SUMMARY
The human default mode network (DMN) is engaged at rest and in cognitive states such as self-directed thoughts. Interconnected homologous cortical areas in primates constitute a network considered as the equivalent. Here, based on a cross-species comparison of the DMN between humans and non-hominoid primates (macaques, marmosets, and mouse lemurs), we report major dissimilarities in connectivity profiles. Most importantly, the medial prefrontal cortex (mPFC) of non-hominoid primates is poorly engaged with the posterior cingulate cortex (PCC), though strong correlated activity between the human PCC and the mPFC is a key feature of the human DMN. Instead, a fronto-temporal resting-state network involving the mPFC was detected consistently across non-hominoid primate species. These common functional features shared between non-hominoid primates but not with humans suggest a substantial gap in the organization of the primate’s DMN and its associated cognitive functions.
In brief
By comparing resting-state networks in humans, macaques, marmosets, and mouse lemurs, Garin et al. identify two networks in non-hominoid primates that include homolog areas of the human default mode network. The mPFC and PCC are tightly connected in the human DMN but poorly connected to each other across non-hominoid primates.
Graphical Abstract

INTRODUCTION
The default mode network (DMN) is unique in supporting internal mental processes in humans with its deactivation being critical for engaging in cognitive functions (Raichle et al., 2001). The anterior part of the DMN is more active during self-directed thoughts and the posterior DMN during passive rest (Davey and Harrison, 2018). The regional features of the human DMN are now well defined and encompass multiple, typical cerebral structures, such as the inferior parietal lobule (PG), the lateral temporal cortex, as well as two core regions: the posterior cingulate cortex (PCC) and the medial prefrontal cortex (mPFC) (Buckner et al., 2008; Raichle et al., 2001). A homologous network to the DMN was proposed in multiple non-human primate species such as chimpanzees (Barks et al., 2015), macaques (Vincent et al., 2007), marmosets (Belcher et al., 2013), and mouse lemurs (Garin et al., 2021). Electrophysiological literature in macaques confirms that PCC firing rates are higher during resting baseline and are suppressed during task performance, consistent with a DMN-like role (Hayden et al., 2009). Despite the similarities of the human and non-human primate DMN, qualitative differences have been reported in some non-human primate studies. For example, absence or weak involvement of the medial prefrontal areas with the DMN has been reported in marmosets (Liu et al., 2019), whereas the mPFC in humans is deemed to support a regulator or “gateway” function of self-representations (Davey et al., 2016). Instead, a connectivity peak in the dorso-lateral prefrontal area A8aD was detected in the marmoset DMN (Liu et al., 2019), though this area is mainly involved in orienting behavior (Germann and Petrides, 2020), which is at odds with the proposed role of the DMN in self-directed thought or introspection. Only the chimpanzee DMN has shown strong equivalence in connectivity patterns with humans (Barks et al., 2015).
We were thus motivated to perform a comprehensive, cross-species study, using comparable methods to characterize the network architecture as well as the connectivity patterns of four primate species: human (Homo sapiens); rhesus macaque (Macaca mulatta), a species of Old World monkey; common marmoset (Callithrix jacchus), a species of New World monkey; and mouse lemur (Microcebus Murinus), a Strepsirrhini that is one of the smallest and most phylogenetically distant primates when compared to humans (Ezran et al., 2017). By comparing the functional organization of these four species, we conclude that the characteristics of high-order networks in all the examined non-hominoid primate species (NHoPs) are closer to each other than to humans. Our interspecies comparison of DMN connectivity offer insights in the cognitive gap between hominoid and non-hominoid primates.
RESULTS
Thirteen anaesthetized mouse lemurs were scanned using an EPI sequence on an 11.7T scanner (Garin et al., 2021). Four common marmosets were scanned awake and anesthetized on a 9.4T scanner (Hori et al., 2020a). Four awake macaques were scanned on a 3T Siemens Magnetom Prisma system scanner, and another thirteen anaesthetized macaques were scanned on a 3T Siemens Magnetom Skyra scanner. Forty healthy human resting-state fMRI images were generated by Castellanos et al. (Castellanos et al., 2009). Images from each species were co-registered to a standardized space (AFNI) for statistical analyses (Cox, 1996). A comprehensive summary table of acquisition parameters for each dataset as well as a discussion of the potential impact of parameter variation on the reported results is provided in Table S1.
Identification of the resting-state network architecture in four primate species
We used group dictionary analysis on the fMRI data of four different primates (humans, macaques, marmosets, and mouse lemurs). Seven components were set for all analyses to extract similar networks as in the functional atlas of Yeo et al. with the exception that we included subcortical regions (Fan et al., 2016; Yeo et al., 2011). For each species, the seven large-scale networks were labeled according to the literature and to their anatomical features (Figures 1 and S1). In humans, we extracted networks identified in the literature as the DMN, fronto-parietal control, dorsal attention, salience, primary and secondary visual, and dorsal somatomotor (Figures 1A and S1).
Figure 1. Dictionary learning statistical map of resting-state large-scale networks in four primate species using seven components.
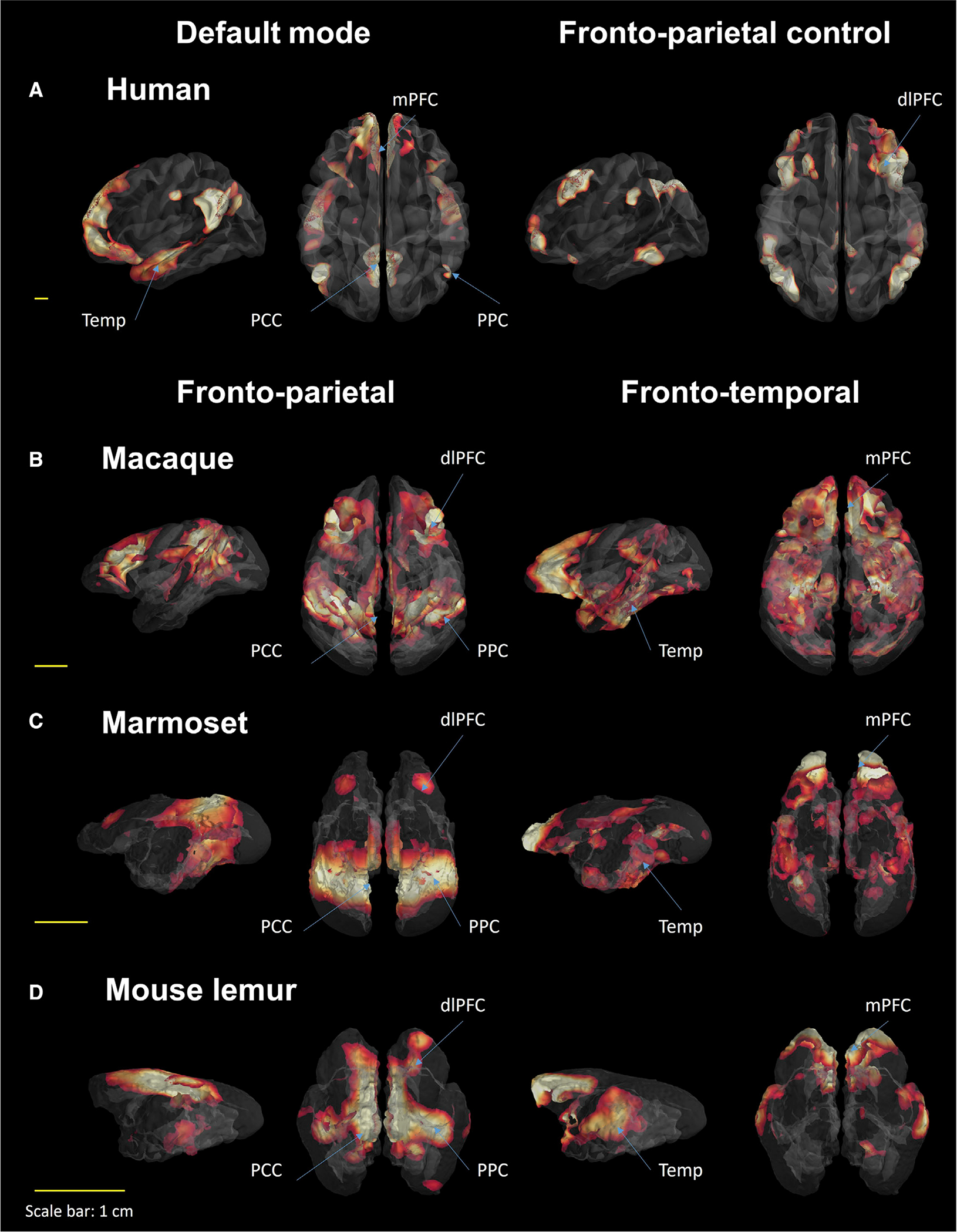
Two high-order networks are illustrated for each species: Human (A): default mode and fronto-parietal control networks. Macaque (B), Marmoset (C), Mouse lemur (D): fronto-parietal and fronto-temporal networks. dlPFC, dorso-lateral prefrontal cluster; mPFC, medial prefrontal cluster; PPC, posterior parietal cluster; Temp, temporal cluster; PCC, posterior cingulate cluster.
In awake macaques, the same analysis produced two distinct networks encompassing areas of the human DMN: First, a cortical network encompassing mainly posterior cingulate cortex (PCC), parietal areas (7a or caudal inferior parietal lobule area and 7m), superior temporal areas (medial superior temporal, middle temporal), visual area V4, and dlPFC areas (area 8, dorso-lateral sulcus upper limb, agranular frontal area F2). We identified this cortical network as the fronto-parietal network (FPN; Figure 1B). Secondly, a cortical network involving mainly the mPFC (areas 9, 32, 13, 10), various areas of the dlPFC, the anterior cingulate cortex and PCC, the hippocampus, areas surrounding the inferior temporal cortex, and the rostrotemporal cortex. We identified this network as the fronto-temporal network (FTN; Figure 1B). The five other networks are identified in Figure S1. Identical analysis performed in anesthetized macaques is displayed in Figure S2.
The same, dual networks were observed in awake and anesthetized marmosets. In awake marmosets, we extracted a component encompassing posterior cingulate areas, parietal areas (parietal area medial, intraparietal area), dlPFC areas (8A, 6 dorsal), and visual areas (1, 3, 6). We identified this component as the FPN (Figure 1C). We also extracted a network that we called FTN encompassing the mPFC (area 9, 10, 14, 8b, 32, 47), temporal areas such as interior temporal area, temporo-parieto association area rostral, auditory parabelt area rostral, temporo polar proisocortex, and some parts of the thalamus and striatum (Figure 1C). The five other networks are identified in Figure S1. The same analysis performed in anesthetized marmoset is displayed in Figure S2.
These findings were also replicated in anesthetized mouse lemurs. We highlighted a cortical network encompassing the PCC, the anterior cingulate cortex, the parietal posterior, and the frontal anterior lateral that was named FPN (Figure 1D). Another component encompassing the mPFC, the frontal anterior lateral, the anterior cingulate cortex, and the middle-temporal cortex was identified as the FTN (Figure 1D). The five other networks are identified in Figure S1 as in Garin et al. (Garin et al., 2021).
Each large-scale network was transformed into a mask and concatenated together, producing a 3D atlas for each species (Figure 2; each atlas can freely be downloaded at http://www.nitrc.org/projects/prim_func_2020/). Cerebral clusters were spatially separated and attributed to a unique label. In this study, we focused on the FTN and FPN (DMN and fronto-parietal control in humans).
Figure 2. Large-scale network functional atlas of the primate brains.
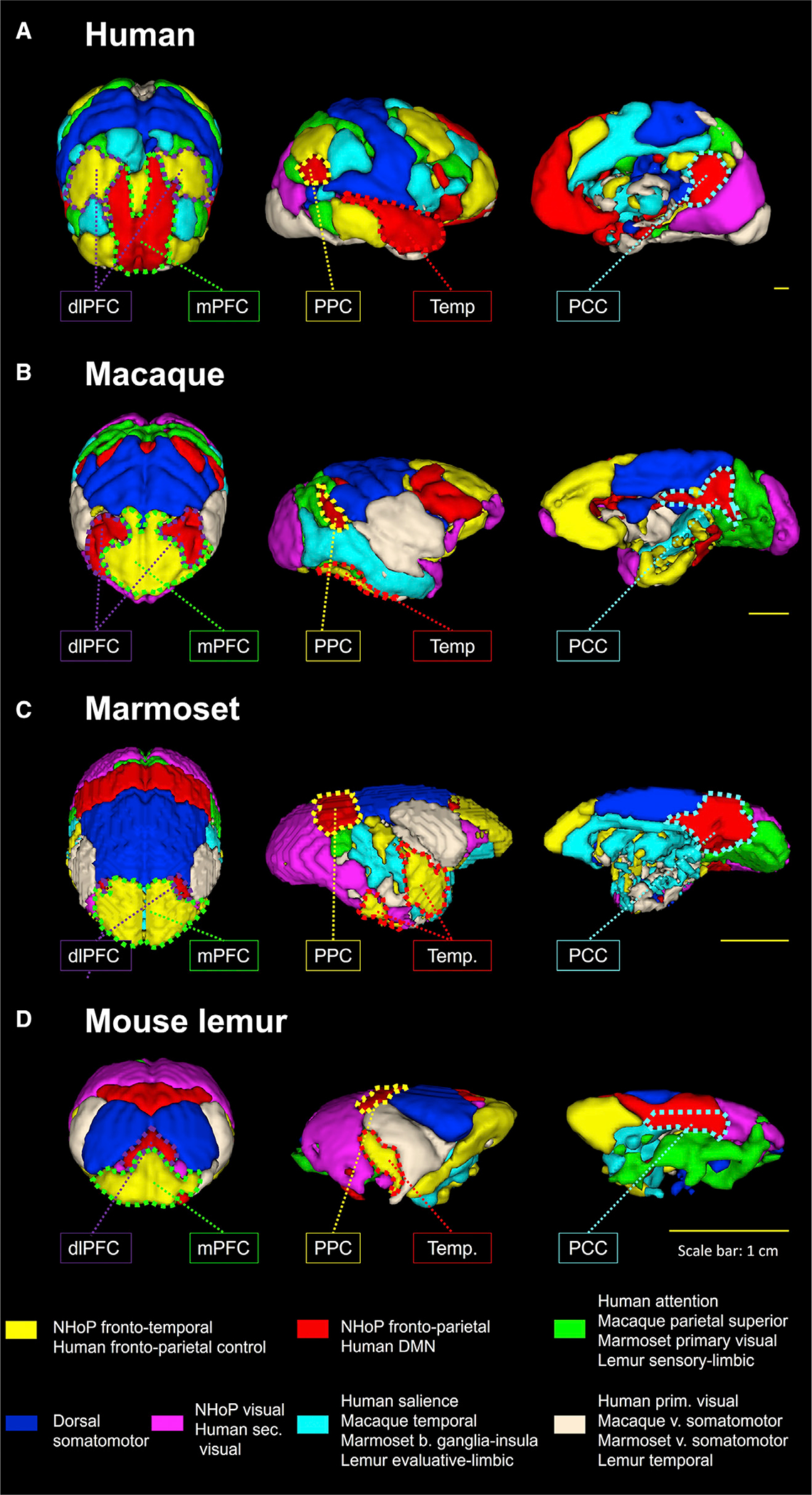
Seven components of the dictionary learning analysis were concatenated and labeled based on their anatomical features in four species: Human functional atlas (A), Macaque (B), Marmoset (C), and Mouse lemur (D). Cerebral clusters were spatially separate (colored dashed line) and were then used to extract their correlation strength with other clusters of interest. dlPFC, dorso-lateral prefrontal cluster; mPFC, medial prefrontal cluster; PPC, posterior parietal cluster; Temp, temporal cluster; PCC, posterior cingulate cluster; prim, primary; sec, secondary, v, ventral, b, basal.
Complementary analyses were performed to test the non-dependency of our networks’ architecture to the statistical analysis. A seed-based analysis was implemented using frontal seeds from the human anatomical atlas of Glasser (Figure S3A) (Glasser et al., 2016), the D99 macaque atlas (Figure S3B) (Reveley et al., 2017), the marmoset atlas of Liu (Liu et al., 2018) (Figures S3C and S3D), and the functional atlas of the mouse lemur brain atlas (Figure S3D). We validated that area 8Ad or its equivalent (8a, dlPFC) in each of the four species leads to the detection of the FPN (or DMN in humans). The DMN anatomical architecture extracted from each of the species shows that PCC voxels are among the most connected to the frontal seed. Finally, an independent component analysis (ICA) with seven components was performed and allowed us to detect similar architecture of the FPN and FTN in macaques (Figures S4A and S4B), marmosets (Figures S4C and S4D), and mouse lemurs (Figures S4E and S4F).
Fingerprint comparison
Fingerprint analysis allows comparison of connectivity between brain clusters across various species. This analysis was performed using normalized average correlation coefficients on key clusters of the fronto-parietal network/DMN in humans, macaques, marmosets, and mouse lemurs. We focused on five clusters identified as the PCC (Figure 3A), mPFC (Figure 3B), dlPFC (Figure 3C), posterior parietal cortex (PPC; Figure 3D), and temporal cortex (Temp; Figure 3E). Visual inspection revealed regions in which connectivity was strong in humans and low in NHoPs (mPFC-PCC, mPFC-Temp, mPFC-PPC, PPC-Temp) and regions with high connectivity in NHoPs and low connectivity in humans (dlPFC-PCC, dlPFC-mPFC, dlPFC-PPC). Cosine similarity can be used as an index indicating the degree of similarity between two fingerprints. Permutation testing was performed on the cosine similarity indexes. A low cosine similarity associated to low p value (p < 0.05) suggests differences in connectivity profiles. We compared the fingerprints using permutation cosine similarity on similar clusters and between every pair of species. These differences with humans were associated to a low cosine similarity score when compared to NHoPs species (Figure 3F). Globally, p values observed between NHoPs were lower than when NHoPs were compared to humans (Figure 3G).
Figure 3. Fingerprint analysis between key functional regions of the fronto-parietal network/DMN in humans, macaques, marmosets, and mouse lemurs.

Average connectivity pattern of five clusters—PCC (A), mPFC (B), dlPFC (C), PPC (D), and Temporal (E)—was extracted and transformed for fingerprint visualization. To allow comparability between species, correlations were normalized between 0 and 1. The analysis reveals regions in which connectivity was strong in humans and low in NHoPs (as for mPFC-PCC) and regions with high connectivity in NHoPs and low connectivity in humans (as for PCC-dlPFC). For each couple of species, cosine similarity between two fingerprints is evaluated and plotted in matrix form (F). Statistical analysis was performed using permutation tests and plotted in matrix form (G). A low cosine similarity associated to low p value (p < 0.05) suggests differences in connectivity profile. Human PCC, PPC, and mPFC clusters display the lowest cosine values when compared with macaques, marmosets, and mouse lemurs. Globally, fewer profile differences were observed between NHoPs than when any NHoPs were compared to humans. dlPFC, dorso-lateral prefrontal cluster; mPFC, medial prefrontal cluster; PPC, posterior parietal cluster; Temp, temporal cluster; PCC, posterior cingulate cluster.
Comparison of pairwise correlations
To further evaluate the relative connections of these key clusters across the four species, we compared pairs of functional clusters of interest (PCC or PPC and dlPFC or mPFC; Figure 4). We first compared the correlation coefficients between PCC-dlPFC and PCC-mPFC. In humans, the connection between PCC and mPFC is known to be a major characteristic of the DMN. By contrast, in marmosets, area 8Ad (dlPFC) displays a strong correlation with PCC (Liu et al., 2019). We established that in macaques, marmosets, and mouse lemurs, the functional connection between PCC-mPFC (x1) was lower than between PCC-dlPFC (x2) (Macaque: , ; pairwise T test; p = 0.0078; Marmoset: , , p = 0.000057; Mouse lemur: , , p = 0.049; Figure 4A). The opposite relationship was observed in humans (Human: , , p = 2.7e-09; Figure 4A).
Figure 4. Functional connectivity between key clusters of the fronto-parietal network/DMN in four different primate species.

Box plots represent median and interquartile range of correlation coefficients computed between different clusters while the whiskers extend to show the rest of the distribution, except for points that are determined to be outliers. Cerebral clusters were segmented using the functional atlas of each primate species (Figure 2). BOLD signal time course was extracted, and correlation coefficient strengths between PCC or PPC and dlPFC or mPFC functional clusters were reported (each line corresponding to one run). Connectivity between PCC and dlPFC was higher than between PCC and mPFC in macaques, marmosets, and mouse lemurs (A, Macaque; Marmoset; Mouse lemur). The opposite relationship was observed in humans (A, Human). We obtained a similar result by replacing PCC by PPC (B, Human; Macaque; Marmoset; Mouse lemur). Homolog regions were extracted from the human atlas (Glasser et al., 2016), the D99 macaque atlas (Reveley et al., 2017), and the marmoset atlas (Liu et al., 2018): BOLD signal time course was extracted and correlation coefficient strengths between 23b or PG(i) (PCC and PPC region) and area 8Ad (dlPFC) or area 9m (dlPFC and mPFC region) were reported. Connectivity between PCC and 8Ad was higher than between PCC and area 9m in macaques and marmosets (C, Macaque; Marmoset). No difference was observed in humans (C, Human). Connectivity between area PG and 8Ad was higher than between area PG and area 9m in macaques and marmosets (D, Macaque; Marmoset). No difference was observed in humans (D, Human). Anesthesia effect on pairwise correlations in marmosets: the differences between area 23b-8A and 23b-9 or PG-8A and PG-9 connections were preserved in anesthetized conditions (E). Direct comparison of the correlation coefficients between 23b and 8A and 23b-9 in awake and anesthetized conditions (F). Isoflurane decreases the connectivity between both anatomical regions (23b-8A, 23b-9). dlPFC, dorso-lateral prefrontal cluster; mPFC, medial prefrontal cluster; PCC, posterior cingulate cluster; PPC, posterior parietal cluster. *p < 0.05; **p < 0.01, ***p < 0.001, ****p < 0.0001.
Similarly, the connection between mPFC and PPC was stronger in humans than NHoPs, relative to the connection between PPC-dlPFC. Correlation coefficients between PPC-mPFC (x1) and PPC-dlPFC (x2) were as follows: Human: , , p = 0.00024; Macaque: , , p = 0.002; Marmoset: , , p = 0.00031; Mouse lemur: , , p = 0.023 (pairwise T test/Wilcoxon; Figure 4B). Extensive results of the statistical analysis are provided in Table S2.
In order to test the dependency of our results to the functional segmentation, we used anatomical atlases of humans (Glasser et al., 2016), macaques (Reveley et al., 2017), and marmosets (Liu et al., 2018) to reproduce the results obtained with functional atlases (the mouse lemur anatomical atlas [Nadkarni et al., 2019] did not identify area 8A). Anatomical regions that were particularly involved in each functional cluster and were analogous across species atlases were identified, such as 8A/8Ad in dlPFC, 9m/9 in mPFC, 23/23b in PCC, and PG/PGi in PPC. We thus compared the connectivity between the anatomical regions 23b-9m (x1) and 23b-8Ad (x2). We obtained similar results as with functional atlases in that posterior cingulate-medial prefrontal connectivity was consistently lower in primates (Macaque: , , p = 0.019; Marmoset: , , p = 3.54e-08, pairwise T test/Wilcoxon, Figure 4C). The only difference in this analysis was that connectivity between these sets of areas did not reach statistical significance in humans (Human: , , p = 0.30; pairwise T test, Figure 4C). Finally, we replicated the PPC-mPFC analysis by comparing PG(i)-9m (x1) and PG(i)-8Ad (x2) connectivity. Correlation coefficients were as follows: Human: , , p = 0.55; Macaque: , , p = 0.00011; Marmoset: , , p = 0.000075 (pairwise T test/Wilcoxon, Figure 4D).
To assess and control for the effects of anesthesia, we compared anaesthetized and awake conditions in marmosets on an exemplar connection of interest (23b-8A and 23b-9 or PG-8A and PG-9). First, we confirmed that the difference between these pairs of connections was preserved under anesthesia (23b-9(x1)/23b-8A(x2): p = 0.0026, , ; PG-9(x1)/PG-8A(x2): p = 0.00079, , ; Figure 4E). By directly comparing the correlation coefficients between 23b and 8A/23b-9 in anesthetized (x1) and awake (x2) conditions, we found that isoflurane decreases the connectivity between both anatomical regions (23b-8A; p = 1.96e-13, , ; 23b-9; p = 0.000007, , ; pairwise T test; Figure 4F).
DISCUSSION
In the current study, the human DMN has been shown to exhibit important architectural differences with NHoPs when extracted with dictionary analysis. We found that whereas the PCC and the mPFC are involved in the human DMN, these two clusters were associated with two different networks (which we termed FTN and FPN, respectively) across all examined NHoPs. Some common characteristics were observed between the NHoP FPN and the human DMN, such as the inclusion of the PCC as well as the involvement of some regions of the PPC. The statistical evaluation of similarities between different species remains challenging, and multiple methods have been proposed in the literature to facilitate interspecies comparisons such as functional finger-printing (Passingham et al., 2002) or cross-species cortical alignment (Eichert et al., 2020). Here, we first used fingerprint analysis in association to permutation cosine similarity to measure the connectivity pattern of various functional regions associated with the DMN, FTN, and FPN. Our results suggested that the functional pattern of these regions is more similar between NHoPs than with humans. In addition, we relied on an analysis that measures the correlation magnitude between three regions of interest. We found that the correlations between PCC and PPC with mPFC were statistically weaker in NHoPs compared with humans.
Non-hominoid primate networks and human DMN
Our results showed 8Ad/8A (dlPFC area) to be a shared cluster of the FPN in NHoPs, which partially overlaps with the human DMN. Moreover, while the PCC is strongly connected with the mPFC in the human DMN, this connection is not as obvious in NHoPs since the frontal peak is localized in area 8A/8Ad. As in humans, the function of the PCC in macaques is oriented toward passive rest function (Davey and Harrison, 2018). In primates, area 8A is well known to be involved in exogenous (visual and auditory) orienting (Germann and Petrides, 2020). Such a function is at odds with some of the identified functions of the human DMN such as self-directed thoughts, though it may account for the finding that in macaques (Arsenault et al., 2018) as well as in humans (Vatansever et al., 2017), the “DMN-like”/DMN network appears to be recruited in attention shifts. In addition, the PCC was associated with spatial perception in humans and macaques, consistent with the function of 8A/8Ad in orienting behavior (Germann and Petrides, 2020).
The FTN that we identified in all three NHoPs was unexpected. It involved mPFC areas in all three NHoPs and comprised various temporal clusters adjacent to the inferior temporal cortex in macaques and marmosets. However, the involvement of the PCC in this network was limited in NHoPs (in macaques a sub-cluster containing parts of PCC was found in the FTN with dictionary learning, which was not the case in other NHoPs). Previous resting-state fMRI studies in macaques identified a homologous network to the FTN using a seed in area 9 (Sallet et al., 2013) or clustering approaches (Hutchison et al., 2012; Xu et al., 2019). Interestingly, Lopez-Persem and colleagues recently showed that hominoid-specific sulcal variability in the mPFC influences the location of the cortical hub of the DMN in mPFC (Lopez-Persem et al., 2019). These results suggest that variation in sulcal morphology may participate in reorganizing the primate DMN. Contrasting our findings, a prior study of the rat DMN shows medial frontal structures connected to the posterior medial structures (Hsu et al., 2016), supporting the idea of frontal connectivity disparities across mammalian species. To our knowledge, the functions of the FTN remain to be determined. Individually, primate area 9 has been implicated in metacognitive judgment in macaques (Miyamoto et al., 2018), whereas inferior temporal and rostrotemporal areas have been implicated in social interaction processing (Froesel et al., 2021; Sallet et al., 2011; Sliwa and Freiwald, 2017).
In humans, a recent study proposed two sub-segmentations of the DMN that are closely juxtaposed: the DMN-A and the DMN-B (Braga and Buckner, 2017). DMN-A includes the posterior inferior parietal lobule, lateral temporal cortex, ventromedial PFC, retrosplenial/ventral posteromedial cortex, and parahippocampal cortex. DMN-B includes the temporoparietal junction, lateral temporal cortex, an inferior region of ventromedial PFC, a dorsal region of anteromedial PFC, and the PCC. Associating DMN-A or DMN-B with the NHoP FTN or FPN remains difficult since both networks have prominent mPFC components. Anatomically, an expansion of PFC gray and white matter volumes could explain the inclusion or reinforcement of the PFC connectivity in the hominoids’ DMN. Interestingly, Donahue et al. found an enlargement of the PFC gray matter volume in humans when compared to non-human primates (up to 1.9-fold larger than in macaques and 1.2-fold larger than in chimpanzees). This enlargement is even more important for subcortical PFC white matter (2.4-fold larger than in macaques and 1.7-fold larger than in chimpanzees; Donahue et al., 2018). However, this expansion remains a matter of debate (Donahue et al., 2019).
Conclusions
In conclusion, only a partial fit could be found between the human DMN and the NHoPs high-order networks (FPN, FTN). Our results suggest that mPFC connectivity to posterior DMN regions (most importantly the PCC) could have been reinforced in the hominoid evolution. In humans, the prefrontal engagement in cognitive tasks is directly involved with suppression of the DMN activity (Greicius et al., 2003). The importance of DMN suppression was confirmed by animal electrophysiology during demanding attention and working memory tasks, and higher firing in the PCC was related with higher error rates and slower task performance (Hayden et al., 2009). This result, also observed in humans (Anticevic et al., 2010), illustrates the importance of top-down versus bottom-up mechanisms in DMN function. We hypothesize that DMN suppression during internally focused cognitive tasks such as mind-wandering or working memory might be more efficient in hominoid species with the strengthening of the coupling mPFC/PCC. This function, necessary for flexible disengagement from various external distracting events, may have provided significant cognitive evolutionary advantages to hominoid species.
Limitations of the study
Evaluating resting-state networks in anesthetized and not in awake animals is an obvious limitation (Grandjean et al., 2014). Isoflurane modifies the functional connectivity at high doses (greater than 1.5%) or after a long exposure (Hutchison et al., 2014; Li and Zhang, 2018), and other effects on the connectivity dynamics in macaques (Barttfeld et al., 2015) as well as a reduction of “functional complexity” (Varley et al., 2020) were observed during unconsciousness. However, several animal studies showed that the DMN architecture can be extracted under anesthesia (Mandino et al., 2019; Vincent et al., 2007). In our study, we compared the connectivity pattern between regions of interest (23b-8A; 23b-9) in marmosets under anesthesia and awake conditions. We observed a weaker correlation most importantly between 23b and 8A when compared with 23b-9. Thus, differences of correlation between 23b and 8A and 23b-9 are underestimated in the anesthetized condition when compared to the awake condition. This analysis was not performed in macaques because awake and anesthetized datasets came from different subjects, with different acquisition methods. In mouse lemurs, awake data were not available. However, under the reasonable assumption that isoflurane has a similar effect on mouse lemurs, more important differences between the frontal cluster (mPFC and dlPFC) are to be expected in awake lemurs. Moreover, this result may explain why previous studies performed under anesthesia were less likely to detect this particular connectivity contrast between mPFC and dlPFC in NHoPs. Another limitation of this study is the variety of resolutions and signal-to-noise ratios in the MRI images, principally due to important differences in brain sizes across primate species. In addition, the blood-oxygen-level-dependent (BOLD) sequences used in this study have a lower scan duration than recommended in macaques or humans (Autio et al., 2021) and may have generated variability in our results. Finally, in the absence of comparison between task and awake state in this study, we cannot exclude either the FTN or the FPN as homologous candidates to the human DMN. Furthermore, if the resting-state condition can be partially assessed in humans, it cannot be controlled in awake NHoPs for obvious reasons. Future studies may address these issues.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources should be directed to Clément Garin (garincle@gmail.com) and will be fulfilled by the lead contact.
Materials availability
Raw MRI data are available upon request following a formal data sharing agreement required by the authors’ institutions.
The atlases generated in this study are available for download in NIfTI-1 format at http://www.nitrc.org/projects/prim_func_2020/.
Data and code availability
This paper does not report standardized data types. All data reported in this paper will be shared by the lead contact upon request.
All original code has been deposited at https://zenodo.org/record/6369417#.YjUe3-rMLIW.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Information on approvals for animal and/or human experiments, and information on sex of animals/subjects can be found in each section below.
METHOD DETAILS
Human dataset
Forty healthy participants generously posted in NITRC site for public use were downloaded (http://fcon_1000.projects.nitrc.org/fcpClassic/FcpTable.html). This dataset was generated by the Milham and Castellanos groups (Castellanos et al., 2009). In the present study, 6 subjects were discarded according to our quality control procedure, including visual inspection of the final images, and automatic rejection by AFNI (afni_proc.py), and 34 subjects (15 males and 19 females ranging from 12.2 to 25.2 years old (mean ± SD: 19.9 ± 4.2)) were kept. Resting state time series (one run per subject) data were acquired using echo planar imaging on a Siemens 3.0 Tesla Allegra (TR = 2000 ms; TE = 25 ms; flip angle = 90, 39 slices, matrix = 64 × 64; FOV = 192mm; acquisition voxel size = 3 × 3 × 3mm; 6.5 min.
Awake macaque preparation and MRI acquisition
The project was authorized by the French Ministry for Higher Education and Research (project no. 2016120910476056 and 2015090114042892) in agreement with the French transposition of Directive 2010/63/UE. This authorization was based on the ethical evaluation by the French Committee on the Ethics of Experiments in Animals C2EA#42.
Subjects and material
4 male rhesus monkeys (Macaca mulatta) participated in the study. They were aged between 10 and 17 years (TO: 17y; EL: 15 y; SC: 10y and SA: 12y).The animals were implanted with a plastic MRI compatible headpost covered by dental acrylic. The anaesthesia during surgery was induced by Zoletil (Tiletamine-Zolazepam, Virbac, 5 mg/kg) and followed by isoflurane (Belamont, 1%–2%). Post-surgery analgesia was ensured thanks to Temgesic (buprenorphine, 0.3 mg/mL, 0.01 mg/kg). During recovery, proper analgesic and antibiotic coverage was provided. The surgical procedures conformed to European and National Institutes of Health Guidelines for the Care and Use of Laboratory Animals.
Scanning procedures
In this study, in-vivo MRI scans were performed on a 3T Magnetom Prisma system (Siemens Healthineers, Erlangen, Germany).
Anatomical MRI acquisitions
Monkeys were first anesthetized with an intramuscular injection of ketamine (10 mg/kg). Then, the subjects were intubated and maintained under 1%–2% of isoflurane. During the scan, animals were placed in a sphinx position in a Kopf MRI-compatible stereotaxic frame (Kopf Instruments, Tujunga, CA). Two L11 coils were placed on each side of the head and a L7 coil was placed on the top of it. T1-weighted anatomical images were acquired for each subject using a magnetization-prepared rapid gradient-echo (MPRAGE) pulse sequence. Spatial resolution was set to 0.5 mm, with TR = 3000 ms, TE = 3.62 ms, Inversion Time (TI) = 1100 ms, flip angle = 8°, bandwidth = 250 Hz/pixel, 144 slices.
Functional MRI acquisitions
Before each scanning session, a contrast agent, composed of monocrystalline iron oxide nanoparticles, Molday ION, was injected into the animal’s saphenous vein (9–11 mg/kg) to increase the signal to noise ratio. We acquired gradient-echoechoplanar images covering the whole brain (TR = 2000 ms TE = 18 ms, 38 sagittal slices, acquisition voxel size = 1.25 × 1.25 × x1.25×1.38 mm anisotropic) with an eight-channel phased-array receive coil; and a saddle-shaped, radial transmit-only surface coil (MRI Coil Laboratory, Laboratory for Neuro- and Psychophysiology, Katholieke Universiteit Leuven, Leuven, Belgium, see Kolster et al., 2014). During the scanning sessions, monkeys sat in a sphinx position in a plastic monkey chair facing a translucent screen placed 60 cm from the eyes. Their head was restrained and equipped with MRI-compatible headphones customized for monkeys (MR Confon GmbH). During the resting state acquisitions, animals were sat the dark and no reward was delivered. They were trained to stay calm in front of the dark screen. No fixation was required.
Eye position (X, Y, pupil size) was recorded thanks to a pupil-corneal reflection tracking system EyeLink at 1000Hz (SR-Research) interfaced with a program for stimulus delivery and experimental control (EventIDE®). Only scan in which the monkeys maintained their eyes open are considered. A total of 40 fMRI run were selected for resting-state analysis (TO: 8; EL: 8; SC: 10 and SA: 14).
Anesthetized macaque preparation and MRI acquisition
All procedures were conducted in compliance with State and Federal laws, standards of the US Department of Health and Human Services, and guidelines established by the Wake Forest University Health Sciences Institutional Animal Care and Use Committee as well as the National Institute of Health Guide for the Care and Use of Laboratory Animals (A20–079; A18–037). Thirteen macaques (9 males and 4 females) ranging from 2.5 to 5.9 years old (mean ± SD: 3.7 ± 1.2) were included in this study. Three macaques were scanned twice. In preparation for the MRI scan, anesthesia was induced using ketamine (5–10mg/kg), dexmedetomidine (0.015mg/kg) and was maintained using isoflurane. The animals were intubated and artificially ventilated at 95–120 breath per minute. Expired CO2 was monitored and maintained between 35 and 45 mmHg. Animals were scanned under isoflurane anaesthesia at 1%–1.5%. Heart rate and oxygen saturation levels were monitored using a pulseoximeter. Body temperature was maintained using warm blankets. The MRI system was a 3 Tesla Siemens MAGNETOM Skyra (Siemens Healthcare, Erlangen, Germany). Anatomical images were acquired using a T1-weighted MPRAGE sequence: TR = 2700 ms, TE = 3.32 ms, inversion time = 880, FOV = 128 × 128 mm, 192 slices of 0.5 mm thickness, resolution = 0.5 mm isotropic. Resting state time series data were acquired using a multiband EPI sequence: TR = 700 ms, TE = 32.0 ms, flip angle = 52°, repetitions = 700, FOV = 128 × 128 mm, 32 slices, resolution = 2 mm isotropic.
Marmoset dataset
Four common marmosets (3 males and 1 females) ranging from 1.6 ± 0.4 years old at the beginning of awake experiments were used in this study. Details of how marmosets were restrained and adapted to the scanner environment can be found in Hori et al. (Hori et al., 2020a, 2020b). Data were acquired using a 9.4-T horizontal bore magnet (Varian/Agilent, Yarnton, UK) and Bruker BioSpec Avance III console with the software package Paravision-6 (Bruker BioSpin Corp, Billerica,MA). The dataset was previously published in Hori et al. (Hori et al., 2020a). Animals were scanned awake or under isoflurane anesthesia at 1.5% in medical air. A T2-weighted anatomical image was acquired for each animal using rapid imaging with refocused echoes (RARE) sequences. Six resting-state time series data (at 600 volumes each) were acquired for each animal, in separate sessions, and also for each condition (anesthetized and awake) resulting in 24 BOLD time series in awake condition and 24 BOLD time series in anesthetized condition. The sequence used was a gradient-echo based single-shot echo-planar imaging sequence (See Hori et al., 2020a for more details).
Mouse lemur dataset
Data from thirteen mouse lemurs previously published in Garin et al. were included in this study (Garin et al., 2021). Eleven animals were used (10 males and 1 females), ranging from 1.3 to 3.1 years old (mean ± SD: 2 ± 0.6). Animals were scanned under isoflurane anesthesia at 1.25%–1.5% in air. The MRI system was an 11.7 Tesla Bruker BioSpec (Bruker, Ettlingen, Germany). Anatomical images were acquired using a T2-weighted multi-slice multi-echo (MSME) sequence and resting state time series data (one run per subject) were acquired using a gradient-echo EPI sequence (See Garin et al. (2021) for more details).
MRI pre-processing
Human data
Spatial pre-processing was performed using AFNI afni_proc.py (Cox, 1996). Anatomical T1 and functional images were registered to a high-resolution template in the Montreal Neurological Institute (MNI) space. fMRI images were corrected for slice timing, motion, detrend, smoothed (4mm) and bandpass filtered (0.01–0.1 Hz). TRs with excessive motion of 0.35mm or where too many voxels were flagged as outliers by 3dToutcount (AFNI) were censored (percentage of censored volumes (mean ± SD: 2.1 ± 3.53)). The first 5 volumes were excluded from analysis to ensure steady-state magnetization.
Awake macaque data
Spatial pre-processing was performed using AFNI afni_proc.py and @animal_warper (Cox, 1996). Anatomical T1 images were registered to the high-resolution NMT template (NIH Macaque Template) using @animal_warper (Seidlitz et al., 2018). Then, afni_proc.py was used for registration of fMRI images to the template and for correction. fMRI images were corrected for slice timing, motion, detrend, smoothed (4mm) and bandpass filtered (0.01–0.1 Hz). TRs with excessive motion of 0.5mm or where too many voxels were flagged as outliers by 3dToutcount (AFNI) were censored (percentage of censored volumes (mean ± SD: 8 ± 4)). 5 volumes were excluded from analysis to ensure steady-state magnetization. Due to an important number of outliers, 18 runs were rejected from the analysis.
Anesthetized macaque data
Spatial pre-processing was performed using AFNI afni_proc.py and @animal_warper (Cox, 1996). Anatomical T1 images were registered to the high-resolution NMT template (NIH Macaque Template) using @animal_warper (Seidlitz et al., 2018). Then, afni_proc.py was used for registration of fMRI images to the template and for correction. fMRI images were corrected for slice timing, motion, detrend, smoothed (4mm) and bandpass filtered (0.01–0.1 Hz). TRs with excessive motion of O.35mm or where too many voxels were flagged as outliers by 3dToutcount (AFNI) were censored (percentage of censored volumes (mean ± SD: 0 ± 0)). Either 5 or 10 volumes were excluded from analysis to ensure steady-state magnetization.
Marmoset data
Spatial pre-processing was performed using AFNI afni_proc.py and @animal_warper (Cox, 1996). Anatomical T2 images were registered to the high-resolution NIH marmoset template (Liu et al., 2018) using @animal_warper. Then, afni_proc.py was used for registration of the fMRI images to the template and for correction. fMRI images were corrected for slice timing, motion, detrend, smoothed (1.5mm) and bandpass filtered (0.01–0.1 Hz). TRs with excessive motion of O.2mm or where too many voxels were flagged as outliers by 3dToutcount (AFNI) were censored (percentage of censored volumes (mean ± SD: 0.5 ± 1.1)). Because of a high number of outliers, two fMRI images were excluded from analysis. Five volumes were excluded from analysis to ensure steady-state magnetization.
Mouse lemur data
Spatial pre-processing was performed using the python module sammba-mri (SmAll MaMmals BrAin MRI; http://sammba-mri.github.io). Anatomical images were registered to a high-resolution anatomical mouse lemur template (Nadkarni et al., 2019), fMRI images were corrected for slice timing (interleaved), B0 distortion, per-slice registration to respective anatomicals were also performed with sammba-mri. As with the other primates, motion, detrend, smoothing (0.9mm) and bandpass filtered 0.01 to 0.1 Hz were performed using AFNI. TRs with excessive motion of 0.07mm or where too many voxels were flagged as outliers by 3dToutcount (AFNI) were censored (percentage of censored volumes (mean ± SD: 2.7 ± 3.2)). Due to an important number of outliers, two animals were excluded from analysis. The first 5 volumes were excluded from analysis to ensure steady-state magnetization.
Identification of large-scale networks
Dictionary learning analysis
Multi-animal dictionary learning statistical analysis was performed with Nilearn (Mensch et al., 2016) with 7 components on our pre-processed EPI images. This number of components was chosen in order to facilitate comparison with the human large-scale network atlas of Yeo et al. (Yeo et al., 2011). Using 17 components did not allow for the identification of additional networks. A mask containing cortical and subcortical areas without the cerebellum was used to restrain analysis space. Independent component analysis (ICA) was also performed with Nilearn (Mensch et al., 2016) using the same mask and number of components.
3D atlas of the primate brain large-scale networks
The large-scale network functional atlas of each primate brain was created using the seven components previously extracted by group dictionary learning statistical analysis. The statistical images were then assigned a threshold based on percentile (98% for marmoset, macaques and humans, 99% for lemurs) using threshold_img from Nilearn, concatenated (for intersecting voxels, the highest statistical score was kept) and excluding regions smaller than 300 mm3 for macaques and humans and 2 mm3 for marmosets and mouse lemurs.
Cluster selection
The primate FTN (Temporal, mPFC) and DMN clusters (PCC, dlPFC, PPC) were defined by dividing our functional atlas into separated regions using Nilearn (connected_label_regions). In marmosets and mouse lemurs, the PPC, the dlPFC (mouse lemur only), and the PCC clusters were separated manually based on current anatomical knowledge (Nadkarni et al., 2019). In order to define homologous clusters across species, different factors were taken into account: i. common anatomical cerebral regions of interest included a cluster (identified with an atlas when available: e.g. area 8Ad, 23b, 9, PG in macaques (Reveley et al., 2017) and humans (Glasser et al., 2016)); ii. the spatial localization of cluster relative to a network or lobe (e.g. in mouse lemurs, the dlPFC and the temporal clusters were respectively at the border of the dorsal somatomotor network and the ventral somatomotor network as in macaques; the posterior parietal cluster was restricted to the parietal area as well as involved in the FPN); iii. the spatial localization of a cluster relative to a sulcus (STS in macaque and humans).
QUANTIFICATION AND STATISTICAL ANALYSIS
Extensive results of the statistical analysis are provided in Table S2.
Fingerprint analysis
The mean connectivity matrix between clusters selected in the previous chapter was calculated. To allow for pattern comparability, the fingerprints were normalized between 0 and 1. Statistical analysis was performed using a permutation test on the cosine similarity values calculated between two given groups. Cosine similarity compares the angle between vectors and provides an index to indicate similarities between the orientation of a set of vectors. In other words, cosine similarity can be used as an index indicating the degree of similarity between two fingerprints numerically defined by a set of vectors. A low cosine similarity associated to low p value (p < 0.05) suggests differences in connectivity profile. Permutation statistical analysis was used to test for differences between fingerprints across the primates’ species (Schaeffer et al., 2020). Permutation tests were performed using in-house code written in Python. Individual fingerprints were randomly divided into two groups, average, and normalized. Pairwise statistical comparisons were then performed by calculating cosine similarity and iterating this process 100,000 times. For calculating p value, a Weibull distribution was fitted to the distribution of null similarity scores using scipy’s weibull_min function (Virtanen et al., 2020).
Connectivity based on the functional clusters of the FPN and the FTN
Correlation magnitude between clusters
Directly comparing the correlation coefficient strength between species is not yet possible because of variations of MRI sequences, brain sizes, artefacts and therefore of signal to noise ratio between species. To counter these limitations, comparisons were only made based on the connection trend and between typical clusters of the networks of interest. The purpose of this relative comparison is to statistically identify any modification of a common pattern. In each species, the correlation coefficients were calculated between functional clusters or anatomical regions in fully pre-processed EPI images using Nilearn (Mensch et al., 2016). Correlation coefficients were computed using the Ledoit and Wolf shrinkage coefficient (Brier et al., 2015; Ledoit and Wolf, 2004; Varoquaux et al., 2012). We first formally tested that there was an interaction across species using Mixed-design ANOVA, including species as a between factor, the correlation coefficient as a within-subject factor (repeated measurements) and the scan or BOLD image from a given subject as a between-subject identifier. Comparison of the different correlation coefficients in humans, macaques, marmosets, and mouse lemurs was calculated using a pairwise T test when the distribution was normal (evaluated using a Shapiro-Wilk test). Otherwise, a Wilcoxon signed-rank test was used (non-parametric version of the paired T test). For macaques and marmosets, multiple sessions were evaluated per subject. In order to take these sessions into account, we performed a linear mixed effects model implemented by statsmodels (Seabold and Perktold, 2010). Correlation strength was used as dependent variable, the regions as the independent variable, and the subject as fixed effects nested in the sessions.
Seed-based analysis
Seed based analysis of the prefrontal cortex was performed using a seed in dlPFC (8Ad, 8a) extracted from the atlas of Glasser (Glasser et al., 2016), D99 (Reveley et al., 2017) and Liu (Liu et al., 2018). In mouse lemurs, the dlPFC area extracted from the functional atlas was used as seed. Correlation coefficients to each seed were calculated for each voxel. Individual correlation maps were z-Fisher transformed and grouped in one statistical map using one-sample t test. An adapted threshold (due to variation on the number of subjects per group, SNR) was applied to each species (Humans: p < 0.0000001, Macaques: p < 0.0000001, Marmosets awake: p < 0.0000001; Macaques anesthetized: p < 0.05, Mouse lemurs: p < 0.01). Cluster sizes associated to each threshold were calculated using AFNI “Clustersim” associated to 3dttest++, with the exception of mouse lemurs, which lacked a sufficient number of subjects (>14) for cluster estimation.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Deposited data | ||
|
| ||
| Human BOLD images | NYU Child Study Center | http://fcon_1000.projects.nitrc.org/fcpClassic/FcpTable.html |
| Functional atlases | Wake Forest University | http://www.nitrc.org/projects/prim_func_2020/ |
| Code (Python) | Wake Forest University | https://doi.org/10.5281/zenodo.6369417 |
|
| ||
| Software and algorithms | ||
|
| ||
| AFNI | National Institutes of Health (NIH) | https://afni.nimh.nih.gov/ |
| SmAll MaMmals BrAin MRI | Commissariat à l’Energie Atomique (CEA) | https://github.com/sammba-mri/sammba-mri |
| FreeSurfer | Athinoula A. Martinos Center for Biomedical Imaging | https://surfer.nmr.mgh.harvard.edu/ |
| Statsmodels | N/A | https://www.statsmodels.org/stable/index.html |
| Pingouin | University of California | https://pingouin-stats.org/ |
| Pandas | N/A | https://pandas.pydata.org/ |
| Seaborn | N/A | https://seaborn.pydata.org/ |
| Numpy | N/A | https://numpy.org/ |
| Matplotlib | N/A | https://matplotlib.org/ |
| Scipy | N/A | https://www.scipy.org/ |
| Nilearn | N/A | https://nilearn.github.io/ |
| ITK-snap | University of Pennsylvania, University of Utah | http://www.itksnap.org/pmwiki/pmwiki.php |
| PySurfer | N/A | https://pysurfer.github.io/ |
|
| ||
| Other | ||
|
| ||
| Siemens 3T | Siemens Healthcare | N/A |
| Varian/Agilent 9.4-T | Agilent Technologies | N/A |
| Bruker 11.7 T | Bruker | N/A |
Highlights.
Resting-state fMRI reveals DMN structure across four primate species
Two distinct networks in non-hominoid primates included homolog areas of the human DMN
The mPFC cluster is poorly connected to the PCC cluster in non-hominoid primates
Functional atlases available for each species
ACKNOWLEDGMENTS
This work was supported by the National Institute of Mental Health of the National Institutes of Health under award numbers R01 MH116675 and R01 MH117996. We wish to thank Russell Jaffe for valuable comments on the manuscript, and Gracie Hilber, Connor Hebert, Austin Lodish, Junda Zhu, Lena Marie Moretz, and Stephanie Rideout for their outstanding assistance in the experiments.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.110669.
REFERENCES
- Anticevic A, Repovs G, Shulman GL, and Barch DM (2010). When less is more: TPJ and default network deactivation during encoding predicts working memory performance. NeuroImage 49, 2638–2648. 10.1016/j.neuroimage.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsenault JT, Caspari N, Vandenberghe R, and Vanduffel W (2018). Attention shifts recruit the monkey default mode network. J. Neurosci. 38, 1202–1217. 10.1523/JNEUROSCI.1111-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autio JA, Zhu Q, Li X, Glasser MF, Schwiedrzik CM, Fair DA, Zimmermann J, Yacoub E, Menon RS, Van Essen DC, et al. (2021). Minimal specifications for non-human primate MRI: challenges in standardizing and harmonizing data collection. NeuroImage 236, 118082. 10.1016/j.neuroimage.2021.118082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barks SK, Parr LA, and Rilling JK (2015). The default mode network in chimpanzees (Pan troglodytes) is similar to that of humans. Cereb. Cortex 25, 538–544. 10.1093/cercor/bht253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barttfeld P, Uhrig L, Sitt JD, Sigman M, Jarraya B, and Dehaene S (2015). Signature of consciousness in the dynamics of resting-state brain activity. Proc. Natl. Acad. Sci. U S A. 112, 887–892. 10.1073/pnas.1418031112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belcher AM, Yen CC, Stepp H, Gu H, Lu H, Yang Y, Silva AC, and Stein EA (2013). Large-scale brain networks in the awake, truly resting marmoset monkey. J. Neurosci. 33, 16796–16804. 10.1523/JNEUROSCI.3146-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braga RM, and Buckner RL (2017). Parallel interdigitated distributed networks within the individual estimated by intrinsic functional connectivity. Neuron 95, 457–471.e5. 10.1016/j.neuron.2017.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brier MR, Mitra A, McCarthy JE, Ances BM, and Snyder AZ (2015). Partial covariance based functional connectivity computation using Ledoit-Wolf covariance regularization. NeuroImage 121, 29–38. 10.1016/j.neuroimage.2015.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner RL, Andrews-Hanna JR, and Schacter DL (2008). The brain’s default network: anatomy, function, and relevance to disease. Ann. N. Y Acad. Sci. 1124, 1–38. 10.1196/annals.1440.011. [DOI] [PubMed] [Google Scholar]
- Castellanos FX, Kelly C, and Milham MP (2009). The restless brain: attention-deficit hyperactivity disorder, resting-state functional connectivity, and intrasubject variability. Can J. Psychiatry 54, 665–672. 10.1177/070674370905401003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox RW (1996). AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput. Biomed. Res. Int. J. 29, 162–173. 10.1006/cbmr.1996.0014. [DOI] [PubMed] [Google Scholar]
- Davey CG, and Harrison BJ (2018). The brain’s center of gravity: how the default mode network helps us to understand the self. World Psychiatry 17, 278–279. 10.1002/wps.20553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey CG, Pujol J, and Harrison BJ (2016). Mapping the self in the brain’s default mode network. NeuroImage 132, 390–397. 10.1016/j.neuroimage.2016.02.022. [DOI] [PubMed] [Google Scholar]
- Donahue CJ, Glasser MF, Preuss TM, Rilling JK, and Van Essen DC (2018). Quantitative assessment of prefrontal cortex in humans relative to nonhuman primates. Proc. Natl. Acad. Sci. U S A. 115, E5183–E5192. 10.1073/pnas.1721653115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue CJ, Glasser MF, Preuss TM, Rilling JK, and Van Essen DC (2019). Reply to Barton and Montgomery: a case for preferential prefrontal cortical expansion. Proc. Natl. Acad. Sci. U S A. 116, 5–6. 10.1073/pnas.1819241116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichert N, Robinson EC, Bryant KL, Jbabdi S, Jenkinson M, Li L, Krug K, Watkins KE, and Mars RB (2020). Cross-species cortical alignment identifies different types of anatomical reorganization in the primate temporal lobe. eLife 9, e53232. 10.7554/eLife.53232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezran C, Karanewsky CJ, Pendleton JL, Sholtz A, Krasnow MR, Willick J, Razafindrakoto A, Zohdy S, Albertelli MA, and Krasnow MA (2017). The mouse Lemur, a genetic model organism for primate biology, behavior, and Health. Genetics 206, 651–664. 10.1534/genetics.116.199448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L, Li H, Zhuo J, Zhang Y, Wang J, Chen L, Yang Z, Chu C, Xie S, Laird AR, et al. (2016). The human brainnetome atlas: a New brain atlas based on connectional architecture. Cereb. Cortex 26, 3508–3526. 10.1093/cercor/bhw157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froesel M, Gacoin M, Clavagnier S, Hauser M, Goudard Q, and Hamed SB (2021). Neural correlates of audio-visual integration of socially meaningful information in macaque monkeys. Preprint at bioRxiv. 10.1101/2021.05.02.442333. [DOI] [Google Scholar]
- Garin CM, Nadkarni NA, Landeau B, Chetelat G, Picq JL, Bougacha S, and Dhenain M (2021). Resting state functional atlas and cerebral networks in mouse lemur primates at 11.7 Tesla. NeuroImage 226, 117589. 10.1016/j.neuroimage.2020.117589. [DOI] [PubMed] [Google Scholar]
- Germann J, and Petrides M (2020). Area 8A within the posterior middle frontal gyrus underlies cognitive selection between competing visual targets. eNeuro 7. 10.1523/ENEURO.0102-20.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser MF, Coalson TS, Robinson EC, Hacker CD, Harwell J, Yacoub E, Ugurbil K, Andersson J, Beckmann CF, Jenkinson M, et al. (2016). A multi-modal parcellation of human cerebral cortex. Nature 536, 171–178. 10.1038/nature18933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandjean J, Schroeter A, Batata I, and Rudin M (2014). Optimization of anesthesia protocol for resting-state fMRI in mice based on differential effects of anesthetics on functional connectivity patterns. NeuroImage 102 Pt 2, 838–847. 10.1016/j.neuroimage.2014.08.043. [DOI] [PubMed] [Google Scholar]
- Greicius MD, Krasnow B, Reiss AL, and Menon V (2003). Functional connectivity in the resting brain: a network analysis of the default mode hypothesis. Proc. Natl. Acad. Sci. U S A. 100, 253–258. 10.1073/pnas.0135058100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden BY, Smith DV, and Platt ML (2009). Electrophysiological correlates of default-mode processing in macaque posterior cingulate cortex. Proc. Natl. Acad. Sci. U S A. 106, 5948–5953. 10.1073/pnas.0812035106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori Y, Schaeffer DJ, Gilbert KM, Hayrynen LK, Clery JC, Gati JS, Menon RS, and Everling S (2020a). Altered resting-state functional connectivity between awake and isoflurane anesthetized marmosets. Cereb. Cortex 30, 5943–5959. 10.1093/cercor/bhaa168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori Y, Schaeffer DJ, Gilbert KM, Hayrynen LK, Clery JC, Gati JS, Menon RS, and Everling S (2020b). Comparison of resting-state functional connectivity in marmosets with tracer-based cellular connectivity. NeuroImage 204, 116241. 10.1016/j.neuroimage.2019.116241. [DOI] [PubMed] [Google Scholar]
- Hsu L-M, Liang X, Gu H, Brynildsen JK, Stark JA, Ash JA, Lin C-P, Lu H, Rapp PR, Stein EA, and Yang Y (2016). Constituents and functional implications of the rat default mode network. Proc. Natl. Acad. Sci. 113, E4541. 10.1073/pnas.1601485113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison RM, Hutchison M, Manning KY, Menon RS, and Everling S (2014). Isoflurane induces dose-dependent alterations in the cortical connectivity profiles and dynamic properties of the brain’s functional architecture. Hum. Brain Mapp. 35, 5754–5775. 10.1002/hbm.22583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison RM, Womelsdorf T, Gati JS, Leung LS, Menon RS, and Everling S (2012). Resting-state connectivity identifies distinct functional networks in macaque cingulate cortex. Cereb. Cortex 22, 1294–1308. 10.1093/cercor/bhr181. [DOI] [PubMed] [Google Scholar]
- Kolster H, Janssens T, Orban GA, and Vanduffel W (2014). The retinotopic organization of macaque occipitotemporal cortex anterior to V4 and caudoventral to the middle temporal (MT) cluster. J. Neurosci. 34, 10168–10191. 10.1523/JNEUROSCI.3288-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledoit O, and Wolf M (2004). A well-conditioned estimator for large-dimensional covariance matrices. J. Multivariate Anal. 88, 365–411. 10.1016/S0047-259x(03)00096-4. [DOI] [Google Scholar]
- Li CX, and Zhang X (2018). Evaluation of prolonged administration of isoflurane on cerebral blood flow and default mode network in macaque monkeys anesthetized with different maintenance doses. Neurosci. Lett. 662, 402–408. 10.1016/j.neulet.2017.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Ye FQ, Yen CC, Newman JD, Glen D, Leopold DA, and Silva AC (2018). A digital 3D atlas of the marmoset brain based on multi-modal MRI. NeuroImage 169, 106–116. 10.1016/j.neuroimage.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Yen CC, Szczupak D, Ye FQ, Leopold DA, and Silva AC (2019). Anatomical and functional investigation of the marmoset default mode network. Nat. Commun. 10, 1975. 10.1038/s41467-019-09813-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Persem A, Verhagen L, Amiez C, Petrides M, and Sallet J (2019). The human ventromedial prefrontal cortex: sulcal morphology and its influence on functional organization. J. Neurosci. 39, 3627. 10.1523/JNEUROSCI.2060-18.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandino F, Cerri DH, Garin CM, Straathof M, van Tilborg GAF, Chakravarty MM, Dhenain M, Dijkhuizen RM, Gozzi A, Hess A, et al. (2019). Animal functional magnetic resonance imaging: trends and path toward standardization. Front. Neuroinform. 13, 78. 10.3389/fninf.2019.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mensch A, Varoquaux G, and Thirion B (2016). Compressed online dictionary learning for fast resting-state fMRI decomposition. In 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI) (IEEE), pp. 1282–1285. [Google Scholar]
- Miyamoto K, Setsuie R, Osada T, and Miyashita Y (2018). Reversible silencing of the frontopolar cortex selectively impairs metacognitive judgment on non-experience in primates. Neuron 97, 980–989.e86. 10.1016/j.neuron.2017.12.040. [DOI] [PubMed] [Google Scholar]
- Nadkarni NA, Bougacha S, Garin C, Dhenain M, and Picq JL (2019). A 3D population-based brain atlas of the mouse lemur primate with examples of applications in aging studies and comparative anatomy. NeuroImage 185, 85–95. 10.1016/j.neuroimage.2018.10.010. [DOI] [PubMed] [Google Scholar]
- Passingham RE, Stephan KE, and Kotter R (2002). The anatomical basis of functional localization in the cortex. Nat. Rev. Neurosci. 3, 606–616. 10.1038/nrn893. [DOI] [PubMed] [Google Scholar]
- Raichle ME, MacLeod AM, Snyder AZ, Powers WJ, Gusnard DA, and Shulman GL (2001). A default mode of brain function. Proc. Natl. Acad. Sci. U S A. 98, 676–682. 10.1073/pnas.98.2.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reveley C, Gruslys A, Ye FQ, Glen D, Samaha J, Brian ER, Saad Z, Anil KS, Leopold DA, and Saleem KS (2017). Three-Dimensional digital template Atlas of the macaque brain. Cereb. Cortex 27, 4463–4477. 10.1093/cercor/bhw248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallet J, Mars RB, Noonan MP, Andersson JL, O’Reilly JX, Jbabdi S, Croxson PL, Jenkinson M, Miller KL, and Rushworth MF (2011). Social network size affects neural circuits in macaques. Science 334, 697–700. 10.1126/science.1210027. [DOI] [PubMed] [Google Scholar]
- Sallet J, Mars RB, Noonan MP, Neubert FX, Jbabdi S, O’Reilly JX, Filippini N, Thomas AG, and Rushworth MF (2013). The organization of dorsal frontal cortex in humans and macaques. J. Neurosci. 33, 12255–12274. 10.1523/JNEUROSCI.5108-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer DJ, Hori Y, Gilbert KM, Gati JS, Menon RS, and Everling S (2020). Divergence of rodent and primate medial frontal cortex functional connectivity. Proc. Natl. Acad. Sci. U S A. 117, 21681–21689. 10.1073/pnas.2003181117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seabold S, and Perktold J (2010). Statsmodels: econometric and statistical modeling with python. In Proceedings of the 9th python in science conference. 10.25080/Majora-92bf1922-011. [DOI] [Google Scholar]
- Seidlitz J, Sponheim C, Glen D, Ye FQ, Saleem KS, Leopold DA, Ungerleider L, and Messinger A (2018). A population MRI brain template and analysis tools for the macaque. NeuroImage 170, 121–131. 10.1016/j.neuroimage.2017.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sliwa J, and Freiwald WA (2017). A dedicated network for social interaction processing in the primate brain. Science 356, 745–749. 10.1126/science.aam6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varley TF, Luppi AI, Pappas I, Naci L, Adapa R, Owen AM, Menon DK, and Stamatakis EA (2020). Consciousness & brain functional complexity in propofol anaesthesia. Sci. Rep. 10, 1018. 10.1038/s41598-020-57695-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varoquaux G, Gramfort A, Poline JB, and Thirion B (2012). Markov models for fMRI correlation structure: is brain functional connectivity small world, or decomposable into networks? J. Physiol. Paris 106, 212–221. 10.1016/j.jphysparis.2012.01.001. [DOI] [PubMed] [Google Scholar]
- Vatansever D, Menon DK, and Stamatakis EA (2017). Default mode contributions to automated information processing. Proc. Natl. Acad. Sci. U S A. 114, 12821–12826. 10.1073/pnas.1710521114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent JL, Patel GH, Fox MD, Snyder AZ, Baker JT, Van Essen DC, Zempel JM, Snyder LH, Corbetta M, and Raichle ME (2007). Intrinsic functional architecture in the anaesthetized monkey brain. Nature 447, 83–86. 10.1038/nature05758. [DOI] [PubMed] [Google Scholar]
- Virtanen P, Gommers R, Oliphant TE, Haberland M, Reddy T, Cournapeau D, Burovski E, Peterson P, Weckesser W, Bright J, et al. (2020). SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods 17, 261–272. 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Sturgeon D, Ramirez JSB, Froudist-Walsh S, Margulies DS, Schroeder CE, Fair DA, and Milham MP (2019). Interindividual variability of functional connectivity in awake and anesthetized rhesus macaque monkeys. Biol. Psychiatry 4, 543–553. 10.1016/j.bpsc.2019.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo BT, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M, Roffman JL, Smoller JW, Zollei L, Polimeni JR, et al. (2011). The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J. Neurophysiol. 106, 1125–1165. 10.1152/jn.00338.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This paper does not report standardized data types. All data reported in this paper will be shared by the lead contact upon request.
All original code has been deposited at https://zenodo.org/record/6369417#.YjUe3-rMLIW.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


