Abstract

Glycosaminoglycans (GAGs), in particular, heparan sulfate and heparin, are found colocalized with Aβ amyloid. They have been shown to enhance fibril formation, suggesting a possible pathological connection. We have investigated heparin’s assembly of the KLVFFA peptide fragment using molecular dynamics simulation, to gain a molecular-level mechanistic understanding of how GAGs enhance fibril formation. The simulations reveal an exquisite process wherein heparin accelerates peptide assembly by first “gathering” the peptide molecules and then assembling them. Heparin does not act as a mere template but is tightly coupled to the peptides, yielding a composite protofilament structure. The strong intermolecular interactions suggest composite formation to be a general feature of heparin’s interaction with peptides. Heparin’s chain flexibility is found to be essential to its fibril promotion activity, and the need for optimal heparin chain length and concentration has been rationalized. These insights yield design rules (flexibility; chain-length) and protocol guidance (heparin:peptide molar ratio) for developing effective heparin mimetics and other functional GAGs.
Introduction
Amyloid deposits are characterized by hierarchically structured fibrils, each comprising helices or bundles of protofilaments composed of stacked β-strands of the protein.1 The formation of amyloid is a general phenomenon being exhibited by some 30 different human proteins.2 Notable examples include the peptide amyloid-β (Aβ) linked with Alzheimer’s disease3 and the islet amyloid polypeptide (IAPP) associated with type II diabetes.4 Amyloid formation is also thought to play a functional role, that is, as a part of an essential physiological response, with evidence suggesting that the process can serve to sequestrate (rogue) peptides/proteins, store hormones, or modulate mechanical properties of cells.5
In vivo, amyloid deposits invariably contain additional “cofactors” that include lipids, nucleic acids, proteoglycans (PG), glycosaminoglycans (GAGs), serum amyloid P component, apolipoprotein E, collagen, and metal ions,6 reflecting the crowded, heterogeneous biological environment.7,8 The role of GAGs in amyloidosis is now considered to be significant, with much attention focusing on heparan sulfate and heparin and how they modulate the mechanistic and kinetic pathways.
Heparin has been shown to enhance fibril formation of many amyloidogenic proteins in vitro including amylin,9 synuclein,10 transthyretin,11 tau protein,12 and the intrinsically low-amyloidogenic peptide (PLB 1-23) acetylated cytoplasmic domain of the phospholamban transmembrane protein.13 Heparan sulfate has been found to accelerate the oligomer formation of native human muscle acylphosphatase (mAcp).14 These enhancement effects can include a reduction in the lag phase of the nucleation step,15 an increase in the rate and/or extent of elongation of the fibrils, and/or an increase in fibril yield,16 the exact nature of the enhancement effect being peptide specific.17
The fibril enhancement role of GAGs offers an additional target for developing therapeutic agents for amyloid-based diseases.18 Inhibition of heparan sulfate biosynthesis in animal models can result in the complete loss of fibril formation and amyloid deposition.19−21 Further, heparin has been shown to convert toxic, soluble peptide oligomers into insoluble stable fibrils that are resistant to proteolysis, suggesting a neuroprotective role.21,11 Indeed, GAG mimetics are being tested clinically for Alzheimer’s disease and as potential diagnostic agents for amyloid.22,23
A molecular-level mechanistic understanding of how GAGs enhance fibril formation is lacking, though essential for the further development of this field. It has been a challenge to develop a coherent mechanistic model given that GAGs can exert their action at multiple levels, from inducing β-sheet structure in monomers, facilitating the assembly of monomers, and/or oligomers,11,19 to enhancing extension of the protofilaments resulting in different fibril morphologies.24 For heparan sulfate and heparin, it has been speculated that they serve as molecular scaffolds to facilitate fibril formation.25 However, a recent study of heparin-facilitated aggregation of the neuropeptide β-endorphin has revealed that heparin does not act as a mere scaffold but is an integral component of the resulting fibril.26 As to whether this could be a general phenomenon (i.e., not just restricted to β-endorphin) requires a better understanding of the nature and strength of the interaction of heparin with peptides.
Heparin is an unbranched, linear polymer of disaccharide units consisting of N-sulfated glucosamine (GlcNS) and iduronic acid linked by a (1–4) glycosidic bond (Figure 1).27 The anionic, sulfate groups give heparin the highest negative charge density of any known biomolecule.28 The sulfates appear to be critical to heparin’s fibril enhancement role, as their removal has been found to result in the loss of heparin’s ability to promote the aggregation of Aβ40 and Aβ42.29,30
Figure 1.
Chemical structure of the disaccharide repeating unit of heparin. The saccharide unit on the left is glucosamine (GlcN), while that on the right is iduronic acid (IdoA).
Proteins and polyelectrolytes such as heparin form complexes primarily due to electrostatic interactions. These complexes vary in stoichiometry and architecture depending on several factors such as pH,31 the charge on the protein,32 ionic concentration of the solution,33 the degree of polymerization (dp),17 hydrophobicity,34 stiffness, and flexibility of the polyelectrolyte.35 Generally, aggregation is enhanced with increasing GAGs chain length, although for some proteins, the effect on fibril assembly plateaus when the chain length becomes sufficiently long (dp ≫ 18, where dp is the number of saccharide units).9,36 Short-length GAGs with dp ≤ 4 are essentially ineffective in accelerating fibrillogenesis. In contrast, medium-length GAGs, dp6–dp12, can significantly reduce the lag phase and accelerate the conversion of oligomeric species into ordered fibrillar assemblies.37,38 With respect to the molar heparin to peptide concentration ratio, a study on the prion-related protein fragment PrP demonstrated that relatively low heparin to peptide ratios ranging from 0.2:1 to 0.5:1 accelerated fibril formation. At even lower molar ratios (0.1:1 and below), the effect is less pronounced. Higher molar ratios (5:1, 2:1, and 1:1) of heparin to peptide were found to inhibit peptide aggregation.39
Here, we present a first clear perspective of how heparin interacts with KLVFFA peptide to enhance fibrillogenesis using unbiased molecular dynamics (MD) simulations. The simulations reveal that heparin does not act as a mere template but is tightly coupled to KLVFFA peptides, yielding a composite protofilament structure. The strong intermolecular interactions suggest composite formation to be a general feature of heparin’s interaction with peptides. Also, heparin’s flexibility is found to be essential to its fibril promotion activity, and we have rationalized the need for optimal chain length and heparin:KLVFFA peptide concentration ratio. These insights will underpin therapeutic approaches including the design of more effective GAG mimetics for chelating and minimizing toxicity of oligomers and for modulating amyloidosis, both its inhibition and promotion.
Methodology
A significant issue with molecular simulation is the limited time scales (at best a few microseconds) that are accessible. Indeed, the self-assembly of multiple units of the full Aβ into an ordered structure is outside the time scales of unbiased MD simulation. Given this, we investigated the heparin-promoted assembly of the KLVFFA fragment peptide (lysine-leucine-valine-phenylalanine-phenylalanine-alanine), which is a stretch of hydrophobic amino acids (residues 16–21) in Aβ, rather than the full Aβ. The KLVFFA peptide is the shortest fragment for which experimental evidence of amyloid formation is available.40,41 Solid-state NMR shows that this fragment acts as a prototype for aggregation, forming antiparallel strands leading to the formation of fibrils.42−45
Nine distinct sets of simulations were carried out: (i) simulation of a single heparin molecule; (ii) the interaction of a single KLVFFA monomer (PDB 2Y2A)46 with a single heparin molecule (flexible and restrained in its extended form); (iii) the self-assembly of the KLVFFA peptides alone, which served as a control; (iv) the assembly of the KLVFFA peptides in the presence of heparin; (v) assembly of the KLVFFA peptides in the presence of a heparin molecule that was restrained in its extended conformation to investigate the role of the flexibility of heparin; (vi) the effect of heparin:peptide concentration on KLVFFA assembly; (vii) the effect of heparin chain length (degree of polymerization) on heparin-promoted KLVFFA assembly; (viii) the self-assembly of the KLVFFA dimers (PDB 3OW9)46 alone; and (ix) the assembly of the KLVFFA dimers in the presence of heparin. The full complement of simulations carried out is listed in Table 1.
Table 1. Details of Simulation Studies Carried Outa.
| study | study details | no. of heparin molecules | no. of KLVFFA peptide(s) |
|---|---|---|---|
| (i) | heparin alone in the water | 1 | – |
| (ii) | heparin’s interaction with single KLVFFA | 1 | 1 |
| (iii) | self-assembly of KLVFFA (alone) in water | – | 20 |
| (iv) | KLVFFA assembly in the presence of heparin | 1 | 20 |
| (v) | KLVFFA assembly in the presence of restrained heparin in an extended conformation | 1 | 1 |
| 1 | 20 | ||
| (vi) | effect of heparin:peptide molar concentration on KLVFFA assembly | 1 | 20 |
| 1 | 100 | ||
| 2 | 20 | ||
| (vii) | effect of heparin chain length on KLVFFA | dp2 (12) | 20 |
| dp4 (6) | 20 | ||
| dp6 (4) | 20 | ||
| dp8 (3) | 20 | ||
| dp24 (1) | 20 | ||
| (viii) | self-assembly of predimerized KLVFFA in water | – | 10 |
| (ix) | assembly of predimerized KLVFFA in the presence of heparin | 1 | 10 |
This entire complement of simulations was repeated in a low ionic strength (system neutralized with counterions) and in a physiological ionic strength aqueous environment (150 mM NaCl). Unless otherwise stated, the heparin chain length in terms of the degree of polymerization was dp = 24.
Given that the heparin molecule is significantly charged, the ionic strength of the solvent environment is likely to be critical as it would modulate the Columbic intermolecular interactions. We, therefore, conducted the above simulation studies in a low ionic strength environment in which just sufficient Na+ counterions were added to balance the net charge of the system and a physiological ionic strength environment that contained additional Na+ and Cl– ions to yield a 150 mM NaCl.
Finally, on noting that preformed Aβ KLVFFA dimers aggregated faster than monomers,47,48 we explored the assembly of preformed KLVFFA peptide dimers alone and in the presence of heparin (simulation studies (xiii) and (ix)). The simulations of the KLVFFA monomers showed that there are kinetic barriers to the monomer organization once the molecules become locked onto heparin. The necessary realignment to get antiparallel β-sheet arrangements become the rate-limiting step, delaying the peptide organization process beyond the time scales of unbiased MD. In contrast, the preformed dimer simulations revealed rapid fibrillation of the peptides to yield partially ordered aggregates, revealing the generic features of the ordering process.
The structure of heparin dp24 (PDB 3IRJ)49 was taken from the protein data bank. The structures of shorter heparin fragments were based on heparin 18-mer (PDB 3IRI)49 with the topologies being obtained from the PRODRG server and reviewed for accuracy. The charges were assigned from literature, which had been calculated using ab initio calculations at 6-31G**.50 Unless otherwise indicated, the heparin molecule comprised 24 saccharide units (dp = 24). In the self-assembly and heparin-facilitated assembly simulations, there were 20 KLVFFA monomers. Each of the assembly simulations were run with 3 replicates. The initial set of coordinates for each was randomized, with the KLVFFA peptide molecules being placed randomly around the heparin molecule to ensure that the process kinetics were not biased by the choice of starting coordinates.
To investigate whether flexibility and bending of the heparin molecule are essential for its assembling role, we simulated the facilitated peptide assembly with the heparin molecule position restrained to remain in the extended form using restraints on all the atoms with a force constant of 500 kJ mol–1 nm2.
Systems investigating the effects of different molar ratios of heparin:peptide on the assembly of KLVFFA peptides contained 1:20, 1:100, and 2:20 heparin:peptide molecules. For the effect of heparin chain length on heparin’s propensity to promote KLVFFA peptide assembly, the heparin fragments investigated included chain lengths of 2, 4, 8, 12, and 24 saccharide units (dp2, dp4, dp8, dp12, and dp24).
All the simulations were carried out at 360 K and pressure of 1 bar using Gromacs 5.0.4.51 The higher temperature serves to accelerate potential conformational and configurational transitions of the peptide assembly, an approach employed by the others to enhance sampling.52 The systems were equilibrated in the NVT ensemble and then in the NPT ensemble using a velocity-rescaling thermostat53 and the Berendsen barostat54 to control the temperature and pressure, respectively. In the production run, the Parrinello–Rahman barostat was used with a coupling time of 2 ps and isothermal compressibility of 4.5 × 10–5 bar–1.55 The time step used was 2.0 fs. PME was used to calculate long-range electrostatics with a grid spacing of 0.16 nm and a real-space cutoff of 1 nm.56 Both van der Waals and neighbor list cut-offs describing short-range interactions were set to 1.0 nm. The simulations were run for up to 200 ns of trajectory unless stated otherwise. Heparin and the KLVFFA fragments monomer (PDB 2Y2A) and dimers (PDB 3OW9) with the ends uncapped were modeled using the Gromos96 53a6 force field as employed and validated by others.57−60 The analyses of the simulation trajectories were performed using Gromacs analysis tools and VMD.61 For example, the gmx clustsize tool was used to follow the process of self-assembly in terms of the number of aggregates/clusters and the size of the largest aggregate present in the solution. The free energy of KLVFFA peptide binding to heparin was estimated using the MM-PBSA method.62
Results and Discussion
KLVFFA Interacts Strongly with Heparin and Is Unable to Move up or down the Heparin Molecule
The simulations reveal that heparin by itself in a low ionic strength environment exists in an extended chain form with an average end-to-end distance of around 9.2 nm, with a limited propensity to flex or coil. This is expected as the charged sulfate and carboxylate moieties are content in the polar aqueous environment and repulsion between the charged groups is also likely to encourage the extended form. In the higher ionic strength environment, this distance reduces slightly to an average of 8.8 nm. It appears that the repulsion between the charged heparin moieties is screened by the increased ionic strength enabling the heparin to adopt a less extended chain (Figure 2a).
Figure 2.
End-to-end distance analysis of heparin chain at two ionic strengths in an aqueous environment. (a) Heparin chain alone. (b) Heparin in the presence of a single KLVFFA peptide.
Now we consider the inclusion of a KLVFFA monomer into both systems (low and high ionic strength), while also investigating the role of the flexibility of the heparin molecule. For both the flexible and the restrained extended form of heparin, the KLVFFA peptide quickly attaches to one of the anionic sites on heparin through its lysine residue. For the restrained (extended) heparin at both ionic strengths, the KLVFFA monomer, once attached, resides essentially at the same anionic site throughout the simulation (Figure SI 1b,d). This confirms that heparin does not act as a track on which a KLVFFA peptide can readily move up or down. For the case of heparin in its fully flexible form, KLVFFA remains localized on the heparin chain at high ionic strength (Figure SI 1c) but can explore a much larger displacement range at low ionic strength (Figure SI 1a). This occurs because of the flexing of the heparin molecule, which enables the KLVFFA molecule to detach from one location to another as another strongly interacting site on the heparin molecule comes in close proximity to the KLVFFA peptide. Thus, it appears that the conformational flexibility of heparin is essential to its facilitation of the assembly of Aβ peptides (see also later discussion). The attachment of the single KLFVAA monomer to heparin does not produce any significant change in heparin’s conformation: The average end-to-end distance reduces from 9.2 to 8.8 nm for the low ionic strength system and from 8.8 to 8.3 nm for the high ionic strength media (Figure 2b).
MM-PBSA calculations show that the binding of the KLVFFA monomer to heparin is strong, where the binding free energy is −122.7 ± 24 kcal/mol in the low ionic strength environment and as expected (due to charge screening) lower −92.1 ± 19 kcal/mol in the high ionic strength system (see a detailed breakdown in Table 2). The major contribution to the total binding energy comes from the electrostatic interactions between the lysine residues of the KLVFFA and negative groups on the heparin (Figure SI 2).
Table 2. Breakdown of the Free Energy of Binding ΔG of KLVFFA Peptide with Heparin in Low and High Ionic Strength (150 mM NaCl) Aqueous Environments Calculated by the MM-PBSA Method.
| energy component | ΔG (kcal mol–1, 0 mM NaCl) | ΔG (kcal mol–1, 150 mM NaCl) |
|---|---|---|
| van der Waals energy | –15.5 | –23.0 |
| electrostatic energy | –254.2 | –204.2 |
| polar solvation energy | 149.42 | 138.5 |
| SASAa energy | –2.5 | –3.5 |
| total binding energy | –122.7 ± 24 | –92.2 ± 19 |
SASA (solvent accessible surface area) used to calculate Gnonopolar energies in the mm-pbsa method.
KLVFFA Peptides Aggregate to Form Ordered Oligomers but not a Fibrillar Structure in 200 ns of MD Simulation
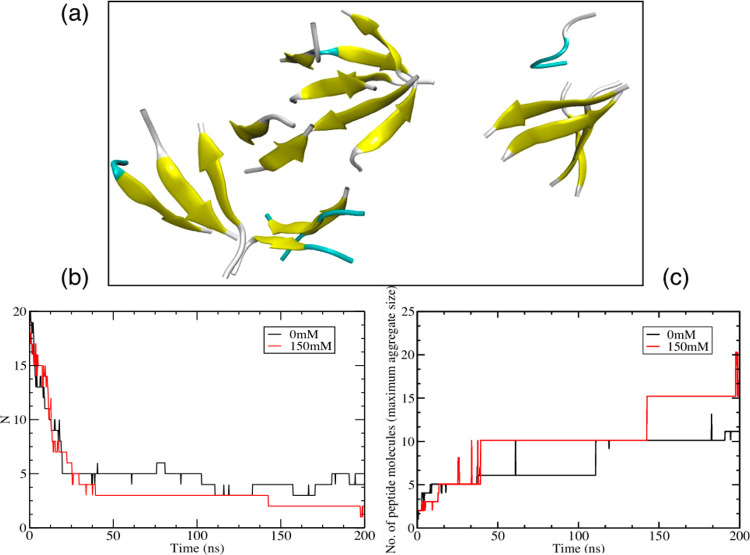
The self-assembly of KLVFFA peptide monomers without heparin in both low and high ionic strength aqueous media results in the formation of 2–3 ordered aggregates but not a contiguous fibrillary structure (Figure 3a). Oligomeric structures form relatively rapidly for about 45 ns and then aggregate further to yield 2–3 larger structures. On formation, the oligomers begin to order internally to form antiparallel β-sheets. The rapid aggregation of the KLVFFA peptides is driven by the strong attraction resulting from the hydrophobic effect. The aggregation is slightly faster (Figure 3b), and the aggregate size is bigger (Figure 3c) in the higher ionic strength system, which would be expected (and has been observed by others)63,64 given that the chemical potential of the hydrophobic KLVFFA would be enhanced in a stronger ionic environment.
Figure 3.
(a) Snapshot of the self-assembly of KLVFFA peptides (monomers) at 200 ns simulation time in 150 mM NaCl in water. (b) A number of peptide aggregates formed during the self-assembly of KLVFFA peptides as a function of time for both the 0 mM and 150 mM ionic strength systems. (c) Maximum aggregate size for the self-assembly of KLVFFA peptides as a function of time for both the 0 mM and 150 mM ionic strength systems. (Cluster criteria: cutoff of 0.35 nm).
Aggregation of KLVFFA Peptides on Heparin Takes Place in Two Steps: Selection and Assembly
Considering heparin-facilitated assembly of KLVFFA peptides, in all six simulations (three at low ionic strength and three at high ionic strength, each beginning from a different starting configuration), we note that there are two distinct stages: a rapid selection and localization of the peptides onto the heparin framework, followed by a cooperative process of heparin-induced assembly of the KLVFFA peptides. The peptide localization onto heparin appears to compete with peptide–peptide interactions with a few of the peptides self-aggregating (mostly as dimers) in isolation, comprising antiparallel β-sheets. In due course, these aggregates also become attached to the heparin. The isolated peptide–peptide aggregates occur mostly in the high-ionic strength simulations, presumably because the high ionic strength screens the charge–charge interaction between the KLVFFA peptides and heparin while also increasing the chemical potential of the hydrophobic KLVFFA peptides, thus favoring peptide–peptide interaction.
The heparin-induced assembly process (second stage) involves bending and unbending of heparin, carrying along with it the adsorbed KLVFFA peptide units that realign and attach to other peptide units, all the while enhancing the development of a more coherent protofilament. The emerging protofilament remains integrated with heparin throughout the process. The heparin-facilitated assembly, in general, is faster relative to the self-assembly of KLVFAA peptides but shows variability that depends on the starting coordinates and is further modulated by the ionic strength of the media.
The aggregation of the peptides in the presence and absence of heparin is compared in Figure 4. At the lower ionic strength, the peptides aggregate onto the heparin to form two intertwined helices of the stacked β-sheets and the heparin, the final structure generally being extended Figure 5. The number of aggregates inclusive of heparin and KLVFFA peptides converges to a single structure within the time period 25–45 ns in all three simulations, reflecting the rapid localization of the KLVFFA molecules onto the heparin. The peptide only clustering (reflecting the development of a more contiguous peptide structure, albeit while integrated with the heparin) is a little slower and is characterized by numerous fluctuations that result from the flexing of the heparin molecule and the attachment/detachment of the peptides in a bid to form a more contiguous structure.
Figure 4.

Evolution of the number of aggregates inclusive of heparin and peptides and peptides only. The plot of peptides and heparin aggregates (black) reflects the initial, rapid association of the peptides with the heparin (within about 45 ns), while the peptides-only aggregates plot (red) reflects the relatively slower, facilitated assembly of the peptides. Note the numerous, repetitive peptide–peptide cluster making and breaking events as the heparin flexes to align and order the emergent protofilament.
Figure 5.

(a) Snapshots of KLVFFA peptide–heparin structures in 0 mM NaCl at 200 ns for three separate simulations each started from a different random configuration. (b) Snapshot of the complex from the second simulation system. (c) Snapshot of the complex from the third simulation but with a “surface” representation to show the formation of a contiguous structure. Heparin and peptides are shown in CPK and cartoon (secondary structure) representation, respectively.
At the higher ionic strength, the resulting peptide–heparin composite shows a greater variation in morphology (Figure 6) ranging from annular to linear structures. In one instance, the heparin molecule with attached KLVFFA peptides folds itself into a ring-like structure. Additionally, in these systems, we also observe the formation of isolated KLVFFA peptide only aggregates, some of which eventually attach to the heparin molecule. As noted earlier, the greater charge screening of the higher ionic strength enables heparin to sample more retracted conformations and also increases the chemical potential of the hydrophobic peptides, thus encouraging peptide–peptide cluster formation alongside localization of the KLVFFA peptides onto the heparin molecule. Here we see both effects playing a role.
Figure 6.

Snapshots of KLVFFA peptide–heparin structures in 150 mM NaCl at 200 ns for three separate simulations (a–c) each started from a different random configuration, showing wide variation in the emergent morphology.
Heparin Flexibility Appears to be Essential for Its Role in Assembling Peptides
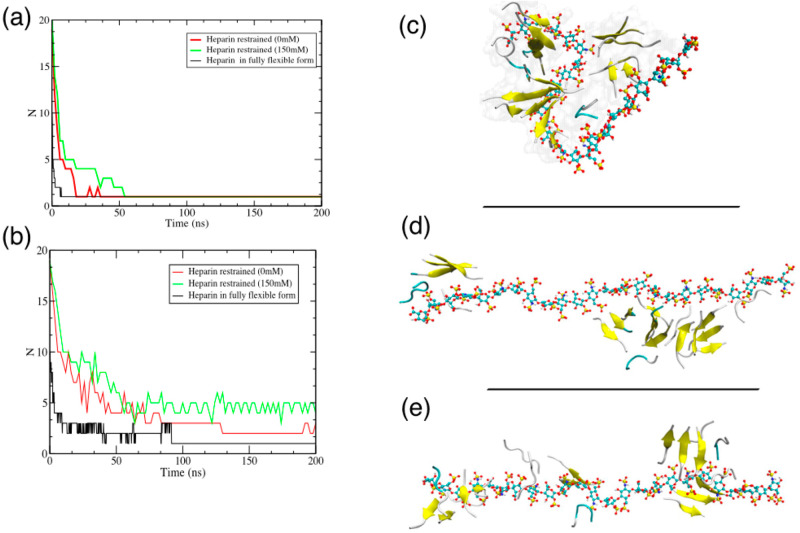
To investigate whether the flexibility and bending of the heparin molecule are essential for its assembling role, we simulated the heparin facilitated KLVFFA peptide assembly with the heparin molecule restrained in its fully extended form. The KLVFFA peptides quickly localized onto the heparin framework (Figure 7a), but there was no subsequent organization or assembly of peptides, only some local ordering of the attached oligomers at a higher ionic concentration (Figure 7b, i.e., peptide only aggregates and Figure 7d,e). This contrasts with the assembly process involving a fully flexible heparin molecule, which yields a more contiguous peptide protofilament albeit integrated with heparin (Figure 7c).
Figure 7.
(a) Total number of aggregates (including KLVFFA peptide aggregates and heparin) as a function of time at both ionic concentrations, reflecting rapid localization of the peptide molecules onto the heparin. (b) The number of peptide-only aggregates as a function of time at both ionic concentrations. The plot shows that peptides assemble into a single structure when heparin is fully flexible within 100 ns while there are still 3–4 loose aggregates when heparin is restrained in its extended state. (c) Snapshots of the heparin–peptide composite for the heparin molecule when fully flexible at 0 mM. (d, e) Heparin restrained in its fully extended form at 0 mM and 150 mM NaCl. For these restrained heparin systems, the lack of a contiguous peptide structure is apparent.
These results clearly illustrate that heparin’s flexibility is essential to its role in facilitating amyloid formation. The peptide–heparin interaction is strong and limits the movement of the peptide once it is adsorbed onto the heparin. The idea of the heparin framework providing a strongly interacting but iso-potential surface along which the peptide molecules can freely translocate (enabling it to form a contiguous structure) is appealing, but unfortunately is not borne out. Instead, the formation of a contiguous protofilament structure comes via peptide detachment/attachment that occurs as the heparin molecule flexes, bringing peptide molecules located at one site of the heparin molecule into the interaction zone of another.
Given that heparin’s preference, when alone, is to be in its extended form, what induces the heparin molecule to flex and bring the attached peptides units together? While the origin of this force is difficult to disentangle, the interactions suggest that it is a combination of a strong, attractive hydrophobic force between the (hydrophobic) peptide units and electrostatics. Unlike valence bonds, electrostatic interactions do not become saturated. Hence, the strong electrostatic interactions responsible for localizing the peptide units onto the heparin are also able to act between an attached peptide and other regions of the heparin. It appears that the combined hydrophobic effect and electrostatic interactions are sufficient to overcome the repulsion between the anionic sulfate moieties on the heparin and any elastic energy penalty associated with its bending.
Heparin–Peptide Interaction Is Not Transitory; Heparin Forms an Integral Part of the Resulting Protofibrils
It is well established that GAGs, and in particular heparan sulfate, are found colocalized with Aβ amyloid.65 This begs the question of whether the GAGs are an integral component of the fibrils or merely just colocalize within the amyloid mass. Indeed, a recent study provides evidence that heparin forms an integral component of the emerging fibrils for the endogenous opiate β-endorphin.26
As noted above, the current simulations also reveal that heparin does not act as a mere template but is tightly coupled to the KLVFFA peptides, yielding a composite protofilament structure consisting of intertwined helices of the stacked β-sheets and the heparin (Figure 8). The binding energy between the two helices (comprising 20 KLVFFA peptides and 1 heparin molecule) was calculated to be around −2000 kcal/mol, which equates to >3500 kβT (thermal energy at T = 310 K). Given this huge value and the intertwining nature of the two helices, there is little or no possibility of their (unaided) disentanglement. However, any unwinding of the helices is likely to occur stepwise, beginning from one of the ends. The interaction energy between a single peptide and heparin ranges from −90 ± 19 kcal/mol to −120 ± 25 kcal/mol depending upon the ionic strength of the solution, while the interpeptide interaction (that between the antiparallel peptide strands) is about −20 kcal/mol. Therefore, the lowest bound for detaching the peptide strand, one residue at a time, from the composite would be −90 kcal/mol and for stripping away altogether an individual peptide molecule from the end would be −110 kcal/mol. These values equate to >150 kT, again pointing to an extremely low probability for the unwinding/detachment process. Furthermore, any stepwise disentanglement would require coordinated molecular displacements.
Figure 8.

KLVFFA peptide–heparin composite results from heparin’s assembly of KLVFFA peptides. Both the formed peptide protofilament and the heparin form helical structures that are intertwined. The heparin helix is characterized by a pitch consisting of four disaccharides with a translation of 2.0–2.1 nm along the axis. (a) Heparin is represented as vdW beads (sulfur in yellow; oxygen in red; carbon in turquoise), while the Aβ peptide protofilament is represented in surface form. (b) Peptides’ secondary structure intertwined against heparin shown in CPK representation.
Given the tendency of the KLVFFA peptides and heparin to form intertwined helices coupled with the strong, electrostatic heparin–peptide interaction, we infer that the formation of composite heparin–peptide protofilaments is a general phenomenon.
Lower Heparin:Peptide Concentration Ratio Is More Effective Because Peptide Monomers Are Not Partitioned among Individual Heparin Molecules
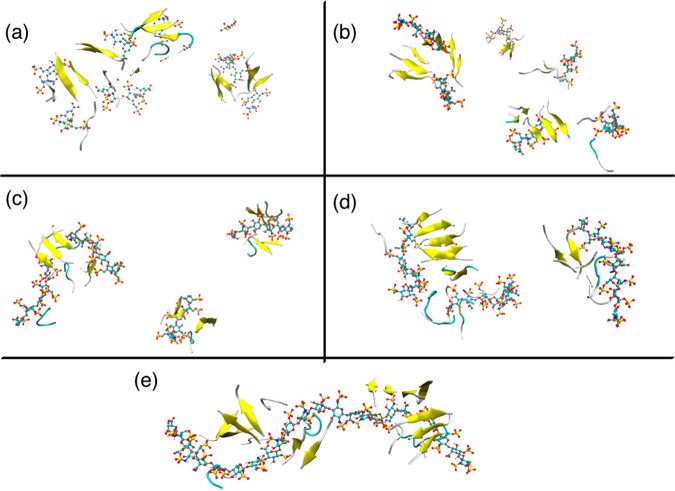
We investigated how the aggregation kinetics depends upon the molar concentration ratio of heparin to KLVFFA peptide. Simulations containing the low molar ratio, that is, 1 heparin molecule:100 KLVFFA peptides (monomers), revealed very fast peptide aggregation and structuring. At low ionic strength, this results in a single dense fibrillar structure that appeared to comprise two protofilaments integrated around the heparin molecule (Figure 9a). At high ionic strength, there were two distinct pathways: the formation of a heparin–peptide structure and ordered filamentous peptide-only structures (Figure 9d). As stated earlier, the high ionic strength enhances the hydrophobic peptide–peptide interactions while screening charge–charge interactions, thereby facilitating the development of peptide-only protofilaments. At a higher molar ratio of 1 heparin:20 KLVFFA peptides, for both ionic strength environments, all of the peptides assembled around the heparin giving rise to an integrated structure (Figure 9b,e). At a still higher molar ratio of 2 heparin molecules:20 KLVFFA peptides (i.e., 1:10), the peptide units apportioned themselves among the 2 heparin molecules, from which they did not dissociate and hence were unable to form a contiguous structure (Figure 9c,f). Each subpopulation attached to a particular heparin molecule, being small in number, can only yield at best a small local oligomeric peptide structure.
Figure 9.
Assembly of KLVFFA peptides at various heparin: peptide molar ratios at 0 mM (left-hand side) and 150 mM NaCl (right-hand side): (a, d) 1:100; (b, e) 1:20; (c, f) 2:20 heparin:peptide molar ratio. The encircled structure in (d) shows the formation of a well-ordered fibril at 150 mM.
These results are consistent with experimental observations and are intuitive.36,39,66 At lower heparin:KLVFFA peptide ratio, as the relative number of peptides is increased, an individual heparin molecule can attract a full complement of peptide units, the number of peptides being commensurate with the available sites on the heparin, and then heparin facilitates their assembly into a protofilament structure. We surmise that at very low heparin:peptide ratios, the relatively few heparin molecules will facilitate peptide assembly, but their effect will be localized and will not impact much the bulk of the solution.
Shorter Heparin Chain Lengths Are Less Effective as They Partition the Peptides to Form Multiple Heparin–Peptide Complexes That Aggregate Only Slowly
The simulations reveal that peptide assembly (toward the development of a contiguous aggregate structure) is facilitated by the longer heparin chain length of dp24 (i.e., 24 saccharide units) at lower ionic strength. Heparin chain lengths of dp4, dp6, and dp8 did not yield a contiguous peptide structure that includes all of the peptide molecules, as the peptide molecules get apportioned among the individual heparin units that then do not in general show any significant tendency to come together (Figure 10b–d). For dp8, the KLVFFA peptides did yield an ordered protofilament that spanned two of the heparin dp8 units.
Figure 10.
Effect of different heparin chain lengths on heparin-facilitated assembly of KLVFF peptides at 0 mM: (a) dp2, (b) dp4, (c) dp6, (d) dp8, and (e) dp24, where dp is the number of saccharide units. Longer heparin chain lengths (dp = 8 and dp = 24) are more effective at facilitating peptide assembly. Shorter heparin chain lengths (dp = 4 and dp = 6) are less effective as they form multiple heparin–peptide complexes that then aggregate only slowly. The disaccharide (dp = 2) complexes show a marginal reversal in trend by aggregating together, courtesy of their faster diffusion rate that enhances complex–complex interactions.
In contrast, the shortest chain length studied, the disaccharide heparin dp2 shows two distinct behaviors: promotion of peptide assembly for the low ionic strength environment (Figure 10a) and little or no effect in the high ionic strength environment. The promotion at low ionic strength, while unexpected, does not contradict any experiments, as dp2 fragments have never been studied experimentally in this context. The heparin dp2–peptide complexes formed quickly and then aggregated to form 1–2 clusters followed by some ordering of the KLVFFA peptides. It appears that being small units, further aggregation of these dp2–peptide complexes can be driven by the complex peptides courtesy of the hydrophobic effect. In principle, such a potential must also exist for the larger heparin units but will be counteracted by their greater inertia due to their size. Indeed, estimated diffusion coefficients of the various heparin chain lengths in pure water from simulation bear this out: D (dp2) = 4.3 ± 1.8 × 10–5 cm2/s; D (dp4) = 4.8 ± 2.0 × 10–5 cm2/s; D (dp6) = 4.0 ± 3.0 × 10–5 cm2/s; D (dp8) = 3.1 ± 1.7 × 10–5 cm2/s; D (dp24) = 0.4 ± 0.3 × 10–5 cm2/s. In the high ionic strength environment, the shorter heparin chains dp2 were ineffective in accelerating the aggregation. While larger chains dp4–dp18 significantly increase the aggregation of peptides giving rise to larger peptide aggregates; dp24 does not enhance further aggregation (Figure SI 3). The extent of the formation of peptide aggregates for the various sized heparin units at both ionic strengths is shown in Figure 11.
Figure 11.
A number of KLVFFA peptide aggregates were observed for the various heparin chain lengths (dp2–dp24) at the end of each simulation. (a) At low ionic concentration, larger chain lengths favor greater KLVFFA peptide aggregation (fewer aggregates), though the shortest chain length dp2 shows distinct behavior and also promotes peptide aggregation. (b) At higher ionic concentration, the aggregation increases (fewer aggregates) with an increase in chain length up to dp8, while dp24 does not enhance further aggregation.
Accessing Longer Time Scale Ordering: Heparin-Induced Assembly of Preformed Peptide Dimers
A limitation of unbiased MD is the relatively short time scales (of the order of a microsecond) that can be accessed. Experiments show that dimers aggregate remarkably faster than the monomers, and there may be some subtle differences in the microstructure of monomers and dimers that are responsible for the marked increase in the aggregation propensity of dimer.47,48 To observe the development of ordered heparin– KLVFFA peptide composite protofilament in a shorter time scale, we simulated heparin-induced assembly of preformed KLVFFA peptide dimers in the form of antiparallel β-sheets. We also looked at the interaction of a single dimer with heparin and carried out the control simulation of the self-assembly of the preformed peptide dimers alone, that is, without heparin.
Assembly of KLVFFA peptide dimers alone led to the formation of ordered oligomers but never a single, ordered protofilament structure over the 200 ns simulation time, as in monomers (Figure 12a). However, the oligomeric structures form more rapidly after about 50 ns into the trajectory (as compared to 145 ns for monomeric KLVFFA) and then rearrange and order but do not associate any further (Figure 12b).
Figure 12.
(a) Snapshot of the self-assembly of preformed dimers of the peptide KLVFFA at 200 ns. (b) Total number of aggregates (peptides only) as a function of simulation time for self-assembly of monomers and preformed peptide dimers. Dimers rapidly come together, while monomer fragments take time to order and form antiparallel β-sheets resulting in slower aggregation.
In the simulations investigating heparin-facilitated assembly of the preformed KLVFFA peptide dimers, the peptides rapidly localize onto the heparin which then displays a choreographic behavior, flexing and unflexing until the attached peptide dimers are assembled into a single stable protofilament, concluding the process just under 200 ns (Figure 13). This causes kinking in the heparin chain and reduces the end-to-end distance up to 4.0 nm (Figure SI 4).
Figure 13.
Heparin-facilitated assembly of preformed dimers of KLVFFA in the presence of one molecule of heparin. The trajectory reveals two distinct stages: rapid association, and localization of the peptides onto the heparin which occurs in about 5 ns, and the heparin-facilitated assembly of the associated peptides into a protofilament, which is concluded well before 200 ns.
The protofilament structure is not “released” but remains integrated with the heparin molecule. As with monomer peptides, the assembly also takes place in two distinct stages. However, in the case of dimers, the process is much faster, with the selection and localization of the peptides onto the heparin framework taking only 5 ns (Figure 14).
Figure 14.

Total number of aggregates (inclusive of heparin and peptides) as a function of simulation time for the heparin-facilitated assembly of peptide monomers and preformed peptide dimers. Dimers are rapidly attracted to the heparin to form a single aggregate (only 5 ns), while monomers attachment is slower.
Mapping Molecular Insights from Heparin’S Assembly of KLVFFA to Larger Peptides and Proteins
The current study looked specifically at the effect of heparin on the assembly of the peptide KLVFFA at two ionic strengths. Nevertheless, many of the simulations results fully rationalize experimental observations with respect to heparin’s interaction with other larger and more complex peptides/proteins, suggesting that the inferences from the results may be generic. Thus, the simulations not only reproduce heparin’s promotion of peptide aggregation and formation of contiguous peptide aggregates (and by inference, protofibrils, and fibrils) at both low and high ionic strengths but also rationalize the effects of variations in heparin chain length as observed, for example, for the 8 kDa fragment of gelsolin (65 residues), human muscle acylphosphatase (about 98 residues),14,25 and a monomer of the transthyretin (127 residues).
Further, the simulations rationalize the effects of the heparin:peptide molar ratio observed for the prion-related fragment PrP.39 Peptide/protein molecular size effects and the nature of the amino acid residues will undoubtedly modulate the mechanistic picture uncovered here for heparin-KLVFFA peptide. The optimum heparin chain length and the molar ratio will depend on the commensurability of the stacked peptides with the interaction sites on the heparin.67 For large peptides, from diffusional inertia considerations, we should not expect the peptides localizing onto the heparin molecule (as observed for KLVFFA here) but rather the heparin (being of a relatively smaller size) being attracted to the larger peptide molecules.
An important question raised in the literature concerns the first step of heparin/peptide interaction, that is,, whether the peptide units localize onto the heparin framework or do they aggregate first to form oligomers before adsorbing onto heparin.14 While KLVFFA shows a tendency to adsorb on heparin’s surface first and then self-assemble due to hydrophobic interactions, this does depend on the ionic strength of the aqueous environment. At 150 mM NaCl ionic strength, self-association of the peptides begins to compete with the peptide–heparin association. What happens for a particular peptide would depend on how the peptide–peptide interaction, particularly the hydrophobicity, fares relative to the peptide–heparin interaction.
Summary and Significance
While there is much experimental data on heparin’s promotion of peptide fibrillogenesis, a molecular-level understanding of the process has been lacking. Here, we have employed MD simulation to provide a first clear perspective of how heparin promotes the initial stages of fibrillogenesis, addressing many of the outstanding questions regarding its activity and mode of action. The simulations corroborate the experimentally observed Aβ fibrillogenesis activity of heparin. Heparin achieves this by first rapidly selecting the peptides from the surrounding solution (in line with earlier inferences from experiments)14 and then by repeated flexing, relocates and assembles the peptide components to yield ordered protofilaments. The heparin–peptide interactions (mostly electrostatic) are strong, and there appears to be no scope for individual peptide molecules to move up and down the heparin framework, as it was a track. This explains the need for the heparin to flex as a means of relocating peptides. We were able to demonstrate that flexing is essential for heparin’s assembling activity. A further consequence of the strong interaction is that the formed protofilament structure is not “released” but remains integrated with the heparin molecule. This provides the basis for earlier experimental evidence for heparin being an integral component of the formed fibrils for the case of β-endorphin26 and suggests GAGs forming composite fibrils to be a more general phenomenon. The simulations rationalize the optimum heparin chain length and the dependency of its aggregation enhancement activity on the heparin:peptide molar concentration ratio. Higher heparin:peptide ratio is less effective, as the peptide molecules (sparsely) are partitioned among the heparin molecules, reducing the probability of assembling a contiguous protofilament.
An important finding of this study is that predimerized peptides aggregate remarkably faster than the monomers as supported by various experimental studies.47,48,68 In our simulations, at the higher peptide concentration, the aggregation of dimers in the presence of heparin is some eight times faster than that of monomers. The monomer–dimer transition for KLVFFA is significant. These findings and insights underpin significant biological and therapeutic implications. In particular, we have identified design rules (flexibility; chain length) and protocol guidance (heparin: peptide molar ratio) for developing effective heparin mimetics and other functional GAGs.
Acknowledgments
Special thanks to Simon Boothroyd for providing structures of smaller chains of heparin. Computational resources supporting this work were provided by the High-End Computing (HEC) Cluster at Lancaster university, UK.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.2c01034.
Additional analysis on the role of the flexibility, chain lengths, and structures of the heparin molecule (PDF)
Author Contributions
⊥ These authors contributed equally to this study.
The authors declare no competing financial interest.
Supplementary Material
References
- Willbold D.; Strodel B.; Schröder G. F.; Hoyer W.; Heise H. Amyloid-Type Protein Aggregation and Prion-like Properties of Amyloids. Chem. Rev. 2021, 121, 8285. 10.1021/acs.chemrev.1c00196. [DOI] [PubMed] [Google Scholar]
- Dobson C. M.; Knowles T. P. J.; Vendruscolo M. The Amyloid Phenomenon and Its Significance in Biology and Medicine. Cold Spring Harb. Perspect. Biol. 2020, 12, a033878. 10.1101/cshperspect.a033878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brothers H. M.; Gosztyla M. L.; Robinson S. R. The Physiological Roles of Amyloid-β Peptide Hint at New Ways to Treat Alzheimer’s Disease. Frontiers in Aging Neuroscience 2018, 10.3389/fnagi.2018.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira S.; Raimundo A.; Menezes R.; Martins I. Islet Amyloid Polypeptide & Amyloid Beta Peptide Roles in Alzheimer’s Disease: Two Triggers, One Disease. Neural Regeneration Research 2021, 16, 1127. 10.4103/1673-5374.300323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley J. E.; Farr S. A.; Nguyen A. D.; Xu F. What Is the Physiological Function of Amyloid-Beta Protein?. Journal of Nutrition, Health and Aging 2019, 23, 225. 10.1007/s12603-019-1162-5. [DOI] [PubMed] [Google Scholar]
- Stewart K. L.; Radford S. E. Amyloid Plaques beyond Aβ: A Survey of the Diverse Modulators of Amyloid Aggregation. Biophys. Rev. 2017, 9 (4), 405–419. 10.1007/s12551-017-0271-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo R.; Ranson N. A.; Radford S. E. Amyloid Structures: Much More than Just a Cross-β Fold. Curr. Opin. Struct. Biol. 2020, 60, 7. 10.1016/j.sbi.2019.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madine J. Cofactor-Mediated Amyloidogenesis. Biosci. Rep. 2019, 10.1042/BSR20190327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha S.; Patil S. M.; Gibson J.; Nelson C. E.; Alder N. N.; Alexandrescu A. T. Mechanism of Amylin Fibrillization Enhancement by Heparin. J. Biol. Chem. 2011, 286 (26), 22894–22904. 10.1074/jbc.M110.215814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohlberg J. A.; Li J.; Uversky V. N.; Fink A. L. Heparin and Other Glycosaminoglycans Stimulate the Formation of Amyloid Fibrils from ??-Synuclein in Vitro. Biochemistry 2002, 41 (5), 1502–1511. 10.1021/bi011711s. [DOI] [PubMed] [Google Scholar]
- Bourgault S.; Solomon J. P.; Reixach N.; Kelly J. W. Sulfated Glycosaminoglycans Accelerate Transthyretin Amyloidogenesis by Quaternary Structural Conversion. Biochemistry 2011, 50 (6), 1001–1015. 10.1021/bi101822y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedert M.; Jakes R.; Spillantini M. G.; Hasegawa M.; Smith M. J.; Crowther R. A. Assembly of Microtubule-Associated Protein Tau into Alzheimer-like Filaments Induced by Sulphated Glycosaminoglycans. Nature 1996, 383 (6600), 550–553. 10.1038/383550a0. [DOI] [PubMed] [Google Scholar]
- Madine J.; Davies H. A.; Hughes E.; Middleton D. A. Heparin Promotes the Rapid Fibrillization of a Peptide with Low Intrinsic Amyloidogenicity. Biochemistry 2013, 52 (50), 8984–8992. 10.1021/bi401231u. [DOI] [PubMed] [Google Scholar]
- Motamedi-Shad N.; Garfagnini T.; Penco A.; Relini A.; Fogolari F.; Corazza A.; Esposito G.; Bemporad F.; Chiti F. Rapid Oligomer Formation of Human Muscle Acylphosphatase Induced by Heparan Sulfate. Nat. Struct. Mol. Biol. 2012, 19 (5), 547–554. 10.1038/nsmb.2286. [DOI] [PubMed] [Google Scholar]
- Vilasi S.; Sarcina R.; Maritato R.; de Simone A.; Irace G.; Sirangelo I. Heparin Induces Harmless Fibril Formation in Amyloidogenic W7FW14F Apomyoglobin and Amyloid Aggregation in Wild-Type Protein In Vitro. PLoS One 2011, 6 (7), e22076. 10.1371/journal.pone.0022076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran G.; Udgaonkar J. B. Understanding the Kinetic Roles of the Inducer Heparin and of Rod-like Protofibrils during Amyloid Fibril Formation by Tau Protein. J. Biol. Chem. 2011, 286 (45), 38948–38959. 10.1074/jbc.M111.271874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iannuzzi C.; Irace G.; Sirangelo I. The Effect of Glycosaminoglycans (GAGs) on Amyloid Aggregation and Toxicity. Molecules 2015, 20 (2), 2510–2528. 10.3390/molecules20022510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naini S. M. A.; Soussi-Yanicostas N. Heparan Sulfate as a Therapeutic Target in Tauopathies: Insights from Zebrafish. Front. Cell Dev. Biol. 2018, 6, 00163. 10.3389/fcell.2018.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suk J. Y.; Zhang F.; Balch W. E.; Linhardt R. J.; Kelly J. W. Heparin Accelerates Gelsolin Amyloidogenesis. Biochemistry 2006, 45 (7), 2234–2242. 10.1021/bi0519295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elimova E.; Kisilevsky R.; Szarek W. A.; Ancsin J. B. Amyloidogenesis Recapitulated in Cell Culture: A Peptide Inhibitor Provides Direct Evidence for the Role of Heparan Sulfate and Suggests a New Treatment Strategy. FASEB J. 2004, 18 (14), 1749–1751. 10.1096/fj.03-1436fje. [DOI] [PubMed] [Google Scholar]
- Maïza A.; Chantepie S.; Vera C.; Fifre A.; Huynh M. B.; Stettler O.; Ouidja M. O.; Papy-Garcia D. The Role of Heparan Sulfates in Protein Aggregation and Their Potential Impact on Neurodegeneration. FEBS Lett. 2018, 592, 3806. 10.1002/1873-3468.13082. [DOI] [PubMed] [Google Scholar]
- Gervais F.; Chalifour R.; Garceau D.; Kong X.; Laurin J.; Mclaughlin R.; Morissette C.; Paquette J. Glycosaminoglycan Mimetics: A Therapeutic Approach to Cerebral Amyloid Angiopathy. Amyloid 2001, 8, 28–35. [PubMed] [Google Scholar]
- Ma Q.; Cornelli U.; Hanin I.; Jeske W. P.; Linhardt R. J.; Walenga J. M.; Fareed J.; Lee J. M. Heparin Oligosaccharides as Potential Therapeutic Agents in Senile Dementia. Curr. Pharm. Des. 2007, 13 (15), 1607–1616. 10.2174/138161207780765918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaurin J.; Franklin T.; Zhang X.; Deng J.; Fraser P. E. Interactions of Alzheimer Amyloid-Beta Peptides with Glycosaminoglycans Effects on Fibril Nucleation and Growth. Eur. J. Biochem. 1999, 266 (3), 1101–1110. 10.1046/j.1432-1327.1999.00957.x. [DOI] [PubMed] [Google Scholar]
- Motamedi-Shad N.; Monsellier E.; Chiti F. Amyloid Formation by the Model Protein Muscle Acylphosphatase Is Accelerated by Heparin and Heparan Sulphate through a Scaffolding-Based Mechanism. J. Biochem. 2009, 146 (6), 805–814. 10.1093/jb/mvp128. [DOI] [PubMed] [Google Scholar]
- Nespovitaya N.; Mahou P.; Laine R. F.; Schierle G. S. K.; Kaminski C. F. Heparin Acts as a Structural Component of β-Endorphin Amyloid Fibrils Rather than a Simple Aggregation Promoter. Chem. Commun. 2017, 53 (7), 1273–1276. 10.1039/C6CC09770G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss J. L.; Regatieri C. V.; Jarrouge T. R.; Cavalheiro R. P.; Sampaio L. O.; Nader H. B. Heparan Sulfate Proteoglycans: Structure, Protein Interactions and Cell Signaling. An. Acad. Bras. Cienc. 2009, 81 (3), 409–429. 10.1590/S0001-37652009000300007. [DOI] [PubMed] [Google Scholar]
- Pita R. EU Regulatory Landscape of Non-Biological Complex Drugs 2015, 20, 357–380. 10.1007/978-3-319-16241-6_11. [DOI] [Google Scholar]
- Castillo G. M.; Lukito W.; Wight T. N.; Snow A. D. The Sulfate Moieties of Glycosaminoglycans Are Critical for the Enhancement of β-Amyloid Protein Fibril Formation. J. Neurochem. 1999, 72 (4), 1681–1687. 10.1046/j.1471-4159.1999.721681.x. [DOI] [PubMed] [Google Scholar]
- Townsend D.; Fullwood N. J.; Yates E. A.; Middleton D. A. Aggregation Kinetics and Filament Structure of a Tau Fragment Are Influenced by the Sulfation Pattern of the Cofactor Heparin. Biochemistry 2020, 59, 4003. 10.1021/acs.biochem.0c00443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y.; Seeman D.; Yan Y.; Sun L.; Post J.; Dubin P. L. Effect of Heparin on Protein Aggregation: Inhibition versus Promotion. Biomacromolecules 2012, 13 (5), 1642–1651. 10.1021/bm3003539. [DOI] [PubMed] [Google Scholar]
- Meneghetti M. C. Z.; Hughes A. J.; Rudd T. R.; Nader H. B.; Powell A. K.; Yates E. A.; Lima M. A. Heparan Sulfate and Heparin Interactions with Proteins. J. R. Soc., Interface 2015, 12, 20150589. 10.1098/rsif.2015.0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyrek E.; Dubin P. L.; Tribet C.; Gamble E. A. Ionic Strength Dependence of Protein-Polyelectrolyte Interactions. Biomacromolecules 2003, 4 (2), 273–282. 10.1021/bm025664a. [DOI] [PubMed] [Google Scholar]
- Sofronova A. A.; Evstafyeva D. B.; Izumrudov V. A.; Muronetz V. I.; Semenyuk P. I. Protein-Polyelectrolyte Complexes: Molecular Dynamics Simulations and Experimental Study. Polymer (Guildf) 2017, 113, 39–45. 10.1016/j.polymer.2017.02.047. [DOI] [Google Scholar]
- Cooper C. L.; Dubin P. L.; Kayitmazer A. B.; Turksen S. Polyelectrolyte-Protein Complexes. Curr. Opin. Colloid Interface Sci. 2005, 10, 52–78. 10.1016/j.cocis.2005.05.007. [DOI] [Google Scholar]
- Takase H.; Tanaka M.; Yamamoto A.; Watanabe S.; Takahashi S.; Nadanaka S.; Kitagawa H.; Yamada T.; Mukai T. Structural Requirements of Glycosaminoglycans for Facilitating Amyloid Fibril Formation of Human Serum Amyloid A. Amyloid 2016, 23 (2), 67–75. 10.3109/13506129.2016.1168292. [DOI] [PubMed] [Google Scholar]
- Fraser P. E.; Darabie A. A.; McLaurin J. Amyloid-Beta Interactions with Chondroitin Sulfate-Derived Monosaccharides and Disaccharides. Implications for drug development. J. Biol. Chem. 2001, 276 (9), 6412–6419. 10.1074/jbc.M008128200. [DOI] [PubMed] [Google Scholar]
- Quittot N.; Sebastiao M.; Bourgault S. Modulation of Amyloid Assembly by Glycosaminoglycans: From Mechanism to Biological Significance. Biochem. Cell Biol. 2017, 95 (3), 329–337. 10.1139/bcb-2016-0236. [DOI] [PubMed] [Google Scholar]
- Bazar E.; Jelinek R. Divergent Heparin-Induced Fibrillation Pathways of a Prion Amyloidogenic Determinant. ChemBioChem. 2010, 11 (14), 1997–2002. 10.1002/cbic.201000207. [DOI] [PubMed] [Google Scholar]
- Antzutkin O. N. Amyloidosis of Alzheimer’s A Beta Peptides: Solid-State Nuclear Magnetic Resonance, Electron Paramagnetic Resonance, Transmission Electron Microscopy, Scanning Transmission Electron Microscopy and Atomic Force Microscopy Studies. Magn. Reson. Chem. 2004, 42 (2), 231–246. 10.1002/mrc.1341. [DOI] [PubMed] [Google Scholar]
- Brown M. R.; Radford S. E.; Hewitt E. W. Modulation of β-Amyloid Fibril Formation in Alzheimer’s Disease by Microglia and Infection. Frontiers in Molecular Neuroscience 2020, 10.3389/fnmol.2020.609073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miravalle L.; Tokuda T.; Chiarle R.; Giaccone G.; Bugiani O.; Tagliavini F.; Frangione B.; Ghiso J. Substitutions at Codon 22 of Alzheimer’s Aβ Peptide Induce Diverse Conformational Changes and Apoptotic Effects Human Cerebral Endothelial Cells. J. Biol. Chem. 2000, 275 (35), 27110–27116. 10.1074/jbc.M003154200. [DOI] [PubMed] [Google Scholar]
- Ma B.; Nussinov R. Stabilities and Conformations of Alzheimer’s β-Amyloid Peptide Oligomers (Aβ16–22, Aβ16–35, and Aβ10–35): Sequence Effects. Proc. Natl. Acad. Sci. U. S. A. 2002, 99, 14126. 10.1073/pnas.212206899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C.; Ni R.; Smith J. E.; Childers W. S.; Mehta A. K.; Lynn D. G. Kinetic Intermediates in Amyloid Assembly. J. Am. Chem. Soc. 2014, 136 (43), 15146–15149. 10.1021/ja508621b. [DOI] [PubMed] [Google Scholar]
- Baumketner A.; Bernstein S. L.; Wyttenbach T.; Bitan G.; Teplow D. B.; Bowers M. T.; Shea J.-E. Amyloid Beta-Protein Monomer Structure: A Computational and Experimental Study. Protein Sci. 2006, 15 (3), 420–428. 10.1110/ps.051762406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colletier J.-P.; Laganowsky A.; Landau M.; Zhao M.; Soriaga A. B.; Goldschmidt L.; Flot D.; Cascio D.; Sawaya M. R.; Eisenberg D. Molecular Basis for Amyloid-Beta Polymorphism. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 16938–16943. 10.1073/pnas.1112600108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portillo A.; Hashemi M.; Zhang Y.; Breydo L.; Uversky V. N.; Lyubchenko Y. L. Role of Monomer Arrangement in the Amyloid Self-Assembly. Biochim. Biophys. Acta - Proteins Proteomics 2015, 1854 (3), 218–228. 10.1016/j.bbapap.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munter L. M.; Voigt P.; Harmeier A.; Kaden D.; Gottschalk K. E.; Weise C.; Pipkorn R.; Schaefer M.; Langosch D.; Multhaup G. GxxxG Motifs within the Amyloid Precursor Protein Transmembrane Sequence Are Critical for the Etiology of Aβ42. EMBO J. 2007, 26 (6), 1702–1712. 10.1038/sj.emboj.7601616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S.; Gor J.; Mulloy B.; Perkins S. J. Semi-Rigid Solution Structures of Heparin by Constrained X-Ray Scattering Modelling: New Insight into Heparin-Protein Complexes. J. Mol. Biol. 2010, 395 (3), 504–521. 10.1016/j.jmb.2009.10.064. [DOI] [PubMed] [Google Scholar]
- Verli H.; Guimarães J. A. Molecular Dynamics Simulation of a Decasaccharide Fragment of Heparin in Aqueous Solution. Carbohydr. Res. 2004, 339 (2), 281–290. 10.1016/j.carres.2003.09.026. [DOI] [PubMed] [Google Scholar]
- Van Der Spoel D.; Lindahl E.; Hess B.; Groenhof G.; Mark A. E.; Berendsen H. J. C. GROMACS: Fast, Flexible, and Free. J. Comput. Chem. 2005, 26, 1701. 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- Cino E. A.; Choy W. Y.; Karttunen M. Comparison of Secondary Structure Formation Using 10 Different Force Fields in Microsecond Molecular Dynamics Simulations. J. Chem. Theory Comput. 2012, 8 (8), 2725–2740. 10.1021/ct300323g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussi G.; Donadio D.; Parrinello M. Canonical Sampling through Velocity Rescaling. J. Chem. Phys. 2007, 126, 014101. 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- Berendsen H. J. C.; Postma J. P. M.; Van Gunsteren W. F.; Dinola A.; Haak J. R. Molecular Dynamics with Coupling to an External Bath. J. Chem. Phys. 1984, 81, 3684. 10.1063/1.448118. [DOI] [Google Scholar]
- Parrinello M.; Rahman A. Polymorphic Transitions in Single Crystals: A New Molecular Dynamics Method. J. Appl. Phys. 1981, 52, 7182. 10.1063/1.328693. [DOI] [Google Scholar]
- Di Pierro M.; Elber R.; Leimkuhler B. A Stochastic Algorithm for the Isobaric-Isothermal Ensemble with Ewald Summations for All Long Range Forces. J. Chem. Theory Comput. 2015, 11, 5624. 10.1021/acs.jctc.5b00648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pol-Fachin L.; Verli H. Depiction of the Forces Participating in the 2-O-Sulfo-α-l-Iduronic Acid Conformational Preference in Heparin Sequences in Aqueous Solutions. Carbohydr. Res. 2008, 343 (9), 1435–1445. 10.1016/j.carres.2008.04.016. [DOI] [PubMed] [Google Scholar]
- Pol-Fachin L.; Rusu V. H.; Verli H.; Lins R. D. GROMOS 53A6 GLYC, an Improved GROMOS Force Field for Hexopyranose-Based Carbohydrates. J. Chem. Theory Comput. 2012, 8 (11), 4681–4690. 10.1021/ct300479h. [DOI] [PubMed] [Google Scholar]
- Pol-Fachin L.; Fernandes C. L.; Verli H. GROMOS96 43a1 Performance on the Characterization of Glycoprotein Conformational Ensembles through Molecular Dynamics Simulations. Carbohydr. Res. 2009, 344, 491. 10.1016/j.carres.2008.12.025. [DOI] [PubMed] [Google Scholar]
- Wei Y.; Li C.; Zhang L.; Xu X. Design of Novel Cell Penetrating Peptides for the Delivery of Trehalose into Mammalian Cells. Biochim. Biophys. Acta - Biomembr. 2014, 1838, 1911. 10.1016/j.bbamem.2014.02.011. [DOI] [PubMed] [Google Scholar]
- Humphrey W.; Dalke A.; Schulten K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33. 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Kumari R.; Kumar R.; Lynn A. G-Mmpbsa -A GROMACS Tool for High-Throughput MM-PBSA Calculations. J. Chem. Inf. Model. 2014, 54 (7), 1951–1962. 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
- Yun S.; Urbanc B.; Cruz L.; Bitan G.; Teplow D. B.; Stanley H. E. Role of Electrostatic Interactions in Amyloid β-Protein (Aβ) Oligomer Formation: A Discrete Molecular Dynamics Study. Biophys. J. 2007, 92 (11), 4064–4077. 10.1529/biophysj.106.097766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyer W.; Antony T.; Cherny D.; Heim G.; Jovin T. M.; Subramaniam V. Dependence of α-Synuclein Aggregate Morphology on Solution Conditions. J. Mol. Biol. 2002, 322 (2), 383–393. 10.1016/S0022-2836(02)00775-1. [DOI] [PubMed] [Google Scholar]
- Zhang G.; Zhang X.; Wang X.; Li J.-P. Towards Understanding the Roles of Heparan Sulfate Proteoglycans in Alzheimer’s Disease. Biomed Res. Int. 2014, 2014, 516028. 10.1155/2014/516028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So M.; Hata Y.; Naiki H.; Goto Y. Heparin-Induced Amyloid Fibrillation of B2-Microglobulin Explained by Solubility and a Supersaturation-Dependent Conformational Phase Diagram. Protein Sci. 2017, 26 (5), 1024–1036. 10.1002/pro.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noborn F.; O’Callaghan P.; Hermansson E.; Zhang X.; Ancsin J. B.; Damas A. M.; Dacklin I.; Presto J.; Johansson J.; Saraiva M. J.; et al. Heparan Sulfate/Heparin Promotes Transthyretin Fibrillization through Selective Binding to a Basic Motif in the Protein. Proc. Natl. Acad. Sci. U. S. A. 2011, 108 (14), 5584–5589. 10.1073/pnas.1101194108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh D. M.; Lomakin a; Benedek G. B.; Condron M. M.; Teplow D. B. Amyloid β-Protein Fibrillogenesis–Detection of a Protofibrillar Intermediate. J. Biol. Chem. 1997, 272 (35), 22364–22372. 10.1074/jbc.272.35.22364. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.