Abstract
BMS-284756 (T-3811ME), a novel des-F(6) quinolone, was tested in the supercoiling inhibition and cleavable complex assays against Escherichia coli DNA gyrase, a target of quinolones. The results suggest that BMS-284756 has the same mechanism of action against DNA gyrase as other quinolones and a similar level of potency.
Quinolones are antibacterial agents known to target DNA gyrase and topoisomerase IV in bacteria (3). For most quinolones, DNA gyrase is the primary target in gram-negative organisms and topoisomerase IV is the primary target in gram-positive organisms (6). Exceptions to this pattern are seen (e.g., sparfloxacin and clinafloxacin), indicating that the primary target of quinolones depends on drug structure (6). DNA gyrase, a member of the type II topoisomerase family, is involved in the control of DNA topology in bacterial cells (2, 13). It consists of two DNA gyrase subunits, A (GyrA) and B (GyrB), which form an A2B2 tetramer (9). GyrA comprises the domains for DNA wrapping and the breakage-reunion activity, while GyrB possesses the ATPase activity (2, 3, 9, 13). The holoenzyme introduces a transient double-stranded break and permits passage of another double helix through the gap (2, 3, 9, 13). Quinolones inhibit DNA gyrase activity by stabilizing the enzyme-DNA complex, termed the cleavable complex (3). The complex is thought to be bactericidal upon release of the cleaved DNA (3).
In this study, we investigated the specific inhibition of DNA gyrase by BMS-284756, a novel des-F(6) quinolone that lacks the classical C-6 fluorine thought to be essential for the enhanced potency of recent fluoroquinolones (11). Unlike the earlier generation of nonfluorinated quinolones such as nalidixic acid and oxolinic acid, which had only moderate activity against gram-negative organisms, BMS-284756 has a broad spectrum of antibacterial activity, including good activity against anaerobes, exceptional activity against gram-positive bacteria (e.g., Streptococcus pneumoniae and the staphylococci), and the potential to cover quinolone-resistant pathogens in the clinic (4, 11). Recently it has been shown that DNA gyrase is the primary target for BMS-284756 in S. pneumoniae and Staphylococcus aureus (5; L. F. Discotto, L. E. Lawrence, K. L. Denbleyker, and J. F. Barrett, Abstr. 101st Gen. Meet. Am. Soc. Microbiol., abstr. A-51, 2001).
The inhibition of DNA gyrase generally correlates with antimicrobial activity (7, 10, 12). Two assays, the supercoiling inhibition assay (SCIA) and cleavable complex assay (CCA), were used to test the inhibitory activity of BMS-284756 and three quinolone comparators against Escherichia coli DNA gyrase. DNA gyrase inhibition can be categorized into at least six major types: specific, mechanism-based inhibition of GyrA, specific, mechanism-based inhibition of GyrB, less specific DNA intercalation, less specific minor groove DNA binding, nonspecific chelation, and nonspecific inhibition caused by environmental conditions (1). The SCIA measures the inhibition of DNA gyrase supercoiling and detects all six types of DNA gyrase inhibitors. The CCA measures the formation of enzyme-DNA complex that is only facilitated by DNA GyrA inhibitors, thus distinguishing the specific GyrA inhibitors from the other five types of gyrase inhibition (including nonspecific inhibition).
(The initial report of the work was presented at the 40th Interscience Conference on Antimicrobial Agents and Chemotherapy, 17 to 20 September, 2000, in Toronto, Canada).
The preparation of DNA gyrase and of relaxed DNA substrate and the SCIA were performed according to the methods of Lawrence et al. (8). The strains used in MIC testing are all clinical isolates, excluding E. coli and Haemophilus influenzae species. E. coli/wild type (wt) (A15119) is the parent strain of E. coli/acrA::kana (A28911), which is an engineered mutant strain that is deficient in efflux via an acrA knockout. H. influenzae/wt (A29354) is the parent strain of H. influenzae/acrB::kana (A29353), which is an engineered mutant strain deficient in efflux.
The DNA gyrase CCA was performed according to the method of Barrett et al. (1). The samples were processed for gel electrophoresis by adding 2 μl of tracking dye (50% glycerol and 0.125% bromophenol blue) and 35 μl of chloroform/isoamyl alcohol (24:1). The mixture was then vortexed and centrifuged briefly, of which 30 μl of the blue upper phase was withdrawn and loaded onto a 0.8% agarose–Tris-borate-EDTA horizontal gel for electrophoresis separation. The gel was stained with 2.5 μg of ethidium bromide/ml for 30 min after electrophoresis. The linear DNA bands were visualized and individually quantified using an AlphaImager 2200 system (AlphaInnotech Corp., San Leandro, Calif.). These data were directly fit for using GraphPad Prism (GraphPad Software, Inc., San Diego, Calif.), generating a nonlinear sigmoidal dose response curve. There was a drug-independent plateau of maximum cleavage with increasing drug concentrations. The maximum amount of cleavage was determined by optimizing the enzyme concentration, time of assay, and ethidium bromide staining; the same conditions were then used for testing all drugs. The CC50 is defined as the amount of drug required to induce 50% cleavage of the maximal amount of linear DNA formed at the standard reaction conditions. For supercoiling inhibition, the intensity of each supercoiled DNA band was directly fit to a sigmoidal dose response curve using GraphPad Prism from which the 50% inhibitory concentration (IC50) was determined.
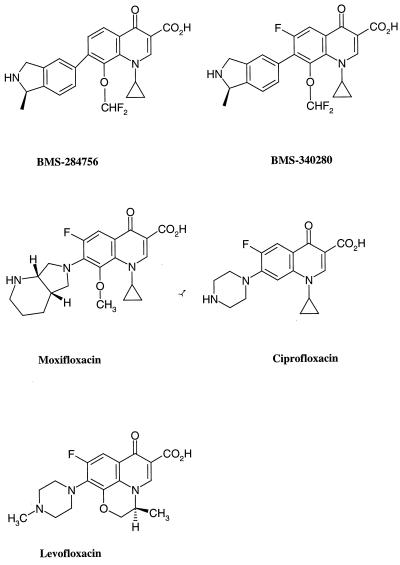
Table 1 shows the MICs of BMS-284756 and three relevant quinolone comparators (Fig. 1) for various gram-negative and gram-positive bacteria. The MICs of BMS-284756 ranged from 0.015 to 2 μg/ml for gram-negative strains and from 0.007 to 0.015 μg/ml for several gram-positive strains; it was generally as active as ciprofloxacin, levofloxacin, and moxifloxacin. Figures 2 and 3 show the concentration-dependent inhibition of BMS-284756 against E. coli DNA gyrase supercoiling activity and facilitation of the cleavable complex, respectively. Both SCIA and CCA were also performed for ciprofloxacin, levofloxacin, moxifloxacin, and BMS-340280, the C-6 fluorinated analog of BMS-284756 (Table 2). The IC50s and CC50s generally correlated well for the quinolones. BMS-284756 was more active than moxifloxacin and levofloxacin in both the SCIA and CCA. BMS-284756 inhibited the supercoiling activity at the same level as ciprofloxacin and had a twofold-higher CC50. The difference in the absolute IC50s and CC50s is possibly caused by differences in enzyme concentrations or other experimental conditions. BMS-340280 has been shown to have MICs equivalent to or within twofold of those of BMS-284756 for a panel of gram-negative and gram-positive bacteria (8). In SCIA and CCA, BMS-340280 exhibited similar inhibition as BMS-284756, indicating that the C-6 fluorine does not affect target activity. Also tested was novobiocin, a known GyrB subunit inhibitor. As expected, it inhibited the supercoiling activity of DNA gyrase with an IC50 of 0.22 μg/ml but did not facilitate the formation of the cleavable complex at a concentration up to 3 mg/ml, indicating that it does not target the E. coli GyrA subunit. Novobiocin serves as a negative control, confirming that the CCA specifically detects GyrA inhibitors. A correlation of MICs and DNA gyrase inhibition for a single bacterium has been observed (7, 10, 11), but other factors such as drug penetration and efflux may affect the whole-cell antibacterial activity.
TABLE 1.
MICs of BMS-284756 and three quinolone comparators against various gram-negative and positive bacteria
| Organism | Strain no. | MIC (μg/ml)
|
|||
|---|---|---|---|---|---|
| BMS-284756 | Ciprofloxacin | Levofloxacin | Moxifloxacin | ||
| E. coli | A15119 | 0.015 | 0.007 | 0.03 | 0.03 |
| E. coli/acrA::kana | A28911 | 0.015 | 0.007 | 0.03 | 0.03 |
| E. coli/wt | A29352 | 0.015 | 0.007 | 0.03 | 0.03 |
| Klebsiella pneumoniae | A9664 | 0.06 | 0.03 | 0.06 | 0.06 |
| Klebsiella oxytoca | A20345 | 0.06 | 0.03 | 0.06 | 0.125 |
| Enterobacter aerogenes | A20985 | 0.06 | 0.03 | 0.06 | 0.06 |
| Enterobacter cloacae | A9656 | 0.06 | 0.03 | 0.06 | 0.06 |
| Enterobacter cloacae (norfloxacin resistant) | A26179 | 2 | 2 | 4 | 2 |
| Proteus mirabilis | A9900 | 0.125 | 0.015 | 0.06 | 0.125 |
| Proteus vulgaris | A21559 | 0.06 | 0.015 | 0.06 | 0.125 |
| Proteus rettgeri | A22424 | 1 | 1 | 1 | 1 |
| Proteus stuartii | A20615 | 1 | 1 | 1 | 1 |
| Morganella morganii | A15153 | 0.25 | 0.03 | 0.06 | 0.125 |
| Serratia marcescens | A20019 | 0.5 | 0.125 | 0.125 | 0.25 |
| Salmonella enteritidis | A9531 | 0.25 | 0.03 | 0.06 | 0.125 |
| Xanthomonas maltophilia | A24258 | 2 | 2 | 1 | 1 |
| Moraxella catarrhalis | A25409 | 0.015 | 0.03 | 0.125 | 0.125 |
| H. influenzae/acrB::kana | A29354 | 0.015 | 0.25 | 0.5 | 0.03 |
| H. influenzae/ATCC 51907/wt | A29353 | 0.003 | 0.007 | 0.015 | 0.007 |
| S. pneumoniae wt | A29417 | 0.015 | 1 | 0.5 | 0.25 |
| S. aureus | A24407 | 0.007 | 0.125 | 0.125 | 0.03 |
| S. aureus (homogeneous, methicillin resistant) | A27223 | 0.007 | 0.06 | 0.125 | 0.03 |
FIG. 1.
Structure of quinolones tested in these studies.
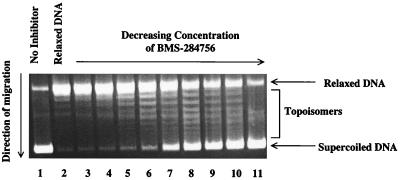
FIG. 2.
Concentration-dependent inhibition of E. coli DNA gyrase supercoiling activity by BMS-284756. Lane 1, no-drug control; lane 2, relaxed pBR322 substrate; lanes 3 through 11, BMS-284756 at 6.25, 3.12, 1.56, 0.78, 0.39, 0.195, 0.098, 0.049, and 0.024 μg/ml, respectively. IC50 = 0.17 μg/ml.
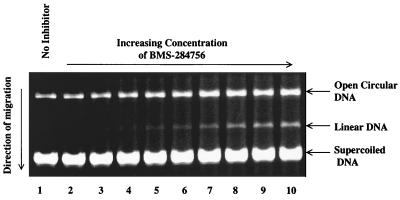
FIG. 3.
Concentration-dependent generation of the cleavable complex of E. coli DNA gyrase by BMS-284756. Lane 1, no-drug control; lanes 2 through 10, BMS-284756 at 0.08, 0.16, 0.32, 0.65, 1.30, 2.60, 5.20, 10.4, and 20.8 μg/ml, respectively. CC50 =2.47 μg/ml.
TABLE 2.
Inhibition of E. coli DNA gyrase activity
| Drug | Results for SCIA
|
Results for CCA
|
||
|---|---|---|---|---|
| IC50 (μg/ml) | SD (μg/ml) | CC50 (μg/ml) | SD (μg/ml) | |
| BMS-284756 | 0.17 | 0.08 | 2.47 | 0.33 |
| Ciprofloxacin | 0.18 | 0.08 | 1.23 | 0.07 |
| Levofloxacin | 0.32 | 0.04 | 6.55 | 1.54 |
| Moxifloxacin | 0.36 | 0.04 | 3.06 | 1.05 |
| BMS-340280 | 0.16 | 0.02 | 2.22 | 0.57 |
| Novobiocin | 0.22 | 0.12 | >3,000 | NDa |
ND, not determined.
BMS-284756 has excellent in vitro activity against both gram-positive and gram-negative bacteria and good intrinsic activity against DNA gyrase. BMS-284756 inhibited DNA gyrase supercoiling activity and facilitated the cleavable complex in a concentration-dependent manner, which is typical of other quinolones. Thus, it is most likely that BMS-284756 appears to have the same mechanism of action against E. coli DNA gyrase as other quinolone comparators, in that it specifically inhibits the GyrA subunit. Further studies such as mutant selection by BMS-284756 in E. coli and biochemical testing of drug sensitivity of GyrA mutant enzyme will be necessary to prove this fact. The definitive determination of the mechanism(s) of antibacterial activity will require additional efforts to demonstrate the involvement of physiological events, such as cellular penetration of drug and inhibition of DNA replication in whole cells. In this study the results from both the SCIA and CCA indicate that the level of inhibition against E. coli DNA gyrase is similar for BMS-284756 and other quinolone comparators, consistent with the similarity of their gram-negative antibacterial activity in vitro. Although the more sensitive target of BMS-284756 has been demonstrated to be DNA gyrase for gram-positive bacteria (5; Discotto et al., Abstr. 101st Gen. Meet. Am. Soc. Microbiol.), whether this is the case for gram-negative bacteria will require further study.
REFERENCES
- 1.Barrett J F, Bernstein J I, Krause H M, Hilliard J J, Ohemeng K A. Testing potential gyrase inhibitors of bacterial DNA gyrase: a comparison of the supercoiling inhibition assay and “cleavable complex” assay. Anal Biochem. 1993;214:313–317. doi: 10.1006/abio.1993.1493. [DOI] [PubMed] [Google Scholar]
- 2.Bates A D, Maxwell A. DNA topology. Oxford, England: IRL Press; 1993. [Google Scholar]
- 3.Drlica K, Zhao X. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol Mol Biol Rev. 1997;61:377–392. doi: 10.1128/mmbr.61.3.377-392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fung-Tomc J C, Minassian B, Kolek B, Huczko E, Aleksunes L, Stickle T, Washo T, Gradelski E, Valera L, Bonner D P. Antibacterial spectrum of a novel des-fluoro(6) quinolone, BMS-284756. Antimicrob Agents Chemother. 2000;44:3351–3356. doi: 10.1128/aac.44.12.3351-3356.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hartman-Neumann S, DenBleyker K, Pelosi L A, Lawrence L E, Barrett J F, Dougherty T J. Selection and genetic characterization of Streptococcus pneumoniae mutants resistant to the des-F(6)-quinolone BMS-284756. Antimicrob Agents Chemother. 2001;45:2865–2870. doi: 10.1128/AAC.45.10.2865-2870.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hooper D C. Mechanisms of fluoroquinolone resistance. Drug Resist Updates. 1999;2:38–55. doi: 10.1054/drup.1998.0068. [DOI] [PubMed] [Google Scholar]
- 7.Hoshino K, Sato K, Akahane K, Yoshida A, Hayakawa I, Sato M, Une T, Osada Y. Significance of the methyl group on the oxazine ring of ofloxacin derivatives in the inhibition of bacterial and mammalian type II topoisomerases. Antimicrob Agents Chemother. 1991;35:309–312. doi: 10.1128/aac.35.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lawrence L E, Wu P, Fan L, Gouveia K E, Card A, Casperson M, Denbleyker K, Barrett J F. The inhibition and selectivity of bacterial topoisomerases by BMS-284756 and its analogues. J Antimicrob Chemother. 2001;48:195–201. doi: 10.1093/jac/48.2.195. [DOI] [PubMed] [Google Scholar]
- 9.Reece R J, Maxell A. DNA gyrase: structure and function. Crit Rev Biochem Mol Biol. 1991;26:335–375. doi: 10.3109/10409239109114072. [DOI] [PubMed] [Google Scholar]
- 10.Saiki A Y C, Shen L L, Chen C-M, Baranowski J, Lerner C G. DNA cleavage activities of Staphylococcus aureus gyrase and topoisomerase IV stimulated by quinolones and 2-pyridones. Antimicrob Agents Chemother. 1999;43:1574–1577. doi: 10.1128/aac.43.7.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takahata M, Mitsuyama J, Yamashiro Y, Yonezawa M, Araki H, Todo Y, Minami S, Watanabe Y, Narita H. In vitro and in vivo antimicrobial activities of T-3811ME, a novel des-F(6)-quinolone. Antimicrob Agents Chemother. 1999;43:1077–1084. doi: 10.1128/aac.43.5.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tanaka M, Sato K, Kimura Y, Hayakawa I, Osada Y, Nishino T. Inhibition by quinolones of DNA gyrase from Staphylococcus aureus. Antimicrob Agents Chemother. 1991;35:1489–1491. doi: 10.1128/aac.35.7.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J. DNA topoisomerases. Annu Rev Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]