Abstract
There has been growing interest in using peptides for the controlled synthesis of nanomaterials. Peptides play a crucial role not only in regulating the nanostructure formation process but also in influencing the resulting properties of the nanomaterials. Leveraging machine learning (ML) in the biomimetic workflow is anticipated to accelerate peptide discovery, make the process more resource-efficient, and unravel associations among attributes that may be useful in peptide design. In this study, a binary ML classifier is formulated that was trained and tested on 1720 peptide examples. The support vector machine classifier uses Kidera factors to categorize peptides into one of two groups based on their binding ability. The classifier exhibits satisfactory performance, as demonstrated by various performance metrics. In addition, key variables that bear a huge impact on the model were identified, such as peptide hydrophobicity. As these trends were derived from a large and diverse dataset, the insights drawn from the data are expected to be generalizable and robust. Thus, the presented ML model is an important step toward the rational and predictive peptide design.
Introduction
Biomimetic synthesis of nanoparticles using peptides is a promising technique of creating highly functional inorganic nanomaterials.1 Through this method, nanostructures that exhibit unique morphologies,2 enhanced properties,3 and controlled composition4 have been created. This method relies on the biomimetic peptide (BMPep) to regulate the nanostructure formation process. The BMPep is able to regulate the growth of the nanomaterial through a capping mechanism, wherein the peptide binds to the surface of the growing nanomaterial, thereby influencing the growth direction.5 Apart from influencing the morphology of the produced nanomaterials, the adsorbed peptide on the nanomaterial surface also impacts the material physicochemical properties, such as catalytic activity.6 As the BMPep plays a central role in the nanostructure formation process, discovering and developing more BMPep is expected to further expand the biomimetic toolkit, which can widen the scope and relevance of this method. A diverse and rich collection of BMPeps will give researchers abundant options to tailor and fine-tune the synthesis process to meet specific requirements or conditions. However, peptide discovery remains to be a bottleneck as currently employed methods are expensive, tedious, and technically demanding, such as the phage display assay.
The integration of machine learning (ML) to biomimetics is a promising approach to address the problems associated with BMPep discovery. ML can minimize trial-and-error by quickly identifying potential sequences that can be used for the biomimetic nanomaterial synthesis. Previous works have reported the creation of ML models that aim to assist the peptide screening process by predicting the peptide binding affinity toward a particular substrate,7 classifying a given sequence if it is a strong binder or not,8,9 and the creation of novel algorithms to predict the peptide binding phenomenon.10 However, these studies have used relatively small datasets, wherein the number of peptides used for training and testing the algorithm is less than 50. Considering that the predictive success of such ML models relies on the quality and quantity of the data used for training,11,12 utilizing a larger dataset for training may lead to the creation of ML models that are more robust and the observed trends more generalizable. In this study, a binary classification model that was trained and tested on more than 1500 peptide sequences is reported. The classification model categorizes peptides based on their ability to bind to gold nanoparticles, which may accelerate the BMPep discovery process and also unravel nonobvious associations that may aid in the rational design of BMPep sequences.
Methodology
The 1720 peptide sequences which are all composed of 10 amino acids and synthesized through solid-phase synthesis employing spot technology and their corresponding intensity values used in this study were obtained from the study of Tanaka et al.13 The paper reported the screening results of peptide-binding assays with gold nanoparticles, as represented by colorimetric intensities. Peptides that exhibited strong binding capability with gold nanoparticles yielded high intensity values. In the categorization of peptides for the dataset to be used for formulating a ML classification model, peptides that reported intensity values greater than 207,500 were designated into class A and others into class B. This threshold value was selected on the basis of the calculated median value for the dataset.
The chemical descriptors for each peptide were calculated using the peptide R package.14 The calculated peptide descriptors were the Blosum indices,15 Cruciani properties,16 factor analysis scale of generalized amino acid information (FASGAI) vectors,17 Kidera factors,18 ProtFP,19 ST-scales,20 T-scales,21 VHSE scales,22 and Z-scales.23 The resulting dataset was then used for the formulation of classification models using the following ML algorithms: generalized linear models in the form of logistic regression (LR), k-nearest neighbor, classification and regression trees, support vector machine (SVM) using the radial basis function, and linear and polynomial kernels. In all classification model formulations, 75% of the dataset was devoted for training, and the remaining 25% served as the test set. A 10-fold cross-validation was likewise conducted for all models during training. The “class A” status was designated as the positive class for the confusion matrix. The default settings of each algorithm which led to the best performance were automatically selected during screening, followed by hyperparameter tuning during model optimization which likewise employed 10-fold cross-validation. The R package caret was used for the creation of the classification models,24 and the IML package was used for feature selection and optimization.25 The feature selection method employed in the IML package involves the stepwise evaluation of the impact of removing a variable in the classification performance. All R packages and their dependents used in the study were executed in R version 4.1.026 running in a MacOS environment. The dataset that consisted of 1720 peptide sequences, color intensity values, and class designation (Table S1), together with the code used in this study, are available in the Supporting Information.
Results and Discussion
This study attempts to overcome the limitations of previous ML models that were applied to metal-binding peptides by utilizing a larger dataset. This aspect is already challenging due to the limited availability and diversity of data. Most papers only report a few BMPeps and their binding affinities or binding constants, wherein a good number of these reported BMPeps are strong binders. However, building ML models also requires the inclusion of weak or nonbinders in order for the model to learn the discriminating attributes between the two classes. The search for more data was widened by considering other types of variables that indirectly relate to peptide binding on to a substrate. Thus, the classification model was built on a dataset that utilized a nonstandard response variable related to nanoparticle binding in the form of color intensity. The first step in creating the binary classifier is identifying the appropriate descriptor and algorithm. This was achieved through the pairwise comparison of the classification accuracy and kappa values for each descriptor–algorithm pair (Figures 1 and 2). Accuracy refers to the proportion of correct classification made over the total number of cases to be classified. Kappa, on the other hand, is a measure of agreement between the predicted and actual classification outcomes. Values for kappa range between −1 to +1, wherein a value closer to +1 is desired. As the heatmaps show, the pair of Kidera factors (KF) and support vector machine using a radial basis function kernel (SVM-R) was the best combination. KF is a class of descriptors specific for peptides and proteins as they are derived from the multivariate analysis of 188 physical properties of the 20 amino acids.18 Following the application of dimension reduction techniques, 10 KFs are obtained, wherein each KF carries more weight on a particular property of the peptide.
Figure 1.
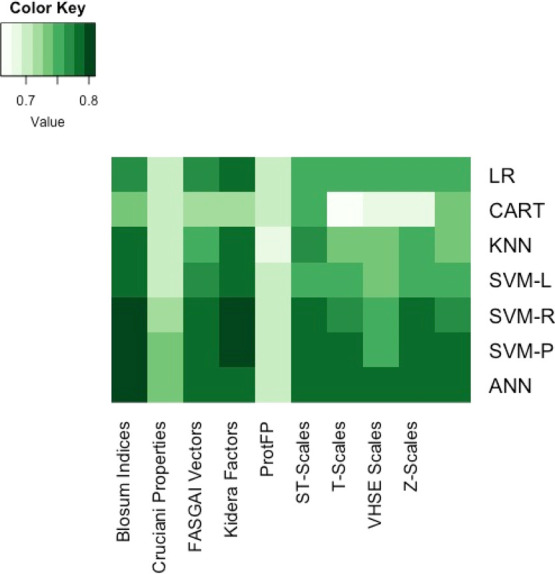
Training performance represented by classification accuracy. Accuracy scores were obtained from training based on n = 1291, followed by 10-fold cross-validation. The darker the color, the higher is the accuracy of the model.
Figure 2.
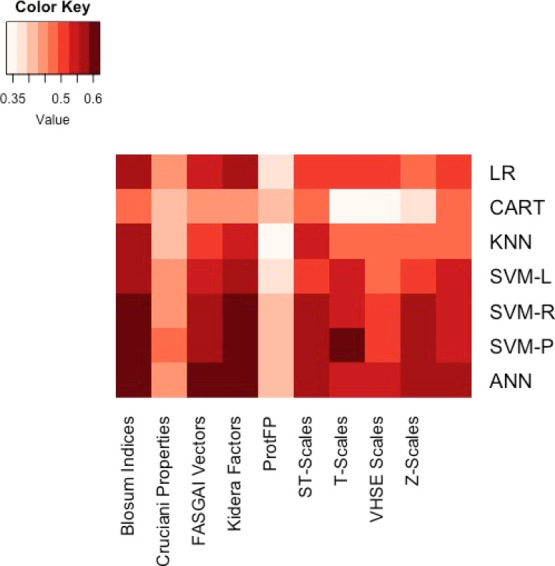
Training performance represented by kappa values. Kappa scores were obtained from training based on n = 1291, followed by 10-fold cross-validation. The darker the color, the higher is the kappa score of the model.
Following the identification of the optimum descriptor–algorithm pair to be used, variable selection was then carried out in order to improve the parsimony of the model. Table 1 shows that KF4 which relates to peptide hydrophobicity had the greatest impact on the model, while KF1 and KF8 which are both related to the peptide helical structure had the least importance. Hence, the classification model was optimized by removing KF1 and KF8, followed by hyperparameter tuning (Figure 3).
Table 1. Variable Importance Scoresa.
| Variable | importance score |
|---|---|
| KF4 (hydrophobicity) | 1.416 |
| KF2 (side chain size) | 1.279 |
| KF3 (extended structure preference) | 1.274 |
| KF9 (pK-C) | 1.252 |
| KF7 (flat extended preference) | 1.102 |
| KF5 (double-bend preference) | 1.084 |
| KF10 (surrounding hydrophobicity) | 1.080 |
| KF6 (partial specific volume) | 1.070 |
| KF8 (occurrence in alpha region) | 1.056 |
| KF1 (helix/bend preference) | 1.027 |
The higher the score, the greater is the impact of the specific variable on the classification model. The importance score is derived from the classification error as a consequence of removing a specific variable.
Figure 3.
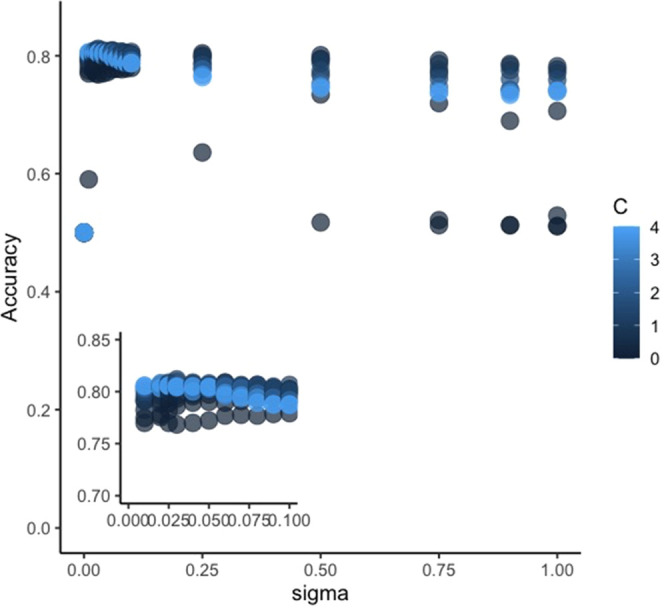
Hyperparameter tuning where the training classification accuracy is monitored while the sigma and C values are varied. Inset graph shows the lower sigma regions where the highest classification accuracy was achieved.
The performance of the final, optimized model that consisted of 8 KF as the classification variables, employing SVM-R as the algorithm using C = 1 and sigma = 0.108 as the hyperparameters, was then evaluated. The test performance is summarized in Table 2 which shows the confusion matrix together with various performance metrics. The nearly identical accuracy scores obtained during the training and testing phases suggest that the model does not exhibit overfitting. Apart from the reported 80.2% classification accuracy for the presented model, 0.604 kappa can be interpreted as a moderate agreement between the predicted and actual outcomes.27 The classifier can also be considered as unbiased as the p-value for the McNemar test is greater than 0.05. This indicates that the proportion of misclassification is statistically the same for each class. Collectively, the performance metrics suggest that the classification model exhibits adequate and satisfactory capability to categorize peptides based on their sequence into either class.
Table 2. Performance of the Optimized Model (SVM with a RBF Kernel, C = 1, and Sigma = 0.108) on the Test Set, as Demonstrated by the Confusion Matrixa.
| class A | class B | |
|---|---|---|
| class A | 182 | 52 |
| class B | 33 | 162 |
Other performance metrics that are derived from the confusion matrix include: accuracy = 0.802, F1 = 0.811, recall = 0.847, sensitivity = 0.847, specificity = 0.757, precision = 0.778, and kappa = 0.604.
External validation is an important component of model building as it tests the optimized model on data that were not part of the training and test phases. For the external validation to be effective, the independent data to be used should come from the same distribution as the training and test data or approximately report the same outcome. This therefore becomes a challenge for the present model due to several factors. First, the definition of a “strong binder” for gold-binding peptides varies from study to study. For example, Du et al.9 used the threshold value of −25 kJ/mol for the binding free energy to differentiate strong from weak binders. In the present study, such categorization was based on the median color intensity values of the dataset. Second, the present ML classifier was developed on a library of 10 mer peptides. Ideally, the peptides to be included in external validation should be of the same length. Aware of these constraints and challenges, the optimized model was subjected to external validation, with the data obtained from different papers,9,28,29 with each paper having different criteria on categorizing strong and weak binders. The external validation dataset is composed of 37 peptides of varying length and is available in the Supporting Information (Table S2). The performance of the optimized model on the external validation is presented and summarized in Table 3. The overall performance of the optimized model on the external validation was lower, as expected. Predictive models tend to perform more poorly on external validation compared to the training and testing sets.30 Taking into consideration the aforementioned limitations and challenges on identifying suitable peptides for external validation, the performance of the optimized model demonstrates its practical utility in BMPep discovery and in the overall biomimetic workflow. The high specificity and precision of the model are valuable in weeding out low-binding peptide sequences as a highly specific model has a low false-positive rate. This is important as the model can conserve time and resources. The excellent ability of the model to reject false positives is also an indication that the model does not overfit.31
Table 3. External Validation of the Optimized Model (SVM with a RBF Kernel, C = 1, and Sigma = 0.108) As Demonstrated by the Confusion Matrixa.
| class A | class B | |
|---|---|---|
| class A | 7 | 1 |
| class B | 9 | 20 |
The dataset used in the external validation is available in the Supporting Information (Table S2). Other performance metrics that are derived from the confusion matrix include: accuracy = 0.73, F1 = 0.583, recall = 0.438, sensitivity = 0.438, specificity = 0.952, precision = 0.875, kappa = 0.415.
Apart from the prospective utility of the classifier for screening BMPeps, the model also has identified variables that have a huge impact in predicting the binding capabilities of peptides. This is fundamentally important and is an incremental advance toward a deeper understanding of the binding phenomenon which may contribute to efforts related to the rational peptide design. Based on current understanding on how BMPeps bind to their substrate, the process is believed to occur through adsorption. Specifically, the peptide-binding process involves expulsion of water molecules that surround the peptide as it approaches the surface. Once bound, the peptide undergoes conformational changes and assembly rearrangement in order to achieve stability.32 Ultimately, these steps in the binding mechanism are influenced by the peptide sequence, and it is envisioned that the presented ML model will be able to capture sequence-based information that are relevant in making the prediction. The identified variables in this study are those related to peptide hydrophobicity (KF4), which was deemed as the most important, followed by extended structure preference (KF3), side chain size (KF2), and pK-C (KF9). The findings presented herein corroborate and reinforce past findings relevant to peptide binding on to gold surface but on a much larger scale as the ML classifier was built on over 1500 peptide examples. The peptide property of hydrophobicity is a known parameter to influence biomolecular interactions. For BMPeps in particular, it was found through experiments and simulations that the binding strength is a delicate balance between interactions with the gold surface and the aqueous environment.33 In addition, interplay between hydrophobicity and peptide conformation exists, and both factors are known to be determinants of how peptides interact with surfaces.32 Previous studies have also identified that the conformation and protonation state of the BMPep influence surface binding.34 In addition, multiple studies have identified that peptide conformation is an important factor that governs surface binding.3,29,32,35−38 The variables identified by the model are all relevant to peptide conformation, an indication that the model is logical and plausible. Finally, the identified variables are also coherent with the phases within the binding mechanism of gold-binding peptides,32 such as the removal of the peptide hydration layer prior to binding (KF4—hydrophobicity and KF9—pKC), and conformation changes and assembly reorganization (KF4—hydrophobicity; KF3—extended structure preference; and KF2—side chain size). Thus, the presented ML model has formalized and generalized the associations of these variables on their role in peptide binding to gold surfaces on a larger scale. These information are valuable for designing surface-binding peptides.
A limitation of the presented model which may curb its application and deployment is the utilization of a nonstandard variable that is associated with peptide binding on to the nanoparticle as the basis for classification. Another limitation is related to the utilization of SVM, which generates a black box model. This limitation is partially addressed by determining variable importance, but the direct association of these variables in the classification process is not clearly known. Despite these limitations, this work is a positive contribution for the wider integration of ML to biomimetic nanomaterial synthesis.
Conclusions
A ML model that was trained and tested on 1720 peptides is presented, wherein the model can categorize peptides into one of the two groups based on peptide binding, as represented by color intensity. The ML model was built using SVM with a radial basis function kernel and Kidera factors as the variables. The ML classifier exhibited satisfactory classification performance, as demonstrated by a test accuracy of 80.2%, among other reported performance metrics. Thus, the formulated model is an enabling tool for the accelerated and more resource-efficient discovery of BMPep.
The ML classifier has also shed light on the significant variables related to BMPep interactions with gold surfaces, wherein it was found that peptide hydrophobicity has the greatest impact on the classification, together with variables related to the structure, side chain, and protonation state of the peptide. As these trends were derived from a large and diverse dataset composed of 1720 peptides, the insights drawn from the data are expected to be generalizable and robust. This is an important step toward the rational and predictive peptide design. Future work should focus on the exploration of other peptide descriptors, conducting peptide motif analysis, and the utilization of interpretable ML methods in order to create a highly generalizable model that can be fully integrated in the materials creation workflow.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.2c00640.
Dataset used for training, testing, and external validation together with the R code to run the model (PDF)
The author declares no competing financial interest.
Supplementary Material
References
- Janairo J. I. B.Peptide-Mediated Biomineralization; Springer: Singapore, 2016. [Google Scholar]
- Mokashi-Punekar S.; Brooks S. C.; Hogan C. D.; Rosi N. L. Leveraging Peptide Sequence Modification to Promote Assembly of Chiral Helical Gold Nanoparticle Superstructures. Biochemistry 2021, 60, 1044–1049. 10.1021/acs.biochem.0c00361. [DOI] [PubMed] [Google Scholar]
- Janairo J. I. B.; Sakaguchi T.; Hara K.; Fukuoka A.; Sakaguchi K. Effects of Biomineralization Peptide Topology on the Structure and Catalytic Activity of Pd Nanomaterials. Chem. Commun. 2014, 50, 9259–9262. 10.1039/c4cc04350b. [DOI] [PubMed] [Google Scholar]
- Ozaki M.; Imai T.; Tsuruoka T.; Sakashita S.; Tomizaki K. ya; Usui K. Elemental Composition Control of Gold-Titania Nanocomposites by Site-Specific Mineralization Using Artificial Peptides and DNA. Chem. Commun. 2021, 4, 1. 10.1038/s42004-020-00440-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppage R.; Slocik J. M.; Briggs B. D.; Frenkel A. I.; Heinz H.; Naik R. R.; Knecht M. R. Crystallographic Recognition Controls Peptide Binding for Bio-Based Nanomaterials. J. Am. Chem. Soc. 2011, 133, 12346–12349. 10.1021/ja203726n. [DOI] [PubMed] [Google Scholar]
- Li Y.; Tang Z.; Prasad P. N.; Knecht M. R.; Swihart M. T. Peptide-Mediated Synthesis of Gold Nanoparticles: Effects of Peptide Sequence and Nature of Binding on Physicochemical Properties. Nanoscale 2014, 6, 3165–3172. 10.1039/c3nr06201e. [DOI] [PubMed] [Google Scholar]
- Janairo J. I. B. Predictive Analytics for Biomineralization Peptide Binding Affinity. Bionanoscience 2019, 9, 74–78. 10.1007/s12668-018-0578-4. [DOI] [Google Scholar]
- Oren E. E.; Tamerler C.; Sahin D.; Hnilova M.; Seker U. O. S.; Sarikaya M.; Samudrala R. A Novel Knowledge-Based Approach to Design Inorganic-Binding Peptides. Bioinformatics 2007, 23, 2816–2822. 10.1093/bioinformatics/btm436. [DOI] [PubMed] [Google Scholar]
- Du N.; Knecht M. R.; Swihart M. T.; Tang Z.; Walsh T. R.; Zhang A. Identifying Affinity Classes of Inorganic Materials Binding Sequences via a Graph-Based Model. IEEE/ACM Trans. Comput. Biol. Bioinf. 2015, 12, 193–204. 10.1109/tcbb.2014.2321158. [DOI] [PubMed] [Google Scholar]
- Janairo J. I. B.; Aviso K. B.; Promentilla M. A. B.; Tan R. R. Enhanced Hyperbox Classifier Model for Nanomaterial Discovery. Ai 2020, 1, 299–311. 10.3390/ai1020020. [DOI] [Google Scholar]
- Hughes Z. E.; Nguyen M. A.; Wang J.; Liu Y.; Swihart M. T.; Poloczek M.; Frazier P. I.; Knecht M. R.; Walsh T. R. Tuning Materials-Binding Peptide Sequences toward Gold- and Silver-Binding Selectivity with Bayesian Optimization. ACS Nano 2021, 15, 18260–18269. 10.1021/acsnano.1c07298. [DOI] [PubMed] [Google Scholar]
- Manavalan B.; Basith S.; Lee G. Comparative Analysis of Machine Learning-Based Approaches for Identifying Therapeutic Peptides Targeting SARS-CoV-2. Briefings Bioinf. 2021, 23, bbab412. 10.1093/bib/bbab412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M.; Hikiba S.; Yamashita K.; Muto M.; Okochi M. Array-Based Functional Peptide Screening and Characterization of Gold Nanoparticle Synthesis. Acta Biomater. 2017, 49, 495–506. 10.1016/j.actbio.2016.11.037. [DOI] [PubMed] [Google Scholar]
- Osorio D.; Rondón-Villarreal P.; Torres R. Peptides: A Package for Data Mining of Antimicrobial Peptides. R J. 2015, 7, 4–14. 10.32614/rj-2015-001. [DOI] [Google Scholar]
- Georgiev A. G. Interpretable Numerical Descriptors of Amino Acid Space. J. Comput. Biol. 2009, 16, 703–723. 10.1089/cmb.2008.0173. [DOI] [PubMed] [Google Scholar]
- Cruciani G.; Baroni M.; Carosati E.; Clementi M.; Valigi R.; Clementi S. Peptide Studies by Means of Principal Properties of Amino Acids Derived from MIF Descriptors. J. Chemom. 2004, 18, 146–155. 10.1002/cem.856. [DOI] [Google Scholar]
- Liang G.; Li Z. Factor Analysis Scale of Generalized Amino Acid Information as the Source of a New Set of Descriptors for Elucidating the Structure and Activity Relationships of Cationic Antimicrobial Peptides. QSAR Comb. Sci. 2007, 26, 754–763. 10.1002/qsar.200630145. [DOI] [Google Scholar]
- Kidera A.; Konishi Y.; Oka M.; Ooi T.; Scheraga H. a. Statistical Analysis of the Physical Properties of the 20 Naturally Occurring Amino Acids. J. Protein Chem. 1985, 4, 23–55. 10.1007/bf01025492. [DOI] [Google Scholar]
- van Westen G. J.; Swier R. F.; Wegner J. K.; IJzerman A. P.; van Vlijmen H. W.; Bender A. Benchmarking of Protein Descriptor Sets in Proteochemometric Modeling (Part 1): Comparative Study of 13 Amino Acid Descriptor Sets. J. Cheminf. 2013, 5, 41. 10.1186/1758-2946-5-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L.; Shu M.; Ma K.; Mei H.; Jiang Y.; Li Z. ST-Scale as a Novel Amino Acid Descriptor and Its Application in QSAM of Peptides and Analogues. Amino Acids 2010, 38, 805–816. 10.1007/s00726-009-0287-y. [DOI] [PubMed] [Google Scholar]
- Tian F.; Zhou P.; Li Z. T-Scale as a Novel Vector of Topological Descriptors for Amino Acids and Its Application in QSARs of Peptides. J. Mol. Struct. 2007, 830, 106–115. 10.1016/j.molstruc.2006.07.004. [DOI] [Google Scholar]
- Mei H.; Liao Z. H.; Zhou Y.; Li S. Z. A New Set of Amino Acid Descriptors and Its Application in Peptide QSARs. Biopolym. - Pept. Sci. Sect. 2005, 80, 775–786. 10.1002/bip.20296. [DOI] [PubMed] [Google Scholar]
- Sandberg M.; Eriksson L.; Jonsson J.; Sjöström M.; Wold S. New Chemical Descriptors Relevant for the Design of Biologically Active Peptides. A Multivariate Characterization of 87 Amino Acids. J. Med. Chem. 2002, 41, 2481–2491. 10.1021/jm9700575. [DOI] [PubMed] [Google Scholar]
- Kuhn M.; Wing J.; Weston S.; Williams A.; Keefer C.; Engelhardt A.; Cooper T.; Mayer Z.; Kenkel B.; Benesty M.; Lescarbeau R.; Ziem A.; Scrucca L.; Tang Y.; Candan C.; Hunt T.. Caret: Classification and Regression Training; 2018.
- Molnar C.; Casalicchio G.; Bischl B. Iml: An R Package for Interpretable Machine Learning. J. Open Source Softw 2018, 3, 786. 10.21105/joss.00786. [DOI] [Google Scholar]
- R Core Team . R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021.
- McHugh M. L. Interrater Reliability: The Kappa Statistic. Biochem. Med. 2012, 22, 276–282. 10.11613/bm.2012.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison E.; Hamilton J. W. J.; Macias-Montero M.; Dixon D. Peptide Functionalized Gold Nanoparticles: The Influence of PH on Binding Efficiency. Nanotechnology 2017, 28, 295602. 10.1088/1361-6528/aa77ac. [DOI] [PubMed] [Google Scholar]
- Lee D. J.; Park H. S.; Koo K.; Lee J. Y.; Nam Y. S.; Lee W.; Yang M. Y. Gold Binding Peptide Identified from Microfluidic Biopanning: An Experimental and Molecular Dynamics Study. Langmuir 2019, 35, 522–528. 10.1021/acs.langmuir.8b02563. [DOI] [PubMed] [Google Scholar]
- Ramspek C. L.; Jager K. J.; Dekker F. W.; Zoccali C.; van Diepen M. External Validation of Prognostic Models: What, Why, How, When and Where?. Clin. Kidney J. 2021, 14, 49–58. 10.1093/ckj/sfaa188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett R.; Jiang S.; White A. D. Classifying Antimicrobial and Multifunctional Peptides with Bayesian Network Models. J. Pept. Sci. 2018, 110, e24079 10.1002/pep2.24079. [DOI] [Google Scholar]
- Seker U. O. S.; Wilson B.; Kulp J. L.; Evans J. S.; Tamerler C.; Sarikaya M. Thermodynamics of Engineered Gold Binding Peptides: Establishing the Structure-Activity Relationships. Biomacromolecules 2014, 15, 2369–2377. 10.1021/bm4019006. [DOI] [PubMed] [Google Scholar]
- Cannon D. A.; Ashkenasy N.; Tuttle T. Influence of Solvent in Controlling Peptide-Surface Interactions. J. Phys. Chem. Lett. 2015, 6, 3944–3949. 10.1021/acs.jpclett.5b01733. [DOI] [PubMed] [Google Scholar]
- Hughes Z. E.; Nguyen M. A.; Li Y.; Swihart M. T.; Walsh T. R.; Knecht M. R. Elucidating the Influence of Materials-Binding Peptide Sequence on Au Surface Interactions and Colloidal Stability of Au Nanoparticles. Nanoscale 2017, 9, 421–432. 10.1039/c6nr07890g. [DOI] [PubMed] [Google Scholar]
- Seker U. O. S.; Wilson B.; Dincer S.; Kim I. W.; Oren E. E.; Evans J. S.; Tamerler C.; Sarikaya M. Adsorption Behavior of Linear and Cyclic Genetically Engineered Platinum Binding Peptides. Langmuir 2007, 23, 7895–7900. 10.1021/la700446g. [DOI] [PubMed] [Google Scholar]
- Hnilova M.; Oren E. E.; Seker U. O. S.; Wilson B. R.; Collino S.; Evans J. S.; Tamerler C.; Sarikaya M. Effect of Molecular Conformations on the Adsorption Behavior of Gold-Binding Peptides. Langmuir 2008, 24, 12440–12445. 10.1021/la801468c. [DOI] [PubMed] [Google Scholar]
- Choi N.; Tan L.; Jang J.-r.; Um Y. M.; Yoo P. J.; Choe W.-S. The Interplay of Peptide Sequence and Local Structure in TiO2 Biomineralization. J. Inorg. Biochem. 2012, 115, 20–27. 10.1016/j.jinorgbio.2012.05.011. [DOI] [PubMed] [Google Scholar]
- Sakaguchi T.; Janairo J. I. B.; Lussier-Price M.; Wada J.; Omichinski J. G.; Sakaguchi K. Oligomerization Enhances the Binding Affinity of a Silver Biomineralization Peptide and Catalyzes Nanostructure Formation. Sci. Rep. 2017, 7, 1400. 10.1038/s41598-017-01442-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


