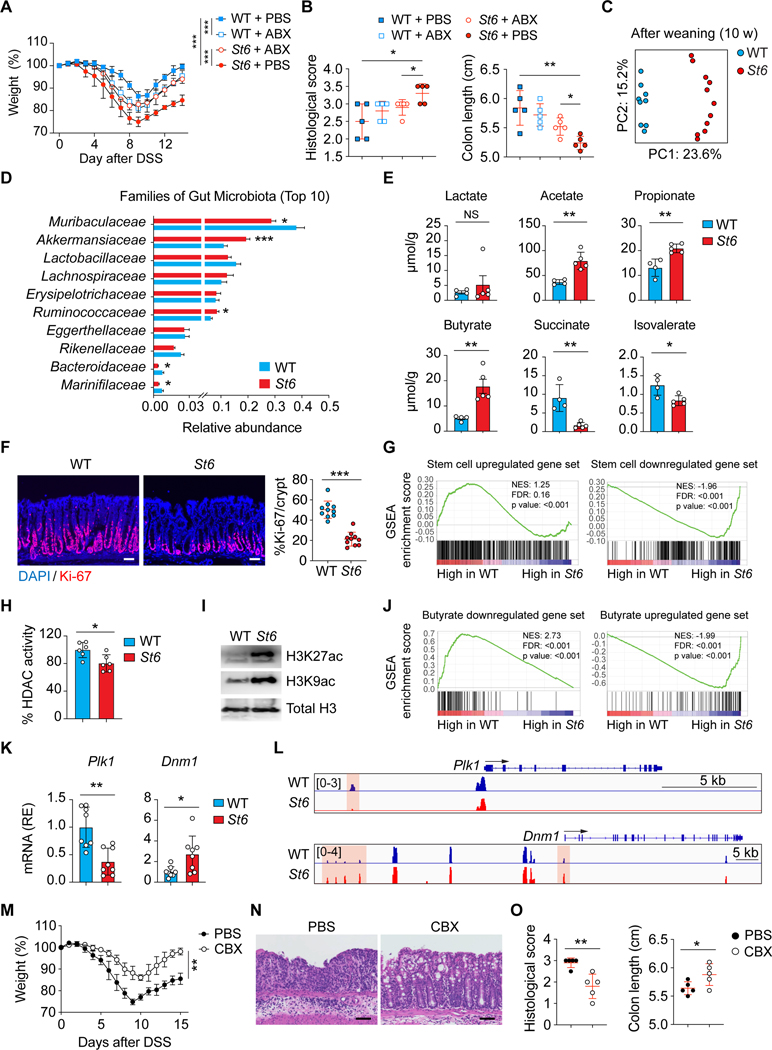
Summary
Intestinal mucus forms the first line of defense against bacterial invasion while providing nutrition to support microbial symbiosis. How the host controls mucus barrier integrity and commensalism is unclear. We show that terminal sialylation of glycans on intestinal mucus by ST6GALNAC1 (ST6), the dominant sialyltransferase specifically expressed in goblet cells and induced by microbial pathogen-associated molecular patterns, is essential for mucus integrity and protecting against excessive bacterial proteolytic degradation. Glycoproteomic profiling and biochemical analysis of ST6 mutations identified in patients show that decreased sialylation causes defective mucus proteins and congenital inflammatory bowel disease (IBD). Mice harboring a patient ST6 mutation have compromised mucus barriers, dysbiosis, and susceptibility to intestinal inflammation. Based on our understanding of the ST6 regulatory network, we show that treatment with sialylated mucin or a Foxo3 inhibitor can ameliorate IBD.
In Brief
Sialylation plays an essential role in protecting mucus barrier integrity from bacterial degradation and is governed by ST6GALNAC1 (ST6), a local sialyltransferase in the gut. ST6-mediated mucus homeostasis controls commensalism to establish intestinal host-microbe symbiosis. ST6 deficiency disrupts this mutualism, increasing susceptibility to intestinal inflammation.
Graphical Abstract
Introduction
Barriers are an innate immune mechanism enforcing host-microbe homeostasis at vulnerable areas of the body. The colonic mucosa is about 2 m2 and apposes a large commensal microbiome (Helander and Fandriks, 2014). The epithelium is comprised of enterocytes, goblet cells (GCs), enteroendocrine cells, Paneth cells, and M cells joined by tight junctions and covered with mucus. Barrier failure can cause inflammatory bowel disease (IBD) (Jeong et al., 2007; Liso et al., 2020; Moehle et al., 2006; Sheng et al., 2012; Tytgat et al., 1996; Van der Sluis et al., 2006; Visschedijk et al., 2016), which enhances the risk of chronic disease and colon cancer (Kvorjak et al., 2020; Molodecky et al., 2012). Epithelial glycans contribute to intestinal mucosal homeostasis and epithelial barrier integrity (Theodoratou et al., 2014; Pickard et al., 2017; Kudelka et al., 2020). They serve as ligands and nutrient sources for the gut microbiota. Abnormal glycosylation disrupts host-microbial interactions, mucin barrier function, and mucosal immunity (Arike et al., 2017; Campbell et al., 2001; Robbe-Masselot et al., 2009; Theodoratou et al., 2014). Understanding GI barriers may improve IBD treatments (Khor et al., 2011; Loftus, 2004).
The GI frontier barrier is intestinal mucus (Johansson et al., 2013). Mucus, a complex gel of mucin glycoproteins, water, immunoglobulin A (IgA), bioactive peptides, and metabolites, is secreted mainly by GCs (Birchenough et al., 2015). Mucus covers the epithelium in 2 layers: an outer loose layer that admits commensal microbiota and an inner tight layer that excludes microbiota (Bergstrom et al., 2020; Theodoratou et al., 2014). The major mucus glycoprotein is Mucin-2 (MUC2) which contains both O-and N-linked glycans (Kudelka et al., 2020). Ulcerative colitis (UC) is associated with a thinner mucus layer (Alipour et al., 2016). Previous studies show that glycosylated MUC2 aids barrier integrity but the key sugar configurations that prevent IBD are unknown (Bergstrom et al., 2020; Theodoratou et al., 2014).
Sialylation is the terminal addition of sialic acid (SA) to glycans and it occurs on mature mucin proteins (Guzman-Aranguez and Argueso, 2010; Kudelka et al., 2020). The main mucin-type glycans have increasing SA from the stomach to the rectum (Kudelka et al., 2020; Robbe et al., 2003). Sialyltransferases (STs) transfer SA from a common donor, cytidine-5’-monophosphate-N-acylneuraminic acid (CMP-Neu5Ac), to acceptor glycans (Harduin-Lepers et al., 2001; Schjoldager et al., 2020). ST proteins have an N-terminal cytoplasmic tail, a transmembrane domain, and a catalytic domain with conserved motifs: L (long) that bind CMP-Neu5Ac, S (short) for substrate binding, and the III (third position in the sequence) and VS (very short) that bind the acceptor substrate (Harduin-Lepers et al., 2001; Patel and Balaji, 2006). ST6GALNAC1 (ST6) is highly conserved in vertebrates and conjugates SA with an a2–6 linkage to N-acetylgalactosamine (GalNAc) glycans generating the tumor-related sialyl-Tn (S-Tn) antigen (Harduin-Lepers et al., 2001; Ikehara et al., 1999; Munkley, 2016; Patel and Balaji, 2006). S-Tn has been associated with ulcerative colitis and various cancers (Kvorjak et al., 2020), but the role of sialylation in intestinal barrier function is unclear.
Results
The ST6 sialyltransferase is highly expressed in GCs
We found ST6 mRNA and protein were selectively expressed in the colon and GI tissues but absent from blood, immune cells, or other organs (Figures 1A–1C, S1A–S1C). Single cell RNA (scRNA) seq data showed high expression of ST6 in GCs and less in (TA) precursor cells, M cells, and enterocytes (Smillie et al., 2019) (Figures 1D and S1D). Immune or stromal cells expressed little ST6 RNA (Figures 1E, S1D, and S1E).
Figure 1. ST6 sialyltransferase is highly expressed in GCs in the colon.
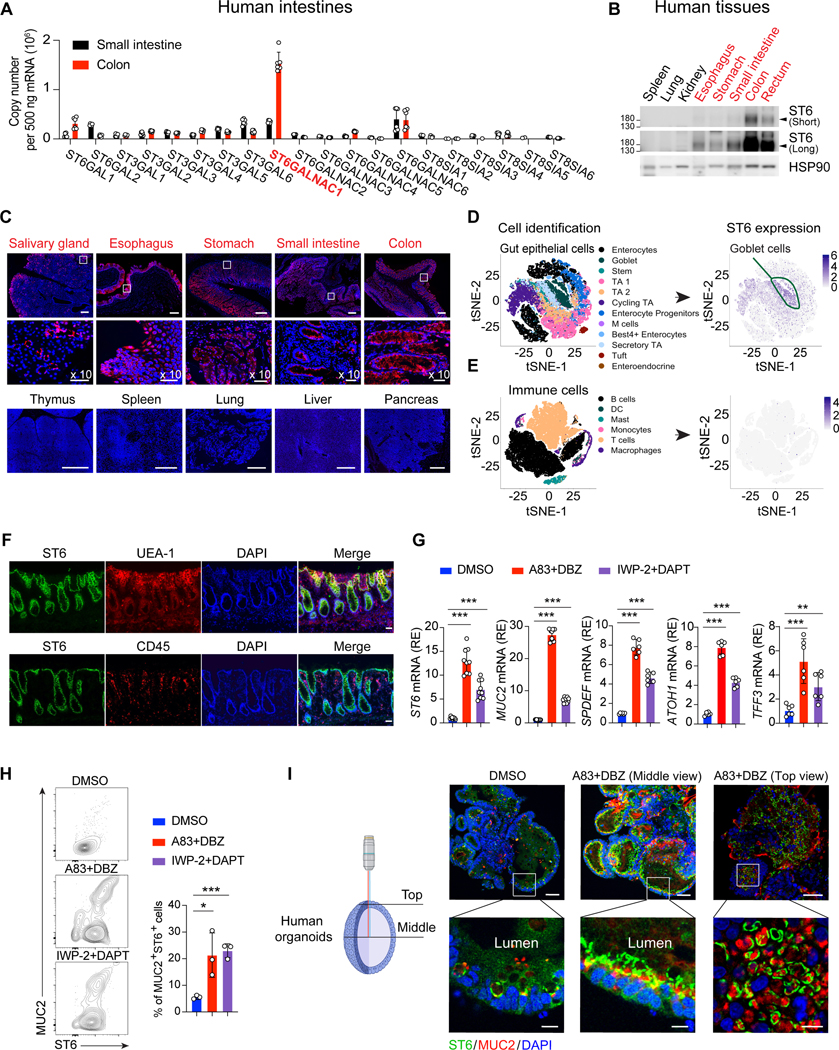
(A) Q-PCR absolute RNA quantification (standard curve method) for the indicated sialyltransferase family genes in human small intestine and colon from healthy donors. ST6GALNAC1 (ST6) in red.
(B) Short and long exposures of ST6 and HSP90 (control) protein immunoblots in healthy donor tissues.
(C) Confocal photomicrographs of human tissues stained for ST6 (red) and DAPI (blue). Scale bar, 300 μm. Middle panels are 10X enlarged, scale bar, 50 μm.
(D, E) t-distributed stochastic neighbor embedding (t-SNE) results of ST6 expression from human colon single cell sequencing obtained from https://doi.org/10.1016/j.cell.2019.06.029. Heat map represents expression level.
(F) Human colon section staining by the indicated antibodies, lectin, or DAPI. UEA-1 is a GC marker and CD45 is a hematopoietic marker. Scale bar, 20 μm.
(G) Q-PCR analysis of indicated genes in human gut organoids after 5-day culturing in DMSO control or two alternative GC differentiation conditions: A83–01, inhibitor of activin receptor-like kinase (ALK) and dibenzazepine (DBZ), or inhibitor of Wnt production-2 (IWP-2) and N-[N-(3,5-Difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl ester (DAPT).
(H) Flow cytometry of MUC2 and ST6 in human gut organoids after 5-day culturing in conditions as in (G) quantitated in the right panel.
(I) Immunofluorescent staining of MUC2, ST6, and DAPI in human gut organoids after 5-day culturing as in (G). Microscopy view depicted at left. Scale bar, 50 μm. Zoom panels (bottom), scale bar, 10 μm. Data represent 2 (A) or 3 (B, C, F-I) experiments. Error bars represent the standard deviation (SD) of samples within a group. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
See also Figure S1.
We next examined GCs in situ by staining healthy human colon tissue. ST6 was mainly expressed in the epithelium in conjunction with GCs, but not immune cells (Figure 1F). Using GC-polarized intestinal organoids derived from healthy human donor induced pluripotent stem cells (iPSCs) (Spence et al., 2011), we observed increased ST6 and GC signature genes (Sato et al., 2011; Yin et al., 2014) (Figure 1G). Flow cytometry confirmed that MUC2+ GCs highly express ST6 (Figure 1H). Confocal microscopy showed increased ST6 and MUC2 protein in GCs bordering the organoid lumen (Figure 1I, middle view), and a map projection showed ST6 surrounding MUC2 (Figure 1I, top view).
ST6 modulates global N-linked glycosylation
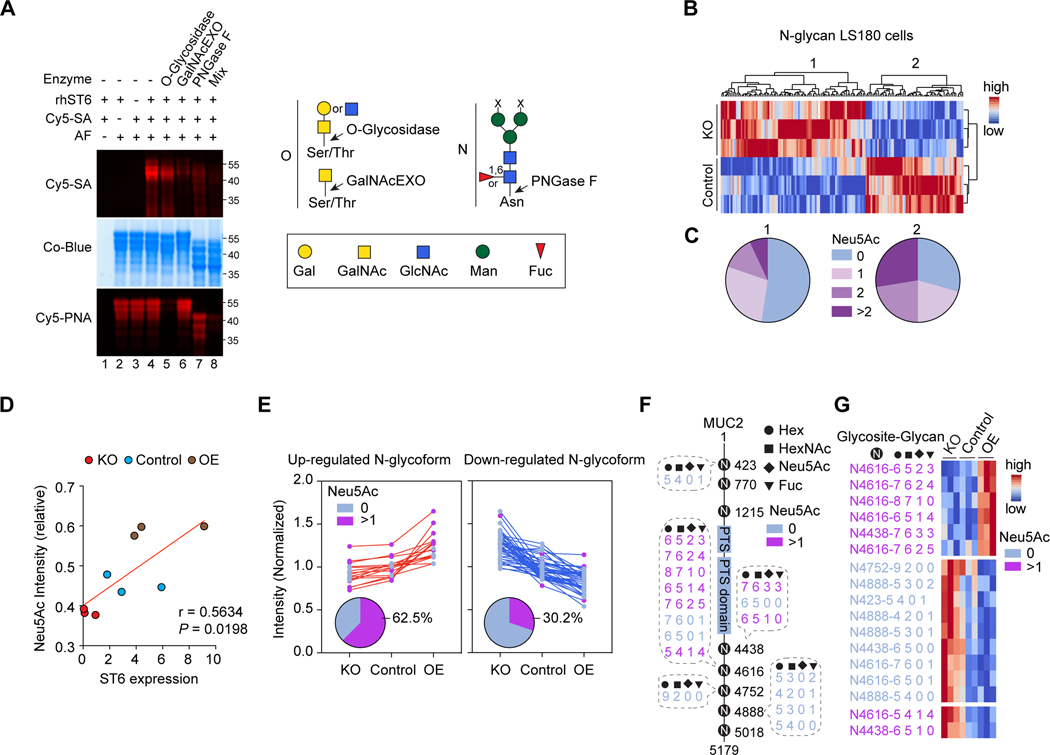
GCs generate intestinal mucus by secreting mucin proteins with extensive O-and N-linked glycosylation (Kudelka et al., 2020). Since ST6 modifies O-linked glycans (Ikehara et al., 1999), we investigated whether ST6 also affects N-linked protein glycosylation. We first established an in vitro fluorescent SA labeling procedure using the common test substrate asialofetuin (AF) (Figure 2A). We found that enzymatically removing either O-or N-glycans reduced the SA signal, showing both promote ST6-mediated SA modification (Figure 2A). We then performed MS-based glycoproteomics to quantitatively profile global N-linked glycans in CRISPR ST6 KO (sgST6) or control (sgSCR) LS180 GC-type cells (Figure S2A). Non-supervised clustering revealed two clusters of differentially expressed N-glycans, which were decreased (cluster 1) or increased (cluster 2) N-glycans in control cells compared to ST6 KO cells (Figure 2B). Of these, cluster 2 N-glycans were significantly enriched for SA decoration (Figure 2C), suggesting more sialylation in control cells. A volcano plot showed more SA on N-glycans in control versus KO cells (Figure S2B).
Figure 2. ST6 modulates N-linked glycosylation.
(A) Fluorescent sialic acid (Cy5-SA, Cy5-Neu5Ac) labeling image (top) and Coomassie blue (Co-Blue) protein stain (middle) of asialofetuin (AF) treated with recombinant human ST6 (rhST6), with or without glycosidase pre-treatments as indicated. Cy5-Peanut agglutinin (PNA) was used as a control (bottom) to detect the enzyme activity of O-glycosidase. (Right) Cartoon of the cleavage sites of O-linked (O) and N-linked (N) glycosidases used.
(B) Non-supervised clustering of differently regulated N-glycans expression level (false discovery rate, FDR = 0.3) detected in control and ST6 knockout (KO) LS180 cells (three biological replicates each). Color scale shows N-glycan intensities.
(C) Pie charts of molar distributions of SA (Neu5Ac)-quantities in N-glycans in clusters 1 and 2 (calculated from three biological replicates) indicated in (B). Pearson’s chi-squared test (p = 0.0009) was used to calculate the p-value of the proportion of glycan status between clusters 1 and 2.
(D) Spearman’s coefficient (r) and significance (P) between ST6 expression level and the relative intensity of SA (Neu5Ac) containing N-glycoforms (FDR = 0.04) on MUC2 detected in LS180 cells with ST6 KO, control, and overexpression (OE).
(E) Normalized intensities of up-and down-regulated MUC2 N-glycoforms detected in LS180 cells as in (D). The distribution of SA (Neu5Ac) non-containing (Neu5Ac 0) and containing (Neu5Ac >1) N-glycoforms are shown in the pie charts.
(F) Visualization of the regulated MUC2 SA (Neu5Ac) non-containing and containing N-glycoforms (FDR = 0.04) in LS180 cells as in (D). Compositions of the N-glycans are indicated. PTS = proline, threonine, and serine rich domains.
(G) Intensity heatmap of the regulated MUC2 N-glycoforms in LS180 cells as in (D). Color scale shows normalized N-glycan intensities. Data represent 3 experiments (A-E, G).
See also Figure S2.
Protein network and cluster enrichment analysis of ST6-affected glycoproteins highlighted a group of mucin proteins, including MUC2, the most abundant mucin in GCs and intestinal mucus (Figures S2C and S2D) (Pelaseyed et al., 2014). So we tested how ST6 contributes to MUC2 sialylation in sgSCR (control), sgST6 (KO), and transduced ST6 overexpressing (OE) LS180 cells (Figure S2E). N-glycoproteomic analysis of MUC2 glycoforms showed a direct correlation between ST6 and sialylation (Figure 2D). As expected, 62.5% of the glycoforms up-regulated with ST6 expression were sialylated, whereas only 30.2% of the down-regulated glycoforms contained SA (Figure 2E). SA addition occurred on numerous MUC2 N-glycosites, covering many glycan compositions (Figures 2F and 2G). Thus, ST6 is a rate-limiting step for sialylation of MUC2.
ST6 determines mucus stability
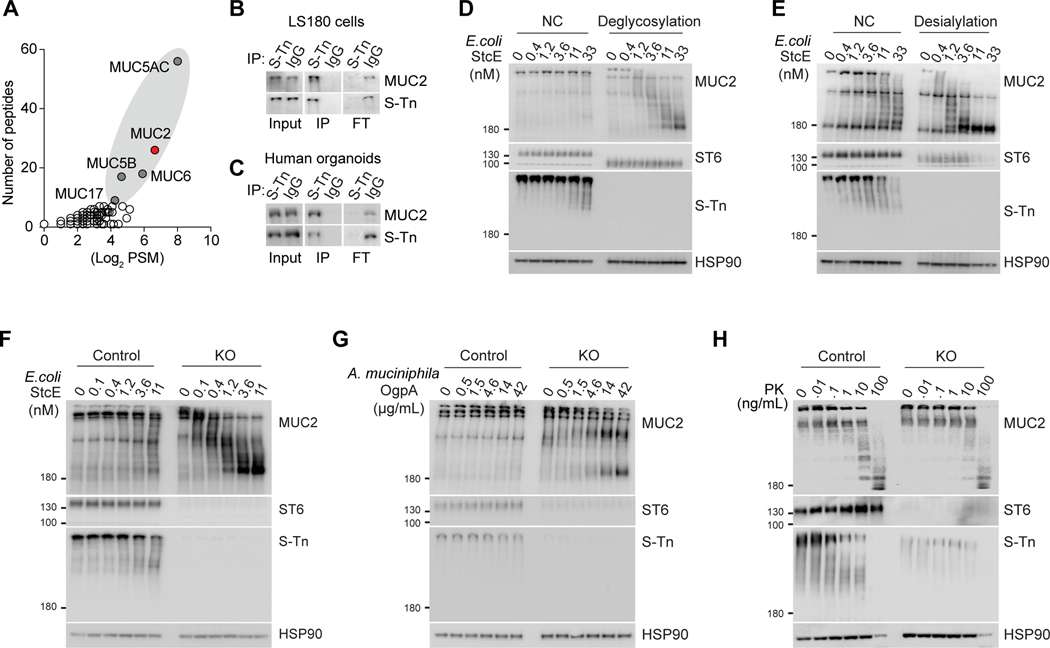
We next investigated how ST6-mediated sialylation regulates mucus homeostasis (Kudelka et al., 2020). The S-Tn antigen, a truncated O-glycan containing SA, is produced specifically by ST6 (Marcos et al., 2004). We used S-Tn as a proxy for ST6 activity and detected sialylation of mucins including MUC2 in LS180 cells (Figure 3A). Immunoprecipitation of LS180 cells and human intestine organoids confirmed that MUC2 bears the S-Tn modification (Figures 3B and 3C). We hypothesized that ST6-mediated sialylation protects MUC2 from bacterial proteolytic degradation to maintain mucus integrity. Mucinases are hydrolytic enzymes produced by gut bacteria that degrade mucins into products that the bacteria consume for growth (Stark et al., 2000). We used the secreted protease of C1 esterase inhibitor (StcE), a well-studied E. coli mucinase (Malaker et al., 2019) and found that MUC2 was relatively resistant to a molar excess of purified StcE but that removal of total glycosylation, and specifically SA, markedly increased MUC2 degradation (Figures 3D and 3E). Moreover, we verified increased sensitivity of MUC2 from ST6 KO LS180 cells to StcE, indicating that an ST6-dependent SA modification protects against MUC2 degradation (Figure 3F). We further used O-glycoprotease (OgpA) from A. muciniphila, which specifically recognizes mucin type O-glycans (Trastoy et al., 2020), and found that MUC2 from ST6 KO cells was more sensitive to degradation (Figure 3G). By contrast, the general protease, proteinase K (PK), equivalently degraded MUC2 from either cell line, indicating that ST6-mediated sialylation preferentially retards digestion by bacterial mucin processing enzymes (Figure 3H).
Figure 3. ST6 regulates MUC2 stability from bacterial degradation.
(A) Mass spectrometry measurement of S-Tn modified proteins by immunoprecipitation for S-Tn antigen or control IgG of proteins from LS180 cells. PSM = peptide spectrum matches.
(B) Immunoblot of MUC2 and S-Tn of immunoprecipitation as in (A). IP = immunoprecipitation, FT = flow-through.
(C) Immunoblot as in (B) using human intestine organoids lysates.
(D) Western blot (WB) analysis of MUC2, ST6, S-Tn, and HSP90 loading control using LS180 cell lysates pre-treated with buffer only (NC) or deglycosylation enzymes (deglycosylation), then incubated with StcE at the indicated concentrations for 3 hours at 37 °C.
(E)WB analysis as in (D) using LS180 cell lysates pre-treated with buffer only (NC) or desialylation enzymes.
(F) WB analysis as in (D) using cell lysates from control and ST6 KO LS180 cells.
(G) WB analysis as in (D) using OgpA protease at the indicated concentrations for 3 hours at 37 °C.
(H) WB analysis as in (D) using proteinase K (PK) at the indicated concentrations for 3 hours at 37 °C. Data represent 2 (A) or 3 (B-H) experiments.
ST6 mutations in IBD patients
Since ST6 could play a protective role by restraining bacterial degradation of mucus, it may be induced during intestinal inflammation. Indeed, we found that colitis patients (Pts) had increased ST6, along with other GC-related genes, in GCs compared to healthy donors (HDs), but not in intestinal immune cells (Figures 4A and S3A). In mice, St6 was specifically expressed in purified intestinal epithelial cells (IECs) but not intraepithelial lymphocytes (IELs) or immune cells in lamina propria (LP) (Figure S3B). Moreover, in the mouse model of dextran sulfate sodium (DSS)-induced colitis, which showed increased 1l6 as a marker gene for inflammation, we also observed elevated St6 mRNA expression in IECs (Figures 4B and S3C).
Figure 4. ST6 deficiency causes early onset colitis in humans.
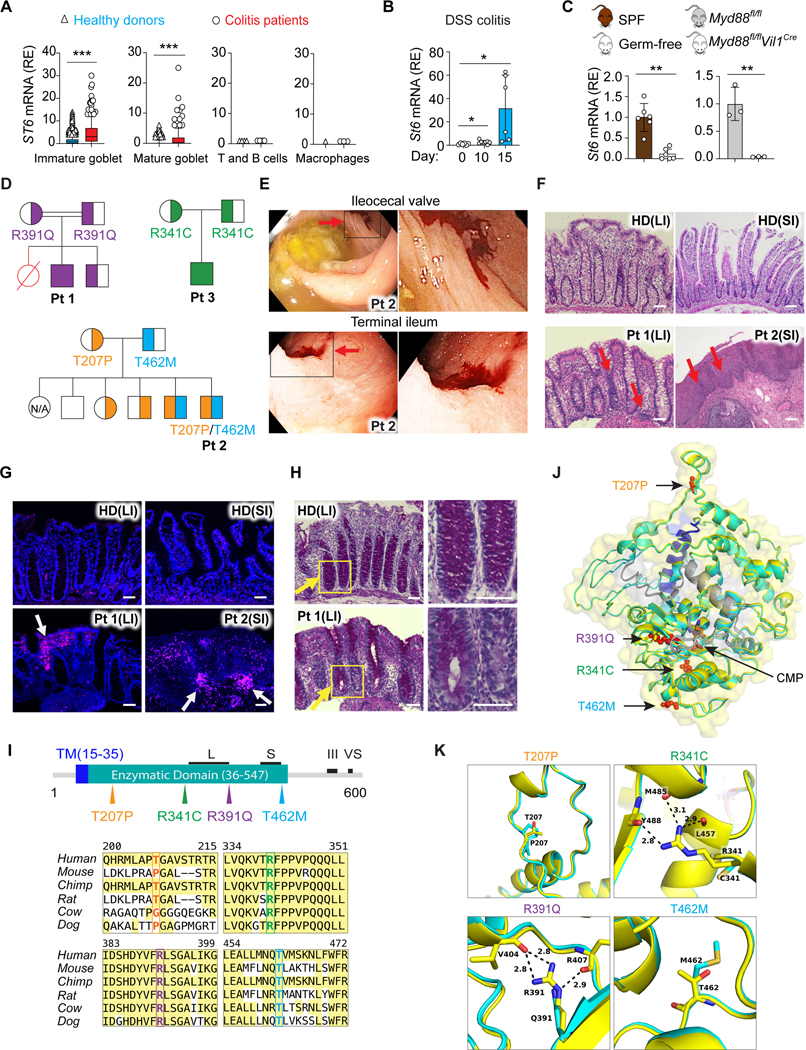
(A) ST6 mRNA in the indicated cell types (Figures 1D and 1E) from healthy donors and colitis patients. Colon single cell sequencing data obtained from https://doi.org/10.1016/j.cell.2019.06.029. Box and whiskers represent the 10th−90th percentiles.
(B) St6 mRNA in IECs from WT mice on the indicated days after initiation of 2.5% DSS water treatment.
(C) St6 mRNA in IECs from specific-pathogen-free (SPF) and germ-free mice (left panel) or Myd88 IEC specific deficient (Myd88fl/fl Vil1Cre) and control (Myd88fl/fl) mice (right panel).
(D) Pedigrees for kindreds 1–3 and patient 1 (Pt 1), Pt 2, and Pt 3, respectively, showing ST6 allele inheritance illustrated by the amino acid substitution. Strikethrough red = deceased individuals; double line = consanguinity.
(E) Colonoscopy photographs of Pt 2 showing mucosal ulceration and hemorrhage (red arrows). (Right) Enlargements of the black box.
(F) Photomicrographs of H&E-stained biopsy sections from large intestine (colon) (LI) and small intestine (SI) from healthy donor (HD), Pt 1, and Pt 2 with mononuclear infiltration and structural damage indicated by red arrows. Scale bar, 50 μm.
(G) Immunofluorescence of biopsies as in (F). White arrows show lymphocyte infiltration. Stains: blue = 4’,6-diamidino-2-phenylindole (DAPI); magenta = CD45. Scale bar, 50 μm.
(H) Periodic Acid Schiff staining of colonic tissues from HD and Pt 1. Yellow arrows show the reduced GC staining in Pt 1 sample. Right panels are enlargements of the yellow box. Scale bar, 50 μm.
(I) (Top) ST6 protein schematic depicting the transmembrane domain (TM), enzymatic domain, and L (long), S (short), III (third position in the sequence), and VS (very short) motif. (Bottom) Amino acid alignment illustrating conservation in ST6 orthologues with mutants in color.
(J, K) Structural model of cytidine 5’-monophosphate (CMP)-bound ST6 color-coded by domains as in (I). The enzymatic domain is shown in yellow, mutant domain in cyan, transmembrane domain (TM) in blue, C terminal domains in grey, and CMP in orange. The Pt amino acid changes are in red (J). Zoomed in models of the amino acids affected by Pt mutations (K). Data represent 3 experiments (B, C, F-H). Error bars represent the SD of samples within a group. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Since colitis involves inappropriate contact of microbiota with intestinal epithelium (Kudelka et al., 2020), we examined whether microbiota drive St6 expression. We found that constitutive St6 expression was significantly higher in specific-pathogen-free (SPF) mice than in germ-free (GF) mice, implying induction by endogenous microbiota (Figure 4C). To determine whether microbiota-associated signals from toll-like receptors (TLRs) conveyed through the “myeloid differentiation primary response 88” (Myd88) signal transducer were important for St6 expression, we analyzed mice conditionally deficient in Myd88 with IECs (Myd8$mVil1cre) and found decreased St6 expression (Figure 4C). Further, we found that lipopolysaccharide (LPS) increased ST6 mRNA, while other TLR ligands had a modest effect (Figure S3D). Loss of TLR4, the LPS receptor upstream of Myd88, significantly reduced St6 expression in IECs (Figure S3E). Using formaldehyde-assisted isolation of regulatory elements (FAIRE) assay, we detected St6 promoter regions with greater eviction of nucleosomes from active chromatin in wildtype (WT) than in Tlr4−/− IECs, indicating that TLR4 signaling governs St6 gene transcription (Figure S3F).
To further investigate the role of ST6 in humans, we screened worldwide IBD cohorts and studied Pts from three independent families with very early onset (< 6 years old) IBD causing episodic diarrhea, abdominal pain, autoimmunity, and failure to thrive, associated with germline ST6 mutations (Figures 4D and S3G; Table S1; Data S1). Endoscopy revealed ulcers and hyperemic, inflamed mucosa and biopsy tissue showed lymphocytic infiltration in the small intestine (SI) and large intestine (LI; colon) (Figures 4E–4G; Table S1). Using periodic acid-Schiff (PAS)-staining for glycosylated mucins, we found disorganized and decreased GCs in Pt 1 colon (Figure 4H; Table S1). Clinical hematological and immune parameters were normal (Table S2). Additionally, CD4+ and CD8+ T cell subsets, natural killer (NK) cells, and B cells, as well as T cell activation, were normal (Figures S3H–S3M). Pt 2 manifested high IgE and increased C-reactive protein (CRP), erythrocyte sedimentation rate (ESR), and antinuclear antibody (ANA) (Table S2). Both Pt 1 and Pt 2 had anti-Saccharomyces cerevisiae antibodies (ASCAs), but no antineutrophil cytoplasmic autoantibodies (ANCAs), as in Crohn’s disease (Table S2).
Using whole exome DNA sequencing to evaluate all rare protein-altering variants, we found biallelic deleterious variants altering conserved amino acids in the ST6 gene (Figures 4D, 4I, and S3N). Pt 1 was consanguineous and homozygous for the rare p.R391Q, Pt 2 was compound heterozygous for p.T462M and p.T207P, and Pt 3 was homozygous for the p.R341C substitution (Figure 4D and 4I). Unaffected siblings and parents were heterozygous or WT except for Pt 2’s brother, who has both disease variants but is currently healthy, indicating incomplete disease penetrance.
We next examined how these variants impact the ST6 protein using a model based on the ST6GalNAc2 structure (PDB 6APL) (Roy et al., 2010). This showed conservation of a single Rossmann-like (glycosyltransferase A variant 2) fold with the p.R391Q, p.R341C, and p.T462M substitutions near the active site where CMP binds (Moremen et al., 2018) (Figure 4J). The p.R391Q substitution likely destabilizes the active site where Arg391 is hydrogen bonded to Val404 and Arg407 (Figure 4K). Similarly, the hydrogen bonds of Arg341 with Tyr488, Met485, and Leu457 are lost with a Cys substitution in the p.R341 C variant (Figure 4K). Thus, p.R391 Q, p.R341C, and p.T462M likely compromise ST6 enzyme activity. T207P is distal to the active site but might disrupt an alpha helix affecting enzyme assembly (Banfield, 2011; Rudresh et al., 2002).
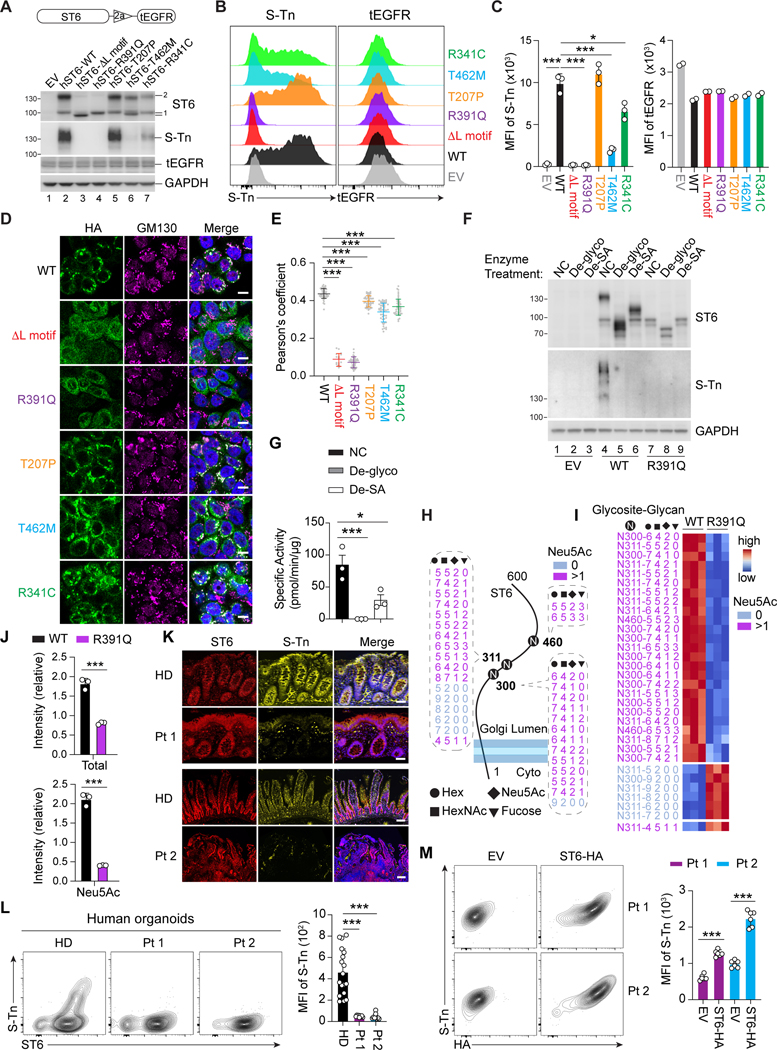
Mutations alter ST6 sialylation and physiological S-Tn expression
We next transduced HT29 human colorectal cells with lentiviral constructs in which full-length ST6 variants were fused by a T2A cleavage motif to a truncated human epidermal growth factor receptor (tEGFR) (Figure 5A, top). Transduction efficiency was measured by EGFR staining. ST6 immunoblotting of the WT sample revealed two bands (100 and 140 kD, respectively) (Figure 5A, ST6 band 1 and 2). Deletion of the L motif (AL), a conserved motif in the ST6 catalytic domain, and all mutations except T207P caused reduction or disappearance of the larger band 2 and variable reductions in sum of the main protein bands (1 + 2) (R391 Q=AL<R341 C<T462M<T207P=WT) (Figure S4A). S-Tn blotting showed that the 140 kD S-Tn band, comigrating with ST6 band 2, was strongest in the WT and T207P lysates, reduced in the T462M and R341C, and absent in the R391Q, EV, and AL samples (Figure 5A). All Pt variants except T207P showed decreased S-Tn levels, indicating that ST6 was auto-sialylated and naturally carried the S-Tn modification. We verified these findings by flow cytometry (Figures 5B and 5C) and immunoprecipitation (data not shown). The same defects were observed in 293T cells (Figures S4B–S4D). We next examined why Pt 2, harboring the active T207P allele and the inactive T462M allele, is susceptible to disease. By co-expressing various combinations of the WT, T207P, and T462M variants in HT29 cells, we found that the WT form paired with T462M could fully rescue S-Tn expression but the T207P form combined with T462M only partially restored ST6 activity (Figures S4E–S4G). Thus, T462M dominantly interferes with T207P function which explains why compound heterozygosity in Pt 2 causes functional ST6 deficiency.
Figure 5. Impaired enzymatic activity of ST6 mutations.
(A) (Top) Lentiviral co-expression vector with truncated Epidermal Growth Factor Receptor (tEGFR). 2a = 2A self-cleaving peptides. (Bottom) Immunoblot analysis of ST6 and Sialyl-Tn (S-Tn) with stable tEGFR lentiviral vector expression of WT and mutant forms of human ST6 (hST6) in HT29 cells. EV = empty vector. tEGFR and GAPDH are transduction and loading controls, respectively.
(B) Flow cytometry analysis of S-Tn and tEGFR in HT29 cells with stable expression of WT and mutant forms of human ST6.
(C) Quantification of S-Tn and tEGFR mean fluorescence intensity (MFI) in (B).
(D) Confocal photomicrographs of HA-tagged WT and ST6 mutants and the Golgi marker GM130 in HT29 cells with stable ST6 expression. Scale bar, 10 μm.
(E) Pearson’s coefficient scores for ST6 and Golgi staining shown visually in (D).
(F)ST6 and S-Tn were detected by immunoblot after deglycosylation enzyme mix (De-glyco) (PNGase F, O-Glycosidase, α2–3,6,8,9 Neuraminidase A, β1-4 Galactosidase S, β-N-acetylhexosaminidasef), or desialylation (De-SA) (α2–3,6,8,9 Neuraminidase A) treatment in HT29 cells with stable ST6-WT and ST6-R391Q expression.
(G) ST6 enzyme activity was quantitated after deglycosylation (De-glyco) and desialylation (De-SA) treatment.
(H) Asparagine (N)-linked glycosylation sites and the modified residues on ST6 in HT29 cells with stable expression of ST6-WT and ST6-R391Q. Glycoforms are shown with their glycan compositions presented as the numbers of Hex, HexNAc, Neu5Ac, and Fucose (dashed). Non-SA (Neu5Ac 0) and SA (Neu5Ac >1)-containing glycoforms are color coded. Cyto = cytoplasm.
(I) Intensity heatmap of the ST6 N-glycoforms as in (H). Color scale shows normalized N-glycoform intensities.
(J) Relative intensity of total or SA (Neu5Ac)-containing N-glycoforms on ST6 from HT29 cells with stable ST6-WT and ST6-R391Q expression.
(K) Immunofluorescent staining of colon biopsies of Pt 1 and HD, and small intestine of Pt 2 and HD showing S-Tn and ST6. Scale bar, 50 μm.
(L) Flow cytometry of S-Tn and ST6 in induced pluripotent stem cell (iPSC)-generated intestinal organoids from Pt 1, Pt 2, and HDs after 5 days of culture in GC-inducing conditions. Statistics for S-Tn MFI shown in right panel.
(M) Flow cytometry of S-T n and HA (ST6-HA) in iPSC-generated intestinal organoids from Pts 1 and 2 after transducing with EV and ST6-HA lentivirus. (Right) Statistics for S-Tn MFI. Data represent 3 experiments (A-G, I-M). Error bars represent the SD of samples within a group. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
See also Figure S4.
ST6 localizes to the Golgi apparatus where it exerts its enzymatic function (Sewell et al., 2006). Interestingly, we found that normal ST6 Golgi localization was compromised for the ΔL and Pt variants (Figures 5D and 5E). Previous studies suggest that glycosylation of glycotransferase proteins themselves affects catalytic activity and Golgi localization (Banfield, 2011; Nagai et al., 1997; Shauchuk et al., 2020). We focused on the R391Q mutation as it exemplified loss of ST6 function (Figure 5A). Electrophoresis showed that R391Q ST6 protein migrated much faster than WT, implying that post-translational modifications were lost (Figure 5F, compare lanes 4 and 7). Deglycosylation treatment shifted the WT ST6 bands near the R391Q protein size and extinguished the S-Tn bands, indicating that WT ST6 is heavily glycosylated (Figure 5F, compare lanes 4, 5, and 7). Using SA glycosidases, we found WT ST6 was modified by sialylation (Figure 5F, compare lanes 4 and 6), which was impaired in R391Q ST6 (Figure 5F, compare lanes 7 and 9). Hence, the R391Q mutation compromises both glycosylation and sialylation. Finally, measuring ST6 ST enzyme activity, we demonstrated that glycosylation and sialylation of ST6 are required for catalysis (Figure 5G).
Since N-glycosylation can strongly affect the activity and Golgi localization of glycosyltransferases (Nagai et al., 1997; Shauchuk et al., 2020), we performed N-glycoproteomic analysis in HT29 cells stably transduced with ST6-WT, empty vector (EV), and ST6-R391Q (Figure S4H). Non-supervised clustering revealed two clusters of differentially expressed N-glycans, which were decreased (cluster 1) or increased (cluster 2) N-glycans in ST6-WT expressing cells compared to EV and ST6-R391Q cells (Figure S4H). Of these, cluster 2 N-glycans were significantly enriched for SA decoration (Figure S4I). A volcano plot showed sialylation overrepresented on the N-glycans that were more abundant in ST6-WT than ST6-R391Q expressing cells (Figure S4J). We detected 32 heterogeneous glycoforms of ST6 at Asn300, Asn311, and to a lesser extent, Asn460 (Figure 5H). Of these, 25 glycoforms, all of which contain SA (Neu5Ac), were reduced by the R391Q mutant (Figure 5I). The intensities of both total and SA (Neu5Ac) containing glycoforms were decreased in R391Q, suggesting impaired N-linked glycosylation and sialylation (Figure 5J). Hence, the R391Q allele impairs Golgi localization and enzymatic function and causes global glycosylation changes.
Lastly, we observed strong S-Tn staining overlapping with ST6 in goblet and epithelial cells in HD colon samples that was lost in tissue from Pts 1 and 2 (Figure 5K), confirming the loss-of-function of these mutations in vivo. We also generated GC-enriched human intestinal organoids from Pts 1 and 2 iPSCs, expressing normal levels of GC signature genes (Gersemann et al., 2009) (Figures S4K–S4M). Pt 1 and 2 organoids stained normal for the ST6 protein but were devoid of S-Tn (Figure 5L). Lentiviral expression of HA-tagged ST6 (ST6-HA) in Pt organoids rescued S-Tn expression (Figure 5M). Hence, S-Tn is a physiological modification in normal intestinal GCs and certain patient ST6 variants have lost catalytic activity.
Defective ST6 predisposes to intestinal inflammation
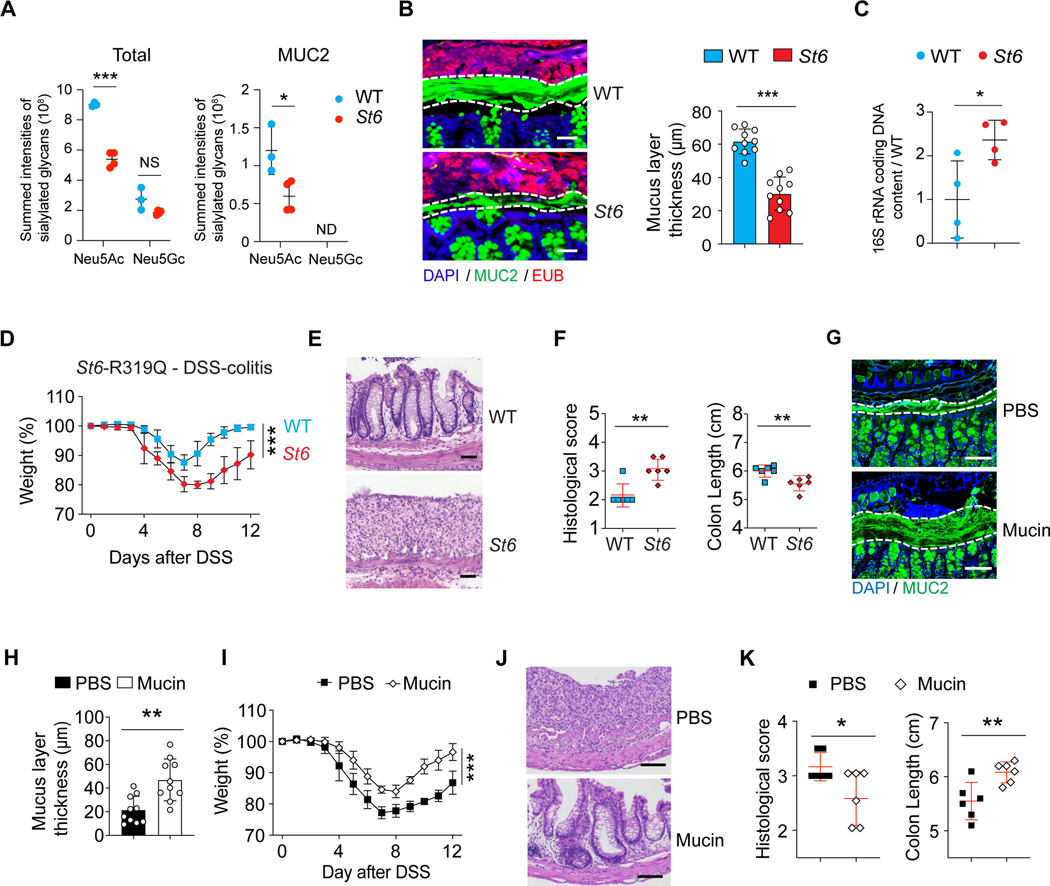
To further study how ST6 governs glycosylation in vivo, we introduced the R319Q substitution (equivalent to patient R391Q) into the mouse germline (St6 mice) (Figure S5A). Under SPF conditions, histopathology revealed no GI abnormalities. GCs, Paneth cells, and tight junction markers as well as immune cells were also normal in St6 mice (Figures S5B–S5M). N-glycoproteomics profiling and protein network analysis of IECs from WT and St6 mice again pinpointed the significant influence of ST6 on mucin glycosylation, including MUC2 (Figure S5N). The summed intensities of N-glycoforms revealed lowered protein sialylation, especially on MUC2, in St6 IECs (Figure 6A). Additionally, St6 mice displayed a marked thinning of the inner mucus layer in the colon (Figure 6B). Bacterial quantification revealed that St6 mice had increased gut microbiota invading the colonic mucus (Figure 6C).
Figure 6. St6 mutant mice are more susceptible to DSS-induced colitis.
(A) Summed intensities of the Neu5Ac and N-glycolylneuraminic acid (Neu5Gc) containing N-glycoforms for total protein and MUC2 in IECs from 12-week-old female WT and St6 mice. FDR = 0.3. NS = not significant. ND = not detected.
(B) Representative confocal micrographs (left) of colon samples stained for cell nuclei (DAPI), mucus (MUC2 antibody), and fluorescence in situ staining for bacteria (EUB) to quantify the thickness of the mucus layer in 10-week-old female WT and St6 mice (right). Scale bar, 100 μm.
(C) Ratio of the quantity of rRNA from infiltrating bacteria isolated from mouse colon mucus of 10-week-old female St6 mice compared to matched WT mice determined by Q-PCR using universal primers for bacterial 16S rRNA genes.
(D-F) Body weight changes (D), representative hematoxylin and eosin staining (E), histological scores (left) and colon length measurements (right) (F) in WT and St6 mice after oral administration at day 10 of 2.5% DSS water. Scale bar, 100 μm.
(G-K) DSS colitis was induced in St6 mice which were treated with either PBS control or mucins. The thickness of the mucus layer was measured before DSS water (G, H) together with analyses of colitis as in (D-F) (I-K). Scale bar, 100 μm. Data represent 3 experiments (A-K). Error bars represent the SD of samples within a group. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
See also Figures S5.
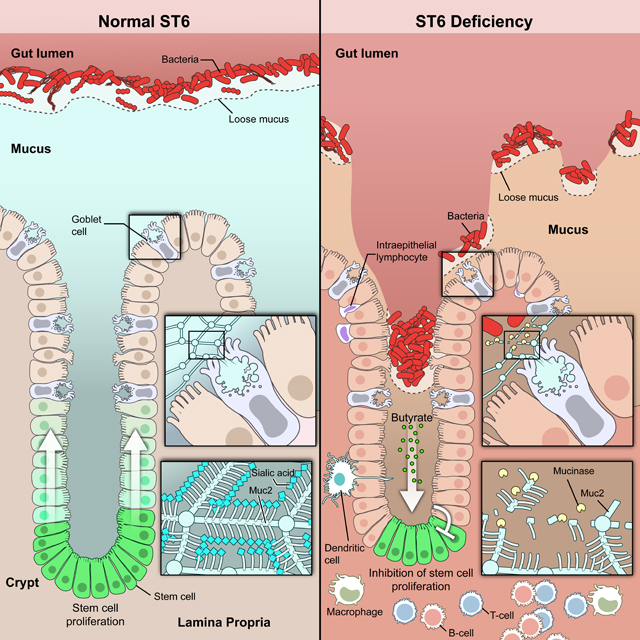
We next found that St6 deficiency led to more severe DSS-induced colitis compared to WT mice (Figures 6D–6F). Given that mucins were altered by St6 deficiency, we conjectured that the vulnerability of St6 mice to DSS-mediated colitis might be reversible by replenishing mucin. We administered mucin purified from WT mice or phosphate-buffered saline (PBS) control by oral gavage to St6 mice (Shan et al., 2013). We observed a thickening of the mucus barrier accompanied by a marked improvement in DSS colitis in mucin-treated mice (Figures 6G–6K). We conclude that ST6 is essential for the mucus barrier that prevents bacterial invasion and inflammation.
ST6 determines intestinal stem cell (ISC) homeostasis via microbiota
Reciprocal interactions between mucus and microbiota significantly contribute to intestinal homeostasis and inflammation (Chen et al., 2021; Kudelka et al., 2020). To elucidate the role of microbiota in colitis, we treated St6 mice with an antibiotic cocktail (ABX) 2 weeks prior to DSS administration. We found that ABX treatment eliminated the difference in severity of DSS colitis between WT and St6 mice (Figures 7A and 7B), suggesting that gut microbiota drive intestinal inflammation due to St6 deficiency.
Figure 7. Microbiota changes and excess butyric acid production in St6 mice impairs ISC proliferation.
(A, B) DSS colitis was induced in 10-week-old female WT and St6 mice treated with or without antibiotics (ABX) and analyzed as in Figures 6D and 6F.
(C) Principal component analyses (PCA) of gut microbiome using 16S rDNA gene sequencing from WT and St6 female mice after weaning (10 weeks). Dots = individual mice.
(C) Ten most abundant (Top 10) gut bacterial families in 10-week-old female WT and St6 mice.
(E) Mass spectrometry analysis of SCFAs as indicated in stool samples from 10-week-old WT and St6 female mice.
(F) DAPI and Ki-67 staining of colon of WT and St6 mice 10 days after DSS colitis (left) and % Ki-67+ cells (right). Scale bar, 100 μm.
(G) Gene set enrichment analysis (GSEA) of murine intestinal crypt stem cell upregulated (left) and downregulated (right) genes from RNA-seq data in Habowski et al., 2020 were compared in WT and St6 mice as in (F). NES = normalized enrichment score. FDR = false discovery rate.
(H) Enzyme-based fluorometric assay of HDAC enzyme activity in purified crypt intestinal stem cells (ISCs) from WT and St6 mice as in (F).
(I) WB analysis of histone H3 acetylation (H3K27ac and H3K9ac) in ISCs from WT and St6 mice as in (F). Total histone H3 = loading control.
(J) GSEA as in (G) of butyrate downregulated (left) and upregulated (right) genes from data in Kaiko et al., 2016 were compared in WT and St6 mice as in (F).
(K) Q-PCR RNA analyses of Polo-like kinase 1 (Plk1) and Dynamin 1 (Dmn1) in samples of crypt ISCs of WT and St6 mice as in (F).
(L) Normalized ATAC-seq sequencing tracks from WT and St6 mice as in (F) at the Plk1 (top) and the Dmn1 (bottom) loci. Orange shows areas of significant difference.
(M-O) DSS colitis induced in St6 mice treated with either PBS or carbenoxolone (CBX) analyzed as in Figures 6D–6F. Scale bar, 100 μm. Data represent 3 experiments (A-F, H, I, K-O). Error bars represent the SD of samples within a group. *, p < 0.05; **, p < 0.01.
See also Figure S6.
We next compared fecal bacteria in WT and St6 mice and found that the microbiota taxa diverged greatly after weaning (Figures 7C and S6A). 16S rDNA sequence analysis showed that adult St6 mice harbored a different microbiome with less diversity compared to WT mice (Figures 7C, S6B, and S6C). Detailed analysis showed reduced Muribaculaceae and increased Akkermansiaceae and Ruminococcaceae taxa as well as broader changes in phylum to genus levels in St6 mice (Figures 7D and S6D).
Previously, Akkermansiaceae and Ruminococcaceae taxa were shown to produce short chain fatty acid (SCFAs) (Koh et al., 2016). Indeed, our metabolomics study indicated disrupted levels of SCFAs in St6 fecal samples with elevated acetate, propionate, and butyrate, and reduced succinate and isovalerate (Figure 7E). Since acetate, propionate, and butyrate are known to repress cell expansion while succinate promotes it (Banerjee et al., 2020; Jan et al., 2002; Kaiko et al., 2016; Kim et al., 2019; Ko et al., 2017; Long et al., 2015; Sahuri-Arisoylu et al., 2021), we examined colon tissues from DSS-treated mice and found reduced ISC proliferation in St6 mice (Figure 7F). We attributed this to the microbiota since ABX treatment restored ISC proliferation (Figures S6E and S6F). Moreover, ATAC-seq and mRNA-seq on crypt ISCs (highly expressing Lgr5) during DSS colitis showed altered profiles in St6 mice (Figures S6G–S6J). We found that St6 deficiency resulted in a loss of ISC signature gene expression (Habowski et al., 2020) (Figure 7G). Additionally, many inflammatory response pathways were concomitantly increased in St6 mice (Figure S6K). Butyrate inhibits HDAC which impairs ISC proliferation and delays wound healing after epithelial injury (Kaiko et al., 2016; Koh et al., 2016). Indeed, we found that colonic crypt ISCs purified from DSS treated St6 mice exhibited decreased histone deacetylase (HDAC) activity and increased acetylation at the histone H3 K27 and K9 sites (Figures 7H and 7I). An mRNA-seq analysis displayed depletion of butyrate-downregulated gene set and enrichment of butyrate-upregulated gene in St6 mice (Kaiko et al., 2016) (Figure 7J). Based on butyrate-targeted gene sets (Kaiko et al., 2016), we validated ST6 deficiency-altered genes for stem/progenitor cell functions from mRNA-seq and ATAC-seq data by q-PCR, including cell proliferation and cell death genes (Plk1, Birc5, Nox1), cell cycle genes (Cdk2, Cdkn1c), cell proliferation and adhesion genes (Cldn2), and vesicle transport and trafficking genes (Dnm1, Kif1a). For example, we found reduced chromatin accessibility and mRNA expression for Plk1 and the converse for Dmn1 consistent with known gene control by butyrate (Figures 7K, 7L, S6L, and S6M). Thus, dysbiosis and excess butyrate impaired ISC proliferation, likely compromising epithelial repair during DSS colitis. Lastly, Foxo3 can mediate butyrate suppression of ISCs (Kaiko et al., 2016). We administered a known glycyrrhetinic acid derivative Foxo3 inhibitor, carbenoxolone (CBX) and found significantly improved disease in St6 mice (Figures 7M–7O) (Salcher et al., 2020).
Discussion
The evolution of 20 human STs that form specific sugar linkages suggests functional specialization, but few studies address SA functions in health and disease (InanlooRahatloo et al., 2014; Li and Chen, 2012; Varki, 2008). The sialylation of many O-and N-glycans was impaired in ST6-deficient cells. The latter may be caused by a failure of direct ST6 modification of N-glycans or, indirectly, by a failure of sialylation of O-glycans that alters activity of N-linked oligosaccharyl transferases. For example, glycoproteomics profiling uncovered STT3B, which is a principal catalytic subunit of the N-linked glycosylation complex, was altered by ST6 deficiency (Shrimal et al., 2015). There are both increases and decreases of new glycoforms because when sialylation is defective, sialylated glycoforms decrease and, by definition, the unsialylated versions increase. However, we observed that glycoform changes in ST6-deficient cells are not only limited to sialylated forms. If sialylation stabilizes the protein, then all glycoforms on the protein, whether sialylated or not, would be increased. There may also be altered sialylation of glycosylation enzymes affecting their modification of substrate proteins. Hence, ST6 deficiency alters glycoforms more globally than expected. Glycosylation, such as FUT2-dependent fucosylation and COSMC-dependent O-glycosylation, is critical for intestinal homeostasis (Kudelka et al., 2020; Pickard et al., 2014). Thus, the orchestration of different glycosyltransferases controls intestinal barrier function, inflammation, and the microbial composition of the GI ecosystem, which all influence IBD.
St6 deficiency not only alters mucus homeostasis but also commensalism. St6- deficient mice, like mucus-deficient mice, manifest dysbiosis with reduced bacterial diversity (Bergstrom et al., 2020; Chen et al., 2021; Wu et al., 2018). Thus, SA modification of glycans is a rate-limiting mechanism that achieves equipoise between the two different functions of mucus vis-à-vis the microbiota: foodstuff vs. barrier. Broader analyses of bacterial taxa showed expansion of Akkermansiaceae and Ruminococcaceae, important producers of SCFAs (Koh et al., 2016), leading to excessive butyrate production in St6 mice. Butyrate reportedly induces regulatory T cells (Tregs) and suppresses proinflammatory cytokines (Arpaia et al., 2013; Atarashi et al., 2013; Chang et al., 2014; Furusawa et al., 2013; Smith et al., 2013). Though proposed as an IBD treatment, no benefit in experimental colitis models or in clinical trials has been achieved (Atarashi et al., 2013; Chang et al., 2014; Furusawa et al., 2013; Hamer et al., 2010; Ji et al., 2016; Mayne and Williams, 2013; Smith et al., 2013; Takayama et al., 2007; Tarrerias et al., 2002). Our study potentially explains this paradox; we found no changes of Tregs or cytokine levels in St6 mice or the patients. Instead, inhibiting the butyrate pathway ameliorated DSS colitis possibly through rescuing stem cells (Eichele and Kharbanda, 2017). Perhaps butyrate may exert its function more potently on IECs during DSS colitis but affect adaptive immunity in the T cell-induced colitis model (Singh et al., 2001).
Previously, the S-Tn antigen was mainly investigated on tumor cells and was believed to be rarely, if at all, present on normal tissues (Prendergast et al., 2017). SA overexpression on malignant cells was reported to promote tumor cell metastases and poor disease outcome (Julien et al., 2012; Munkley, 2016). We find that S-Tn expression in humans is exclusively due to ST6 in GCs, implying that it functions ectopically in tumors. Further, we show that S-Tn is part of the mucosal barrier. Besides mucins, our mass spectrometry results show many proteins modified by ST6. Therefore, using specific S-Tn neutralizing antibodies for cancer immunotherapy may interfere with the physiological functions of S-Tn modification on normal proteins (Prendergast et al., 2017).
In the classic textbook conception, the intestine is the largest immunological organ where most T cells in the body reside and IBD reflects mainly immune dysregulation. Our study offers another perspective based on a growing body of evidence that IBD pathogenesis may also stem from changes in the biochemical composition of the mucus barrier, specifically glycan sialylation, and epithelium, so that anti-inflammatory or other immune-based treatments may not be sufficient. Rather, treatments that improve gut barriers, epithelial repair, or the microbial community should be considered. Mucin administration may be a relatively benign and broadly applicable intervention. Treatment with the FDA-approved drug CBX or antibiotics that affect glycosylation, ST6 function, or the gut microbiota may re-establish the GI barrier integrity.
Limitations of the study
The current studies on ST6 modulating intestinal bacteria are limited to in vitro cell-based assays and in vivo mouse model studies. The patients harbor rare genetic variants with limited patient access and material for experimental analysis. We found many more GI proteins besides MUC2 affected by ST6 deficiency and clarifying their biological significance requires further investigation.
STAR★Methods
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Michael J. Lenardo: (lenardo@nih.gov).
Materials availability
Reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and code availability
The human exome and genome sequencing data reported in this paper has been deposited at dbGaP. mRNA-seq and ATAC-seq in this paper are available at GEO. The mass spectrometry-based glycoproteomics data has been deposited to the ProteomeXchange Consortium via the PRIDE partner repository. Data are publicly available as of the date of publication. These accession numbers for the datasets are listed in the Key resources table.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-human ST6GALNAC1 | Sigma | Cat# HPA014975; RRID: AB 1857529 |
| Rabbit monoclonal recombinant anti-human MUC2 | Abcam | Cat# ab133555 |
| Rabbit monoclonal anti-HSP90 | Ceii Signaling Technology | Cat# 4877; RRID: AB 2233307 |
| Mouse monoclonal anti-human Sialyl Tn (S-Tn) [BRST3, B72.3] | BioLegend | Cat# 915206; RRID: AB 2616965 |
| Goat polyclonal anti-human EGFR | R&D | Cat# AF231; RRID: AB 355220 |
| Rabbit monoclonal anti-HA-tag | Ceii Signaling Technology | Cat# 3724; RRID: AB 1549585 |
| Rabbit monoclonal anti-GAPDH | Ceii Signaling Technology | Cat# 2118; RRID: AB 561053 |
| Rabbit polyclonal anti-H3K27ac | Abcam | Cat# ab4729; RRID: AB 2118291 |
| Rabbit monoclonal anti-H3K9ac | Ceii Signaling Technology | Cat# 9649; RRID: AB 823528 |
| Rabbit monoclonal anti-histone H3 | Ceii Signaling Technology | Cat# 4499; RRID: AB 10544537 |
| Mouse monoclonal anti-MUC2 | Santa Cruz Biotechnology | Cat# sc-515032; RRID: AB 2815005 |
| Mouse monoclonal anti-HA epitope tag | Covance | Cat# MMS-101P; RRID: AB 2314672 |
| Rabbit monoclonal recombinant anti-GM130 | Abcam | Cat# ab52649; RRID: AB 880266 |
| Rabbit polyclonal anti-MUC2 | GeneTex | Cat# GTX100664; RRID:AB 1950958 |
| Mouse monoclonal anti-Sialyl Tn | Abcam | Cat# ab115957; RRID: AB 10901541 |
| Rabbit polyclonal anti-CD45 | Abcam | Cat# ab10558; RRID: AB 442810 |
| Rabbit polyclonal anti-REG3G | Abgent | Cat# AP5606c; RRID: AB 10817069 |
| Goat polyclonal anti-rabbit IgG (H+L) highly crossadsorbed Alexa Fluor Plus 647 | Thermo Fisher Scientific | Cat# A32733; RRID: AB 2633282 |
| Goat polyclonal anti-mouse IgG (H+L) highly crossadsorbed Alexa Fluor Plus 647 | Thermo Fisher Scientific | Cat# A32728; RRID: AB 2633277 |
| Goat polyclonal anti-rabbit IgG (H+L) highly crossadsorbed Alexa Fluor Plus 555 | Thermo Fisher Scientific | Cat# A32732; RRID: AB 2633281 |
| Goat recombinant polyclonal anti-mouse IgG (H+L) Superclonal™ Alexa Fluor 555 | Thermo Fisher Scientific | Cat# A28180; RRID: AB 2536164 |
| Goat recombinant polyclonal anti-rabbit IgG (H+L) Superclonal™ Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A27034; RRID: AB 2536097 |
| Goat polyclonal anti-mouse IgG (H+L) highly crossadsorbed Alexa Fluor Plus 488 | Thermo Fisher Scientific | Cat# A32723; RRID: AB 2633275 |
| Mouse monoclonal Ultra-LEAF™ purified anti-human CD3 | BioLegend | Cat# 300334; RRID: AB 2616670 |
| Mouse monoclonal LEAF™ purified anti-human CD28 | BioLegend | Cat# 302914; RRID: AB 314316 |
| Human IgG recombinant monoclonal human EGFR (Research Grade Cetuximab Biosimilar) Alexa Fluor® 488-conjugated | R&D | Cat# FAB9577G-100 |
| Mouse anti-mouse-IgG-control-human | Santa Cruz Biotechnology | Cat# sc-2025; RRID: AB 737182 |
| Mouse monoclonal FITC anti-human CD57 | BioLegend | Cat# 322306; RRID: AB 535992 |
| Mouse monoclonal Brilliant Violet 421™ anti-human CD197 (CCR7) | BioLegend | Cat# 353208; RRID: AB 11203894 |
| Mouse monoclonal PerCP/Cyanine5.5 anti-human CD4 | BioLegend | Cat# 344608; RRID: AB 1953236 |
| Mouse monoclonal PE/Cyanine7 anti-human CD45RA | BioLegend | Cat# 304126; RRID: AB 10708879 |
| Armenian hamster monoclonal APC antimouse/rat/human CD27 | BioLegend | Cat# 124212; RRID: AB 2073425 |
| Mouse monoclonal Brilliant Violet 605™ anti-human CD8a | BioLegend | Cat# 301040; RRID: AB 2563185 |
| Mouse monoclonal PE anti-human CD28 | BioLegend | Cat# 302940; RRID: AB 2564147 |
| Mouse monoclonal Brilliant Violet 605™ anti-human CD3 | BioLegend | Cat# 317321; RRID: AB 11126166 |
| Mouse monoclonal Alexa Fluor® 488 antimouse/rat/human FOXP3 | BioLegend | Cat# 320012; RRID: AB 439748 |
| Mouse monoclonal PE anti-human CD25 | BioLegend | Cat# 302606; RRID: AB 314276 |
| Mouse monoclonal APC anti-human CD127 (IL-7Ra) | BioLegend | Cat# 351316; RRID: AB 10900804 |
| Mouse monoclonal PE/Cyanine7 anti-human CD194 (CCR4) | BioLegend | Cat# 359410; RRID: AB 2562431 |
| Mouse monoclonal Pacific Blue™ anti-human CD45RO | BioLegend | Cat# 304216; RRID: AB 493659 |
| Mouse monoclonal PE anti-human CD24 | BioLegend | Cat# 311106; RRID: AB 314855 |
| Mouse BV786 anti-Human IgM | BD Biosciences | Cat# 740998; RRID: AB 2740621 |
| Mouse monoclonal PE-Cy™5 anti-human CD20 | BD Biosciences | Cat# 561761; RRID: AB 10898182 |
| Mouse monoclonal FITC anti-human CD19 | BD Biosciences | Cat# 555412; RRID: AB 395812 |
| Mouse monoclonal PerCP anti-human CD27 | BioLegend | Cat# 302817; RRID: AB 893294 |
| Mouse monoclonal APC anti-human CD38 | BioLegend | Cat# 356606; RRID: AB 2561902 |
| Mouse monoclonal Brilliant Violet 421™ anti-human CD3 | BioLegend | Cat# 344834; RRID: AB 2565675 |
| Mouse monoclonal Brilliant Violet 510™ anti-human IgD | BioLegend | Cat# 348220; RRID: AB 2561945 |
| Mouse monoclonal PE anti-human CD56 (NCAM) | BioLegend | Cat# 362507; RRID: AB 2563924 |
| Mouse monoclonal APC anti-human CD16 | BioLegend | Cat# 302011; RRID: AB 314211 |
| Mouse monoclonal FITC eBioscience™ anti-human CD14 | Thermo Fisher Scientific | Cat# 11–0149-42; RRID: AB 10597597 |
| Mouse monoclonal Brilliant Violet 421™ anti-human HLA-DR | BioLegend | Cat# 307636; RRID: AB 2561831 |
| Mouse monoclonal PE/Cyanine7 anti-human CD11c | BioLegend | Cat# 371507; RRID: AB 2650779 |
| Mouse monoclonal PerCP anti-human CD19 | BioLegend | Cat# 302228; RRID: AB 893272 |
| Rat monoclonal Brilliant Violet 605™ anti-mouse/human CD44 | BioLegend | Cat# 103047; RRID: AB 2562451 |
| Mouse monoclonal Alexa Fluor® 700 anti-human CD3 | BioLegend | Cat# 344821; RRID: AB 2563419 |
| Mouse monoclonal PerCP anti-human CD8a | BioLegend | Cat# 300922; RRID: AB 1575072 |
| Mouse monoclonal APC anti-human CD4 | BioLegend | Cat# 300552; RRID: AB 2564153 |
| Mouse monoclonal Brilliant Violet 510™ anti-human CD183 (CXCR3) | BioLegend | Cat# 353725; RRID: AB 2562066 |
| Mouse monoclonal Brilliant Violet 421™ anti-human CD184 (CXCR4) | BioLegend | Cat# 306517; RRID: AB 10901163 |
| Mouse monoclonal FITC anti-human CD196 (CCR6) | BioLegend | Cat# 353411; RRID: AB 10915473 |
| Mouse monoclonal PE/Cyanine7 anti-human CD194 (CCR4) | BioLegend | Cat# 359409; RRID: AB 2562430 |
| Armenian hamster monoclonal PE anti-human CCR10 | BioLegend | Cat# 341503; RRID: AB 1595542 |
| Mouse monoclonal FITC anti-human CD8a | BioLegend | Cat# 300906; RRID: AB 314110 |
| Mouse monoclonal APC anti-human CD69 | BioLegend | Cat# 310910; RRID: AB 314845 |
| Rat monoclonal Brilliant Violet 711 ™ anti-mouse CD45 | BioLegend | Cat# 103147; RRID: AB 2564383 |
| Armenian hamster monoclonal FITC anti-mouse TCR p chain | BioLegend | Cat# 109206; RRID: AB 313429 |
| Rat monoclonal PerCP/Cyanine5.5 anti-mouse CD4 | BioLegend | Cat# 100434; RRID: AB 893324 |
| Armenian hamster monoclonal Alexa Fluor® 647 antimouse TCR y/6 | BioLegend | Cat# 118133; RRID: AB 2566406 |
| Syrian hamster monoclonal Alexa Fluor® 700 antimouse CD3e | Biolegend | Cat# 152316; RRID: AB 2632713 |
| Rat monoclonal PE anti-mouse CD8a | BD Biosciences | Cat# 553033; RRID: AB 394571 |
| Mouse monoclonal PE-Cyanine7 eBioscience™ antihuman CD8b | Thermo Fisher Scientific | Cat# 25–5273-41; RRID: AB 11217487 |
| Rat monoclonal Brilliant Violet 510™ anti-mouse/human CD45R/B220 | BioLegend | Cat# 103248; RRID: AB 2561394 |
| Rat monoclonal FITC anti-mouse CD8a | BioLegend | Cat# 100706; RRID: AB 312745 |
| Rat monoclonal PerCP anti-mouse/human CD44 | BioLegend | Cat# 103035; RRID: AB 10639933 |
| Rat monoclonal APC anti-mouse CD62L | BioLegend | Cat# 104412; RRID: AB 313099 |
| Rat monoclonal PE anti-mouse CD4 | BioLegend | Cat# 100407; RRID: AB 312692 |
| Mouse monoclonal PE-Cy™7 anti-mouse NK-1.1 | BD Biosciences | Cat# 552878; RRID: AB 394507 |
| Mouse monoclonal BV650 anti-mouse NK-1.1 | BD Biosciences | Cat# 564143; RRID: AB 2738617 |
| Rat monoclonal EOMES (Dan11 mag), eFluor 450, eBioscience™ | Thermo Fisher Scientific | Cat# 48–4875-82; RRID: AB 2574062 |
| Rat monoclonal Alexa Fluor® 488 anti-mouse CD3 | BioLegend | Cat# 100212; RRID: AB 493530 |
| Rat monoclonal Alexa Fluor® 488 anti-mouse CD19 | BioLegend | Cat# 115524; RRID: AB 493339 |
| Rat monoclonal Alexa Fluor® 488 anti-mouse Ter119 | BioLegend | Cat# 116215; RRID: AB 493402 |
| Rat monoclonal Alexa Fluor® 488 anti-mouse Gr-1 | BioLegend | Cat# 108419; RRID: AB 493480 |
| Rat monoclonal Alexa Fluor® 488 anti-mouse CD11 b | BioLegend | Cat# 101219; RRID: AB 493545 |
| Hamster monoclonal Alexa Fluor® 488 anti-mouse CD11c | BioLegend | Cat# 117313; RRID: AB 492849 |
| Hamster monoclonal Alexa Fluor® 488 anti-mouse FceRIa | BioLegend | Cat# 134329; RRID: AB 2687238 |
| Mouse monoclonal BUV395 anti-mouse CD45.2 | BD Biosciences | Cat# 565448; RRID: AB 2738867 |
| Rat monoclonal BUV737 anti-mouse CD335 (NKp46) | BD Biosciences | Cat# 612805; RRID: AB 2870131 |
| Mouse monoclonal Alexa Fluor® 647 anti-T-bet | BD Biosciences | Cat# 561267; RRID: AB 10564093 |
| Armenian hamster monoclonal PE anti-mouse CD196 (CCR6) | BioLegend | Cat# 129804; RRID: AB 1279137 |
| Mouse monoclonal PE-CF594 anti-mouse RORYt | BD Biosciences | Cat# 562684; RRID: AB 2651150 |
| Rat monoclonal PE/Cyanine7 anti-mouse CD127 (IL-7Ra) | BioLegend | Cat# 135014; RRID: AB 1937265 |
| Rat monoclonal Brilliant Violet 421 ™ anti-mouse Ly-6C | BioLegend | Cat# 128031; RRID: AB 2562177 |
| Rat monoclonal anti-CD11 b mouse | BD Biosciences | Cat# 553310; RRID: AB 394774 |
| Rat monoclonal PerCP/Cyanine5.5 anti-mouse I-A/I-E | BioLegend | Cat# 107625; RRID: AB 2191072 |
| Rat monoclonal Alexa Fluor® 647 anti-mouse F4/80 | BioLegend | Cat# 123121; RRID: AB 893492 |
| Rat monoclonal PE anti-mouse Ly-6G | BioLegend | Cat# 127608; RRID: AB 1186099 |
| Armenian hamster monoclonal PE/Cyanine7 anti-mouse CD11c | BioLegend | Cat# 117318; RRID: AB 493568 |
| Rat monoclonal V450 anti-mouse Foxp3 | BD Biosciences | Cat# 561293; RRID: AB 10611728 |
| Rat monoclonal APC-R700 anti-mouse CD25 | BD Biosciences | Cat# 565134; RRID: AB 2744344 |
| Bacterial and virus strains | ||
| Stellar™ Competent Cells | Takara | Cat# 636766 |
| One Shot™ Stbl3™ Chemically Competent E. coli | ThermoFisher Scientific |
Cat# C737303 |
| BL21(DE3) Competent Cells | ThermoFisher Scientific |
Cat# EC0114 |
| Sendai virus (from Cytotune 2.0 kit) | ThermoFisher Scientific |
Cat# A16517 |
| Biological samples | ||
| Healthy human PBMCs | This study | NIH biood bank |
| Patient and family PBMCs (Pt 1) | This study | Marmara University School of Medicine |
| Patient and family PBMCs (Pt 2) | This study | Schneider Children’s Medical Center of Israel |
| Human Colon Total RNA | Zyagen | Cat# HR-311 |
| Human Small Intestine Total RNA | Zyagen | Cat# HR-306 |
| Human Spleen Paraffin Sections | Zyagen | Cat# HP-701 |
| Human Thymus Paraffin Sections | Zyagen | Cat# HP-702 |
| Human Lung Paraffin Sections | Zyagen | Cat# HP-601 |
| Human Colon Paraffin Sections | Zyagen | Cat# HP-311 |
| Human Salivary Gland Paraffin Sections | Zyagen | Cat# HP-317 |
| Human Lung Total Protein | zyagen | Cat# HT-601 |
| Human Rectum Total Protein | zyagen | Cat# HT-312 |
| Human Small Intestine Total Protein | zyagen | Cat# HT-306 |
| Human Spleen Total Protein | zyagen | Cat# HT-701 |
| Human Stomach Total Protein | zyagen | Cat# HT-302 |
| Human Liver Total Protein | zyagen | Cat# HT-314 |
| Human Brain Total Protein | zyagen | Cat# HT-201 |
| Human Esophagus Whole Tissue Lysate | novusbio | Cat# NB820–59214 |
| Human Kidney Total Protein | zyagen | Cat# HT-901 |
| Human Small Intestine, Duodenum Paraffin Sections | zyagen | Cat# HP-307 |
| Human Small Intestine, Ileum Paraffin Sections | zyagen | Cat# HP-309 |
| Human Esophagus Paraffin Sections | zyagen | Cat# HP-301 |
| Human Stomach Paraffin Sections | zyagen | Cat# HP-302 |
| Human Multi-tissue Tissue MicroArray | novusbio | Cat# NBP2–78067 |
| Chemicals, peptides, and recombinant proteins | ||
| HILIC amphion | Welch | Lot# 7301.26 |
| Sera-Mag SpeedBeads | GE Healthcare | Cat# 45152101010250 Cat# 65152105050250 |
| Corning® Matrigel® Growth Factor Reduced (GFR) Basement Membrane Matrix, LDEV-free, 10 mL | Corning Life Science | Cat# 354230 |
| CryoStor® CS10 Freeze Media | Biolifesolutions | Cat# 210374 |
| A 83–01 | Cayman | Cat# 9001799 |
| DBZ | Cayman | Cat# 14627 |
| STEMdiff™ Intestinal Organoid Growth Medium | Stemcell | Cat# 05145 |
| IntestiCult™ Organoid Growth Medium (Mouse) | Stemcell | Cat# 06005 |
| mEGF | PeproTech | Cat# 315–09 |
| mWnt3a | PeproTech | Cat# 315–20 |
| Essential 8™ Medium | ThermoFisher Scientific |
Cat# A1517001 |
| Opti-MEM™ I Reduced Serum Medium | ThermoFisher Scientific |
Cat# 31985062 |
| Guanidine hydrochloride | Sigma | Cat# G3272–25G |
| Iodoacetamide | Sigma | Cat# I1149–5G |
| Dextran Sulfate Sodium Salt | MPbio | Cat# SKU 0216011050 |
| BsmBI-v2 | NEB | Cat# R0739S |
| STEMdiff™ Intestinal Organoid Kit | Stemceii | Cat# 05140 |
| T4 Polynucleotide Kinase | NEB | Cat# M0201s |
| T4 DNA Ligase | NEB | Cat# M0202L |
| Human TruStain FcX™ (Fc Receptor Blocking Solution) | Bioiegend | Cat# 422301 |
| Protein Deglycosylation Mix II | NEB | Cat# P6044S |
| a2–3,6,8,9 Neuraminidase A | NEB | Cat# P0722S |
| PNGase F (Glycerol-free), Recombinant | NEB | Cat# P0709S |
| O-Glycosidase | NEB | Cat# P0733S |
| Recombinant Human ST6GALNAC1 Protein | R&D | Cat# 9154-GI-020 |
| Manganese(II) chloride | Sigma | Cat# 244589–10G |
| Asialofetuin from fetal calf serum | Sigma | Cat# A4781–50MG |
| CMP-Cy5-Sialic Acid | R&D | Cat# ES302–050 |
| Corning® Matrigel® Growth Factor Reduced (GFR) Basement Membrane Matrix, LDEV-free, 10 mL | Corning Life Science | Cat# 354230 |
| TrypLE™ Express Enzyme (1X), no phenol red | Thermo Fisher Scientific | Cat# 12604013 |
| BSA | Sigma | Cat# A7030 |
| EDTA | Thermo Fisher Scientific | Cat# AM9261 |
| StcE | From Dr. Bertozzi’s lab | N/A |
| OgpA | From Dr. Lawrence Tabak’s lab, NIDCR | N/A |
| polybrene | Sigma-Aldrich | Cat# TR-1003 |
| Blasticidin S HCl | Thermo Fisher Scientific | Cat# A1113903 |
| Puromycin | Thermo Fisher Scientific | Cat# A1113803 |
| SCF | R&D | Cat# 7466-SC |
| StemSpan™SFEM II medium | StemCell | Cat# 09605 |
| IL-3 | R&D | Cat# 203-IL |
| EPO | R&D | Cat# 287-TC |
| IGF-1 | R&D | Cat# 201-G1 |
| Dexamethasone | Sigma | Cat# D4902 |
| holo-transferrin | Sigma | Cat# I0665 |
| IWP2 | Sigma | Cat# I0536 |
| DAPT | Sigma | Cat# D5942 |
| Y27632 | Cayman | Cat# 10005583 |
| DAPI Fluoromount-G | SouthernBiotech | Cat# 0100–20 |
| GalNAcEXO | From Dr. Lawrence Tabak’s lab, NIDCR | N/A |
| Dithiothreitol | Sigma | Cat# 10197777001 |
| Trifluoroacetic acid (TFA) | Sigma | Cat# I6508–1L |
| Urea | Sigma | Cat# U0631–1 KG |
| Formic acid | Thermo Fisher Scientific | Cat# A117–50 |
| Trypsin | Promega | Cat# V5111 |
| Tris-(2-Carboxyethyl)phosphine, Hydrochloride (TCEP) | Thermo Fisher Scientific | Cat# I2556 |
| Acetonitrile (ACN) | Thermo Fisher Scientific | Cat# A9554 |
| Guanidine hydrochloride solution | Sigma | Cat# SRE0066– 100ML |
| carbenoxolone (CBX) | Selleckchem | Cat# S4368 |
| Ampicillin sodium salt | Sigma | Cat# A9518 |
| Metronidazole | Sigma | Cat# M3761 |
| Vancomycin hydrochloride from Streptomyces orientalis | Sigma | Cat# V2002 |
| Neomycin trisulfate salt hydrate | Sigma | Cat# N1876 |
| Liberase | Sigma | Cat# 5401020001 |
| DNase I | Sigma | Cat# 4716728001 |
| Collagenase D | Sigma | Cat# 11088858001 |
| phenol-chloroform-isoamyl alcohol (25:24:1) | Sigma | Cat# P2069–100ML |
| SCFA standards | Protein Characterization Core, NCI |
N/A |
| Percoll Plus | GE Healthcare | Cat# 17544501 |
| Monensin Solution | Biolegend | Cat# 420701 |
| EcoR I-HF | NEB | Cat# R3101L |
| MluI-HF | NEB | Cat# R3198S |
| Lipo3000 transfection reagent | ThermoFisher Scientific |
Cat# L3000075 |
| Sodium Pyruvate (100 mM) | ThermoFisher Scientific |
Cat# 11360070 |
| Percoll | Sigma | Cat# GE17–0891-01 |
| UEA 1 fluorescin | Vector Laboratories | Cat# FL-1061 |
| Cy5-Peanut agglutinin (PNA) | Vectorlabs | Cat# CL-1075–1 |
| Fixable Viability Dye eFluor™ 780 | Thermo Fisher Scientific | Cat# 65–0865-18 |
| β-mercaptoethanol | Gibco | Cat# 21985 |
| L-Glutamine | Gibco | Cat# 25030 |
| NEAA | Gibco | Cat# 11140 |
| Penicillin and streptomycin | Gibco | Cat# 15140 |
| Critical commercial assays | ||
| TMT10plex™ Isobaric Labeling Reagent Set | ThermoFisher Scientific |
Cat# 90110 |
| Pierce BCA Protein Assay Kit | ThermoFisher Scientific |
Cat# 23225 |
| XBridge Peptide BEH C18 Column, 300Ä, 3.5 μm, 1 mm × 150 mm | Waters | Cat# SKU: 186003606 |
| Histone Deacetylase (HDAC) Activity Assay Kit (Fluorometric) (ab156064) | Abcam | Cat# ab156064 |
| Human Pan T cell isolation kit | Miltenyi | Cat# 130–096-535 |
| Dynabeads™ Human T-Activator CD3/CD28 for T Cell Expansion and Activation | Thermo Fisher Scientific | Cat#11131D |
| Dynabeads™ Protein G for Immunoprecipitation | ThermoFisher Scientific |
Cat# 10004D |
| Lenti-X™ Concentrator | Takara | Cat# 631231 |
| PrimeScript™ RT Reagent Kit (Perfect Real Time) | Takara | Cat# RR037A |
| In-Fusion® HD Cloning Kit | Takara | Cat# 639650 |
| Lipofectamine™ 3000 Transfection Reagent | ThermoFisher Scientific |
Cat# L3000075 |
| NuPAGE™ 4–12% Bis-Tris Protein Gels, 1.5 mm, 15-well | ThermoFisher Scientific |
Cat# NP0336B0X |
| NuPAGE™ 3 to 8%, Tris-Acetate, 1.5 mm, Mini Protein Gel, 15-well | ThermoFisher Scientific |
Cat# EA03785BOX |
| Foxp3 / Transcription Factor Staining Buffer Set | Thermo Fisher Scientific, | Cat# 00–5523-00 |
| BCA protein assay kit | Thermo Fisher Scientific, | Cat# 23225 |
| Sialyltransferase Activity Kit | R&D | Cat# EA002 |
| In-Fusion® HD Cloning Plus CE kit | Iakara | Cat# 638919 |
| Anti-Adherence Rinsing solution | Stemceii | Cat# 07010 |
| ATAC-Seq Kit | Active motif | Cat# 53150 |
| LabSafe GEL Blue | G-biosciences | Cat# 786–35 |
| C18 cartridge | Seq-Pak C18 | Cat# WAI054955 |
| Sera-Mag SpeedBeads with a hydrophilic surface | GE Healthcare | Cat# 45152101010250 |
| Sera-Mag SpeedBeads with a hydrophobic surface | GE Healthcare | Cat# 65152105050250 |
| Halt™ Protease Inhibitor Cocktail, EDTA-Free | Thermo Fisher Scientific | Cat# 78439 |
| Halt™ PhosphataseInhibitor Cocktail | Thermo Fisher Scientific | Cat# 78427 |
| EndoTrap red | Biovendor | Cat# LEI0001 |
| ToxinSensor™ Chromogenic LAL Endotoxin Assay Kit | Genescript | Cat# L00350 |
| DNeasy Blood & Tissue Kit | Qiagen | Cat# 69506 |
| Cytotune 2.0 kit | ThermoFisher Scientific |
Cat# A16517 |
| Deposited data | ||
| Site-specific N-glycoproteome MS raw and searched data | This paper | https://www.ebi.ac.uk/pride/archiveidentifierPXD028239 |
| mRNA-seq and ATAC-seq data | This paper | GEO ID: GSE183821 |
| whole exome sequencing (WES) (Pt 1) | This paper | dbGaP ID: phs002598.v1.p1 |
| Experimental models: Celi lines | ||
| HI-29 | AICC | Cat# HTB-38 |
| LS 180 | AICC | Cat# CL-187 |
| Lenti-X™ 293T Cells | Iakara | Cat# 632180 |
| HEK293I | AICC | Cat# CRL-3216 |
| Experimental models: Organisms/strains | ||
| Mouse: Myd88flox/flox | The Jackson Laboratory | Cat# 008888 |
| Mouse: Vil1cre | The Jackson Laboratory | Cat# 21504 |
| Mouse: Tlr4−/− | The Jackson Laboratory | Cat# 29015 |
| Mouse: St6 (St6galnac1R319Q) | This paper | N/A |
| Oligonucleotides | ||
| A full list is provided in Table S3 | ||
| Recombinant DNA | ||
| pLV-EFIa-IRES-Puro | Hayer et al., 2016 | Addgene, Cat# 85132 |
| psPAX2 | A gift from Didier Trono | Addgene, Cat# 12260 |
| pMD2.G | A gift from Didier Trono | Addgene, Cat# 12259 |
| lentiGuide-Puro | Sanjana et al., 2014 | Addgene, Cat# 52963 |
| lentiCas9-Blast | Sanjana et al., 2014 | Addgene, Cat# 52962 |
| lentiCRISPRv2 | Sanjana et al., 2014 | Addgene, Cat# 52961 |
| pLV-EF1a-2a-tEGFR | This paper | N/A |
| Software and algorithms | ||
| ImageJ | Schneider et al., 2012 | https://imagej.nih.gov/ij/ |
| Graphpad Prism | https://www.graphpad.com/scientific-software/prism | version 9.0 |
| QIIME2 | Bolyen et al., 2019 | version 2019.10 |
| DADA2 method | Callahan et al., 2016 | https://benjjneb.github.io/dada2/ |
| Thermo Xcalibur software | https://www.thermofisher.com/order/catalog/product/0PT0N-30965 | Cat# 0PT0N-30965 |
| HOMER | http://homer.ucsd.edu/homer/interactions/ | version 4.11.1 |
| Python | https://www.python.org/ | Version 3.6–3.8 |
| Macs | https://pypi.org/project/MACS/ | version 1.4.3 |
| Bedtools | https://bedtools.readthedocs.io/en/latest/ | version 2.30.0 |
| HTSeq | https://pypi.org/project/HTSeq/ | version 0.11.4 |
| DESeq2 | version 1.30.1 | |
| RStudio | https://www.rstudio.com/ | version 1.4.1106 |
| R | https://www.r-project.org | version 4.0.4 |
| RColorBrewer | http://colorbrewer2.org/ | version 1.1–2 |
| Samtools | http://www.htslib.org/ | version 1.12 |
| Deeptools | https://deeptools.readthedocs.io/en/develop/ | version 3.5.0 |
| Enrichr | Kuleshov at el., 2016 | https://maayanlab.cloud/Enrichr/ |
| FastQC | https://www.bioinformatics.babraham.ac.uk | version 0.11.9 |
| Bowtie | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml | version 1.3.0 |
| IGV | https://software.broadinstitute.org/software/igv/ | version 2.10.0 |
| Benchling algorithm | https://www.benchling.com/crispr/ | https://www.benchling.com |
| PyMOL | https://pymol.org/2/ | version 2.3.1 |
| I-TASSER | Roy et al., 2010 | https://zhanggroup.org/I-TASSER/ |
| pFind3.0 | Chi et al., 2018 | https://github.com/pFindStudio/pFind3 |
| FlowJo | https://www.flowjo.com/ | version 10.6.2 |
| GlycoBinder | Fang et al., 2020 | https://github.com/IvanSilbern/GlycoBinder |
| IPA | https://digitalinsights.qiagen.com/products-overview/discovery-insights-portfolio/analysis-and-visualization/qiagen-ipa/ | version 01–20-04 |
| Imaris | https://imaris.oxinst.com/ | version 9.8.1 |
| Snapgene | https://www.snapgene.com/ | version 6.0 |
| Cytoscape | https://cytoscape.org/ | version 3.8.2 |
| Other | ||
| Agilent 1100 series HPLC | Agilent Technologies | https://www.agilent.com/en/products/liquid-chromatography |
| Orbitrap Fusion™ Lumos™ Tribrid™ Mass Spectrometer | ThermoFisher Scientific |
https://www.thermofisher.com/order/catalog/product/IQLAAEGAAPFADBMBHQ |
| Fluorescent imager Azure 500 | Azure biosystems | https://www.azurebiosystems.com/imaging-systems/azure-500/ |
Experimental model and subject details
Human subjects
All enrolled subjects provided written informed consent and were collected through protocols following local ethics and IRB recommendations. Pt 1 patient was enrolled on NIH Protocol 06-I-0015 (clinicaltrials.gov NCT00246857). IRB approval was obtained from the NIH IRB 02/16/2021. Pt 2 patient consented to protocol SMC-3312–16 by Sheba Medical Center. Pt 3 patient consented to a protocol approved at Hospital for Sick Children (REB 1000024905). All experiments were carried out with the approval of the Research Ethics Board at the Hospital for Sick Children, Toronto, Canada. Informed consent to participate in research was obtained from all participants. Blood from healthy donors was obtained at the NIH Clinical Center under approved protocols.
Mice
All protocols were approved by the NIAID Animal Care and Use Committee and followed NIH and DHSS guidelines. Myd88flox/flox, Vil1cre and Tlr4−/− mice were obtained from Jackson Laboratories. IEC-specific Myd88-deficient (Myd88fl/flV/V1cre) mice were generated by crossing Myd88flox/flox with Vil1cre mice. St6 (St6galnac1R319Q) C57BL6/N mice were generated by Transgenic Core in National Heart, Lung, and Blood Institute (NHLBI). Briefly, the St6 knock in mouse line was generated using CRISPR/Cas9 (Wang et al., 2013). A sgRNA (GCAACGGAGGGAGAACATAC) was designed to cut near the mutation site and was made using ThermoFisher’s custom in vitro transcription service. A donor oligonucleotide (TGGGCAACGGGGGCATCCTGAATGATTCACGTGTTGGCCGGGAGATAGACAGCCATGACTATGTTTTCCAGTATGTTCTCATCCTCCGTTGCTTGTAGATATGGGCTATAATGGGCGACCCGATAAGTGTCCTGTGCTGGTTTGAATTGGAATGGCCC) was purchased from IDT. The sgRNA (20 ng/μL) and donor oligonucleotides (100 ng/μL) were co-microinjected with Cas9 mRNA (20 ng/μL, purchased from Trilink Biotechnologies) into the cytoplasm of zygotes collected from C57BL6/N mice. Injected embryos were cultured in M16 medium (Millipore Sigma) overnight in a 37 °C incubator with 6% CO2. The next morning, embryos that reached 2-cell stage of development were implanted into the oviducts of pseudopregnant surrogate mothers. Offspring born to foster mothers were genotyped by PCR and Sanger DNA sequencing. Experiments used true littermates from an St6+/− father and an St6+/− mother or St6+/− father and harem held St6+/− mothers. After weaning, the male and female mice were separated and same genotype mice were kept in the same cage. Gender-matched 10-to 12-week-old female mice were used for most experiments. Otherwise, more specific usage is indicated in figure legends.
Method details
Exome and whole genome sequencing analysis
Genomic DNA was isolated from peripheral blood mononuclear cells (PBMCs) of proband, parents, and healthy relatives from each pedigree. Exome sequencing was carried out using the IDT XGen exome target reagent or the Agilent SureSelect Human All Exon 50Mb Kit (Agilent Technologies) followed by next-generation short read sequencing on an Illumina HiSeq 2500 instrument. Whole genome sequencing (WGS) was performed based on Standard Coverage Human WGS from Broad Institutes. For individual samples, whole exome sequencing (WES) produced ~50–100X sequence coverage for targeted regions and WGS produced 60X coverage for proband and 30X coverage for family members. Short-read sequence data was mapped and aligned to the reference human genome assembly (build 19) using the Burrows-Wheeler Aligner (BWA) with default parameters. Variants and genotypes were called using the Genome Analysis ToolKit (GATK). Variants were then annotated according to their functional impact on encoded proteins and prioritized based on frequency, conservation, deleteriousness, and potential disease-causing genetic models. Variants with minor allele frequency > 0.1% in the dbSNP (version 137), 1000 Genomes (1094 subjects of various ethnicities; May 2011 data release), Exome Sequencing Project (ESP, 4300 European and 2203 African-American subjects; last accessed August 2016), the Genome Aggregation Consortium (gnomAD) database, or Yale internal database (2500 European subjects including 1894 Turkish exomes) were filtered out. All possible inheritance modes and variants including de novo, recessive, and hemizygous were considered and analyzed. Autosomal-recessive inheritance was investigated and genes with rare homozygous or compound heterozygous variants were prioritized.
Human peripheral blood mononuclear cells (PBMCs) isolation and primary T cell culture
Human PBMCs were isolated by Ficoll-Paque PLUS (GE Healthcare) density gradient centrifugation, washed twice in phosphate-buffered saline (PBS), and resuspended at 106/mL in complete RPMI 1640 (cRPMI) medium containing 10% fetal bovine serum, 2 mM glutamine, and 100 U/mL each of penicillin and streptomycin (Invitrogen). For T cell activation, human primary T cells were isolated by MACS beads (MACS, 130–096-535) and activated with 1 μg/mL anti-CD3 (clone HIT3a, BioLegend) and 1 μg/mL anti-CD28 (clone CD28.2, BioLegend) or with Dynabeads® Human T-Activator CD3/CD28 (Thermo Fisher Scientific).
Flow cytometry
For standard surface staining, PBMCs and T cell blasts were washed once in fluorescence activated cell sorting (FACS) buffer (PBS containing 1% BSA and 2 mM EDTA) and stained in 50 μL of FACS buffer containing indicated fluorochrome-labeled antibodies and Fixable Viability Dye (eBioscience™ Fixable Viability Dye eFIuor™ 780, Thermo Fisher Scientific, cat: 65–0865-18) for 30 min. Cells were then washed three times in FACS buffer and fixed before acquisition on either a LSR II or LSRFortessa (BD Biosciences). Cells were washed three times in PBS before acquisition. For intracellular Foxp3 staining, cells were fixed with eBioscience™ Foxp3 / Transcription Factor Staining Buffer Set (Thermo Fisher Scientific, cat: 00–5523-00) by following the manuals. For S-Tn staining, cells were detached from the plates with cell scraper without trypsin digestion and resuspended in FACS buffer. After washing three times with FACS buffer, cells were stained in 50μL of FACS buffer containing Fixable Viability Dye, S-Tn antibodies (Purified anti-BRST-3 Antibody, clone: B72.3, Biolegend, cat: 915206), or (Anti-Sialyl Tn antibody [STn 219], abcam, cat: ab115957) (Loureiro et al., 2018) (Prendergast et al., 2017) and Human EGFR (Cetuximab) Alexa Fluor 488-conjugated Antibody (R&D, cat: FAB9577G-100) for 30 min. Cells were then washed three times in FACS buffer, and anti-Mouse IgG APC secondary antibody staining for 30 min followed. Cells were washed three times in PBS before acquisition. Data were analyzed using FlowJo v. 10.6.2.
Single cell RNA-Seq analysis
Single cell data used in this study are from Smillie et al. (Smillie et al., 2019). A total of 366650 cells from the colon mucosa of 18 ulcerative colitis (UC) Pts and 12 healthy individuals were downloaded as RData for epithelial, stromal, and immune cells (https://www.dropbox.com/sh/dn4gwdww8pmfebf/AACXYu8rda5LoLwuCZ8aZXfma?dl=0). Data processing including batch correction, doublet removal, cell clustering, partitioning cells into epithelial, stromal, and immune compartments are described by Smillie et al. (Smillie et al., 2019). Seurat R toolkit (Stuart et al., 2019) was used to run the Barnes-Hut t-Distributed Stochastic Neighbor Embedding (t-SNE) analysis using 1 to 30 PCs to produce two-dimensional embeddings of the data for visualization. Expression of ST6 gene in each of the cell subsets are visualized in violin plots.
ST6 structure modeling and analysis
Full-length ST6 model was predicted by I-TASSER (Roy et al., 2010). The mutation ST6 (blue) was done by using PyMOL 2.3.1, compared with wildtype (WT) ST6 (yellow).
Immunoblotting
Proteins were solubilized in 1% NP-40 buffer containing 50 mM Tris-Cl (pH 8.0), 1 mM EDTA, 1% NP-40, and 150 mM NaCl supplemented with Halt™ Protease Inhibitor Cocktail, EDTA-Free (Thermo Fisher Scientific, cat: 78439) plus Halt™ Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific, cat: 78427). After 30 min incubation on ice, the lysates were spun down at 14000 rpm at 70 °C for 20 min. The supernatant was transferred to new tubes and proteins were quantified with the BCA protein assay kit (Thermo Fisher Scientific, cat: 23225). Equal amounts of lysate were mixed with NuPAGE™ LDS Sample Buffer (4X) (Thermo Fisher Scientific, cat: NP0008) and boiled for 10 min at 70 °C before being subjected to electrophoresis in NuPAGE™ 4–12% Bis-Tris Protein Gels (Thermo Fisher Scientific, cat: NP0336BOX). Cleavage of ST6 modifications were achieved with Protein Deglycosylation Mix II (New England BioLabs, cat: P6044S) or a2–3,6,8,9 Neuraminidase A (New England BioLabs, cat: P0722S) in accordance with the manufacturer’s procedures. Samples were analyzed using immunoblotting techniques. For MUC2 Western blot, NuPAGE™ 3 – 8% Tris-Acetate Gels (Thermo Fisher Scientific, cat: EA03785BOX) were used to separate the high molecular weight proteins. Human organ samples are from Zyagen. The following antibodies were used for immunoblotting: rabbit anti-human ST6 Ab (Sigma, HPA014975), rabbit anti-MUC2 Ab (abcam, ab133555), rabbit anti-HSP90 Ab (CST, #2118), mouse anti-Sialyl Tn (S-Tn) Ab [BRST3, B72.3] (Biolegend, 915206), goat anti-EGFR Ab (R&D, AF231), rabbit anti-HA-Tag (C29F4) Ab (CST, #3724), rabbit anti-GAPDH Ab (CST, #2118), rabbit anti-HSP90 Ab (CST, #4877), anti-H3K27ac Ab (abcam, ab4729), anti-H3K9ac Ab (CST, #9649), and anti-H3 Ab (CST, #4499).
Deglycosylation, desialylation treatment and ST6 sialyltransferase activity measurement
HT29 cells with stable expression of WT and mutant form of human ST6 were digested with Deglycosylation Mix II (for removing total glycosylation, NEB, cat: P6044S), PNGase F (for removing N-linked glycosylation, NEB, cat: P0709S), O-Glycosidase (NEB, P0733S) plus a2–3,6,8,9 Neuraminidase A (NEB, cat: P0722S) (for removing O-linked glycosylation), or a2–3,6,8,9 Neuraminidase A (for removing sialic acid, NEB, cat: P0722S) at 37 °C for 24 hours in accordance with the manufacturer’s procedures. The digested lysate was mixed with NuPAGE™ LDS Sample Buffer (4X) and boiled for 10 min at 70 °C before being subjected to electrophoresis in NuPAGE™ 4–12% Bis-Tris Protein Gels. For ST6 sialyltransferase activity measurement, recombinant human ST6 (R&D, 9154-GT-020) was digested with Deglycosylation Mix II or a2–3,6,8,9 Neuraminidase A at 37 °C for 1 hour. The ST6 sialyltransferase activity was then quantified using Sialyltransferase Activity Kit (R&D, cat: EA002) according to the manufacturer’s instructions.
StcE and OgpA digestion assay
StcE was kindly provided from Dr. Bertozzi’s lab, and the purification was described in a previous study (Malaker et al., 2019). OgpA was kindly provided by WeiMing Yang from Dr. Lawrence Tabak’s lab, NIDCR and the purification was performed as below. DNA for recombinant enzymes of OgpA (B2UR60) without signal peptide sequences were synthesized and cloned into pET28a vector (Trastoy et al., 2020). The plasmids were transformed into BL21(DE) competent cells (ThermoFisher Scientific). The bacteria were cultured, and enzyme expression was induced by IPTG (final concentration 0.5 mM) for 4 hours. The bacteria were pelleted and lysed using sonication. Bacteria debris was pelleted, and the supernatant with the enzyme was mixed with HisPur™ Ni-NTA Resin (ThermoFisher Scientific) for 1 hour at room temperature. The resin was then washed, and enzymes were eluted using imidazole (final concentration 250 mM). The enzymes were further purified through the 10 KDa cutoff Amicon Centrifugal Filter Unit (Sigma). Cell pellets from LS180 cells were solubilized in lysate buffer containing 50 mM Tris-Cl (pH 8.0), 1% NP-40, 0.3% Triton X-100, 3% glycerol, and 150 mM NaCl on ice for 30 min. After spinning down at 14000 rpm for 20 min, the supernatant was aliquoted and digested in a total volume of 20 μL of 50 mM ammonium bicarbonate with StcE, OgpA or protein K (Qiagen) at the indicated dose for 3 hours at 37 °C. Afterwards, 7 μL of NuPAGE™ LDS Sample Buffer (4X) was added and boiled for 10 min at 70 °C before being subjected to electrophoresis in NuPAGE™ 3 to 8% Tris-Acetate Gels. For de-glycosylation and de-sialylation samples, the supernatant from LS180 cells was digested with Deglycosylation Mix II (New England BioLabs, cat: P6044S) or a2–3,6,8,9 Neuraminidase A (New England BioLabs, cat: P0722S) for 3 hours at 37 °C in accordance with the manufacturer’s procedures. Afterward, the samples were mixed with different doses of StcE or OgpA for 3 hours at 37 °C before electrophoresis.
Lentivirus production and infection
On day 0, Lenti-X™ 293T Cells (Takara, cat: 632180) were seeded in 10 cm dishes or 6 well plates. On day 1, the cells were transfected with Lipofectamine 3000 Reagent (Thermo Fisher Scientific, cat: L3000075) according to the online transfection protocol for Lipofectamine 3000. The cells were 60–70% confluent at the time of transfection. For example, 3rd generation Lentivirus backbone: psPAX2: pMD2.G = 1250 ng: 1250 ng: 125 ng were used for 6 well plate per well and 11000 ng: 11000 ng: 1100 ng were used for 10 cm dishes. All plasmids were amplified in Stbl3™ Chemically Competent (Thermo Fisher Scientific, cat: C737303) and purified with endotoxin-free NucleoBond® Xtra Maxiprep EF kit (Clontech, 740424.10). Cells were incubated in cell incubator at 37 °C for 48 hours and the lentivirus-containing supernatants were collected. The supernatants were centrifuged briefly (500 × g for 10 min) or filtered through a 0.45 jm filter. Clarified supernatant was transferred to a sterile container and 1 volume of Lenti-X Concentrator (Takara, cat: 631232) was combined with 3 volumes of clarified supernatant. Gentle inversion was used to mix. The mixture incubated at 4 °C overnight. The next day, samples were centrifuged at 1500 × g for 45 min at 4 °C. The supernatant was carefully removed, and the residual supernatant was removed by brief centrifugation at 1500 × g. After centrifugation, an off-white pellet was visible. The pellet was gently resuspended in 1/20 of the original volume using complete DMEM or RPMI medium. Samples were stored at −80 °C in single-use aliquots.
CRISPR/Cas9-mediated gene knockout in cell line
sgRNA vector lentiGuide-Puro (Addgene, cat: 52963), was obtained from Addgene (Sanjana et al., 2014). sgRNAs targeting ST6 were designed with the Benchling algorithm (https://www.benchling.com) and subsequently cloned into lentiGuide-Puro. For cloning, the lentiGuide-Puro vector was gel purified after digestion with BsmBI. sgRNA oligos from IDT were annealed through incubation of 1μL of 100μM forward oligo, 1μL of 100 jM reverse oligo, 1μL 10X T4 Ligation Buffer, 0.5μL T4 PNK, and 6.5μL nuclease -free water at 37 °C for 30 min and cooled to 25 °C at 5 °C/min. Annealed oligos were ligated into the digested lentiGuide-Puro vector by combining 1μL of 20X diluted annealed oligo, 50 ng digested lentiGuide-Puro, 1μL 10X T4 Ligation Buffer, and 1μL NEB T4 DNA ligase (NEB M0202), and brought to a total volume of 10μL with nuclease-free water, and incubated at room temperature for 18 hours. The plasmids were verified by Sanger sequencing from Genewiz. Sequences of all sgRNAs are listed in the Key resources table. For infection, lentiCas9-Blast (Addgene, cat: 52962) and lentiGuide-Puro-sgRNA lentiviral particles were produced in Lenti-X™ 293T Cell Line as described in Lentivirus production and infection. Concentrated virus lentiCas9-Blast was added to HT29 or LS180 cells to generate Cas9 stable expressing cells first, and cells were incubated overnight in growing media plus virus and polybrene (10 μg/mL, Sigma-Aldrich, TR-1003). The following day, the virus-containing media was removed and replaced with growing media, and cells were selected with Blasticidin S HCl (Thermo Fisher Scientific, cat: A1113903) at 5 jg/mL beginning 72 hours after infection. The lentiGuide-Puro-sgRNA lentiviral particles were added as described above and selected with puromycin (thermo Fisher Scientific, cat: A1113803) at 2 jg/mL beginning 72 hours after infection to generate the ST6 deficient cells.
Induced pluripotent stem cell (iPSC) generation and culturing
Briefly, PBMCs isolated from whole blood were expanded for 9 days using StemSpan™SFEM II medium with added erythroid expansion supplement (100 ng/m L SCF, 10 ng/mL IL-3, 2 U/mL EPO, 40 ng/mL IGF-1, 1μM Dexamethasone, and 100μg/mL holo-transferrin). 200,000 expanded erythroblasts were infected by 20 μL mixed Sendai Virus from Cytotune 2.0 kit using centrifugation and then plated onto Matrigel (Corning, 354277)-coated 48-well plates on day 2–3 post-infection by a serial dilution of 20–15000 cells/well. Single iPSC clones were picked and then passaged by EDTA method. Established iPSC lines were maintained in E8 medium.
Organoid generation, culturing, and differentiation
The differentiation from iPSCs into intestinal organoids were performed with STEMdiff Intestinal Organoid Kit (STEMCELL) according to the manufacturer’s instructions. The human intestinal organoids were cultured in STEMdiff Intestinal Organoid Growth Medium (STEMCELL) in Matrigel. The organoids were differentiated into GCs with the addition of IWP2 (1.5 mM; Sigma Aldrich, cat: I0536) and DAPT (10 mM, Sigma Aldrich, cat: D5942) or A83–01 (1 mM; Cayman, cat: 9001799) and DBZ (10 mM; Cayman, cat: 14627) in Organoid Growth Medium.
Organoid lentivirus transduction and analysis
The WT and mutant version of ST6 were cloned into pLV-EF1a-IRES-Puro (Addgene, cat: 85132) or pLV-EF1a-2a-tEGFR backbone. pLV-EF1a-2a-tEGFR was generated in our lab; briefly, human T2A-EGFR extracellular part (tEGFR) was cloned into pLV-EF1a-IRES-Puro by cutting with EcoR I and TspM I. For cloning, the EF1a-IRES-Puro or EF1a-2a-tEGFR vector was gel purified after digestion with EcoR I and Mlu I. gBlock of WT and mutant versions of ST6 from IDT were cloned into the digested vector using In-Fusion® HD Cloning Plus CE kit (Takara, cat: 638919) by following the online User Manual. The plasmids were verified by Sanger sequencing from Genewiz. For generating ST6-stable expressing organoids, the organoids were harvested by pipetting the Matrigel and medium up and down, thereby disrupting the mixture with Anti-Adherence Rinsing Solution (STEMCELL, cat: 07010) pre-treated p1000 micropipette. The mixture was placed in a 15 mL tube and centrifuged for 5 min at 300 × g to form a pellet. The supernatant was removed, and the pellet resuspended with 5 mL DMEM/F12 medium. The organoids were washed with DMEM/F12 medium twice and dissociated with TrypLE Express (thermo Fisher Scientific, cat: 12604013) for 5–10 min at 37 °C to prepare single cell suspensions. Samples were centrifuged for 5 min at 500 × g to remove the supernatant, and the pellet was resuspended with STEMdiff™ Intestinal Organoid Growth Medium from STEMCELL (cat: 05145) for human organoids and IntestiCult™ Organoid Growth Medium (Mouse) from STEMCELL (cat: 06005) containing 30 μg/mL mouse EGF (PeproTech, cat: 315–09) and 30μg/mL mouse Wnt3a (PeproTech, cat: 315–20) for mouse organoids. High titer lentivirus was added in organoids culture medium containing 10μg/mL Polybrene. Organoid-virus mixture was placed in culture incubator and incubated for 1 hour at 37 °C to allow transduction. Afterwards, lentivirus spin down infection was performed by putting the 24 well plate containing organoids in a pre-warmed centrifuge at 32 °C and rotating at 2000 rpm for 90 min. Organoid-virus mixture was placed in culture incubator and incubated for 3 hours at 37 °C to enhance transduction efficiency. The organoids were then transferred into a microcentrifuge tube and centrifuged for 5 min at 300 × g for pelleting. The supernatant was removed and the organoids were resuspended in 30 μL of Matrigel (Corning Life Science, cat: 354230). The cells were mixed by pipetting up and down several times. The droplet was put in the middle of each well in a 24 wells plate and incubated in culture incubator at 37 °C for 15–30 min to solidify. Afterwards, 500 μL of organoid culture medium supplemented with 10 μM Y27632 was carefully added. The organoid culture medium without Y27632 was replaced on the second day of spin down infection. Small disrupted organoid fragments formed into small cystic organoids within 24–48 hours. 4 jg/mL puromycin was added to the organoid culture medium on day 3 to remove the uninfected organoid cells.
Confocal microscopy and immunofluorescence staining
WT and mutant versions of ST6 stable expressing HT29 cells were plated onto coverslips in 24 well plates overnight before addition of 500 μL 4% paraformaldehyde for 15 min at room temperature. After washing the coverslips twice with PBS, samples were permeabilized for 15 min in PBS buffer containing 1% Triton X-100 and 5% BSA, washed, and then blocked with 5% BSA in PBST (PBS + 0.05% Tween-20) for an additional 1 hour at room temperature. Samples were then incubated with primary antibodies diluted in PBS buffer containing 1% Triton X-100 and 5% BSA at 4 °C overnight. After washing three times, the samples were treated with fluorochrome-conjugated secondary antibodies (Invitrogen) diluted in PBS buffer containing 1% Triton X-100 and 5% BSA for 1 hour at room temperature in the dark. Following three washes with PBS, coverslips were mounted with DAPI Fluoromount-G® (SouthernBiotech) onto slides and left to dry overnight. For human intestine organoids, the organoids were spun down at 100 × g for 5 min and gently resuspended with 4% paraformaldehyde in PBS for 15 min at room temperature. After washing the organoids twice with PBS, the organoids were permeabilized for 30 min in PBS buffer containing 1% Triton X-100 and 5% BSA, and then spun down and changed the same buffer with primary antibodies at 4 °C for 24 hours. Afterwards, samples were washed three times and treated with fluorochrome-conjugated secondary antibodies (Invitrogen) diluted in PBS buffer containing 1% Triton X-100 and 5% BSA at 4 °C overnight in the dark. The primary antibodies were used as below: rabbit anti-human ST6 Ab (Sigma, HPA014975, 1:200), mouse anti-MUC2 Ab (Santa cruz, sc-515032, 1:50), mouse anti-HA-Tag Ab (Covance, MMS-101P, 1:200), and rabbit anti-GM130 Ab (Abcam, ab52649, 1:100). Confocal images were acquired on a Leica SP5 X-WLL microscope and analysis was performed using Imaris and ImageJ software (Schneider et al., 2012). For mouse intestinal samples, the tissue was fixed in 10% neutral formalin and stored in 70% ethanol. Different human organ samples from healthy donors were obtained from Novusbio (NBP2–78067) and Zyagen. Patient and mouse intestinal samples were paraffin-embedded and cut into 10 μm longitudinal sections, with the H&E and PAS staining performed by Histoserv Inc. (Germantown, MD). GCs were counted and calculated from 10 crypts in each slide. For immunofluorescence staining, slides were deparaffinized by xylene and antigen retrieval was conducted for 20 min in a 95 °C water bath in 10 mM sodium citrate, pH 6.0 followed by a 15 min incubation to cool down to room temperature. Slides were washed, blocked in 5% BSA, and stained with the primary antibodies rabbit anti-human ST6 Ab (Sigma, HPA014975, 1:200), mouse anti-MUC2 Ab (GeneTex, GTX100664,1:100), mouse Anti-Sialyl Tn (S-Tn) antibody Ab (Abcam, ab115957, 1:100), mouse anti-CD45 Ab (Biolegend, 304002, 1:100), anti-Ki-67 Ab (Thermo, 12–5698-82, 1:100), anti-Lysozyme Ab (Abcam, ab108508, 1:100), anti-RegIII-gamma Ab (Abgent, AP5606c, 1: 100) and secondary antibodies conjugated to Alexa fluor 488, 633, or 594 (Thermo Fisher) or UEA-1fluorescein (Vector lab, FL-1061, 1:100), EUB probe (EUB 338, 5’-GCTGCCTCCCGTAGGAGT-3’ conjugated with Alexa546 (Thermo) 1:100). Slides were mounted in Fluoromount-G (Thermo Fisher), and 3–15 images were taken per slide at 20X or 40X magnification along transections of the intestinal crypts for each biological replicate (Zeiss).
Direct fluorescent SA (Neu5Ac) labeling
Direct fluorescent SA (Cy5-SA) labeling was performed according to previously established protocols with modifications (Wu et al., 2019). 5–10 μg target protein Asialofetuin (AF) in 20 μL buffer of 25 mM Tris pH 7.5 was treated with de-glycosylation enzymes O-Glycosidase (NEB, P0733S), GalNAcEXO (from WeiMing Yang from Dr. Lawrence Tabak’s lab, NIDCR, NIH, 0. μg), PNGase F (NEB, P0709S) or mixed with all the above de-glycosylation enzymes overnight at 37 °C. Samples with enzymes were then heat inactivated at 75 °C for 10 min. Recombinant human ST6 protein (rhST6) (R&D, 9154-GT-020, 0.2μg) and 10 mM MnCl2 were added to the reactions with 0.2 nmol Cy5-conjugated CMP-SA (R&D, ES302–050) at 37 °C for 1–2 hours. The reactions were then analyzed on a 4–12% Tris-Bis NuPAGE gel. Cy5-SA and Cy5-Peanut agglutinin (PNA) (vectorlabs, CL-1075–1) signaling was imaged by a fluorescent imager Azure 500. Protein was stained with LabSafe GEL Blue (G-biosciences, 786–35) and imaged using scanner.
S-Tn antibody pull down and mass spectrometry analysis
For S-Tn carrying protein LS180 IP-MS experiments, cells from two T175 Flasks (around 400 mg cell pellets) were used per IP. Cell pellets were lysed in 800 μL lysis buffer (Tris-HCl 30mM, pH 8.0, 1 % NP-40, 150 mM NaCl, 1 % Triton X-100 and 2.5% Glycerol) on ice for 30 min. After spinning down, the supernatant was transferred to a new tube and Dynabeads™ Protein G were added at 4 °C for 2 hours for pre-clearing. After removing the Dynabeads by magnet and dividing into two tubes, samples were rotated overnight in cold room with 20 μg anti-S-Tn antibody (Biolegend, 915206) or mouse IgG. Afterwards, 200 uL Dynabeads™ Protein G was added to each tube for an additional 2 hours. IP production was isolated by magnet and resuspended with elution buffer (8 M urea, 200 mM Tris-HCl (pH 8.0)). The samples were then sequentially treated with 5 mM dithiothreitol (DTT) and 10 mM iodoacetamide (IAM) and incubated for 1 hour at 37 °C and 40 min in the dark at room temperature (RT, 20–25 °C), respectively. The samples were diluted at least five times using Tris-HCl (200 mM, pH 8.0) and treated overnight with trypsin using an enzyme-to-protein ratio of 1:40 (wt/wt) at RT. After the samples were acidified with trifluoroacetic acid (TFA) to a final concentration of 1 % (vol/vol), the digested peptides were purified using a 100 mg C18 cartridge (Seq-Pak C18, cat: WAT054955) and vacuum dried. The peptides were then dissolved in 0.1% TFA and analyzed on a Fusion Lumos mass spectrometer coupled with an Dionex U3000 nanoHPLC system (Thermo Fisher Scientific). Injected peptides were separated on a homemade C18 column (ReproSil-Pur 120 C18-AQ, 1.9 jm pore size, 75 jm inner diameter, Dr. Maisch GmbH) using a gradient composed of 0.1% FA in water (A) and 0.1% FA/80% acetonitrile (B) at a flow rate of 300 nL/min -7.5% B for 5 min, 7.5–50% B for 154 min, 50–90% B for 10 min. MS analysis was performed using a spray voltage of 1.8 kV. MS1 survey scans (AGC target 4 × 105 and maximum injection time (maxIT) of 50 ms) were collected from 375 to 1800 m/z at a resolution of 60 K and followed by data-dependent Top15 HCD MS/MS scans (at a resolution of 50 K, normalized collision energy of 30, intensity threshold of 5 × 104, and maxIT of 86 ms) with an isolation window of 1.2 m/z. Charge-state screening was enabled to reject unassigned, single, and more than six protonated ions. Fixed first mass was 110 m/z. A dynamic exclusion time of 45 sec was used to discriminate against previously selected ions. MS data were searched against a reviewed human protein database (UniProt, downloaded on July-02–2021) using pFind3.0 (Chi et al., 2018) with the following settings: tryptic digestion, up to two missed cleavages, oxidation (M) as variable modifications and carbamidomethyl(C) as fixed modification.
Preparation of TMT-labelled peptides, glycopeptides enrichment, MS analysis and database search
Site-specific N-glycoproteomics analyses of HT29, LS180, and mice IECs were performed as previously described (Fang et al., 2020). Briefly, cell pellets were lysed in 4% SDS (w/v), 50 mM HEPES (pH 7.5) and sonicated for 20 min (15 sec on, 15 sec off) using Bioruptor at 4 °C. After centrifugation at 14000 × g for 15 min, the supernatants were collected, and the protein concentrations were determined using Pierce BCA Protein Assay Kit (Thermo Scientific). All proteins were reduced and alkylated with 10 mM TCEP (500 mM stock, Thermo Scientific) and 20 mM IAM at 37 °C for 60 min in the dark. A mixture of Sera-Mag SpeedBeads with a hydrophilic surface (GE Healthcare, cat: 45152101010250) and Sera-Mag SpeedBeads with a hydrophobic surface (GE Healthcare, cat: 65152105050250) were rinsed twice with water and then added to protein lysates at the working ratio of 10:1 (wt/wt, beads to proteins). After adding acetonitrile (ACN) to a final percentage of 70% (vol/vol) and removing the supernatant, the beads were washed three times with 95% (vol/vol) ACN, resuspended in 50 mM HEPES (pH 8.0) containing sequencing grade modified trypsin (1:50 of enzyme-to-protein ratio), and incubated at 37 °C overnight in a ThermoMixer with mixing at 1000 rpm. The resulting peptides in the supernatant were collected and labeled with TMT reagents according to the manufacturer’s instruction (Thermo Scientific).
TMT-labeled glycopeptides were enriched using spin-columns self-packed with the Ultimate Hydrophilic Interaction Liquid Chromatography (HILIC) Amphion II beads (5μm, Welch, Lot. no. 7301.26). Briefly, samples were dissolved in loading buffer (75% (vol/vol) ACN and 1% TFA) and loaded five times to the HILIC columns followed by three times washing with loading buffer. The retained TMT-labeled glycopeptides were eluted with 100 μL 0.1% TFA and 1% TFA and dried in a SpeedVac concentrator. Subsequently, the enriched glycopeptides were fractionated using an XBridge C18 column (3.5μm particles, 1.0 mm × 150 mm, Waters) installed on an Agilent 1100 series High-performance liquid chromatography (HPLC) system. In total, 58 fractions were collected and pooled into 6 concatenated fractions. LC-MS/MS analysis was performed on a Orbitrap Fusion Lumos as described above with the following settings: MS1 settings: Orbitrap Resolution-120 k, Mass Range (m/z)-350–2000, MaxIT-50 ms, AGC target-5e5, RF Lens-60%, Precursor selection range (m/z)-700–2000; MS2 settings: Isolation window-2 m/z, First mass-132, Activation type-HCD, Collision energy-25, Detector type-Orbitrap, Orbitrap resolution-15 K, MaxIT-150 ms, AGC target-5e5; MS3 settings: Precursor selection range-700–2000, Number of Notches-10, Isolation window (m/z)-2, MS2 isolation window (m/z)-2, First mass (m/z)-120, Activation type-HCD, Collision energy (%)-35, Orbitrap resolution-60 K, MaxIT-350 ms, AGC target-5e5.
All raw files were processed by GlycoBinder (Fang et al., 2020). Main parameters used for GlycoBinder include fully specific trypsin digestion with maximal two missed cleavages and mass tolerance for precursors and fragment ions of 10 and 20 ppm, respectively. Cysteine carbamidomethylation and TMT10 on peptide N-termini and lysine residues were set as fixed modifications and methionine oxidation was set as a variable modification. The reviewed human protein database was downloaded from Swiss-Prot (October 2020, human, 20370 entries). GlycoBinder propagates all glycopeptide-to-spectra matches and directly reported the quantification of glycosylation sites, glycan compositions, and glycoforms.
Co-immunoprecipitations
LS180 cells with 80–90% density from T175 flask were washed with PBS three times and collected using cell scraper to 15 mL tubes. The cells were spun down at 300 × g at 4 °C for 5 min. The cell pellet was lysed in 1% NP-40 buffer containing 50 mM Tris-Cl (pH 8.0), 1 mM EDTA, 1% NP-40, and 150 mM NaCl supplemented with Halt™ Protease Inhibitor Cocktail, EDTA-Free (Thermo Fisher Scientific, cat: 78439) plus Halt™ Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific, cat: 78427) and kept on ice for 30 min. After 30 min incubation, the lysates were spun down at 14000 rpm at 4 °C for 20 min. The supernatant was transferred to new tubes and proteins were quantified with the BCA protein assay kit (Thermo Fisher Scientific, cat: 23225). Supernatant was aliquoted at 5 mg per tube and cell lysates were precleared with unconjugated Dynabeads Protein G (Thermo Fisher Scientific, cat: 10003D) for 2 hours with rotation at 4 °C. Primary antibodies or IgG control (mouse IgG, Santa Cruz, cat: sc-2025) were then added to the precleared lysates and incubated with rotation overnight at 4 °C. Dynabeads Protein G were added and incubated for an additional 2 hours at 4 °C. Immunoprecipitated proteins were eluted from the Dynabeads in 4×LDS sample loading buffer for 10 min at 70 °C resolved by electrophoresis and analyzed by immunoblotting.
RT-PCR and real time PCR
Total RNA was extracted from in vitro-cultured cells using TRIzol reagent (Invitrogen) or RNeasy Plus Mini Kit (Qiagen, cat: 74136). Complementary DNA was synthesized with PrimeScript™ RT Reagent Kit (Takara, cat: RR037A) at 37 °C for 15 min and de-activated at 85 °C for 5 sec. PCR with reverse transcription was performed using PowerUp™ SYBR™ Green Master Mix (Thermo Fisher Scientific, A25777) in a StepOnePlus detection system (Applied Biosystems). The fold difference in mRNA expression between treatment groups was determined using the ΔΔCt method. β-actin, GAPDH, or HSP90 was used as a human internal control and β-actin or Rpl13a was used as a mouse internal control. For absolute RNA quantification Q-PCR, the standards for each sialyltransferase gene production were ordered from IDT. The copy number for each gene was calculated by using the following formula: copy number = (amount × 6.022 × 1023) ÷ (length × 1× 109 × 650), where amount is the DNA concentration obtained in ng/μL and length is the total length of the standards in bp. The copy number was obtained as copies/μL. After generating the standard curve with 5-fold dilution of standard productions and amplification cycles from Q-PCR, the copy number for each sialyltransferase gene in human small intestine and colon was calculated based on their individual amplification cycles and standard curve. The primer pair sequences of individual genes are listed in Key resources table.
Transfection
HEK293T cells were seeded to 50–70% density in cDMEM. Tagged plasmids were diluted in Opti-MEM reduced serum media (Thermo Fisher Scientific) and combined with Lipofectamine 3000 Transfection Reagent (Thermo Fisher Scientific) for transfection according to the manufacture’s protocol. Cell lysates were made 24–48 hours later for immunoblotting and co-immunoprecipitation. Full-length ST6 mutants were cloned into the pLV-EF1a-IRES-Puro (Addgene, cat: 85132) (Hayer et al., 2016) or pLV-EF1a-2a-tEGFR (our lab) backbones.
ATAC-seq and mRNA-seq
We followed the Omni-ATAC protocol (Corces et al., 2017). Briefly, 50,000 cells were washed and lysed in lysis buffer (10 mM Tris-HCl (pH 7.4), 10 mM NaCl, 3mM MgCl2) containing 0.1% NP-40, 0.1% Tween-20, and 0.01% Digitonin for 10min on ice to prepare the nuclei. Immediately after lysis, nuclei were spun at 500 × g for 10 min to remove the supernatant. Nuclei were then incubated with Tn5 transposome and tagmentation buffer (20 mM Tris-HCl (pH 7.6), 10 mM MgCl2, 20% Dimethylformamide) containing 0.1% Tween-20 and 0.005% Digitonin at 37 °C for 30 min. Stop buffer was added directly into the reaction to end tagmentation. PCR was performed to amplify the library. Libraries were then purified with SPRI (Beckman) beads and deep sequencing was performed by Sequencing Facility in the National Cancer Institute (NCI). For mRNA-seq, the mRNA from fresh isolated 2–5 million colon crypt ISCs was isolated using RNeasy Plus Micro Kit (Qiagen, cat: 74034) and the concentration and QC was determined by Agilent technologies. The library generation and sequencing were performed by Sequencing Facility in the NCI.
For mRNA-seq analysis, reads of the samples were trimmed for adapters and low-quality bases using Cutadapt before alignment with the reference genome (mm10) and the annotated transcripts using STAR. Read counts were generated using HTSeq (version 0.11.4) with parameters -m union, --stranded=reverse, and the mm10 genes.gtf file from UCSC. Differential gene expression analysis was performed using DESeq2 (version 1.30.1) in RStudio (version 1.4.1106) using R (version 4.0.4). Any genes with summed counts less than 10 across all samples were excluded. PCA plots were generated using DESeq2’s rlog function. Normalization was performed using DESeq2 default parameters. The DESeq2 results were generated using default parameters and transformed using lfcShrink function. Volcano plots were generated using EnhancedVolcano (version 1.8.0). Heatmaps were generated with pheatmap (version 1.0.12) and RColorBrewer (version 1.1–2), using normalized count data generated with DESeq2. For generation of bigwig files, bam files were indexed using samtools (version 1.12) with default parameters. Bigwig files were created using deeptools’ (version 3.5.0) function bamCoverage with default bin size and the forward RNA strand. For pathway analysis, genes were selected that had p-adjusted < 0.05 and log2FC > 1 or < −1. The upregulated and downregulated were separately input into Enrichr (https://maayanlab.doud/Enrichr/) (Kuleshov et al., 2016).
For ATAC-seq analysis, paired-end sequencing fastq files were checked for quality using FastQC (version 0.11.9), then aligned to the mm10 genome using Bowtie (version 1.3.0) with parameters -v 2 -m 1 --pairtries 200. Bowtie output was converted to bed files using the homemade bash script. Tag directories and bigwig files were created using HOMER (version 4.11.1); parameter -sspe was used for tag directory creation, otherwise default parameters were used. Peaks were called using Macs1 (version 1.4.3) with a p-value cutoff of 1E-5. Peak files were merged for each condition using homer’s mergePeaks command with parameter -d given. Peak files were converted to bed format using HOMER’s pos2bed.pl function and compared using bedtools (version 2.30.0). Only regions called in both replicates were used in downstream analysis. A master list of peaks was generated using HOMER’s mergePeaks function with -d given, and annotated using HOMER’s annotatePeaks.pl function, with reference genome mm10, -size given, and tag counts from tag directories created previously.
Mouse mucin isolation
Mouse mucin isolation was performed and defined by following previously described procedures (Johansson et al., 2009; Shan et al., 2013). Briefly, the lumen surface of colon of WT B6 mice were exposed by longitudinal dissection and washed twice with 1× Dulbecco’s phosphate-buffered saline (DPBS). Mucus was gently scraped off with a glass slide and collected into 1.7-mL tubes together with an equal volume of ice-cold 1× DPBS containing protease inhibitors (Thermo). After centrifuging at 14000 rpm for 30 min at 4 °C, the pellet was resuspended with six volumes of 6 M guanidinium-hydrochloride buffer (6 M GuHCl, 0.1 M Tris pH 8.0, 1 mM EDTA) and gently stirred overnight at 4 °C. The samples were centrifuged at 14000 rpm for 30 min at 4 °C the next day. Supernatants were collected and the insoluble fraction (the pellet) was resuspended in 6 M guanidinium-hydrochloride buffer and centrifuged again. Reducing agent DTT was added to the insoluble and soluble fractions to a final concentration of 50 mM and samples were stirred for 1.5 hours at 37 °C. Samples were then incubated with 125 mM iodoacetamin overnight in the dark at room temperature. Samples were transferred to Slide-A-Lyzer MINI dialysis cups and dialyzed against water at 4 °C overnight. LPS was removed using EndoTrap red (Hyglos). Pooled soluble and insoluble fractions were concentrated with a SpeedVac apparatus, resuspended in 1× DPBS, and analyzed by Agarose-Acrylamide Composite Gel Electrophoresis (Ag-PAGE) gel (Johansson and Hansson, 2012) and mass spectrometry. The endotoxin levels in isolated mucus samples were checked by ToxinSensor™ Chromogenic LAL Endotoxin Assay Kit from Lonza (Genescript) according to the manufacturer’s instruction.
Murine colitis model
Experimental colitis was initiated by treatment of mice with 2.5% dextran sulfate sodium (DSS) (36000–50000 M.Wt, MP Biomedicals) in drinking water for 6 days. DSS was then replaced by normal water for another 6~7 days. Body weight was monitored daily. For carbenoxolone (CBX) (Selleckchem, cat: S4368) treatment, mice were intraperitoneally injected with CBX (12.5 mg/kg body weight) once a day from day 4 to 6. For antibiotics treatment, mice were given an antibiotic cocktail (ABX; 1 g/L Ampicillin, 1 g/L Neomycin, 1 g/L Metronidazole, 0.5 g/L Vancomycin, all from Sigma) in drinking water for 3 weeks. After mouse stool samples were confirmed as sterile in agar plate incubation, mice received 2.5% DSS plus antibiotic cocktail in drinking water for 6 days and then were switched to antibiotic cocktail until the end of the experiment. For mucin administration, mice were treated with antibiotic cocktail (1 g/L Ampicillin, 1 g/L Neomycin, 1 g/L Metronidazole, 0.5 g/L Vancomycin) in drinking water for 3 weeks. The mice were then given normal water and gavaged with 200 μL concentrated mucin at 1 mg/mL in PBS every other day for 10 days. Mice were then administered 2.5% DSS water.
Intestinal epithelial cells (IECs), intraepithelial lymphocytes (IELs), lamina propria cells, and colon crypt cell isolation
For isolation of lamina propria cells, small intestines and colon from euthanized mice were emptied of contents, excised of Peyer’s patches in small intestines, opened longitudinally, and cut into 0.5–1 cm pieces. The intestinal epithelial cells (IECs) and intraepithelial lymphocytes (IELs) were dissociated from the intestine fragments by first shacking the fragments for 30 min at 37 °C in HBSS buffer containing 1% FBS, 5 mM EDTA, and 1 mM dithiothreitol, vortexed for 30 sec and passed intestine through a gray-mesh (100 micron). Intestine pieces on the gray-mesh were returned to the original 50 mL tube, washed with same HBSS buffer twice, and combined with the supernatant in 50 mL tube. The supernatant containing IELs and IECs was spun down at 300 × g for 5 min. The IELs (Live CD45+epcam-) and IECs (Live CD45-epcam+) were further purified by staining with sorting via FACS Aria III (BD Biosciences). To isolate lamina propria cells, the remaining fragments were minced and digested at 37 °C for 30 min in RPMI-1640 medium containing 0.1 mg/mL Liberase (Roche) and 10 U/mL DNase I (Roche). The digestion suspension was then filtered through a 40-mm cell strainer and centrifuged at 300 × g for 5 min. Cell pellets were washed twice and spun down at 300 × g for 5 min. The pellets were resuspended with 40% Percoll in RPMI medium, and the 40% Percoll was overlayed onto 70% Percoll RPMI medium in 15 mL clear falcon tube. Samples were centrifuged for 20 min at 1500 rpm at 20 °C with the descending rate 5. Cells were collected from 40–70% Percoll interface and transferred to 10 mL tubes. To purify the lymphocytes (Live CD45+epcam-) from lamina propria, the cells were further sorted via FACS Aria III (BD Biosciences). For colon crypt cell isolation, colon was taken carefully and put into 10 cm dish containing cold PBS, and associated fat tissue was removed carefully. The lumen of the intestine was flushed with cold PBS. The colon was cut longitudinally with scissors and placed in a 50 mL tube with 25 mL cold PBS, inverted 10–15 times, and then the PBS was removed (repeated until the supernatant no longer contained any visible debris). The colon was cut into around 5 mm pieces and placed into cold 10 mL 5 mM EDTA-PBS. The fragments were vigorously triturated by pipetting up and down into a 10 mL pipette 15 times, then the fragments settled by gravity for 30 sec and the supernatant was aspirated (with care to avoid the intestinal fragments). Supernatant was replaced with 10 mL 5 mM EDTA-PBS and samples were placed at 4 °C on roller for 15 min. EDTA-PBS was aspirated from the tube. Samples were washed with PBS twice and 3 mL Basal media (RPMI 1640) containing 500U/mL Collagenase Type IV were added. A 5 mL pipette was used to pipette up and down 5 times. Tubes were placed in 37 °C water bath for 30–40 min. 10 mL cold PBS were added and 10 mL pipette was used to vigorously vortex samples 10 times. The 10 mL supernatant fraction was collected in a separate tube. The cold PBS/vortex was repeated 2 more times, and the 2nd and 3rd round washes were combined into 50 mL tube to gather crypt cells.
Formaldehyde-assisted isolation of regulatory elements (FAIRE) assay
IECs were crosslinked and sonicated as described for the ChIP assay. Briefly, the fresh isolated IECs from WT and Tlr4−/− mice were weighed. 25 mg of cell pellets were used for each IP to be performed. Cells were transferred to a 15 mL conical tube and 1 mL of PBS + protein inhibitor cocktail (PIC) was added per 25 mg cell pellets. To crosslink proteins to DNA, 45 μL of 37% formaldehyde were added per 1 mL of PBS + PIC to a final concentration of 1.5%. Samples were rocked at room temp for 20 min. 100 μL of 10X Glycine were added per 1 mL of PBS + PIC to stop cross-linking and samples were mixed for 5 min at room temperature. Tissue was centrifuged at 500 × g in a benchtop centrifuge for 5 min at 4 °C. Supernatant was removed, and samples were washed one time with 1 mL PBS + PIC per 25 mg cell pellet. Centrifugation was repeated at 500 × g for 5 min at 4 °C. Supernatant was removed and tissue was resuspended in 1 mL PBS + PIC per 25 mg tissue and stored on ice. Cell pellets were disaggregated into single-cell suspension using a Medimachine or Dounce homogenizer. Samples were centrifuged and the aqueous phase was isolated by three consecutive extractions with phenol-chloroform-isoamyl alcohol (25:24:1). The DNA was precipitated by ethanol. After the final extraction, the FAIRE samples were purified and reverse crosslinked as described for the ChIP assay. The enrichment of fragmented genomic DNA in the FAIRE samples relative to each input DNA was measured by quantitative PCR. The primers are listed in the Key resources table.
Metabolite and sample preparation
For all LCMS methods, LCMS grade solvents were used. Intestinal samples were excised and immediately submerged in 0.5 mL of ice-cold methanol. Fecal samples were transferred directly to methanol. Samples were pulverized with a micropestle. 0.5 mL of water and 0.5 mL of chloroform was added to each sample. Samples were agitated for 30 min at 4 °C and subsequently centrifuged at 16000 × g for 20 min to induce layering. 400 μL of the top (aqueous) layer was collected. The aqueous layer was diluted 5× in 50% methanol in water and prepared for LCMS injection.
Short Chain Fatty Acid (SCFA) derivatization and measurement
The fecal samples were sent to the Protein Characterization Core at the NCI. Briefly, Fecal samples were homogenized in 70% isopropyl alcohol (35 mg/mL) using the Bead Rupter (Omni International). The SCFA calibration standards and the isotopic internal standard (IS) solution were prepared in 50% acetonitrile. EDC [1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride] was prepared as 120 mM solution in 50% acetonitrile containing 6% pyridine. 3-NPH (3-nitrophenylhydrazine hydrochloride) was prepared as 200 mM solution in 50% acetonitrile. SCFA were determined by LC/MS/MS after derivatization with 3-NPH according to previous established protocols with slight modification (Han et al., 2015). In a 500μL Eppendorf vial, 20μL each of the fecal supernatant (or standard), IS, EDC, and 3-NPH were mixed and reacted for 30 min at 40 °C. After centrifugation, the reaction mixtures were transferred to injection vials. LC were performed using a Shimadzu 20AC-XR system at 60°C with a 2.1 × 100 mm, 2.7 jm Cortecs C18 column (Waters). The injection volume was 2μL. Mobile phase A was 0.01 % formic acid in water and mobile phase B was 0.01 % formic acid in acetonitrile. The flow rate was 300 μL/min and the SCFA derivatives were eluted with a gradient (0– 0.2min/5%B; 11min/35%B; 11.1–13min/95%B; 13.1–15min/5%B). MS/MS was performed with a TSQ Quantiva triple quadrupole mass spectrometer (Thermo Fisher Scientific) operating in negative SRM mode. The derivatized SCFA were detected using the following m/z precursor > product ions: lactate/lactate-13C3 (224>152/227>152); acetate/acetate-D3 (194>152/197>137); propionate/propionate-13C3 (208>137/211>137); butyrate/butyrate-13C4 (222>137/226>137); succinate/succinate-13C4 (387>234/391 >238); isovalerate/isovalerate-D9 (236>137/245>137). The SCFA in the samples were determined by linear calibration curves with 1/x weighting generated by the Thermo Xcalibur software. The curves were constructed by plotting the peak area ratios vs. standard concentrations. The peak area ratios were calculated by dividing the peak areas of the SCFA by the peak area of the corresponding IS.
Microbiota sequence and data analysis
Fecal samples were collected from live mice, snap-frozen, and stored at −80°C. DNA was extracted and amplified using barcoded V4 region primers targeting bacterial 16S rRNA gene and sequenced using a Mi-Seq Illumina sequencer by NCI Genetics and Microbiome core. The microbial community analysis was performed by using QIIME2 software (version 2019.10) (Bolyen et al., 2019). Demultiplexed paired-end raw reads were filtered (quality score>20) and then denoised by DADA2 method (Callahan et al., 2016). Taxonomy was assigned by feature-classifier (Bokulich et al., 2018) that high-quality sequences were against to the Greengenes reference database, and the sequence identity cutoff was set at 99%. For the quantification of total 16s rDNA in the mucus, 0.5 ng DNA extracted from each scraped mucin samples were used for Q-PCR with the universal bacterial primers targeting the 16S rRNA gene (16S-rRNA-f and 16S-rRNA-r). The plasmid containing the PCR products was used for standard curve. The results were calculated according to the weight of each mucin sample.
Histone deacetylase (HDAC) activity measurement
The level of HDAC activity in crypts of small and large intestine was quantified using the enzyme-based fluorometric HDAC Activity Assay Kit (Abcam, cat: 156064) according to manufacturer’s instructions.
Quantification and statistical analysis
Data were analyzed using Graphpad Prism 9.0 (Graph Pad software, La Jolla, CA, USA). Depending on experimental design, statistical significance was tested via either two-tailed unpaired or paired Student’s t test or by two-way ANOVA with Geisser-Greenhouse correction. The difference in gut bacteria between two groups was assessed by unpaired t-test. P-values < 0.05 were considered significant (*p<0.05; **p<0.01; ***p<0.001), p > 0.05 non-significant (NS). Details on the test used can be found in the respective figure legends.
Supplementary Material
(A) Principal component analysis (PCA) of gut microbiome by 16S rDNA gene sequencing from WT and St6 female mice before (3 weeks) and after (6 weeks) weaning. Dots represent individual mice.
(B) Modified volcano plot showing the significant changes of gut bacterial families in 10-week-old female WT vs. St6 mice.
(C) Gut bacterial community composition (family level) of 10-week-old female WT and St6 mice. Red rectangle with asterisk indicates significantly changed families.
(D) Relative abundance of the indicated bacteria in feces from 10-week-old female WT and St6 mice.
(E) Confocal photomicrographs of Ki-67 (red) and DAPI (blue) stained colon sections from WT and St6 mice 10 days after DSS colitis with antibiotic (ABX) treatment. Scale bar, 100 μm
(F) Quantification of the results from (E). NS = no significance.
(G) Q-PCR analysis of Lgr5 using RNA from IECs and crypt intestinal stem cells (ISCs) of WT and St6 mice.
(H) PCA of transcriptomes (RNA-seq) of ISCs from WT and St6 mice 10 days after DSS colitis.
(I) PCA of assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq) using purified ISCs from WT and St6 mice as in (H).
(J) Venn diagram showing the numbers of differentially expressed genes in WT and St6 mice in (H) and (I).
(K) Gene Ontology (GO) term analysis of RNA-seq of ISCs from WT and St6 mice as in (H).
(L) Q-PCR analysis of ISC functional genes using RNA from purified crypt ISCs from WT and St6 mice as in (H).
(M) Normalized ATAC-seq sequencing tracks from WT and St6 mice for the indicated loci as in (L). Data represent 3 experiments (A, D-I, L, M). Error bars represent the SD of samples within a group. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
(A) Schematic of St6 mice prepared by CRISPR knock-in of a single base substitution (G > A) encoding the single amino acid change from Arg (R) to Gln (Q) corresponding to the Pt 1 mutation. Sanger sequencing of St6 mutants in IECs from WT and St6 mice are shown in the right panel.
(B) Photomicrographs of representative hematoxylin and eosin-stained sections of small intestine (SI) and colon from 8–10-week-old female WT and St6 mice. Scale bar, 50 μm.
(C) Photomicrographs of representative colon sections from 8–10-week-old female WT and St6 mice stained with Periodic Acid-Schiff (PAS) (left). Statistical results of mucin area in each cell and GC number in each crypt (right). Scale bar, 50 μm.
(D) Q-PCR analysis of Muc2 mRNA in SI and colon IECs from naive WT and St6 mice.
(E) Q-PCR analysis of indicated GC genes from IEC RNA as in (D).
(F) Confocal microscopic images of colon from WT and St6 mice stained as indicated Paneth cell genes. Scale bar, 100 μm.
(G) Q-PCR analysis of antimicrobial genes in IECs as in (D).
(H) Q-PCR analysis of tight junction genes in IECs as in (D).
(I-M) Quantification of flow cytometry analyses of mouse ILC2, ILC3 (I), intraepithelial lymphocytes (IELs) (J), B cells, T cells, NK cells (K), macrophages, neutrophils (L), and Tregs (M) in SI and colon from WT and St6 mice.
(N) The changed N-glycoform carrying proteins in St6 mice were analyzed by protein interaction network and cluster enrichment by STRING database and Cytoscape. Mucin proteins are circled in red. Data represent 3 experiments (A-M). Error bars represent the SD of samples within a group.
(A) Densitometric quantification of Figure 5A for the ratio of ST6 upper (modified by sialylation)/lower (unmodified protein) bands, total ST6 (upper + lower bands)/GAPDH, and total ST6 (upper + lower bands)/tEGFR. ND = not determined.
(B) WB analysis of ST6, S-Tn, and GAPDH loading control using lysates from 293T cells overexpressing WT and mutant forms of human ST6. EV = empty vector control. × = nonspecific bands.
(C) Flow cytometry analysis of S-Tn and tEGFR in 293T cells as in (B).
(D) Quantification and statistical graphs of (C).
(E) Flow cytometry analysis as in (C) in HT29 cells after lentiviral co-transduction of T462M and T207P or T462M and WT human ST6 at the indicated ratios (T462M/T207P or T462M/WT).
(F) Quantification of S-Tn and tEGFR mean fluorescence intensity (MFI) in (E).
(G) Sanger sequencing of T462M ST6 mutants to indicate the ratio of T462M/WT or T462M/T207P after lentiviral co-transduction in HT29 cells as in (E).
(H) Non-supervised clustering of differently regulated N-glycans expression level (False Discovery Rate, FDR = 0.3) detected in HT29 cells with stable ST6-WT, EV, and ST6-R391Q expression (three biological replicates each). N-glycan intensities are shown with a color scale.
(I) Pie charts showing the molar distributions of SA (Neu5Ac)-quantities in N-glycans in clusters 1 and 2 (calculated from three biological replicates) indicated in (H). Pearson’s chi-squared test (p <0.0001) was used to calculate the p-value regarding the proportion of glycan status between clusters 1 and 2.
(J) Volcano plot of quantified N-glycans in ST6-WT compared to that in ST6-R391Q stable expressing HT29 cells. Glycan compositions are depicted by the numbers of monosaccharide moieties (top to bottom: Hex, HexNAc, Neu5Ac, Fucose) and color coded for molar amount of SA (Neu5Ac) residues. Examples of most significantly changed SA (Neu5Ac)-containing N-glycan structures are shown.
(K) Phase contrast photomicrographs of induced pluripotent stem cell (iPSC)-generated intestinal organoids from Pt 1, Pt 2, and healthy donors (HDs). Scale bar, 100 μm.
(L, M) Sanger sequencing confirming ST6 mutants (L) and Q-PCR analysis of GC differentiation genes (M) of the organoids from Pt 1, Pt 2, and HDs as in (K). Data represent 3 experiments (A-M). Error bars represent the SD of samples within a group. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
(A) Relative expression (RE) of mRNA of GC-related genes Mucin-2 (MUC2), Trefoil Factor 3 (TFF3), and Kruppel-Like Factor 4 (KLF4) compared to GAPDH in the indicated cell types (Figures 1D and 1E) from healthy donors and colitis patients. Colon single cell sequencing data obtained from https://doi.org/10.1016/Lcell.2019.06.029. Box and whiskers represent the 10th−90th percentiles.
(B) Q-PCR analysis of indicated genes using RNA prepared from flow cytometry-sorted mouse colon cells from lamina propria (LP), intraepithelial lymphocytes (IELs), and intestinal epithelial cells (IECs).
(C) Q-PCR analysis of interleukin-6 (Il6) mRNA in IECs from WT mice at the indicated days after 2.5% DSS water treatment.
(D) Q-PCR analysis of ST6 in human IEC HT29 cells after toll like receptor ligands treatment for 8 hours.
(E) Q-PCR analysis of Tlr4 and St6 in IECs from WT and Tlr4−/− mice.
(F) (Top) Formaldehyde-assisted isolation of regulatory elements (FAIRE)-Q-PCR analyses of chromatin accessibility at 8 tandem sites (x-axis) in the St6 promoter in WT or Tlr4−/− mouse IECs, y-axis presented enrichment percentage relative to input.(Bottom) Q-PCR primer sites (small horizontal bars) in the St6 locus (black arrow, transcription start site).
(G) Extended pedigree of family 1 showing consanguinity and ST6 allele (filled) inheritance. Deceased individuals are indicated by strikethrough and consanguinity is indicated by a double line. ND = whole exome DNA sequencing not determined.
(H) Fraction of cells determined by flow cytometry of patients (Pts 1 and 2) compared to healthy donor (HD) blood samples stained with antibodies as indicated showing CD57+, central memory (TCM) (CCR7+CD45RA-), naive (TN) (CCR7+CD45RA+), terminally differentiated effector memory (TEMRA) (CCR7-CD45RA+), and effector memory (TEM) (CCR7-CD45RA-) CD4+ and CD8+ T cells. Each dot represents an individual.
(I) Percentage of CD69+ CD4+ and CD8+ T cells 24 hours after activation of purified cells using CD3 and CD28 activation beads as in (H).
(J-M) Specific analyses for T regulatory (Treg), B, natural killer (NK), monocyte, and dendritic cell (DC) subsets as in (H).
(N) Table showing bioinformatic information for ST6 variants. Data represent 3 experiments (B-F). Error bars represent the SD of samples within a group. *, p < 0.05;**, p < 0.01; ***, p < 0.001.
(A) Schematic of Glycan (glycan structures linked to any glycosite), Glycoform (specific glycosite with specific glycan structure on specific protein), Glycosite (specific modification site on specific protein), and Sialylation (addition of sialic acid (SA, Neu5A(G)c) to the terminal end of glycoproteins).
(B) Volcano plot of quantified N-glycans in ST6 wildtype (WT) (control) compared to that in ST6 knockout (KO) LS180 cells. FDR = false discovery rate. Glycan compositions are depicted by the numbers of monosaccharide moieties (top to bottom: Hex, HexNAc, Neu5Ac, Fucose) and color coded for molar amount of SA (Neu5Ac) residues. Examples of most significantly changed SA (Neu5Ac)-containing N-glycan structures are shown.
(C) Protein interaction network and cluster enrichment analyzed by STRING and Cytoscape of the proteins carrying regulated N-glycoforms in ST6 KO LS180 cells with FDR = 0.05. Mucus proteins are encircled in red.
(D) MUC2 average expression level and percent expression of MUC2 and mucin members in the identified epithelial, stromal, glia, and immune cell types from human colon single cell sequencing obtained from https://doi.org/10.1016/j.cell.2019.06.029. Heat map represents expression level. Arrow shows goblet cell expression of MUC2.
(E) Western blot (WB) analysis of ST6 in the indicated genetically modified LS180 cells. OE = overexpression. Densitometry of the ST6 band was normalized to the HSP90 band and ratios are shown below each lane. Data represent 3 experiments (B, E).
(A) RNA expression of SA transferase family genes in the indicated human tissues displayed in the order of Enzyme Commission (EC) numbers (right). GI tract and ST6GALNAC1 (ST6) are shown in red. Data processing and normalization by GTEx portal. TPM = transcripts per million.
(B) RNA expression of ST6 in the indicated human tissues. Red indicates GI tract tissues. Data processing and normalization by GTEx portal.
(C) ST6 protein level displayed in descending order of abundance in indicated human tissues analyzed by mass spectrometry. Red indicates GI tract tissues. Data are from https://doi.org/10.15252/msb.20188503.
(D) ST6 average expression level and percent expression of ST6 and sialyltransferase family members as in (A) in individual cell types. Red arrows = high expression.
(E) t-SNE-1 plots showing ST6 expression (bottom) in the indicated stromal and glia cell types (top) from human colon single cell sequencing obtained from https://doi.org/10.1016Zj.cell.2019.06.029. Heat map represents expression level.
Highlights.
A local sialyltransferase dominantly controls intestinal protein sialylation
Sialylation protects intestinal mucus integrity from bacterial degradation
Mucus sialylation is critical for commensalism and bacterial metabolite homeostasis
Treatment with sialylated mucins ameliorates gut inflammation
ACKNOWLEDGMENTS
We thank the participating physicians and families. We thank Jizhong Zou from the National Heart, Lung, and Blood Institute (NHLBI) for iPSC cell generation. We thank Ronald Germain and Jeff Zhu for critically reading the manuscript; Ryan Kissinger, NIAID for illustrations; and acknowledge support from the Regeneron Genetics Center. This research was supported by the Intramural Research Program of the NIAID, NIH. C.R.B. was funded by National Cancer Institute (NCI) Grant (R01CA200423), H.Y.S. by the National Eye Institute (NEI) Intramural Research Program (ZIAEY000569-01), and K.T.P. by the LOEWE Center Frankfurt Cancer Institute (FCI), Hessen State Ministry for Higher Education, Research, and the Arts [III L 5-519/03/03.001-(0015)]. We thank Dr. Yan Wang of NIDCR Mass Spectrometry Facility for mass spectrometry of S-Tn carrying proteins. The work with NIDCR Mass Spectrometry Facility is supported by the Division of Intramural Research, NIDCR/NIH (ZIA DE000751) and the Intramural Research Program of the NIDCR, NIH (ZIA-DE000739-05) with thanks to Lawrence A. Tabak. This work was supported by National Multiple Sclerosis Society Career Transition Award (TA 3059-A-2 to C.W.). This work utilized the NIH HPC Biowulf cluster. A.M.M and C.K were funded by the Leona M. and Harry B. Helmsley Charitable Trust and NIH (RC2DK122532) and A.M.M by a Canada Research Chair (Tier 1) in Pediatric IBD, CIHR Foundation Grant, and NIH (RC2DK118640).
Footnotes
Declaration of interests
The authors declare no competing interests.
Supplemental information
Document S1. Tables S1 and S2, related to Figure 4.
Table S3. Oligonucleotides used in this paper, related to STAR Methods.
Data S1. Detailed patient clinical features, related to Figure 4.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alipour M, Zaidi D, Valcheva R, Jovel J, Martinez I, Sergi C, Walter J, Mason AL, Wong GK, Dieleman LA, et al. (2016). Mucosal Barrier Depletion and Loss of Bacterial Diversity are Primary Abnormalities in Paediatric Ulcerative Colitis. Journal of Crohn’s & colitis 10, 462–471. 10.1093/ecco-jcc/jjv223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arike L, Holmen-Larsson J, and Hansson GC (2017). Intestinal Muc2 mucin O-glycosylation is affected by microbiota and regulated by differential expression of glycosyltranferases. Glycobiology 27, 318–328. 10.1093/glycob/cww134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, and Rudensky AY (2013). Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504, 451–455. 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, et al. (2013). Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 500, 232–236. 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- Banerjee A, Herring CA, Chen B, Kim H, Simmons AJ, Southard-Smith AN, Allaman MM, White JR, Macedonia MC, McKinley ET, et al. (2020). Succinate Produced by Intestinal Microbes Promotes Specification of Tuft Cells to Suppress Ileal Inflammation. Gastroenterology 159, 2101–2115 e2105. 10.1053/j.gastro.2020.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banfield DK (2011). Mechanisms of protein retention in the Golgi. Cold Spring Harbor perspectives in biology 3, a005264. 10.1101/cshperspect.a005264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstrom K, Shan X, Casero D, Batushansky A, Lagishetty V, Jacobs JP, Hoover C, Kondo Y, Shao B, Gao L, et al. (2020). Proximal colon-derived O-glycosylated mucus encapsulates and modulates the microbiota. Science 370, 467–472. 10.1126/science.aay7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchenough GM, Johansson ME, Gustafsson JK, Bergstrom JH, and Hansson GC (2015). New developments in goblet cell mucus secretion and function. Mucosal Immunol 8, 712–719. 10.1038/mi.2015.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokulich NA, Kaehler BD, Rideout JR, Dillon M, Bolyen E, Knight R, Huttley GA, and Gregory Caporaso J. (2018). Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6, 90. 10.1186/s40168-018-0470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37, 852–857. 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, and Holmes SP (2016).DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 13, 5811609 583. 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell BJ, Yu LG, and Rhodes JM (2001). Altered glycosylation in inflammatory bowel disease: a possible role in cancer development. Glycoconj J 18, 851–858. 10.1023/a:1022240107040. [DOI] [PubMed] [Google Scholar]
- Chang PV, Hao L, Offermanns S, and Medzhitov R. (2014). The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci U S A 111, 2247–2252. 10.1073/pnas.1322269111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Luo J, Li J, Kim G, Chen ES, Xiao S, Snapper SB, Bao B, An D, Blumberg RS, et al. (2021). Foxo1 controls gut homeostasis and commensalism by regulating mucussecretion. The Journal of experimental medicine 218. 10.1084/jem.20210324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi H, Liu C, Yang H, Zeng WF, Wu L, Zhou WJ, Wang RM, Niu XN, Ding YH,Zhang Y, et al. (2018). Comprehensive identification of peptides in tandem mass spectra using an efficient open search engine. Nat Biotechnol. 10.1038/nbt.4236. [DOI] [PubMed] [Google Scholar]
- Corces MR, Trevino AE, Hamilton EG, Greenside PG, Sinnott-Armstrong NA, Vesuna S, Satpathy AT, Rubin AJ, Montine KS, Wu B, et al. (2017). An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. NatMethods 14, 959–962. 10.1038/nmeth.4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichele DD, and Kharbanda KK (2017). Dextran sodium sulfate colitis murine model: An indispensable tool for advancing our understanding of inflammatory bowel diseases pathogenesis. World J Gastroenterol 23, 6016–6029. 10.3748/wjg.v23.i33.6016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang P, Ji Y, Silbern I, Doebele C, Ninov M, Lenz C, Oellerich T, Pan KT, and Urlaub H. (2020). A streamlined pipeline for multiplexed quantitative site-specific N-glycoproteomics. Nat Commun 11, 5268. 10.1038/s41467-020-19052-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, et al. (2013). Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504, 446–450. 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- Gersemann M, Becker S, Kubler I, Koslowski M, Wang G, Herrlinger KR, Griger J, Fritz P, Fellermann K, Schwab M, et al. (2009). Differences in goblet cell differentiation between Crohn’s disease and ulcerative colitis. Differentiation 77, 84–94. 10.1016/j.diff.2008.09.008. [DOI] [PubMed] [Google Scholar]
- Guzman-Aranguez A, and Argueso P. (2010). Structure and biological roles of mucin-type O-glycans at the ocular surface. Ocul Surf 8, 8–17. 10.1016/s1542-0124(12)70213-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habowski AN, Flesher JL, Bates JM, Tsai CF, Martin K, Zhao R, Ganesan AK,Edwards RA, Shi T, Wiley HS, et al. (2020). Transcriptomic and proteomic signatures of stemness and differentiation in the colon crypt. Commun Biol 3, 453. 10.1038/s42003-020-01181-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer HM, Jonkers DM, Vanhoutvin SA, Troost FJ, Rijkers G, de Bruine A, Bast A, Venema K, and Brummer RJ (2010). Effect of butyrate enemas on inflammation and antioxidant status in the colonic mucosa of patients with ulcerative colitis in remission. Clin Nutr 29, 738–744. 10.1016/j.clnu.2010.04.002. [DOI] [PubMed] [Google Scholar]
- Han J, Lin K, Sequeira C, and Borchers CH (2015). An isotope-labeled chemical derivatization method for the quantitation of short-chain fatty acids in human feces by liquid chromatography-tandem mass spectrometry. Anal Chim Acta 854, 86–94. 10.1016/j.aca.2014.11.015. [DOI] [PubMed] [Google Scholar]
- Harduin-Lepers A, Vallejo-Ruiz V, Krzewinski-Recchi MA, Samyn-Petit B, Julien S, and Delannoy P. (2001). The human sialyltransferase family. Biochimie 83, 727–737. 10.1016/s0300-9084(01)01301-3. [DOI] [PubMed] [Google Scholar]
- Hayer A, Shao L, Chung M, Joubert LM, Yang HW, Tsai FC, Bisaria A, Betzig E, and Meyer T. (2016). Engulfed cadherin fingers are polarized junctional structures between collectively migrating endothelial cells. Nat Cell Biol 18, 1311–1323. 10.1038/ncb3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helander HF, and Fandriks L. (2014). Surface area of the digestive tract -revisited. Scand J Gastroenterol 49, 681–689. 10.3109/00365521.2014.898326. [DOI] [PubMed] [Google Scholar]
- Ikehara Y, Kojima N, Kurosawa N, Kudo T, Kono M, Nishihara S, Issiki S, Morozumi K, Itzkowitz S, Tsuda T, et al. (1999). Cloning and expression of a human gene encoding an N-acetylgalactosamine-alpha2,6-sialyltransferase (ST6GalNAc I): a candidate for synthesis of cancer-associated sialyl-Tn antigens. Glycobiology 9, 1213–1224. 10.1093/glycob/9.11.1213. [DOI] [PubMed] [Google Scholar]
- InanlooRahatloo K, Parsa AF, Huse K, Rasooli P, Davaran S, Platzer M, Kramer M, Fan JB, Turk C, Amini S, et al. (2014). Mutation in ST6GALNAC5 identified in family with coronary artery disease. Sci Rep 4, 3595. 10.1038/srep03595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan G, Belzacq AS, Haouzi D, Rouault A, Metivier D, Kroemer G, and Brenner C. (2002). Propionibacteria induce apoptosis of colorectal carcinoma cells via short-chain fatty acids acting on mitochondria. Cell Death Differ 9, 179–188. 10.1038/sj.cdd.4400935. [DOI] [PubMed] [Google Scholar]
- Jeong YH, Kim MC, Ahn EK, Seol SY, Do EJ, Choi HJ, Chu IS, Kim WJ, Kim WJ, Sunwoo Y, and Leem SH (2007). Rare exonic minisatellite alleles in MUC2 influence susceptibility to gastric carcinoma. PLoS One 2, e1163. 10.1371/journal.pone.0001163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji J, Shu D, Zheng M, Wang J, Luo C, Wang Y, Guo F, Zou X, Lv X, Li Y, et al. (2016). Microbial metabolite butyrate facilitates M2 macrophage polarization and function. Sci Rep 6, 24838. 10.1038/srep24838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson ME, and Hansson GC (2012). Analysis of assembly of secreted mucins. Methods Mol Biol 842, 109–121. 10.1007/978-1-61779-513-8_6. [DOI] [PubMed] [Google Scholar]
- Johansson ME, Sjovall H, and Hansson GC (2013). The gastrointestinal mucus system in health and disease. Nature reviews. Gastroenterology & hepatology 10, 352–361. 10.1038/nrgastro.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson ME, Thomsson KA, and Hansson GC (2009). Proteomic analyses of the two mucus layers of the colon barrier reveal that their main component, the Muc2 mucin, is strongly bound to the Fcgbp protein. J Proteome Res 8, 3549–3557. 10.1021/pr9002504. [DOI] [PubMed] [Google Scholar]
- Julien S, Videira PA, and Delannoy P. (2012). Sialyl-tn in cancer: (how) did we miss the target? Biomolecules 2, 435–466. 10.3390/biom2040435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiko GE, Ryu SH, Koues OI, Collins PL, Solnica-Krezel L, Pearce EJ, Pearce EL, Oltz EM, and Stappenbeck TS (2016). The Colonic Crypt Protects Stem Cells from Microbiota-Derived Metabolites. Cell 165, 1708–1720. 10.1016/j.cell.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khor B, Gardet A, and Xavier RJ (2011). Genetics and pathogenesis of inflammatory bowel disease. Nature 474, 307–317. 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Kwon O, Ryu TY, Jung CR, Kim J, Min JK, Kim DS, Son MY, and Cho HS (2019). Propionate of a microbiota metabolite induces cell apoptosis and cell cycle arrest in lung cancer. Mol Med Rep 20, 1569–1574. 10.3892/mmr.2019.10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko SH, Choi GE, Oh JY, Lee HJ, Kim JS, Chae CW, Choi D, and Han HJ (2017). Succinate promotes stem cell migration through the GPR91-dependent regulation of DRP1-mediated mitochondrial fission. Sci Rep 7, 12582. 10.1038/s41598-017-12692-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh A, De Vadder F, Kovatcheva-Datchary P, and Backhed F. (2016). From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell 165, 1332–1345. 10.1016/jcell.2016.05.041. [DOI] [PubMed] [Google Scholar]
- Kudelka MR, Stowell SR, Cummings RD, and Neish AS (2020). Intestinal epithelial glycosylation in homeostasis and gut microbiota interactions in IBD. Nature reviews. Gastroenterology & hepatology 17, 597–617. 10.1038/s41575-020-0331-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S,Jenkins SL, Jagodnik KM, Lachmann A, et al. (2016). Enrichr: a comprehensive gene setenrichment analysis web server 2016 update. Nucleic Acids Res 44, W90–97. 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvorjak M, Ahmed Y, Miller ML, Sriram R, Coronnello C, Hashash JG, Hartman DJ,Telmer CA, Miskov-Zivanov N, Finn OJ, and Cascio S. (2020). Cross-talk between ColonCells and Macrophages Increases ST6GALNAC1 and MUC1-sTn Expression in UlcerativeColitis and Colitis-Associated Colon Cancer. Cancer immunology research 8, 167–178. 10.1158/2326-6066.CIR-19-0514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, and Chen X. (2012). Sialic acid metabolism and sialyltransferases: natural functions and applications. Appl Microbiol Biotechnol 94, 887–905. 10.1007/s00253-012-4040-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liso M, De Santis S, Verna G, Dicarlo M, Calasso M, Santino A, Gigante I, Eri R, Raveenthiraraj S, Sobolewski A, et al. (2020). A Specific Mutation in Muc2 Determines Early Dysbiosis in Colitis-Prone Winnie Mice. Inflamm Bowel Dis 26, 546–556. 10.1093/ibd/izz279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus EV Jr. (2004). Clinical epidemiology of inflammatory bowel disease: Incidence, prevalence, and environmental influences. Gastroenterology 126, 1504–1517. 10.1053/j.gastro.2004.01.063. [DOI] [PubMed] [Google Scholar]
- Long PM, Tighe SW, Driscoll HE, Fortner KA, Viapiano MS, and Jaworski DM (2015). Acetate supplementation as a means of inducing glioblastoma stem-like cell growth arrest. J Cell Physiol 230, 1929–1943. 10.1002/jcp.24927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loureiro LR, Sousa DP, Ferreira D, Chai W, Lima L, Pereira C, Lopes CB, Correia VG, Silva LM, Li C, et al. (2018). Novel monoclonal antibody L2A5 specifically targeting sialyl-Tn and short glycans terminated by alpha-2–6 sialic acids. Sci Rep 8, 12196. 10.1038/s41598-018-30421-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malaker SA, Pedram K, Ferracane MJ, Bensing BA, Krishnan V, Pett C, Yu J, Woods EC, Kramer JR, Westerlind U, et al. (2019). The mucin-selective protease StcE enables molecular and functional analysis of human cancer-associated mucins. Proc Natl Acad Sci U S A 116, 7278–7287. 10.1073/pnas.1813020116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcos NT, Pinho S, Grandela C, Cruz A, Samyn-Petit B, Harduin-Lepers A, Almeida R, Silva F, Morais V, Costa J, et al. (2004). Role of the human ST6GalNAc-I andST6GalNAc-II in the synthesis of the cancer-associated sialyl-Tn antigen. Cancer Res 64, 7050–7057. 10.1158/0008-5472.CAN-04-1921. [DOI] [PubMed] [Google Scholar]
- Mayne CG, and Williams CB (2013). Induced and natural regulatory T cells in thedevelopment of inflammatory bowel disease. Inflamm Bowel Dis 19, 1772–1788. 10.1097/MIB.0b013e318281f5a3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moehle C, Ackermann N, Langmann T, Aslanidis C, Kel A, Kel-Margoulis O, Schmitz-Madry A, Zahn A, Stremmel W, and Schmitz G. (2006). Aberrant intestinal expression andallelic variants of mucin genes associated with inflammatory bowel disease. J Mol Med (Berl) 84, 1055–1066. 10.1007/s00109-006-0100-2. [DOI] [PubMed] [Google Scholar]
- Molodecky NA, Soon IS, Rabi DM, Ghali WA, Ferris M, Chernoff G, Benchimol EI,Panaccione R, Ghosh S, Barkema HW, and Kaplan GG (2012). Increasing incidence andprevalence of the inflammatory bowel diseases with time, based on systematic review.Gastroenterology 142, 46–54 e42; quiz e30. 10.1053/j.gastro.2011.10.001. [DOI] [PubMed] [Google Scholar]
- Moremen KW, Ramiah A, Stuart M, Steel J, Meng L, Forouhar F, Moniz HA, Gahlay G, Gao Z, Chapla D, et al. (2018). Expression system for structural and functional studies ofhuman glycosylation enzymes. Nat Chem Biol 14, 156–162. 10.1038/nchembio.2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munkley J. (2016). The Role of Sialyl-Tn in Cancer. Int J Mol Sci 17, 275. 10.3390/ijms17030275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai K, Ihara Y, Wada Y, and Taniguchi N. (1997). N-glycosylation is requisite for theenzyme activity and Golgi retention of N-acetylglucosaminyltransferase III. Glycobiology 7,769–776. 10.1093/glycob/7.6.769. [DOI] [PubMed] [Google Scholar]
- Patel RY, and Balaji PV (2006). Identification of linkage-specific sequence motifs insialyltransferases. Glycobiology 16, 108–116. 10.1093/glycob/cwj046. [DOI] [PubMed] [Google Scholar]
- Pelaseyed T, Bergstrom JH, Gustafsson JK, Ermund A, Birchenough GM, Schutte A, van der Post S, Svensson F, Rodriguez-Pineiro AM, Nystrom EE, et al. (2014). The mucus and mucins of the goblet cells and enterocytes provide the first defense line of the gastrointestinal tract and interact with the immune system. Immunol Rev 260, 8–20. 10.1111/imr.12182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickard JM, Maurice CF, Kinnebrew MA, Abt MC, Schenten D, Golovkina TV, Bogatyrev SR, Ismagilov RF, Pamer EG, Turnbaugh PJ, and Chervonsky AV (2014). Rapid fucosylation of intestinal epithelium sustains host-commensal symbiosis in sickness. Nature 514, 638–641. 10.1038/nature13823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickard JM, Zeng MY, Caruso R, and Nunez G. (2017). Gut microbiota: Role in pathogen colonization, immune responses, and inflammatory disease. Immunol Rev 279, 70–89. 10.1111/imr.12567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendergast JM, Galvao da Silva AP, Eavarone DA, Ghaderi D, Zhang M, Brady D, Wicks J, DeSander J, Behrens J, and Rueda BR (2017). Novel anti-Sialyl-Tn monoclonal antibodies and antibody-drug conjugates demonstrate tumor specificity and anti-tumor activity. MAbs 9, 615–627. 10.1080/19420862.2017.1290752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbe C, Capon C, Maes E, Rousset M, Zweibaum A, Zanetta JP, and Michalski JC (2003). Evidence of regio-specific glycosylation in human intestinal mucins: presence of an acidic gradient along the intestinal tract. J Biol Chem 278, 46337–46348. 10.1074/jbc.M302529200. [DOI] [PubMed] [Google Scholar]
- Robbe-Masselot C, Maes E, Rousset M, Michalski JC, and Capon C. (2009). Glycosylation of human fetal mucins: a similar repertoire of O-glycans along the intestinal tract. Glycoconj J 26, 397–413. 10.1007/s10719-008-9186-9. [DOI] [PubMed] [Google Scholar]
- Roy A, Kucukural A, and Zhang Y. (2010). I-TASSER: a unified platform for automated protein structure and function prediction. Nature protocols 5, 725–738. 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudresh, Jain R, Dani V, Mitra A, Srivastava S, Sarma SP, Varadarajan R, and Ramakumar S. (2002). Structural consequences of replacement of an alpha-helical Pro residue in Escherichia coli thioredoxin. Protein Eng 15, 627–633. 10.1093/protein/15.8.627. [DOI] [PubMed] [Google Scholar]
- Sahuri-Arisoylu M, Mould RR, Shinjyo N, Bligh SWA, Nunn AVW, Guy GW, Thomas EL, and Bell JD (2021). Acetate Induces Growth Arrest in Colon Cancer Cells Through Modulation of Mitochondrial Function. Front Nutr 8, 588466. 10.3389/fnut.2021.588466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salcher S, Spoden G, Hagenbuchner J, Fuhrer S, Kaserer T, Tollinger M, Huber-Cantonati P, Gruber T, Schuster D, Gust R, et al. (2020). A drug library screen identifies Carbenoxolone as novel FOXO inhibitor that overcomes FOXO3-mediated chemoprotection in high-stage neuroblastoma. Oncogene 39, 1080–1097. 10.1038/s41388-019-1044-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, and Zhang F. (2014). Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11, 783–784. 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S, Van Houdt WJ, Pronk A, Van Gorp J, Siersema PD, and Clevers H. (2011). Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 141, 1762–1772. 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- Schjoldager KT, Narimatsu Y, Joshi HJ, and Clausen H. (2020). Global view of human protein glycosylation pathways and functions. Nature reviews. Molecular cell biology 21, 729–1801749. 10.1038/s41580-020-00294-x. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9, 671–675. 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewell R, Backstrom M, Dalziel M, Gschmeissner S, Karlsson H, Noll T, Gatgens J, Clausen H, Hansson GC, Burchell J, and Taylor-Papadimitriou J. (2006). The ST6GalNAc-I sialyltransferase localizes throughout the Golgi and is responsible for the synthesis of the tumor-associated sialyl-Tn O-glycan in human breast cancer. The Journal of biological chemistry 281,3586–3594. 10.1074/jbc.M511826200. [DOI] [PubMed] [Google Scholar]
- Shan M, Gentile M, Yeiser JR, Walland AC, Bornstein VU, Chen K, He B, Cassis L, Bigas A, Cols M, et al. (2013). Mucus enhances gut homeostasis and oral tolerance by delivering immunoregulatory signals. Science 342, 447–453. 10.1126/science.1237910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shauchuk A, Szulc B, Maszczak-Seneczko D, Wiertelak W, Skurska E, and Olczak M. (2020). N-glycosylation of the human beta1,4-galactosyltransferase 4 is crucial for its activity and Golgi localization. Glycoconj J 37, 577–588. 10.1007/s10719-020-09941-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng YH, Hasnain SZ, Florin TH, and McGuckin MA (2012). Mucins in inflammatory bowel diseases and colorectal cancer. J Gastroenterol Hepatol 27, 28–38. 10.1111/j.1440-1746.2011.06909.x. [DOI] [PubMed] [Google Scholar]
- Shrimal S, Cherepanova NA, and Gilmore R. (2015). Cotranslational and posttranslocational N-glycosylation of proteins in the endoplasmic reticulum. Semin Cell Dev Biol 41, 71–78. 10.1016/j.semcdb.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh B, Read S, Asseman C, Malmstrom V, Mottet C, Stephens LA, Stepankova R, Tlaskalova H, and Powrie F. (2001). Control of intestinal inflammation by regulatory T cells. Immunological reviews 182, 190–200. 10.1034/j.1600-065x.2001.1820115.x. [DOI] [PubMed] [Google Scholar]
- Smillie CS, Biton M, Ordovas-Montanes J, Sullivan KM, Burgin G, Graham DB,Herbst RH, Rogel N, Slyper M, Waldman J, et al. (2019). Intra-and Inter-cellular Rewiring of the Human Colon during Ulcerative Colitis. Cell 178, 714–730 e722. 10.1016/j.cell.2019.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, Glickman JN, and Garrett WS (2013). The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341, 569–573. 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence JR, Mayhew CN, Rankin SA, Kuhar MF, Vallance JE, Tolle K, Hoskins EE, Kalinichenko VV, Wells SI, Zorn AM, et al. (2011). Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 470, 105–109. 10.1038/nature09691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark RM, Wiggins R, Walley E, Hicks SJ, Gill GA, Carrington SD, and Corfield AP (2000). Mucinase activity. Methods Mol Biol 125, 385–392. 10.1385/1-59259-048-9:385. [DOI] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, and Satija R. (2019). Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902 e1821. 10.1016/jcell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama N, Igarashi O, Kweon MN, and Kiyono H. (2007). Regulatory role of Peyer’s patches for the inhibition of OVA-induced allergic diarrhea. Clin Immunol 123, 199–208. 10.1016/j.clim.2007.01.007. [DOI] [PubMed] [Google Scholar]
- Tarrerias AL, Millecamps M, Alloui A, Beaughard C, Kemeny JL, Bourdu S, Bommelaer G, Eschalier A, Dapoigny M, and Ardid D. (2002). Short-chain fatty acid enemas fail to decrease colonic hypersensitivity and inflammation in TNBS-induced colonic inflammation in rats. Pain 100, 91–97. 10.1016/s0304-3959(02)00234-8. [DOI] [PubMed] [Google Scholar]
- Theodoratou E, Campbell H, Ventham NT, Kolarich D, Pucic-Bakovic M, Zoldos V, Fernandes D, Pemberton IK, Rudan I, Kennedy NA, et al. (2014). The role of glycosylation in IBD. Nature reviews. Gastroenterology & hepatology 11, 588–600. 10.1038/nrgastro.2014.78. [DOI] [PubMed] [Google Scholar]
- Trastoy B, Naegeli A, Anso I, Sjogren J, and Guerin ME (2020). Structural basis of mammalian mucin processing by the human gut O-glycopeptidase OgpA from Akkermansia muciniphila. Nat Commun 11, 4844. 10.1038/s41467-020-18696-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tytgat KM, Opdam FJ, Einerhand AW, Buller HA, and Dekker J. (1996). MUC2 is the prominent colonic mucin expressed in ulcerative colitis. Gut 38, 554–563. 10.1136/gut.38.4.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Sluis M, De Koning BA, De Bruijn AC, Velcich A, Meijerink JP, Van Goudoever JB, Buller HA, Dekker J, Van Seuningen I, Renes IB, and Einerhand AW (2006). Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology 131, 117–129. 10.1053/j.gastro.2006.04.020. [DOI] [PubMed] [Google Scholar]
- Varki A. (2008). Sialic acids in human health and disease. Trends Mol Med 14, 351–360. 10.1016/j.molmed.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visschedijk MC, Alberts R, Mucha S, Deelen P, de Jong DJ, Pierik M, Spekhorst LM, Imhann F, van der Meulen-de Jong AE, van der Woude CJ, et al. (2016). Pooled Resequencing of 122 Ulcerative Colitis Genes in a Large Dutch Cohort Suggests Population-Specific Associations of Rare Variants in MUC2. PLoS One 11, e0159609. 10.1371/journal.pone.0159609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, and Jaenisch R. (2013). One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–918. 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Wu Y, Li J, Bao Y, Guo Y, and Yang W. (2018). The Dynamic Changes of Gut Microbiota in Muc2 Deficient Mice. Int J Mol Sci 19. 10.3390/ijms19092809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu ZL, Person AD, Burton AJ, Singh R, Burroughs B, Fryxell D, Tatge TJ, Manning T, Wu G, Swift KAD, and Kalabokis V. (2019). Direct fluorescent glycan labeling with recombinant sialyltransferases. Glycobiology 29, 750–754. 10.1093/glycob/cwz058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, Farin HF, van Es JH, Clevers H, Langer R, and Karp JM (2014). Niche-independent high-purity cultures of Lgr5+ intestinal stem cells and their progeny. Nat Methods 11, 106–112. 10.1038/nmeth.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Principal component analysis (PCA) of gut microbiome by 16S rDNA gene sequencing from WT and St6 female mice before (3 weeks) and after (6 weeks) weaning. Dots represent individual mice.
(B) Modified volcano plot showing the significant changes of gut bacterial families in 10-week-old female WT vs. St6 mice.
(C) Gut bacterial community composition (family level) of 10-week-old female WT and St6 mice. Red rectangle with asterisk indicates significantly changed families.
(D) Relative abundance of the indicated bacteria in feces from 10-week-old female WT and St6 mice.
(E) Confocal photomicrographs of Ki-67 (red) and DAPI (blue) stained colon sections from WT and St6 mice 10 days after DSS colitis with antibiotic (ABX) treatment. Scale bar, 100 μm
(F) Quantification of the results from (E). NS = no significance.
(G) Q-PCR analysis of Lgr5 using RNA from IECs and crypt intestinal stem cells (ISCs) of WT and St6 mice.
(H) PCA of transcriptomes (RNA-seq) of ISCs from WT and St6 mice 10 days after DSS colitis.
(I) PCA of assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq) using purified ISCs from WT and St6 mice as in (H).
(J) Venn diagram showing the numbers of differentially expressed genes in WT and St6 mice in (H) and (I).
(K) Gene Ontology (GO) term analysis of RNA-seq of ISCs from WT and St6 mice as in (H).
(L) Q-PCR analysis of ISC functional genes using RNA from purified crypt ISCs from WT and St6 mice as in (H).
(M) Normalized ATAC-seq sequencing tracks from WT and St6 mice for the indicated loci as in (L). Data represent 3 experiments (A, D-I, L, M). Error bars represent the SD of samples within a group. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
(A) Schematic of St6 mice prepared by CRISPR knock-in of a single base substitution (G > A) encoding the single amino acid change from Arg (R) to Gln (Q) corresponding to the Pt 1 mutation. Sanger sequencing of St6 mutants in IECs from WT and St6 mice are shown in the right panel.
(B) Photomicrographs of representative hematoxylin and eosin-stained sections of small intestine (SI) and colon from 8–10-week-old female WT and St6 mice. Scale bar, 50 μm.
(C) Photomicrographs of representative colon sections from 8–10-week-old female WT and St6 mice stained with Periodic Acid-Schiff (PAS) (left). Statistical results of mucin area in each cell and GC number in each crypt (right). Scale bar, 50 μm.
(D) Q-PCR analysis of Muc2 mRNA in SI and colon IECs from naive WT and St6 mice.
(E) Q-PCR analysis of indicated GC genes from IEC RNA as in (D).
(F) Confocal microscopic images of colon from WT and St6 mice stained as indicated Paneth cell genes. Scale bar, 100 μm.
(G) Q-PCR analysis of antimicrobial genes in IECs as in (D).
(H) Q-PCR analysis of tight junction genes in IECs as in (D).
(I-M) Quantification of flow cytometry analyses of mouse ILC2, ILC3 (I), intraepithelial lymphocytes (IELs) (J), B cells, T cells, NK cells (K), macrophages, neutrophils (L), and Tregs (M) in SI and colon from WT and St6 mice.
(N) The changed N-glycoform carrying proteins in St6 mice were analyzed by protein interaction network and cluster enrichment by STRING database and Cytoscape. Mucin proteins are circled in red. Data represent 3 experiments (A-M). Error bars represent the SD of samples within a group.
(A) Densitometric quantification of Figure 5A for the ratio of ST6 upper (modified by sialylation)/lower (unmodified protein) bands, total ST6 (upper + lower bands)/GAPDH, and total ST6 (upper + lower bands)/tEGFR. ND = not determined.
(B) WB analysis of ST6, S-Tn, and GAPDH loading control using lysates from 293T cells overexpressing WT and mutant forms of human ST6. EV = empty vector control. × = nonspecific bands.
(C) Flow cytometry analysis of S-Tn and tEGFR in 293T cells as in (B).
(D) Quantification and statistical graphs of (C).
(E) Flow cytometry analysis as in (C) in HT29 cells after lentiviral co-transduction of T462M and T207P or T462M and WT human ST6 at the indicated ratios (T462M/T207P or T462M/WT).
(F) Quantification of S-Tn and tEGFR mean fluorescence intensity (MFI) in (E).
(G) Sanger sequencing of T462M ST6 mutants to indicate the ratio of T462M/WT or T462M/T207P after lentiviral co-transduction in HT29 cells as in (E).
(H) Non-supervised clustering of differently regulated N-glycans expression level (False Discovery Rate, FDR = 0.3) detected in HT29 cells with stable ST6-WT, EV, and ST6-R391Q expression (three biological replicates each). N-glycan intensities are shown with a color scale.
(I) Pie charts showing the molar distributions of SA (Neu5Ac)-quantities in N-glycans in clusters 1 and 2 (calculated from three biological replicates) indicated in (H). Pearson’s chi-squared test (p <0.0001) was used to calculate the p-value regarding the proportion of glycan status between clusters 1 and 2.
(J) Volcano plot of quantified N-glycans in ST6-WT compared to that in ST6-R391Q stable expressing HT29 cells. Glycan compositions are depicted by the numbers of monosaccharide moieties (top to bottom: Hex, HexNAc, Neu5Ac, Fucose) and color coded for molar amount of SA (Neu5Ac) residues. Examples of most significantly changed SA (Neu5Ac)-containing N-glycan structures are shown.
(K) Phase contrast photomicrographs of induced pluripotent stem cell (iPSC)-generated intestinal organoids from Pt 1, Pt 2, and healthy donors (HDs). Scale bar, 100 μm.
(L, M) Sanger sequencing confirming ST6 mutants (L) and Q-PCR analysis of GC differentiation genes (M) of the organoids from Pt 1, Pt 2, and HDs as in (K). Data represent 3 experiments (A-M). Error bars represent the SD of samples within a group. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
(A) Relative expression (RE) of mRNA of GC-related genes Mucin-2 (MUC2), Trefoil Factor 3 (TFF3), and Kruppel-Like Factor 4 (KLF4) compared to GAPDH in the indicated cell types (Figures 1D and 1E) from healthy donors and colitis patients. Colon single cell sequencing data obtained from https://doi.org/10.1016/Lcell.2019.06.029. Box and whiskers represent the 10th−90th percentiles.
(B) Q-PCR analysis of indicated genes using RNA prepared from flow cytometry-sorted mouse colon cells from lamina propria (LP), intraepithelial lymphocytes (IELs), and intestinal epithelial cells (IECs).
(C) Q-PCR analysis of interleukin-6 (Il6) mRNA in IECs from WT mice at the indicated days after 2.5% DSS water treatment.
(D) Q-PCR analysis of ST6 in human IEC HT29 cells after toll like receptor ligands treatment for 8 hours.
(E) Q-PCR analysis of Tlr4 and St6 in IECs from WT and Tlr4−/− mice.
(F) (Top) Formaldehyde-assisted isolation of regulatory elements (FAIRE)-Q-PCR analyses of chromatin accessibility at 8 tandem sites (x-axis) in the St6 promoter in WT or Tlr4−/− mouse IECs, y-axis presented enrichment percentage relative to input.(Bottom) Q-PCR primer sites (small horizontal bars) in the St6 locus (black arrow, transcription start site).
(G) Extended pedigree of family 1 showing consanguinity and ST6 allele (filled) inheritance. Deceased individuals are indicated by strikethrough and consanguinity is indicated by a double line. ND = whole exome DNA sequencing not determined.
(H) Fraction of cells determined by flow cytometry of patients (Pts 1 and 2) compared to healthy donor (HD) blood samples stained with antibodies as indicated showing CD57+, central memory (TCM) (CCR7+CD45RA-), naive (TN) (CCR7+CD45RA+), terminally differentiated effector memory (TEMRA) (CCR7-CD45RA+), and effector memory (TEM) (CCR7-CD45RA-) CD4+ and CD8+ T cells. Each dot represents an individual.
(I) Percentage of CD69+ CD4+ and CD8+ T cells 24 hours after activation of purified cells using CD3 and CD28 activation beads as in (H).
(J-M) Specific analyses for T regulatory (Treg), B, natural killer (NK), monocyte, and dendritic cell (DC) subsets as in (H).
(N) Table showing bioinformatic information for ST6 variants. Data represent 3 experiments (B-F). Error bars represent the SD of samples within a group. *, p < 0.05;**, p < 0.01; ***, p < 0.001.
(A) Schematic of Glycan (glycan structures linked to any glycosite), Glycoform (specific glycosite with specific glycan structure on specific protein), Glycosite (specific modification site on specific protein), and Sialylation (addition of sialic acid (SA, Neu5A(G)c) to the terminal end of glycoproteins).
(B) Volcano plot of quantified N-glycans in ST6 wildtype (WT) (control) compared to that in ST6 knockout (KO) LS180 cells. FDR = false discovery rate. Glycan compositions are depicted by the numbers of monosaccharide moieties (top to bottom: Hex, HexNAc, Neu5Ac, Fucose) and color coded for molar amount of SA (Neu5Ac) residues. Examples of most significantly changed SA (Neu5Ac)-containing N-glycan structures are shown.
(C) Protein interaction network and cluster enrichment analyzed by STRING and Cytoscape of the proteins carrying regulated N-glycoforms in ST6 KO LS180 cells with FDR = 0.05. Mucus proteins are encircled in red.
(D) MUC2 average expression level and percent expression of MUC2 and mucin members in the identified epithelial, stromal, glia, and immune cell types from human colon single cell sequencing obtained from https://doi.org/10.1016/j.cell.2019.06.029. Heat map represents expression level. Arrow shows goblet cell expression of MUC2.
(E) Western blot (WB) analysis of ST6 in the indicated genetically modified LS180 cells. OE = overexpression. Densitometry of the ST6 band was normalized to the HSP90 band and ratios are shown below each lane. Data represent 3 experiments (B, E).
(A) RNA expression of SA transferase family genes in the indicated human tissues displayed in the order of Enzyme Commission (EC) numbers (right). GI tract and ST6GALNAC1 (ST6) are shown in red. Data processing and normalization by GTEx portal. TPM = transcripts per million.
(B) RNA expression of ST6 in the indicated human tissues. Red indicates GI tract tissues. Data processing and normalization by GTEx portal.
(C) ST6 protein level displayed in descending order of abundance in indicated human tissues analyzed by mass spectrometry. Red indicates GI tract tissues. Data are from https://doi.org/10.15252/msb.20188503.
(D) ST6 average expression level and percent expression of ST6 and sialyltransferase family members as in (A) in individual cell types. Red arrows = high expression.
(E) t-SNE-1 plots showing ST6 expression (bottom) in the indicated stromal and glia cell types (top) from human colon single cell sequencing obtained from https://doi.org/10.1016Zj.cell.2019.06.029. Heat map represents expression level.
Data Availability Statement
The human exome and genome sequencing data reported in this paper has been deposited at dbGaP. mRNA-seq and ATAC-seq in this paper are available at GEO. The mass spectrometry-based glycoproteomics data has been deposited to the ProteomeXchange Consortium via the PRIDE partner repository. Data are publicly available as of the date of publication. These accession numbers for the datasets are listed in the Key resources table.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-human ST6GALNAC1 | Sigma | Cat# HPA014975; RRID: AB 1857529 |
| Rabbit monoclonal recombinant anti-human MUC2 | Abcam | Cat# ab133555 |
| Rabbit monoclonal anti-HSP90 | Ceii Signaling Technology | Cat# 4877; RRID: AB 2233307 |
| Mouse monoclonal anti-human Sialyl Tn (S-Tn) [BRST3, B72.3] | BioLegend | Cat# 915206; RRID: AB 2616965 |
| Goat polyclonal anti-human EGFR | R&D | Cat# AF231; RRID: AB 355220 |
| Rabbit monoclonal anti-HA-tag | Ceii Signaling Technology | Cat# 3724; RRID: AB 1549585 |
| Rabbit monoclonal anti-GAPDH | Ceii Signaling Technology | Cat# 2118; RRID: AB 561053 |
| Rabbit polyclonal anti-H3K27ac | Abcam | Cat# ab4729; RRID: AB 2118291 |
| Rabbit monoclonal anti-H3K9ac | Ceii Signaling Technology | Cat# 9649; RRID: AB 823528 |
| Rabbit monoclonal anti-histone H3 | Ceii Signaling Technology | Cat# 4499; RRID: AB 10544537 |
| Mouse monoclonal anti-MUC2 | Santa Cruz Biotechnology | Cat# sc-515032; RRID: AB 2815005 |
| Mouse monoclonal anti-HA epitope tag | Covance | Cat# MMS-101P; RRID: AB 2314672 |
| Rabbit monoclonal recombinant anti-GM130 | Abcam | Cat# ab52649; RRID: AB 880266 |
| Rabbit polyclonal anti-MUC2 | GeneTex | Cat# GTX100664; RRID:AB 1950958 |
| Mouse monoclonal anti-Sialyl Tn | Abcam | Cat# ab115957; RRID: AB 10901541 |
| Rabbit polyclonal anti-CD45 | Abcam | Cat# ab10558; RRID: AB 442810 |
| Rabbit polyclonal anti-REG3G | Abgent | Cat# AP5606c; RRID: AB 10817069 |
| Goat polyclonal anti-rabbit IgG (H+L) highly crossadsorbed Alexa Fluor Plus 647 | Thermo Fisher Scientific | Cat# A32733; RRID: AB 2633282 |
| Goat polyclonal anti-mouse IgG (H+L) highly crossadsorbed Alexa Fluor Plus 647 | Thermo Fisher Scientific | Cat# A32728; RRID: AB 2633277 |
| Goat polyclonal anti-rabbit IgG (H+L) highly crossadsorbed Alexa Fluor Plus 555 | Thermo Fisher Scientific | Cat# A32732; RRID: AB 2633281 |
| Goat recombinant polyclonal anti-mouse IgG (H+L) Superclonal™ Alexa Fluor 555 | Thermo Fisher Scientific | Cat# A28180; RRID: AB 2536164 |
| Goat recombinant polyclonal anti-rabbit IgG (H+L) Superclonal™ Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A27034; RRID: AB 2536097 |
| Goat polyclonal anti-mouse IgG (H+L) highly crossadsorbed Alexa Fluor Plus 488 | Thermo Fisher Scientific | Cat# A32723; RRID: AB 2633275 |
| Mouse monoclonal Ultra-LEAF™ purified anti-human CD3 | BioLegend | Cat# 300334; RRID: AB 2616670 |
| Mouse monoclonal LEAF™ purified anti-human CD28 | BioLegend | Cat# 302914; RRID: AB 314316 |
| Human IgG recombinant monoclonal human EGFR (Research Grade Cetuximab Biosimilar) Alexa Fluor® 488-conjugated | R&D | Cat# FAB9577G-100 |
| Mouse anti-mouse-IgG-control-human | Santa Cruz Biotechnology | Cat# sc-2025; RRID: AB 737182 |
| Mouse monoclonal FITC anti-human CD57 | BioLegend | Cat# 322306; RRID: AB 535992 |
| Mouse monoclonal Brilliant Violet 421™ anti-human CD197 (CCR7) | BioLegend | Cat# 353208; RRID: AB 11203894 |
| Mouse monoclonal PerCP/Cyanine5.5 anti-human CD4 | BioLegend | Cat# 344608; RRID: AB 1953236 |
| Mouse monoclonal PE/Cyanine7 anti-human CD45RA | BioLegend | Cat# 304126; RRID: AB 10708879 |
| Armenian hamster monoclonal APC antimouse/rat/human CD27 | BioLegend | Cat# 124212; RRID: AB 2073425 |
| Mouse monoclonal Brilliant Violet 605™ anti-human CD8a | BioLegend | Cat# 301040; RRID: AB 2563185 |
| Mouse monoclonal PE anti-human CD28 | BioLegend | Cat# 302940; RRID: AB 2564147 |
| Mouse monoclonal Brilliant Violet 605™ anti-human CD3 | BioLegend | Cat# 317321; RRID: AB 11126166 |
| Mouse monoclonal Alexa Fluor® 488 antimouse/rat/human FOXP3 | BioLegend | Cat# 320012; RRID: AB 439748 |
| Mouse monoclonal PE anti-human CD25 | BioLegend | Cat# 302606; RRID: AB 314276 |
| Mouse monoclonal APC anti-human CD127 (IL-7Ra) | BioLegend | Cat# 351316; RRID: AB 10900804 |
| Mouse monoclonal PE/Cyanine7 anti-human CD194 (CCR4) | BioLegend | Cat# 359410; RRID: AB 2562431 |
| Mouse monoclonal Pacific Blue™ anti-human CD45RO | BioLegend | Cat# 304216; RRID: AB 493659 |
| Mouse monoclonal PE anti-human CD24 | BioLegend | Cat# 311106; RRID: AB 314855 |
| Mouse BV786 anti-Human IgM | BD Biosciences | Cat# 740998; RRID: AB 2740621 |
| Mouse monoclonal PE-Cy™5 anti-human CD20 | BD Biosciences | Cat# 561761; RRID: AB 10898182 |
| Mouse monoclonal FITC anti-human CD19 | BD Biosciences | Cat# 555412; RRID: AB 395812 |
| Mouse monoclonal PerCP anti-human CD27 | BioLegend | Cat# 302817; RRID: AB 893294 |
| Mouse monoclonal APC anti-human CD38 | BioLegend | Cat# 356606; RRID: AB 2561902 |
| Mouse monoclonal Brilliant Violet 421™ anti-human CD3 | BioLegend | Cat# 344834; RRID: AB 2565675 |
| Mouse monoclonal Brilliant Violet 510™ anti-human IgD | BioLegend | Cat# 348220; RRID: AB 2561945 |
| Mouse monoclonal PE anti-human CD56 (NCAM) | BioLegend | Cat# 362507; RRID: AB 2563924 |
| Mouse monoclonal APC anti-human CD16 | BioLegend | Cat# 302011; RRID: AB 314211 |
| Mouse monoclonal FITC eBioscience™ anti-human CD14 | Thermo Fisher Scientific | Cat# 11–0149-42; RRID: AB 10597597 |
| Mouse monoclonal Brilliant Violet 421™ anti-human HLA-DR | BioLegend | Cat# 307636; RRID: AB 2561831 |
| Mouse monoclonal PE/Cyanine7 anti-human CD11c | BioLegend | Cat# 371507; RRID: AB 2650779 |
| Mouse monoclonal PerCP anti-human CD19 | BioLegend | Cat# 302228; RRID: AB 893272 |
| Rat monoclonal Brilliant Violet 605™ anti-mouse/human CD44 | BioLegend | Cat# 103047; RRID: AB 2562451 |
| Mouse monoclonal Alexa Fluor® 700 anti-human CD3 | BioLegend | Cat# 344821; RRID: AB 2563419 |
| Mouse monoclonal PerCP anti-human CD8a | BioLegend | Cat# 300922; RRID: AB 1575072 |
| Mouse monoclonal APC anti-human CD4 | BioLegend | Cat# 300552; RRID: AB 2564153 |
| Mouse monoclonal Brilliant Violet 510™ anti-human CD183 (CXCR3) | BioLegend | Cat# 353725; RRID: AB 2562066 |
| Mouse monoclonal Brilliant Violet 421™ anti-human CD184 (CXCR4) | BioLegend | Cat# 306517; RRID: AB 10901163 |
| Mouse monoclonal FITC anti-human CD196 (CCR6) | BioLegend | Cat# 353411; RRID: AB 10915473 |
| Mouse monoclonal PE/Cyanine7 anti-human CD194 (CCR4) | BioLegend | Cat# 359409; RRID: AB 2562430 |
| Armenian hamster monoclonal PE anti-human CCR10 | BioLegend | Cat# 341503; RRID: AB 1595542 |
| Mouse monoclonal FITC anti-human CD8a | BioLegend | Cat# 300906; RRID: AB 314110 |
| Mouse monoclonal APC anti-human CD69 | BioLegend | Cat# 310910; RRID: AB 314845 |
| Rat monoclonal Brilliant Violet 711 ™ anti-mouse CD45 | BioLegend | Cat# 103147; RRID: AB 2564383 |
| Armenian hamster monoclonal FITC anti-mouse TCR p chain | BioLegend | Cat# 109206; RRID: AB 313429 |
| Rat monoclonal PerCP/Cyanine5.5 anti-mouse CD4 | BioLegend | Cat# 100434; RRID: AB 893324 |
| Armenian hamster monoclonal Alexa Fluor® 647 antimouse TCR y/6 | BioLegend | Cat# 118133; RRID: AB 2566406 |
| Syrian hamster monoclonal Alexa Fluor® 700 antimouse CD3e | Biolegend | Cat# 152316; RRID: AB 2632713 |
| Rat monoclonal PE anti-mouse CD8a | BD Biosciences | Cat# 553033; RRID: AB 394571 |
| Mouse monoclonal PE-Cyanine7 eBioscience™ antihuman CD8b | Thermo Fisher Scientific | Cat# 25–5273-41; RRID: AB 11217487 |
| Rat monoclonal Brilliant Violet 510™ anti-mouse/human CD45R/B220 | BioLegend | Cat# 103248; RRID: AB 2561394 |
| Rat monoclonal FITC anti-mouse CD8a | BioLegend | Cat# 100706; RRID: AB 312745 |
| Rat monoclonal PerCP anti-mouse/human CD44 | BioLegend | Cat# 103035; RRID: AB 10639933 |
| Rat monoclonal APC anti-mouse CD62L | BioLegend | Cat# 104412; RRID: AB 313099 |
| Rat monoclonal PE anti-mouse CD4 | BioLegend | Cat# 100407; RRID: AB 312692 |
| Mouse monoclonal PE-Cy™7 anti-mouse NK-1.1 | BD Biosciences | Cat# 552878; RRID: AB 394507 |
| Mouse monoclonal BV650 anti-mouse NK-1.1 | BD Biosciences | Cat# 564143; RRID: AB 2738617 |
| Rat monoclonal EOMES (Dan11 mag), eFluor 450, eBioscience™ | Thermo Fisher Scientific | Cat# 48–4875-82; RRID: AB 2574062 |
| Rat monoclonal Alexa Fluor® 488 anti-mouse CD3 | BioLegend | Cat# 100212; RRID: AB 493530 |
| Rat monoclonal Alexa Fluor® 488 anti-mouse CD19 | BioLegend | Cat# 115524; RRID: AB 493339 |
| Rat monoclonal Alexa Fluor® 488 anti-mouse Ter119 | BioLegend | Cat# 116215; RRID: AB 493402 |
| Rat monoclonal Alexa Fluor® 488 anti-mouse Gr-1 | BioLegend | Cat# 108419; RRID: AB 493480 |
| Rat monoclonal Alexa Fluor® 488 anti-mouse CD11 b | BioLegend | Cat# 101219; RRID: AB 493545 |
| Hamster monoclonal Alexa Fluor® 488 anti-mouse CD11c | BioLegend | Cat# 117313; RRID: AB 492849 |
| Hamster monoclonal Alexa Fluor® 488 anti-mouse FceRIa | BioLegend | Cat# 134329; RRID: AB 2687238 |
| Mouse monoclonal BUV395 anti-mouse CD45.2 | BD Biosciences | Cat# 565448; RRID: AB 2738867 |
| Rat monoclonal BUV737 anti-mouse CD335 (NKp46) | BD Biosciences | Cat# 612805; RRID: AB 2870131 |
| Mouse monoclonal Alexa Fluor® 647 anti-T-bet | BD Biosciences | Cat# 561267; RRID: AB 10564093 |
| Armenian hamster monoclonal PE anti-mouse CD196 (CCR6) | BioLegend | Cat# 129804; RRID: AB 1279137 |
| Mouse monoclonal PE-CF594 anti-mouse RORYt | BD Biosciences | Cat# 562684; RRID: AB 2651150 |
| Rat monoclonal PE/Cyanine7 anti-mouse CD127 (IL-7Ra) | BioLegend | Cat# 135014; RRID: AB 1937265 |
| Rat monoclonal Brilliant Violet 421 ™ anti-mouse Ly-6C | BioLegend | Cat# 128031; RRID: AB 2562177 |
| Rat monoclonal anti-CD11 b mouse | BD Biosciences | Cat# 553310; RRID: AB 394774 |
| Rat monoclonal PerCP/Cyanine5.5 anti-mouse I-A/I-E | BioLegend | Cat# 107625; RRID: AB 2191072 |
| Rat monoclonal Alexa Fluor® 647 anti-mouse F4/80 | BioLegend | Cat# 123121; RRID: AB 893492 |
| Rat monoclonal PE anti-mouse Ly-6G | BioLegend | Cat# 127608; RRID: AB 1186099 |
| Armenian hamster monoclonal PE/Cyanine7 anti-mouse CD11c | BioLegend | Cat# 117318; RRID: AB 493568 |
| Rat monoclonal V450 anti-mouse Foxp3 | BD Biosciences | Cat# 561293; RRID: AB 10611728 |
| Rat monoclonal APC-R700 anti-mouse CD25 | BD Biosciences | Cat# 565134; RRID: AB 2744344 |
| Bacterial and virus strains | ||
| Stellar™ Competent Cells | Takara | Cat# 636766 |
| One Shot™ Stbl3™ Chemically Competent E. coli | ThermoFisher Scientific |
Cat# C737303 |
| BL21(DE3) Competent Cells | ThermoFisher Scientific |
Cat# EC0114 |
| Sendai virus (from Cytotune 2.0 kit) | ThermoFisher Scientific |
Cat# A16517 |
| Biological samples | ||
| Healthy human PBMCs | This study | NIH biood bank |
| Patient and family PBMCs (Pt 1) | This study | Marmara University School of Medicine |
| Patient and family PBMCs (Pt 2) | This study | Schneider Children’s Medical Center of Israel |
| Human Colon Total RNA | Zyagen | Cat# HR-311 |
| Human Small Intestine Total RNA | Zyagen | Cat# HR-306 |
| Human Spleen Paraffin Sections | Zyagen | Cat# HP-701 |
| Human Thymus Paraffin Sections | Zyagen | Cat# HP-702 |
| Human Lung Paraffin Sections | Zyagen | Cat# HP-601 |
| Human Colon Paraffin Sections | Zyagen | Cat# HP-311 |
| Human Salivary Gland Paraffin Sections | Zyagen | Cat# HP-317 |
| Human Lung Total Protein | zyagen | Cat# HT-601 |
| Human Rectum Total Protein | zyagen | Cat# HT-312 |
| Human Small Intestine Total Protein | zyagen | Cat# HT-306 |
| Human Spleen Total Protein | zyagen | Cat# HT-701 |
| Human Stomach Total Protein | zyagen | Cat# HT-302 |
| Human Liver Total Protein | zyagen | Cat# HT-314 |
| Human Brain Total Protein | zyagen | Cat# HT-201 |
| Human Esophagus Whole Tissue Lysate | novusbio | Cat# NB820–59214 |
| Human Kidney Total Protein | zyagen | Cat# HT-901 |
| Human Small Intestine, Duodenum Paraffin Sections | zyagen | Cat# HP-307 |
| Human Small Intestine, Ileum Paraffin Sections | zyagen | Cat# HP-309 |
| Human Esophagus Paraffin Sections | zyagen | Cat# HP-301 |
| Human Stomach Paraffin Sections | zyagen | Cat# HP-302 |
| Human Multi-tissue Tissue MicroArray | novusbio | Cat# NBP2–78067 |
| Chemicals, peptides, and recombinant proteins | ||
| HILIC amphion | Welch | Lot# 7301.26 |
| Sera-Mag SpeedBeads | GE Healthcare | Cat# 45152101010250 Cat# 65152105050250 |
| Corning® Matrigel® Growth Factor Reduced (GFR) Basement Membrane Matrix, LDEV-free, 10 mL | Corning Life Science | Cat# 354230 |
| CryoStor® CS10 Freeze Media | Biolifesolutions | Cat# 210374 |
| A 83–01 | Cayman | Cat# 9001799 |
| DBZ | Cayman | Cat# 14627 |
| STEMdiff™ Intestinal Organoid Growth Medium | Stemcell | Cat# 05145 |
| IntestiCult™ Organoid Growth Medium (Mouse) | Stemcell | Cat# 06005 |
| mEGF | PeproTech | Cat# 315–09 |
| mWnt3a | PeproTech | Cat# 315–20 |
| Essential 8™ Medium | ThermoFisher Scientific |
Cat# A1517001 |
| Opti-MEM™ I Reduced Serum Medium | ThermoFisher Scientific |
Cat# 31985062 |
| Guanidine hydrochloride | Sigma | Cat# G3272–25G |
| Iodoacetamide | Sigma | Cat# I1149–5G |
| Dextran Sulfate Sodium Salt | MPbio | Cat# SKU 0216011050 |
| BsmBI-v2 | NEB | Cat# R0739S |
| STEMdiff™ Intestinal Organoid Kit | Stemceii | Cat# 05140 |
| T4 Polynucleotide Kinase | NEB | Cat# M0201s |
| T4 DNA Ligase | NEB | Cat# M0202L |
| Human TruStain FcX™ (Fc Receptor Blocking Solution) | Bioiegend | Cat# 422301 |
| Protein Deglycosylation Mix II | NEB | Cat# P6044S |
| a2–3,6,8,9 Neuraminidase A | NEB | Cat# P0722S |
| PNGase F (Glycerol-free), Recombinant | NEB | Cat# P0709S |
| O-Glycosidase | NEB | Cat# P0733S |
| Recombinant Human ST6GALNAC1 Protein | R&D | Cat# 9154-GI-020 |
| Manganese(II) chloride | Sigma | Cat# 244589–10G |
| Asialofetuin from fetal calf serum | Sigma | Cat# A4781–50MG |
| CMP-Cy5-Sialic Acid | R&D | Cat# ES302–050 |
| Corning® Matrigel® Growth Factor Reduced (GFR) Basement Membrane Matrix, LDEV-free, 10 mL | Corning Life Science | Cat# 354230 |
| TrypLE™ Express Enzyme (1X), no phenol red | Thermo Fisher Scientific | Cat# 12604013 |
| BSA | Sigma | Cat# A7030 |
| EDTA | Thermo Fisher Scientific | Cat# AM9261 |
| StcE | From Dr. Bertozzi’s lab | N/A |
| OgpA | From Dr. Lawrence Tabak’s lab, NIDCR | N/A |
| polybrene | Sigma-Aldrich | Cat# TR-1003 |
| Blasticidin S HCl | Thermo Fisher Scientific | Cat# A1113903 |
| Puromycin | Thermo Fisher Scientific | Cat# A1113803 |
| SCF | R&D | Cat# 7466-SC |
| StemSpan™SFEM II medium | StemCell | Cat# 09605 |
| IL-3 | R&D | Cat# 203-IL |
| EPO | R&D | Cat# 287-TC |
| IGF-1 | R&D | Cat# 201-G1 |
| Dexamethasone | Sigma | Cat# D4902 |
| holo-transferrin | Sigma | Cat# I0665 |
| IWP2 | Sigma | Cat# I0536 |
| DAPT | Sigma | Cat# D5942 |
| Y27632 | Cayman | Cat# 10005583 |
| DAPI Fluoromount-G | SouthernBiotech | Cat# 0100–20 |
| GalNAcEXO | From Dr. Lawrence Tabak’s lab, NIDCR | N/A |
| Dithiothreitol | Sigma | Cat# 10197777001 |
| Trifluoroacetic acid (TFA) | Sigma | Cat# I6508–1L |
| Urea | Sigma | Cat# U0631–1 KG |
| Formic acid | Thermo Fisher Scientific | Cat# A117–50 |
| Trypsin | Promega | Cat# V5111 |
| Tris-(2-Carboxyethyl)phosphine, Hydrochloride (TCEP) | Thermo Fisher Scientific | Cat# I2556 |
| Acetonitrile (ACN) | Thermo Fisher Scientific | Cat# A9554 |
| Guanidine hydrochloride solution | Sigma | Cat# SRE0066– 100ML |
| carbenoxolone (CBX) | Selleckchem | Cat# S4368 |
| Ampicillin sodium salt | Sigma | Cat# A9518 |
| Metronidazole | Sigma | Cat# M3761 |
| Vancomycin hydrochloride from Streptomyces orientalis | Sigma | Cat# V2002 |
| Neomycin trisulfate salt hydrate | Sigma | Cat# N1876 |
| Liberase | Sigma | Cat# 5401020001 |
| DNase I | Sigma | Cat# 4716728001 |
| Collagenase D | Sigma | Cat# 11088858001 |
| phenol-chloroform-isoamyl alcohol (25:24:1) | Sigma | Cat# P2069–100ML |
| SCFA standards | Protein Characterization Core, NCI |
N/A |
| Percoll Plus | GE Healthcare | Cat# 17544501 |
| Monensin Solution | Biolegend | Cat# 420701 |
| EcoR I-HF | NEB | Cat# R3101L |
| MluI-HF | NEB | Cat# R3198S |
| Lipo3000 transfection reagent | ThermoFisher Scientific |
Cat# L3000075 |
| Sodium Pyruvate (100 mM) | ThermoFisher Scientific |
Cat# 11360070 |
| Percoll | Sigma | Cat# GE17–0891-01 |
| UEA 1 fluorescin | Vector Laboratories | Cat# FL-1061 |
| Cy5-Peanut agglutinin (PNA) | Vectorlabs | Cat# CL-1075–1 |
| Fixable Viability Dye eFluor™ 780 | Thermo Fisher Scientific | Cat# 65–0865-18 |
| β-mercaptoethanol | Gibco | Cat# 21985 |
| L-Glutamine | Gibco | Cat# 25030 |
| NEAA | Gibco | Cat# 11140 |
| Penicillin and streptomycin | Gibco | Cat# 15140 |
| Critical commercial assays | ||
| TMT10plex™ Isobaric Labeling Reagent Set | ThermoFisher Scientific |
Cat# 90110 |
| Pierce BCA Protein Assay Kit | ThermoFisher Scientific |
Cat# 23225 |
| XBridge Peptide BEH C18 Column, 300Ä, 3.5 μm, 1 mm × 150 mm | Waters | Cat# SKU: 186003606 |
| Histone Deacetylase (HDAC) Activity Assay Kit (Fluorometric) (ab156064) | Abcam | Cat# ab156064 |
| Human Pan T cell isolation kit | Miltenyi | Cat# 130–096-535 |
| Dynabeads™ Human T-Activator CD3/CD28 for T Cell Expansion and Activation | Thermo Fisher Scientific | Cat#11131D |
| Dynabeads™ Protein G for Immunoprecipitation | ThermoFisher Scientific |
Cat# 10004D |
| Lenti-X™ Concentrator | Takara | Cat# 631231 |
| PrimeScript™ RT Reagent Kit (Perfect Real Time) | Takara | Cat# RR037A |
| In-Fusion® HD Cloning Kit | Takara | Cat# 639650 |
| Lipofectamine™ 3000 Transfection Reagent | ThermoFisher Scientific |
Cat# L3000075 |
| NuPAGE™ 4–12% Bis-Tris Protein Gels, 1.5 mm, 15-well | ThermoFisher Scientific |
Cat# NP0336B0X |
| NuPAGE™ 3 to 8%, Tris-Acetate, 1.5 mm, Mini Protein Gel, 15-well | ThermoFisher Scientific |
Cat# EA03785BOX |
| Foxp3 / Transcription Factor Staining Buffer Set | Thermo Fisher Scientific, | Cat# 00–5523-00 |
| BCA protein assay kit | Thermo Fisher Scientific, | Cat# 23225 |
| Sialyltransferase Activity Kit | R&D | Cat# EA002 |
| In-Fusion® HD Cloning Plus CE kit | Iakara | Cat# 638919 |
| Anti-Adherence Rinsing solution | Stemceii | Cat# 07010 |
| ATAC-Seq Kit | Active motif | Cat# 53150 |
| LabSafe GEL Blue | G-biosciences | Cat# 786–35 |
| C18 cartridge | Seq-Pak C18 | Cat# WAI054955 |
| Sera-Mag SpeedBeads with a hydrophilic surface | GE Healthcare | Cat# 45152101010250 |
| Sera-Mag SpeedBeads with a hydrophobic surface | GE Healthcare | Cat# 65152105050250 |
| Halt™ Protease Inhibitor Cocktail, EDTA-Free | Thermo Fisher Scientific | Cat# 78439 |
| Halt™ PhosphataseInhibitor Cocktail | Thermo Fisher Scientific | Cat# 78427 |
| EndoTrap red | Biovendor | Cat# LEI0001 |
| ToxinSensor™ Chromogenic LAL Endotoxin Assay Kit | Genescript | Cat# L00350 |
| DNeasy Blood & Tissue Kit | Qiagen | Cat# 69506 |
| Cytotune 2.0 kit | ThermoFisher Scientific |
Cat# A16517 |
| Deposited data | ||
| Site-specific N-glycoproteome MS raw and searched data | This paper | https://www.ebi.ac.uk/pride/archiveidentifierPXD028239 |
| mRNA-seq and ATAC-seq data | This paper | GEO ID: GSE183821 |
| whole exome sequencing (WES) (Pt 1) | This paper | dbGaP ID: phs002598.v1.p1 |
| Experimental models: Celi lines | ||
| HI-29 | AICC | Cat# HTB-38 |
| LS 180 | AICC | Cat# CL-187 |
| Lenti-X™ 293T Cells | Iakara | Cat# 632180 |
| HEK293I | AICC | Cat# CRL-3216 |
| Experimental models: Organisms/strains | ||
| Mouse: Myd88flox/flox | The Jackson Laboratory | Cat# 008888 |
| Mouse: Vil1cre | The Jackson Laboratory | Cat# 21504 |
| Mouse: Tlr4−/− | The Jackson Laboratory | Cat# 29015 |
| Mouse: St6 (St6galnac1R319Q) | This paper | N/A |
| Oligonucleotides | ||
| A full list is provided in Table S3 | ||
| Recombinant DNA | ||
| pLV-EFIa-IRES-Puro | Hayer et al., 2016 | Addgene, Cat# 85132 |
| psPAX2 | A gift from Didier Trono | Addgene, Cat# 12260 |
| pMD2.G | A gift from Didier Trono | Addgene, Cat# 12259 |
| lentiGuide-Puro | Sanjana et al., 2014 | Addgene, Cat# 52963 |
| lentiCas9-Blast | Sanjana et al., 2014 | Addgene, Cat# 52962 |
| lentiCRISPRv2 | Sanjana et al., 2014 | Addgene, Cat# 52961 |
| pLV-EF1a-2a-tEGFR | This paper | N/A |
| Software and algorithms | ||
| ImageJ | Schneider et al., 2012 | https://imagej.nih.gov/ij/ |
| Graphpad Prism | https://www.graphpad.com/scientific-software/prism | version 9.0 |
| QIIME2 | Bolyen et al., 2019 | version 2019.10 |
| DADA2 method | Callahan et al., 2016 | https://benjjneb.github.io/dada2/ |
| Thermo Xcalibur software | https://www.thermofisher.com/order/catalog/product/0PT0N-30965 | Cat# 0PT0N-30965 |
| HOMER | http://homer.ucsd.edu/homer/interactions/ | version 4.11.1 |
| Python | https://www.python.org/ | Version 3.6–3.8 |
| Macs | https://pypi.org/project/MACS/ | version 1.4.3 |
| Bedtools | https://bedtools.readthedocs.io/en/latest/ | version 2.30.0 |
| HTSeq | https://pypi.org/project/HTSeq/ | version 0.11.4 |
| DESeq2 | version 1.30.1 | |
| RStudio | https://www.rstudio.com/ | version 1.4.1106 |
| R | https://www.r-project.org | version 4.0.4 |
| RColorBrewer | http://colorbrewer2.org/ | version 1.1–2 |
| Samtools | http://www.htslib.org/ | version 1.12 |
| Deeptools | https://deeptools.readthedocs.io/en/develop/ | version 3.5.0 |
| Enrichr | Kuleshov at el., 2016 | https://maayanlab.cloud/Enrichr/ |
| FastQC | https://www.bioinformatics.babraham.ac.uk | version 0.11.9 |
| Bowtie | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml | version 1.3.0 |
| IGV | https://software.broadinstitute.org/software/igv/ | version 2.10.0 |
| Benchling algorithm | https://www.benchling.com/crispr/ | https://www.benchling.com |
| PyMOL | https://pymol.org/2/ | version 2.3.1 |
| I-TASSER | Roy et al., 2010 | https://zhanggroup.org/I-TASSER/ |
| pFind3.0 | Chi et al., 2018 | https://github.com/pFindStudio/pFind3 |
| FlowJo | https://www.flowjo.com/ | version 10.6.2 |
| GlycoBinder | Fang et al., 2020 | https://github.com/IvanSilbern/GlycoBinder |
| IPA | https://digitalinsights.qiagen.com/products-overview/discovery-insights-portfolio/analysis-and-visualization/qiagen-ipa/ | version 01–20-04 |
| Imaris | https://imaris.oxinst.com/ | version 9.8.1 |
| Snapgene | https://www.snapgene.com/ | version 6.0 |
| Cytoscape | https://cytoscape.org/ | version 3.8.2 |
| Other | ||
| Agilent 1100 series HPLC | Agilent Technologies | https://www.agilent.com/en/products/liquid-chromatography |
| Orbitrap Fusion™ Lumos™ Tribrid™ Mass Spectrometer | ThermoFisher Scientific |
https://www.thermofisher.com/order/catalog/product/IQLAAEGAAPFADBMBHQ |
| Fluorescent imager Azure 500 | Azure biosystems | https://www.azurebiosystems.com/imaging-systems/azure-500/ |