Abstract
Background and Aims
Histone deacetylase inhibitors [HDACi] exert potent anti-inflammatory effects. Because of the ubiquitous expression of HDACs, clinical utility of HDACi is limited by off-target effects. Esterase-sensitive motif [ESM] technology aims to deliver ESM-conjugated compounds to human mononuclear myeloid cells, based on their expression of carboxylesterase 1 [CES1]. This study aims to investigate utility of an ESM-tagged HDACi in inflammatory bowel disease [IBD].
Methods
CES1 expression was assessed in human blood, in vitro differentiated macrophage and dendritic cells, and Crohn’s disease [CD] colon mucosa, by mass cytometry, quantitative polymerase chain reaction [PCR], and immunofluorescence staining, respectively. ESM-HDAC528 intracellular retention was evaluated by mass spectrometry. Clinical efficacy of ESM-HDAC528 was tested in dextran sulphate sodium [DSS]-induced colitis and T cell transfer colitis models using transgenic mice expressing human CES1 under the CD68 promoter.
Results
CES1 mRNA was highly expressed in human blood CD14+ monocytes, in vitro differentiated and lipopolysaccharide [LPS]-stimulated macrophages, and dendritic cells. Specific hydrolysis and intracellular retention of ESM-HDAC528 in CES1+ cells was demonstrated. ESM-HDAC528 inhibited LPS-stimulated IL-6 and TNF-α production 1000 times more potently than its control, HDAC800, in CES1high monocytes. In healthy donor peripheral blood, CES1 expression was significantly higher in CD14++CD16- monocytes compared with CD14+CD16++ monocytes. In CD-inflamed colon, a higher number of mucosal CD68+ macrophages expressed CES1 compared with non-inflamed mucosa. In vivo, ESM-HDAC528 reduced monocyte differentiation in the colon and significantly improved colitis in a T cell transfer model, while having limited potential in ameliorating DSS-induced colitis.
Conclusions
We demonstrate that monocytes and inflammatory macrophages specifically express CES1, and can be preferentially targeted by ESM-HDAC528 to achieve therapeutic benefit in IBD.
Keywords: HDAC inhibitor, CES1, IBD
1. Introduction
Histone deacetylases [HDAC] are a family of 18 epigenetic enzymes that regulate histone and non-histone acetylation by erasing acetyl groups from lysine residues, leading to a chromatin remodelling and altered gene expression.1 Several studies report a fundamental role of HDAC in regulating cell survival and inflammatory response,2 and therefore various HDAC inhibitors have been developed and investigated in, for instance, fields of cancer and inflammatory diseases. Several randomised clinical trials have been conducted in the oncology field, and four HDACi have been Food and Drug Administration [FDA]-approved for some malignancies like multiple melanoma, cutaneous T cell lymphoma, and peripheral T cell lymphoma.3,4 Also, however, trial outcomes revealed a wide range of adverse events associated with HDACi5 with treatment discontinuation as a result.6
In immune-related diseases, HDACi treatment inhibits inflammatory responses both in vitro and in vivo.7,8 Furthermore, in patients receiving HDACi, low plasma levels of pro-inflammatory cytokines were reported, and isolated immune cells from these patients were also less responsive to inflammatory stimuli.9–11 In preclinical models of several inflammatory diseases including IBD, HDACi have demonstrated promising therapeutic benefits.12–18 However, clinical translation is limited due to their off-target activity and wide toxicity profile, including life-threatening cardiac arrhythmias, bone marrow suppression effects, and gastrointestinal and hepatic toxicities.19–22
As histone acetylation drives gene expression in a cell-specific manner, selective targeting strategies of relevant immune cells is a highly attractive approach. In this regard, a targeted drug delivery technology has been developed to selectively retain inhibitors tagged with an esterase-sensitive motif [ESM] in mononuclear myeloid cells, based on their expression of the carboxylesterase 1 [CES1] enzyme.23 This technology is of interest in immune-related diseases where these cell subsets play a role in driving the pathology, and such an approach can minimise off-target effects. Murine models are designed to investigate this approach through transgenic human CES1 expression, which allow human CES1 to be expressed predominantly in mononuclear myeloid cells, driven by CD68 promoter. These models have shown promising results; in an arthritis model, ESM-HDAC528 has improved the outcome of the disease at a 100-fold lower dose compared with non-targeted compound, SAHA.23
We reasoned that this approach is likely to have a therapeutic benefit in inflammatory bowel disease [IBD]. First, HDACi ameliorates colitis in preclinical models and second, IBD is an immune-driven pathology where mononuclear myeloid cells, including inflammatory macrophages, are enriched in Crohn’s disease [CD] colon mucosa24 and are believed to perpetuate mucosal inflammation.25 Furthermore, a recent report suggests that a unique mononuclear phagocyte cytokine/chemokine network is linked to anti-TNF-α resistance in CD.26
In the current study, we demonstrate anti-inflammatory effects of ESM-HDAC528 in monocytes and macrophages, reflecting their differential CES1 expression. In preclinical models of IBD, ESM-HDAC528 showed clinical efficacy in the T cell transfer colitis model, and in the dextran sulphate sodium [DSS]-induced colitis, ESM-HDAC528 attenuated monocytes-to-macrophages maturation in the colon and blunted response to inflammatory stimuli in peritoneal macrophages.
2. Materials and Methods
Detailed information on the materials, methods, and associated references can be found in the SI Appendix, available as Supplementary data at ECCO-JCC online.
2.1. Compounds
ESM-HDAC528 and its non-hydrolysable HDAC800 control were provided by GlaxoSmithKline [Stevenage, UK].
2.2. Animals
The human CES1 transgenic mouse [CES1/Es1elo] was generated as described earlier.23,27 The transgenic mouse [CES1/Es1elo/Rag-/-] was generated by cross-breeding human CES1 transgenic mouse [CES1/Es1elo] with the immunodeficient RAG−/− mouse. All animal studies were ethically reviewed and carried out in accordance with European Directive 2010/63/EEC, the guidelines of the Ethical Animal Research Committee of the University of Amsterdam, and the GSK Policy on the Care, Welfare and Treatment of Animals.
2.3. Murine colitis models
For DSS-induced colitis, CES1/Es1elo mice were given 2% dextran sulphate sodium [DSS; TdB Consultancy] for 7 days, followed by 2 days of normal drinking water. Simultaneously, mice received daily intraperitoneal [IP] injections of 1 or 3 or 10 mg/kg of ESM-HDAC528 or vehicle until sacrifice. For T cell transfer colitis; CES1/Es1elo/Rag-/- mice received IP injection of CD4+CD45Rbhigh cells, isolated from spleens of C57BL/6 WT mice. Three weeks later; mice received daily IP injections of 3 mg/kg of ESM-HDAC528 or vehicle for another 4 weeks until sacrifice.
2.4. Human clinical samples
The human biological samples were sourced ethically and their research use was in accord with the terms of the informed consents under an IRB/EC approved protocol or approval of the accredited Medical Ethics Committee at the Amsterdam UMC, University of Amsterdam.
2.5. Cytokine measurement
Cytokines were measured using either mouse inflammation CBA kit [BD Bioscience], Meso Scale Discovery [MSD] plates or enzyme-linked immunosorbent assay [ELISA] kits [R&D systems], according to the manufacturer’s protocol.
2.6. Flow cytometry analysis
All samples were acquired using a FACS Fortessa [BD Biosciences] and analysed using FlowJo software [Treestar Inc., Ashland, OR].
2.7. Mass cytometry analysis
Samples were acquired on a CyTOF Helios mass cytometer. Data were normalised using bead normalisation.28 Deconvolution of pooled samples was performed by processing flow cytometry standard [FCS] files with the standard single-cell debarcoding algorithm for CyTOF data.29 Analysis were performed using R Studio. Clusters of phenotypically similar cells were identified using the FlowSOM-package.30
2.8. Quantitative real-time polymerase chain reaction
RNA was isolated using RNeasy mini kit [Qiagen] and cDNA was synthesised using cDNA synthesis kit [Qiagen] following the manufacturer’s protocol. Quantitative polymerase chain reaction [PCR] was performed on a QuantStudio Flex 7 [Applied Biosystems] or a LightCycler 480 II [Roche Applied Science].
2.9. Immunofluorescence
Paraffin sections, prepared from surgically resected colons of CD patients undergoing colectomy at Amsterdam University Medical Center, were stained for CD68 [clone PG-M1, Dako], CES1 [polyclonal, Novus Biologicals], and DAPI.
2.10. In vitro human monocyte and macrophage assays
Freshly isolated CD14+ monocytes or GM-CSF differentiated macrophages were pre-incubated for 1 h with ESM-HDAC528 or HDAC800, then stimulated with 1 ng/mL lipopolysaccharide [LPS] for 24 h. Supernatants were collected for cytokine analysis and cells were used for ATP bioluminescence assay.
2.11. Mass spectrometry assay
Cell lysate samples were extracted using protein precipitation and directly injected onto the HPLC-MS/MS system. Analysis was conducted by reverse-phase HPLC-MS/MS. Nominal MRM transitions for HDAC800, hydrolysed ESM-HDAC528, and parent ESM-HDAC528 were 391 to 178, 335 to 178, and 403 to 178, respectively.
2.12. Statistical analysis
The significance of the differences was analysed using Student’s t test, a Mann-Whitney U test, and one-way or two-way analysis of variance [ANOVA], as indicated; p-values <0.05 were considered significant.
3. Results
3.1. ESM-HDAC528 is hydrolysed by CES1 and accumulates in human blood-derived CD14+ monocytes
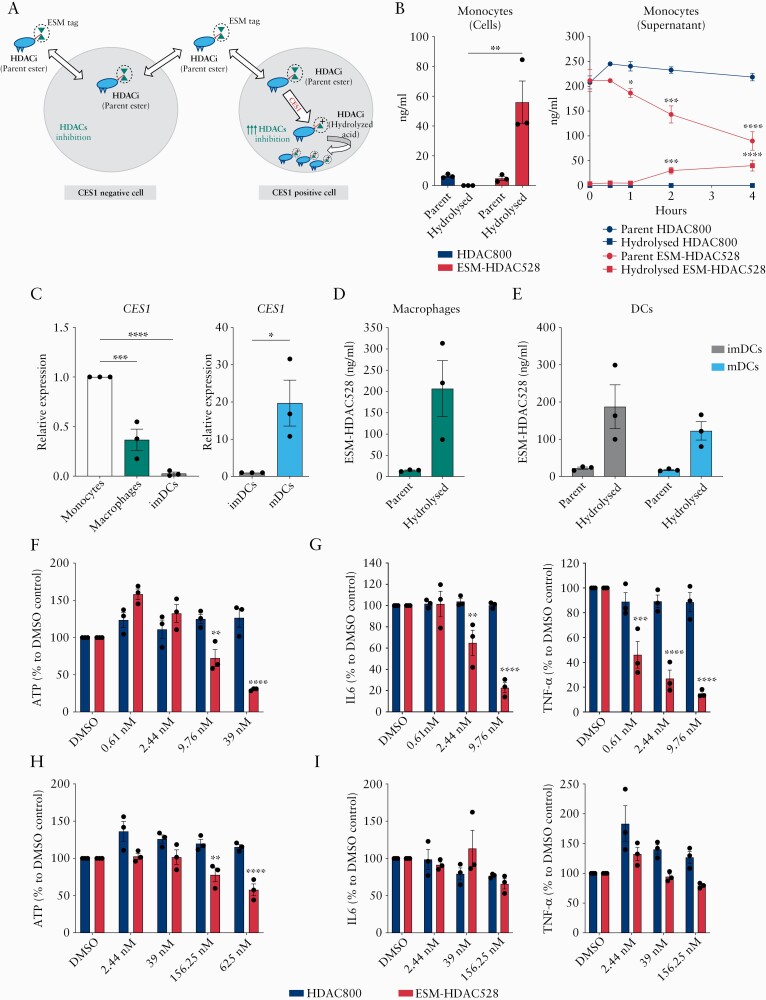
ESM-targeted drug delivery technology depends on the expression and hydrolysing activity of CES1 in target cells, where an ESM-tagged compound is hydrolysed and retained23 [Figure 1A]. First, we aimed to validate this technology using human monocytes as a model of CES1-expressing cells. Monocytes were incubated with ESM-HDAC528 or non-hydrolysable HDAC800 control compound; then both parent ester and hydrolysed acid of the compounds were measured intracellularly and in the supernatant [Figure 1B]. Monocytes retained little HDAC800, with no hydrolysed acid formed, whereas ESM-HDAC528 was retained more strongly, with efficient hydrolysed acid generation intracellularly. Concurrently, ESM-HDAC528 parent ester was more consumed over time by monocytes compared with HDAC800 parent ester, as measured in the supernatant, reflecting the enhanced take-up and consumption of the ESM-tagged compound. ESM-HDAC528 hydrolysed acid was gradually detected as well in the supernatant over time, indicating some active efflux activity [Figure 1B].
Figure 1.
Profiling ESM-HDAC528 accumulation and anti-inflammatory effect in mononuclear myeloid cell subsets. [A] Schematic diagram of ESM technology for CES1-based targeted drug delivery to mononuclear myeloid cells; ESM-tagged HDACi [parent ester] can freely move in and out CES1- cells but, once it enters CES1+ cells, it is hydrolysed by CES1 to the ESM-HDACi [hydrolysed acid] form of the compound which cannot leave the cells and is retained intracellularly, causing augmented HDACs inhibition. [B] Monocytes were incubated for 4 h with ESM-HDAC528 or non-hydrolysable HDAC800; the parent ester and hydrolysed acid of both compounds were measured both intracellularly and in the supernatant by LC-MS/MS. [C] CES1 mRNA expression in CD14+ monocytes, monocyte-differentiated macrophages and dendritic cells [imDCs], and LPS-polarised dendritic cells [mDCs]. [D and E] Macrophages imDCs or mDCs were all incubated with ESM-HDAC528 for 4 h; the intracellular parent ester and its hydrolysed acid concentrations were measured by LC-MS/MS. [F and G] CD14+ isolated monocytes or [H and I] macrophages differentiated from the same donors were pre-incubated with ESM-HDAC528 or HDAC800 and stimulated for 1 day with LPS, and then ATP production, IL-6, and TNF-α secretion were measured. Data are represented as mean with SEM of three donors, two technical replicates for each. In [B, right panel], parent or hydrolysed forms of HDAC800 and ESM-HDAC528 were compared. In [F to I], similar doses of ESM-HDAC528 and HDAC800 treatment were compared. Statistical testing was performed using two-way ANOVA test [B,E,F,G,H,I] or one-way ANOVA or Student’s t test [C]; *p ≤0.05, **p ≤0.01, *** p ≤0.001, **** p ≤0.0001. SEM, standard error of the mean.
3.2. In vitro differentiated inflammatory macrophages and dendritic cells show high CES1 expression
Next we aimed to profile CES1 gene expression within in vitro monocytes-derived macrophages and dendritic cells [DCs] subsets, to address their potential to retain ESM-HADC528. CES1 was found to be highly expressed in human monocytes, and its expression reduced upon differentiation into macrophages or immature DCs [imDCs] [Figure 1C]. However, an upregulation of CES1 expression was observed in mature DCs [mDCs] upon LPS-primed maturation [Figure 1C].
Furthermore, we aimed to validate the ability of macrophages and DC subsets to generate and retain the hydrolysed acids from ESM parent ester. Both macrophages and DCs showed efficient ESM-HDAC528 acid hydrolysis and intracellular retention following incubation with ESM-HDAC528 [Figure 1D and E]. Interestingly, mDCs did not show better ability to hydrolyse and retain ESM-HDAC528 compared with the imDCs [Figure 1E], despite the upregulation of CES1 mRNA levels seen upon LPS maturation of DCs [Figure 1C]. These data demonstrate the efficient CES1-mediated ESM-HDAC528 retention in mononuclear myeloid cell subsets, as well as the differential CES1 gene expression among these cell populations.
3.3. ESM-HDAC528 shows an enhanced anti-inflammatory effect in human blood monocytes compared with monocytes-derived macrophages
We next hypothesised that the observed differences in CES1 expression among mononuclear myeloid cell subsets might be reflected in differential ESM-HDAC528 anti-inflammatory potency. Both human blood CD14+ monocytes and in vitro differentiated macrophages from matched donors were incubated with serial dilutions of ESM-HDAC528 or HDAC800, and then stimulated with LPS. ESM-HDAC528 compromised adenosine triohosphate [ATP] production in monocytes culture at 9.7 nM compared with 156.2 nM in macrophages [Figure 1F–H]. Meanwhile, the same doses of HDAC800 treatment did not affect ATP production either in monocytes or in macrophages [Figure 1F–H]. In monocytes, ESM-HDAC528 strongly downregulated LPS-induced IL-6 and TNF-α production at low doses compared with HDAC800 control compound [Figure 1G]. This enhanced anti-inflammatory potency of ESM-HDAC528 was also observed in macrophages [Figure 1I] but to a much lesser extent compared with donor-matched monocytes. The enhanced potency of ESM-HDAC528 over HDAC800 confirms the augmented effect of the ESM-HDAC528, mediated by CES1 activity.
3.4. CES1 expression profiles in peripheral blood cells of healthy donors demonstrates higher expression in classical CD14++CD16- monocytes compared with non-classical CD14+CD16++ monocytes
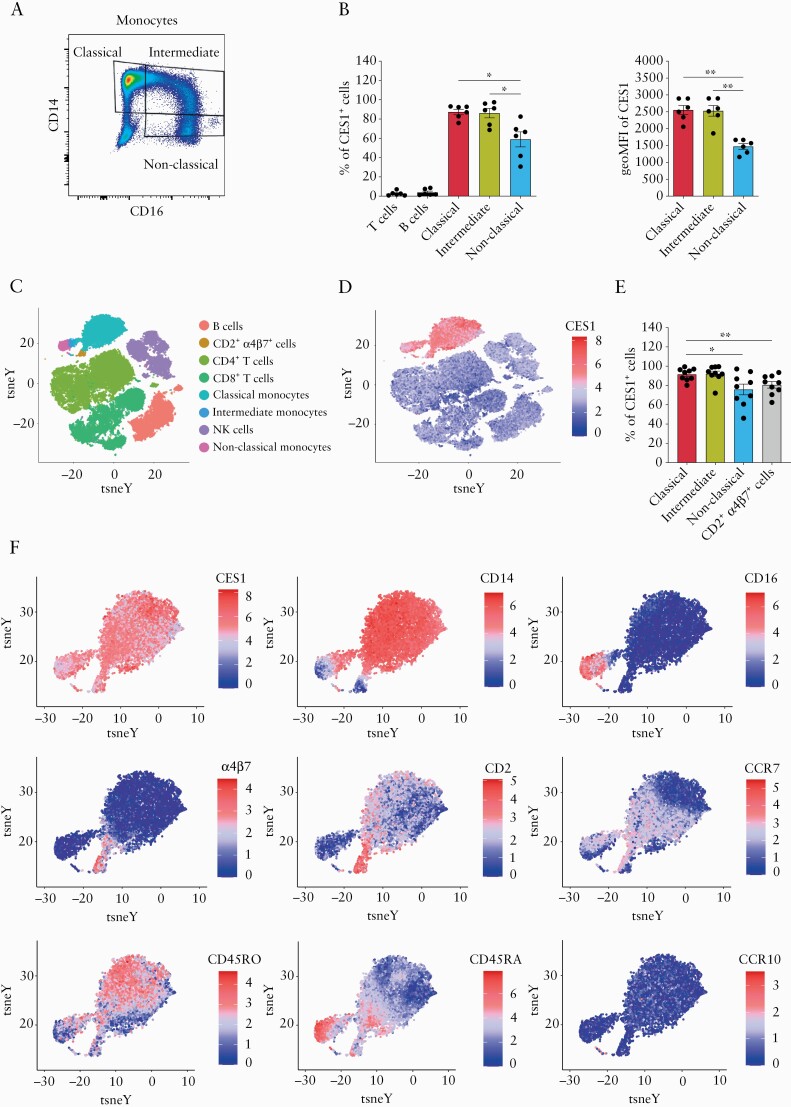
We next aimed to explore CES1 expression in peripheral blood. Peripheral blood mononuclear cells [PBMCs] were isolated from healthy donors and analysed by flow cytometry for CES1 expression [Figure S1, available as Supplementary data at ECCO-JCC online]. Monocytes subsets were further characterised by CD14 and CD16 expression pattern [Figure 2A] into classical monocytes [CD14++CD16-], intermediate monocytes [CD14++ CD16+], and non-classical monocytes [CD14+ CD16++]. CES1 was specifically and highly expressed in monocytes, with no CES1 expression detected in CD3+ T and CD19+ B cells [Figure 2B]. In the monocyte population, non-classical monocytes showed significantly less CES1 expression compared with both classical and intermediate monocytes as identified by both frequency of CES1+ cells and geometric mean of intensity of CES1 expression [Figure 2B].
Figure 2.
CES1 expression profiling in peripheral blood monocyte populations reveals predominant expression in classical and intermediate monocytes. [A] Flow cytometry analysis of healthy donors PBMCs [n = 6] for CES1 expression within monocyte subsets, plots shown are gated on monocytes, identified by high FSC-A/ SSC-A; monocyte subsets are further distinguished by CD14/CD16 expression into; classical monocytes [CD14++CD16-], intermediate monocytes [CD14++CD16+], and non-classical monocytes [CD14+ CD16++]. [B] Frequency of CES1+ cells among CD3+ T cells; CD19+ B cells and monocyte subsets are shown, geoMFI of CES1 among monocytes subsets are plotted. [C] tSNE plots show immune cell subsets in healthy donor PBMCs [n = 9]. [D] CES1 expression among clustered populations as analysed by mass cytometry. [E] Frequencies of CES1+ cells among identified CES1-expressing immune cell subsets are shown. [F] Monocyte diversity tSNE plots are generated to demonstrate expression levels of monocyte-related markers [CD14 – CD16 – CD2 – α4β7 – CD45RO – CD45RA – CCR7 – CCR10] and CES1 among monocyte populations. The events identified as classical monocytes, intermediate monocytes, non-classical monocytes, and the CD2+a4b7+ myeloid cells were shown in the annotation of FlowSOM clusters. Data are represented as mean with SEM of six to nine patients, one technical replicate each. Statistical testing was performed using one-way ANOVA test or Student’s t test; *p ≤0.05, **p ≤0.01. SEM, standard error of the mean; PBMC, peripheral blood mononuclear cells.
Next, using mass cytometry, we further investigated differential CES1 expression in healthy donor PBMCs in more detail. Clusters of phenotypically similar cells were identified and shown in tSNE plots [Figure 2C], the CES1 expression pattern was shown to localise among monocyte populations clusters [Figure 2D]. The heterogeneity of monocyte populations was further dissected, showing the expression pattern of monocyte-related markers among the monocyte population [Figure 2F]. Interestingly, we could identify a CD14-CD16- population that expressed CD2 and α4β7 markers that have previously been linked to a DC precursor population.31,32 This CD2+ α4β7+ DC precursor population expressed a similar level of CES1 as did non-classical monocytes, and both express significantly less CES1 compared with classical and intermediate monocytes [Figure 2E]. In conclusion, CES1 profiling in peripheral blood shows predominantly high CES1 expression among classical and intermediate monocyte populations and relatively less expression in non-classical monocytes.
3.5. CES1-expressing macrophages are enriched in inflamed CD intestinal mucosa
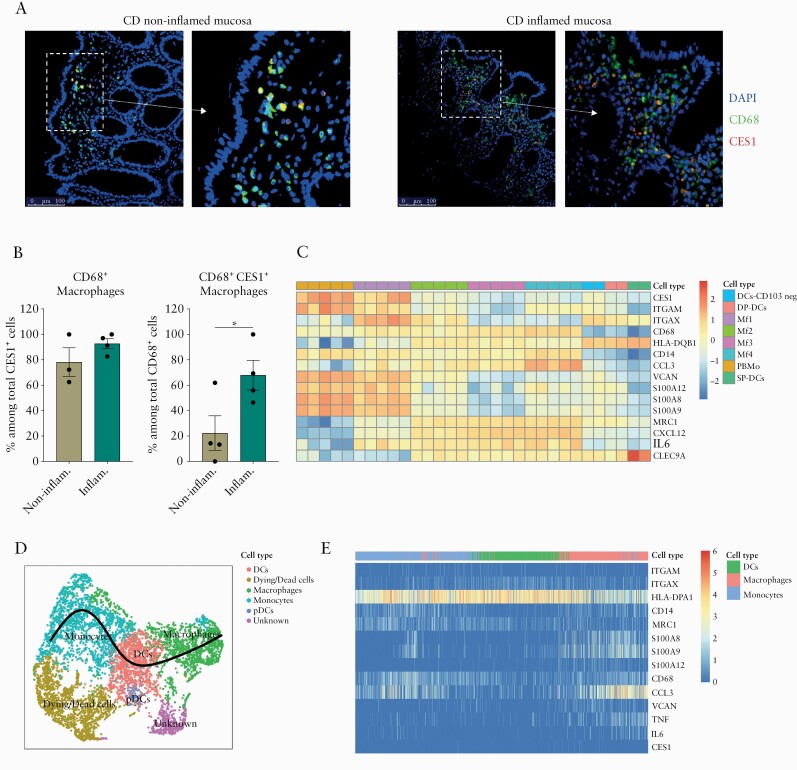
Next, we addressed the CES1 expression in intestinal mucosa in both healthy and inflamed conditions. In CD colon mucosa, immunofluorescent staining for CES1 and the pan-macrophage marker CD68 was performed in macroscopically inflamed and non-inflamed areas. CES1 was found to be expressed in a proportion of CD68+ cells in both inflamed and non-inflamed mucosa [Figure 3A]. The frequency of colon mucosal macrophages, as defined by CD68 expression, which express CES1 was higher in inflamed mucosa compared with non-inflamed mucosa [Figure 3B]. In line with earlier reports, CES1 was mainly expressed in CD68+ macrophages as quantified by percentage of CD68+ macrophages among CES1+ cells, confirming restricted CES1 expression among mucosal CD68+ mononuclear myeloid cells [Figure 3B].
Figure 3.
Profiling CES1 expression in non-inflamed and inflamed intestinal mucosa. [A] Immunofluorescence staining for CES1, CD68, and DAPI is shown within paraffin-embedded sections from surgically resected colons of CD patients [n = 4]. [B] Quantification of CD68+ macrophages among CES1+ cells and CES1+ CD68+ macrophages among total CD68+ macrophages is shown. [C] Heat map of CES1 gene expression along with some other differentially expressed myeloid cell-related genes expression is shown among peripheral blood monocytes [PBMo] and small intestinal macrophage and DCs subsets; data are retrieved from Bujko et al. 2018. RNAseq data, cells are flow cytometry sorted from small intestine of patients undergoing the Whipple procedure [n = 2-5]. [D] Developmental trajectory analysis of intestinal mononuclear phagocytes [MNPs] is conducted on [Martin et al. 2019] a scRNAseq dataset of CD patient intestinal biopsies [n = 11]. [E] Gene expression of CES1 and myeloid cell-related genes along the MNPs trajectory are demonstrated. Data are represented as mean with SEM of four patients, one technical replicate each. RNAseq expression values [log2] were median-centred by transcript. Statistical testing was performed using Student’s t test; *p ≤0.05. SEM, standard error of the mean; CD, Crohn’s disease.
Furthermore, RNAseq data retrieved from Bujko et al. 201833 were re-examined for CES1 mRNA expression, along with other myeloid cell-related genes, in peripheral blood monocytes [PBMo] and flow cytometry-sorted macrophages and dendritic cell subsets from small intestinal mucosa of patients undergoing a Whipple surgical procedure. CES1 showed higher expression in blood monocytes [PBMo] and intestinal immature macrophages [MF1: CD14+CD11c+HLA-DRint] queued, whereas more mature intestinal macrophage subsets [MF2: CD14+CD11c+HLA-DRhi], [MF3: CD14+CD11c−CD11b−,and [MF4: CD14hiCD11c−CD11b+], as well as DC subsets [SP-DCs: CD103+SIRPα −], [DP-DCs: CD103+SIRPα +], and [CD103− DCs: CD103−SIRPα +], showed less CES1 expression [Figure 3C]. This observation was consistent with CES1 mRNA expression dynamics during in vitro monocyte differentiation [Figure 1C]. Interestingly we could observe ITGAX [CD11c], S100A8, S100A9, and S100A12 genes to be enriched in high CES1-expressing intestinal macrophages [MF1].
To examine CES1 expression development in intestinal mononuclear phagocytes [MNPs] of CD patients, a developmental trajectory analysis was conducted on scRNAseq data retrieved from Martin et al. 201926 [Figure 3D]. There was lower CES1 mRNA transcript expression in MNPs retrieved from this dataset as compared with Bujko et al.’s 201833 dataset or protein expression as analysed by mass cytometry in PBMCs [Figure 2D]. Irrespectively, by analysing a developmental trajectory analysis for CD intestinal MNPs, CES1 was relatively enriched in differentiated macrophages along with S100A8, S100A9, and S100A12 genes that all follow the same pattern [Figure 3E].
3.6. ESM-HDAC528 attenuates colon monocytes to macrophage differentiation and peritoneal macrophage reactivity in DSS-induced colitis
We next aimed to evaluate the therapeutic potential of ESM-HDAC528 in a mouse model. To achieve CES1 expression in mice, we made use of CES1/Es1elo transgenic mice that express human CES1 (hCES1) predominantly in a monocyte-macrophage lineage-selective manner, driven by a human CD68 promoter known from its expression pattern to reflect macrophage populations in the intestinal mucosa.34 The ability of ESM-HDAC528 to accumulate in CES1/Es1elo mouse mononuclear myeloid cells was demonstrated both in vitro [Figure S2A and B, available as Supplementary data at ECCO-JCC online] and in vivo [Figure S2E] by means of flow cytometry analysis of acetylated lysine expression as indirect measure of ESM-HDAC528 accumulation. Gene expression levels of hCES1 were shown to be similar across differently polarised CES1/Es1elo mononuclear myeloid cells [Figure S2C]. However, enhanced acetylated lysine expression upon ESM-HDAC528 [50nM] treatment of these cells showed higher activity in bone marrow-derived dendritic cells [BMDCs] compared with bone marrow-derived macrophages [BMDM] [Figure S2D].
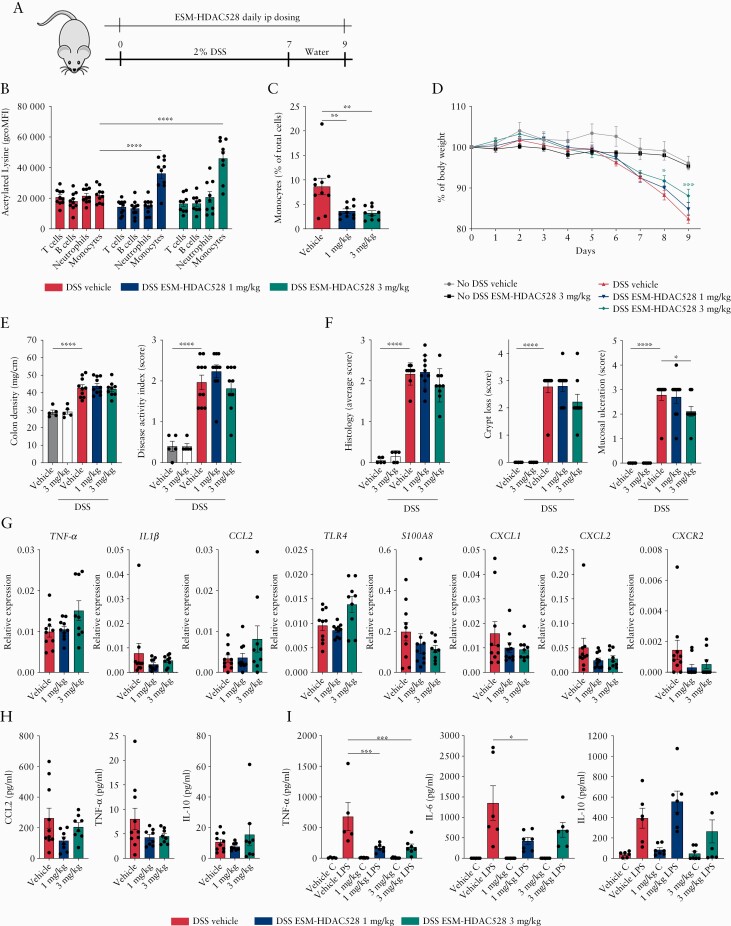
To evaluate the clinical relevance of the mononuclear myeloid cell-targeting approach by ESM-HDAC528, DSS colitis was induced in CES1/Es1elo mice using 2% DSS in drinking water. At the same time mice started to receive daily intraperitoneal injections of 1 or 3 mg/kg ESM-HDAC528 or vehicle [Figure 4A]. Both doses showed efficient targeting of ESM-HDAC528 to blood monocytes compared with other immune cells. The global acetylated lysine expression was particularly enhanced in blood monocytes [Figure 4B], and a reduced frequency of monocytes was observed [Figure 4C] when assessed 3 h after last injection at both tested doses. Despite efficient monocyte targeting of ESM-HDAC528, clinical improvement of colitis was limited to a number of clinical and biochemical parameters. Weight loss was significantly improved in a dose-dependent manner [Figure 4D], although no significant effect was observed in disease clinical activity index or colon density [weight/length ratio] [Figure 4E]. Colon inflammation score was improved in mice dosed at 3 mg/kg, reflecting reduced mucosal ulceration and crypt loss [Figure 4F]. Colon inflammatory biomarkers [TNF-α, IL1β, CCL2, TLR4, S100A8, CXCL1, CXCL2, CXCR2] mRNA expression were measured. S100A8, CXCL1, CXCL2, and CXCR2 mRNA levels tended to be reduced at both doses, whereas CCL2 and TLR4 were upregulated in ESM-HDAC528-treated mice only at the 3 mg/kg dose; but those trends did not meet significance in the groups tested. No change was detected in TNF-α or IL1β [Figure 4G]. However, serum levels of TNF-α, IL-10 and CCL2 were not significantly changed [Figure 4H]. In ESM-HDAC528-treated mice, peritoneal resident macrophages isolated at the end of the study showed a significantly reduced response to LPS [as assessed by IL-6 and TNF-α secretion] when cultured ex vivo [Figure 4I].
Figure 4.
ESM-HDAC528 improves clinical parameters in DSS-induced colitis and reduces peritoneal macrophage response to inflammatory stimulus. [A] A scheme of the acute DSS colitis model experimental design. [B] Flow cytometry analysis of acetylated lysine expression within blood immune cells, 3 h after IP dosing, along with [C] frequency of blood monocytes across all groups. [D] Weight changes are indicated as percentage of initial body weight. [E] Colon density [weight/length ratio] and average disease activity index [DAI] were measured at sacrifice. DAI consisted of average scores of oedema [0–3], diarrhoea [0–3], and the presence of blood in the stool [0–3], with a maximal DAI of 3 points. [F] Colon histopathology scores were graded from 0 to 4 points as indicated in Table S1, available as Supplementary data at ECCO-JCC online. Crypt loss and mucosal ulceration scores are highlighted. [G] Colon mRNA expression of inflammation biomarkers are shown. [H] Serum CCL2, TNF-α, and IL-10 are demonstrated. [I] Ex vivo retrieved peritoneal macrophages after mouse sacrifice were stimulated with LPS. TNF-α, IL-6, and IL-10 were quantified. Data are represented as mean with SEM, DSS groups [n = 10] and no DSS groups [n = 5], one technical replicate each. The DSS vehicle group is compared with the DSS compound-treated groups. In [E and F], thw no-DSS vehicle group is compared with the DSS-vehicle group as well. Statistical testing was performed using two-way ANOVA test [B, D] or otherwise one-way ANOVA test; *p ≤0.05, **p ≤0.01, *** p ≤0.001, **** p ≤0.0001. SEM, standard error of the mean; DSS, dextran sulphate sodium.
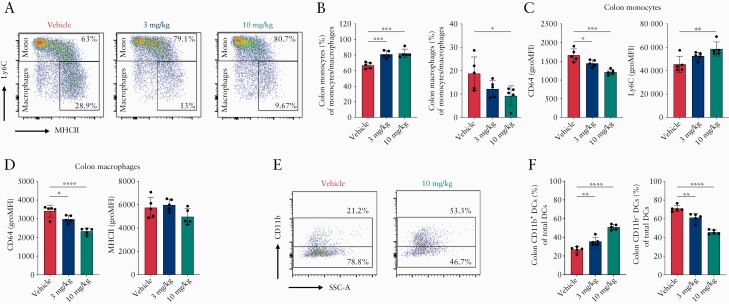
Next, in a repeat experiment, mice were dosed at 3 and 10 mg/kg ESM-HDAC528 or vehicle, following analyses of colon cell dynamics through flow cytometry. CD45+ live immune cells were identified and further analysed to define CD3+ T cells and Ly6G+ neutrophils. Monocytes and macrophages were defined as CD64+CD11b+CD11c- within CD3-Ly6G- gating and were differentiated by pattern of Ly6C and MHCII expression, with monocytes being Ly6Chigh/MHCIIlow and macrophages Ly6Clow/MHCIIhigh. DCs were defined as CD11c+ MHCII+ among CD3-Ly6G-CD64-CD11b- gating, and two subsets of DCs were further identified as CD11b+ and CD11b- DCs [Figure S3A, available as Supplementary data at ECCO-JCC online]. ESM-HDAC528 treatment did not interfere with recruitment of total immune cells to the colon during inflammation [Figure S3B], or with monocyte-macrophage population [CD64+CD11b+] recruitment in particular [data not shown]. However, ESM-HDAC528 attenuated colon monocyte to macrophage tissue differentiation in a dose-dependent manner [Figure 5A and B]. Monocytes found in the colon [Ly6Chigh MHCIIlow] exhibited a less mature phenotype, with higher Ly6C expression and lower CD64 protein expression in ESM-HDAC528-treated groups [Figure 5C]. Colon macrophages [Ly6Clow MHCIIhigh] exhibited reduced CD64 but no impact on MHCII expression [Figure 5D]. Total DC frequency was not affected, but the DCs subset distribution was modulated, with enrichment of the CD11b+ subset compared with the CD11b- subset in a dose-dependent manner, but with no impact on MHCII expression in DCs [Figure 5E and F]. With regard to clinical outcome of the colitis, the 10 mg/kg dose did not lead to any additional improvement in disease activity or inflammation score compared with the 1 and 3 mg doses [data not shown].
Figure 5.
ESM-HDAC528 modulates colon mononuclear myeloid cell populations during DSS-induced colitis. Acute DSS colitis was induced as described earlier; mice were simultaneously treated with vehicle or ESM-HDAC528 [3 or10 mg/kg] and colon mononuclear myeloid cells were analysed by flow cytometry. [A] Representative expression of MHCII and Ly6C among colon monocyte and macrophage populations were compared across treatment groups. [B] Frequencies of colon monocytes and macrophages among parent population [CD11b+CD64+CD11c-] are shown. [C] GeoMFI of CD64 and Ly6C in colon monocytes are quantified. [D] GeoMFI of CD64 and MHCII in colon macrophages are quantified. [E] CD11b+ and CD11b- dendritic cell [DCs] subsets are identified among colon DCs and shown across treatment groups. [F] Frequencies of colon DC subsets among total colon DCs population are compared across vehicle and 10 mg/kg groups. Data are represented as mean with SEM, DSS groups [n = 10] and no DSS groups [n = 5], two or three mouse colon were combined per sample. The DSS vehicle group is compared with DSS compound-treated groups. Statistical testing was performed using one-way ANOVA test; *p ≤0.05, **p ≤0.01, *** p ≤0.001, **** p ≤0.0001. SEM, standard error of the mean; DSS, dextran sulphate sodium.
3.7. ESM-HDAC528 significantly improves colon inflammation in a T cell transfer colitis model
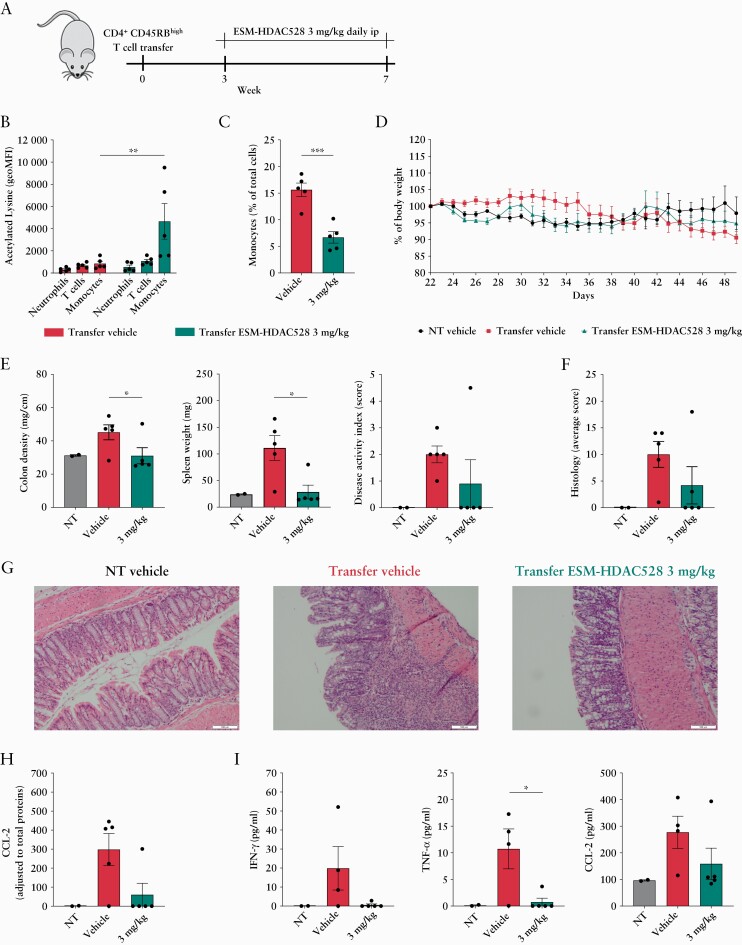
Next, we tested the potential of 3 mg/kg ESM-HDAC528 to reduce colitis in a T cell transfer colitis model. To this end, we generated transgenic CES1/Es1elo/Rag-/- mice to transfer CD45Rbhigh T cells into a RAG-/- host overexpressing the hCES1 gene. Three weeks following transfer of CD4+45Rbhigh T cells, mice started to receive daily intraperitoneal injections of 3 mg/kg ESM-HDAC528 or vehicle [Figure 6A]. Efficient targeting of blood monocytes was observed, as assessed by enhanced global acetylated lysine expression in blood monocytes compared with other blood immune cells [Figure 6B] along with reduced frequency of blood monocytes [Figure 6C] when assessed 3 h after IP injection. Clinical improvement of colitis was apparent in ESM-HDAC528-treated mice: weight loss was reduced in the ESM-HDAC528-treated group [Figure 6D], and colon density and spleen weight were significantly improved compared with the vehicle-treated group. Additionally, the disease activity index [DAI] showed a trend towards improvement in ESM-HDAC528-treated mice [Figure 6E] and a reduced colon inflammation histological score was noted after ESM-HDAC528 as compared with controls [Figure 6F]. As shown in representative images of haematoxylin-eosin stained colon histology sections, ESM-HDAC528-treated mice exhibited preserved crypt architecture along with reduced mucosal immune cell infiltration compared with colitis controls [Figure 6G]. A reduction in colon homogenate CCL2 protein expression was observed in ESM-HDAC528-treated mice [Figure 6H]. Finally, serum levels of IFN-γ, TNF-α, and CCL2 were reduced in ESM-HDAC528-treated mice, but only the reduction of TNF-α reached significance [Figure 6I].
Figure 6.
ESM-HDAC528 improves colon inflammation in a T cell transfer colitis model. [A] A schematic of the T cell transfer colitis model experimental design. [B] Flow cytometry analysis of acetylated lysine expression within blood immune cells, 3 h after ip dosing, along with [C] frequency of blood monocytes across indicated groups. [D] Weight changes are indicated as percentage of body weight on the first day of treatment. [E] Colon density [weight/length ratio], spleen weight [mg], and total Disease Activity Index [DAI] were measured at sacrifice. DAI; consisted of total scores of oedema [0–3], diarrhoea [0–3], and the presence of blood in the stool [0–3], with a maximal DAI of 9 points. [F] Colon histopathology scores were graded from 0 to 4 points, as indicated in Table S2, available as Supplementary data at ECCO-JCC online. Total scores are calculated according to this formula [total score = goblet cell loss score + 2 x crypt loss score + 2 x crypt hyperplasia score + 3 x submucosal inflammation score]. [G] Representative images of colon haematoxylin and eosin staining are shown. [H] Colon CCL2 and [I] serum IFN-γ, TNF-α, and CCL2 protein expression are shown. Data are represented as mean with SEM, transfer groups [n = 5] and no transfer group [n = 2]: the transfer vehicle group is compared with transfer compound-treated groups. Statistical testing was performed using two-way ANOVA test [B, D] or otherwise Student’s t test; *p ≤0.05, **p ≤0.01, *** p ≤0.001. SEM, standard error of the mean.
4. Discussion
As the ESM drug delivery technology is based on the ability of CES1 to hydrolyse and retain the tagged compound in the targeted cells, we aimed to demonstrate that the ESM-HDAC528 is superior over non-ESM tagged HDAC800 to target mononuclear myeloid cells. Monocytes – as CES1 high expressing cells – showed efficient ability to hydrolyse and retain ESM-HDAC528. In comparison, non-hydrolysable HDAC800 control was minimally retained intracellularly. This was reflected in enhanced anti-inflammatory effect of ESM-HDAC528 in monocytes and to lesser extent, in macrophages. ESM-HDAC528 potently inhibited IL-6 and TNF-α at doses 1000 times lower than non-hydrolysable HDAC800 control. Notably, only at higher dose ranges, ESM-HDAC528 also reduced intracellular ATP levels in both monocytes and macrophages. In this context, intracellular ATP levels were reported to positively correlate with cell proliferation and were widely used as a method to assess cell proliferation and cytotoxicity.35 The anti-inflammatory and anti-proliferative effects of ESM-HDAC528 were all absent with HDAC800 treatment at similar doses, despite having the same chemical potency, as a consequence of the CES1-mediated accumulation of ESM-HDAC528 in monocytes and macrophages. Therefore administering low doses of ESM-HDAC528 is expected to affect the CES1-expressing myeloid cells only. This feature will improve the tolerability and safety of HDACi therapeutic application through selectively targeting CES1-expressing inflammatory myeloid cells.
In the ex-vivo setting, differential CES1 expression was observed in both peripheral blood and intestinal mucosa of the healthy and the IBD environments. We defined populations of CD14+ and CD16+ monocytes in healthy donor peripheral blood expressing CES1 at single cell resolution using mass cytometry, and observed that CD14++CD16- classical monocytes showed higher CES1 expression compared with CD14+CD16++ non-classical monocytes. CES1 expression is also detected in CD2+α4β7+ DCs precursors at lower levels compared with CD14++CD16- classical monocytes, indicating that CES1 is expressed in mononuclear myeloid cells largely but not per se restricted to monocytes only.
In CD colonic mucosa, CES1 expression is largely seen in CD68+ tissue macrophages. This confirms earlier observations that show CES1 expression confined to the monocyte-macrophage population but not infiltrating DCs.36 The CES1-expressing macrophages were more abundant in inflamed mucosa compared with non-inflamed mucosa. This may be explained by upregulation of CES1 expression in macrophages in response to local inflammatory cues, as CES1 is shown to be regulated by inflammatory NF-κB signalling.37 Another explanation could be the abundance of recently recruited CES1high classical monocytes to the inflamed colon and expansion of CES1high immature pro-inflammatory macrophages within inflamed colon. In line with that, CES1 mRNA expression – in the non-inflamed intestinal environment – is highest in the CD14+CD11c+HLA-DRint immature macrophage population [MF1], a population that corresponds to the recently recruited monocytes in early stages of macrophage differentiation, in contrast to mature macrophages that show much reduced CES1 expression.33 Interestingly the CES1high MF1 population highly co-expressed S100A12, a previously reported reliable IBD biomarker that highly correlates with inflammation severity.38 S100A12 is also enriched in inflammatory macrophages and neutrophils within intestinal mucosa and upon TLR2/1 ligand polarisation.39,40 Concurrently, a developmental trajectory analysis of inflamed intestinal MNPs reveals relative enrichment of CES1 mRNA expression in intestinal macrophages, along with S100A12. Next to S100A8 and S100A9 [also known as calprotectin], S100A12 is specifically found in human inflammatory IBD tissue macrophages and neutrophils41 and is known to correspond to disease severity.42 Such S100A proteins can endogenously activate TLR4 and subsequently induce the NF-κB signalling pathway,43,44 which in turn upregulates CES1 expression in response to inflammatory stimuli.37 This can explain the correlation between CES1 and S100A protein expression in mononuclear myeloid cells.
Mononuclear myeloid cells play a major role in murine colitis models, driving both colon inflammation and healing.35 In DSS-induced colitis, an innate immune cell driven model, inflammation is largely driven by recruitment of Ly6Chigh blood monocytes to colon and enrichment of pro-inflammatory signals (IL1β) in colon macrophages and DCs. Alternatively, CX3CR1high resident macrophages are essential to maintain colon homeostasis and tolerance.45 In T cell transfer colitis, a T cell-driven colitis model, mononuclear phagocytes are essential to process antigens and induce T cell activation and expansion in the colon.46 Whereas the CD103- DCs subset promotes IFN-γ producing T cell differentiation, the CD103+ DCs subset is essential for Treg-mediated protective effect.47
In both murine colitis models, ESM-HDAC528 demonstrated specific blood monocyte targeting at tested doses, as indicated by preferential enhanced lysine acetylation in monocytes This specific targeting reflects the restricted pattern of human CES1 expression in our transgenic CD68 promoter-driven human CES1 mice, used in our in vivo studies, which results in expression of human CES1 in mononuclear myeloid cells and subsequent ESM-HDAC528 accumulation.23 In this setting, a reduction in blood monocytes was observed. This is consistent with earlier reports showing loss of PU.1 expression in murine macrophages and subsequently myeloid cell markers like CD11b and CD115 [c-fsm] following HDAC inhibition.48 Given that PU.1 is a critical transcription factor to maintain monocyte-macrophage cell lineages,49 this effect may well explain the notable loss of circulating blood monocytes following ESM-HDAC528 treatment.
When applied in vivo, ESM-HDAC528 partially reduced some endpoints of DSS-induced colitis and concurrently affected mononuclear myeloid cell differentiation in the colon. In conjunction, peritoneal macrophages showed less LPS-induced cytokine responses. Similar effects were reported with the same compound in a peritonitis model.27 Some colon inflammation biomarkers of mRNA expression were improved; this included CXCL1 and CXCL2, known targets of HDAC inhibition.50 The increased CCL2 and TL4 expression in the colon might be a compensatory mechanism for impaired monocyte-macrophage responses due to the specific compound targeting. However, the clinical outcome of colitis was unexpectedly moderate, with alleviation of weight loss, colonic mucosal ulceration, and crypt loss. The observed effect of ESM-HDAC528 is likely mediated through a combination of reduced chemokine secretion as well as reduced tissue monocyte differentiation and macrophage reactivity, as, in earlier studies in DSS-induced colitis, blocking monocyte recruitment to the colon ameliorated colitis.51,52 ESM-HDAC528 targeted monocytes likely attain a reduced ability to induce gene expression allowing macrophages differentiation. Additionally, ESM-HDAC528 significantly improved multiple key clinical outcomes of T cell transfer colitis; attenuating colon inflammation and reducing serum IFN-γ, TNF-α, and CCL2. A reduced colon CCL2 level was observed, a key inflammatory biomarker in this model53 indicative of less severe colonic inflammation. HDAC inhibitors are reported to modulate DC functions, compromising T cell stimulatory capacity54 and secretion of IL-12,54,55 a key Th1 polarising cytokine. HDAC inhibitor-treated DCs are less able to support Th1 cell skewing, resulting in less T cell IFN-γ secretion,56 while supporting type 1 T regulatory cell polarisation,57 in agreement with our observations. The observed discrepancy between the outcome of the two colitis models, may be attributed to their different predominant mechanisms driving inflammation.58 Earlier studies show that macrophage-DC depletion strategies to aggravate DSS induced colitis largely, mediated by increased colon CXCL1 expression and neutrophils infiltration.59,60 Several studies have addressed targeting DCs and macrophages in multiple murine colitis models, with conflicting outcomes.52,61,62 This is largely attributed to the dual protective and inflammatory roles that these cell populations play in pathogenesis of colitis, depending on their phenotype or predominant subset.
Our data suggest that CES1 expression is more enriched in inflammatory subsets of mononuclear myeloid cells in the human setting. In contrast to that, transgenic CD68 promoter-driven human CES1 expression in the murine system [CES1/ES1lo mice] shows similar CES1 expression and activity among different mononuclear myeloid cell subsets. Therefore, due to discrepancy between CES1 regulation in human vs transgenic murine systems, translating in vivo finding from murine studies to human should be done with care. A potential explanation of less efficacy in DSS-induced colitis model could be the universal targeting of all macrophages and DC subsets including anti-inflammatory subsets that play a role to limit colon inflammation in the DSS model; therefore any beneficial effect from targeting monocyte and pro-inflammatory macrophage subsets can be mitigated by dampening anti-inflammatory subsets activity. Unlike in humans where CES1 seems to be more expressed in inflamed setting, a beneficial therapeutic effect can be potentially achieved by refining treatment dosing.
Together, we addressed CES1 expression in a variety of IBD intestinal tissue and established high CES1 expression in pro-inflammatory cells. Given their high sensitivity to HDACi, and strong potential to drive excess inflammatory pathology and tissue damage, our findings warrant further application of ESM-based small molecule delivery to specific target cells in IBD.
Supplementary Material
Contributor Information
Ahmed M I Elfiky, Tytgat Institute for Liver and Intestinal Research, Amsterdam Gastroenterology & Metabolism, Amsterdam University Medical Centers, Amsterdam, The Netherlands; Immunology Research Unit, GSK Medicines Research Centre, Stevenage, UK.
Mohammed Ghiboub, Tytgat Institute for Liver and Intestinal Research, Amsterdam Gastroenterology & Metabolism, Amsterdam University Medical Centers, Amsterdam, The Netherlands; Immunology Research Unit, GSK Medicines Research Centre, Stevenage, UK.
Andrew Y F Li Yim, Tytgat Institute for Liver and Intestinal Research, Amsterdam Gastroenterology & Metabolism, Amsterdam University Medical Centers, Amsterdam, The Netherlands; Immunology Research Unit, GSK Medicines Research Centre, Stevenage, UK; Department of Clinical Genetics, Amsterdam Reproduction & Development, Amsterdam University Medical Centers, Amsterdam, The Netherlands.
Ishtu L Hageman, Tytgat Institute for Liver and Intestinal Research, Amsterdam Gastroenterology & Metabolism, Amsterdam University Medical Centers, Amsterdam, The Netherlands.
Jan Verhoeff, Department of Molecular Cell Biology & Immunology, Amsterdam Infection & Immunity Institute and Cancer Center Amsterdam, Amsterdam University Medical Centers, Amsterdam, The Netherlands.
Manon de Krijger, Tytgat Institute for Liver and Intestinal Research, Amsterdam Gastroenterology & Metabolism, Amsterdam University Medical Centers, Amsterdam, The Netherlands.
Patricia H P van Hamersveld, Tytgat Institute for Liver and Intestinal Research, Amsterdam Gastroenterology & Metabolism, Amsterdam University Medical Centers, Amsterdam, The Netherlands.
Olaf Welting, Tytgat Institute for Liver and Intestinal Research, Amsterdam Gastroenterology & Metabolism, Amsterdam University Medical Centers, Amsterdam, The Netherlands.
Iris Admiraal, Tytgat Institute for Liver and Intestinal Research, Amsterdam Gastroenterology & Metabolism, Amsterdam University Medical Centers, Amsterdam, The Netherlands.
Shafaque Rahman, Tytgat Institute for Liver and Intestinal Research, Amsterdam Gastroenterology & Metabolism, Amsterdam University Medical Centers, Amsterdam, The Netherlands.
Juan J Garcia-Vallejo, Department of Molecular Cell Biology & Immunology, Amsterdam Infection & Immunity Institute and Cancer Center Amsterdam, Amsterdam University Medical Centers, Amsterdam, The Netherlands.
Manon E Wildenberg, Tytgat Institute for Liver and Intestinal Research, Amsterdam Gastroenterology & Metabolism, Amsterdam University Medical Centers, Amsterdam, The Netherlands.
Laura Tomlinson, Discovery DMPK, IVIVT, GSK Medicines Research Centre, Stevenage, UK.
Richard Gregory, Discovery DMPK, IVIVT, GSK Medicines Research Centre, Stevenage, UK.
Inmaculada Rioja, Immunology Research Unit, GSK Medicines Research Centre, Stevenage, UK.
Rab K Prinjha, Immunology Research Unit, GSK Medicines Research Centre, Stevenage, UK.
Rebecca C Furze, Immunology Research Unit, GSK Medicines Research Centre, Stevenage, UK.
Huw D Lewis, Immunology Research Unit, GSK Medicines Research Centre, Stevenage, UK.
Palwinder K Mander, Immunology Research Unit, GSK Medicines Research Centre, Stevenage, UK.
Sigrid E M Heinsbroek, Tytgat Institute for Liver and Intestinal Research, Amsterdam Gastroenterology & Metabolism, Amsterdam University Medical Centers, Amsterdam, The Netherlands.
Matthew J Bell, Immunology Research Unit, GSK Medicines Research Centre, Stevenage, UK.
Wouter J de Jonge, Tytgat Institute for Liver and Intestinal Research, Amsterdam Gastroenterology & Metabolism, Amsterdam University Medical Centers, Amsterdam, The Netherlands; Department of Surgery, University of Bonn, Bonn, Germany.
Funding
This project is funded by the European Union’s Horizon 2020 research and innovation programme under Grant Agreement No. ITN-2014-EID-641665. PH, OW, IA, SR SH, and WJ are funded by a grant from Dutch Economic Affairs Top Sector Life Sciences & Health (LSH) - Top Consortia for Knowledge and Innovation’s (TKI), grants no. TKI-LSH T2017, and European Crohn’s and Colitis Organization (ECCO) Pioneer Grant, 2018.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. LT, RG, IR, RP, RF, HL, PM, and MB were employed by GSK at the time of conducting this study. AE, MG, MK, IH, PH, OW, IA, SR, SH, JV, JG, and WJ were employed by Amsterdam University Medical Centers at the time of conducting this study.
Author Contributions
Conduct of the study, laboratory work, and writing of the manuscript: AE; study design: AE, WJ, MB and SH; mass cytometry analyses: JV and JJG; bioinformatics analysis: AY; mass spectrometry analyses: LT and RG; patient sample collection: MK and IH; supervision: WJ, MB, and SH; reviewing and editing: WJ, MG, MB, RF, HL, PM, IR, and RB; in vivo studies and technical support: AE, OW, PH, IA, SR, MG, and SH. All authors have read and agreed to the published version of the manuscript.
References
- 1. Gregoretti IV, Lee YM, Goodson HV. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J Mol Biol 2004;338:17–31. [DOI] [PubMed] [Google Scholar]
- 2. Sweet MJ, Shakespear MR, Kamal NA, Fairlie DP. HDAC inhibitors: modulating leukocyte differentiation, survival, proliferation and inflammation. Immunol Cell Biol 2012;90:14–22. [DOI] [PubMed] [Google Scholar]
- 3. Singh AK, Bishayee A, Pandey AK. Targeting histone deacetylases with natural and synthetic agents: an emerging anticancer strategy. Nutrients 2018;10:731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li Y, Wang F, Chen X, et al. Zinc-dependent Deacetylase [HDAC] inhibitors with different zinc binding groups. Curr Top Med Chem 2019;19:223–41. [DOI] [PubMed] [Google Scholar]
- 5. Subramanian S, Bates SE, Wright JJ, Espinoza-Delgado I, Piekarz RL. Clinical toxicities of histone deacetylase inhibitors. Pharmaceuticals 2010;3:2751–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Van Veggel M, Westerman E, Hamberg P. Clinical pharmacokinetics and pharmacodynamics of panobinostat. Clin Pharmacokinet 2018;57:21–9. [DOI] [PubMed] [Google Scholar]
- 7. Das Gupta K, Shakespear MR, Iyer A, Fairlie DP, Sweet MJ. Histone deacetylases in monocyte/macrophage development, activation and metabolism: refining HDAC targets for inflammatory and infectious diseases. Clin Transl Immunology 2016;5:e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nijhuis L, Peeters JGC, Vastert SJ, van Loosdregt J. Restoring T cell tolerance, exploring the potential of histone deacetylase inhibitors for the treatment of juvenile idiopathic arthritis. Front Immunol 2019;10:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Høgh Kølbæk Kjær AS, Brinkmann CR, Dinarello CA, et al. The histone deacetylase inhibitor panobinostat lowers biomarkers of cardiovascular risk and inflammation in HIV patients. AIDS 2015;29:1195–200. [DOI] [PubMed] [Google Scholar]
- 10. Brinkmann CR, Højen JF, Rasmussen TA, et al. Treatment of HIV-infected individuals with the histone deacetylase inhibitor panobinostat results in increased numbers of regulatory T cells and limits ex vivo lipopolysaccharide-induced inflammatory responses. mSphere 2018;3:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Choi SW, Gatza E, Hou G, et al. Histone deacetylase inhibition regulates inflammation and enhances Tregs after allogeneic hematopoietic cell transplantation in humans. Blood 2015;125:815–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Glauben R, Batra A, Fedke I, et al. Histone hyperacetylation is associated with amelioration of experimental colitis in mice. J Immunol 2006;176:5015–22. [DOI] [PubMed] [Google Scholar]
- 13. Glauben R, Siegmund B. Molecular basis of histone deacetylase inhibitors as new drugs for the treatment of inflammatory diseases and cancer. Methods Mol Biol 2009;512:365–76. [DOI] [PubMed] [Google Scholar]
- 14. Joosten LA, Leoni F, Meghji S, Mascagni P. Inhibition of HDAC activity by ITF2357 ameliorates joint inflammation and prevents cartilage and bone destruction in experimental arthritis. Mol Med 2011;17:391–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bombardo M, Saponara E, Malagola E, et al. Class I histone deacetylase inhibition improves pancreatitis outcome by limiting leukocyte recruitment and acinar-to-ductal metaplasia. Br J Pharmacol 2017;174:3865–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Friedrich M, Gerbeth L, Gerling M, et al. HDAC inhibitors promote intestinal epithelial regeneration via autocrine TGFβ1 signalling in inflammation. Mucosal Immunol 2019;12:656–67. [DOI] [PubMed] [Google Scholar]
- 17. Cui SN, Chen ZY, Yang XB, et al. Trichostatin A modulates the macrophage phenotype by enhancing autophagy to reduce inflammation during polymicrobial sepsis. Int Immunopharmacol 2019;77:105973. [DOI] [PubMed] [Google Scholar]
- 18. Ali MN, Choijookhuu N, Takagi H, et al. The HDAC inhibitor, SAHA, prevents colonic inflammation by suppressing pro-inflammatory cytokines and chemokines in DSS-induced colitis. Acta Histochem Cytochem 2018;51:33–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rius M, Lyko F. Epigenetic cancer therapy: rationales, targets and drugs. Oncogene 2012;31:4257–65. [DOI] [PubMed] [Google Scholar]
- 20. Hadley M, Noonepalle S, Banik D, Villagra A. Functional analysis of HDACs in tumorigenesis. In: Brosh RM Jr, editor. Protein Acetylation: Methods and Protocols. New York, NY: Springer; 2019: 279–307. [DOI] [PubMed] [Google Scholar]
- 21. Suraweera A, O’Byrne KJ, Richard DJ. Combination therapy with histone deacetylase inhibitors [HDACi] for the treatment of cancer: achieving the full therapeutic potential of HDACi. Front Oncol 2018;8:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shah RR. Safety and tolerability of histone deacetylase [HDAC] inhibitors in oncology. Drug Saf 2019;42:235–45. [DOI] [PubMed] [Google Scholar]
- 23. Needham LA, Davidson AH, Bawden LJ, et al. Drug targeting to monocytes and macrophages using esterase-sensitive chemical motifs. J Pharmacol Exp Ther 2011;339:132–42. [DOI] [PubMed] [Google Scholar]
- 24. Jones GR, Bain CC, Fenton TM, et al. Dynamics of colon monocyte and macrophage activation during colitis. Front Immunol 2018;9:2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kamada N, Hisamatsu T, Okamoto S, et al. Unique CD14 intestinal macrophages contribute to the pathogenesis of Crohn disease via IL-23/IFN-gamma axis. J Clin Invest 2008;118:2269–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Martin JC, Chang C, Boschetti G, et al. Single-cell analysis of Crohn’s disease lesions identifies a pathogenic cellular module associated with resistance to anti-TNF therapy. Cell 2019;178:1493–508.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Luque-Martin R, Van den Bossche J, Furze RC, et al. Targeting histone deacetylases in myeloid cells inhibits their maturation and inflammatory function with limited effects on atherosclerosis. Front Pharmacol 2019;10:1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Finck R, Simonds EF, Jager A, et al. Normalization of mass cytometry data with bead standards. Cytometry A 2013;83:483–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zunder ER, Finck R, Behbehani GK, et al. Palladium-based mass tag cell barcoding with a doublet-filtering scheme and single-cell deconvolution algorithm. Nat Protoc 2015;10:316–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Van Gassen S, Callebaut B, Van Helden MJ, et al. FlowSOM: using self-organizing maps for visualization and interpretation of cytometry data. Cytometry A 2015;87:636–45. [DOI] [PubMed] [Google Scholar]
- 31. Crawford K, Gabuzda D, Pantazopoulos V, et al. Circulating CD2+ monocytes are dendritic cells. J Immunol 1999;163:5920–8. [PubMed] [Google Scholar]
- 32. Di Pucchio T, Lapenta C, Santini SM, Logozzi M, Parlato S, Belardelli F. CD2+/CD14+ monocytes rapidly differentiate into CD83+ dendritic cells. Eur J Immunol 2003;33:358–67. [DOI] [PubMed] [Google Scholar]
- 33. Bujko A, Atlasy N, Landsverk OJB, et al. Transcriptional and functional profiling defines human small intestinal macrophage subsets. J Exp Med 2018;215:441–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gough PJ, Gordon S, Greaves DR. The use of human CD68 transcriptional regulatory sequences to direct high-level expression of class A scavenger receptor in macrophages in vitro and in vivo. Immunology 2001;103:351–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Crouch SP, Kozlowski R, Slater KJ, Fletcher J. The use of ATP bioluminescence as a measure of cell proliferation and cytotoxicity. J Immunol Methods 1993;160:81–8. [DOI] [PubMed] [Google Scholar]
- 36. Satoh T, Hemmerlein B, Zschunke F, Radzun HJ. In situ detection of human monocyte/macrophage serine esterase-1 mRNA expression in human tissues. Pathobiology 1999;67:158–62. [DOI] [PubMed] [Google Scholar]
- 37. Capece D, D’Andrea D, Begalli F, et al. Enhanced triacylglycerol catabolism by carboxylesterase 1 promotes aggressive colorectal carcinoma. J Clin Invest 2021;131:e137845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sidler MA, Leach ST, Day AS. Fecal S100A12 and fecal calprotectin as noninvasive markers for inflammatory bowel disease in children. Inflamm Bowel Dis 2008;14:359–66. [DOI] [PubMed] [Google Scholar]
- 39. Realegeno S, Kelly-Scumpia KM, Dang AT, et al. S100A12 is part of the antimicrobial network against Mycobacterium leprae in human macrophages. PLoS Pathog 2016;12:e1005705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Heilmann RM, Nestler J, Schwarz J, et al. Mucosal expression of S100A12 [calgranulin C] and S100A8/A9 [calprotectin] and correlation with serum and fecal concentrations in dogs with chronic inflammatory enteropathy. Vet Immunol Immunopathol 2019;211:64–74. [DOI] [PubMed] [Google Scholar]
- 41. Däbritz J, Musci J, Foell D. Diagnostic utility of faecal biomarkers in patients with irritable bowel syndrome. World J Gastroenterol 2014;20:363–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Planell N, Masamunt MC, Leal RF, et al. Usefulness of transcriptional blood biomarkers as a non-invasive surrogate marker of mucosal healing and endoscopic response in ulcerative colitis. J Crohns Colitis 2017;11:1335–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Foell D, Wittkowski H, Kessel C, et al. Proinflammatory S100A12 can activate human monocytes via Toll-like receptor 4. Am J Respir Crit Care Med 2013;187:1324–34. [DOI] [PubMed] [Google Scholar]
- 44. Loes AN, Bridgham JT, Harms MJ. Coevolution of the toll-like receptor 4 complex with calgranulins and lipopolysaccharide. Front Immunol 2018;9:304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Grainger JR, Konkel JE, Zangerle-Murray T, Shaw TN. Macrophages in gastrointestinal homeostasis and inflammation. Pflugers Arch 2017;469:527–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rossini V, Zhurina D, Radulovic K, et al. CX3CR1+ cells facilitate the activation of CD4 T cells in the colonic lamina propria during antigen-driven colitis. Mucosal Immunol 2014;7:533–48. [DOI] [PubMed] [Google Scholar]
- 47. Toribio-Fernández R, Herrero-Fernandez B, Zorita V, et al. Lamin A/C deficiency in CD4+ T-cells enhances regulatory T-cells and prevents inflammatory bowel disease. J Pathol 2019;249:509–22. [DOI] [PubMed] [Google Scholar]
- 48. Laribee RN, Klemsz MJ. Loss of PU.1 expression following inhibition of histone deacetylases. J Immunol 2001;167:5160–6. [DOI] [PubMed] [Google Scholar]
- 49. Rosa A, Ballarino M, Sorrentino A, et al. The interplay between the master transcription factor PU.1 and miR-424 regulates human monocyte/macrophage differentiation. Proc Natl Acad Sci U S A 2007;104:19849–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gatla HR, Muniraj N, Thevkar P, Yavvari S, Sukhavasi S, Makena MR. Regulation of chemokines and cytokines by histone deacetylases and an update on histone decetylase inhibitors in human diseases. Int J Mol Sci 2019;20:1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zigmond E, Varol C, Farache J, et al. Ly6C hi monocytes in the inflamed colon give rise to proinflammatory effector cells and migratory antigen-presenting cells. Immunity 2012;37:1076–90. [DOI] [PubMed] [Google Scholar]
- 52. Becker F, Kurmaeva E, Gavins FN, et al. A critical role for monocytes/macrophages during intestinal inflammation-associated lymphangiogenesis. Inflamm Bowel Dis 2016;22:1326–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Scheerens H, Hessel E, de Waal-Malefyt R, Leach MW, Rennick D. Characterization of chemokines and chemokine receptors in two murine models of inflammatory bowel disease: IL-10-/- mice and Rag-2-/- mice reconstituted with CD4+CD45RBhigh T cells. Eur J Immunol 2001;31:1465–74. [DOI] [PubMed] [Google Scholar]
- 54. Misaki K, Morinobu A, Saegusa J, et al. Histone deacetylase inhibition alters dendritic cells to assume a tolerogenic phenotype and ameliorates arthritis in SKG mice. Arthritis Res Ther 2011;13:R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jiang H, Zhang S, Song T, Guan X, Zhang R, Chen X. Trichostatin a protects dendritic cells against oxygen-glucose deprivation via the SRSF3/PKM2/glycolytic pathway. Front Pharmacol 2018;9:612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Brogdon JL, Xu Y, Szabo SJ, et al. Histone deacetylase activities are required for innate immune cell control of Th1 but not Th2 effector cell function. Blood 2007;109:1123–30. [DOI] [PubMed] [Google Scholar]
- 57. Kaisar MMM, Pelgrom LR, van der Ham AJ, Yazdanbakhsh M, Everts B. Butyrate conditions human dendritic cells to prime type 1 regulatory T cells via both histone deacetylase inhibition and G protein-coupled receptor 109A signaling. Front Immunol 2017;8:1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kiesler P, Fuss IJ, Strober W. Experimental models of inflammatory bowel diseases [Internet]. Cell Mol Gastroenterol Hepatol 2001;59:241–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Qualls JE, Tuna H, Kaplan AM, Cohen DA. Suppression of experimental colitis in mice by CD11c+ dendritic cells. Inflamm Bowel Dis 2009;15:236–47. [DOI] [PubMed] [Google Scholar]
- 60. Qualls JE, Kaplan AM, van Rooijen N, Cohen DA. Suppression of experimental colitis by intestinal mononuclear phagocytes. J Leukoc Biol 2006;80:802–15. [DOI] [PubMed] [Google Scholar]
- 61. Muzaki AR, Tetlak P, Sheng J, et al. Intestinal CD103[+]CD11b[-] dendritic cells restrain colitis via IFN-γ-induced anti-inflammatory response in epithelial cells. Mucosal Immunol 2016;9:336–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Steinbach EC, Plevy SE. The role of macrophages and dendritic cells in the initiation of inflammation in IBD. Inflamm Bowel Dis 2014;20:166–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.