA-approved products, are hydrated through strong electrostatic interactions that lead to lower protein adsorption during systemic circulation since there is no gain in free energy in displacing the water molecules via protein binding [11]. As a consequence, zwitterionic PMPC/siRNA polyplexes showed a five-fold increase in circulation half-life compared to polyplexes containing a 5 kDa PEG corona, and significantly increased tumor cell uptake. Nanoparticles containing zwitterionic coronas have shown longer circulation and greater accumulation in cancer cells and tumor tissues compared to the traditional PEG counterparts [12, 13].
Our goal was to investigate if the above effects of increased siRNA transfection are also applicable to PMPC-DB/DNA polyplexes. We seek to apply this polyplex chemistry in the setting of glioblastoma, which is the most refractory of all cancers with a median survival time of these patients less than 15 months despite receiving standard-of-care therapies such as surgery, chemotherapy and radiation therapy [14]. Phosphatidylcholine (PC) is a natural component of cell membranes that makes up nearly 50% of the total membrane lipid content and is required for membrane structural integrity and function [15]. Clinical samples collected from patients with grade II/III glioblastoma contained higher lipid levels, specifically phosphatidylcholine (PC), compared to normal brain tissues [16]. Rapid metabolism, cell proliferation, and increased lipogenesis rate in malignant cells overexpressing choline transporters [17], a precursor of PC, provides a unique opportunity to target abnormal lipid metabolism as an anticancer therapeutic strategy [18]. Jiang et al. investigated a choline derivative for magnetic resonance imaging of glioblastoma tumors synthesis [19] and as a targeting ligand to deliver combination anticancer therapies such as the human tumor necrosis factor-alpha ligand gene and doxorubicin to U-87MG glioblastoma cells and mice bearing U-87MG xenografts [17]. The above observations compelled us to choose glioma cell lines as model systems to test the delivery efficiency of PMPC-DB/DNA polyplexes.
IFN-β has clinically demonstrated anti-cancer effects by enhancing Temozolomide sensitivity via downregulation of O(6)-methylguanine DNA methyltransferase [20]. IFN-β has also exhibited anti-angiogenic activity in glioblastoma multiforme, restricted cell growth, and differentiation, blocked oncogene expression, and can act as an adjuvant for immunotherapies by activating T lymphocytes, natural killer cells and macrophages [20, 21]. The demonstrated clinical safety, stability, bioavailability, and multi-faceted anticancer effects of IFN-β compared to IFN-α [22] encouraged us to select IFN-β as a therapeutic plasmid. IFN-β interacts with the IFNAR-2 ligand that facilitates the dimerization with IFNAR-1 and activates the Tyk-2/JAK-1 receptor kinases through tyrosine phosphorylation of the cytosolic receptor domains [22]. Subsequently, the secondary signaling cascade activates the transcription factors of the Signal Transducer and Activator of Transcription family to initiate the IFN-β protein synthesis [23]. We tested IFN-β1 pDNA delivery using PMPC-DB polyplexes to determine its suitability to deliver a therapeutic gene.
To summarize, we explored the capability of PMPC-DB, a well-studied siRNA carrier, for the delivery of pDNA to glioblastoma cells. We studied the physicochemical characteristics of PMPC/DB-DNA polyplexes, determined its cytocompatibility and transfection activity in U-87MG and U-138MG glioblastoma cell lines. The transfection efficiency of PMPC-DB/DNA polyplexes was compared to polyethyleneimine (PEI)/DNA polyplexes. While PEI cytotoxicity is a concern, it arguably remains as the gold standard and one of the most efficient synthetic polycations [24]. We demonstrated the capability of PMPC-DB/DNA polyplexes to deliver a therapeutic gene, IFN-β1, and confirmed its anti-proliferative, apoptotic, and inhibitory effects on cell migration in vitro.
MATERIALS AND METHODS
Materials
gWiz plasmid containing luciferase (gWiz-Luc), or Green Fluorescence Protein reporter gene (gWiz-GFP) were purchased from Aldevron (Fargo, ND) as 5 mg/ml aqueous solution and used without further purification. Plasmid DNA encoding the human interferon-beta1 (IFN-β1) inserted into a pCMV6-XL4 vector was purchased from OriGene Technologies Inc. (Rockville, MD) and used without further purification. Polyethylenimine branched (PEI, molecular weight 25,000 Da) was purchased from Sigma-Aldrich (St. Louis, MO). Ethidium Bromide: 1% solution, and agarose were procured from Fisher Bioreagents (Fair Lawn, NJ). GeneRuler 1 kb (DNA ladder) and 6x DNA loading dye were purchased from Thermo Scientific (Vilnius, Lithuania). Cell Titer Glo 2.0 reagent and cell culture lysis 5x reagent were from Promega (Madison, WI). ATP, Lucifer Yellow CH dilithium, and heparin sodium salt were purchased from MP Biomedicals (Illkirch, France). Pierce BCA protein assay kit for total protein measurement was purchased from Thermo Scientific (Rockford, IL). Annexin V Alexa Fluor 647 conjugate and e-Bioscience 7-AAD viability staining solution were procured from Thermo Fisher Scientific (Carlsbad, CA). All the chemicals used for the experiments were obtained as molecular biology grade, DNase, RNase, and protease-free materials.
Cell Lines and Cell Culture
U-87 MG (ATCC HTB-14) and U-138 MG (ATCC HTB-16) cell lines were purchased from ATCC (Manassas, VA). All cells were maintained in the complete growth media containing glutamine-supplemented Modified Eagle Medium (MEM (1x) + Glutamax, gibco, Carlsbad, CA)) with an added 10% fetal bovine serum (FBS, HyClone, Logan, UT). The cells were maintained at 37 ± 0.5°C and 5% CO2 in a humidified incubator (Isotemp, Thermo Fisher Scientific, USA). Notably, the U-87 MG cell line was used between passage numbers (P) 127 to P133, whereas the U-138 MG cell line was used from P178 to P183. The cell lines were received from ATCC at comparable passage numbers. Prior to passage, the cells were washed using 1x Phosphate Buffered Saline (PBS, Hyclone, Logan, UT), and were dissociated using 1x TrypLE Express Enzyme (gibco, Denmark) except for the apoptosis assay where the cells were dissociated using 1x enzyme-free Cell Dissociation Buffer containing ethylenediaminetetraacetic acid (EMD Millipore Corp, MA).
Methods
Polymer Synthesis
We RAFT polymerized a pH-responsive block comprising a random copolymer of dimethylaminoethyl methacrylate (DMAEMA) and butyl methacrylate (BMA). This macro-CTA was subsequently extended using RAFT to polymerize 20 kDa zwitterionic poly methacryloyloxyethyl phosphorylcholine (PMPC) [5]. The copolymer is referred to as PMPC-DB throughout the manuscript. The molecular mass of the diblock copolymer measured using gel permeation chromatography was 44,901 g/mol. The degree of the poly(DMAEMA-co-BMA) block polymerization was 81 with 50% of each monomer. For our N:P calculations, we assumed that the polymer is 50% protonated, so we used 40.5 cationic charges per mole of the copolymer resulting in a mass/charge value of 1108.67 Da.
Ethidium Bromide Exclusion Assay
The ability of PMPC-DB to condense plasmid DNA was confirmed by a standard ethidium bromide exclusion assay by measuring the changes in ethidium bromide/DNA fluorescence upon gradual addition of pre-calculated volumes of PMPC-DB to achieve a range of N/P ratios. DNA (gWiz-Luc) solution at a concentration of 20 μg/ml in 10 mM citrate buffer, pH 4.0 was mixed with ethidium bromide (EtBr, 1 μg/ml). The fluorescence of DNA/ethidium bromide solution was measured using a Fluoromax-4 spectrofluorometer (HORIBA Scientific, NJ, USA) at 545 nm excitation and 595 nm emission, a 3 nm slit width, and set to 100%. The background fluorescence was set to 0% using 1 μg/ml EtBr solution. The mass per charge (m/z) ratio of PMPC-DB is 1108.67 Da, whereas the m/z of pDNA is 325 Da.
Preparation of PMPC-DB/DNA Polyplexes
We used a rapid titration procedure described previously to prepare the DNA polyplexes [25]. Briefly, plasmid DNA (1 mg/ml) was diluted in 10 mM citrate buffer (pH 4.0), the DNA solution was vortexed for 30 s at low speeds on a benchtop vortexer (Fisher Analog Vortex, 120 V, Fisher Scientific, USA) and was allowed to stand for 10 min at room temperature (RT). We define the molar mixing ratio (N/P) as a ratio of the mass of polymer/mass per charge of the charged amines in the copolymer (1108.67 g/mol) to that of the mass of pDNA added/mass per charge of pDNA (325 g/mol). Polyplexes were prepared by adding precalculated volumes of the polymer solution slowly along the walls of the microtube and the polymer and DNA solution were allowed to come into contact while vortexing the tube at maximum speeds for 10 s. The polyplexes were initially formed at pH 4 to allow protonation of the tertiary amines in DMAEMA in the DMAEMA-BMA (DB) polymer chains and allow effective electrostatic binding to the pDNA [9]. After mixing, the solution was spun down using a benchtop mini centrifuge (VWR, USA/China). The polyplexes were allowed to stand for 30 min at RT. Finally, we added a 5x volume of 10 mM phosphate buffer (pH 8.0) to the polyplexes prior to use in experiments. For 24-well plate transfections, the final concentration of pDNA in the polyplexes was maintained at 20, 30, and 40 μg/ml for 1, 1.5, and 2 μg/well pDNA dose, respectively. PEI polyplexes were prepared in 10 mM HEPES buffer, pH 7.4 using the above described rapid titration method.
Gel Electrophoresis
The formation of PMPC-DB/DNA polyplexes was further confirmed using a standard agarose gel electrophoresis assay. PMPC-DB or PEI DNA polyplexes were prepared at different N/P ratios as described earlier. Polyplexes containing 0.4 μg DNA in a 20 μl volume were mixed with a 6x loading dye prior to loading. The samples were loaded in a 0.8% agarose gel containing 0.5 μg/ml EtBr prepared in 0.5x Tris-borate EDTA buffer and were subjected to electrophoresis using a gel assembly (OWL EASYCAST B2, Thermo scientific) at 120 V, 100 mA for 75 min. The gel was exposed to a UV chamber using a Protein simple red imaging system (Santa Clara, CA, USA).
Particle Diameter and Zeta Potential
Z-Average particle diameter and zeta potential of DNA polyplexes were measured using Dynamic Light Scattering (DLS). DNA polyplexes containing 20 μg/ml gWiz-Luc pDNA were prepared at various N/P ratios as described earlier. DNA polyplexes were prepared in 10 mM phosphate buffer or 10 mM HEPES buffer, pH 7.4, and the measurements were done on a Zetasizer Nano ZS90 (Malvern Panalytical Inc., Westborough, PA). Each sample was run thrice and the data reported are representative of three independent experiments. The standard error of measurement was <5%.
Time-Dependent Colloidal Stability of DNA Polyplexes
We studied the time-dependent colloidal stability in [1] a low ionic strength, 10 mM phosphate buffer; pH 7.4, [2] 10 mM phosphate buffer containing 0.15 M NaCl; defined here as high salt conditions, [3] cell culture medium supplemented with serum at concentrations ranging from 0 to 50% and [4] also determined the stability of polyplexes challenged with different concentrations of an anionic competitor, heparin. The stability of PMPC-DB/pDNA polyplexes was compared with PEI/pDNA polyplexes. The polyplexes were stored at room temperature (RT) and 2–8°C for seven days. The physicochemical characteristics such as particle size distribution (PSD) and polydispersity index (PdI) of the polyplexes were measured using the dynamic light scattering, whereas the ability of polyplexes to retain pDNA during the storage period was evaluated using agarose gel electrophoresis at 0, 1, 4, and 7 days. The details about the stability studies in 10 mM phosphate buffer containing 0.15 M NaCl, serum-containing cell culture medium and the heparin competition assay were mentioned in the supplementary file.
Cytocompatibility
The effect of PMPC-DB and PEI polymers, or its DNA polyplexes was tested in U-87MG and U-138MG cell lines using a CellTiter-Glo Luminescent Cell Viability Assay that correlates intracellular ATP levels to metabolic cell viability. The ATP assay is a sensitive technique compared to the widely used MTS assay because the luminescent ATP readout is a more accurate representation of the metabolically viable cell numbers [26]. Polyethylenimine (PEI) was used as a positive control. U-87MG or U138MG cells were seeded in a 96-well plate at a seeding density of 16,500 cells/well and were cultured for 48 h in the complete growth medium until about 80% confluency. Cells were incubated with [1] PEI from 2.5 to 75 μg/ml in an isotonic buffer containing 0.15 M NaCl; 10 mM HEPES buffer, pH 7.4 and PMPC-DB from 5 to 100 μg/ml in isotonic 10 mM citrate buffer pH 4.0 further diluted using 10 mM phosphate buffer pH 8.0, and [2] PMPC-DB/pDNA polyplexes from N/P 2 to 10 and naked pDNA at a concentration of 2.8 μg DNA/ml. Prior to treating the cells, the polymer solution and DNA polyplexes were diluted in 60 μl of MEM/FBS. The treatment mixture was removed after 4 h and replaced with 200 μl fresh MEM/FBS medium. After overnight incubation in a humidified CO2 incubator, the medium was replaced by 60 μl fresh MEM/FBS medium and 60 μl ATP assay reagent. After shaking the plate for 15 min on a Benchtop Orbital Shaker in dark at room temperature, 60 μl of each sample was transferred into a 96-well white opaque polystyrene microplate. The luminescence was measured with a 1 s integration time using a SpectraMax i3 multimode plate reader (Molecular Devices, Sunnyvale, CA). The percent (%) cell viability was calculated using this formula: (luminescence of transfected cells/luminescence of control, untreated cells) × 100%. Data presented are average ± S.D. (n = 6) and are representative of three independent experiments.
Transfection Efficiency In Vitro
Transfection activity of DNA polyplexes containing gWiz-Luc prepared at different N/P ratios was measured using a standard luciferase assay [27]. Briefly, U-87MG cells were seeded in 48-well plates at a seeding density of 50,000 cells/well and plated 48 h before transfection. On a day of transfection, the cells were incubated for 4 h with DNA polyplexes (25 μl) in 150 μl of complete medium resulting in a DNA concentration of 2.85 μg/ml and a pDNA dose of 0.5 μg/well. After the 4 h incubation, the transfection mixture was removed and cells were cultured for an additional 24 h in fresh complete medium prior to lysing cells for luciferase measurement. The culture medium was removed, the cells were washed once with prewarmed 1x PBS, and 100 μl of 1x cell lysis reagent was added to each well. To measure the luciferase protein, 20 μl of the cell lysate was mixed with 100 μl of luciferase assay buffer (20 mM glycylglycine (pH 8), 1 mM MgCl2, 0.1 mM EDTA, 3.5 mM DTT, 0.5 mM ATP, 0.27 mM coenzyme A) and the luminescence was immediately measured over 10 s integration using a GloMax 20/20 Luminometer. The total cellular protein in the cell lysate was determined using a Pierce BCA protein assay using a calibration curve constructed with standard bovine serum albumin solutions. The transfection results are expressed as average Relative Light Units (RLU)/mg of cellular protein ± S.D. (n = 4) and are representative of at least three independent experiments.
Antiproliferative effects of PMPC-DB/interferon-β1 DNA polyplexes In Vitro
The transfection and subsequent antiproliferative effects of PMPC-DB/plasmid interferon -β1 DNA (pIFN-β1) polyplexes were determined using the ATP assay. Two key parameters, N/P ratio of polyplexes and pIFN-β1 DNA dose, were optimized in U-87MG and U-138MG cell lines. U-87MG or U-138MG cells were seeded at a density of 16,500 cells/well in a 96-well plate and cultured for 48 h so that the cells were 70–80% confluent at the time of transfection. For N/P optimization, pIFN-β1 polyplexes using either PMPC-DB or PEI at N/P ratios from 2 to 10 were prepared using the same protocol described earlier and cells were incubated with a transfection mixture containing 2.85 μg/ml IFN-β1 diluted in complete medium. The pDNA concentration in the transfection mixture was 2.85 μg/ml for a dose of 1 μg/well/0.35 ml treatment volume and the concentrations were correspondingly scaled-up for higher doses. For formulation optimization, cells were transfected with PMPC-DB/pIFN-β1 polyplexes using a full factorial combination of [1] N/P 8, 10 and 12, and [2] DNA doses at 1, 1.5, and 2 μg/well. PEI at 40 and 60 μg/ml were used as a positive controls. Cells were also incubated with naked pDNA or free polymer solution at an amount equivalent to the N/P ratios used in DNA polyplexes. Untreated cells were used as a negative control. Cells were transfected for 4 h, the transfection mixture was removed, and the cells were cultured with 200 μL complete medium for 48 h prior to determining cell viability using the ATP assay as described in cytotoxicity studies above. Data presented are average ± S.D. (n = 6) and are representative of at least two independent experiments.
Apoptosis Assay Using Flow Cytometry
The ability of IFN-β1 pDNA polyplexes to induce apoptosis was investigated using flow cytometry. Cellular apoptosis was evaluated using the Annexin-V AF647 conjugate and 7-AAD double-staining assay. U-87MG cells were seeded at a density of 100,000 cells/well in a 24-well plate and cultured for 48 h so that the cells were 70–80% confluent at the time of transfection. IFN-β1 pDNA polyplexes were prepared using the same protocol described earlier and cells were incubated with a transfection mixture containing 1.0 μg/well IFN-β1 diluted in complete medium. Cells were transfected for 4 h, the transfection mixture was removed, and the cells were cultured for 48 h prior to harvesting them using a non-enzymatic cell dissociation buffer for flow cytometry analysis. Cells were washed once with ice-cold 1x PBS and the supernatant was discarded. The pellet was resuspended and diluted in annexin-binding buffer (10 mM HEPES, 140 mM NaCl, and 2.5 mM calcium chloride, pH 7.4) to adjust the concentration to ~1 × 106 cells/ml. Per assay, a 100 μl aliquot of the cell suspension was incubated with 5 μl annexin V conjugate at room temperature in dark for 10 min and then 5 μl 7-AAD was added for 5 min to stain the dead cells. The mixture was diluted to 500 μl using the annexin-binding buffer and the samples were immediately analyzed using Attune NxT Acoustic Focusing Cytometer (Invitrogen, Singapore) equipped with Attune NxT software. The fluorescence intensity of annexin V conjugate-stained cells was detected at 670/40 nm, while 7-AAD positive cells were detected using a 574/26 nm detector. In each experiment, about 50,000 events from a 150 μl sample volume were recorded. The percentage of viable (annexin V conjugate-negative and 7-AAD-negative), early apoptotic (annexin V-positive and 7-AAD-negative), and late apoptotic and necrotic cells (annexin V-positive and 7-AAD-positive) were analyzed and are presented as histograms obtained from the Attune NxT software.
Scratch Assay
The antiproliferative effect of pIFN-β1 DNA polyplexes in vitro was evaluated using a scratch or wound assay [28]. U-87MG cells were seeded at a density of 100,000 cells/well in a 24-well plate and incubated at 37°C in a humidified 5% CO2 incubator for 72 h to reach about 90% confluency. Prior to treatment, the media was removed, and a scratch was made vertically near the center of the well using a 10 μl pipet tip. The scratch was noted underside of the plate with a marker dot followed by washing the cells with pre-warmed 250 μl 1x PBS buffer. The wash was discarded, and cells were transfected with PMPC-DB/pIFN-β1 DNA polyplexes at N/P 8 (preparation described earlier) diluted in a complete growth media at 1 ml/well containing a dose of 2.0 μg pDNA/well. Cells were also incubated with naked pDNA or an equivalent amount of PMPC-DB polymer used in the polyplexes. All samples were diluted in complete medium prior to adding to the cells. Cells incubated with complete medium alone was used as a control. Cells were transfected for 4 h, and the treatment mixture was replaced with fresh complete medium. The initial image at 0 h was taken using an EVOS microscope under transmitted light settings and 4x magnification. Additional images were taken at 24 and 48 h post-transfection. An arbitrary boundary enclosing the scratch was made and the width of this boundary was calibrated to be a constant in all groups. As cells continued to proliferate or otherwise as an effect of the treatment, they filled this boundary space, and the width of the “unfilled” boundary was measured at three different positions along the length: the top, middle, and bottom. The width of the unfilled boundary is defined here as the wound width. The scale bar of the images was set at 1000 μm using Image J software (NIH).
GFP Gene Expression Assay
Transfection activity and gene expression efficiency of PMPC-DB/DNA polyplexes were confirmed using fluorescence microscopy. The transfection experiments using gWiz-GFP pDNA were conducted as follows. U-87MG cells were seeded at a density of 100,000 cells/well in a 24-well plate and cultured for 48 h so that the cells were 70–80% confluent at the time of transfection. Plasmid gWiz-GFP DNA polyplexes were prepared using the same protocol described earlier and cells were incubated with a transfection mixture containing 2 μg pDNA/well diluted in complete medium. Cells were transfected for 4 h and the transfection mixture was replaced with fresh complete medium. The images of the cells were acquired at 24 and 48 h post-transfection using an EVOS microscope under transmitted light and GFP channel settings (excitation 470/22 nm and emission 510/12 nm) at 20x magnification. The images were compared with the control (untreated cells), cells treated with naked gWiz-GFP, or those treated with an equivalent quantity of free PMPC-DB polymer used in the polyplexes.
Statistical Analysis
Data in figures indicate mean ± standard deviation and ‘n’ values indicate the number of replicates in each group. GraphPad Prism 8.4.1 and JMP Pro 14 software (SAS Institute Inc., NC, USA) were used for statistical analysis. Mean comparison between control and treatment groups in the EtBr assay, cytocompatibility studies, luciferase gene expression assays, anti-proliferative, and apoptosis studies were performed using one-way ANOVA followed by Bonferroni’s Multiple Comparison Test. p < 0.05 was considered statistically significant. DOE study was statistically analyzed using JMP Pro 14 software. The scale bar of microscopic images was calibrated and the wound width was quantified using Image J software (NIH). The intensity of the transmitted, GFP channel, and overlay images for GFP expression studies were adjusted at a constant setting using Image J (NIH) software.
RESULTS
DNA Condensation
The ability of PMPC-DB to condense pDNA was studied using the ethidium bromide (EtBr) exclusion and agarose gel electrophoresis assays (Fig. 1). The gradual addition of PMPC-DB to a solution of pDNA containing EtBr displaced the dye as a result of polymer binding to the pDNA, resulting in a gradual decrease of fluorescence values. The relative fluorescence unit (RFU) value at N/P 0 was considered 100%, and percent RFU (%RFU) for N/P 0.05 to 10 was calculated in comparison to N/P 0. A significant (p < 0.05) drop in RFU was observed at N/P 0.2 (Fig. 1a). The %RFU value decreased steeply from N/P 0.2 (81.9 ± 1.1%) to N/P 1 (24.0 ± 2.5%) and stayed constant up to N/P 10 at about 20%. The reduction in %RFU from N/P 0.4 to 10 was statistically significant (p < 0.0001). We used agarose gel retardation assay as an orthogonal technique to confirm the formation of DNA polyplexes. The EtBr exclusion assay allowed us to follow the real-time formation of DNA polyplexes by monitoring the decrease in EtBr fluorescence whereas the agarose gel retardation is a static assay that allowed us to compare differences in pre-made polyplexes at different N/P ratios. The intense band under UV light indicates the presence of DNA intercalated with EtBr. The DNA bands in the loading wells were considered a complete condensation of DNA in the polyplexes. DNA polyplexes of poly(ethylenimine) (PEI) prepared at N/P ratios 0.5 to 5 were used for comparison. PEI/pDNA at N/P 2 showed condensation of DNA into polyplexes (Fig. 1b) accompanied by a strong fluorescence signal near the loading wells. Increased PEI amounts in the N/P 3 and 5 polyplexes showed a complete condensation of pDNA and less intense bands in the loading wells suggesting complete complexation at these N/P ratios. PMPC-DB/DNA polyplexes showed a migration pattern that was fairly consistent with the EtBr data. As expected, we noted DNA migration from the cathode to anode in the samples prepared at N/P ratios ≤0.8–1.0 whereas the polyplexes at N/P 0.8–1.0 remained in the wells (Fig. 1b). Interestingly, we noted a smear-like pattern migrating towards the anode at N/P ratios 2.0 and 2.5. The N/P 3.0 sample showed an interesting pattern where the DNA migrated a little towards the anode, and a noticeable band appeared to migrate in a direction towards the cathode. Polyplexes prepared at N/P ratios 4, 6, and 10 showed a similar pattern where some of the DNA stayed in the wells and the remaining pDNA migrated away from the wells towards the cathode. In summary, both EtBr exclusion and agarose gel electrophoresis data demonstrated that PMPC-DB copolymer condensed pDNA into nanoparticles at N/P ≥ 3.
Fig. 1.
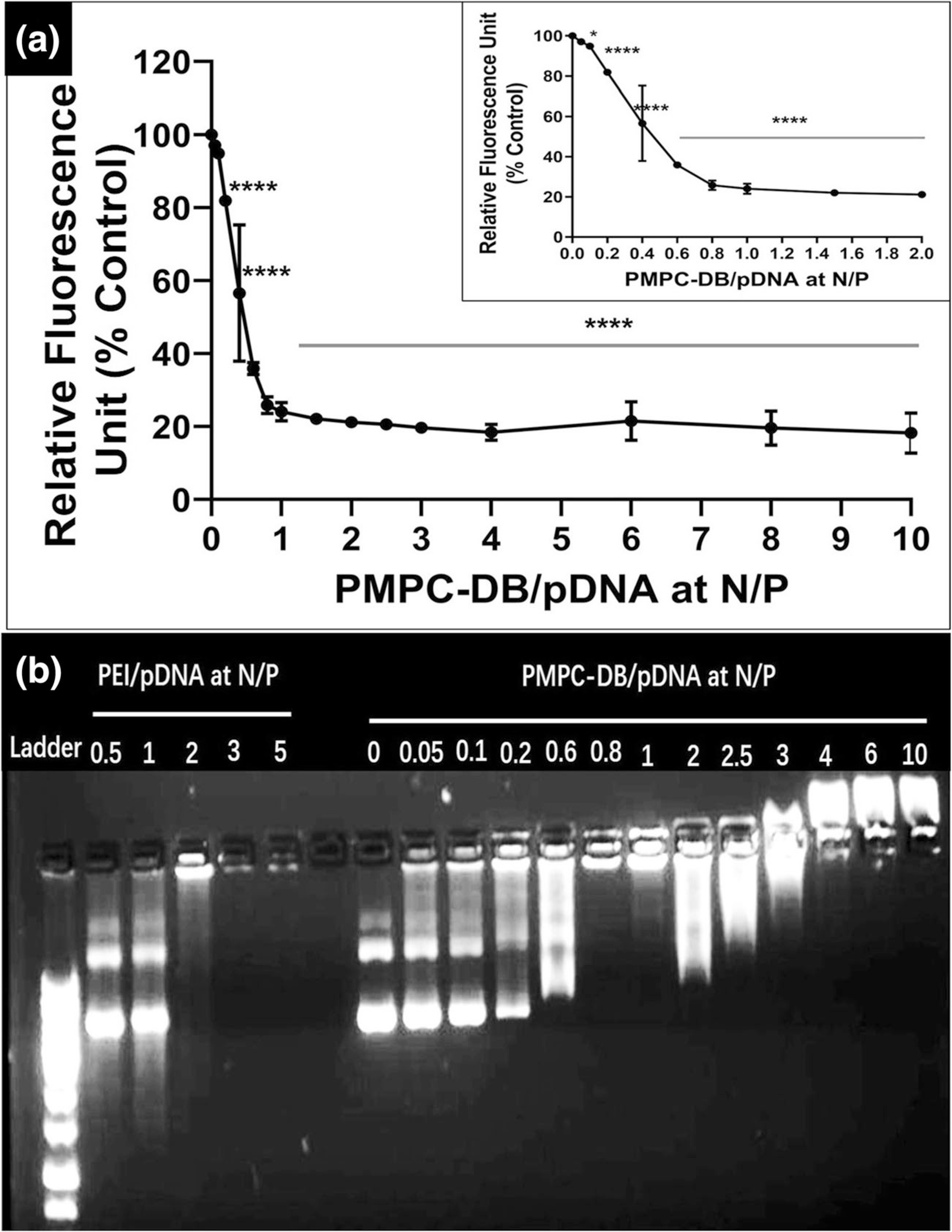
DNA condensation by PMPC-DB polymer at different N/P ratios confirmed using (a) ethidium bromide exclusion assay and (b) agarose gel electrophoresis. (a) Precalculated volumes of a PMPC-DB stock (3 mg/ml) solution was added to 20 μg/ml pDNA containing 1 μg/ml ethidium bromide in 10 mM citrate buffer pH 4.0 (N/P 0; baseline) to obtain the specified N/P ratios. The relative fluorescence unit (%) of PMPC-DB/pDNA polyplexes was calculated against the baseline. The RFU values were measured on a Fluoromax-4 spectrofluorometer. The data is presented as mean ± standard deviation (SD) from three independent experiments. One-way ANOVA was performed using GraphPad Prism 8.4.1. Asterisks indicate significant differences (****p < 0.0001, * p < 0.05). (b) Polyplexes containing 0.4 μg pDNA were loaded into a 0.8% w/v agarose gel. The gel was run at 120 V and 100 mA for 75 min. The gel was scanned under UV using a ProteinSimple red imaging system.
Characterization of DNA Polyplexes Using Dynamic Light Scattering (DLS)
Physicochemical characteristics like particle size and surface charge are important determinants of the biological activity of DNA polyplexes. So, we measured the Z-average particle diameter and ζ-potential using dynamic light scattering, the gold standard tool to characterize these parameters (Table I). Plasmid DNA solution at N/P 0 showed an average diameter of about 100 nm, a broad size distribution (PdI) of 0.4 ± 0.2, and a negative zeta potential of −30.7 ± 7.3 mV. DNA polyplexes at N/P ratio 0.5 showed a larger particle size of ca. 216 nm (PdI 0.2). DNA polyplexes formed at N/P ratios 1 to 8 showed particle diameters ranging from ca. 100–150 nm with narrow polydispersity indices (PdI) ranging from 0.15–0.20 and showed a unimodal distribution on the intensity vs. particle diameter plots (SL Fig. 1). DNA polyplexes at N/P 10 showed a larger particle diameter of ca. 211 nm with a broader PdI of ca. 0.3. The measured zeta-potentials across N/P ratios from 0 to 10 were suggestive of DNA polyplex formation. Though the N/P 0.5 polyplexes showed a negative zeta potential of ca. −25.4 mV, the net surface charge shifted towards electroneutral values as the PMPC-DB amounts increased at N/P values ranging from 1 to 10. PEI/DNA polyplexes showed particle diameters >200 nm at N/P 1 and 2 with a PdI > 0.2. PEI/pDNA at N/P 10 showed an average particle size of ca. 131 nm and a narrow PdI of 0.14. As expected, the zeta potential shifted from ca. −34 mV at N/P 1 to ca. +25 mV at N/P 10.
Table I.
Particle size distribution, polydispersity index, and zeta potential of PMPC-DB/pDNA or PEI polyplexes measured using dynamic light scattering. PMPC-DB/pDNA polyplexes containing 20 μg/ml pDNA were initially prepared in 10 mM citrate buffer and further diluted 5× using 10 mM phosphate buffer pH 8.0, whereas PEI/pDNA polyplexes were prepared in 10 mM HEPES buffer pH 7.4 for size, PdI, and zeta potential measurements on a Malvern ZS90 Zetasizer
| Polyplexes | N/P | Z-avg. (nm) | PdI | Zeta Potential (mV) |
|---|---|---|---|---|
|
| ||||
| PMPC-DB/pDNA | 0 | 96.8 ± 15.9 | 0.44 ± 0.23 | −30.7 ± 7.3 |
| 0.5 | 215.6 ± 0.9 | 0.21 ± 0.01 | −25.4 ± 3.8 | |
| 1 | 144.6 ± 1.3 | 0.19 ± 0.01 | −0.3 ± 0.2 | |
| 2 | 131.3 ± 1.4 | 0.18 ± 0.02 | 0.1 ± 0.2 | |
| 3 | 123.8 ± 1.2 | 0.20 ± 0.00 | 0.7 ± 0.1 | |
| 4 | 99.1 ± 0.6 | 0.14 ± 0.01 | 0.2 ± 0.2 | |
| 6 | 103.1 ± 0.5 | 0.19 ± 0.02 | 0.4 ± 0.2 | |
| 8 | 102.0 ± 0.6 | 0.15 ± 0.01 | 0.5 ± 0.1 | |
| 10 | 211.0 ± 3.7 | 0.32 ± 0.01 | 0.5 ± 0.1 | |
| PEI/pDNA | 1 | 190.9 ± 3.7 | 0.19 ± 0.00 | −33.9 ± 0.6 |
| 2 | 366.0 ± 34.8 | 0.62 ± 0.06 | 9.4 ± 0.4 | |
| 10 | 131.2 ± 1.2 | 0.14 ± 0.01 | 24.3 ± 0.8 | |
Cytocompatibility of PMPC-DB Polyplexes
We measured the cytocompatibility of free polymer and PMPC-DB/DNA polyplexes in two different glioma cell lines derived from malignant adult tumors, U-87MG and U-138MG. We used U-87MG and U-138MG cell lines since these cells express both the IFN-β receptors, IFNAR1 and IFNAR2, that dimerize for its functional activity [29]. The Cell Titer Glo based-ATP Assay was used as a high throughput cytotoxicity measurement method due to the rapid and sensitive nature of this assay [26] for evaluating the cell viability of polymer and DNA polyplex-treated cells. The relative luminescence unit (RLU) readout in this assay is directly proportional to the intracellular ATP levels. The RLU levels of the untreated cells were normalized as 100% cell viability and thus, the measurements allowed us to determine the cytocompatibility of the polymer or polyplexes based on the measured ATP levels. First, we evaluated the cell viability of U-87 MG cells treated with PEI at concentrations ranging from 2.5–75 μg/ml (Fig. 2a). PEI at 10 μg/ml showed a significant (p < 0.001) reduction in U-87 MG cell viability. There was a linear and significant (p < 0.001) reduction in cell viability from about 56% at 10 μg/ml to about 3% at 75 μg/ml. The IC50 of PEI for U-87MG cells was ca. 15 μg/ml. Second, the cytocom-patibility of isotonic PMPC-DB polymer solution (Fig. 2b) was evaluated in U-87MG and U-138MG cells. Our data showed that U-87MG and U-138MG cells incubated with PMPC-DB copolymer at concentrations ranging from 5 to 60 μg/ml showed an average cell viability of ≥80% following a 4 h exposure (Fig. 2b). Third, we also determined the cytocompatibility of PMPC-DB/DNA polyplexes with U-87MG (Fig. 2c) and U-138MG cells (Fig. 2d) using the ATP assay. We formed polyplexes at N/P ratios varying from 2 to 10 and our data demonstrated that all the formulations were safe resulting in cell viabilities of ca. 100% in both cell lines (Fig. 2c and d).
Fig. 2.
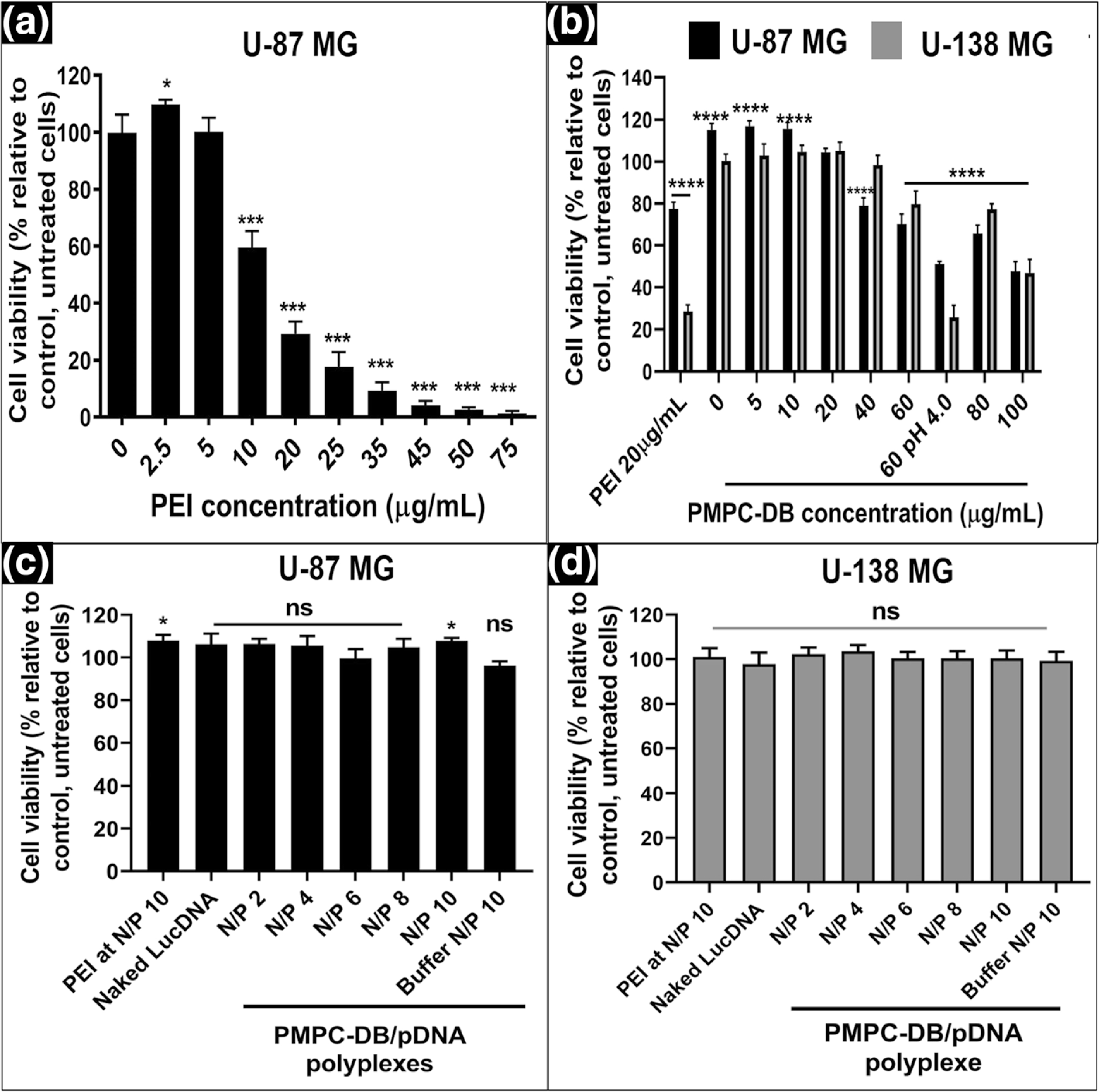
Cytocompatibility of PMPC-DB polymer and PMPC-DB/pDNA polyplexes with U-87MG and U-138MG cells using the ATP assay. U-87MG (P130) cells were seeded in 96-well plates at 16,500 cells/well. At about 80% confluency, U-87MG cells were incubated with (a) an isotonic mixture of PEI diluted in MEM/FBS medium for 4 h. U-87MG and U-138MG (P179) were (b) incubated for 4 h with PMPC-DB polymer initially dissolved in citrate buffer and 5x diluted in 10 mM phosphate buffer pH 8.0 buffer. (c-d) transfected with naked DNA, PEI/pDNA polyplexes, and PMPC-DB/pDNA at the indicated N/P ratios for 4 h. The pDNA concentration in each group was 2.8 μg/ml in a 60 μL volume/well. For ATP assays in a-d, the treatment media was replaced with fresh complete culture media and incubated for 24 h before the ATP assay. Cell Titer Glo 2.0 reagent was added at an equal volume to that of cell culture medium. The luminescence was measured at 1 s integration time using a luminometer. The % cell viability of the treated cells was calculated using the following equation: ((relative luminescence unit (RLU) of treated cells/RLU of untreated cells) × 100. The significance of treated groups was compared against control using a student’s unpaired t-test compared to control or one-way ANOVA (Bonferroni’s Multiple Comparison test) and the significance levels are indicated as *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 (ns = non-significant)
Transfection Activity of PMPC-DB/Luc pDNA Polyplexes
We formed PMPC-DB/pDNA polyplexes using luciferase pDNA and measured their transfection activity in the glioma cell lines. We normalized luciferase gene expression levels to the total cellular protein because this allowed us to use the protein concentration values as an indirect readout of potential transfection-related toxicity. This also allowed us to confirm that the observed changes in transfection activity were not due to polyplex toxicity. In all transfection experiments, cells were microscopically examined at various stages during the transfection experiment: prior to adding polyplexes, after the 4 h transfection, and prior to lysing cells for luciferase gene expression measurement 24 h post-transfection. The cells appeared healthy similar to the control, untreated cells during those observations. We prepared polyplexes at N/P ratios ranging from 2 to 10 and used PEI/DNA polyplexes as a positive control. A previous study tested the effect of the N/P ratio on the luciferase transgene expression of PEI/DNA polyplexes and determined that polyplexes at N/P 10 mediated the highest level of expression [30]. Thus, we used PEI polyplexes at N/P ratio 10 as our positive control. As expected, naked pDNA did not mediate measurable levels of luciferase expression, and the readout was similar to untreated U-87MG cells (Fig. 3a). We observed a bell-shaped profile in the case of PMPC-DB/pDNA polyplexes at N/P ratios ranging from 2 to 10. Polyplexes at N/P 2 mediated a 21-fold increase in luciferase expression compared to cells exposed to naked pDNA whereas polyplexes prepared at N/P 4, 6, 8 and 10 mediated increases that were 308-, 38–11- and 6-fold higher compared to polyplexes at N/P 2. This bell-shaped profile related to the effect of PMPC-DB N/P ratios was reproducibly verified in independent transfection experiments. Notably, PMPC-DB/pDNA polyplexes at N/P 4 showed an 18-fold increase in luciferase transfection compared to the gold standard PEI/DNA polyplexes.
Fig. 3.
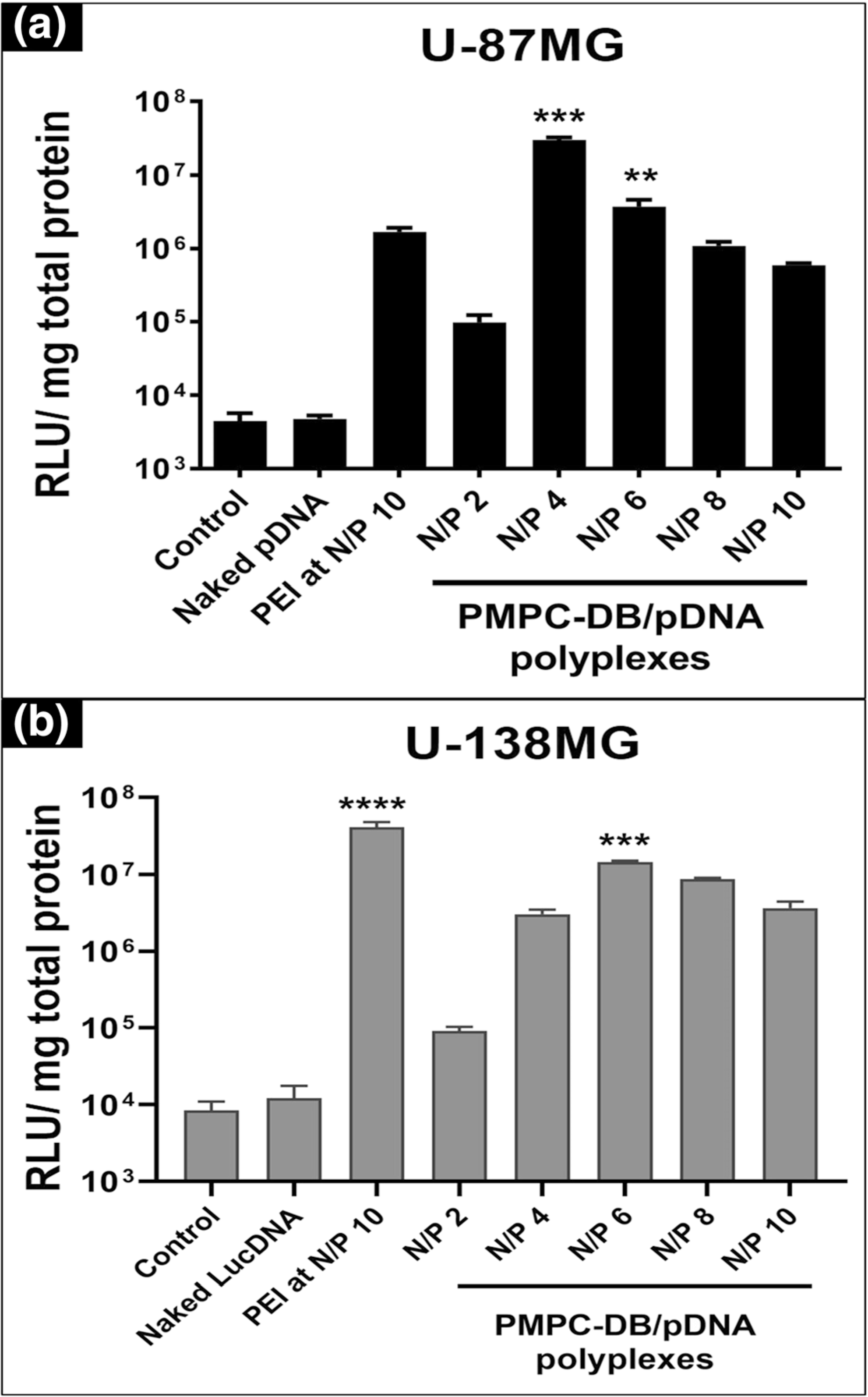
Transfection activity of PMPC-DB/DNA polyplexes in U-87MG and U-138MG cells. (a) U-87MG (P133) and (b) U-138MG (P178) were seeded at 50,000 cells/well in 48-well plates. At about 80% confluency, cells were transfected with the indicated samples for 4 h and the luciferase expression was measured 24 h post-transfection using a luminometer. The luciferase expression was normalized to the total protein content of the cell lysate. The significance of treated groups was compared against control using one-way ANOVA (Bonferroni’s Multiple Comparison test) and the significance levels are indicated as *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001
We conducted the same luciferase transfection experiment in the U-138MG cell line and observed a similar trend as noted in the U-87MG cell line (Fig. 3b). The transfection levels of polyplexes at N/P 4, 6, 8, and 10 were 37-, 161-, 94- and 40-fold greater than that of PMPC-DB/pDNA polyplexes at N/P 2.We noticed the same bell-shaped profile related to the effect of the N/P ratio on the transfection activity of PMPC-DB/pDNA polyplexes. In the U-138MG cell line, the highest transfection activity was observed at N/P 6 instead of at N/P 4 as observed in the U-87MG cell line (Fig. 3b). PEI/pDNA polyplexes showed a small, three-fold increase in the transfection compared to the best-forming PMPC/DB-pDNA N/P 6 polyplexes. Overall, PMPC-DB polyplexes showed the highest luciferase expression at N/P 4 in U-87MG cells, whereas N/P 6 was the better performing composition in U-138MG cells.
Antiproliferative Activity and DNA Dose Optimization of PMPC-DB Interferon-β1 DNA Polyplexes In Vitro
PMPC-DB/pDNA polyplexes increased the transfection efficacy of luciferase and GFP transgene (Fig. 9) in the glioma cell lines. We then wanted to determine if the PMPC-DB copolymer can also deliver a therapeutic plasmid, interferon-beta1 (pIFN-β1) to kill glioma cells in culture. A plasmid encoding the human IFN-β1 gene inserted into a pCMV6-XL4 vector (pIFN-β1) was custom-synthesized and its purity and size were verified by agarose gel electrophoresis (SL Fig. 2). The anticancer activity of pIFN-β1 delivered via PMPC-DB polyplexes was optimized and evaluated using three different assays.
Fig. 9.
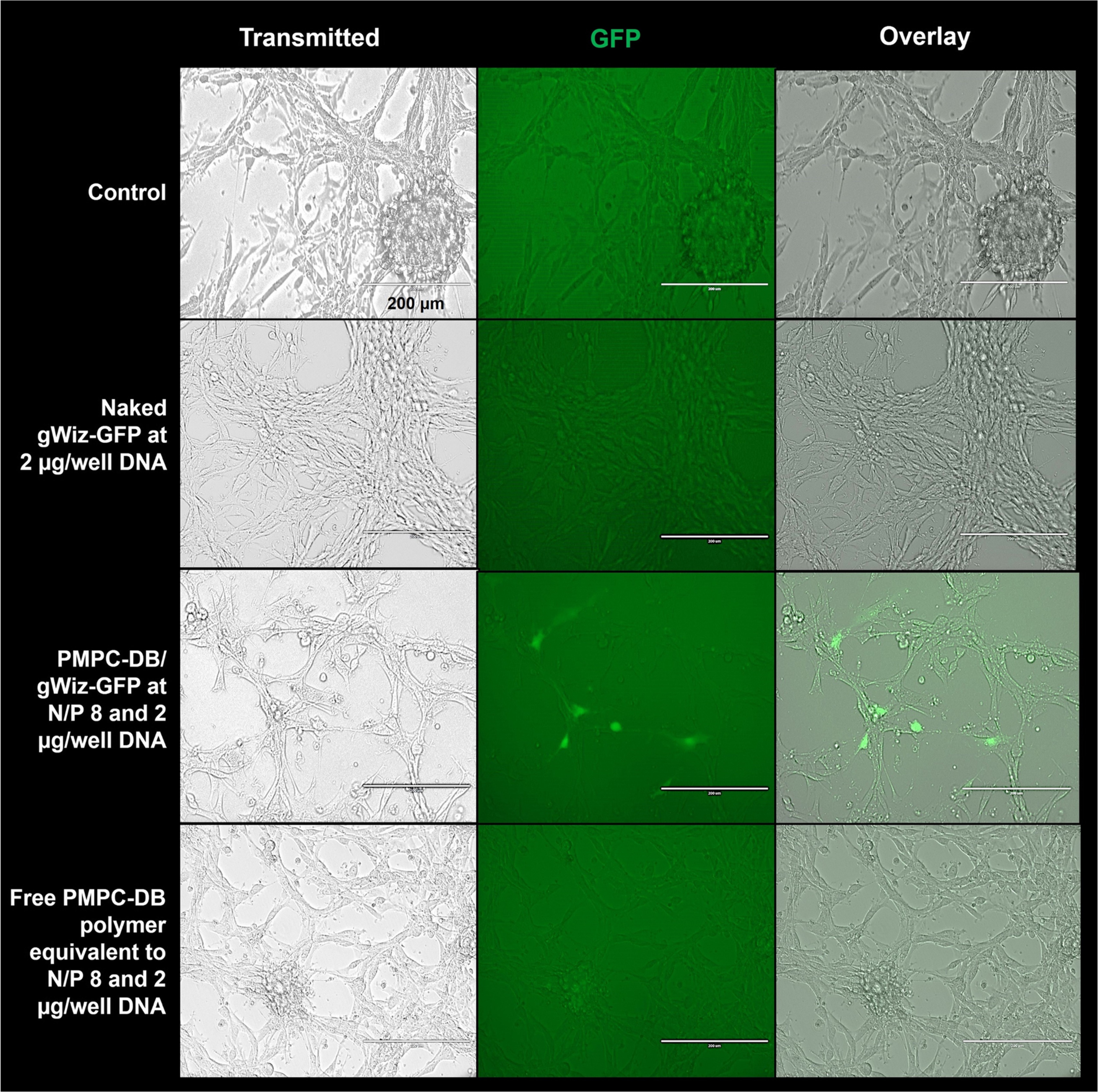
Gene expression efficiency of PMPC-DB/GFP pDNA polyplexes 48 h post-transfection in vitro. U-87MG cells were transfected with PMPC-DB/gWiz-GFP DNA at N/P 8 with 2 μg/well DNA for 4 h. Cell treated with naked gWiz-GFP DNA at 2 μg/well and an equivalent amount of free PMPC-DB copolymer were used as negative controls. Control, untreated cells were incubated with fresh MEM/FBS medium. GFP expression was observed under a fluorescence microscope 48 h post-transfection. The images were acquired at 20x magnification under the transmitted and GFP channel settings. The overlay images were produced using Image J software (NIH) at the same intensity settings for each image. Scale bar: 200 μm
First, the transfection efficiency of PMPC-DB/pIFN-β1 DNA polyplexes in U-87MG and U-138MG cells were determined by screening different N/P ratios at a constant pDNA concentration in the transfection mixture; 2.85 μg/ml. We transfected U-87MG cells using PMPC-DB/pDNA polyplexes prepared at the same range of N/P ratios as earlier; 2–10 and determined its efficacy to inhibit cell proliferation using the ATP assay (Fig. 4a). We used the following samples as negative controls to study the IFN-β1-specific cell killing activity: the best-performing PMPC-DB/Luc pDNA N/P 4 polyplexes, free PMPC-DB copolymer at N/P ratios equivalent to 4, 8, 10 and free luciferase pDNA. As described earlier, untreated cells were set to 100% viability, and the effects of IFN-β1 transfection were expressed as % cell viability relative to untreated cells. We observed a N/P ratio-dependent gradual decrease in viability in cells transfected with PMPC-DB/IFN-β1 DNA polyplexes (Fig. 4a). The average cell viability significantly (p < 0.0001) decreased from about 74% at N/P 2 to nearly 65% at N/P 10. Interestingly, we saw no effect of the N/P ratio on the extent of cell growth inhibition at N/P ratios >8 (Fig. 4a). We compared the activity of PMPC-DB/IFN-β1 polyplexes with PEI/IFN-β1 polyplexes at the same N/P ratios. PEI polyplexes at N/P 4 mediated a 65% reduction in cell viability that was similar to the effects noted in cells treated with PMPC-DB/IFN-β1 polyplexes at N/P 10. Notably, an equivalent quantity of free PEI polymer induced about a 15% reduction in cell viability and appeared to be toxic to the cells. Cells treated with equivalent amounts of free copolymer at N/P ratios 4, 8, and 10 showed an average cell viability of 113%, suggesting that the PMPC-DB may have a small nourishing effect via its choline groups, an essential nutrient [31]. Cells treated with PMPC-DB/Luc pDNA polyplexes showed a cell viability of 95% indicating that IFN-β1-mediated inhibition in cell proliferation is specific to the delivered gene and not due to carrier toxicity. Noteworthy, while the PEI polyplexes appeared to be equally efficient to the PMPC-DB/IFN-β1 polyplexes, the extent of cell killing to some degree was mediated by non-specific, carrier-mediated toxicity whereas the effects mediated by PMPC-DB/IFN-β1 polyplexes were specific to the delivered gene. Naked IFN-β1 DNA did not inhibit cell proliferation and in fact, the cell viability was >100%. This suggests that the anionic naked pDNA failed to enter cells in measurable amounts. Cells treated with free Luc pDNA also showed no inhibition in cell proliferation. Notwithstanding the effects we observed in the U-87MG cell line, our data did not show any significant reduction in U-138MG cell viability transfected for 4 h with PMPC-DB/IFN-β1 polyplexes across the entire range of the tested compositions from N/P 2–10 (Fig. 4b). We speculate that the lack of effects in U-138MG could be due to a cell line-dependent phenomenon on transfection activity [32, 33]. PEI/IFN-β1 polyplexes showed ca. 17% reduction in U-138MG cell viability, which was likely due to the toxic effects of the free PEI polymer that alone reduced the cell viability by ca. 19%. Due to a lack of activity in the U-138MG cell line, we focused all our subsequent studies in the U-87MG cell line.
Fig. 4.
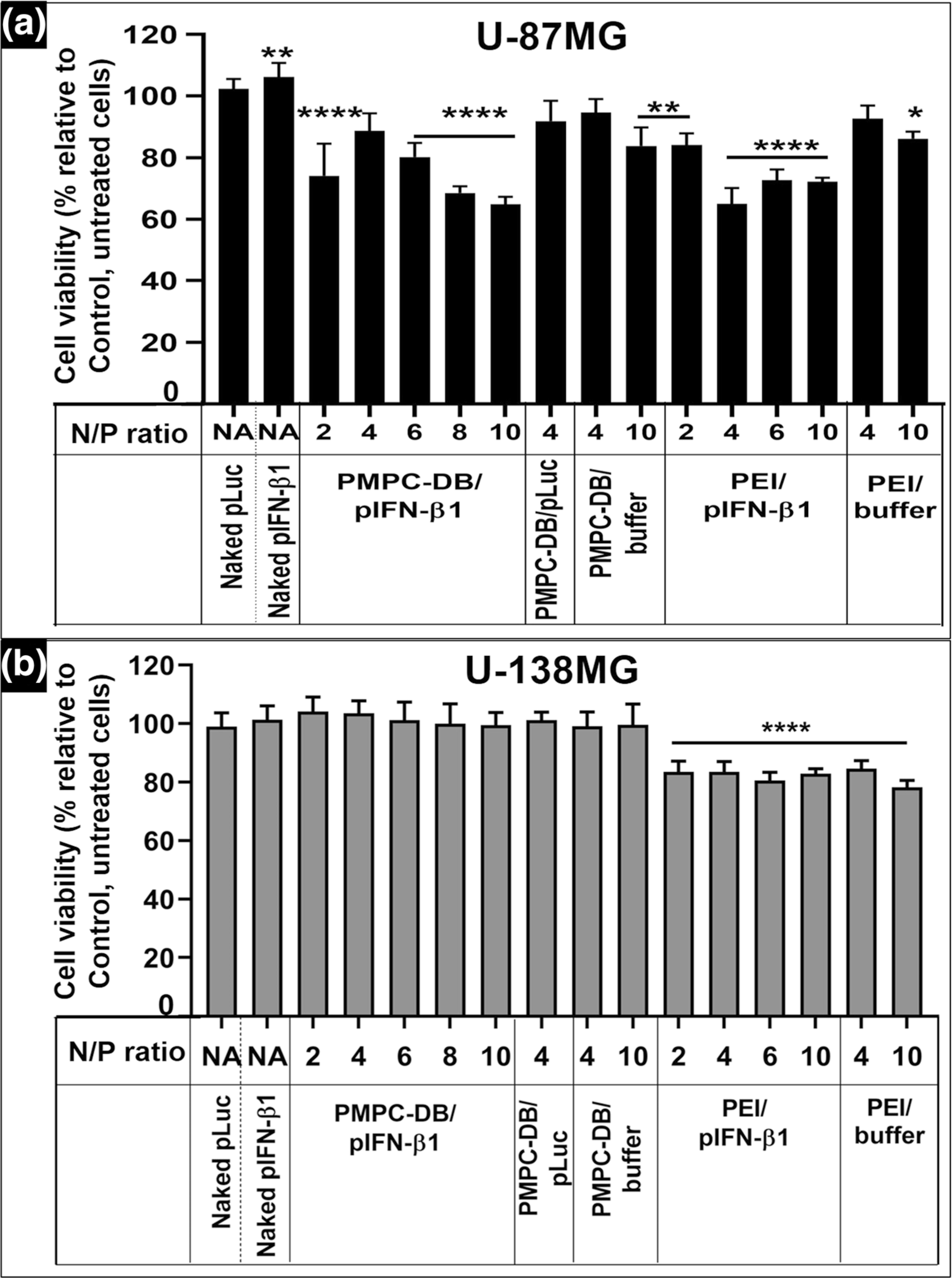
Anti-proliferative effects of IFN-β1 polyplexes on glioblastoma cells in vitro at varying N/P ratios and a constant DNA dose. (a) U-87MG (P132) cells or (b) U-138MG cells (P179) were seeded at 16,500 cells/well in 96-well plates. At about 80% confluency, cells were transfected for 4 h with the indicated treatment groups (naked DNA or DNA polyplexes) at 0.17 μg/well pDNA dose. Untreated cells were considered as a control. After 4 h transfection, the treatment mixture was replaced with fresh complete culture media, and the cells were allowed to regenerate for 48 h. As indicated, cells were also treated with equivalent amounts of the free polymer used to form polyplexes at the indicated N/P ratios. The cell viability was determined using the ATP assay. Statistical comparisons between the treated groups and the control were done using one-way ANOVA (Bonferroni’s Multiple Comparison test) and the significance levels are indicated as *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. NA: not applicable
Second, we optimized the polyplex N/P ratio and IFN-β1 pDNA dose to evaluate the individual and possible interactive effects of N/P ratio and DNA dose on the cell viability of U87MG cells. We tested two factors (N/P ratio and pDNA dose) and developed a two-three level (2 × 3) full factorial Design of Experiments (DOE) with two center points to evaluate the effect of factors on the response: percent cell viability. N/P ratio was studied at two levels, 8 and 12, and the DNA dose was studied at three levels: 1, 1.5, and 2 μg/well. The center point of the design was a combination of N/P 10 and a DNA dose; 1.5 μg/well. The center point was repeated (n = 2, hence two center points) for measuring the pure error by estimating the variance of the responses for the identical independent variables. The lack of fit or goodness of fit test, calculated using the pure error indicates whether the model fits the data well. Free PEI polymer at 40 and 60 μg/ml were used as positive controls. We used the following samples as negative controls to study the IFN-β1-specific cell killing activity: naked IFN-β1 at the highest tested dose of 2 μg/well, equivalent amounts of free PMPC-DB copolymer at N/P ratios 8, 10 and 12 for a DNA dose of 1.5 μg/well, and PEI polyplexes at N/P 12 with or without 1.5 μg IFN-β1 DNA (Fig. 5a). A stepwise statistical analysis was performed to evaluate the individual and interactive effects of independent variables; N/P ratios and DNA dose on the response, % cell viability. The p value at α= 0.05 from the lack of fit test suggests that the lack of fit is non-significant (p > 0.05) and the model fits the data well. The F-test ANOVA table in Fig. 5b demonstrated that the N/P ratio and the interaction term between N/P ratio and DNA dose are statistically significant with p-values of 0.007 and 0.03 at α = 0.05 respectively, whereas the DNA dose alone did not have a significant effect (p > 0.05) on cell viability. A significant interaction between the N/P ratio and DNA dose indicates that the effect of the polyplex N/P ratio on the cell viability is different at different values of the pDNA dose. The bar graph in Fig. 5a and the interaction profiles in Fig. 5b demonstrated that the cell viability was significantly (p < 0.0001, 48.2%) reduced in cells treated with N/P 8 polyplexes irrespective of the tested pDNA dose. Despite a significant (p < 0.0001) reduction in cell viability at N/P 10 and 1.5 μg/well pDNA dose, the cell viability was about 10% higher than the cells treated with N/P 8 polyplexes. Unexpectedly, the cell viability in N/P 12 polyplex-treated cells steeply increased from 56.9% at 1 μg/well to 85.8% at 2 μg/well DNA dose. Naked pIFN-β1 DNA at 2 μg/well dose showed ca. 20% decrease in cell viability, whereas PEI/IFN-β1 at N/P 12 with 1.5 μg DNA did not show any significant reduction in cell viability. U-87MG cells treated with an equivalent quantity of free PMPC-DB polymer (N/P 12; 1.5 μg pDNA) appeared healthy and showed no effects on cell viability.
Fig. 5.
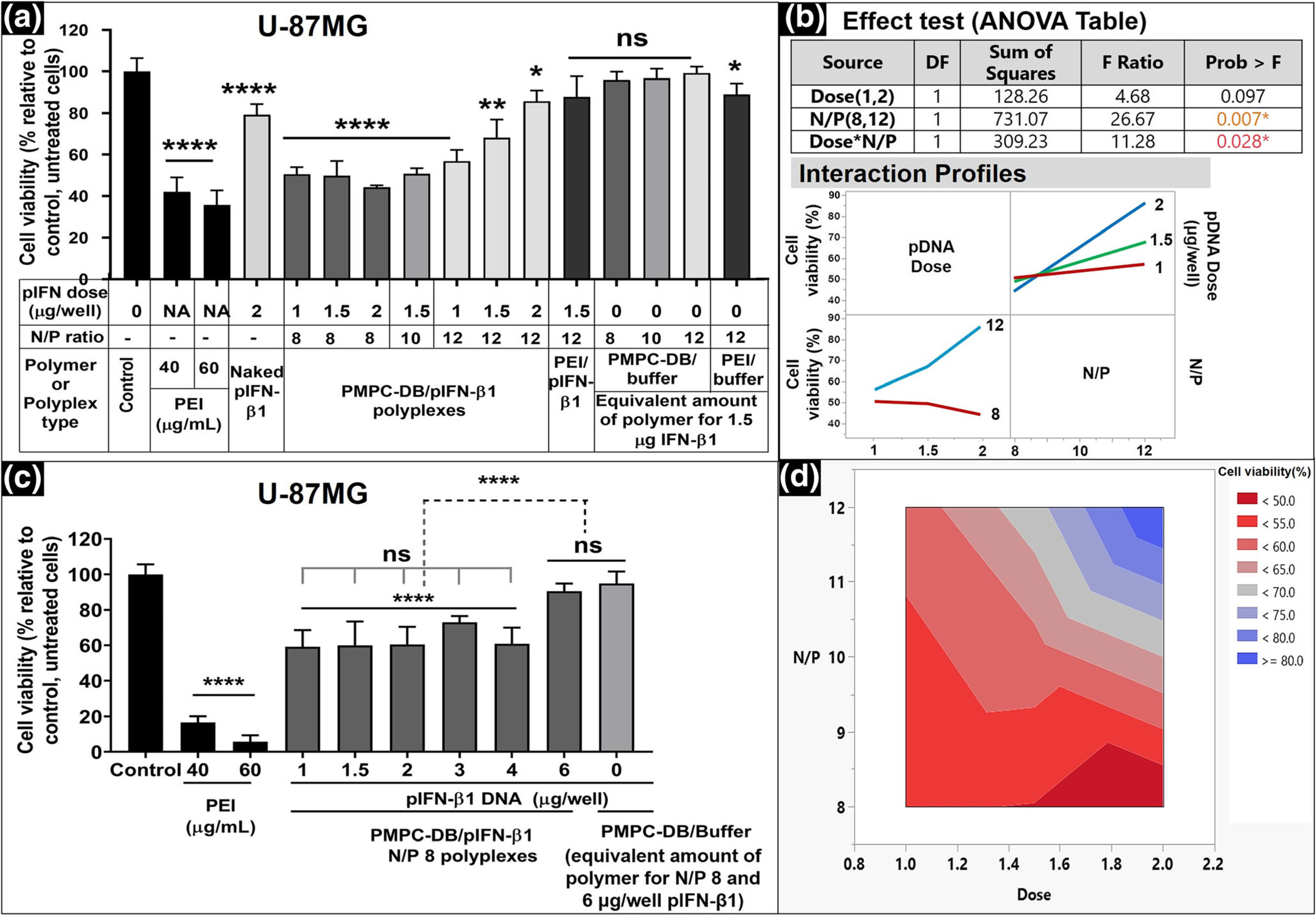
Effect of DNA dose and polyplex N/P ratio on the activity of IFN-β1 in U-87MG cells. (a) U-87MG cells were transfected for 4 h with the indicated samples. Naked DNA at 2 μg/well, PEI/IFN-β1 polyplexes at N/P 12, and an equivalent amount of free polymer at each N/P were used as negative controls. PEI at 40 and 60 μg/ml was used as positive controls. Untreated cells were used as the baseline control. The cell viability was measured using the ATP assay 48 h posttransfection, and % relative cell viability was calculated relative to the control. (b) The statistical analysis of the effect of DNA dose and N/P ratios on cell viability was studied using JMP Pro 14 software. The significance of individual factors and their interaction on cell viability was evaluated using the Effect test (ANOVA table). Interaction profiles showed the effect of polyplex N/P ratios on the cell viability at different pDNA doses. (c) The best -performing PMPC-DB/IFN-β1 polyplex composition, N/P 8, that yielded the lowest post-treatment cell survival was evaluated at different DNA doses ranging from 1 to 6 μg/well. The cell viability of U-87MG cells transfected for 4 h with DNA polyplexes was measured at 48 h post-transfection using the ATP assay. (d) The Contour plot for cell viability at different DNA doses and polyplex N/P ratios derived using the JMP Pro 14 software. The gradient map represents the predicted combined effect of the DNA doses and polyplex N/P ratios on % cell viability. Statistical analysis between specified groups in a and c was performed using GraphPad Prism 8.4.1. Asterisks represent statistically significant differences and the significance levels are indicated as *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. Data are presented as mean ± SD of n = 5 samples.
The contour plot in Fig. 5d showed that N/P 8 polyplexes at the dose higher than 1.5 μg pIFN-β1 showed a maximum <55% reduction in cell viability. Therefore, we tested the cell viability at higher pIFN-β1 doses using the best performing PMPC-DB-DNA N/P 8 polyplexes. There was a significant (p < 0.0001) reduction in cell viability across the tested range of DNA doses: 1–6 μg/well (Fig. 5c) compared to the control, untreated cells. Interestingly, at a 6 μg/well pIFN-β1 DNA dose, no reduction in cell viability was observed and in fact, the cell viability value was similar to control, untreated cells. A possible reason could be that there exists a maximum threshold of cell growth inhibition that is likely specific to the delivered transgene (pIFN-β1, in this case) and the cell line or a combination of both. Overall, our results suggest that the best-performing PMPC-DB/pIFN-β1 polyplexes inhibited glioma cell proliferation by nearly 40%.
PMPC-DB/IFN-β1 Polyplexes Induced Apoptosis of Glioblastoma Cells In Vitro
Numerous in vitro and in vivo studies have demonstrated that the chemotherapeutic effect of IFN-β1 is associated with its direct cytotoxic effect and/or induction of apoptosis in malignant cells [34–36]. We studied PMPC-DB/IFN-β1 polyplex-induced cellular apoptosis in U-87MG cells via Annexin V/7-AAD double staining and analyzed the cells using flow cytometry (Figs. 6, 7 and SL Fig. 3). 7-AAD is a dead cell marker and was used in the conjugation with the apoptosis marker Annexin V to determine if the polyplex-treated cells were viable(7-AAD−−/Ann V−−), early apoptotic (7-AAD−−/Ann V+), or were late apoptotic/necrotic cells (7-AAD+/Ann V+). U-87MG cells were transfected for 4 h with PMPC-DB/IFN-β1 polyplexes containing 1.5 μg/well DNA either at N/P 8, 10, or 12. Negative controls included naked IFN-β1 at a dose of 2 μg/well and free PMPC-DB copolymer at a quantity equivalent to N/P 12 polyplexes containing 1.5 μg pDNA. Cells were analyzed using flow cytometry 48 h post-transfection. Moghimi et al. demonstrated that branched PEI (25 kD) induces time and concentration-dependent membrane damage and apoptosis in three human cell lines including Jurkat T, umbilical vein endothelial, and THLE3 hepatocyte-like cells [37]. Therefore, we used PEI as a positive control in these studies.
Fig. 6.
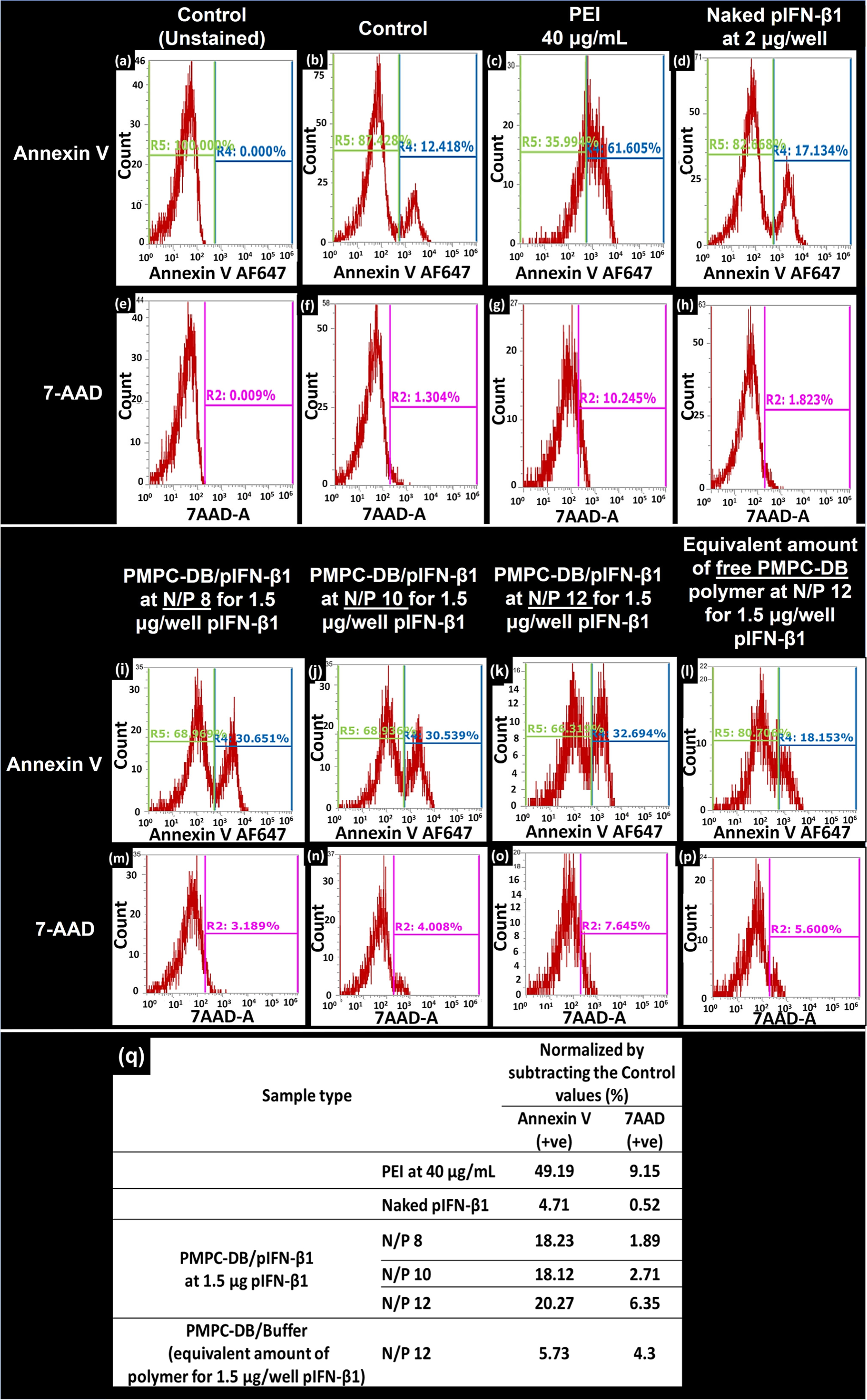
Flow cytometry analysis demonstrating the effect of PMPC-DB/IFN-β1 polyplexes on the induction of glioblastoma cell apoptosis. Cells were transfected for 4 h with using the indicated samples. PEI polymer at 40 μg/ml was used as a positive control. Naked IFN-β1 and an equivalent amount of the free PMPC-DB polymer at the indicated N/P ratios were used as negative controls. Post-transfection, cells were washed once with warm PBS and detached using a non-enzymatic cell dissociation buffer. Cells from each treatment group (n = 3) were pooled into the same tube and a single-cell suspension was stained using an annexin-binding buffer with Annexin v/7-AAD conjugates. Data are presented as histograms of cell count vs. fluorescence intensity for Annexin V-positive (+ve) and 7-AAD + ve cells. (a, e) Control, unstained sample was used to manually gate the Annexin V and 7-AAD + ve cells on histograms. The R4 region on histograms represents the %AnnexinV+ve cells, whereas the R2 gate represents the % 7-AAD + ve cells. (b-d, i-l) % of annexinV+ve cells in the indicated samples. (f-h, m-p) % 7-AAD positive cells for the groups described above. (q) Normalized % annexin V and 7-AAD + ve cells for each treatment group was calculated by subtracting the values corresponding to the stained, control untreated cells. The presented data are representative of two independent experiments.
Fig. 7.
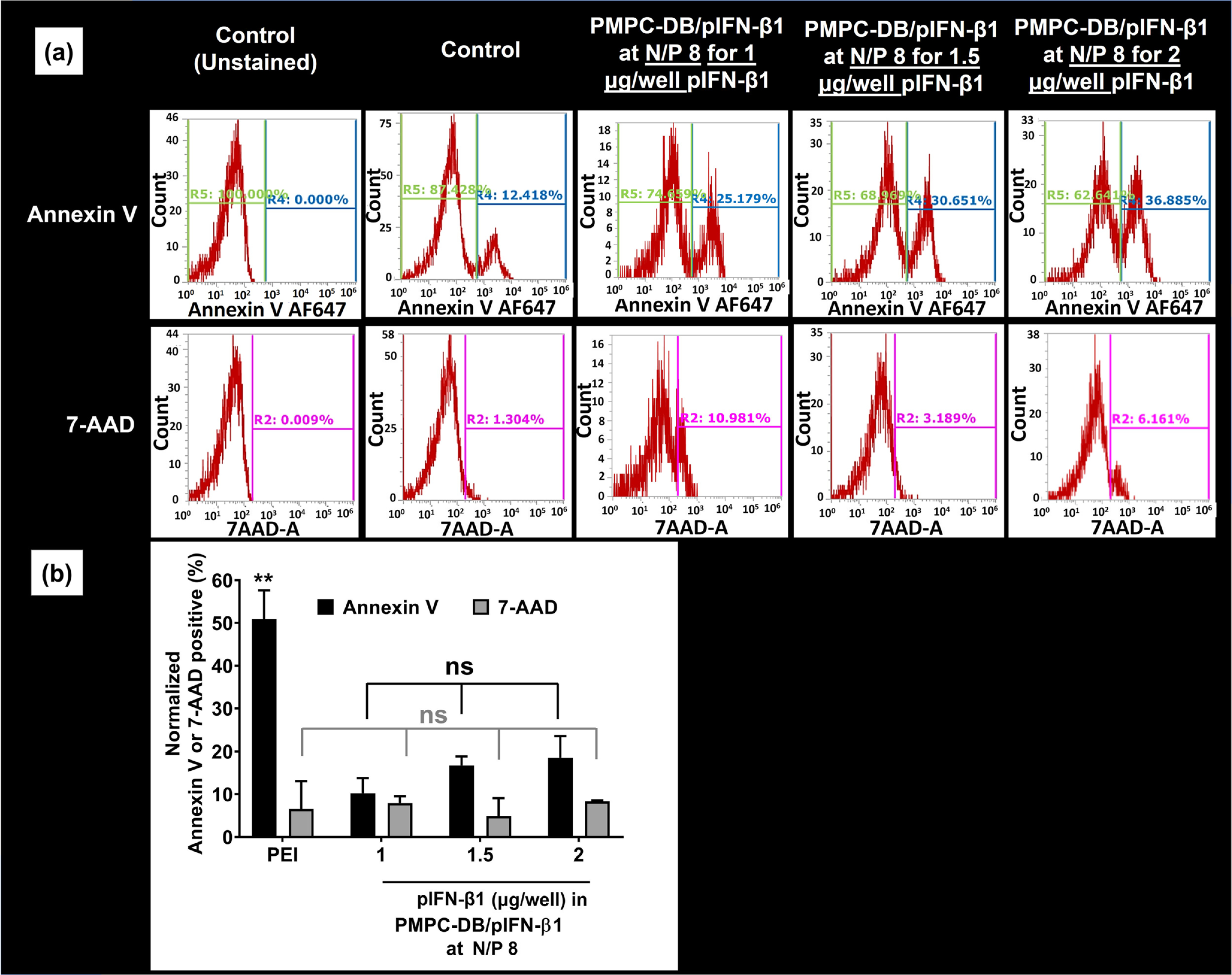
The effect of DNA dose on PMPC-DB/IFN-β1 polyplex transfection in glioblastoma cells. (a) IFN-β1 DNA at three different doses 1, 1.5, and 2 μg/well were transfected using PMPC-DB polyplexes at N/P 8 for 4 h, and the % annexin V and 7-AAD positive cells were analyzed using flow cytometry. A control, unstained sample was used to manually gate the Annexin V and 7-AAD positive cells on histograms. The R4 gate on histograms represents the % Annexin V + ve cells, whereas the R2 gate represents the % 7-AAD + ve cells. (b) Normalized % annexin V and 7-AAD + ve cells for each treatment group was obtained by subtracting the control annexin V and 7-AAD percent values. Data are presented as mean ± SD of n = 3 samples from independent experiments. Statistical analysis between specified groups was performed using GraphPad Prism 8.4.1. ** indicates a p value of <0.01 and ns indicates non-significant.
We optimized the measurement parameters such as voltage settings for forward scattering (FSC), side scattering (SSC), and the compensation matrix of Annexin V detected at 670/40 nm and 7-AAD detected at 574/26 nm double staining to eliminate light spillover to both detectors. For each sample, 50,000 events of U-87MG cells were recorded in the FSC/SSC dot plot (SL Fig. 3). We captured all events (R8 gate, SL Fig. 3A), and eliminated the dead cells or debris that showed reduced forward and side scattering in the FSC/SSC plot by a manual gating (R1 gate, SL Fig. 3A) process. SL Fig. 3A shows the representative gating strategy in control, unstained cells, and was consistently applied to all samples. The events captured in the R1 gate were analyzed further for determining the %viable, early, or late apoptotic cells.
Figure 6 demonstrates the histograms of Annexin V and 7-AAD positive cells in the control and the treated groups. A control, unstained sample was used for Annexin V (R4 gate) and 7-AAD (R2 gate) gating on histograms. As shown in Fig. 6a, the R4 gate for measuring Annexin V positive cells was manually created using signal intensity spanning from 103 to 106 arbitrary fluorescence units, whereas the R2 gate for 7-AAD-positive cells spanned from 2 × 102 to 106 arbitrary fluorescence units. Annexin V- or 7-AAD-positive events out of the total events are represented as a percent (%) Annexin V or 7-AAD, respectively. The double-stained control group showed about 12.5% Annexin V-positive cells (Fig. 6b) and 1.3% necrotic cells (Fig. 6f). Subsequently, Annexin V and 7-AAD-positive cells were measured in groups treated with PEI (Fig. 6c and g), naked IFN-β1 (Fig. 6d and h), PMPC-DB/IFN-β1 polyplexes at N/P 8, 9, and 10 each containing 1.5 μg DNA (Fig. 6i–k and m-o), and free PMPC-DB copolymer (Fig. 6l and p). The absolute percent of Annexin V-positive cells calculated by subtracting the values obtained from the double-stained control sample was demonstrated in Fig. 6q. The PMPC-DB/IFN-β1polyplexes showed ca. 20% Annexin V positive cells that were ca. 3.8-fold higher than cells treated with naked IFN-β1 DNA. No measurable differences were observed between the different N/P ratios. The density plots of Annexin V and 7-AAD double-staining in control U-87MG cells and cells treated with PMPC-DB polyplexes, naked DNA, and free polymers were demonstrated in SL Fig. 3B. The cells were differentiated as viable (7-AAD−ve/Ann V−ve), early apoptotic (7-AAD−ve/Ann V +ve), or late apoptotic and necrotic cells (7-AAD + ve/Ann V +ve). The percent of viable, early apoptotic, or late apoptotic and necrotic cells were calculated from the total cellular events captured in the R1 gate and were in agreement with the histogram data shown in Fig. 6.
We further investigated the effect of DNA dose on apoptosis using PMPC-DB/IFN-β1 polyplexes at N/P 8 (Fig. 7). The histograms of Annexin V and 7-AAD positive cells treated with 1, 1.5, and 2 μg/well IFN-β1 DNA was depicted in Fig. 7a. On an average, we noticed that 10.2%, 16.7%, and 18.6% of the cells were Annexin V-positive at DNA doses of 1, 1.5, and 2 μg/well, respectively (Fig. 7b). However, this gradual increase in % apoptotic cells at increasing DNA doses was not statistically significant. The overall trend was consistent with the ATP assay data that demonstrated that the maximum reduction in cell viability was observed at N/P 8 and 2 μg/well pDNA dose. To summarize, PMPC-DB/IFN-β1 polyplexes demonstrated a modest ca. 20% apoptosis that was 3.8-fold higher compared to cells treated with naked IFN-β1 DNA.
Anti-Proliferative Efficacy of PMPC-DB/IFN-β1 Polyplexes
It has been reported that endogenously secreted IFN-β from melanoma cells suppresses their proliferation in an autocrine manner [38]. The efficacy of the delivered IFN-β1 pDNA to retard the migration of U-87MG cells was evaluated using a simple and cost-effective wound closure assay [28]. An artificial gap (scratch or wound) created on a confluent U-87MG monolayer enabled the cells to migrate towards the gap and fill the space until complete closure of the wound. The comparison of images captured immediately after the scratch and those captured at regular intervals post-scratch enabled us to determine the effect of the treatment on the extent of cell migration and the efficiency of wound closure. In our study, a scratch was made in confluent U-87MG monolayers cultured in a 24-well plate, and cells were transfected with N/P 8 PMPC-DB/IFN-β1 polyplexes containing 2 μg DNA, naked IFN-β1 DNA, or an equivalent amount of free PMPC-DB polymer for 4 h. Control, untreated cells were incubated with fresh MEM/FBS medium. The images of the scratch within each group were acquired immediately after transfection (time 0 h), 24, and 48 h post-transfection (Fig. 8). The average distance between the two parallel lines of the scratch at 0 h was calibrated at 500 μm using the Image J software. As shown in Fig. 8a, the wound was partially filled with new cells at 24 h, and almost completely filled with new cells at 48 h post-transfection in the untreated cells, naked IFN-β1-, and free polymer-treated groups. Whereas, the wound remained unfilled (in other words, cells did not migrate to close the wound) in the cells treated with PMPC-DB/IFN-β1 polyplexes 24 h post-transfection. There were cell growth and migration showing wound closure at 48 h post-transfection. The wound closure via the migration of new cells was quantified for each treatment at the indicated time points (Fig. 8b). The control, untreated cells, and negative control groups showed ca. 70% and then nearly 90% wound closure at 24 and 48 h post-transfection, respectively. Whereas, cells transfected with PMPC-DB/IFN-β1 polyplexes showed only 0.29% and 46.5% wound closure at 24 and 48 h post-transfection respectively. The significant (p < 0.0001) inhibition in cell growth and migration in PMPC-DB/IFN-β1 polyplex-transfected cells further confirmed the anti-proliferative effects of the DNA polyplexes in glioblastoma cells. The cell migration inhibitory effects of PMPC-DB IFN-β1 polyplexes were reproducibly verified in an independent transfection experiment (SL Fig. 4).
Fig. 8.
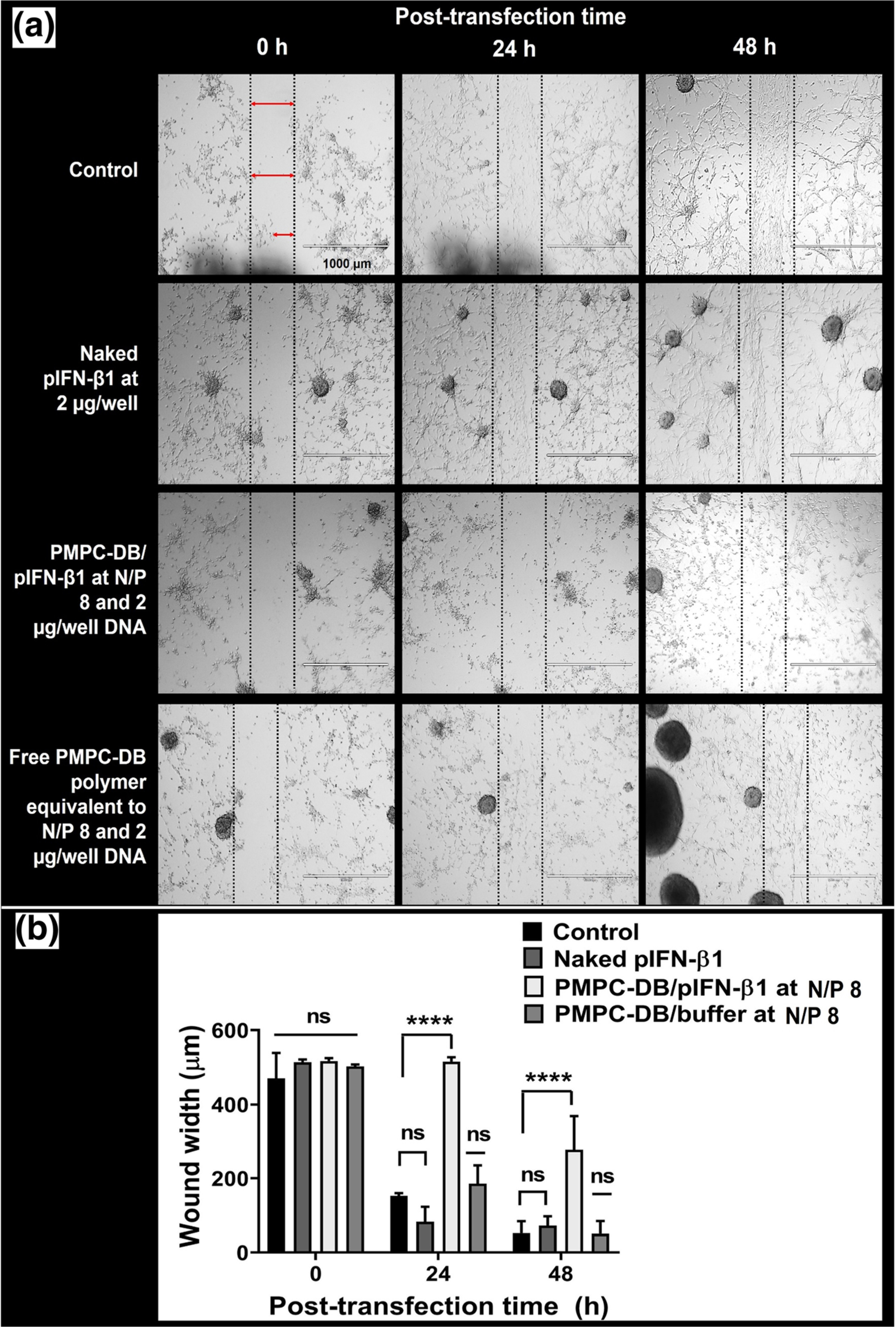
Anti-proliferative and cell migratory inhibition effect of PMPC-DB/IFN-β1 polyplexes in U-87MG cells analyzed using a scratch assay. (a) An artificial scratch was made on confluent U-87MG cells cultured in a 24-well plate. Cells were transfected with naked IFN-β1 DNA, PMPC-DB/IFN-β1 at N/P 8 polyplexes containing 2 μg DNA, and an equivalent amount of free PMPC-DB polymer for 4 h. Control, untreated cells were incubated with fresh MEM/FBS. The images of the scratch within each group were acquired at 0, 24, and 48 h post-transfection. As described earlier, the width of the unfilled boundary (wound width) was measured using the Image J software after calibrating the image scale bar. (b) The wound width was measured and plotted at 0, 24, and 48 h for control, naked IFN-β1, IFN-β1 polyplexes, and free PMPC-DB polymer. Data are presented as mean ± SD of n = 3 measurements at different locations for each wound. Statistical analysis between specified groups was performed using GraphPad Prism 8.4.1. The significance of treated groups was compared against control using one-way ANOVA (Bonferroni’s Multiple Comparison test) where **** indicates p < 0.0001. ns = non-significant. Scale bar: 1000 μm
Gene Expression Efficiency of Optimized PMPC-DB/GFP pDNA Polyplexes In Vitro
We visually confirmed the DNA transfection and subsequent gene expression of the optimized PMPC-DB/DNA polyplexes using gWiz-GFP pDNA. U-87MG cells were transfected with PMPC-DB/gWiz-GFP DNA polyplexes at N/P 8 containing 2 μg DNA, naked gWiz-GFP DNA, and an equivalent amount of free PMPC-DB polymer for 4 h. The images were acquired 48 h post-transfection under bright field and green fluorescence channels and were then processed as overlay images using a fluorescence microscope (Fig. 9). U-87MG cells transfected with PMPC-DB/GFP DNA polyplexes showed cytoplasmic green fluorescence indicating the transfection and expression of GFP 48 h (Fig. 9) post-transfection. Whereas, the lack of green fluorescence in U-87MG cells treated with naked gWiz-GFP DNA 48 h post-transfection suggested inefficient transfection of the naked DNA confirming our previous observations from the luciferase and IFN-β1 transfections. As expected, untreated cells and cells incubated with the free polymer did not express GFP at 48 h post-transfection (Fig. 9). The images acquired 24 h post-transfection did not show any green fluorescence in polyplex-treated or control, untreated cells suggesting that the delivered GFP transgene requires at least 48 h for protein expression (SL Fig. 5). Overall, N/P 8 PMPC-DB/GFP DNA polyplexes containing 2 μg DNA transfected cells and subsequently expressed the encoded GFP protein.
Time-Dependent Colloidal Stability in Low Ionic Strength 10 mM Phosphate Buffer, pH 7.4
We determined the colloidal stability of PMPC-DB/pDNA and PEI/pDNA polyplexes over a period of 7 days in a low ionic strength, 10 mM phosphate buffer using DLS (Fig. 10 and SL Fig. 6). PMPC-DB/pDNA polyplexes at N/P 4 and 8 showed average particle diameters ranging from 78 to 111 nm with a narrow PdI of <0.25 throughout the 7 d storage at RT and 4°C (Fig. 10a–d). However, the average particle diameter of PMPC/pDNA polyplexes at N/P 10 gradually increased from 211 nm initially to above 500 nm with broader PdI >0.3 upon 4 and 7 d storage at RT and 4°C respectively. PEI/pDNA polyplexes at N/P 2 showed a larger particle diameter of >300 nm with a broader PdI of >0.5 during storage at RT and 4°C (Fig. 10a and c). PEI/pDNA at N/P 10 showed Z-average below 150 nm with a narrow PdI of <0.17 during the storage period. The particle size distribution of PMPC-DB/pDNA polyplexes at N/P 4, 8, and PEI/pDNA at N/P 10 showed a unimodal distribution throughout the storage period, whereas PMPC-DB/pDNA at N/P 10 and PEI/pDNA at N/P 2 showed bi−/ multimodal distribution (SL Fig. 6).
Fig. 10.
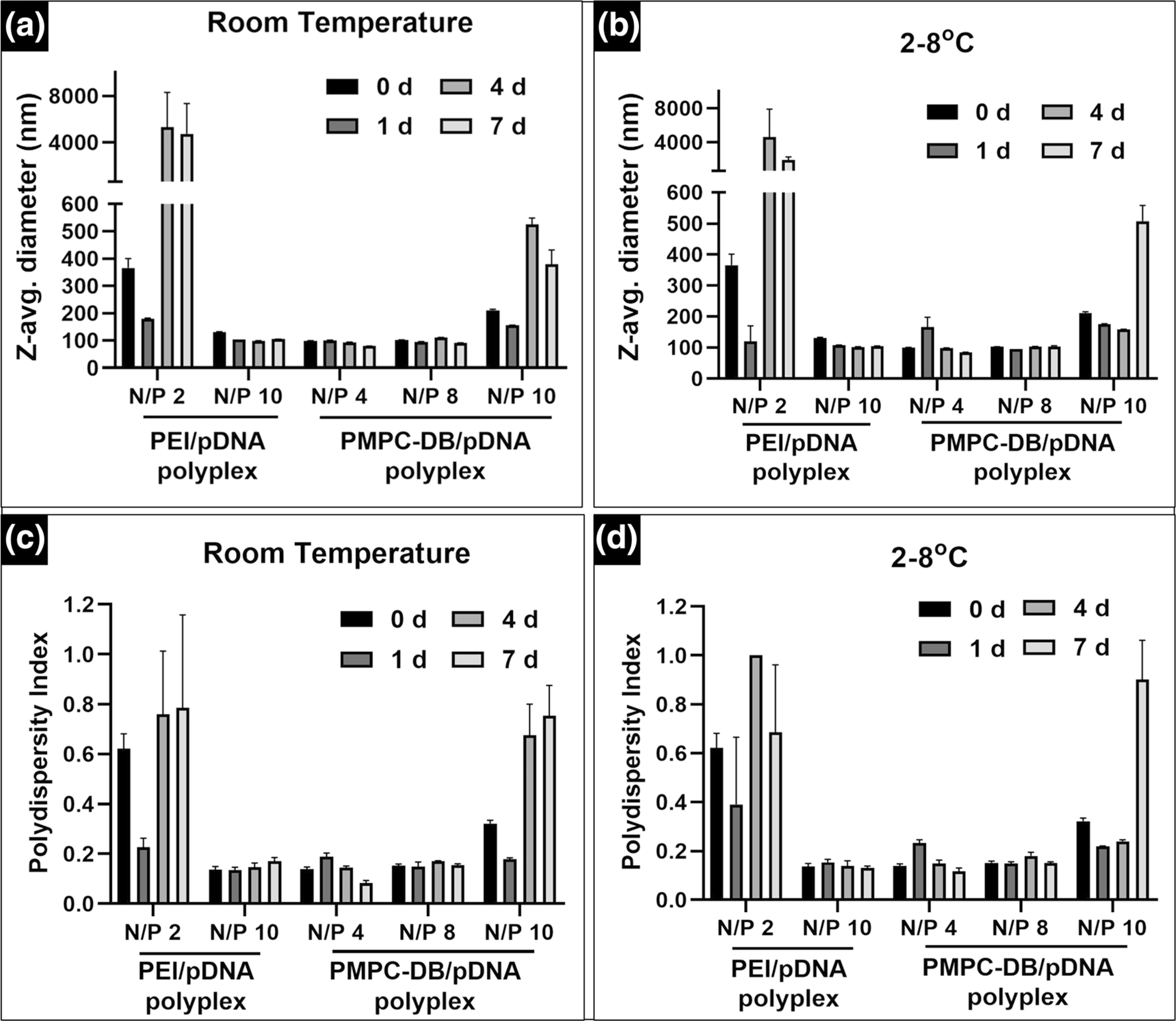
Time-dependent colloidal stability of PMPC-DB/pDNA or PEI polyplexes measured using dynamic light scattering. PMPC-DB/pDNA polyplexes containing 20 μg/ml pDNA were initially prepared in 10 mM citrate buffer and further diluted 5x using 10 mM phosphate buffer pH 8.0, whereas PEI/pDNA polyplexes were prepared in 10 mM HEPES buffer pH 7.4. The samples were stored at room temperature (a and c) and 2–8°C (b and d). Z-average particle diameters (a and b) and polydispersity indices (c and d) were measured either immediately after preparation or 2, 4, and 7 days post-preparation on a Malvern ZS90 Zetasizer. Data are presented as mean ± SD of n = 3 measurements.
PMPC-DB/pDNA at N/P 4, 8, and 10 showed a complete DNA condensation during 7 days of storage at RT and 4°C characterized by a strong fluorescence signal near the loading wells (Fig. 11). PEI/pDNA at N/P 2 showed DNA condensation with less intense fluorescence band whereas PEI/pDNA at N/P 5 and 10 showed an absence of DNA bands suggesting a complete condensation of DNA at these N/P ratios. DNA remained condensed by PEI during the 7 d storage at RT and 4°C. Naked pDNA migrated towards the anode upon electrophoresis, however, the intense bands observed at all time points suggested that pDNA did not degrade for 7 d when stored at either RT or 4°C. The discussion of the stability studies in 10 mM phosphate buffer containing 0.15 M NaCl, cell culture medium supplemented with serum at concentrations ranging from 0 to 50% and a heparin competition assay were presented in SL Figs. 7 SL 8, and SL 9, respectively in the supplementary file.
Fig. 11.
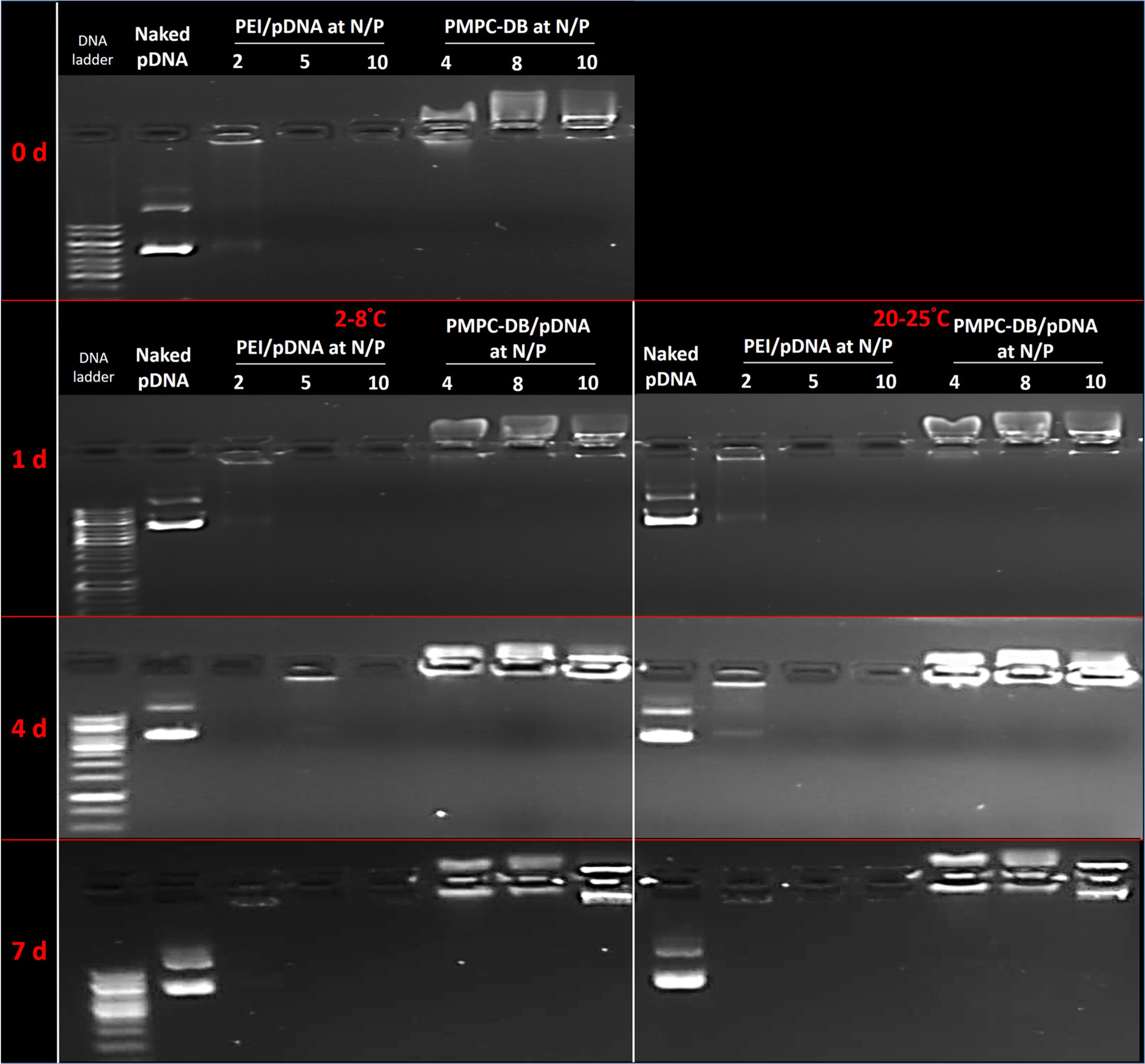
DNA retention by polyplexes at different N/P ratios stored at room temperature and 2–8°C for 7 days studied using agarose gel electrophoresis. PMPC-DB/pDNA polyplexes containing 20 μg/ml pDNA at N/P 4, 8, and 10 were prepared in 10 mM citrate buffer and further diluted 5x using 10 mM phosphate buffer pH 8.0, whereas PEI/pDNA polyplexes were prepared in 10 mM HEPES buffer pH 7.4. Naked pDNA was diluted in 10 mM citrate buffer followed by 5x dilution with 10 mM phosphate buffer. The samples were stored at room temperature and in a 2–8°C fridge. Polyplexes containing 0.4 μg pDNA were electrophoresed immediately after preparation or 1, 4, and 7 days post-preparation on a 0.8% w/v agarose gel containing 0.5 μg/mL ethidium bromide.
DISCUSSION
In the present study, we utilized PMPC-DB copolymer to form DNA polyplexes that contain a core consisting of charge neutralized DB polymer-DNA chains and a zwitterionic PMPC shell. The positively charged tertiary amines from the choline group and negatively charged phosphate in 2-Methacryloxyethyl phosphorylcholine (MPC)-based copolymers result in a zwitterionic moiety that confers advantageous characteristics including hydrophilicity, antifouling surfaces, reduced protein adsorption and improved physiological and colloidal stability of the MPC-containing polyplexes [5, 6]. Lam et al. reported that optimized dimethylaminoethyl methacrylate (DMAEMA)-MPC copolymers demonstrated stable DNA condensation and showed very little non-specific cellular association [6]. Under an acidic pH (~4.0–4.5), the tertiary amine of DMAEMA moiety is nearly 100% protonated [39] and shows strong electrostatic interactions with the negatively charged phosphate group of pDNA whereas, at a physiological pH (pH 7.2–7.4), DMAEMA has about 50% protonated amines [39]. Therefore, the pH-dependent changes in charge density can reduce the non-specific association of the DNA polyplexes with the extracellular serum components at the physiological pH while the increased degree of protonation at the acidic, endosomal pH plays a key role in the release of polyplexes from endosomes resulting in higher transfection efficiency. It should be noted that the colloidal stability at the physiological pH is also translated to improved blood serum stability in vivo [5, 8, 9].
In an earlier study, our colleagues demonstrated improved pharmacokinetics and biodistribution of PMPC-DB/siRNA polyplexes resulting in efficient target gene silencing in vivo [5]. In the present study, we hypothesized that PMPC-DB/DNA polyplexes can transfect plasmid DNA into glioblastoma cell lines and facilitate the subsequent protein expression. The objectives of this study were to [1] study the physicochemical characteristics of PMPC-DB/DNA polyplexes, [2] determine their cytocompatibility with glioblastoma cell lines, [3] screen polyplexes at various N/P ratios to determine the optimal composition that yields the highest transfection efficiency, and [4] use optimized polyplex compositions to determine the transfection efficiency of a therapeutic gene, IFN-β1.
The EtBr and agarose condensation assay data confirmed the formation of polyplexes between the cationic DMAEMA moiety and anionic DNA (Fig. 1a). The excess polycation in polyplexes prepared at N/P > 1 surrounds the stoichiometric polyplexes allowing a combination of the hydrophobic BMA core-mediated internal stabilization and PMPC corona-based steric stabilization [8, 40]. The average particle diameters ranged from 100 to 140 nm at N/P ratios 1 to 8 (Table I) supporting the stabilization effects noted above. It has been reported that polyplexes with a diameter of ~150 nm are most favorable for the classical nuclear uptake pathway in glioma cells [41] indicating that the formed polyplexes are suitable for nuclear entry. The net surface charge of the polyplexes correlated with the N/P ratio of PMPC-DB and DNA. A gradual swift was noted from the negative surface charge of free pDNA (N/P 0) to a near-neutral charge at N/P 1 that remained consistent up to N/P 10. The electroneutral surface charge of polyplexes due to the presence of the PMPC corona is a critical feature that prevents colloidal aggregation, facilitates in vitro stability, blocks serum protein adsorption, and enables longer circulation which all lead to better tumor delivery [5]. This electroneutral surface charge is also an essential characteristic for choline transporter-mediated enhanced glioblastoma cellular uptake [17, 18, 42].
Cytocompatibility and low off-target toxicity are important aspects of the novel polymeric materials that otherwise limit their use for gene delivery. We chose malignant glioma cell lines, U-87MG and U-138 MG, for the biological studies since these cells express both the IFN-β receptors, IFNAR1 and IFNAR2, that dimerize for its functional activity [29]. We determined the effects of acute exposure of free PMPC-DB copolymer and their polyplexes using a reporter luciferase gene in both cell lines (Fig. 2). We compared the cytocompatibility results with branched PEI, the gold standard synthetic polymer that has been well studied for gene transfection in a variety of cell culture and in vivo models [43]. Boeckle et al. demonstrated that excess free PEI required to increase the transfection efficiency both in vitro and in vivo caused extreme cellular metabolic toxicity resulting in cell viabilities as low as 10% [44]. Janjira et al. compared the physicochemical characteristics, in vitro, and in vivo transfection efficiency of linear and branched PEI/DNA complexes [45]. The authors reported that branched PEI/DNA complexes showed a higher zeta potential and smaller particle size compared to linear PEI due to stronger electrostatic interactions resulting in greater compaction of the branched PEI/DNA complexes. Further, branched PEI/DNA complexes prepared at N/P ratios ≥10 resulted in significantly higher luciferase gene expression compared to linear PEI polyplexes in HeLa, HEK293, COS7, and HepG2 cell lines [45]. Seib et al. compared the endocytic properties of linear and branched PEI polymer, and demonstrated a five-fold increase in extracellular binding of branched PEI compared to the linear counterpart [46]. Branched PEI polymer is endocytosed via cholesterol-dependent pathways increasing the likelihood of uptake by cellular membranes, whereas endocytosis of linear PEI is independent of clathrin and cholesterol pathways [46]. In contrast to the observed lower in vitro transfection efficiencies, polyplexes of linear PEI showed a greater in vivo transfection efficiency compared to branched PEI polyplexes [45]. The strong packaging between branched PEI and DNA results in less efficient disassembly of the complexes that retards the release of DNA and eventually reduces transfection efficiency [45] whereas linear PEI/DNA complexes dissociate efficiently in the presence of physiological biomolecules such as heparin [45]. Collectively, branched PEI is a highly efficient non-viral vector for in vitro DNA transfection due to higher polyplex zeta potential, smaller particle size, and enhanced cellular uptake and endosomal release whilst linear PEI demonstrated efficient gene transfection in vivo. In the present studies, we evaluated the transfection efficiency of PMPC-DB polyplexes in vitro in glioblastoma cell lines, and therefore, we selected branched PEI/DNA polyplexes as a positive control. Our results demonstrated that PMPC-DB copolymer at concentrations below 60 μg/ml (Fig. 2b) and its DNA polyplexes are well-tolerated by both cell lines (Fig. 2c–d), confirming previous observations [5]. Therefore, PMPC-DB polyplexes at N/P≤ 10 are cytocompatible and did not show carrier-mediated non-specific toxicity to cancer cells.
We transfected luciferase transgene into the glioma cell lines to determine the transfection activity of PMPC-DB/pDNA polyplexes (Fig. 3a–b). The simple readout and its dynamic range of measurement allowed us to screen various polyplex compositions to determine optimal N/P ratios for achieving the highest transgene expression. Transfection in both U-87MG (Fig. 3a) and U-138MG (Fig. 3b) cell lines revealed a bell-shaped curve for polyplex gene expression at N/P ratios 2–10. A decrease in gene expression at higher N/P ratios is expected if the polycation is toxic at higher concentrations. Noteworthy, in our experiments, cells transfected with polyplexes at N/P ratios 2–10 appeared healthy 24 h post-transfection and did not show significant differences in total cellular protein content relative to untreated cells. Thus, we conclude that the observed decrease in N/P 6–10 polyplex gene expression (in the so-called “curving down portion” of the bell-shaped profile) is not due to transfection-related toxicity.
Once we confirmed efficient luciferase gene transfection in glioblastoma cell lines, we then evaluated the transfection efficiency of PMPC-DB polyplexes to deliver a therapeutic gene, pDNA encoding interferon-β (IFN-β1). We evaluated the activity of the delivered pIFN-β1 on the cell viability, induction of apoptosis, and migration inhibitory effects in U-87MG cells using the ATP assay (Figs. 4–5), flow cytometry analysis of cells stained with Annexin V/7-AAD (Figs. 6–7), and a scratch assay (Fig. 8) respectively. We noticed a significant effect of the polyplex N/P ratio on the transfection activity of the IFN-β1 gene (Fig. 4) as we did in the case of the luciferase transfections. The cell viability gradually decreased with increasing polyplex N/P ratios up to N/P 8 and remained constant till N/P 12 at a 1 μg/well pDNA dose (Figs. 4a and 5a). Optimization of polyplex compositions showed that PMPC-DB/IFN-β1 polyplexes at N/P 8.0 containing 2 μg DNA/well inhibited glioma cell proliferation by nearly 40% (Figs. 4–5). Interestingly, we observed a significant interaction between the N/P ratio and the pDNA dose on the cell viability (Fig. 5b). There is a minimal effect of pDNA dose on the cell viability at the lower polyplex N/P ratio 8, whereas, at a higher N/P ratio of 12, the cell viability significantly increased with increasing pDNA doses. Our results suggest that polyplex compositions at a lower polyplex N/P ratio containing higher pDNA dose result in the maximum efficacy. We used negative controls including reporter DNA polyplexes, naked IFN-β1, and an equivalent free PMPC-DB copolymer at the indicated N/P ratios to confirm that the observed cell growth inhibition effects are specific to the delivered IFN-β1 gene and not the carrier itself. Importantly, the best PMPC-DB/IFN-β1 polyplexes appeared to be just as efficient as the gold standard PEI polyplexes but did not show non-specific toxicity as PEI did. Their improved safety profile is a significant advantage encouraging further exploration of these systems.
Upon incubation with IFN-β protein, tumor cells show apoptotic features such as cell shrinkage, membrane blebbing, chromatin condensation, and DNA fragmentation. Dedoni et al. demonstrated that the IFN-β treatment-mediated suppression of the activation of the PI3K/AKT survival signaling pathway led to apoptosis in neuroblastoma cells [35, 47]. Zhang et al. reported that the apoptosis of IFN-β treated U-87MG cells occurred due to reduced activity of the NF-κB survival pathway [35, 48]. The loss of plasma membrane symmetry and the resulting exposure of phosphatidylserine (PS) residue at the outer plasma membrane is known to occur early during cell apoptosis and was measured using an Annexin V binding assay since Annexin V has a strong and specific binding affinity to PS [49]. Our data showed that at 48 h post-transfection, cells transfected with PMPC-DB/IFN-β1 polyplexes showed a modest fraction of 20% Annexin V-positive cells that was 3.8-fold higher than naked IFN-β1 DNA transfected cells (Figs. 6–7, and SL Fig. 2). The tested range of N/P ratios and DNA doses did not show any considerable difference in the extent of early apoptosis as measured using Annexin V staining.
The anti-proliferative or cell migration efficacy of IFN-β1 polyplex-treated U-87MG cells was evaluated using a scratch assay. It allows visual confirmation of the cell migration effect and also to some extent mimics the migration of cells in vivo [28]. Cell migration and invasion are critical processes in cancer initiation, progression, and subsequent metastasis when tumor cells migrate from the primary location to distant organs [50]. Hence, it is important to study the ability of anti-cancer drugs to inhibit cell migration. Therefore, the lack of cell migration and subsequent wound closure is an expected therapeutic outcome for IFN-β1 exposure in the scratch assay. IFN-β1 polyplexes reproducibly demonstrated a statistically significant (p < 0.0001) lack of wound closure compared to control, naked IFN-β1, and free polymer-treated cells 48 h post-transfection (Fig. 8 and SL Fig. 3). Only 45% of the wound closed in IFN-β1 polyplex-treated cells compared to cells treated with naked IFN-β1 DNA (~86%) confirming its anti-proliferative and inhibitory effects on cell migration. The transfection efficiency of PMPC-DB/DNA polyplexes was microscopically confirmed using gWiz-GFP DNA. Only IFN-β1 polyplex-treated U-87MG cells showed intracellular GFP expression at 48 h post-transfection while naked GFP DNA-treated cells did not (Fig. 9).
PMPC-DB/pDNA at N/P 4 and 8 showed consistent particle size distribution of ca. 100 nm with a narrow PdI of ca. 0.15 throughout the 7 day storage period at RT and 4°C. PMPC-DB/pDNA polyplexes at N/P 4 and 8 showed a unimodal size distribution and completely retained the DNA payload during the storage time. Our results align with the previously reported results using PMPC-DB/siRNA polyplexes where the 20k PMPC corona-containing siRNA polyplexes appeared most resistant to higher salt concentration-mediated destabilization [5]. Our data further supports the previous findings that PMPC coronas improve polyplex stability in higher ionic strength conditions. The likely reason for the stability of PMPC-DB polyplexes in high salt conditions is due to the steric stabilization effects of the zwitterionic PMPC corona whereas PEI polyplexes are stabilized by electrostatic forces alone, and therefore they aggregate more rapidly under high salt conditions [8]. PMPC-DB/pDNA polyplexes showed considerable DNA complexation for 4 h when incubated in a serum-free, cell culture medium, and 50% serum-containing medium—a condition that somewhat simulates an in vivo environment [51]. Our data is in agreement with the previously reported serum stability observations by our coworkers, Jackson et al. who demonstrated that PMPC-DB/siRNA polyplexes were stable in 10% and 30% FBS for 100 min [5]. Naked pDNA rapidly degraded in 10% FBS/DMEM in less than 4 h, also in agreement with previous reports [51, 52]. At the same N/P ratios, PMPC-DB polyplexes showed greater resistance to charge-induced destabilization compared to PEI polyplexes confirmed by heparin competition assay. Importantly, we observed a correlation between the physicochemical parameters in the stability studies with the transfection efficiency of PMPC-DB/pDNA polyplexes in cell culture models. It should be noted that the PMPC-DB/pDNA polyplexes at N/P 4–8 that demonstrated consistent stability in low ionic strength-, high salt concentration- and in high serum- conditions, also showed the maximum luciferase expression in U-87 MG and U-138 MG cells as well as anti-proliferative, apoptotic and migration inhibitory effects in case of the PMPC-DB/pIFN-β1 DNA polyplexes.
Our previous studies demonstrated that the PMPC surface chemistry makes this carrier more amenable to intravenous systemic delivery and showed high uptake by tumor cells at least in a breast cancer model [5]. The neutral zeta potential is especially important for longer circulation time which is typically associated with better tumor delivery compared to conventional PEI-based high zeta potential carriers. Our data on PMPC-DB/pDNA polyplexes encourage further optimization and exploration of these systems for in vivo gene delivery. In our future work, we plan to study the in vivo pharmacokinetics of the polyplexes after intravenous administration in experimental mice. We will determine the following pharmacokinetic parameters: area under the curve, Cmax, Tmax, mean residence time, half-life, and clearance. For the biodistribution studies, animals will be treated with polyplexes and the organs will be harvested at selected time points post-injection [5, 53]. Plasmid DNA in blood and homogenized tissue samples will be measured using RT-PCR using a previously reported method [53]. In our future work addressing the mechanism of action, we will evaluate the anti-angiogenic effects of IFN-β that is known to function as a potent inhibitor of angiogenesis in glioma cells [20, 54, 55]. Takano et al. investigated the anti-angiogenic effect of IFN-β in glioblastoma cells in vitro and in vivo and demonstrated that IFN-β inhibited VEGF protein production and induced interferon-inducible protein 10 (IP10) that collectively resulted in inhibition of human umbilical vein endothelial cells (HUVEC) migration-mediated angiogenesis [20]. Therefore, we will evaluate the anti-angiogenic effect of IFN-β1 in PMPC-DB/pIFN-β1 polyplex-transfected U-87 MG cells. Further, IFN-β1 mediated up-regulation of IP10 and reduction in VEGF mRNA and protein levels in U-87 MG cells will be evaluated using RT-PCR and western blotting. We further plan to study the anti-angiogenic effect of PMPC-DB/IFN-β1DNA polyplexes in U-87 MG, HUVEC, and human fibrosarcoma U3A cell lines as the HUVEC and U3A cells express INFR1, VEGF, and IP10 proteins [20, 54]. We will evaluate the modulation in IFN-β1, VEGF, and IP10 mRNA and protein expression levels using quantitative RT-PCR and western blot analysis in the PMPC-DB/IFN-β1-transfected U-87 MG, HUVEC, and U3A cells. In the tumor microenvironment, the absence of endogenous IFN-β increases the synthesis and release of VEGF proteins resulting in HUVEC migration-mediated angiogenesis, and as a result, an increase in IFN-β1-dependent IP10 expression leads to the downregulation of VEGF and HUVEC cell migration. Therefore, we will evaluate cell migration and the anti-proliferative effects of PMPC-DB/IFN-β1 polyplexes in HUVEC cells using the wound closure assay and cytotoxicity study (ATP assay) that we have used in the current study.
CONCLUSIONS
We developed and characterized PMPC-DB polyplexes for the delivery of plasmid DNA to glioblastoma cells. We demonstrated the transfection efficiency of PMPC-DB polyplexes using three different plasmid DNA constructs including gWiz-Luc, gWiz-GFP, and pCMV6-XL4-IFN-β1. PMPC-DB/Luc-DNA polyplexes were safe and showed an 18-fold increase in luciferase transgene expression compared to the gold-standard PEI polyplexes in U-87MG cells. Complementary to the results observed with the luciferase reporter gene, PMPC-DB/gWiz-GFP DNA polyplexes showed GFP expression 48 and 72 h post-transfection in U-87MG cells. The DOE screen revealed that polyplex compositions at a lower polyplex N/P ratio containing higher pDNA doses result in the maximum efficacy of the delivered IFN-β gene. We demonstrated anti-proliferative, apoptosis-inducing, and inhibitory effects on cell migration in IFN-β1 DNA polyplex -transfected glioma cells. The results described herein should serve to guide the future optimization of PMPC-DB/DNA delivery systems for glioblastoma therapy.
Supplementary Material
ACKNOWLEDGMENTS AND DISCLOSURES
The study was supported using start-up funds for the Manickam laboratory from the School of Pharmacy and a Faculty Development Fund award to the PI from Duquesne University (DU). The authors thank Mr. Suyang Wu (DU) for providing Annexin V AF647 and 7-AAD dyes for the flow cytometry studies. The authors express their deep appreciation to Ms. Manisha Chandwani, Ms. Yashika Kamte, and Dr. Lauren O’Donnell (DU) for flow cytometry support. The authors are thankful to Ms. Akshita Bhatt and Dr. Jane Cavanaugh (DU) for fluorescence microscopy support. The authors are thankful to Drs. Joel Gillespie (University of Pittsburgh) and Jelena Janjic (DU) for allowing the use of Malvern ZS90 Zetasizer for the DLS studies. The authors declare no conflicts of interest.
ABBREVIATIONS
- DLS
Dynamic light scattering
- DMAEMA-BMA
Dimethylaminoethyl methacrylate and butyl methacrylate
- GFP
Green fluorescence protein
- IFN-β1
Interferon beta-1
- Luc
Luciferase
- PC
Phosphatidylcholine
- Pdna
Plasmid deoxyribonucleic acid
- PEG
Polyethylene glycol
- PMPC
Poly (methacryloyloxyethyl phosphorylcholine)
- PMPC-DB
Poly (methacryloyloxyethyl phosphorylcholine) conjugated with DMAEMA-BMA
- siRNA
Small interfering ribonucleic acid
- U-138MG
Uppsala 138 malignant glioma
- U-87MG
Uppsala 87 malignant glioma
Footnotes
Electronic supplementary material The online version of this article (https://doi.org/10.1007/s11095-020-02899-5) contains supplementary material, which is available to authorized users.
Publisher’s Note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
REFERENCES
- 1.Christie R, Nishiyama N, Kataoka K. Minireview: delivering the code: Polyplex carriers for deoxyribonucleic acid and ribonucleic acid interference therapies. Endocrinology. 2010;151:466–73. [DOI] [PubMed] [Google Scholar]
- 2.Patil S, Rhodes D, Burgess D. DNA-based therapeutics and DNA delivery systems: a comprehensive review. AAPS J. 2005;7:E61–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lopes A, Vandermeulen G, Préat V. Cancer DNA vaccines: current preclinical and clinical developments and future perspectives. J Exp Clin Cancer Res. 2019;38(1):146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kang HC, Huh KM, Bae YH. Polymeric nucleic acid carriers: current issues and novel design approaches. J Control Release. 2012;164(3):256–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jackson MA, Werfel TA, Curvino EJ, Yu F, Kavanaugh TE, Sarett SM, et al. Zwitterionic Nanocarrier surface chemistry improves siRNA tumor delivery and silencing activity relative to polyethylene glycol. ACS Nano. 2017;11(6):5680–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lam JKW, Armes SP, Lewis AL, Stolnik S. Folate conjugated phosphorylcholine-based polycations for specific targeting in nucleic acids delivery. J Drug Target. 2009;17(7):512–23. [DOI] [PubMed] [Google Scholar]
- 7.Jackson MA, Bedingfield SK, Yu F, Stokan ME, Miles RE, Curvino EJ, et al. Dual carrier-cargo hydrophobization and charge ratio optimization improve the systemic circulation and safety of zwitterionic nano-polyplexes. Biomaterials. 2019;192:245–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adolph EJ, Nelson CE, Werfel TA, Guo R, Davidson JM, Guelcher SA, et al. Enhanced performance of plasmid DNA polyplexes stabilized by a combination of core hydrophobicity and surface PEGylation. J Mater Chem B. 2014;2(46):8154–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nelson CE, Kintzing JR, Hanna A, Shannon JM, Gupta MK, Duvall CL. Balancing cationic and hydrophobic content of PEGylated siRNA polyplexes enhances endosome escape, stability, blood circulation time, and bioactivity in vivo. ACS Nano. 2013;7(10):8870–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kilchrist KV, Dimobi SC, Jackson MA, Evans BC, Werfel TA, Dailing EA, et al. Gal8 visualization of endosome disruption predicts carrier-mediated biologic drug intracellular bioavailability. ACS Nano. 2019;13(2):1136–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schlenoff JB. Zwitteration: coating surfaces with Zwitterionic functionality to reduce nonspecific adsorption. Langmuir. 2014;30(32): 9625–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu X, Li H, Chen Y, Jin Q, Ren K, Ji J. Mixed-charge nano-particles for long circulation, low Reticuloendothelial system clearance, and high tumor accumulation. Advanced HealthCare Mater. 2014;3(9):1439–47. [DOI] [PubMed] [Google Scholar]
- 13.Wang J, Yuan S, Zhang Y, Wu W, Hu Y, Jiang X. The effects of poly(zwitterions)s versus poly(ethylene glycol) surface coatings on the biodistribution of protein nanoparticles. Biomater Sci. 2016;4(9):1351–60. [DOI] [PubMed] [Google Scholar]
- 14.Wen PY, Kesari S. Malignant Gliomas in Adults. N Engl J Med. 2008;359(5):492–507. [DOI] [PubMed] [Google Scholar]
- 15.Michel V, Yuan Z, Ramsubir S, Bakovic M. Choline transport for phospholipid synthesis. Exp Biol Med (Maywood). 2006;231(5): 490–504. [DOI] [PubMed] [Google Scholar]
- 16.Guo D, Bell EH, Chakravarti A. Lipid metabolism emerges as a promising target for malignant glioma therapy. CNS Oncol. 2013;2(3):289–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li J, Guo Y, Kuang Y, An S, Ma H, Jiang C. Choline transporter-targeting and co-delivery system for glioma therapy. Biomaterials. 2013;34(36):9142–8. [DOI] [PubMed] [Google Scholar]
- 18.Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer. 2007;7:763–77. [DOI] [PubMed] [Google Scholar]
- 19.Li J, Huang S, Shao K, Liu Y, An S, Kuang Y, et al. A choline derivate-modified nanoprobe for glioma diagnosis using MRI. Scientific Reports. 2013;3:1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takano S, Ishikawa E, Matsuda M, Yamamoto T, Matsumura A. Interferon-β inhibits glioma angiogenesis through downregulation of vascular endothelial growth factor and upregulation of interferon inducible protein 10. Int J Oncol. 2014;45(5):1837–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolpert F, Happold C, Reifenberger G, Florea A-M, Deenen R, Roth P, et al. Interferon-β modulates the innate immune response against Glioblastoma initiating cells. PLoS One. 2015;10(10): e0139603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dicitore A, Grassi ES, Borghi MO, Gelmini G, Cantone MC, Gaudenzi G, et al. Antitumor activity of interferon-beta1a in hormone refractory prostate cancer with neuroendocrine differentiation. J Endocrinol Investig. 2017;40(7):761–70. [DOI] [PubMed] [Google Scholar]
- 23.Borden EC. Interferons α and β in cancer: therapeutic opportunities from new insights. Nat Rev Drug Discov. 2019;18(3):219–34. [DOI] [PubMed] [Google Scholar]
- 24.Patnaik S, Gupta KC. Novel polyethylenimine-derived nanoparticles for in vivo gene delivery. Expert Opinion on Drug Delivery. 2013;10(2):215–28. [DOI] [PubMed] [Google Scholar]
- 25.Oupicky D, Konak C, Ulbrich K, Wolfert MA, Seymour LW. DNA delivery systems based on complexes of DNA with synthetic polycations and their copolymers. J Control Release. 2000;65(1–2):149–71. [DOI] [PubMed] [Google Scholar]
- 26.Dave KM, Ali L, Manickam DS. Characterization of the SIM-A9 cell line as a model of activated microglia in the context of neuropathic pain. PLoS One. 2020;15(4):e0231597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soundara Manickam D, Bisht HS, Wan L, Mao G, Oupicky D. Influence of TAT-peptide polymerization on properties and transfection activity of TAT/DNA polyplexes. J Control Release. 2005;102(1):293–306. [DOI] [PubMed] [Google Scholar]
- 28.Liang C-C, Park AY, Guan J-L. In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat Protoc. 2007;2(2):329–33. [DOI] [PubMed] [Google Scholar]
- 29.Happold C, Roth P, Silginer M, Florea A-M, Lamszus K, Frei K, et al. Interferon-β induces loss of Spherogenicity and overcomes therapy resistance of Glioblastoma stem cells. Mol Cancer Ther. 2014;13(4):948–61. [DOI] [PubMed] [Google Scholar]
- 30.Manickam DS, Oupicky D. Multiblock reducible copolypeptides containing histidine-rich and nuclear localization sequences for gene delivery. Bioconjug Chem. 2006;17(6):1395–403. [DOI] [PubMed] [Google Scholar]
- 31.Zeisel SH, da Costa K-A. Choline: an essential nutrient for public health. Nutr Rev. 2009;67(11):615–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raup A, Jérôme V, Freitag R, Synatschke CV, Müller AHE. Promoter, transgene, and cell line effects in the transfection of mammalian cells using PDMAEMA-based nano-stars. Biotechnology Reports. 2016;11:53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.von Gersdorff K, Sanders NN, Vandenbroucke R, De Smedt SC, Wagner E, Ogris M. The internalization route resulting in successful gene expression depends on both cell line and polyethylenimine polyplex type. Mol Ther. 2006;14(5):745–53. [DOI] [PubMed] [Google Scholar]
- 34.Makowska A, Wahab L, Braunschweig T, Kapetanakis N-I, Vokuhl C, Denecke B, et al. Interferon beta induces apoptosis in nasopharyngeal carcinoma cells via the TRAIL-signaling pathway. Oncotarget. 2018;9(18):14228–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kotredes KP, Gamero AM. Interferons as inducers of apoptosis in malignant cells. J Interf Cytokine Res. 2013;33(4):162–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thyrell L, Erickson S, Zhivotovsky B, Pokrovskaja K, Sangfelt O, Castro J, et al. Mechanisms of interferon-alpha induced apoptosis in malignant cells. Oncogene. 2002;21(8):1251–62. [DOI] [PubMed] [Google Scholar]
- 37.Moghimi SM, Symonds P, Murray JC, Hunter AC, Debska G, Szewczyk A. A two-stage poly(ethylenimine)-mediated cytotoxicity: implications for gene transfer/therapy. Mol Ther. 2005;11(6):990–5. [DOI] [PubMed] [Google Scholar]
- 38.Satomi H, Wang B, Fujisawa H, Otsuka F. Interferon-beta from melanoma cells suppresses the proliferations of melanoma cells in an autocrine manner. Cytokine. 2002;18(2):108–15. [DOI] [PubMed] [Google Scholar]
- 39.Zhou J, Horev B, Hwang G, Klein MI, Koo H, Benoit DSW. Characterization and optimization of pH-responsive polymer nanoparticles for drug delivery to oral biofilms. J Mater Chem B. 2016;4(18):3075–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kabanov AV, Kabanov VA. DNA complexes with Polycations for the delivery of genetic material into cells. Bioconjug Chem. 1995;6(1):7–20. [DOI] [PubMed] [Google Scholar]
- 41.Tammam SN, Azzazy HME, Lamprecht A. The effect of nanoparticle size and NLS density on nuclear targeting in cancer and normal cells; impaired nuclear import and aberrant nanoparticle intracellular trafficking in glioma. J Control Release. 2017;253:30–6. [DOI] [PubMed] [Google Scholar]
- 42.Boeckle S, Fahrmeir J, Roedl W, Ogris M, Wagner E. Melittin analogs with high lytic activity at endosomal pH enhance transfection with purified targeted PEI polyplexes. J Control Release. 2006;112(2):240–8. [DOI] [PubMed] [Google Scholar]
- 43.Boussif O, Lezoualc’h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, et al. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci U S A. 1995;92(16):7297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boeckle S, von Gersdorff K, van der Piepen S, Culmsee C, Wagner E, Ogris M. Purification of polyethylenimine polyplexes highlights the role of free polycations in gene transfer. J Gene Med. 2004;6(10):1102–11. [DOI] [PubMed] [Google Scholar]
- 45.Intra J, Salem AK. Characterization of the transgene expression generated by branched and linear polyethylenimine-plasmid DNA nanoparticles in vitro and after intraperitoneal injection in vivo. J Control Release. 2008;130(2):129–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seib FP, Jones AT, Duncan R. Comparison of the endocytic properties of linear and branched PEIs, and cationic PAMAM dendrimers in B16f10 melanoma cells. J Control Release. 2007;117(3):291–300. [DOI] [PubMed] [Google Scholar]
- 47.Dedoni S, Olianas MC, Onali P. Interferon-beta induces apoptosis in human SH-SY5Y neuroblastoma cells through activation of JAK-STAT signaling and down-regulation of PI3K/Akt pathway. J Neurochem. 2010;115(6):1421–33. [DOI] [PubMed] [Google Scholar]
- 48.Zhang R, Banik NL, Ray SK. Combination of all-trans retinoic acid and interferon-gamma suppressed PI3K/Akt survival pathway in glioblastoma T98G cells whereas NF-kappaB survival signaling in glioblastoma U87MG cells for induction of apoptosis. Neurochem Res. 2007;32(12):2194–202. [DOI] [PubMed] [Google Scholar]
- 49.van Engeland M, Nieland LJ, Ramaekers FC, Schutte B, Reutelingsperger CP. Annexin V-affinity assay: a review on an apoptosis detection system based on phosphatidylserine exposure. Cytometry. 1998;31(1):1–9. [DOI] [PubMed] [Google Scholar]
- 50.Pijuan J, Barceló C, Moreno DF, Maiques O, Sisó P, Marti RM, et al. In vitro Cell Migration, Invasion, and Adhesion Assays: From Cell Imaging to Data Analysis. Frontiers in Cell and Developmental Biology. 2019;7(107). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gwak SJ, Macks C, Bae S, Cecil N, Lee JS. Physicochemical stability and transfection efficiency of cationic amphiphilic copolymer/pDNA polyplexes for spinal cord injury repair. Sci Rep. 2017;7(1):11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Frickhofen N, Müller E, Sandherr M, Binder T, Bangerter M, Wiest C, et al. Rearranged Ig heavy chain DNA is detectable in cell-free blood samples of patients with B-cell Neoplasia. Blood. 1997;90(12):4953–60. [PubMed] [Google Scholar]
- 53.Zhou Q-H, Wu C, Manickam DS, Oupický D. Evaluation of pharmacokinetics of bioreducible gene delivery vectors by real-time PCR. Pharm Res. 2009;26(7):1581–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng H, Qian J, Carbone CJ, Leu NA, Baker DP, Fuchs SY. Vascular endothelial growth factor-induced elimination of the type 1 interferon receptor is required for efficient angiogenesis. Blood. 2011;118(14):4003–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sagar SM, Yance D, Wong RK. Natural health products that inhibit angiogenesis: a potential source for investigational new agents to treat cancer-Part 1. Current oncology (Toronto, Ont). 2006;13(1):14–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


