Abstract
Polystyrene is a chemically inert synthetic aromatic polymer. This widely used form of plastic is recalcitrant to biodegradation. The exponential production and consumption of polystyrene in various sectors has presented a great environment risk and raised the problem of waste management. Biodegradation by bacteria has previously shown great potential against various xenobiotics but there are only a few reports concerning polyolefins. By screening wetland microbes, we found two bacterial species – Exiguobacterium sibiricum strain DR11 and Exiguobacterium undae strain DR14 which showed promising biodegradation potential against polystyrene. In this study, we report the degradation of non-irradiated solid polystyrene material after incubation with these isolates. Growth studies suggested that the Exiguobacterium strains utilize polystyrene as a carbon source. Moreover, our data suggest that polymer degradation was initiated by biofilm formation over the PS surface leading to alteration in the physical properties of the material. Surface property analysis by AFM revealed significantly enhanced roughness resulting in reduced surface hydrophobicity of polystyrene. Fourier-transfer infrared (FT-IR) spectroscopic analysis showed breakdown of polystyrene backbone by oxidation. The extent of deterioration was further determined by percent weight reduction of polystyrene after incubation with bacteria. Our data support the fact that strains of extremophile bacterium Exiguobacterium are capable of degrading polystyrene and can be further used to mitigate the environmental pollution caused by plastics.
Isolation and characterization Exiguobacterium sp. DR11 and DR14 from wetlands able to establish biofilm and alter polystyrene surface properties.
1. Introduction
Over the years plastic or synthetic polymers have been deeply incorporated into human life. Their high durability, resistance to change in temperature, light weight and low cost have made plastic a material of choice in each sector.1,2 Among the most widely used forms of polyolefins, polyethylene (PE), polypropylene (PP) and polystyrene (PS) are majorly used in the packaging industry.3 PS is a synthetic aromatic polymer synthesized by polymerization of styrene units. The expanded form of PS is styrofoam, and it is commonly used as a packaging material and for manufacturing disposable cups and cutlery, containers and laboratory consumables.4 Because of the extensive use and increasing demand, annual production of PS has crossed the scale of several million tonnes.5 The recalcitrant nature of plastic poses a severe environmental hazard. PS is also a major terrestrial and marine pollutant.6–9
Polymer degradation is defined as the conversion of polymer into smaller subunits by breaking bonds through physical, chemical or biological processes leading to alteration in the physico-chemical properties of the polymer. A few alternative abiotic processes are also known to play a role in polymer degradation. These include thermal, photo-oxidative, ozone-based degradation, catalytic and mechanochemical methods.10 Exposure to sunlight/UV-radiation has been reported to enhance the chain breakage in PS structure initiating degradation process. As an alternative to the conventional methods of polymer degradation, biodegradation presents itself as an economic and environment-friendly approach. Biodegradation is a complex process of physico-chemical transformation of polymers into smaller units by the action of microorganisms. It is a relatively slow process in which cleavage of polymer backbone by bond scission leads to generation of carbonyl groups (oxidation) which are considered to be utilized by the microorganism as a source of carbon and energy.11–13 There are a few reports showing biodegradation of synthetic polymers including polyethylene, polyurethane, polyvinyl chloride14–20 and their microplastic forms.21–23 PS also belongs to a non-biodegradable class of synthetic polymers12 but very limited data is available on its biodegradation. The monomeric form, styrene, has been studied and has shown degradable properties.24–27 It is considered that to initiate the process of PS biodegradation, prior exposure to photo-oxidation and thermal induction is required. This, in turn, causes bond scission and formation of new functional groups.28,29
In nature, bacteria exist in sessile communities known as biofilms. Biofilms provide protection from harsh environmental conditions and resistance to chemical stress.30,31 Studies show that biodegradation by microbial biofilms is more favourable than planktonic bacteria.32 Initial adherence of bacteria on the surface of plastics is followed by the colonization of the exposed plastic surface. It is proposed that initiation of biofilms enhances carbon utilization from non-soluble substrates such as PS and PE. Biodegradation of polymer is achieved by two types of enzymes intracellular and extracellular depolymerases. The extracellular depolymerases secreted from microorganisms convert the polymers into monomers, dimers or oligomers which can permeate through bacterial cell membrane and are utilized as a source of carbon and energy.
Biodiversity and occurrence of polymer-degrading microorganisms show variation depending on the ecological habitat, such as soil, water, compost, or sludge. Wetlands are one of the richest habitats of microbial diversity as they provide complex as well as distinct environmental conditions.33 These ecosystems are ideal grounds for isolating bacteria with novel enzymes. Members of Exiguobacterium genus are considered extremophiles as they possess extreme tolerance to temperature (−12 °C to 55 °C), halo tolerance (up to 13%) and can grow in a wide range of pH (5–11). These firmicutes thrive in most extreme environments such as ancient Siberian permafrost and hot springs to freshwater ecosystem.34–36 Yet, only one study reported the role of Exiguobacterium sp. strain YT2 in PS degradation which was isolated from the gut of a mealworm.37 In this study, we are reporting initiation of PS degradation by Exiguobacterium sp. strains DR11 and DR14 isolated from Dadri wetlands, India. PS degradation by Exiguobacterium sp. strains DR11 and DR14 was verified by analysis of various parameters such as FT-IR analysis, hydrophobicity test and AFM imaging. We are also reporting biofilm formation over the PS surface by the Exiguobacterium strains.
2. Materials and methods
2.1. Sample collection
Water samples were collected in sterile glass bottles from Dadri wetland, Uttar Pradesh, India (coordinates – 28°31′30.7′′N77°34′40.1′′E) in April 2014. Collected samples were taken immediately for further processing which included serial dilution and filtration through Whatman filter sheets, 0.22 μm-pore size (GE Healthcare, India). Filter papers having concentrated sample were kept over different media plates including Luria–Bertani (LB) agar, tryptic soy broth (TSB) agar and nutrient agar (NA) plates in triplicates. After 2 h, filter papers were removed and plates were incubated overnight at 30 °C. The next day, yellow and orange coloured colonies were selected and streaked on fresh media plates. All media were procured from Himedia Pvt. Ltd., India.
2.2. Bacterial strains and culture conditions
For initial screening and isolation NB, LB and TSB were selected. Nutrient agar (NA) and skim milk agar (SMA) (Himedia Pvt. Ltd., India) was used for motility and proteolytic assays respectively. After identification for subsequent experiments Exiguobacterium sp. strain DR11 and Exiguobacterium sp. strain DR14 were grown in nutrient broth (NB) on a shaking incubator (200 rpm) at 30 °C. To detect the activity of PS degradation, liquid carbon-free basal medium (LCFBM) was used containing (g l−1): 0.7 g of KH2PO4, 0.7 g of K2HPO4, 0.7 g of MgSO4·7H2O, 1.0 g of NH4NO3, 0.005 g of NaCl, 0.002 g of FeSO4·7H2O, 0.002 g of ZnSO4·7H2O, and 0.001 g of MnSO4·H2O.37
2.3. Identification of bacterial species
For identification, DNA was isolated conventionally and then 16S rRNA gene sequence was amplified by PCR using universal primers: 27F – (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492R – (5′-TACGGYTACCTTGTTACGACTT-3′). PCR reaction mixture containing Taq polymerase (NEB) was set under the following cycling conditions: 94 °C for 10 min, (94 °C for 40 s, 56 °C for 1 min, 74 °C for 2 min) X30 cycles, 74 °C for 10 min using Eppendorf thermal master cycler (Eppendorf, Hamburg, Germany). PCR amplified products were purified using PCR purification kit (Qiagen India Pvt Ltd) and sent for sequencing (SciGenom Labs Pvt Ltd, Kerala, India). The sequence similarity was verified by using basic local alignment search tool (BLAST) on NCBI (http://www.ncbi.nlm.nih.gov/). Sequences of both the isolates, Exiguobacterium sp. strain DR11 and DR14, have been submitted in the NCBI (Genbank accession ID numbers – MG645229, MG645233 respectively). Both the strains are publically deposited in Microbial Type Culture Collection (MTCC 12887, 12888), Chandigarh, India and are available for public use.
2.4. Polystyrene material
Two types of PS material were used for this study including (1 × 1) cm cut pieces of optically clear, sterile polystyrene Petri plates, 90 mm (Himedia Lab Pvt Ltd, Mumbai, India) and PS beads, atactic (Alfa Aesar, Massachusetts, USA).
2.5. Polystyrene biodegradation assay
Biodegradation test was performed with PS chips which were weighed, sterilized (70% ethanol) for an hour, dried inside biosafety cabinet and then added to the Erlenmeyer flasks (150 ml capacity) containing 40 ml LCFBM. Overnight culture grown in LB broth was washed twice with saline water to remove residual medium then cells were collected after centrifugation (8000 rpm, 5 min). After that, the pellet was resuspended in saline water to maintain the cell concentration approximately 108 cells per ml for inoculation. An uninoculated flask containing growth medium was used as control. Flasks were incubated for 30 days at 30 °C. Similarly, the experiment was executed with PS beads. At the end of 30 days of incubation, PS chips were removed, washed and dried for further analysis. All experiments were performed independently three times.
2.6. Growth assessment of bacterial strains
Exiguobacterium sp. strain DR11 and DR14 were inoculated (108 cells per ml, ∼0.1 OD600) separately in LCFBM with glucose (0.2%) and sterilized PS beads (1.0 g). The growth among cultures was compared by observing the absorbance over 45 days of incubation at 30 °C under shaking conditions. The growth curve was plotted using OriginPro 8 software.
2.7. Biofilm assay
Biofilm formation of the strains over PS plates was assessed initially using a range of media (LB, NB and TSB). Standard biofilm formation assay was followed with slight modifications.38 Briefly, cultures of Exiguobacterium sp. strain DR11 and DR14 were grown for 18 h in TSB medium at 30 °C. The next day, cultures were re-inoculated (1 : 100 dilution) and seeded in 24-well plates (non-coated, flat bottom, Himedia Labs Pvt. Ltd., India). To study multi-species biofilm formation by both strains DR11 and DR14, equal volume of diluted cultures (1 : 100) were seeded in 6-well plates. In co-culture experiment, cell numbers were kept equivalent to the mono-species biofilm experiment. After 48 h of incubation at 30 °C (without agitation), cultures were removed and washed twice with deionized water. For staining, 1 ml of 0.1% solution of crystal violet was added per well and incubated at room temperature for 15 minutes then rinsed thrice as described previously. After drying the plates overnight, biofilms were quantified by adding 1 ml 30% glacial acetic acid per well. Absorbance (OD550 nm) was measured at 550 nm. Similarly, biofilm formation was tested on PS pieces kept inside 6-well plates. Experiments were done in triplicate and repeated independently three times. Bright field microscopy was performed using Leica-DM IL LED model (Leica Microsystems) to image biofilm. Images were taken at 40× magnification.
2.8. Live–dead staining
Biofilms were grown in 35 mm non-coated polystyrene Petri dishes (Thermo Scientific) after 48 h of incubation in TSB growth medium at 30 °C. After the incubation period, media containing suspension cells were removed and the biofilm was washed twice with 1× PBS. Biofilms were treated with 1 : 1 ratio of SYTO9 : propidium iodide mixture (final concentration of SYTO9 = 6 μM; PI = 30 μM) using LIVE/DEAD BacLight bacterial viability kit (Invitrogen-Molecular probes). Biofilms were incubated at room temperature in the dark for 15 min followed by a single wash with 1× PBS. Biofilms were observed using a 40× objective and Nikon Eclipse Ti microscope with 485 nm and 535 nm excitation wavelengths and 498 nm, 617 nm emission filters (green fluorescence represents SYTO-9 and red is depicted by propidium iodide).
2.9. Evaluation of polystyrene hydrophobicity
Water contact angle (WCA) was used to analyze the changes in surface hydrophobicity. For complete removal of bacteria from PS surface, samples were treated with 2% SDS followed by multiple washes with sterile distilled water. The hydrophobicity of PS samples inoculated with and without Exiguobacterium strains were analyzed by measuring contact angle (Krüss GmbH, DSA-25) through software-controlled hanging drop method. On the surface of PS material, 2 μl of deionized water was dropped and images were taken immediately by an attached CCD camera having magnifying lens. CA readings were computed after multiple recordings to verify repeatability.
2.10. FT-IR spectroscopy of polystyrene material
Changes in the PS structure with subsequent incubation with bacteria were analyzed by Fourier-transfer infrared (FT-IR) spectroscopy (Nicolet iS5 spectrometer assembled with iD5-ATR accessory) in the range of 3500–500 cm−1. Three different sets of PS samples – untreated (medium without bacteria) and treated with Exiguobacterium sp. strain DR11 and DR14 were analyzed after 30 days of incubation as described previously.
2.11. Atomic force microscopy
AFM (PARK, XE-007) imaging was performed to observe any changes in the surface structure and smoothness of bacterial treated PS material. After 30 days of incubation, PS pieces were treated with 2% sodium-dodecyl sulfate (SDS) to remove bacteria from the surface of PS followed by multiple washing with Milli-Q water, dried overnight inside the incubator and analyzed thereafter. Images were taken in non-contact mode with the scan speed of 0.4 Hz and resolution of 256 × 256 pixels.
2.12. Determination of percent weight reduction
To determine the ability of Exiguobacterium strain DR11 and DR14 to reduce the weight of PS the following method was employed. The bacterial strains were grown overnight in LB medium and next day washed thrice with saline water to remove any traces of previous medium. The cells were inoculated (108 cells per ml) in 50 ml LCFBM in 150 ml Erlenmeyer conical flasks with PS material. After 30 days of incubation, broth was filtered to recover PS pieces. Samples were rinsed in 70% ethanol, washed three times with MilliQ water and dried for 2–4 h inside biosafety cabinet. The reduction in polymer weight was measured and compared with respect to the control incubated with PS in the absence of bacteria. The extent of polymer degradation was estimated by calculating the percent weight loss using the following formula: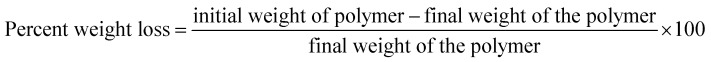
3. Results and discussion
3.1. Isolation and identification of bacteria
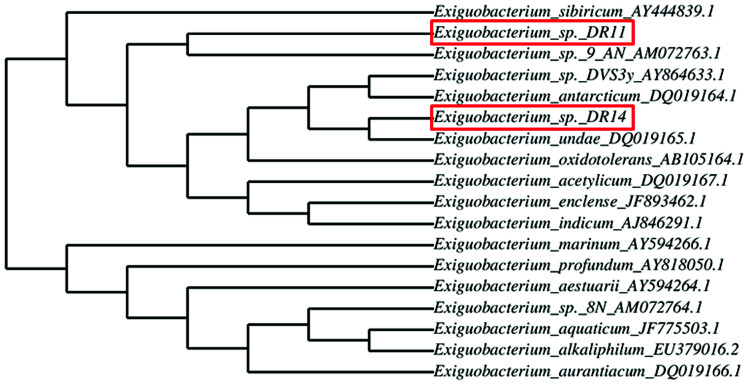
Exiguobacterium strains were isolated from the wetland by serially diluting the collected soil and water samples followed by incubation at 30 °C within various media such as LB, NB, and TSB. After 24 h of incubation, yellow and orange coloured colonies were re-streaked. Two Gram-positive, motile rods were selected and further analysed. After DNA isolation, 16S rRNA gene was amplified using universal primers and sent for sequencing. BLAST analysis of 16S rRNA gene sequencing of isolates DR11 and DR14 showed 97% and 94% similarity to two species of genus Exiguobacterium – E. sibiricum and E. undae respectively. To display the degree of relatedness amongst isolated strains and known Exiguobacterium species, a phylogenetic tree was constructed using phylogeny.fr software (Fig. 1). It was observed that strain DR11 was closely related to Exiguobacterium sibiricum (Accession number – AY444839.1) whereas isolate DR14 has shown highest similarity to Exiguobacterium undae (Accession no. DQ719164.1). Next, strains were morphologically characterized (Fig. S1a and S1b†) and optimum culture conditions were determined. Further characterization showed that the strains were psychrotrophic and display high proteolytic activity as indicated by the formation of zone of clearance on skim-milk agar plates after overnight incubation at 30 °C (Fig. S1c and S1d†).
Fig. 1. Dendrogram showing relatedness between Exiguobacterium species type strains and strains isolated from the wetland. The relation between isolated strains (highlighted in red box) with the known Exiguobacterium species was analyzed by comparing their 16S rRNA gene sequences using the phylogeny.fr program. Accession numbers are indicated prior to the species name.
3.2. Analysis of biofilm formation on polystyrene surface
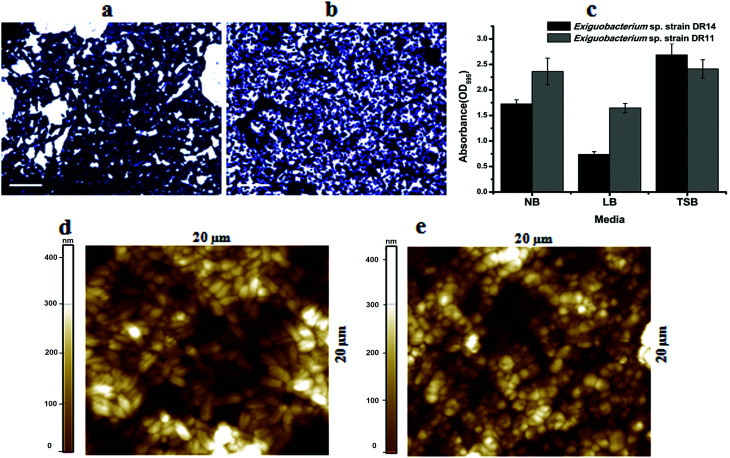
Exiguobacterium species are involved in biofilm formation and were isolated previously from natural biofilm communities, such as ship hull39 and water pipelines in the Gulf of Mexico.40Exiguobacterium antarcticum B7 was isolated from biofilms in Ginger Lake, Antarctica.41 Studies also observed Exiguobacterium sp. biofilms on synthetic polymer surfaces.37,42 Based on these findings we tested whether Exiguobacterium strains isolated in this study can establish mono-species biofilm by performing static biofilm assay in 96-well plates. After crystal violet staining, it was found that both strains were able to establish biofilms. We found that although these psychrotrophic isolates can grow in various growth media they make robust biofilms only in high nutrient conditions. In a comparative study among three nutrient-rich growth media – LB, NB and TSB, biofilm growth was more robust in TSB for both the strains. Although, strain DR11 formed equivalent biofilm in NB and TSB, both strains showed reduced biofilm formation in LB medium (Fig. 2a–c).
Fig. 2. Biofilm formation by Exiguobacterium species on PS surface. Crystal violet stained biofilm images of (a) Exiguobacterium sp. strain DR14, (b) Exiguobacterium sp. strain DR11 and (c) quantification with various growth media formed in 24-well plates after 48 h incubation at 30 °C without agitation. Scale bar is equivalent to 10 μm. Atomic force microscopy images showing biofilm formation over the PS surface by Exiguobacterium sp. strain DR14 (d) and Exiguobacterium sp. strain DR11 (e) under similar conditions. Colour bar indicates thickness of biofilm over the surface.
It is proposed that bacteria in biofilms display more potential for biodegradation. Because of the hydrophobic nature of PS surface, for adhesion, bacterial surface should also possess similar property. Both strains DR11 and DR14 were able to colonize PS chips and crystal violet staining and bright field imaging displayed robust biofilm adhered to the PS surface (Fig. 2a–c). Biofilms initiated on the PS chips were further analyzed by AFM imaging (Fig. 2d, e and S2†). AFM imaging revealed that cell morphology was altered in these biofilms. Exiguobacterium sp. DR14 cells in biofilms were short oval-shaped rods similar to coccobacilli as compared to rod-shaped cells observed in planktonic growth (Fig. 2d, e and S2†). Exiguobacterium sp. DR11 showed elongated rod-shaped cells within biofilms, while it displayed small rod morphology in exponential phase culture (Fig. 2).34 Alteration in cell shape has been reported previously for Exiguobacterium where E. undae, similar to strain DR14, reduce in size in stationary phase. E. sibiricum having similarity with isolated strain DR11 also alters cell shape, forming elongated cells with the change in temperature.34,43
Yang Y. et al.37 reported degradation of pliable PS films by Exiguobacterium strains both in cell suspension and biofilm cultures. In our study we used hard PS chips or PS beads as substrate to mimic the hard plastic materials. Even on hard PS chips both Exiguobacterium sp. DR11 and Exiguobacterium sp. DR14 were able to establish biofilms (Fig. 2). Bacterial growth as biofilms on solid substrates is proposed to enhance carbon availability. Yet, hydrophobicity of PS material acts as a deterrent for biofilm establishment. Thus, microorganisms to be used as potential biodegradation agents should be able to colonize PS surfaces. Both isolates DR11 and DR14 were able to overcome the hydrophobicity of PS and establish robust biofilms. Our results suggest that Exiguobacterium biofilm establishment on PS does not require the addition of a surfactant in the growth medium.
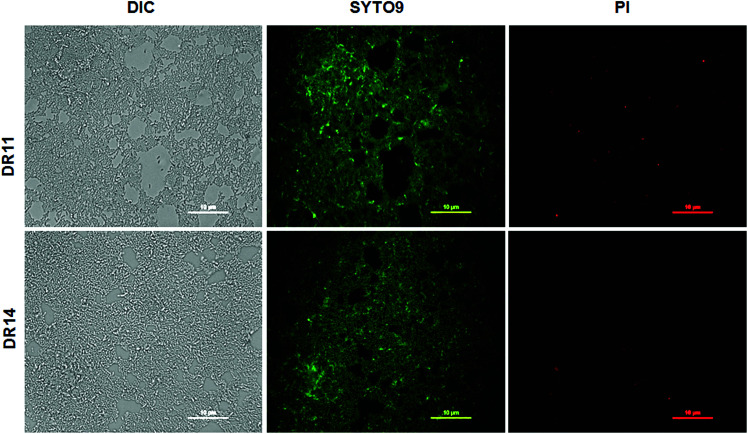
In nature bacteria establish multi-species biofilm communities. These multi-species biofilms are influenced by both cooperative and antagonistic interaction between the microbes in the community.44,45 Since both Exiguobacterium sp. were isolated from aquatic habitats we probed whether these bacterial strains can establish biofilms in co-cultivation systems. Under co-culturing conditions both Exiguobacterium sp. DR11 and Exiguobacterium sp. DR14 are able to colonize the plastic surfaces establishing a dense multi-species biofilm (Fig. S3a–c†) The co-culture biofilm was robust and displayed significantly higher cell density when compared mono-species biofilms formed by either of the Exiguobacterium strains (Fig. S3d†). These results suggest that both Exiguobacterium sp. DR11 and Exiguobacterium sp. DR14 probably exhibit cooperative interaction in mixed biofilm communities. Exiguobacterium sp. DR11 and Exiguobacterium sp. DR14 are able to establish biofilms in 48 h on PS surfaces in static culture conditions. To test whether cell viability is compromised during the incubation period, live–dead staining on biofilms established on PS surfaces was performed as described in material and methods. As seen in Fig. 3 dense aggregation of live cells was observed in biofilms formed by both Exiguobacterium strains (Fig. 3). Overall our data shows that both Exiguobacterium sp. DR11 and Exiguobacterium sp. DR14 can colonize PS surfaces and establish both mono and co-culture biofilms.
Fig. 3. Viability of Exiguobacterium sp. DR11 and DR14 biofilms. Upper panel represents biofilm formed by Exiguobacterium sp. DR11 and the lower panel show biofilm formed by strain DR14. Biofilms were stained with LIVE/DEAD BacLight bacterial viability kit (Invitrogen-Molecular probes) after 48 h of incubation at 30 °C. Green fluorescence signifies live cells stained by SYTO9 dye whereas red dye indicates dead cells stained by propidium iodide. Scale bar equals to 10 μm.
3.3. Exiguobacterium strain can utilize PS substrates for growth
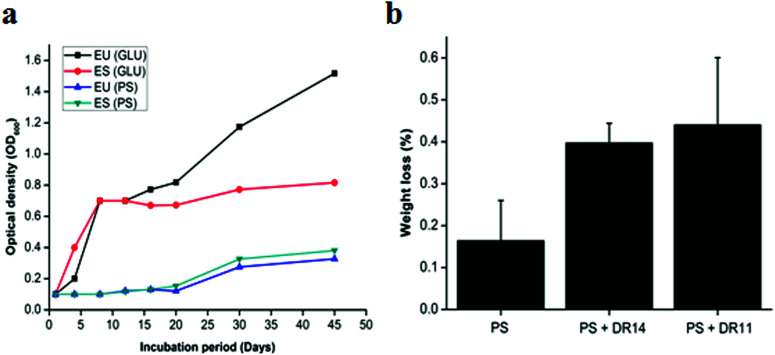
Bacteria having biodegradation properties are able to utilize PS as a carbon source for growth. To the best of our knowledge no reports were available whether Exiguobacterium sp. can utilize PS as sole carbon source and to investigate this we propagated Exiguobacterium sp. in LCFBM in absence of glucose supplemented PS beads. While the control cells displayed rapid growth in presence of glucose, Exiguobacterium cells growing with PS beads showed a longer lag phase and slow growth (Fig. 4a). However, after 20 days there was steady increase in optical density of both Exiguobacterium sibiricum strain DR11 and Exiguobacterium undae strain DR14, with OD reaching ∼0.4 in 45 days (Fig. 4a). These results indicate that both Exiguobacterium strains are able to sustain growth on PS substrates. Moreover, our data shows that both Exiguobacterium sibiricum strain DR11 and Exiguobacterium undae strain DR14 can utilize PS substrates for growth further enhancing their potential for biodegradation (Fig. 4a).
Fig. 4. (a) Exiguobacterium strains growth with glucose and polystyrene as a carbon source. Strains incubated in LCFBM medium with additional glucose and polystyrene beads in separate flasks for 45 days at 30 °C under shaking conditions. Growth was monitored at regular intervals. Advancement in growth was seen in the cultures incubated with polystyrene after 20 days of incubation suggesting that Exiguobacterium strains started utilizing PS as a carbon source. ES and EU represents Exiguobacterium sibiricum strain DR11 and Exiguobacterium undae strain DR14 respectively. (b) Percent weight loss of polystyrene by Exiguobacterium strains. E. sibiricum DR11 and strain DR14 cells were inoculated in LCBF medium (108 cells per ml) with polystyrene pieces and incubated for 30 days at 30 °C. Control was represented by LCFBM with PS without bacteria.
To further examine the PS degradation potential of Exiguobacterium strain DR11 and DR14 weight reduction of PS chips was measured after prolonged incubation with bacterial isolates (Fig. 4b). The cells were inoculated in LCFBM with PS material. After 30 days of incubation, broth was filtered to recover PS pieces. After ethanol wash the PS pieces were dried and weight was measured. The reduction in polymer weight was measured and compared with respect to the control incubated with PS in the absence of bacteria and percentage reduction was calculated as described in material and method. Both strain DR11 and DR14 caused weight reduction of the PS chip. Weight reduction was 0.4 and 0.44% for DR11 and DR14 respectively in 50 ml LCFBM (PS weight = 250 mg/50 ml) which is equivalent to 8% (DR11) and 8.8% (DR14) weight loss per liter medium (Fig. 4b). This was significantly higher than the reported weight reduction of 0.8% by Rhodococcus ruber C208 (ref. 46) and similar to the previous report by Yang Y. et al.37 where they showed 7.4% weight loss of PS by Exiguobacterium sp. YT2 after 60 days of incubation.
Pre-treatment of PE or PS by UV photooxidation, thermal or chemical oxidation methods enhances biodegradation. This synergistic effect of photooxidation is proposed to increase the availability of carbonyl groups on the surface of the plastic material which can be utilized by bacterial cells. In our study, non-irradiated PS chips were employed as a substrate for biodegradation. Surprisingly, both strain DR11 and DR14 were able to not only form biofilm on non-irradiated PS, but cause weight reduction indicating that Exiguobacterium species are capable of degrading unmodified PS. Since not many bacteria can initiate PS degradation in absence of photooxidation, these Exiguobacterium strains could be exploited as early initiators in PS biodegradation process.
3.4. Surface property analysis of degraded polystyrene material
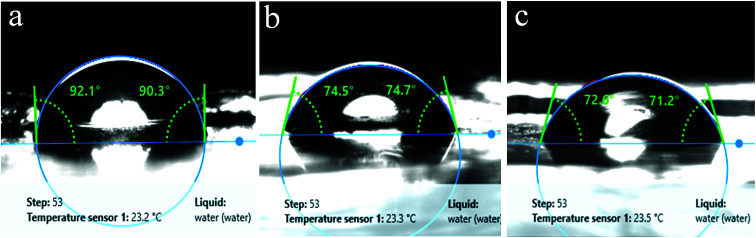
In a previous study, Exiguobacterium sp. isolated from the gut of meal worm were found to be capable of degrading PS.37 In this study, we tested the potential of our strains obtained from a wetland for PS degradation. Exiguobacterium isolates were grown in growth medium having PS chips/beads as sole carbon source as described in material and methods section. After 30 days of incubation, the PS chips were analysed for changes in surface properties. It was seen that due to biodegradation the surface hydrophobicity of the PS chips was altered. Water contact analysis of bacteria treated and untreated PS material was measured by calculating the reduction in contact angle of water droplets over the surface. In comparison to untreated PS, PS chips treated with Exiguobacterium strains had significantly reduced water contact angle (WCA) showing alteration in the surface property of PS (Fig. 5 and Table S1†). The WCA of PS chip inoculated with strain DR11 was 73.2 ± 1.6 and strain DR14 was 74.03 ± 2.63 as compared to the WCA of uninoculated control chip (95.7 ± 1). Taken together our data indicates that bacterial action had altered the surface properties of PS chips. A decrease in hydrophobicity of PS material is an indication of surface alteration and could also lead to better colonization of solid substrate by bacterium, providing enhanced carbon utilization. Both strain DR11 and DR14 were able to decrease the hydrophobicity of the PS chips. WCA was reduced 1.5 times more than the previous published report (Table S1†).
Fig. 5. Surface hydrophobicity analysis of polystyrene surface. Water contact angles were measured for PS pieces incubated for 30 days in LCFBM medium at 30 °C, under shaking conditions. (a) Control having PS (medium without bacteria), (b) PS incubated with Exiguobacterium sp. strain DR14 and (c) Exiguobacterium sp. strain DR11.
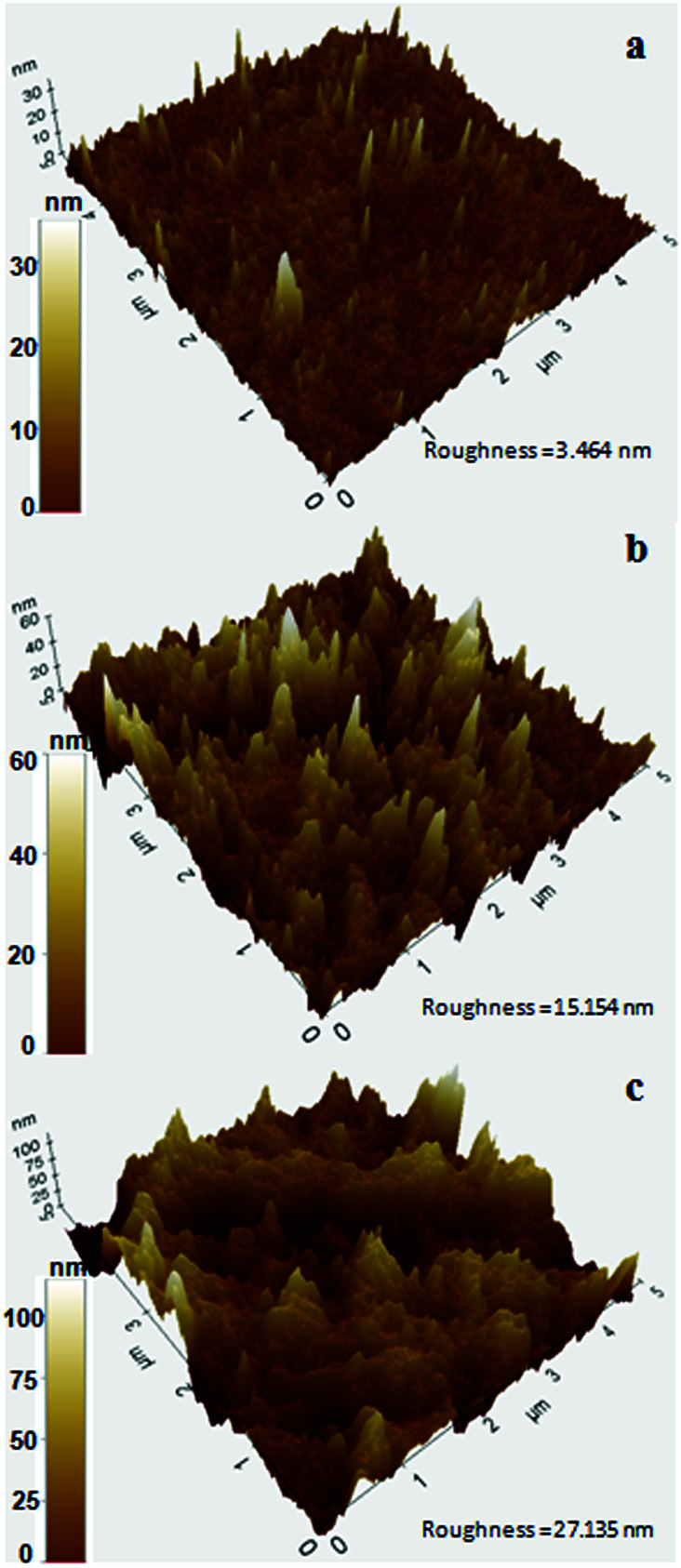
To further analyze the PS surface after bacterial treatment, AFM imaging of the bacterial treated and untreated PS chips was performed. A significant change in surface morphology of PS chips was observed after the bacterial treatment. A clear picture of surface alteration and formation of cavities and grooves of the PS after incubation with DR11 and DR14 could be seen (Fig. 6) which suggests that the surface of PS was eroded by bacterial growth led enzymatic action (Fig. 2). PS chips treated with Exiguobacterium sp. strain DR11 and DR14 displayed enhanced roughness value (27.13 nm and 15.15 nm respectively) compared to untreated PS chips (3.464 nm) and displayed uneven surface with pits and cavities.
Fig. 6. Atomic force microscopic images showing 3D surface topography of PS pieces after bacterial incubation. (a) PS pieces incubated for 30 days without bacterial inoculum (control), (b) after incubation with Exiguobacterium sp. strain DR14, and (c) Exiguobacterium sp. strain DR11. Color bar indicates Z-range.
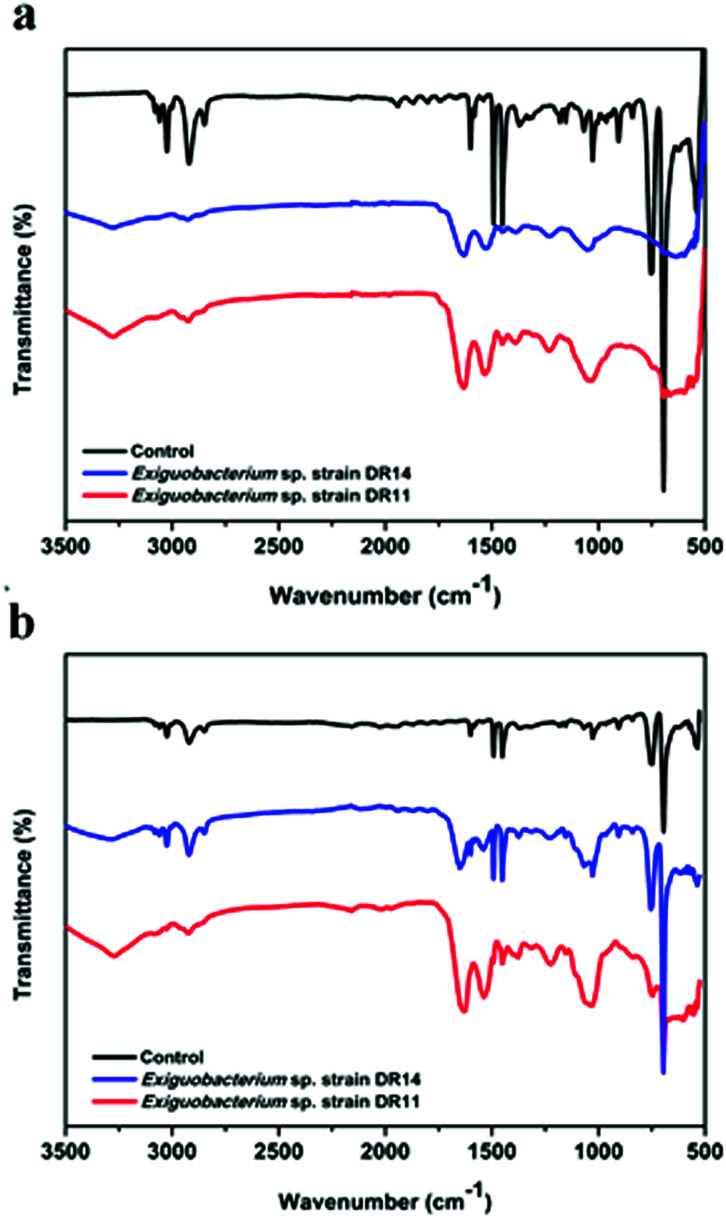
PS degradation was also analysed by the appearance of certain chemicals and functional groups using FT-IR method. PS chips and PS beads incubated with bacteria had hydroxyl peaks (3280 to 3270 cm−1 respectively) due to the vibrations in the stretching of the O–H bond in alcohols and phenols (Fig. 7). In contrast, the PS chips incubated in media without bacterial cells showed no peak of O–H bond ranging from 3500–3200 cm−1. After incubation of PS chips or PS beads with DR11 and DR14 for 30 days, the carbonyl index enhanced in comparison to the untreated PS (Fig. 7). The increase in carbonyl index indicates oxidation of polymer after incubation with bacteria and suggests that Exiguobacterium species are capable of degrading non-irradiated PS. Prior pre-treatment of PE or PS by UV photooxidation, thermal or chemical oxidation methods enhances biodegradation. This synergistic effect of photooxidation is proposed to increase the availability of carbonyl groups on the surface of the plastic material which can be utilized by bacteria. In our study, non-irradiated PS chips were employed as a substrate for biodegradation. Surprisingly, both strain DR11 and DR14 were able to generate carbonyl groups upon incubation of non-irradiated PS, indicating that Exiguobacterium species are capable of degrading unmodified PS. Our studies along with the previous published report suggest that Exiguobacterium species could be a suitable candidate for biodegradation of PS and future research in this direction would be beneficial.
Fig. 7. Fourier-transform infrared (FT-IR) spectroscopy analysis of degraded polystyrene. The plot shows a comparison between untreated and bacterial treated PS samples. Incubation with bacterial strains for 30 days in LCFBM medium at 30 °C leads to the generation of carbonyl group peaks (1632–1644 cm−1) shown with two different PS material – (a) PS pieces (1 × 1 cm), and (b) PS beads. Control indicates PS material incubated and agitated in media without bacteria.
4. Conclusions
In this study, we have shown that Exiguobacterium strain DR11 and DR14, isolated from a wetland, can degrade PS material under given conditions. Exiguobacterium species are known to be extremophiles and previously have shown various potential applications. Exiguobacterium species are known to survive in nutrient-deprived conditions35 and can utilize variable substrates as a source of energy. In our study, we investigated the potential of the isolated species in PS deterioration. We are reporting that strains DR11 and DR14 can establish robust biofilms over PS material and can also utilize PS as a carbon source. It was concluded from the microscopic analysis and quantification after crystal violet staining that these strains can adhere and colonize PS surface as mono-species biofilm. Our data also suggests that both Exiguobacterium sp. DR11 and Exiguobacterium sp. DR14 can co-colonize PS surface and establish viable biofilms. After prolonged incubation, for 30 days, it was observed by AFM analysis that bacteria formed pits and grooves on the PS surface. This indicates that hydrolyzing enzymes produced by the bacterial cell probably initiated the process of depolymerization. In comparison to bacterial untreated PS samples, treated samples showed significantly enhanced roughness. Water contact angle measurement of PS samples incubated with Exiguobacterium cells manifested reduced hydrophobicity of the material in comparison to control. FT-IR analysis further verified the disintegration of PS. The formation of prominent carbonyl peaks of treated samples in comparison to untreated controls showed the breakdown of polymer chains by the oxidation process. Together, our data indicate towards the promising utilization of Exiguobacterium strains for polymer degradation. Our study also emphasizes on further investigation and plausible use of environmental isolates specifically extremophiles for the biodegradation of polyolefins.
Conflicts of interest
The authors declare that they have no conflict of interest.
Supplementary Material
Acknowledgments
We thank Dr Aloke Kanjilal for allowing us to use contact angle instrument and Dr CP Saini for his assistance. We are grateful to Dr Ravikant Upadhyay for his initial suggestions in the study. DC is supported by doctoral research fellowship and GA is supported in part by opportunities for undergraduate research (OUR) fellowship from Shiv Nadar University.
Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ra06448b
References
- Brook M. A. and Organic O., Polymer Chemistry, Wiley, New York, 2000 [Google Scholar]
- Thompson R. C. Swan S. H. Moore C. J. Vom Saal F. S. Philos. Trans. R. Soc., B. 2009;364:2153–2166. doi: 10.1098/rstb.2009.0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olabisi O. and Adewale K., Handbook of thermoplastics, CRC press, 2016 [Google Scholar]
- Maul J., Frushour B. G., Kontoff J. R., Eichenauer H. and Ott K. H., Ullmann's encyclopedia of industrial chemistry, 2000 [Google Scholar]
- Jambeck J. R. Geyer R. Wilcox C. Siegler T. R. Perryman M. Andrady A. Narayan R. Law K. L. Science. 2015;347:768–771. doi: 10.1126/science.1260352. [DOI] [PubMed] [Google Scholar]
- Cózar A. Echevarría F. González-Gordillo J. I. Irigoien X. Úbeda B. Hernández-León S. Palma Á. T. Navarro S. García-de-Lomas J. Ruiz A. Proc. Natl. Acad. Sci. U. S. A. 2014;111:10239–10244. doi: 10.1073/pnas.1314705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrelly T. A. and Shaw I. C., in Household Hazardous Waste Management, InTech, 2017 [Google Scholar]
- Cózar A. Martí E. Duarte C. M. García-de-Lomas J. Van Sebille E. Ballatore T. J. Eguíluz V. M. González-Gordillo J. I. Pedrotti M. L. Echevarría F. Troublè R. Sci. Adv. 2017;3(4):e1600582. doi: 10.1126/sciadv.1600582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochman C. M. Browne M. A. Halpern B. S. Hentschel B. T. Hoh E. Karapanagioti H. K. Rios-Mendoza L. M. Takada H. Teh S. Thompson R. C. Nature. 2013;494:169. doi: 10.1038/494169a. [DOI] [PubMed] [Google Scholar]
- Singh B. Sharma N. Polym. Degrad. Stab. 2008;93:561–584. doi: 10.1016/j.polymdegradstab.2007.11.008. [DOI] [Google Scholar]
- Kyrikou I. Briassoulis D. J. Polym. Environ. 2007;15:125–150. doi: 10.1007/s10924-007-0053-8. [DOI] [Google Scholar]
- Shah A. A. Hasan F. Hameed A. Ahmed S. Biotechnol. Adv. 2008;26:246–265. doi: 10.1016/j.biotechadv.2007.12.005. [DOI] [PubMed] [Google Scholar]
- Sivan A. Curr. Opin. Biotechnol. 2011;22:422–426. doi: 10.1016/j.copbio.2011.01.013. [DOI] [PubMed] [Google Scholar]
- Akutsu Y. Nakajima-Kambe T. Nomura N. Nakahara T. Appl. Environ. Microbiol. 1998;64:62–67. doi: 10.1128/aem.64.1.62-67.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danko A. S. Luo M. Bagwell C. E. Brigmon R. L. Freedman D. L. Appl. Environ. Microbiol. 2004;70:6092–6097. doi: 10.1128/AEM.70.10.6092-6097.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadad D. Geresh S. Sivan A. J. Appl. Microbiol. 2005;98:1093–1100. doi: 10.1111/j.1365-2672.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Howard G. T. Int. Biodeterior. Biodegrad. 2002;49:245–252. doi: 10.1016/S0964-8305(02)00051-3. [DOI] [Google Scholar]
- Orr I. G. Hadar Y. Sivan A. Appl. Microbiol. Biotechnol. 2004;65:97–104. doi: 10.1007/s00253-004-1584-8. [DOI] [PubMed] [Google Scholar]
- Sivan A. Szanto M. Pavlov V. Appl. Microbiol. Biotechnol. 2006;72:346–352. doi: 10.1007/s00253-005-0259-4. [DOI] [PubMed] [Google Scholar]
- Webb J. S. Nixon M. Eastwood I. M. Greenhalgh M. Robson G. D. Handley P. S. Appl. Environ. Microbiol. 2000;66:3194–3200. doi: 10.1128/AEM.66.8.3194-3200.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auta H. Emenike C. Fauziah S. Environ. Pollut. 2017;231:1552–1559. doi: 10.1016/j.envpol.2017.09.043. [DOI] [PubMed] [Google Scholar]
- Khan S. Nadir S. Shah Z. U. Shah A. A. Karunarathna S. C. Xu J. Khan A. Munir S. Hasan F. Environ. Pollut. 2017;225:469–480. doi: 10.1016/j.envpol.2017.03.012. [DOI] [PubMed] [Google Scholar]
- Paço A. Duarte K. da Costa J. P. Santos P. S. Pereira R. Pereira M. Freitas A. C. Duarte A. C. Rocha-Santos T. A. Sci. Total Environ. 2017;586:10–15. doi: 10.1016/j.scitotenv.2017.02.017. [DOI] [PubMed] [Google Scholar]
- Atiq N. Ahmed S. Ali M. I. Ahmad B. Robson G. Afr. J. Microbiol. Res. 2010;4:1537–1541. [Google Scholar]
- Hartmans S. Smits J. Van der Werf M. Volkering F. De Bont J. Appl. Environ. Microbiol. 1989;55:2850–2855. doi: 10.1128/aem.55.11.2850-2855.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmans S. Van der Werf M. de Bont J. Appl. Environ. Microbiol. 1990;56:1347–1351. doi: 10.1128/aem.56.5.1347-1351.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warhurst A. Fewson C. J. Appl. Microbiol. 1994;77:597–606. doi: 10.1111/j.1365-2672.1994.tb02807.x. [DOI] [PubMed] [Google Scholar]
- Ghaffar A. Scott A. Scott G. Eur. Polym. J. 1975;11:271–275. doi: 10.1016/0014-3057(75)90075-0. [DOI] [Google Scholar]
- Teare D. Emmison N. Ton-That C. Bradley R. Langmuir. 2000;16:2818–2824. doi: 10.1021/la9907533. [DOI] [Google Scholar]
- Flemming H.-C. Wingender J. Nat. Rev. Microbiol. 2010;8:623. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- O'Toole G. Kaplan H. B. Kolter R. Annu. Rev. Microbiol. 2000;54:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- Zettler E. R. Mincer T. J. Amaral-Zettler L. A. Environ. Sci. Technol. 2013;47:7137–7146. doi: 10.1021/es401288x. [DOI] [PubMed] [Google Scholar]
- Wang Y. Sheng H.-F. He Y. Wu J.-Y. Jiang Y.-X. Tam N. F.-Y. Zhou H.-W. Appl. Environ. Microbiol. 2012;78:8264–8271. doi: 10.1128/AEM.01821-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues D. F. Goris J. Vishnivetskaya T. Gilichinsky D. Thomashow M. F. Tiedje J. M. Extremophiles. 2006;10:285–294. doi: 10.1007/s00792-005-0497-5. [DOI] [PubMed] [Google Scholar]
- Vishnivetskaya T. A. Kathariou S. Tiedje J. M. Extremophiles. 2009;13:541–555. doi: 10.1007/s00792-009-0243-5. [DOI] [PubMed] [Google Scholar]
- White R. A. Grassa C. J. Suttle C. A. Genome Announc. 2013;1:e00597–e00513. doi: 10.1128/genomeA.00597-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y. Yang J. Wu W.-M. Zhao J. Song Y. Gao L. Yang R. Jiang L. Environ. Sci. Technol. 2015;49:12087–12093. doi: 10.1021/acs.est.5b02663. [DOI] [PubMed] [Google Scholar]
- O'Toole G. A. J. Visualized Exp. 2011:47. doi: 10.3791/2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inbakandan D. Sriyutha Murthy P. Venkatesan R. Ajmal Khan S. Biofouling. 2010;26:893–899. doi: 10.1080/08927014.2010.530347. [DOI] [PubMed] [Google Scholar]
- López M. A. Díaz de la Serna F. J. Z. Jan-Roblero J. Romero J. M. Hernández-Rodríguez C. FEMS Microbiol. Ecol. 2006;58:145–154. doi: 10.1111/j.1574-6941.2006.00137.x. [DOI] [PubMed] [Google Scholar]
- Carneiro A. R. Ramos R. T. J. Dall'Agnol H. Pinto A. C. de Castro Soares S. Santos A. R. Guimarães L. C. Almeida S. S. Baraúna R. A. das Graças D. A. J. Bacteriol. 2012;194:6689–6690. doi: 10.1128/JB.01791-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthukumar T. Aravinthana A. Dineshram R. Venkatesan R. Doble M. J. Microb. Biochem. Technol. 2014;6:116–122. [Google Scholar]
- Frühling A. Schumann P. Hippe H. Sträubler B. Stackebrandt E. Int. J. Syst. Evol. Microbiol. 2002;52:1171–1176. doi: 10.1099/00207713-52-4-1171. [DOI] [PubMed] [Google Scholar]
- Rendueles O. Ghigo J.-M. FEMS Microbiol. Rev. 2012;36:972–989. doi: 10.1111/j.1574-6976.2012.00328.x. [DOI] [PubMed] [Google Scholar]
- Rickard A. H. Gilbert P. High N. J. Kolenbrander P. E. Handley P. S. Trends Microbiol. 2003;11:94–100. doi: 10.1016/S0966-842X(02)00034-3. [DOI] [PubMed] [Google Scholar]
- Mor R. Sivan A. Biodegradation. 2008;19:851–858. doi: 10.1007/s10532-008-9188-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.