Abstract
Speedy and on-time detection of coronavirus disease 2019 (COVID-19) is of high importance to control the pandemic effectively and stop its disastrous consequences. A widely available, reliable, label-free, and rapid test that can recognize tiny amounts of specific biomarkers might be the solution. Nanobiosensors are one of the most attractive candidates for this purpose. Integration of graphene with biosensing devices shifts the performance of these systems to an incomparable level. Between the various arrangements using this wonder material, field-effect transistors (FETs) display a precise detection even in complex samples. The emergence of pioneering biosensors for detecting a wide range of diseases especially COVID-19 created the incentive to prepare a review of the recent graphene-FET biosensing platforms. However, the graphene fabrication and transfer to the surface of the device is an imperative factor for researchers to take into account. Therefore, we also reviewed the common methods of manufacturing graphene for biosensing applications and discuss their advantages and disadvantages. One of the most recent synthesizing techniques - laser-induced graphene (LIG) - is attracting attention owing to its extraordinary benefits which are thoroughly explained in this article. Finally, a conclusion highlighting the current challenges is presented.
Keywords: COVID-19, FET biosensors, graphene, LIG, SARS-CoV-2
I. Introduction
Over 120 million people have been infected by COVID-19 and the death toll has reached 3 million in 1 year by March 2021 [1]. It has a high reproduction number, and superfast transmission rate in comparison to the other emerging viral diseases such as SARS and Ebola. Thus it caused an unprecedented health crisis in the past 200 years compared to any other pandemic [2]. This lethal and fast-spreading infection is caused by a new strand of coronaviruses named Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) [3]. This crown-like single-stranded RNA virus is a member of the Coronaviridae subfamily. Its genome encodes a polyprotein, non-structural, structural, and accessory proteins [4]. The majority of the non-structural proteins are necessary for the replication of the virus, whereas the structural ones like the membrane (M), envelope (E), and nucleocapsid (N) proteins are vital for the assembly process [5]. The spike (S) protein is responsible for the cell membrane fusion as it binds to the receptor molecules on the host cell. It links to the angiotensin-converting enzyme 2 (ACE2) and facilitates cell entry [6]. Between the aforementioned molecules in the structure of SARS-CoV-2, E protein which plays critical roles during the life cycle of the virus, is the most antigenic one which can act as an important target in COVID-19 biosensing, vaccine, and drug discovery [7]. Since this lethal malady is highly contagious and can transmit from symptomatic or asymptomatic individuals, it is essential to identify it in its early phases in order to curb its transmission rate and control it more efficiently [8]. The most widespread detection method is Reverse Transcriptase Polymerase Chain Reaction (RT-PCR), which identifies the existence of SARS-CoV-2’s genetic material in nasopharyngeal swab samples [9]. It has disadvantages like being time-consuming and expensive. Additionally, it cannot spot the virus during the incubation period and after the inception of symptoms [10]. Since it takes time to vaccinate the majority of the people all around the globe, an alternate technique is needed to enable facile management of the disease and relax the strict quarantine rules [11]. Moreover, this new methodology should be accessible, easy-to-use, affordable, and speedy to expedite massive on-site testing, especially in airports and public places [12]–[14]. Biosensing platforms based on field-effect transistors (BioFETs) are one of the most appropriate choices for coronavirus detection as they follow electro-analysis of charged biomolecules like virus-related biomarkers [15]. Attributable to their advantages like being compact, rapid, label-free, real-time, simple to fabricate and use, reliable, and compatible with state-of-the-art micro-and nanofabrication technologies, they have the potential to replace the presently used diagnostic methods [16]. Furthermore, a FET-based biosensor can be used for multiplexed detection of COVID-19 related biomarkers in human biofluids without or with minimal sample preparation steps [17]. However, it is necessary to choose a suitable substrate for covering the channel region of a FET device in order to have an optimized surface functionalization and biorecognition element (BRE) immobilization process [18]. One of the commonly used and competent materials is graphene which offers several benefits as a transducer in BioFETs [19]. It has outstanding electrical and thermal conductivity, a large surface-to-volume ratio, high capacitance, low contact resistance, and adjustable ambipolar field-effect behaviors [20]. The superior electrical conductivity of graphene accelerates the electron shuttle and thus minimizes the response time of the biosensor [21]. Besides, graphene-based FETs (GFETs) are highly scalable, miniaturized, and do not require sophisticated laboratory equipment and personnel [22]. Therefore, they are broadly utilized for point-of-care (POC) measurement of various biomarkers such as viral and bacterial particles, oligonucleotides, oncobiomarkers, hormones, and other biomolecules [20]. POC application of BioGFET demands a suitable method to develop its miniaturized parts like source, drain, and gate terminals, and also the graphene channel with the immobilized BREs on top of it [23], [24]. Conventionally, chemical vapor deposition (CVD) [25], mask photolithography techniques [26] along proper etching and transfer processes are used for this purpose [27]. Despite the homogenous, impermeable, pure, and well-structured final product of the CVD, several toxic gases are produced as the by-products which are the unwanted consequences of this expensive procedure. Additionally, it easily gets affected by the changes of related parameters [28]. These multiple-step methodologies increase the fabrication cost of the device and make the device labor-intensive which hinder the frequent use of the BioGFET in large scale viral and bacterial detection. Therefore, there is an urgent need for an alternate technique to prepare BioGFET in a rapid and facile manner. Laser-induced graphene (LIG) is a strong candidate due to its ability to quickly generate different graphene-based patterns in single step [29], [30]. Additionally, LIGs have successfully been functionalized with different BREs such as antibodies, nucleic acids, and aptamers, which is a crucial part of any viral detection. Therefore, it is considered an efficient strategy in constructing GFET biosensors especially for novel coronavirus detection. This review article discusses different techniques of synthesizing graphene including top-down methodologies (mechanical and chemical exfoliation), bottom-up approaches (Epitaxial growth (EG) and CVD), and the most recent technology of LIG for being used in the structure of GFET-based biosensing devices. It also reviews the recently proposed biosensors for detecting various biomolecules such as, viruses, bacteria, nucleic acids, oncobiomarkers, hormones, etc.
II. Graphene Synthesis
Theoretically, graphene is an infinite two-dimensional monolayer of the honeycomb structure of sp 2 hybridized carbon atoms [31]. Geim and Novoselov first discovered graphene in 2004 during a repeated peeling-off exfoliation experiment on graphite [32], as shown in Fig. 1. Graphite material is composed of graphene sheets connected by weak Van der forces [33]. Exfoliation and cleavage processes can separate these sheets apart. Although, this first discovered graphene method is a time-consuming process but paved the way for the application of other exfoliation processes [34]. Till to date, a large number of graphene synthesis methods have been proposed that can be categories into top-bottom and bottom-up approaches, as depicted in Fig. 1. The first approach involves the split-up of graphite material into graphene layers, and the second involves building-up graphene layers on a substrate [36].
Fig. 1.
Schematic of the conventional fabrication methods of graphene fabrication. (Reproduced with permission from [35]).
A. Top-Down Approach
1). Mechanical Exfoliation:
Mechanical exfoliation follows the application of a suitable mechanical process on a piece of high-quality graphite to separate weakly attached single or few layers of graphene one by one. These processes typically include cleavage during the scotch tape, lathe-like, ball-milling experiments where shear force separates the sheets like three-ball mill, dry and wet ball milling experiments, etc. [37]. This technique is low-yielding, time-consuming, and challenging to scale-up for the industry. After each cleavage process conductivity of the remaining sample decreases. For example, after 12 hours of mechanical exfoliation by a three-roll-mill experiment, a sample of few graphene layers exhibited 7500 S/m conductivity than the original graphite’s conductivity of 25000 S/m before the start of the process [38].
2). Chemical Exfoliation:
Instead of using mechanical processes, different chemical methods have successfully been employed on graphite for its large-scale exfoliation into graphene sheets, like Brodie, Staudenmaier, and Hummer’s methods, etc. [39]. It is one of the low-cost methods. However, the use of strong acids and oxidizers during chemical exfoliation processes creates defects in produced sp 2 hybridized sheets of the graphene. Therefore rather than pristine graphene, these methods provide graphene oxide (GO) sheets that exhibit low conductivity. Hummer’s method is one of the most popular methods used for graphene production and many types of graphene have been seen like three-dimensional graphene, sponge-like graphene, etc. [40]. But this method requires a subsequent reduction method to restore the graphene’s conductivity, called reduced graphene oxide (RGO) [41]. Different methodologies have been employed to reduce GO like chemical reduction, thermal, plasma, and solvothermal reduction [42]. The application of different reduction methods reports different electrical conductivities. For example, the chemical reduction of Zn/HCl reported 15,000 S/m conductivity [43], whereas thermal treatment of C2H2 at 1000°C exhibited 143,000 S/m conductivity [44]. Moreover, the use of RGO as active material in a device demands a suitable experiment-design that involves a patterning process as well [40].
B. Bottom-Up Approach
1). Epitaxial Growth (EG) of Graphene:
Another method that retains high crystallinity is the epitaxial growth (EG) of graphene on silicon carbide (SiC) substrate. In this process, carbon atoms rearrange under high vacuum and form mono or multilayers of high-quality graphene over the substrate [45], [46]. This method is common for the preparation of a miniaturized semiconductor-based electrical biosensor. This process exhibits very low sheet resistance (
 /sq) [47] and it is very suitable for fundamental study on the laboratory scale. However, SiC substrate cost, high temperature >1000°C, size of the substrate, transfer of graphene to another substrate, and control over graphene thickness pose challenges in device preparation. Various other substrates have also been reported using epitaxial growth techniques like Pt, Ru, TiC, and Cu [48].
/sq) [47] and it is very suitable for fundamental study on the laboratory scale. However, SiC substrate cost, high temperature >1000°C, size of the substrate, transfer of graphene to another substrate, and control over graphene thickness pose challenges in device preparation. Various other substrates have also been reported using epitaxial growth techniques like Pt, Ru, TiC, and Cu [48].
2). Chemical Vapor Deposition (CVD):
The other technique that allows the successful transfer of generated high-quality graphene from one substrate to another is the carbon’s CVD. This process follows the diffusion of thermally decomposed carbon atoms from hydrocarbons into the metal surface placed inside a furnace tube’s controlled environment. The graphene formation occurs when metal substrate containing metal-carbon solid solution cools down and allows carbon atoms to precipitate and segregate over the surface [49]. Controlling various CVD parameters like growth temperature, duration, and changing metal substrate and types of hydrocarbons, different graphene layers, and their sizes have been seen. CVD is the most commonly used method nowadays for the fabrication of GFET biosensors [50]. The graphene sheet is grown on a metal substrate (commonly Cu or Ni) by CVD and transfer to another separate substrate. TABLE I summarises conventional methods of graphene fabrication and their electrical performances.
TABLE I. Graphene Synthesis Methods.
| Synthesis Methods | Advantages | Disadvantages | Process | Electronic Properties | Ref | |
|---|---|---|---|---|---|---|
Conductivity S/m Or Sheet resistance
 /sq Or resistance /sq Or resistance
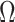
| ||||||
| Top-to-bottom | Mechanical Exfoliation | Graphene layers formation. | Large scale production, time-consuming. | Peel-of | ~2m
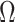 at Vg=0 at Vg=0 |
[32] |
| Wet ball milling | 1200 S/m | [58] | ||||
| Three-roll-mill | 7500 S/m | [38] | ||||
| Chemical Exfoliation | Large scale production. | Surface oxygen, reduction method. | GO reduction by Zn/HCl. | 15,000 S/m | [43] | |
| GO reduction by NH3-BH3 | 20,300 S/m | [59] | ||||
| GO reduction under H2 at 1000°C for 1h. | 76,000 S/m | [60] | ||||
| GO reduction under C2H2 at 1000°C for 30 min. | 143,000 S/m | [44] | ||||
| Bottom-to-top | Epitaxial Growth | High-quality graphene is suitable for device fabrication. | Transfer method, expensive SiC substrate. | Graphene on SiC |
 /sq /sq |
[61] |
| Graphene on SiC |
 /sq /sq |
[62] | ||||
| Graphene on SiC |
 /sq /sq |
[47] | ||||
| Chemical Vapor Deposition | High-quality graphene is suitable for device fabrication. | Transfer method | Plasma CVD |
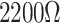 /sq /sq |
[63] | |
| CVD graphene on Cu |
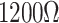 /sq /sq |
[64] | ||||
| CVD graphene on Cu |
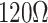 /sq /sq |
[65] | ||||
| Laser-induced graphene | Pattern graphene, single step. | Low resolution. | LIG |
 /sq /sq |
[66] | |
| LIG |
 /sq /sq |
[67] | ||||
| LIG |
 /sq /sq |
[68] | ||||
Preparation of functional GFET demands an efficient manufacturing technique for creating its various parts, including graphene-based FET channel, source, drain, and gate terminals. CVD is the most extensively used technique for the fabrication of GFET based biosensors because produced graphene offers high conductivity and surface area by produced defect-free sp 2 hybridized carbon structure. In a typical GFET device fabrication by CVD, few graphene layers are first produced on a metallic planar substrate (for example, Cu or Ni), transfer to another substrate, followed by the generation of source, drain, and gate electrodes by photolithography. The photolithography technique is an advanced form of lithography commonly employed to develop small-scale electrodes, which calls for photoresist, patterning the photoresist, and controlled etching process. For example, Wu et al. used CVD to develop few graphene layers and coated the graphene with a photoresist for the photolithography process. It followed careful attachment of a tiny featured-mask over the photoresist surface and exposure to ultraviolet light for some time. Multiple etching and metal coating processes were applied in a careful sequence to develop multiple contacts with few tenths of nanometer-wide graphene [51]. J Tu et al. synthesized graphene thin film over the copper foil by CVD, capped with thermally grown SiO2 and polymethylmethacrylate (PMMA), followed by etching copper foil and PMMA by acid and acetone, respectively [52]. After transferring graphene to SiO2 substrate, standard photolithography was employed two times to pattern the graphene channel region and a few nanometer source and drain contacts, as shown in Fig. 2(a). Prepared GFET biosensor detected mercury contaminants based on single-stranded DNA aptamer. Following similar one-time photolithography Z Gao et al. developed in plan two gate Au-electrodes on graphene and source and drain on the surface of SiO2 and used the design as GFET based DNA biosensor [53]. A similar approach is being followed by W Yue et al. to prepare GFET based biosensor for the detection of binding-kinetics of DNA hybridization [54]. There are many other reports about the use of this technique to develop GFET based biosensor [55]–[57].
Fig. 2.

(a) Schematic of the GFET fabrication process (reproduced with permission from [52]), and (b) LIG patterning.
Although photolithography is a very suitable bottom-up technique of the graphene synthesis but involved careful etching of the metallic substrate during the transfer process, coating of photoresist, and photolithography techniques make the whole process lengthy and complex. Moreover, produced defect-free CVD graphene is difficult to functionalize, which is sometimes required for specific biosensing applications.
Therefore, an alternative method is earnestly required to generate channel material more straightforwardly than CVD, which can be further functionalized easily for GFET related biosensing applications.
C. Laser-Induced Graphene (LIG)
The LIG technique is a recently discovered strategy for generating patterned graphene with several advantages. It is simple, fast, environmentally-friendly, and produced material can easily be functionalized and decorated, as shown in Fig. 2(b). Since its first discovery in 2014 [69], different types of. graphene materials have been produced and employed as active materials in many biosensing platforms. The detailed process involves surface interaction between laser and substrate material of some carbon source which triggers localized heating or thermal ablation and photochemical reactions. As a result, temperature in a tiny surface area increases. These reactions derive decomposition of the surface carbon, and as a result, sp3 hybridized carbon molecules of the source rearrange into sp2 hybridized carbon of graphene with some gas molecules’ evolution. The lowest sheet resistance of ~
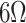 /sq has been seen by one of the LIG materials [70]. This localized surface heat process is also known as laser irradiation, engraving, and laser writing. Different lasering parameters like laser carrier speed, pulse width, wavelength, and power of the used laser light can control the heat. Surface-generated heat plays a vital role in the formation of graphene structures. Hence, many morphologies of different thicknesses like hierarchical porous graphene, fibrous graphene, graphene sheets, and their different physio-chemical properties along with surface functionalization have been seen [78], [79]. Furthermore, graphene patterns of different feature sizes and resolutions have been prepared. Owing to mask-free patterning, non-toxic nature, and process flexibility, it has attracted attention. Developed laser graphene-based architectures have been used in a wide range of advanced graphene-based devices as active materials [78], [80]. This method has also been employed in the biosensor field to design a wide variety of in-plan graphene-based electrodes for various biosensing applications [81], [82]. For example, Guo et al. reported successful LIG-based FET preparation in which only back gate was fabricated using laser writing technique over graphene oxide [76]. He et al. reported the preparation of source, drain, and top gates using laser writing over graphene oxide surface [77]. On-chip application of small channel material of LIG demands successful formation of source and drain for complete preparation of a FET-based biosensor which demands more research in the field. Numerous studies are focusing on designing tailored detection systems employing specific BRE-functionalized LIG. For instance, A R. Cardoso et al. successfully employed LIG on the surface of polyimide to fabricate LIG-based active area of working, counter, and a reference electrode in a one-step engraving process, followed by molecularly imprinted polymerization [71]. Electrochemical polymerization and incubation processes were used along with the electrodes’ passivation to obtain a molecularly-imprinted polymer. This imprinted polymerization was followed by electropolymerization of the functional monomer in chloramphenicol’s presence. The detection process followed electrochemical test performed in a buffer solution after removing chloramphenicol, and enhanced removal is noticed compared with other carbon electrodes, as shown in Fig. 3(a). C. Fenzl et al. reported successful LIG functionalization of the aptamer for thrombin detection. A pyrene butyric acid-treated LIG electrode resulted in the formation of COOH-group on the surface of porous LIG, which attracted aptamers and hence thrombin as shown in Fig. 3(b)
[72]. As a result, aptamer-functionalized LIG electrode of porous morphology blocked (Fe(CN)
/sq has been seen by one of the LIG materials [70]. This localized surface heat process is also known as laser irradiation, engraving, and laser writing. Different lasering parameters like laser carrier speed, pulse width, wavelength, and power of the used laser light can control the heat. Surface-generated heat plays a vital role in the formation of graphene structures. Hence, many morphologies of different thicknesses like hierarchical porous graphene, fibrous graphene, graphene sheets, and their different physio-chemical properties along with surface functionalization have been seen [78], [79]. Furthermore, graphene patterns of different feature sizes and resolutions have been prepared. Owing to mask-free patterning, non-toxic nature, and process flexibility, it has attracted attention. Developed laser graphene-based architectures have been used in a wide range of advanced graphene-based devices as active materials [78], [80]. This method has also been employed in the biosensor field to design a wide variety of in-plan graphene-based electrodes for various biosensing applications [81], [82]. For example, Guo et al. reported successful LIG-based FET preparation in which only back gate was fabricated using laser writing technique over graphene oxide [76]. He et al. reported the preparation of source, drain, and top gates using laser writing over graphene oxide surface [77]. On-chip application of small channel material of LIG demands successful formation of source and drain for complete preparation of a FET-based biosensor which demands more research in the field. Numerous studies are focusing on designing tailored detection systems employing specific BRE-functionalized LIG. For instance, A R. Cardoso et al. successfully employed LIG on the surface of polyimide to fabricate LIG-based active area of working, counter, and a reference electrode in a one-step engraving process, followed by molecularly imprinted polymerization [71]. Electrochemical polymerization and incubation processes were used along with the electrodes’ passivation to obtain a molecularly-imprinted polymer. This imprinted polymerization was followed by electropolymerization of the functional monomer in chloramphenicol’s presence. The detection process followed electrochemical test performed in a buffer solution after removing chloramphenicol, and enhanced removal is noticed compared with other carbon electrodes, as shown in Fig. 3(a). C. Fenzl et al. reported successful LIG functionalization of the aptamer for thrombin detection. A pyrene butyric acid-treated LIG electrode resulted in the formation of COOH-group on the surface of porous LIG, which attracted aptamers and hence thrombin as shown in Fig. 3(b)
[72]. As a result, aptamer-functionalized LIG electrode of porous morphology blocked (Fe(CN)
 ions’ electrochemical activity when electrochemically tested in K3(Fe(CN)6) added buffer solution of phosphorous. Hence, a reduced redox current is reported. This aptamer-labeling electrochemical biosensing system successfully detected a small amount of 1 pM in a buffer solution and 5 pM in the serum’s complex matrix. A. K. Yagati et al. reported thrombin detection with an improved low limit of detection of 0.12 pM in phosphorous buffer solution. In which the surface of the pores of interdigitated LIG electrodes was chemically modified with polymer-contained nanoparticles after the attachment of COOH-groups, as shown in Fig. 3(c)
[73]. This interdigitated design of modified LIG electrodes showed a linear current-concentration response of 0.01 to 1000 nM and an exceptionally low detection limit because of the quick change in impedimetric capacitance calculated in an electrolyte of buffer solution. Furthermore, this study showed a comparison between the label and label-free functionalization of LIG. This approach to quantifying the concentration of thrombin in blood serum was found amazingly effective compared with other expensive thrombin detection methods that can help in the early detection of related human diseases. R. R. S. Soares et al. showed successful loading of anti-Salmonella antibody on LIG material by chemical treatment and reported low-limit detection of Salmonella in chicken broth (as shown in Fig. 3(d)) [74]. Lacquer passivation was used to cover LIG strand and expose circular shape active area for the chemical loading of anti-Salmonella antibody. The modified electrode as a working electrode in a three-electrode electrochemical cell exhibited an electron transfer rate of 0.0146 cm/s and detection of a low concentration of 13 cfu mL−1 of Salmonella in chicken broth. These results of the LIG-based Salmonella biosensor are comparable to other sensors prepared by laborious and expensive CVD and inkjet printing methods. D.C. Vanegas et al. reported the preparation of a LIG-based biogenic amine biosensor prepared by diamine oxidase functionalization due to electrodeposition of copper nano-composite on the surface of LIG pores (as shown in Fig. 3(e)). This biosensor detected a low biogenic amine concentration of
ions’ electrochemical activity when electrochemically tested in K3(Fe(CN)6) added buffer solution of phosphorous. Hence, a reduced redox current is reported. This aptamer-labeling electrochemical biosensing system successfully detected a small amount of 1 pM in a buffer solution and 5 pM in the serum’s complex matrix. A. K. Yagati et al. reported thrombin detection with an improved low limit of detection of 0.12 pM in phosphorous buffer solution. In which the surface of the pores of interdigitated LIG electrodes was chemically modified with polymer-contained nanoparticles after the attachment of COOH-groups, as shown in Fig. 3(c)
[73]. This interdigitated design of modified LIG electrodes showed a linear current-concentration response of 0.01 to 1000 nM and an exceptionally low detection limit because of the quick change in impedimetric capacitance calculated in an electrolyte of buffer solution. Furthermore, this study showed a comparison between the label and label-free functionalization of LIG. This approach to quantifying the concentration of thrombin in blood serum was found amazingly effective compared with other expensive thrombin detection methods that can help in the early detection of related human diseases. R. R. S. Soares et al. showed successful loading of anti-Salmonella antibody on LIG material by chemical treatment and reported low-limit detection of Salmonella in chicken broth (as shown in Fig. 3(d)) [74]. Lacquer passivation was used to cover LIG strand and expose circular shape active area for the chemical loading of anti-Salmonella antibody. The modified electrode as a working electrode in a three-electrode electrochemical cell exhibited an electron transfer rate of 0.0146 cm/s and detection of a low concentration of 13 cfu mL−1 of Salmonella in chicken broth. These results of the LIG-based Salmonella biosensor are comparable to other sensors prepared by laborious and expensive CVD and inkjet printing methods. D.C. Vanegas et al. reported the preparation of a LIG-based biogenic amine biosensor prepared by diamine oxidase functionalization due to electrodeposition of copper nano-composite on the surface of LIG pores (as shown in Fig. 3(e)). This biosensor detected a low biogenic amine concentration of
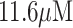 in fish paste samples subjected to fermentation with lactic acid bacteria.
in fish paste samples subjected to fermentation with lactic acid bacteria.
Fig. 3.

(a) Schematic representation of the workflow employed for the LIG electrodes’ preparation and functionalization for the detection of chloramphenicol [71]. Illustration of Thrombin detection by aptamer modified LIG by (b) COOH groups and (c) nanoparticle plus polymer. (d) Depiction of anti-Salmonella antibody attachment with LIG for the detection of Salmonella. Illustration of detection of biogenic amine by (e) diamine oxidase modification of LIG. (f) schematic depiction of bottom-gate GO FET (top) and scheme of the experimental procedures for LIG-based FET (bottom). Reproduced with permissions from [71]–[77].
These successful research studies exhibit the immense potential of LIG-based biosensors for diagnostic applications. They can be a potent candidate for early detection of the COVID-19. Seo et al. fabricated GFET to detect novel coronavirus successfully using antibody attachment within the graphene channel [17]. GFET biosensors are suffering from high cost because they involved complex and expensive fabrication techniques for designing their separate parts (e.g., photolithography and CVD, etc.). In the COVID-19 scenario considering the execution of many daily tests, the biosensor cost is a significant concern. Therefore, the low-cost LIG material’s fabrication in small features makes this technique an ideal choice for the biosensors’ preparation for COVID-19. However, on-chip application of small channel material of LIG demands successful formation of source and drain for complete preparation of a FET-based biosensor which requires more research in the field.
III. FET Biosensors for Detecting Biomolecules
Biosensors based on FET have been evolving in recent decades [83]. They are being used for detecting a wide range of biomolecules sensitively and selectively [84]. Since FETs are sensitive to surface charge and most of the biological particles are charged in physiological circumstances, they are desirable for conducting a rapid, real-time, and label-free identification of numerous biological analytes such as viruses [16]. For this purpose, their surfaces must be modified with biocompatible and conductive materials to be desirable for detecting a biological element and also amplify the generated electrical signal [85]. Graphene is one of the suitable candidates which can act as an efficient interface between the electrical and biological departments. Thus, GFETs are attracting ever-increasing attention in the field of early-stage diagnosis [22]. As it is summarized in TABLE II, s variety of target biomolecules such as viruses, nucleic acids, oncobiomarkers, hormones, etc. have been studied using GFET devices functionalized with specific bioreceptors (antibody, DNA, RNA, aptamer, etc.) [86]. A list of the most recent research works can be seen in Table II. The detection plan of these systems can be easily adopted in designing GFET-based devices for early recognition of COVID-19-related biomarkers (whole virus, antigen, antibody, RNA).
TABLE II. Recently Developed GFET-Based Biosensors.
| Application | Target | RE | Linker | Surface | LOD | Sample | Ref. |
|---|---|---|---|---|---|---|---|
| COVID-19 diagnosis | S protein and whole virus | Ab | PBASE | Graphene |
 2 copies/ml 2 copies/ml |
Clinical samples | [17] |
| COVID-19 diagnosis | S protein | Ab | Graphene | 0.2 pM | Buffer | [92] | |
| JEV and AIV detection | Whole virus | Ab | carboxy | Graphene | 1 fM and 10 fM | Buffer | [93] |
| IV detection | H1N1 virus | Aptamer | EDC/NHS | Graphene | 1 ng/ml | [94] | |
| Virus detection | Whole virus (VSV, HIV, MLV) | Ab | PBASE | Graphene | 47.8 aM | Buffer | [95] |
| Rota-virus detection | Rota-virus | Ab | pyrene-NHS | rGO | 1 nM | Buffer | [96] |
| HPV detection | HPV-16 E7 protein | RNA aptamer Sc5-c3 | EDC/NHS | rGO | 1.75 nM | Saliva | [97] |
| EVD detection | Ebola glycoprotein | Ab | rGO | 1 ng/ml | buffer, serum, and plasma | [98] | |
| Bacteria detection | E. coli K12 | Ab | PBASE ETA | Graphene | Buffer | [103] | |
| E. coli detection | Escherichia coli O157:H7 | Ab | ETA | Graphene | Buffer | [104] | |
| E. coli detection | E. coli | Ab | EDC/NHS/AuNPs | rGO/Al2O3 | 104 CFU/ mL | River water | [105] |
| DNA detection | Target DNA | Probe DNA | PASE ETA | PS/Graphene/PMMA/Silicone rubber | 20 aM | Serum | [110] |
| DNA detection | magnetically labeled ssDNA | Aptamer | PBASE | Graphene | 1 pM | Buffer | [132] |
| SNP detection | SNP | DNA-tweezers | Graphene | Femtomolar level | [133] | ||
| DNA hybridization detection | ssDNA | DNA | Graphene | [134] | |||
| DNA hybridization detection | DNA | DNA | Ar plasma | Ar plasma-treated graphene | 10 aM | [135] | |
| DNA detection | DNA | hairpin probe DNA | PBASE | Graphene | subfM | Buffer | [136] |
| RNA detection | RNA | ssDNA | PBASE | Graphene | 0.1 fM | Buffer | [111] |
| PSA detection | PSA | DNA aptamers | ETA EDC/NHS | Graphene/PYCOOH/PEG | 1 nM | Buffer | [114] |
| AFP detection | AFP | Ab | PBASE | Graphene | 12.9 ng.mL−1 and 0.1 ng.mL−1 | Serum and buffer | [115] |
| CEA detection | CEA | Ab | nano-dBSA EDC/Sulfo-NHS | Graphene | 337.58 fg Ml−1 | Buffer | [116] |
| Exosome detection | Exosome | anti-CD63 | PBASE | Graphene |
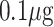 /mL /mL |
Buffer | [118] |
| CA1 detection | CA1 | RNA aptamers | PBASE NHS | Graphene | 70 pM | Human saliva | [117] |
| TSH detection | TSH | Anti-TSH Ab fragment | EDC/NHS | Graphene/PBA,PyMal/PEG | 10−16 M | Serum | [121] |
| hCG detection | hCG | Ab | PBSE ETA Pyr-NHS | Graphene | ~1 pg.mL−1 | Buffer | [122] |
| Cortisol detection | cortisol | C-Mab | EDC/NHS | Graphene | 10 pg/ml | Human tear | [123] |
| ADH detection | ADH peptide | Aptamer | APTES GA | Graphene | 3.55 ag/mL | Serum and buffer | [124] |
| ID detection | ferritin | anti-ferritin | PASE | Graphene | 10 fM | Buffer | [125] |
| Biotin detection | Biotin | Avidin | PBASE | Graphene | 0.37 pM | Buffer | [126] |
| IgG detection | IgG | anti-IgG | AuNPs | TrGO, metal nitride/graphene nanohybrid | 0.2 ng/mL | Buffer | [137] |
| IgG detection | IgG | Ab | AuNPs | VG | 13 pM | Buffer | [138] |
| Methyl vanillate detection | Methyl vanillate | OBP14 | PBSE | rGO | 100 nm | Buffer | [128] |
| odorant molecules detection | odorant molecules | membrane bound receptors | Pyrene NTA, pyrene lipids | rGO | micromole | Buffer | [129] |
| Odorant detection | Eugenol | OBP14 | PBSE | rGO | 100 pM | Buffer | [127] |
| H2O2 detection | H2O2 | rGO/MoS2 | 1 pM | Buffer | [130] | ||
| lead and potassium ions detection | K+ and Pb2+ | TBA | MB | Graphene |
 |
Standard sample | [131] |
A. Virus
As discussed previously, GFET-based sensing systems are one of the beneficial alternatives for the early detection of viral infections like the ongoing universal pandemic [22], [87], [88]. There are four strategies for identifying viruses without using labels. In the first approach, the whole virus is being studied using virus-specific antibodies as the capturing probes. While the target in the second approach is viral antigens like its surface proteins and the probe is the antibodies against them. The third approach targets the viral genomic material utilizing its complementary nucleotides and the fourth is designed to capture the antibodies produced in the host body as a response to the virus employing the matching antigens as BREs [89]–[91]. Overall, the sensing scheme is founded on measuring the electrical characteristics of the GFET’s gate after the occurrence of biological reactions [86]. Recently, several studies have focused on detecting viruses such as COVID-19, Encephalitis Virus (JEV), Avian Influenza Virus (AIV), Vesicular Stomatitis Indiana Virus (VSV), Rotavirus, Human Papillomavirus (HPV), and Ebola exploiting GFET. For example, an innovative GFET-based immunosensing system was developed for the early identification of SARS-CoV-2 in nasopharyngeal swab samples. The graphene-modified surface of the sensor was functionalized by antibodies against the spike protein of the virus. The LOD of this device was reported
 2 copies/mL in biological specimens which is a promising achievement in early detection of this viral infection [17]. Or In a recent study, a GFET was utilized to sense the interaction between COVID-19 spike protein S1- and its specific antibody in less than two minutes. They reached a limit of detection of 0.2 pM [92]. Another example of GFET biosensor for virus detection can be seen in Roberts et al. study (See Fig. 4). They designed a miniaturized and easy-to-use device for recognizing JEV and AIV. The Si/SiO2 surface of the FET was coated with graphene which itself was decorated with carboxy groups. These functional groups facilitated the immobilization of virus-specific antibodies through covalent bonding. Because of the interaction between the antibody and the target antigen, a variation in the resistance occurred and LODs down to 1 fM and 10 fM for JEV and AIV were recorded, respectively [93]. Chen and colleagues constructed an aptamer-modified GFET by Micro-electromechanical system (MEMS) for detecting influenza virus (IV). This system was able to sense as low as 1 ng/ml of the target analyte [94]. In another study, a GFET-based immunosensor was fabricated for VSV detection. The graphene was functionalized with antibodies for capturing the target viral particles using 1-pyrenebutanoic acid succinimidyl ester (PASE) as the linker. This molecule is commonly employed on graphene-based surfaces, since it attaches via
2 copies/mL in biological specimens which is a promising achievement in early detection of this viral infection [17]. Or In a recent study, a GFET was utilized to sense the interaction between COVID-19 spike protein S1- and its specific antibody in less than two minutes. They reached a limit of detection of 0.2 pM [92]. Another example of GFET biosensor for virus detection can be seen in Roberts et al. study (See Fig. 4). They designed a miniaturized and easy-to-use device for recognizing JEV and AIV. The Si/SiO2 surface of the FET was coated with graphene which itself was decorated with carboxy groups. These functional groups facilitated the immobilization of virus-specific antibodies through covalent bonding. Because of the interaction between the antibody and the target antigen, a variation in the resistance occurred and LODs down to 1 fM and 10 fM for JEV and AIV were recorded, respectively [93]. Chen and colleagues constructed an aptamer-modified GFET by Micro-electromechanical system (MEMS) for detecting influenza virus (IV). This system was able to sense as low as 1 ng/ml of the target analyte [94]. In another study, a GFET-based immunosensor was fabricated for VSV detection. The graphene was functionalized with antibodies for capturing the target viral particles using 1-pyrenebutanoic acid succinimidyl ester (PASE) as the linker. This molecule is commonly employed on graphene-based surfaces, since it attaches via
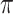 -
-
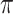 interactions and covalent linkage to the graphene substrate and antibody’s primary amine, respectively. This device was successful enough to identify down to 47.8 aM of the virus [95]. Pant and coworkers designed an rGO-modified FET biosensing platform for Rota-virus recognition. The employment of graphene on the sensor’s surface enabled an efficient antibody immobilization through a linking molecule. Pyrene-NHS which is commonly used in functionalizing graphene-based surfaces was utilized to link the probes to the surface of the sensing site. After the generation of the antibody-antigen complex, a change in conductance was recorded which validated the success of detection [96]. The detection of HPV has also been studied by rGO-FET biosensors. Aspermair et al. modified the surface with RNA aptamers using pyrene and reached a LOD of 1.75 nM in saliva samples [97]. The next example study is done by Chen et al. for detecting the Ebola virus. They fabricated an immunosensor based on rGO-FET and identified down to 1 nM of Ebola glycoprotein in PBS, human serum, and plasma samples [98]. These achievements represent the applicability of graphene-modified FET biosensors for virus detection diagnosis especially COVID-19.
interactions and covalent linkage to the graphene substrate and antibody’s primary amine, respectively. This device was successful enough to identify down to 47.8 aM of the virus [95]. Pant and coworkers designed an rGO-modified FET biosensing platform for Rota-virus recognition. The employment of graphene on the sensor’s surface enabled an efficient antibody immobilization through a linking molecule. Pyrene-NHS which is commonly used in functionalizing graphene-based surfaces was utilized to link the probes to the surface of the sensing site. After the generation of the antibody-antigen complex, a change in conductance was recorded which validated the success of detection [96]. The detection of HPV has also been studied by rGO-FET biosensors. Aspermair et al. modified the surface with RNA aptamers using pyrene and reached a LOD of 1.75 nM in saliva samples [97]. The next example study is done by Chen et al. for detecting the Ebola virus. They fabricated an immunosensor based on rGO-FET and identified down to 1 nM of Ebola glycoprotein in PBS, human serum, and plasma samples [98]. These achievements represent the applicability of graphene-modified FET biosensors for virus detection diagnosis especially COVID-19.
Fig. 4.
(A) Illustration of modifying the graphene surface with virus-specific antibody including, exfoliated graphene, EDC-NHS activated surface which contains carboxylic groups, and antibody-immobilization via its amine groups, (B) SEM images graphene and graphene-Ab. (Reconstructed from [93]).
B. Bacteria
The severe disorders caused by toxins released from different strains of bacteria can lead to lethal consequences [99]. The postponement of the therapeutic process after the initiation of toxicity symptoms may result in the worsening of the patient’s health status [100]. Therefore, it is imperative to identify the infection in its early stages in order to prevent the progress and control it [101]. The currently used techniques for this purpose like polymerase chain reaction (PCR) and colony-counting methodology are time-consuming, expensive, not sensitive, and selective enough [102]. GFETs are capable of conquering these limitations since they are highly scalable, accurate, reliable, economical, and simple. For instance, a pioneering GFET-based immunosensor was developed for bacteria detection to control food and water sanitary. Bacteria-specific antibodies were immobilized using PBASE which could successfully recognize the target biomolecules. Furthermore, they integrated the biosensor with a signal display device to show the variations of the bias current
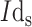 which increased after the attachment of the bacteria [103]. As it can be seen in Fig. 5, another GFET immunosensing platform was developed for Escherichia coli O157:H7 detection. It was reported that the conductance of the sensor’s surface increases after the target antigen attaches to the surface probes. Because of the p-type operation of the GFET and the concentration of the negatively-charged bacteria on the sensing region, an increase in the conductance occurred which augmented the current [104]. Thakur and coworkers reported the use of an rGO-functionalized FET immunosensor for label-free identification of E.coli bacteria. A thin layer of Al2O3 was coated on top of the rGO in order to facilitate easy immobilization of antibody-anchored gold nanoparticles. Attributable to the negatively-charged nature of the target species, the density of charge carriers on the rGO layer increased which enabled rapid (50 s) quantification of E. coli in river water samples [105]. The success of these works validate the adoptability of their strategies in redesigning these devices for recognizing COVID-19 in biological fluids. For example, by replacing the bacteria-specific antibodies by SARS-CoV-2-selective antibodies, antigens, oligonucleotides, or aptamers, innovative platforms can be fabricated for early detection of this disastrous viral infection.
which increased after the attachment of the bacteria [103]. As it can be seen in Fig. 5, another GFET immunosensing platform was developed for Escherichia coli O157:H7 detection. It was reported that the conductance of the sensor’s surface increases after the target antigen attaches to the surface probes. Because of the p-type operation of the GFET and the concentration of the negatively-charged bacteria on the sensing region, an increase in the conductance occurred which augmented the current [104]. Thakur and coworkers reported the use of an rGO-functionalized FET immunosensor for label-free identification of E.coli bacteria. A thin layer of Al2O3 was coated on top of the rGO in order to facilitate easy immobilization of antibody-anchored gold nanoparticles. Attributable to the negatively-charged nature of the target species, the density of charge carriers on the rGO layer increased which enabled rapid (50 s) quantification of E. coli in river water samples [105]. The success of these works validate the adoptability of their strategies in redesigning these devices for recognizing COVID-19 in biological fluids. For example, by replacing the bacteria-specific antibodies by SARS-CoV-2-selective antibodies, antigens, oligonucleotides, or aptamers, innovative platforms can be fabricated for early detection of this disastrous viral infection.
Fig. 5.
Configuration of GFET biosensor for detection of E. coli bacteria. (Reconstructed from [104]).
C. Nucleic Acid
Swift and reliable nucleic acid quantification are vital for biomedical diagnostics which can replace the conventional screening techniques [106], [107]. Owing to the superior biocompatibility, high conductivity, chemical, and mechanical stamina, and large available surface area, graphene-modified bioFETs are considered promising methods for single-molecule detection of nucleic acids [108]. By functionalizing the surface of these miniaturized devices with complementary oligonucleotides or aptamers, the target biomolecules can be captured readily [109]. In recent years, several studies have highlighted the importance of designing such platforms for DNA/RNA measurement. As an example, Hwang et al. employed a unique GFET for nucleic acid identification. Fig. 6 illustrates that a malformed single layer of graphene in the channel region was employed which demonstrated superior performance in comparison to conventional flat designs. It could sense down to 600 zM and 20 aM of the target in the buffer and human serum specimens. It was reported that the novel configuration of the graphene resulted in ultrasensitive detection owing to the reduction in charge screening at the indented sites. Additionally, it displayed an extraordinary shift in the source-drain current which shows the capability of this structure in detecting small biomolecules in millimeter-scale devices [110]. The other example is a biosensing platform for RNA detection based on graphene-modified FET. This device demonstrated superior performance and identified down to 0.1 fM of the analyte. It also differentiated the target RNA from other molecules efficaciously [111]. If the probes of theses developed devices are replaced with COVID-19-specific biorecognition elements like the complementary RNA of this virus or the related aptamer, it can be detected at its incubation time. In this way, the infection can be controlled and better treated.
Fig. 6.
Fabrication of GFETs on flat (A) and crumpled graphene (B), (C) SEM image of crumpled graphene (The scale bar is 500 nm). (Reconstructed from [110]).
D. Cancer Biomarker
Ultra-sensitive measurement of cancer-associated biomolecules in biological fluids such as proteins, enzymes, nucleic acids, and exosomes is essential in the early detection and monitoring of cancer [112]. Although the sample containing these tiny molecules is complex and their concentration is very low especially in early-stage cancer patients, nano-bioelectronic devices have the potential to sense them accurately [113]. The advancement of GFET in recent years paved the way for the emergence of pioneering nano biosensors for accomplishing this goal. As an instance, a novel GFET device was introduced for detecting prostate-specific antigen (PSA). This aptasensor was modified with polyethylene glycol (PEG) in order to ease the DNA aptamers’ immobilization process and reached a LOD of 1 nM (See Fig. 7) [114]. Alpha-fetoprotein (AFP) – known as the hepatocellular carcinoma (HCC) biomarker - is the other biomolecule that has been measured by a tailor-made GFET device. The anti-AFP antibodies were immobilized on the surface of the graphene through PBASE and detected as low as 12.9 ng.mL−1 and 0.1 ng.mL−1 of the target analyte in HCC patients’ serum and buffer, respectively [115]. Zhou and colleagues introduced an accurate and easy-to-use GFET device for recognizing carcinoembryonic antigen (CEA). The surface functionalization was done using nano-denatured bovine serum albumin (nano-dBSA) to both facilitate the anti-CEA immobilization on the EDC and sulfo-NHS-activated graphene channel and protect the sensing site against contamination. This arrangement could detect 337.58 fg mL−1 of the target and validated the functionality of this surface modification strategy in designing biosensing systems [116]. A GFET biosensor was arranged for recognizing human carbonic anhydrase 1 (CA1) using RNA aptamers as capturing probes. PBASE was the linking molecule for aptamer immobilization which bonded with graphene through
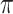 -
-
 interactions. This methodology was capable of detecting low concentrations (70 pM) of the target oncobiomarker which in could act as an alternative technique in the early detection of diseases related to variations in CA1 level [117]. A similar strategy was used to functionalize the surface of a GFET with anti-CD63 antibodies for detecting exosomes. They integrated the sensor with a microfluidic channel and left a section of the graphene surface uncovered to become exposed to the sample. In the presence of exosomes,
interactions. This methodology was capable of detecting low concentrations (70 pM) of the target oncobiomarker which in could act as an alternative technique in the early detection of diseases related to variations in CA1 level [117]. A similar strategy was used to functionalize the surface of a GFET with anti-CD63 antibodies for detecting exosomes. They integrated the sensor with a microfluidic channel and left a section of the graphene surface uncovered to become exposed to the sample. In the presence of exosomes,
 shifted with time which represented the detection of target biomolecules [118]. Such simple and accurate devices have the potential to substitute the conventional screening techniques for the early detection of critical disorders like cancer and critical infectious diseases.
shifted with time which represented the detection of target biomolecules [118]. Such simple and accurate devices have the potential to substitute the conventional screening techniques for the early detection of critical disorders like cancer and critical infectious diseases.
Fig. 7.
(A) Schematic of a GFET device for sensing the biological analyte. (B) The image of a chip-based system on which a PDMS microfluidic channel is mounted. (Reconstructed from [114]).
E. Hormone
Measuring the level of hormones is critical in the prevention, detection, and monitoring of some diseases [119], [120]. During the last few years, GFET biosensors have also been used for this purpose. For instance, a GFET-based immunosensor was fabricated to detect thyroid-stimulating hormone (TSH) in a complicated biological sample. The surface modification was done using polyethylene glycol (PEG) and TSH-specific antibody fragments. This arrangement was able to detect as low as
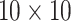 −15 M in serum specimens [121]. Another graphene-based electrolyte-gated FET biosensor for hormone detection was used for quantifying Human Chorionic Gonadotrophin (hCG). The surface was decorated with pyrene-NHS to maintain the sp2 structure and easily immobilize the antibodies. The reported LOD was ~1 pg.mL−1 which demonstrates the potential of graphene-based POC devices for medical applications [122]. In another study, a GFET-based cortisol sensing platform was constructed that can sense down to 10 pg/ml of the target in human tears. This real-time detection was integrated with a smartphone in order to ease the process. The reliability, repeatability, and biocompatibility of this design were validated using live rabbit and human pilot tests [123]. Fig. 8 presents anti-diuretic hormone (ADH) detection utilizing a GFET-based aptasensor. As a result of the captured target peptides by the immobilized aptamers, the density of charge carriers changed which showed the occurrence of detection, and a LOD of 3.55 ag/mL was recorded [124]. By substituting the biorecognition elements of these tailored devices, it is feasible to design a specific biosensing system for spotting SARS-CoV-2-related biomarkers.
−15 M in serum specimens [121]. Another graphene-based electrolyte-gated FET biosensor for hormone detection was used for quantifying Human Chorionic Gonadotrophin (hCG). The surface was decorated with pyrene-NHS to maintain the sp2 structure and easily immobilize the antibodies. The reported LOD was ~1 pg.mL−1 which demonstrates the potential of graphene-based POC devices for medical applications [122]. In another study, a GFET-based cortisol sensing platform was constructed that can sense down to 10 pg/ml of the target in human tears. This real-time detection was integrated with a smartphone in order to ease the process. The reliability, repeatability, and biocompatibility of this design were validated using live rabbit and human pilot tests [123]. Fig. 8 presents anti-diuretic hormone (ADH) detection utilizing a GFET-based aptasensor. As a result of the captured target peptides by the immobilized aptamers, the density of charge carriers changed which showed the occurrence of detection, and a LOD of 3.55 ag/mL was recorded [124]. By substituting the biorecognition elements of these tailored devices, it is feasible to design a specific biosensing system for spotting SARS-CoV-2-related biomarkers.
Fig. 8.
Illustration of the surface functionalization process of a GFET biosensor. 1. KOH activation, 2.APTES, and 3. Glutaraldehyde (GA) treatment, and 4. ADH-specific aptamer immobilization. (Reconstructed from [124]).
F. Other
GFET sensing devices are being utilized for identifying other types of biomolecules as well. For example, Ferritin which is known as a biomarker for early detection of iron deficiency (ID) has been studied employing a GFET immunosensor. This PBASE-anchored graphene-based sensing system was able to detect as low as 10 fM of the ferritin antigen [125]. The other GFET device was designed by Wang et al. to detect biotin. The sensing site of this system was decorated with PBASE cross-linkers to facilitate the attachment of avidin. This immobilization was based on the affinity between the lysine group of avidin and the N-hydroxysuccinimide ester group of PBASE. After anchoring the probes on the monolayer of graphene and introducing the target-containing sample, the current variations were monitored in a real-time manner. This novel structure was capable of selectively capturing the desired target biomolecules with a 0.37 pM sensitivity [126]. Smell sensors are the other category of sensing platforms designed utilizing GFET technology. The target analyte of these devices is a tiny and lipophilic structure like homovanillic acid, eugenol, and methyl vanillate. In a recent research work, an odorant-binding protein-modified GFET biosensor was introduced by Rozman et al. which was able to spot down to 100pM of the target analyte [127]. In another similar study, an rGO-coated FET biosensor was developed for identifying odorants in water-based solutions. Odorant-binding protein 14 (OBP14) was used as the capturing probe that can bind to the hydroxyl group of the target aromatic molecules. Analyzing the electrical measurement results indicated that all the probe proteins were at their optimum functionality such that they can identify the target sensitively and selectively [128]. In a patented work, an odorant biosensor was constructed based on a ligand binding protein-anchored GFET. The use of a lipid bilayer on the first graphene layer of this structure made the immobilization process easier [129]. Zheng and coworkers presented a precise FET biosensor for measuring hydrogen peroxide (H2O2). They used molybdenum disulfide (MoS2) and rGO to modify the sensing area. This methodology demonstrated a sensitive and selective detection in a complex sample. Additionally, it could directly sense the H2O2 produced by cancer cells which is a prominent improvement in monitoring H2O2-associated malfunctions [130]. In another investigation, the possibility of sensitive measurement of lead and potassium ions by a GFET aptasensor was examined. A methylene blue (MB) molecule was attached to the terminal end of the probes. The binding of target ions to the specific aptamers resulted in a shape change and accordingly increased the proximity of the MB to the surface. as a result, an electron was donated and the concentration of the target analyte was quantified by measuring the current [131].
The success of these novel platforms shows the functionality of GFET biosensors for detecting COVID-19-related biomarkers such as surface proteins (spike (S), nucleocapsid (N), membrane (M), and envelope (E)), viral RNA, and host antibodies. For this purpose, the sensing region of the GFET biosensor should be functionalized with capture probes to identify the target analytes. For example, tailor-made antibodies against the viral antigens, complementary strands of an oligonucleotide against the genomic material of the virus, and selective aptamers can be designed and immobilized on the surface of LIG which has many binding sites. Decorating the graphene surface of these devices with bioreceptors can lead to the detection of COVID-19- specific biomarkers even in the most complex samples.
IV. Conclusion
To conclude, the huge potential of GFET-based biosensors for early detection of different biomarkers, including COVID-19-related biomolecules specified in this review paper. Besides, the importance of choosing an efficient graphene fabrication methodology which is an essential parameter in designing a GFET biosensing device was discussed. Among the diverse methods of graphene fabrication, LIG is an advantageous technique since it provides thermal stability, wide available surface area, high electrical conductivity, and cost-effectiveness. These attributions turn it into a popular and appropriate candidate for biosensing applications. Even though these compact, scalable, and ultra-sensitive GFET biosensors are attracting the attention of researchers in the field of early-phase disease detection especially during the past few years, there is still a burdensome track in front of its widespread use in biomedical fields. For instance, synthesizing graphene sheets with uniform properties is arduous and a minor discrepancy in their structure may cause a major change in the performance of the device. Furthermore, the baseline drift in the aqueous milieu is a common issue that these sensing platforms often experience. It complicates the process of response analysis which is not a desirable feature while using a biosensor. Another point that needs to be taken into account is the reusability of the GFET biosensor, particularly when it is utilized as a POC device. For a biosensor to be used over and over again it should minimize the probability of cross-contamination. One simple solution for this problem is cartridge-type GFET biosensors. The disposability of the cartridge and the reusability of the readout system enable using the device several times. However, currently developed GFET Biosystems lack this feature. Thus, constructing such a device would be beneficial in terms of COVID-19 detection, since it can be used numerous times and examine a large number of people. One other point is the sample treatment and delivery to the sensing region which can be done through integrating microfluidics with GFET technology. Innovative designs can be created to easily deliver the sample to be tested with microfluidic-based GFETs. To reach this ultimate goal, effective collaboration between scientists from different fields of study such as electrical engineering, chemistry, physics, nanotechnology, and medicine is required. All in all, LIG is a unique technique that can tackle the current limitations in graphene fabrication and transfer to the sensing area of a biosensing system, particularly GFETs. Thus LIG-based GFET biosensors hold great potential in sensing COVID-19-specific biomolecules in biological samples. They are expected to be one of the most preferred and widely used detection techniques in the near future after further improvements.
Contributor Information
Deniz Sadighbayan, Email: denizsdg@yorku.ca.
Aamir Minhas-Khan, Email: aminhask@yorku.ca.
Ebrahim Ghafar-Zadeh, Sr., Email: egz@yorku.ca.
References
- [1].Worldometer, COVID-19 Coronavirus Pandemic. [Online]. Available: https://www.worldometers.info/coronavirus/?utm_campaign=homeAdvegas1?
- [2].Wilder-Smith A., “COVID-19 in comparison with other emerging viral diseases: Risk of geographic spread via travel,” Tropical Diseases, Travel Med. Vaccines, vol. 7, no. 1, pp. 1–11, Jan. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Li H., Liu S.-M., Yu X.-H., Tang S.-L., and Tang C.-K., “Coronavirus disease 2019 (COVID-19): Current status and future perspectives,” Int. J. Antimicrobial Agents, vol. 55, no. 5, May 2020, Art. no. 105951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Mousavizadeh L. and Ghasemi S., “Genotype and phenotype of COVID-19: Their roles in pathogenesis,” J. Microbiol., Immunol. Infection, vol. 54, pp. 159–163, Mar. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Mittal A., Manjunath K., Ranjan R. K., Kaushik S., Kumar S., and Verma V., “COVID-19 pandemic: Insights into structure, function, and hACE2 receptor recognition by SARS-CoV-2,” PLoS Pathogens, vol. 16, no. 8, Aug. 2020, Art. no. e1008762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Sternberg A. and Naujokat C., “Structural features of coronavirus SARS-CoV-2 spike protein: Targets for vaccination,” Life Sci., vol. 257, Sep. 2020, Art. no. 118056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Walls A. C., Park Y. J., Tortorici M. A., Wall A., McGuire A. T., and Veesler D., “Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein,” Cell, vol. 181, no. 2, pp. 281–292, Apr. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Xu G.et al. , “Clinical pathway for early diagnosis of COVID-19: Updates from experience to evidence-based practice,” Clin. Rev. Allergy Immunol., vol. 59, no. 1, pp. 89–100, Aug. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Udugama B.et al. , “Diagnosing COVID-19: The disease and tools for detection,” ACS Nano, vol. 14, no. 4, pp. 3822–3835, Apr. 2020. [DOI] [PubMed] [Google Scholar]
- [10].Afzal A., “Molecular diagnostic technologies for COVID-19: Limitations and challenges,” J. Adv. Res., vol. 26, pp. 149–159, Nov. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Umakanthan S., Chattu V. K., Ranade A. V., Das D., Basavarajegowda A., and Bukelo M., “A rapid review of recent advances in diagnosis, treatment and vaccination for COVID-19,” AIMS Public Health, vol. 8, no. 1, pp. 137–153, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Parihar A., Ranjan P., Sanghi S. K., Srivastava A. K., and Khan R., “Point-of-care biosensor-based diagnosis of COVID-19 holds promise to combat current and future pandemics,” ACS Appl. Bio Mater., vol. 3, no. 11, pp. 7326–7343, Nov. 2020. [DOI] [PubMed] [Google Scholar]
- [13].Xu L., Li D., Ramadan S., Li Y., and Klein N., “Facile biosensors for rapid detection of COVID-19,” Biosensors Bioelectron., vol. 170, Dec. 2020, Art. no. 112673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Sadighbayan D. and Ghafar-Zadeh E., “Portable sensing devices for detection of COVID-19: A review,” IEEE Sensors J., vol. 21, no. 9, pp. 10219–10230, May 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Poghossian A., Jablonski M., Molinnus D., Wege C., and Schöning M. J., “Field-effect sensors for virus detection: From ebola to SARS-CoV-2 and plant viral enhancers,” Frontiers Plant Sci., vol. 11, Nov. 2020, Art. no. 598103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Vu C. A. and Chen W. Y., “Field-effect transistor biosensors for biomedical applications: Recent advances and future prospects,” Sensors, vol. 19, no. 19, p. 4214, Sep. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Seo G.et al. , “Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor,” ACS Nano, vol. 14, no. 4, pp. 5135–5142, 2020. [DOI] [PubMed] [Google Scholar]
- [18].Bhalla N., Jolly P., Formisano N., and Estrela P., “Introduction to biosensors,” Essays Biochem., vol. 60, pp. 1–8, May 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Forsyth R., Devadoss A., and Guy O., “Graphene field effect transistors for biomedical applications: Current status and future prospects,” Diagnostics, vol. 7, no. 3, p. 45, Jul. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Bai Y., Xu T., and Zhang X., “Graphene-based biosensors for detection of biomarkers,” Micromachines, vol. 11, no. 1, p. 60, Jan. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Szunerits S. and Boukherroub R., “Graphene-based biosensors,” Interface Focus, vol. 8, no. 3, Jun. 2018, Art. no. 20160132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Sengupta J. and Hussain C. M., “Graphene-based field-effect transistor biosensors for the rapid detection and analysis of viruses: A perspective in view of COVID-19,” Carbon Trends, vol. 2, Jan. 2021, Art. no. 100011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Viswanathan S.et al. , “Graphene–protein field effect biosensors: Glucose sensing,” Mater. Today, vol. 18, no. 9, pp. 513–522, 2015. [Google Scholar]
- [24].Choi J. R., “Development of point-of-care biosensors for COVID-19,” Frontiers Chem., vol. 8, p. 517, May 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Huang Y., Dong X., Shi Y., Li C. M., Li L. J., and Chen P., “Nanoelectronic biosensors based on CVD grown graphene,” Nanoscale, vol. 2, no. 8, pp. 1485–1488, Aug. 2010. [DOI] [PubMed] [Google Scholar]
- [26].Park S., Shehzad M. A., Choi W., Kwon Y. H., Eom J., and Seo Y., “Simple photolithography process for a graphene-based device with edge contact,” Nanosci. Nanotechnol. Lett., vol. 10, no. 8, pp. 1072–1079, Aug. 2018. [Google Scholar]
- [27].Cabral P. D.et al. , “Clean-room lithographical processes for the fabrication of graphene biosensors,” Materials, vol. 13, no. 24, p. 5728, Dec. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Saeed M., Alshammari Y., Majeed S. A., and Al-Nasrallah E., “Chemical vapour deposition of graphene—Synthesis, characterisation, and applications: A review,” Molecules, vol. 25, no. 17, p. 3856, Aug. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Shi X., Zhou F., Peng J., Wu R., Wu Z., and Bao X., “One-step scalable fabrication of graphene-integrated micro-supercapacitors with remarkable flexibility and exceptional performance uniformity,” Adv. Funct. Mater., vol. 29, no. 50, Dec. 2019, Art. no. 1902860. [Google Scholar]
- [30].Wang Y., Wang Y., Zhang P., Liu F., and Luo S., “Laser-induced freestanding graphene papers: A new route of scalable fabrication with tunable morphologies and properties for multifunctional devices and structures,” Small, vol. 14, no. 36, Sep. 2018, Art. no. 1802350. [DOI] [PubMed] [Google Scholar]
- [31].Zhen Z. and Zhu H., “Structure and properties of graphene,” Graphene, pp. 1–12, 2018.
- [32].Novoselov K. S.et al. , “Electric field effect in atomically thin carbon films,” Science, vol. 306, no. 5696, pp. 666–669, 2004. [DOI] [PubMed] [Google Scholar]
- [33].Yang G., Li L., Lee W. B., and Ng M. C., “Structure of graphene and its disorders: A review,” Sci. Technol. Adv. Mater., vol. 19, no. 1, pp. 613–648, Dec. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Whitener K. E. and Sheehan P. E., “Graphene synthesis,” Diamond Rel. Mater., vol. 46, pp. 25–34, Jun. 2014. [Google Scholar]
- [35].Wang X.-Y., Narita A., and Müllen K., “Precision synthesis versus bulk-scale fabrication of graphenes,” Nature Rev. Chem., vol. 2, no. 1, pp. 1–10, 2017. [Google Scholar]
- [36].Chauhan N., Maekawa T., and Kumar D. N. S., “Graphene based biosensors—Accelerating medical diagnostics to new-dimensions,” J. Mater. Res., vol. 32, no. 15, pp. 2860–2882, Aug. 2017. [Google Scholar]
- [37].Yi M. and Shen Z., “A review on mechanical exfoliation for the scalable production of graphene,” J. Mater. Chem. A, vol. 3, no. 22, pp. 11700–11715, 2015. [Google Scholar]
- [38].Chen J., Duan M., and Chen G., “Continuous mechanical exfoliation of graphene sheets via three-roll mill,” J. Mater. Chem., vol. 22, no. 37, p. 19625, 2012. [Google Scholar]
- [39].Dreyer D. R., Park S., Bielawski C. W., and Ruoff R. S., “The chemistry of graphene oxide,” Chem. Soc. Rev., vol. 39, no. 1, pp. 228–240, 2010. [DOI] [PubMed] [Google Scholar]
- [40].Qiao Y.et al. , “Graphene-based wearable sensors,” Nanoscale, vol. 11, no. 41, pp. 18923–18945, Nov. 2019. [DOI] [PubMed] [Google Scholar]
- [41].Terse-Thakoor T., Badhulika S., and Mulchandani A., “Graphene based biosensors for healthcare,” J. Mater. Res., vol. 32, no. 15, pp. 2905–2929, Aug. 2017. [Google Scholar]
- [42].Parvez K., Yang S., Feng X., and Müllen K., “Exfoliation of graphene via wet chemical routes,” Synth. Met., vol. 210, pp. 123–132, Dec. 2015. [Google Scholar]
- [43].Mei X. and Ouyang J., “Ultrasonication-assisted ultrafast reduction of graphene oxide by zinc powder at room temperature,” Carbon, vol. 49, no. 15, pp. 5389–5397, 2011. [Google Scholar]
- [44].Liang Y.et al. , “Transparent, highly conductive graphene electrodes from acetylene-assisted thermolysis of graphite oxide sheets and nanographene molecules,” Nanotechnology, vol. 20, no. 43, Oct. 2009, Art. no. 434007. [DOI] [PubMed] [Google Scholar]
- [45].Berger C.et al. , “Electronic confinement and coherence in patterned epitaxial graphene,” Science, vol. 312, no. 5777, pp. 1191–1196, Apr. 2006. [DOI] [PubMed] [Google Scholar]
- [46].Hass J., de Heer W. A., and Conrad E. H., “The growth and morphology of epitaxial multilayer graphene,” J. Phys., Condens. Matter, vol. 20, no. 32, Aug. 2008, Art. no. 323202. [Google Scholar]
- [47].Liu Z.et al. , “Laser-induced growth of large-area epitaxial graphene with low sheet resistance on 4H-SiC (0001),” Appl. Surf. Sci., vol. 514, Jun. 2020, Art. no. 145938. [Google Scholar]
- [48].Sutter P. W., Flege J. I., and Sutter E. A., “Epitaxial graphene on ruthenium,” Nature Mater., vol. 7, no. 5, pp. 406–411, May 2008. [DOI] [PubMed] [Google Scholar]
- [49].Tran T.-T. and Mulchandani A., “Carbon nanotubes and graphene nano field-effect transistor-based biosensors,” TrAC Trends Anal. Chem., vol. 79, pp. 222–232, May 2016. [Google Scholar]
- [50].Novodchuk I., Bajcsy M., and Yavuz M., “Graphene-based field effect transistor biosensors for breast cancer detection: A review on biosensing strategies,” Carbon, vol. 172, pp. 431–453, Feb. 2021. [Google Scholar]
- [51].Wu Y.et al. , “200 GHz maximum oscillation frequency in CVD graphene radio frequency transistors,” ACS Appl. Mater. Interfaces, vol. 8, no. 39, pp. 25645–25649, Sep. 2016. [DOI] [PubMed] [Google Scholar]
- [52].Tu J.et al. , “Graphene FET array biosensor based on ssDNA aptamer for ultrasensitive Hg2+ detection in environmental pollutants,” Frontiers Chem., vol. 6, p. 333, Aug. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Gao Z.et al. , “Scalable production of sensor arrays based on high-mobility hybrid graphene field effect transistors,” ACS Appl. Mater. Interfaces, vol. 8, no. 41, pp. 27546–27552, Oct. 2016. [DOI] [PubMed] [Google Scholar]
- [54].Yue W.et al. , “An electricity-fluorescence double-checking biosensor based on graphene for detection of binding kinetics of DNA hybridization,” RSC Adv., vol. 7, no. 70, pp. 44559–44567, 2017. [Google Scholar]
- [55].Xu S.et al. , “Ultrasensitive label-free detection of DNA hybridization by sapphire-based graphene field-effect transistor biosensor,” Appl. Surf. Sci., vol. 427, pp. 1114–1119, Jan. 2018. [Google Scholar]
- [56].Zheng C., Huang L., Zhang H., Sun Z., Zhang Z., and Zhang G.-J., “Fabrication of ultrasensitive field-effect transistor DNA biosensors by a directional transfer technique based on CVD-grown graphene,” ACS Appl. Mater. Interfaces, vol. 7, no. 31, pp. 16953–16959, Aug. 2015. [DOI] [PubMed] [Google Scholar]
- [57].Chen S., Sun Y., Xia Y., Lv K., Man B., and Yang C., “Donor effect dominated molybdenum disulfide/graphene nanostructure-based field-effect transistor for ultrasensitive DNA detection,” Biosensors Bioelectron., vol. 156, May 2020, Art. no. 112128. [DOI] [PubMed] [Google Scholar]
- [58].Zhao W., Fang M., Wu F., Wu H., Wang L., and Chen G., “Preparation of graphene by exfoliation of graphite using wet ball milling,” J. Mater. Chem., vol. 20, no. 28, p. 5817, 2010. [Google Scholar]
- [59].Pham V. H., Hur S. H., Kim E. J., Kim B. S., and Chung J. S., “Highly efficient reduction of graphene oxide using ammonia borane,” Chem. Commun., vol. 49, no. 59, pp. 6665–6667, Jul. 2013. [DOI] [PubMed] [Google Scholar]
- [60].Wang S., Ang P. K., Wang Z., Tang A. L. L., Thong J. T. L., and Loh K. P., “High mobility, printable, and solution-processed graphene electronics,” Nano Lett., vol. 10, no. 1, pp. 92–98, Jan. 2010. [DOI] [PubMed] [Google Scholar]
- [61].Ouerghi A.et al. , “Epitaxial graphene on cubic SiC(111)/Si(111) substrate,” Appl. Phys. Lett., vol. 96, no. 19, May 2010, Art. no. 191910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Ferrari A. C.et al. , “Raman spectrum of graphene and graphene layers,” Phys. Rev. Lett., vol. 97, no. 18, Oct. 2006, Art. no. 187401. [DOI] [PubMed] [Google Scholar]
- [63].Kim J., Ishihara M., Koga Y., Tsugawa K., Hasegawa M., and Iijima S., “Low-temperature synthesis of large-area graphene-based transparent conductive films using surface wave plasma chemical vapor deposition,” Appl. Phys. Lett., vol. 98, no. 9, Feb. 2011, Art. no. 091502. [Google Scholar]
- [64].Baraket M.et al. , “Aminated graphene for DNA attachment produced via plasma functionalization,” Appl. Phys. Lett., vol. 100, no. 23, Jun. 2012, Art. no. 233123. [Google Scholar]
- [65].Le Quang T.et al. , “Epitaxial electrical contact to graphene on SiC,” Carbon, vol. 121, pp. 48–55, Sep. 2017. [Google Scholar]
- [66].Vanegas D.et al. , “Laser scribed graphene biosensor for detection of biogenic amines in food samples using locally sourced materials,” Biosensors, vol. 8, no. 2, p. 42, Apr. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Li X.et al. , “High-voltage flexible microsupercapacitors based on laser-induced graphene,” ACS Appl. Mater. Interfaces, vol. 10, no. 31, pp. 26357–26364, 2018. [DOI] [PubMed] [Google Scholar]
- [68].Marques A. C., Cardoso A. R., Martins R., Sales M. G. F., and Fortunato E., “Laser-induced graphene-based platforms for dual biorecognition of molecules,” ACS Appl. Nano Mater., vol. 3, no. 3, pp. 2795–2803, Mar. 2020. [Google Scholar]
- [69].Lin J.et al. , “Laser-induced porous graphene films from commercial polymers,” Nature Commun., vol. 5, no. 1, pp. 1–8, Dec. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Minhas-Khan A., Nambi S., and Grau G., “Low-resistance laser-induced graphitic carbon by maximizing energy delivery and pulse overlap,” Carbon, vol. 181, pp. 310–322, Aug. 2021. [Google Scholar]
- [71].Cardoso A. R.et al. , “Molecularly-imprinted chloramphenicol sensor with laser-induced graphene electrodes,” Biosensors Bioelectron., vols. 124–125, pp. 167–175, Jan. 2019. [DOI] [PubMed] [Google Scholar]
- [72].Fenzl C., Nayak P., Hirsch T., Wolfbeis O. S., Alshareef H. N., and Baeumner A. J., “Laser-scribed graphene electrodes for aptamer-based biosensing,” ACS Sensors, vol. 2, no. 5, pp. 616–620, May 2017. [DOI] [PubMed] [Google Scholar]
- [73].Yagati A. K.et al. , “Laser-induced graphene interdigitated electrodes for label-free or nanolabel-enhanced highly sensitive capacitive aptamer-based biosensors,” Biosensors Bioelectron., vol. 164, Sep. 2020, Art. no. 112272. [DOI] [PubMed] [Google Scholar]
- [74].Soares R. R. A.et al. , “Laser-induced graphene electrochemical immunosensors for rapid and label-free monitoring of Salmonella enterica in chicken broth,” ACS Sensors, vol. 5, no. 7, pp. 1900–1911, Jul. 2020. [DOI] [PubMed] [Google Scholar]
- [75].Vanegas D. C., Gomes C. L., Cavallaro N. D., Giraldo-Escobar D., and McLamore E. S., “Emerging biorecognition and transduction schemes for rapid detection of pathogenic bacteria in food,” Comprehensive Rev. Food Sci. Food Saf., vol. 16, no. 6, pp. 1188–1205, Nov. 2017. [DOI] [PubMed] [Google Scholar]
- [76].Guo L.et al. , “Bandgap tailoring and synchronous microdevices patterning of graphene oxides,” J. Phys. Chem. C, vol. 116, pp. 3594–3599, Feb. 2012. [Google Scholar]
- [77].He Y.et al. , “Femtosecond laser direct writing of flexible all-reduced graphene oxide FET,” IEEE Photon. Technol. Lett., vol. 28, no. 18, pp. 1996–1999, Sep. 15, 2016. [Google Scholar]
- [78].Ye R., James D. K., and Tour J. M., “Laser-induced graphene: From discovery to translation,” Adv. Mater., vol. 31, no. 1, Jan. 2019, Art. no. 1803621. [DOI] [PubMed] [Google Scholar]
- [79].Ye R., James D. K., and Tour J. M., “Laser-induced graphene,” Accounts Chem. Res., vol. 51, no. 7, pp. 1609–1620, 2018. [DOI] [PubMed] [Google Scholar]
- [80].You R., Liu Y. Q., Hao Y. L., Han D. D., Zhang Y. L., and You Z., “Laser fabrication of graphene-based flexible electronics,” Adv Mater, vol. 32, no. 15, Apr. 2020, Art. no. e1901981. [DOI] [PubMed] [Google Scholar]
- [81].Wan Z., Nguyen N.-T., Gao Y., and Li Q., “Laser induced graphene for biosensors,” Sustain. Mater. Technol., vol. 25, Jul. 2020, Art. no. e00205. [Google Scholar]
- [82].Ghanam A., Lahcen A. A., Beduk T., Alshareef H. N., Amine A., and Salama K. N., “Laser scribed graphene: A novel platform for highly sensitive detection of electroactive biomolecules,” Biosensors Bioelectron, vol. 168, Nov. 2020, Art. no. 112509. [DOI] [PubMed] [Google Scholar]
- [83].Sadighbayan D., Hasanzadeh M., and Ghafar-Zadeh E., “Biosensing based on field-effect transistors (FET): Recent progress and challenges,” TrAC Trends Anal. Chem., vol. 133, Dec. 2020, Art. no. 116067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Syu Y.-C., Hsu W.-E., and Lin C.-T., “Review—Field-effect transistor biosensing: Devices and clinical applications,” ECS J. Solid State Sci. Technol., vol. 7, no. 7, pp. Q3196–Q3207, 2018. [Google Scholar]
- [85].Kimura J. and Kuriyama T., “FET biosensors,” J. Biotechnol., vol. 15, no. 3, pp. 239–254, Aug. 1990. [DOI] [PubMed] [Google Scholar]
- [86].Béraud A., Sauvage M., Bazán C. M., Tie M., Bencherif A., and Bouilly D., “Graphene field-effect transistors as bioanalytical sensors: Design, operation and performance,” Analyst, vol. 146, no. 2, pp. 403–428, Jan. 2021. [DOI] [PubMed] [Google Scholar]
- [87].Matsumoto K., Hayashi R., Kanai Y., Inoue K., and Ono T., “Graphene field-effect transistor for biosensor,” in Proc. 23rd Int. Workshop Active-Matrix Flatpanel Displays Devices, pp. 45–46, 2016. [Google Scholar]
- [88].Cordaro A., Neri G., Sciortino M. T., Scala A., and Piperno A., “Graphene-based strategies in liquid biopsy and in viral diseases diagnosis,” Nanomaterials, vol. 10, no. 6, p. 1014, May 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Saylan Y., Erdem Ö., Ünal S., and Denizli A., “An alternative medical diagnosis method: Biosensors for virus detection,” Biosensors, vol. 9, no. 2, p. 65, May 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Ribeiro B. V., Cordeiro T. A. R., Oliveira e Freitas G. R., Ferreira L. F., and Franco D. L., “Biosensors for the detection of respiratory viruses: A review,” Talanta Open, vol. 2, Dec. 2020, Art. no. 100007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Aznakayev E., Aznakayeva D., and Kim J., “Label-free biosensor for viruses and bacteria detection,” Proc. SPIE, vol. 11378, Apr. 2020, Art. no. 113781G. [Google Scholar]
- [92].Zhang X.et al. , “Electrical probing of COVID-19 spike protein receptor binding domain via a graphene field-effect transistor,” 2020, arXiv:2003.12529. [Online]. Available: http://arxiv.org/abs/2003.12529
- [93].Roberts A.et al. , “Graphene functionalized field-effect transistors for ultrasensitive detection of Japanese encephalitis and avian influenza virus,” Sci. Rep., vol. 10, no. 1, p. 14546, Sep. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Chen Y.-J., Chiang C.-L., and Huang J.-T., “Wireless portable graphene-FET biosensor for detecting H1N1 virus,” in Graphene. Barcelona, Spain, 2017. [Google Scholar]
- [95].Kim J. W., Kim S., Jang Y.-H., Lim K.-I., and Lee W. H., “Attomolar detection of virus by liquid coplanar-gate graphene transistor on plastic,” Nanotechnology, vol. 30, no. 34, Aug. 2019, Art. no. 345502. [DOI] [PubMed] [Google Scholar]
- [96].Pant M., Kharkwa H., Singh K. P., and Joshi H. C., “Detection of rota virus with the help of nanomaterial based field effect transistor (BIO-FET),” Biosensors J., vol. 6, no. 2, 2017, Art. no. 1000148. [Google Scholar]
- [97].Aspermair P.et al. , “Reduced graphene oxide–based field effect transistors for the detection of E7 protein of human papillomavirus in saliva,” Anal. Bioanal. Chem., vol. 413, no. 3, pp. 779–787, Jan. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Chen Y.et al. , “Field-effect transistor biosensor for rapid detection of ebola antigen,” Sci. Rep., vol. 7, no. 1, p. 10974, Sep. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Doron S. and Gorbach S. L., “Bacterial infections: Overview,” Int. Encyclopedia Public Health, pp. 273–282, 2008.
- [100].Tsalik E. L., Bonomo R. A., and Fowler V. G., “New molecular diagnostic approaches to bacterial infections and antibacterial resistance,” Annu. Rev. Med., vol. 69, pp. 379–394, Jan. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Ahmed A., Rushworth J. V., Hirst N. A., and Millner P. A., “Biosensors for whole-cell bacterial detection,” Clin. Microbiol. Rev., vol. 27, no. 3, pp. 631–646, Jul. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Alahi M. and Mukhopadhyay S., “Detection methodologies for pathogen and toxins: A review,” Sensors, vol. 17, no. 8, p. 1885, Aug. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Zhu J., Niu F., Zhu C., Yang J., and Xi N., “Graphene-based FET detector for E. coli K12 real-time monitoring and its theoretical analysis,” J. Sensors, vol. 2016, pp. 1–9, Jan. 2016. [Google Scholar]
- [104].Pourasl A. H., Ahmadi M. T., Rahmani M., and Ismail R., “Graphene based biosensor model for Escherichia coli bacteria detection,” J. Nanosci. Nanotechnol., vol. 17, no. 1, pp. 601–605, Jan. 2017. [DOI] [PubMed] [Google Scholar]
- [105].Thakur B.et al. , “Rapid detection of single E. coli bacteria using a graphene-based field-effect transistor device,” Biosensors Bioelectron., vol. 110, pp. 16–22, Jul. 2018. [DOI] [PubMed] [Google Scholar]
- [106].Teles F. and Fonseca L., “Trends in DNA biosensors,” Talanta, vol. 77, no. 2, pp. 606–623, Dec. 2008. [Google Scholar]
- [107].Fu Z., Lu Y. C., and Lai J. J., “Recent advances in biosensors for nucleic acid and exosome detection,” Chonnam Med. J., vol. 55, no. 2, pp. 86–98, May 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Suvarnaphaet P. and Pechprasarn S., “Graphene-based materials for biosensors: A review,” Sensors, vol. 17, no. 10, p. 2161, Sep. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Paniel N., Baudart J., Hayat A., and Barthelmebs L., “Aptasensor and genosensor methods for detection of microbes in real world samples,” Methods, vol. 64, no. 3, pp. 229–240, Dec. 2013. [DOI] [PubMed] [Google Scholar]
- [110].Hwang M. T.et al. , “Ultrasensitive detection of nucleic acids using deformed graphene channel field effect biosensors,” Nature Commun., vol. 11, no. 1, pp. 1–11, Dec. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Tian M.et al. , “RNA detection based on graphene field-effect transistor biosensor,” Adv. Condens. Matter Phys., vol. 2018, pp. 1–6, Jun. 2018. [Google Scholar]
- [112].Jayanthi V. S. P. K. S. A., Das A. B., and Saxena U., “Recent advances in biosensor development for the detection of cancer biomarkers,” Biosensors Bioelectron., vol. 91, pp. 15–23, May 2017. [DOI] [PubMed] [Google Scholar]
- [113].Zhang Y., Li M., Gao X., Chen Y., and Liu T., “Nanotechnology in cancer diagnosis: Progress, challenges and opportunities,” J. Hematol. Oncol., vol. 12, no. 1, p. 137, Dec. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Gao N.et al. , “Specific detection of biomolecules in physiological solutions using graphene transistor biosensors,” Proc. Nat. Acad. Sci. USA, vol. 113, no. 51, pp. 14633–14638, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Kim D.et al. , “Detection of alpha-fetoprotein in hepatocellular carcinoma patient plasma with graphene field-effect transistor,” Sensors, vol. 18, no. 11, p. 4032, Nov. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Zhou L.et al. , “Novel graphene biosensor based on the functionalization of multifunctional nano-bovine serum albumin for the highly sensitive detection of cancer biomarkers,” Nano-Micro Lett., vol. 11, no. 1, pp. 1–13, Dec. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Kumar N.et al. , “Detection of a multi-disease biomarker in saliva with graphene field effect transistors,” Med. Devices Sensors, vol. 3, no. 6, Dec. 2020, Art. no. e10121. [Google Scholar]
- [118].Kwong Hong Tsang D.et al. , “Chemically functionalised graphene FET biosensor for the label-free sensing of exosomes,” Sci. Rep., vol. 9, no. 1, p. 13946, Sep. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Honour J. W., “Diagnosis of diseases of steroid hormone production, metabolism and action,” J. Clin. Res. Pediatric Endocrinol., vol. 1, no. 5, pp. 209–226, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Golden S. H., Robinson K. A., Saldanha I., Anton B., and Ladenson P. W., “Clinical review: Prevalence and incidence of endocrine and metabolic disorders in the United States: A comprehensive review,” J. Clin. Endocrinol. Metab., vol. 94, no. 6, pp. 1853–1878, Jun. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Andoy N. M., Filipiak M. S., Vetter D., Gutiérrez-Sanz Ó., and Tarasov A., “Graphene-based electronic immunosensor with femtomolar detection limit in whole serum,” Adv. Mater. Technol., vol. 3, no. 12, Dec. 2018, Art. no. 1800186. [Google Scholar]
- [122].Islam K., Suhail A., and Pan G., “A label-free and ultrasensitive immunosensor for detection of human chorionic gonadotrophin based on graphene FETs,” Biosensors, vol. 7, no. 3, p. 27, Jul. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Ku M.et al. , “Smart, soft contact lens for wireless immunosensing of cortisol,” Sci. Adv., vol. 6, no. 28, Jul. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Selvarajan R. S., Rahim R. A., Majlis B. Y., Gopinath S. C. B., and Hamzah A. A., “Ultrasensitive and highly selective graphene-based field-effect transistor biosensor for anti-diuretic hormone detection,” Sensors, vol. 20, no. 9, May 2020, Art. no. eabb2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Oshin O., Kireev D., Hlukhova H., Idachaba F., Akinwande D., and Atayero A., “Graphene-based biosensor for early detection of iron deficiency,” Sensors, vol. 20, no. 13, p. 3688, Jul. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Wang S.et al. , “Graphene field-effect transistor biosensor for detection of biotin with ultrahigh sensitivity and specificity,” Biosensors Bioelectron., vol. 165, Oct. 2020, Art. no. 112363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Reiner-Rozman C., Kotlowski C., and Knoll W., “Electronic biosensing with functionalized rGO FETs,” Biosensors, vol. 6, no. 2, p. 17, Apr. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Larisika M.et al. , “Electronic olfactory sensor based on A. mellifera odorant-binding protein 14 on a reduced graphene oxide field-effect transistor,” Angew. Chem. Int. Ed., vol. 54, no. 45, pp. 13245–13248, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Nowak C., “Graphene FET-based biosensor,” U.S. Patent EP 2 848 929 A1, Mar. 18, 2015.
- [130].Zheng C.et al. , “Sensitive molybdenum disulfide based field effect transistor sensor for real-time monitoring of hydrogen peroxide,” Sci. Rep., vol. 9, no. 1, p. 759, Jan. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Xu K., Meshik X., Nichols B. M., Zakar E., Dutta M., and Stroscio M. A., “Graphene- and aptamer-based electrochemical biosensor,” Nanotechnology, vol. 25, no. 20, May 2014, Art. no. 205501. [DOI] [PubMed] [Google Scholar]
- [132].Sun J.et al. , “Magnetic graphene field-effect transistor biosensor for single-strand DNA detection,” Nanosc. Res. Lett., vol. 14, no. 1, p. 248, Jul. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Hwang M. T.et al. , “DNA nanotweezers and graphene transistor enable label-free genotyping,” Adv. Mater., vol. 30, no. 34, Aug. 2018, Art. no. 1802440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Manoharan A. K., Chinnathambi S., Jayavel R., and Hanagata N., “Simplified detection of the hybridized DNA using a graphene field effect transistor,” Sci. Technol. Adv. Mater., vol. 18, no. 1, pp. 43–50, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Xia Y.et al. , “Plasma treated graphene FET sensor for the DNA hybridization detection,” Talanta, vol. 223, Feb. 2021, Art. no. 121766. [DOI] [PubMed] [Google Scholar]
- [136].Gao Z.et al. , “Detection of sub-fM DNA with target recycling and self-assembly amplification on graphene field-effect biosensors,” Nano Lett., vol. 18, no. 6, pp. 3509–3515, Jun. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Chen J., Mao S., and Lu G., “Graphene-based field-effect transistor bosensors,” UWM Research Foundation, U.S. Patent 9 676 621, Jun. 14, 2017.
- [138].Mao S., Yu K., Chang J., Steeber D. A., Ocola L. E., and Chen J., “Direct growth of vertically-oriented graphene for field-effect transistor biosensor,” Sci. Rep., vol. 3, no. 1, p. 1696, Dec. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]








