Abstract
The ability of the dehydrogenase and ring cleavage dioxygenase of the naphthalene degradation pathway to transform 3,4-dihydroxylated biphenyl metabolites was investigated. 1,2-Dihydro-1,2-dihydroxynaphthalene dehydrogenase was expressed as a histidine-tagged protein. The purified enzyme transformed 2,3-dihydro-2,3-dihydroxybiphenyl, 3,4-dihydro-3,4-dihydroxybiphenyl, and 3,4-dihydro-3,4-dihydroxy-2,2′,5,5′-tetrachlorobiphenyl to 2,3-dihydroxybiphenyl, 3,4-dihydroxybiphenyl (3,4-DHB), and 3,4-dihydroxy-2,2′,5,5′-tetrachlorobiphenyl (3,4-DH-2,2′,5,5′-TCB), respectively. Our data also suggested that purified 1,2-dihydroxynaphthalene dioxygenase catalyzed the meta cleavage of 3,4-DHB in both the 2,3 and 4,5 positions. This enzyme cleaved 3,4-DH-2,2′,5,5′-TCB and 3,4-DHB at similar rates. These results demonstrate the utility of the naphthalene catabolic enzymes in expanding the ability of the bph pathway to degrade polychlorinated biphenyls.
Oxidative cometabolism of polychlorinated biphenyls (PCBs) by the biphenyl degradation (bph) pathway has been intensively investigated as a means to remove these persistent pollutants from environmental samples (9). This pathway comprises four enzymatic activities which sequentially transform PCBs to chlorobenzoates. The first step in this transformation, 2,3-dihydroxylation of the biphenyl, is catalyzed by a three-component biphenyl dioxygenase (BPH dox). It has been established that the abilities of different bacterial strains to preferentially transform different PCB congeners are specified by structural determinants of the oxygenase component of BPH dox (17, 27). In contrast to the burgeoning literature establishing the importance of the substrate selectivity of BPH dox in the degradation of PCBs, very little has been reported on the substrate specificities of the three subsequent enzymes of the bph pathway.
One of the numerous PCB-degrading strains that have been described, Burkholderia cepacia LB400 (also referred as Pseudomonas sp. strain LB400), transforms a particularly broad range of PCB congeners (3, 7). This microorganism has the rare ability to attack congeners such as 2,2′,5,5′-tetrachlorobiphenyl (2,2′,5,5′-TCB), in which no free adjacent ortho-meta positions are available for 2,3-dihydroxylation by LB400 BPH dox. The ability of B. cepacia LB400 to transform these congeners resides in the ability of LB400 BPH dox to catalyze 3,4-dihydroxylation of these compounds. In the case of 2,2′,5,5′-TCB, this gives rise to 3,4-dihydro-3,4-dihydroxy-2,2′,5,5′-TCB (3,4-DD-2,2′,5,5′-TCB). Nadim et al. (28) reported that cell extracts of biphenyl-induced B. cepacia LB400 were inactive against 3,4-DD-2,2′,5,5′-TCB. This observation suggests that in LB400 the second enzyme of the bph pathway, 2,3-dihydro-2,3-dihydroxybiphenyl dehydrogenase (BphB) is not able to catalyze conversion of this metabolite to 3,4-dihydroxy-2,2′,5,5′-TCB (3,4-DH-2,2′,5,5′-TCB). Furthermore, it has been reported that the third enzyme of the B. cepacia LB400 bph pathway, 2,3-dihydroxybiphenyl dioxygenase (BphC), is not able to catalyze extradiol cleavage of 3,4-dihydroxybiphenyl (3,4-DHB) (6). Similar observations have been reported for the BphC of other bacteria (13, 33). Complete mineralization of PCB congeners whose degradation is initiated by 3,4-dihydroxylation thus requires the presence of enzymes that are able to transform these compounds.
The initial enzymatic steps in aerobic degradation of many aromatic compounds are very similar. Enzymes homologous to BPH dox, BphB, and BphC are found in pathways responsible for degradation of naphthalene, toluene, and benzoate, respectively. Moreover, despite the evolutionary adaptation of the aryl-degrading enzymes for specific substrates, the enzymes of a particular pathway generally catalyze the transformation of a range of aromatic compounds, albeit less specifically. For example, the first three enzymes of the bph pathway can sequentially transform toluene, whereas the first three enzymes of the toluene pathway can sequentially transform biphenyl (8). Furthermore, the enzymes of the naphthalene degradation pathway can transform several polycyclic aromatic hydrocarbons (18), indigo (21), and biphenyl (18). In particular, 1,2-dihydro-1,2-dihydroxynaphthalene dehydrogenase oxidizes 1,2-dihydro-1,2-dihydroxyphenanthrene, 1,2-dihydro-1,2-dihydroxytoluene, and 2,3-dihydroxy-2,3-dihydroxybiphenyl (2,3-DDB) to the corresponding catechols (29). Moreover, 1,2-dihydroxynaphthalene dioxygenase catalyzes the meta fission of 2,3-dihydroxybiphenyl (2,3-DHB), as well as the meta fission of 3,4-DHB (20).
The low specificity of BphB and BphC for 3,4-dihydroxylated PCB metabolites raises the question of whether the corresponding enzymes of the naphthalene degradation pathway can be recruited for the transformation of such compounds. We investigated the ability of purified preparations of the nahB-encoded 1,2-dihydro-1,2-dihydroxynaphthalene dehydrogenase (NahB) of Pseudomonas putida G7 (5) and the doxG-encoded 1,2-dihydroxynaphthalene dioxygenase (DoxG) of Pseudomonas sp. strain C18 (4) to transform metabolites of 2,2′,5,5′-TCB. The reaction products were identified. The implications of the results for engineering strains for degradation of PCBs are discussed.
MATERIALS AND METHODS
Bacterial strains, culture media, and general protocols.
The bacterial strains used in this study were Escherichia coli M15(pREP4) and SG13009(pREP4) (both obtained from QIAGEN Inc., Chatsworth, Calif.), P. putida G7 (5), and Pseudomonas sp. strain C18 (4). The media used were Luria-Bertani broth and H-plates (31). DNA was manipulated by using protocols described by Sambrook et al. (31) unless otherwise noted. PCR to amplify B. cepacia LB400 BPH dox genes and P. putida G7 naphthalene dioxygenase (NAH dox) genes plus nahB were performed by using Pwo DNA polymerase and procedures described previously (32).
Production and purification of enzymes.
His-tagged (ht) LB400 BPH dox components and ht-NAH dox components from P. putida G7 were expressed and purified essentially as ht-BPH dox from Comamonas testosteroni B-356 was (15, 16) and will be described elsewhere. Purified G7 ht-NahB was prepared from E. coli M15(pREP4) by using the QIAGEN expression system, as described previously for strain B-356 ht-BphB (32). The following two oligonucleotides used to amplify nahB from the genomic DNA of P. putida G7 by PCR were based on the nucleotide sequence of nahB (unpublished data): oligonucleotide I (5′-GCGGGATCCGGGCAATCAACAAGTCG-3′) and oligonucleotide II (5′-CGACGGTACCTCACTTGCGACCGAGC-3′). These primers introduced BamHI and KpnI restriction sites at the 5′ and 3′ ends, respectively, of the amplified product, thus facilitating subsequent cloning of the gene.
Expression and anaerobic purification of DoxG will be described elsewhere. Even when preparations of DoxG were handled anaerobically, they were found to contain only 0.35 molar equivalent of iron. For this reason, the activities of these preparations were calculated as functions of their iron contents and not as functions of their protein contents. Exogenous iron was anaerobically removed from samples of DoxG prior to kinetic experiments by gel filtration chromatography. Typically, 100 to 200 μl of purified DoxG (10 mg/ml) was thawed in an anaerobic glove box and applied to a Bio-Gel P6 DG column (0.7 by 5 cm; Bio-Rad, Mississauga, Ontario, Canada) equilibrated with 20 mM HEPPS (N-[2-hydroxyethyl]piperazine-N’-3-propanesulfonic acid)–80 mM NaCl (pH 8.0) containing 10% glycerol and 2 mM dithiothreitol. Prior to use, the enzyme was stored in an inert atmosphere on ice to maximize its activity. The iron concentrations of DoxG preparations were determined colorimetrically by using Ferene S (11).
Protein characterization.
The purity and Mr of the protein were evaluated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as described by Laemmli (22). The gels contained 12% acrylamide and were stained with Coomassie brilliant blue (31). Promega’s midrange markers were used as Mr standards. Protein concentrations were estimated by the method of Lowry et al. (24) by using bovine serum albumin as a standard.
Production of dihydroxylated compounds.
3,4-DD-2,2′,5,5′-TCB was produced enzymatically from 2,2′,5,5′-TCB (ULTRA Scientific, North Kingstown, R.I.) by using reconstituted LB400 ht-BPH dox. The transformation mixture contained 75 nmol of each LB400 ht-BPH dox component (reductase, ferredoxin, and oxygenase), 20 μmol of NADH, and 12 nmol of FeSO4 in 10 ml (total volume) of 50 mM morpholineethanesulfonic acid (MES) buffer (pH 6.0). The transformation was initiated by adding 10 μmol of 2,2′,5,5′-TCB dissolved in 200 μl of acetone, and the preparation was incubated for 10 min at 37°C. 3,4-DD-2,2′,5,5′-TCB was purified from the mixture by reverse-phase chromatography by using an octyldecyl silane Hypersil II (5 μm) column (4 mm by 25 cm), and its identity was verified by performing a gas chromatography (GC)-mass spectrometry (MS) analysis of its butylboronate derivatives by using the protocols described previously for purification and identification of 2,3-DDB (32).
Production of 3,4-DD-2,2′,5,5′-TCB was routinely monitored by performing an analysis with a high-performance liquid chromatograph coupled to a Perkin-Elmer model LC95 UV-visible detector. The detector was set at 283 nm, which was determined to be the maximal wavelength of 3,4-DD-2,2′,5,5′-TCB by Haddock et al. (10). 3,4-DD-2,2′,5,5′-TCB was quantified by using a standard curve in which the increase in the amount of 3,4-DD-2,2′,5,5′-TCB produced in the transformation reaction at different time points was equated to the depletion of 2,2′,5,5′-TCB.
A mixture of 2,3-DDB and 3,4-dihydro-3,4-dihydroxybiphenyl (3,4-DDB) was produced enzymatically by using reconstituted strain G7 ht-NAH dox. The transformation mixture contained 0.6 nmol of each G7 ht-NAH dox component, 100 nmol of NADH, and 1.2 nmol of FeSO4 in 2 ml (total volume) of 50 mM MES buffer (pH 6.0). The transformation was initiated by adding 100 nmol of biphenyl (Aldrich Chemicals, Milwaukee, Wis.) dissolved in 2 μl of acetone, and the preparation was incubated for 10 min at 37°C. The ethyl acetate extract of the reaction mixture contained essentially pure 2,3-DDB and 3,4-DDB.
NahB assay.
The activity of ht-NahB was measured in 50 mM bicine (pH 9.0) at 37°C. Each reaction mixture (total volume, 200 μl) contained 1.0 mM NAD+ and 100 μg of the purified enzyme. The reaction was initiated by adding different amounts of 3,4-DD-2,2′,5,5′-TCB dissolved in 2 μl of acetone and was stopped after 1 min by adding 400 μl of acetonitrile. Each mixture was vortexed and then centrifuged for 30 s. Fifty microliters of the supernatant was injected onto an octyldecyl silane Hypersil II high-performance liquid chromatography column as described above, and the amount of substrate remaining was evaluated at 283 nm.
The ability of NahB to oxidize 2,3-DDB and 3,4-DDB was verified by using the NAH dox reaction mixture described above. This reaction mixture was incubated for 10 min at 37°C, after which 25 μg of ht-NahB and 1 mM NAD+ were added and the mixture was incubated for an additional 10 min at 37°C. The products of the reaction were extracted with ethyl acetate and subjected to a GC-MS analysis.
DoxG assay.
DoxG activity was measured by monitoring the consumption of dioxygen with a Clark type of polarographic oxygen electrode (model 5301; Yellow Springs Instruments). Reactions were performed in a thermojacketed model RC1 respiration chamber (Cameron Instrument Co., Port Aransas, Tex.) equipped with a Lauda circulating water bath. The electrode signal was amplified with a Cameron Instrument model OM200 oxygen meter before it was recorded with a microcomputer equipped with a PC-LPM-16 multifunction board and Virtual Bench Data Logger (National Instruments, Austin, Tex.). Data were recorded every 0.1 s. Initial velocities were determined from progress curves by analyzing the data with Excel (Microsoft, Redmond, Wash.). The slopes calculated in this study had correlation coefficients of at least 0.998.
Activity assays were performed at 25.0 ± 0.1°C in 1.45 ml (total volume) of air-saturated 20 mM HEPPS–80 mM NaCl (pH 8.0) containing 100 μg of ht-NahB per ml and 1 mM NAD+. Approximately 280 nmol of 3,4-DD-2,2′,5,5′-TCB dissolved in 17 μl of acetone was injected into the reaction chamber and incubated for 2 min. This amount was sufficient to convert 3,4-DD-2,2′,5,5′-TCB to 3,4-DH-2,2′,5,5′-TCB essentially quantitatively. The ring cleavage reaction was initiated by injecting between 10 and 25 μl of an appropriate dilution of a DoxG preparation into the reaction chamber. For assays involving 2,3-DHB (Waco Chemicals, Dallas, Tex.) or 3,4-DHB (ULTRA Scientific), the reaction mixture contained no ht-NahB or NAD+. Buffers and stock solutions were prepared fresh daily, and the stock solutions were stored under argon at 4°C. The oxygen electrode was calibrated on each day that kinetic assays were performed by using standard concentrations of 2,3-DHB and an excess of LB400 BphC. One unit of enzymatic activity was defined as the quantity of enzyme required to consume 1 μmol of dioxygen per min.
Analysis of the reaction products.
The metabolites were extracted with ethyl acetate, treated with N,O-bis-trimethylsilyl trifluoroacetamide or n-butylboronic acid, and analyzed by GC-MS as described previously (26, 32).
RESULTS AND DISCUSSION
Preliminary work in our laboratory performed with cell extracts of naphthalene-grown P. putida G7 carrying the NAH plasmid confirmed previously reported data (20) which showed that 3,4-DHB is converted by 1,2-dihydroxynaphthalene dioxygenase to a yellow metabolite which exhibits maximum absorbance at 380 nm at pH 7.5. In the presence of purified components of LB400 BPH dox, the same cell extract also transformed 2,2′,5,5′-TCB into a colored metabolite. These observations suggested that NahB could catalyze dehydrogenation of 3,4-DD-2,2′,5,5′-TCB to the corresponding catechol, which could then be cleaved by 1,2-dihydroxynaphthalene dioxygenase.
To further investigate this transformation, the individual enzymatic steps were investigated with purified 3,4-DD-2,2′,5,5′-TCB and two purified enzymes, G7 ht-NahB and DoxG of Pseudomonas sp. strain C18. The sequence of DoxG is 97% identical to the sequence of G7 1,2-dihydroxynaphthalene dioxygenase (NahC) (12).
Production, purification, and characterization of G7 ht-NahB.
Amplification of the nahB gene from the genomic DNA of P. putida G7 yielded a DNA fragment that was approximately 850 bp long. Ligation of the BamHI-KpnI-digested fragment into appropriately digested plasmid pQE31 produced plasmid pQE31::NahB. The nucleotide sequence of the nahB gene in this construction was verified. In this construction the nahB gene was immediately downstream of a sequence encoding a polyhistidine tag, which added 13 amino acids (MRGSHHHHHHTDP) to the N terminus of G7 NahB, producing G7 ht-NahB.
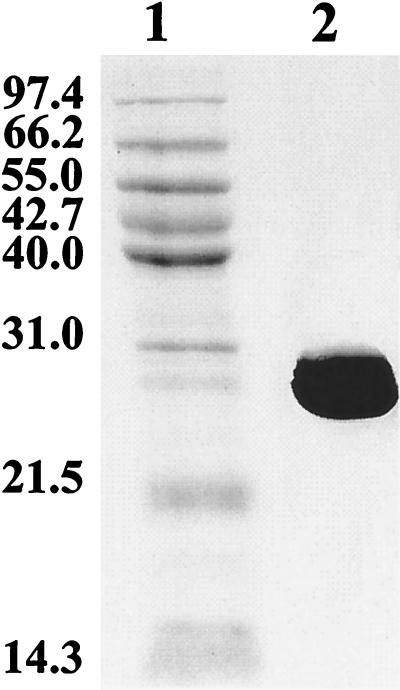
Approximately 30 mg of purified G7 ht-NahB was obtained from 1 liter of induced E. coli M15 harboring pQE31::NahB. The protein was estimated to be more than 99% pure by using Coomassie brilliant blue-stained SDS-PAGE gels (Fig. 1). The Mr of the G7 ht-NahB polypeptide was estimated to be 29,000 based on its migration in SDS-PAGE gels. The enzyme could be stored for days at 4°C with no detectable loss of activity. G7 ht-NahB had an absolute requirement for NAD+. The optimal pH of G7 ht-NahB was found to be 9.0, but the enzyme was fairly active at pH 7.0, as well as pH 6.0. These biochemical features of G7 ht-NahB are similar to those reported for NahB purified from P. putida NP (30).
FIG. 1.
Coomassie brilliant blue-stained SDS-PAGE gel of G7 ht-NahB. The gel was loaded with 6 μg of protein eluted from the Ni2+-nitrilotriacetic acid resin (lane 1) and the Promega midrange Mr standard (lane 2). The separating gel contained 12% acrylamide.
Production of 3,4-DDB and 3,4-DD-2,2′,5,5′-TCB.
Purified G7 ht-NAH dox produced two dihydrodiol metabolites from biphenyl, which were detected by GC-MS as their butylboronate derivatives (data not shown). The major metabolite, which accounted for 97% of the total product, was identified as 2,3-DDB by comparison with spectra of the authentic compound. The MS spectra of the minor metabolite, which accounted for the remaining 3% of the product, had the main features of the spectra of dihydro-dihydroxybiphenyls bearing hydroxyl groups on vicinal carbons. On the basis of results described below we identified this minor product as 3,4-DDB.
Under the reaction conditions described in Materials and Methods, 10 μmol of 2,2′,5,5′-TCB was transformed essentially quantitatively to 3,4-DD-2,2′,5,5′-TCB by LB400 BPH dox within 10 min. 3,4-DD-2,2′,5,5′-TCB was stable for hours in 50 mM phosphate (pH 7.0), as well as in 50 mM bicine (pH 9.0).
Transformation of 3,4-DDB and 3,4-DD-2,2′,5,5′-TCB by G7 ht-NahB.
3,4-DDB is not available commercially, so the ability of G7 ht-NahB to catalyze dehydrogenation of this compound was investigated by using an enzymatically produced mixture of 2,3-DDB and 3,4-DDB. When the NAH dox reaction was coupled to the NahB reaction as described in Materials and Methods, the two dihydro-dihydroxybiphenyls produced were quantitatively converted to the respective catechols. The two catechols produced were identified as 2,3-DHB and 3,4-DHB by comparing their GC retention times and MS spectra (data not shown) with the GC retention times and MS spectra of commercially available compounds. These data showed that dihydroxylation of biphenyl by G7 ht-NAH dox results in production of 2,3-DDB and 3,4-DDB. More importantly, they demonstrated that G7 ht-NahB dehydrogenates 2,3-DDB and 3,4-DDB to 2,3-DHB and 3,4-DHB, respectively.
G7 ht-NahB transformed enzymatically produced, purified 3,4-DD-2,2′,5,5′-TCB into a single metabolite. This metabolite was identified as 3,4-DH-2,2′,5,5′-TCB based on its molecular mass and the similarity of its MS spectra to the spectra of authentic 3,4-DHB (data not shown). The reaction was dependent on the presence of G7 ht-NahB and NAD+ (results not shown). The rate of 3,4-DD-2,2′,5,5′-TCB consumption was determined to be 149 nmol min−1 mg of protein−1 when the reaction was performed as described in Materials and Methods and was initiated with 150 μM 3,4-DD-2,2′,5,5′-TCB.
Transformation of 3,4-DHB by DoxG.
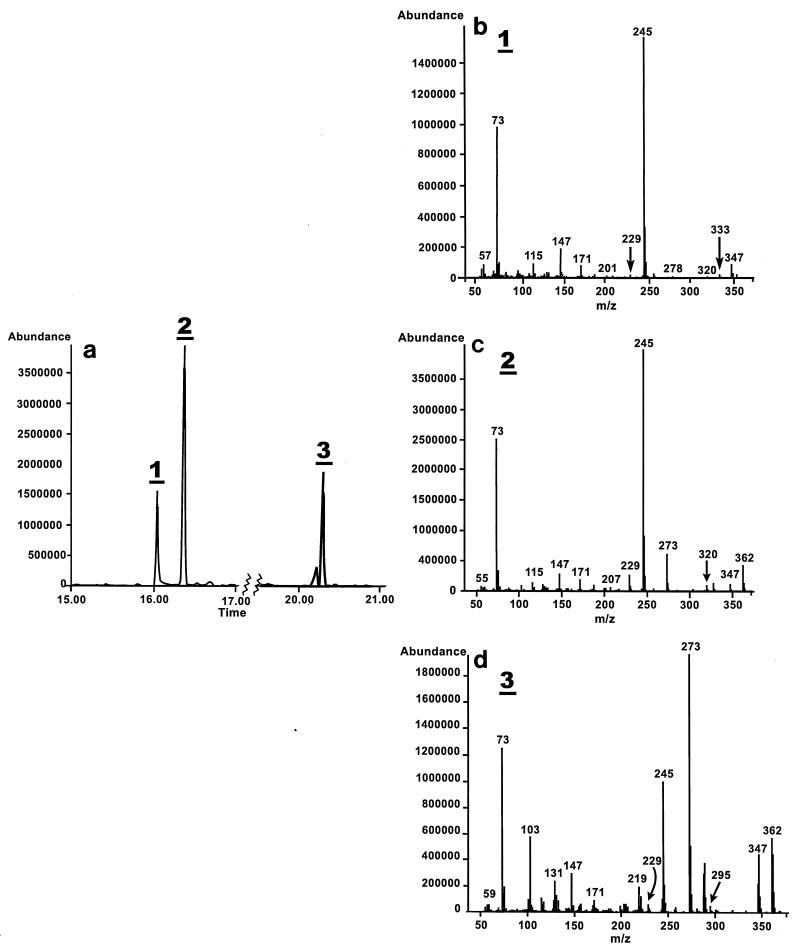
DoxG catalyzed the meta fission of 3,4-DHB, as has been reported previously for other 1,2-dihydroxynaphthalene dioxygenases (20). GC-MS analysis of the trimethylsilyl (TMS) derivatives of the reaction product revealed the presence of three metabolites (Fig. 2) at a ratio of approximately 1:2.6:1.3. These metabolites had similar ion fragmentation patterns. Although the fragmentation pattern of metabolite 1 lacked an ion at molecular mass M+ (m/z 362), all three metabolites yielded two diagnostically important ions, an ion at M-15 (m/z 347) and an ion at m/z 245 (M-COOTMS). The latter ion is characteristic of acidic compounds.
FIG. 2.
Transformation of 3,4-DHB by DoxG. (a) GC elution profile of the metabolites produced by DoxG-catalyzed meta cleavage of 3,4-DHB. (b through d) Mass spectra of the meta-cleavage products 1 through 3. The experimental conditions are described in Materials and Methods.
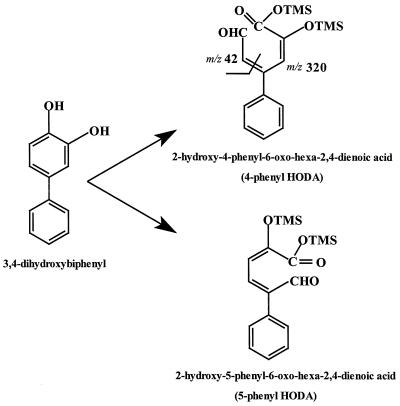
As shown in Fig. 3, there are two ways in which 3,4-DHB can be cleaved in a meta fashion; cleavage between C-2 and C-3 yields 2-hydroxy-5-phenyl-6-oxo-hexa-2,4-dienoic acid (5-phenyl HODA), whereas cleavage between C-4 and C-5 yields 2-hydroxy-4-phenyl-6-oxo-hexa-2,4-dienoic acid (4-phenyl HODA). These two 2-hydroxy-6-oxo-hexa-2,4-dienoic acids (HODAs) have identical predicted molecular masses, which correspond to the molecular masses of the three observed metabolites. However, due to the different positions of the phenyl substituents, we predicted that only 4-phenyl HODA should yield an ion at m/z 320 (M-42), as shown in Fig. 3. Metabolites 1 and 2 had similar retention times, and their fragmentation patterns revealed an ion at m/z 320. These metabolites were thus tentatively identified as isomers of 4-phenyl HODA. It is not clear whether the two isomers of 4-phenyl HODA were cis-trans isomers that arose after the meta-cleavage reaction or whether they were produced during the reaction with N,O-bis-trimethylsilyl trifluoroacetamide which generated the TMS derivatives.
FIG. 3.
Possible meta-cleavage products of 3,4-DHB. Two ions that are predicted to specifically arise from fragmentation of TMS-derivatized 4-phenyl HODA are indicated.
Metabolite 3, which did not yield an ion at m/z 320, was either 5-phenyl HODA or a tautomer of 4-phenyl HODA. While the HODAs produced by meta cleavage of catechols are generally depicted as dienols, their tautomerization to their keto forms has not been well studied. The meta-cleavage metabolite of phenylpropionic acid exists almost exclusively in the dienol form (23). Interestingly, it has been proposed that the hydrolase that catalyzes hydrolysis of the phenylpropionic acid ring fission metabolite catalyzes the tautomerization of HODAs prior to C-C bond hydrolysis (14). A GC-MS analysis after meta cleavage of 2,3-DHB by 2,3-dihydroxybiphenyl dioxygenase of C. testosteroni B-356 also revealed two products having identical molecular masses (26). At this time, however, there is no documented evidence of spontaneous tautomerization of phenyl HODAs. Therefore, metabolite 3 is likely to be 5-phenyl HODA. Although the exact nature of the metabolites produced by meta cleavage of 3,4-DHB by DoxG has yet to be determined unequivocably, our results suggest that this enzyme can open the ring on either side of the vicinal hydroxyl groups. The engineering of strains to enhance degradation of meta-para hydroxylated PCB metabolites will have to accommodate the production of these two types of meta-cleavage products.
As the cleavage of 3,4-DHB by DoxG gave rise to more than one product, the ring cleavage of 3,4-DHB was monitored by measuring oxygen uptake. The assay mixtures typically contained 2.5 μM iron, which corresponded to 80 μg of active DoxG per ml. When 258 μM 3,4-DHB was used, an initial rate of O2 consumption of 21.8 U/μmol of Fe was obtained. Interestingly, a similar concentration of 3,4-DH-2,2′,5,5′-TCB generated in situ from 3,4-DD-2,2′,5,5′-TCB by G7 ht-NahB was cleaved at a rate comparable to the rate of cleavage of the unchlorinated compound. Thus, the initial rate of O2 consumption in the presence of 202 μM 3,4-DD-2,2′,5,5′-TCB was 17.6 μmol min−1 μmol of iron−1 in the coupled G7 NahB-DoxG assay, compared to 21.8 μmol min−1 μmol of iron−1 in the presence of 258 μM 3,4-DHB in the DoxG assay. In both cases, O2 consumption was dependent on the presence of DoxG. In addition, O2 consumption in the presence of 3,4-DD-2,2′,5,5′-TCB was observed only when G7 ht-NahB and NAD+ were present in the reaction mixture.
Unfortunately, the amount of metabolite produced by meta cleavage of 3,4-DH-2,2′,5,5′-TCB by DoxG was too small for structural analyses. However, it is interesting to note that whether the ring is cleaved at position 2,3 or 4,5, a 3-acyl chloride is expected to be produced. Such compounds have been found to be potent irreversible inhibitors of extradiol dioxygenases (1, 19). While DoxG cleaved 3,4-DH-2,2′,5,5′-TCB and 3,4-DHB at comparable rates, it will be interesting to determine whether the 3-acyl chlorides are produced by cleavage of 3,4-DH-2,2′,5,5′-TCB or whether these compounds undergo concomitant dechlorination, as appears to be the case for meta cleavage of 3-chlorocatechol by chlorocatechol 2,3-dioxygenase (25).
In the current work we investigated transformation of 3,4-dihydroxylated biphenyl metabolites by the naphthalene-degrading enzymes. A small number of PCB degraders, including B. cepacia LB400 and Alcaligenes eutrophus H850, dihydroxylate certain congeners in the 3,4 position instead of the usual 2,3 position. While this ability resides in the activity of the BPH dox, previous work has indicated that the resultant 3,4-hydroxylated metabolites are not further degraded by subsequent biphenyl catabolic enzymes (28). It has been suggested that in the natural environment, these dead-end metabolites polymerize and combine with humic acids found in soils, which decreases their bioavailability (2). Utilization of bacteria for remediation of PCB-contaminated soil will require the development of microorganisms that can completely mineralize all PCB congeners.
One possible way to engineer microorganisms to degrade PCBs is to recruit catabolic enzymes whose specificities complement those of the enzymes of the biphenyl degradation pathway. In this work, we clearly showed that NahB and DoxG are enzymes that could potentially serve this purpose as they can metabolize both chlorinated and unchlorinated metabolites of 3,4-dihydroxylated biphenyls. Although the observed levels of activity are somewhat low, particularly in the case of oxidative meta cleavage of 3,4-DD-2,2′,5,5′-TCB, these enzymes represent a good starting point for development of variant enzymes that exhibit increased activity towards the meta-para hydroxylated PCB metabolites. Thus, the enzymes of the naphthalene degradation pathway appear to be useful for engineering bacteria with enhanced PCB-degrading activities.
ACKNOWLEDGMENTS
This work was supported by grant STP0193182 from the Natural Sciences and Engineering Research Council of Canada.
We thank Alain Charlebois for technical assistance with the GC-MS analysis and Nathalie Drouin for skilled technical assistance with the DoxG assays.
REFERENCES
- 1.Bartels I, Knackmuss H-J, Reineke W. Suicide inactivation of catechol 2,3-dioxygenase from Pseudomonas putida mt-2 by 3-halocatechols. Appl Environ Microbiol. 1984;47:500–505. doi: 10.1128/aem.47.3.500-505.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baxter R M. Bacterial formation of humus-like materials from polychlorinated biphenyls (PCBs) Water Pollut Res Can. 1986;21:1–7. [Google Scholar]
- 3.Bedard D L, Unterman R, Bopp L H, Brennan M J, Haberl M L, Johnson C. Rapid assay for screening and characterizing microorganisms for the ability to degrade polychlorinated biphenyls. Appl Environ Microbiol. 1986;51:761–768. doi: 10.1128/aem.51.4.761-768.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Denome S A, Stanley D C, Olson E S, Young K D. Metabolism of dibenzothiophene and naphthalene in Pseudomonas strains: complete DNA sequence of an upper naphthalene catabolic pathway. J Bacteriol. 1993;175:6890–6901. doi: 10.1128/jb.175.21.6890-6901.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunn N W, Gunsalus I C. Transmissible plasmids coding early enzymes of naphthalene oxidation in Pseudomonas putida. J Bacteriol. 1973;114:974–979. doi: 10.1128/jb.114.3.974-979.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eltis L D, Hofmann B, Hecht H J, Lunsdorf H, Timmis K N. Purification and crystallization of 2,3-dihydroxybiphenyl 1,2-dioxygenase. J Biol Chem. 1993;268:2727–2732. [PubMed] [Google Scholar]
- 7.Erickson B D, Mondello F J. Enhanced biodegradation of polychlorinated biphenyl after site-directed mutagenesis of a biphenyl dioxygenase gene. Appl Environ Microbiol. 1993;59:3858–3862. doi: 10.1128/aem.59.11.3858-3862.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Furukawa K, Hirose J, Suyama A, Zaiki T, Hayashida S. Gene components responsible for discrete substrate specificity in the metabolism of biphenyl (bph operon) and toluene (tod operon) J Bacteriol. 1993;175:5224–5232. doi: 10.1128/jb.175.16.5224-5232.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furukawa K, Tonomura K, Kamibayashi A. Effect of chlorine substitution on the biodegradability of polychlorinated biphenyls. Appl Environ Microbiol. 1978;35:223–227. doi: 10.1128/aem.35.2.223-227.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haddock J D, Horton J R, Gibson D T. Dihydroxylation and dechlorination of chlorinated biphenyls by purified biphenyl 2,3-dioxygenase from Pseudomonas sp. strain LB400. J Bacteriol. 1995;177:20–26. doi: 10.1128/jb.177.1.20-26.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haigler B, Gibson D T. Purification and properties of NADH-ferredoxinNAP reductase, a component of naphthalene dioxygenase from Pseudomonas sp. strain NCIB 9816. J Bacteriol. 1990;172:457–464. doi: 10.1128/jb.172.1.457-464.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harayama S, Rekik M. Bacterial aromatic ring-cleavage enzymes are classified into two different gene families. J Biol Chem. 1989;264:15238–15333. [PubMed] [Google Scholar]
- 13.Hein P, Powlowski J, Barriault D, Hurtubise Y, Ahmad D, Sylvestre M. Biphenyl-associated meta-cleavage dioxygenases from Comamonas testosteroni B-356. Can J Microbiol. 1998;44:42–49. [PubMed] [Google Scholar]
- 14.Henderson I M J, Bugg T D H. Pre-steady-state kinetic analysis of 2-hydroxy-6-keto-nona-2,4-diene-1,9-dioic acid 5,6-hydrolase: kinetic evidence for enol/keto tautomerization. Biochemistry. 1997;36:12252–12258. doi: 10.1021/bi971116j. [DOI] [PubMed] [Google Scholar]
- 15.Hurtubise Y, Barriault D, Sylvestre M. Characterization of active recombinant His-tagged oxygenase component of Comamonas testosteroni B-356 biphenyl dioxygenase. J Biol Chem. 1996;271:8152–8156. doi: 10.1074/jbc.271.14.8152. [DOI] [PubMed] [Google Scholar]
- 16.Hurtubise Y, Barriault D, Powlowski J, Sylvestre M. Purification and characterization of the Comamonas testosteroni B-356 biphenyl dioxygenase components. J Bacteriol. 1995;177:6610–6618. doi: 10.1128/jb.177.22.6610-6618.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kimura N, Nishi A, Goto M, Furukawa K. Functional analysis of a variety of chimeric dioxygenases constructed from two biphenyl dioxygenases that are similar structurally but different functionally. J Bacteriol. 1997;179:3936–3943. doi: 10.1128/jb.179.12.3936-3943.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiyohara H, Torigoe S, Kaida N, Asaki T, Iida T, Hayashi H, Takizawa N. Cloning and characterization of a chromosomal gene cluster, pah, that encodes the upper pathway for phenanthrene and naphthalene utilization by Pseudomonas putida OUS82. J Bacteriol. 1994;176:2439–2443. doi: 10.1128/jb.176.8.2439-2443.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klecka G M, Gibson D T. Inhibition of catechol 2,3-dioxygenase from Pseudomonas putida by 3-chlorocatechol. Appl Environ Microbiol. 1981;41:1159–1165. doi: 10.1128/aem.41.5.1159-1165.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuhm A E, Stolz A, Ngai K L, Knackmuss H J. Purification and characterization of a 1,2-dihydroxynaphthalene dioxygenase from a bacterium that degrades naphthalenesulfonic acids. J Bacteriol. 1991;173:3795–3802. doi: 10.1128/jb.173.12.3795-3802.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kurkela S, Lehvaslaiho H, Palva E T, Teeri T H. Cloning, nucleotide sequence and characterization of genes encoding naphthalene dioxygenase of Pseudomonas putida strain NCIB9816. Gene. 1988;73:355–362. doi: 10.1016/0378-1119(88)90500-8. [DOI] [PubMed] [Google Scholar]
- 22.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Lam W W Y, Bugg T D H. Purification, characterization, and stereochemical analysis of a C-C hydrolase: 2-hydroxy-6-keto-nona-2,4-diene-1,9-dioic acid 5,6-hydrolase. Biochemistry. 1997;36:12242–12251. doi: 10.1021/bi971115r. [DOI] [PubMed] [Google Scholar]
- 24.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 25.Mars A E, Kasberg T, Kaschabek S R, Vanagteren M H, Janssen D B, Reineke W. Microbial degradation of chloroaromatics: use of the meta-cleavage pathway for mineralization of chlorobenzene. J Bacteriol. 1997;179:4530–4537. doi: 10.1128/jb.179.14.4530-4537.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Massé R, Messier F, Ayotte C, Lévesque M-F, Sylvestre M. A comprehensive chromatographic/mass spectrometric analysis of 4-chlorobiphenyl bacterial degradation products. Biomed Environ Mass Spectr. 1989;18:27–47. [Google Scholar]
- 27.Mondello F J, Turcich M P, Lobos J H, Erickson B D. Identification and modification of biphenyl dioxygenase sequences that determine the specificity of polychlorinated biphenyl degradation. Appl Environ Microbiol. 1997;63:3096–3103. doi: 10.1128/aem.63.8.3096-3103.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nadim L M, Schocken M J, Higson F K, Gibson D T, Bedard L D, Bopp L H, Mondello F J. Proceedings of the US EPA Thirteenth Annual Research Symposium on Land Disposal, Remedial Action, Incineration, and Treatment of Hazardous Waste. Cincinnati, Ohio: Environmental Protection Agency; 1987. Bacterial oxidation of polychlorinated biphenyls; pp. 395–402. [Google Scholar]
- 29.Patel T R, Gibson D T. Bacterial cis-dihydrodiol dehydrogenases: comparison of physicochemical and immunological properties. J Bacteriol. 1976;128:842–850. doi: 10.1128/jb.128.3.842-850.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patel T R, Gibson D T. Purification and properties of (+)-cis-naphthalene dihydrodiol dehydrogenase of Pseudomonas putida. J Bacteriol. 1974;119:879–888. doi: 10.1128/jb.119.3.879-888.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 32.Sylvestre M, Hurtubise Y, Barriault D, Bergeron J, Ahmad D. Characterization of active recombinant 2,3-dihydro-2,3-dihydroxybiphenyl dehydrogenase from Comamonas testosteroni B-356 sequence of the encoding gene (bphB) Appl Environ Microbiol. 1996;62:2710–2715. doi: 10.1128/aem.62.8.2710-2715.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taira K, Hayase N, Arimura N, Yamashita S, Miyazaki T, Furukawa K. Cloning and nucleotide sequence of the 2,3-dihydroxybiphenyl dioxygenase gene from the PCB-degrading strain of Pseudomonas paucimobilis Q1. Biochemistry. 1988;27:3990–3996. doi: 10.1021/bi00411a015. [DOI] [PubMed] [Google Scholar]