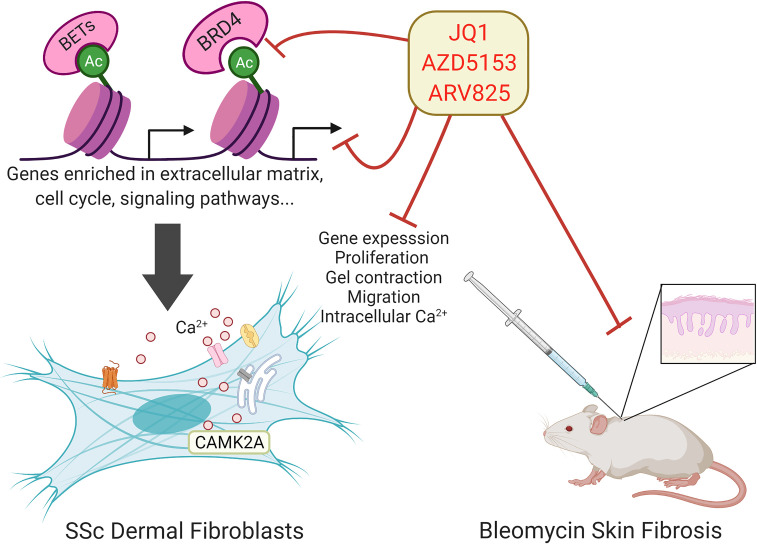
Abstract
Binding of the bromodomain and extraterminal domain proteins (BETs) to acetylated histone residues is critical for gene transcription. We sought to determine the antifibrotic efficacy and potential mechanisms of BET inhibition in systemic sclerosis (SSc). Blockade of BETs was done using a pan-BET inhibitor, JQ1; BRD2 inhibitor, BIC1; or BRD4 inhibitors AZD5153 or ARV825. BET inhibition, specifically BRD4 blockade, showed antifibrotic effects in an animal model of SSc and in patient-derived diffuse cutaneous SSc (dcSSc) fibroblasts. Transcriptome analysis of JQ1-treated dcSSc fibroblasts revealed differentially expressed genes related to extracellular matrix, cell cycle, and calcium (Ca2+) signaling. The antifibrotic effect of BRD4 inhibition was mediated at least in part by downregulation of Ca2+/calmodulin–dependent protein kinase II α and reduction of intracellular Ca2+ concentrations. On the basis of these results, we propose targeting Ca2+ pathways or BRD4 as potentially novel therapeutic approaches for progressive tissue fibrosis.
Keywords: Autoimmunity
Keywords: Autoimmune diseases, Epigenetics, Fibrosis
Introduction
Systemic sclerosis (SSc; also called scleroderma) is an autoimmune disease characterized by vascular dysfunction as well as excessive synthesis and deposition of extracellular matrix (ECM) in affected organs. Activation of the immune system and vasculopathy precede fibrosis, in which fibroblast activation and subsequent myofibroblast transdifferentiation are necessary events. At present, SSc is not curable; current treatments focus on managing disease manifestations in an effort to ease the progression of tissue fibrosis.
Although the exact etiology of the disease is not known, a growing body of literature has been pointing to the critical involvement of epigenetic mechanisms in SSc pathogenesis (1). Epigenetic regulation affects chromatin dynamics and thereby modulates gene transcription. In addition to DNA methylation and noncoding RNAs, histone changes are implicated in SSc fibroblast activation (2–4). Acetylation of histones on lysine residues is one of the most common histone modifications that relaxes the chromatin structure by loosening the histone–DNA interaction, which results in increased chromatin accessibility for transcription. Dysregulation of histone acetylation could result in aberrant gene expression, leading to pathogenic consequences. These histone marks, therefore, are tightly regulated by a set of histone acetyltransferases and histone deacetylases. They are also controlled by proteins containing the bromodomain module, such as the bromodomain extraterminal domain (BET) family proteins. The 4 members of the BETs — BRD2, BRD3, BRD4, and the testis-specific BRD-t — share a common domain consisting of 2 N-terminal bromodomains, BD1 and BD2, that bind to acetylated lysine residues on histones. These histone readers provide scaffolds to attract components of the transcriptional machinery to histone acetylation marks. Pharmacological inhibition of BET proteins results in repression of downstream gene expression, thereby modulating various physiological conditions. Indeed, prototype BET inhibitors such as JQ1 or I-BET attenuate various types of cancer and tissue fibrosis (5–9).
Considering the potential antifibrotic properties of BET inhibition, we investigated whether the BET inhibitor JQ1 can modulate fibrogenesis in an animal model of SSc, as well as in dermal fibroblasts isolated from patients with diffuse cutaneous SSc (dcSSc). We posit that BET inhibition impedes the expression of profibrotic genes and blocks myofibroblast differentiation in SSc, thereby improving fibrosis. Through whole transcriptomic analysis, we revealed a mechanism by which BET inhibition exerts its antifibrotic effect in SSc. Furthermore, we demonstrated the involvement of BRD4 in mediating the profibrotic effect of BETs and identified BRD4 as a therapeutic target for this disease. We believe the novelty of our study is that we performed a comprehensive analysis by using both functional assays and transcriptomic analysis, which allowed us to pinpoint a mechanism whereby BET inhibition affects intracellular calcium (Ca2+) and related signaling in SSc fibroblasts. We also show the antifibrotic properties of 2 BRD4 inhibitors, 1 of which, AZD5153, is being evaluated in a clinical trial (ClinicalTrials.gov NCT03205176). To our knowledge, the effect of these inhibitors has not been examined in other fibrotic models before.
Results
The antifibrotic effects of JQ1 in bleomycin-treated mice.
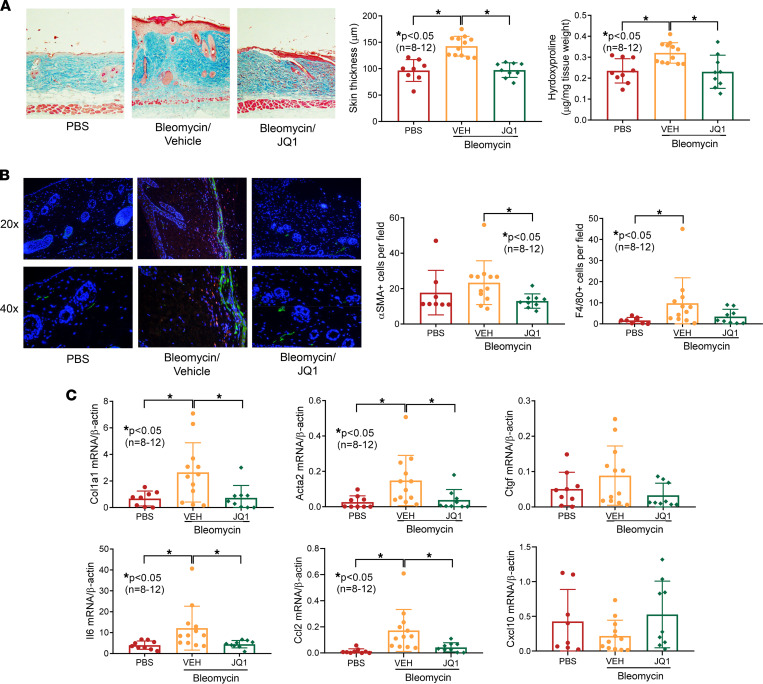
To investigate the antifibrotic effect of BET inhibition, we used a pan-BET inhibitor, JQ1, in an animal model of fibrosis. We observed an approximately 3.5% weight reduction in the bleomycin/vehicle group and a 11.4% weight reduction in the bleomycin/JQ1 group. All animals were active without apparent distress throughout the course of the experiment. Daily injections of bleomycin in a defined area in the back of mice increased dermal thickness and collagen accumulation (Figure 1A). Daily JQ1 oral gavage prevented skin fibrosis in bleomycin-treated mice; importantly, attenuation of dermal thickness and collagen was observed. In addition, immunofluorescent staining revealed increased α-SMA– and F4/80-positive cells in bleomycin-treated mice, with F4/80 staining reaching statistical significance (Figure 1B). JQ1 treatment reduced α-SMA– and F4/80-positive cells, with α-SMA staining reaching statistical significance. These results were also reflected at the mRNA level, as profibrotic genes, including Acta2 and Col1a1, whose expression was significantly elevated in bleomycin-treated mice, were downregulated in the presence of JQ1 (Figure 1C). Bleomycin also induced a significant increase in Il6 and Ccl2 expression, and both were significantly downregulated with JQ1 treatment. In contrast, Ctgf and Cxcl10 expression were not significantly altered by bleomycin or JQ1.
Figure 1. BET inhibition by JQ1 prevents bleomycin-induced skin fibrosis in mice.
(A) Bleomycin-treated mice showed increased dermal thickness and hydroxyproline content in skin, and JQ1 (50 mg/kg) efficiently prevented skin fibrosis in these mice. Original magnification, ×100. (B) Immunofluorescence staining of skin sections showed that JQ1 significantly decreased α-SMA–positive cells (green) in bleomycin-treated mice compared with the vehicle (VEH) group. Similar results were found quantifying F4/80-positive cells (red). Nuclei were stained with DAPI (blue). Original magnification, ×200–400. (C) Acta2, Col1a1, Il6, and Ccl2 were significantly elevated in bleomycin-treated mice and significantly reduced when JQ1 was given. (PBS, n = 8; VEH, n = 12; JQ1, n = 9). Results are expressed as mean ± SD and P < 0.05 was considered significant. Significance was determined by 1-way ANOVA (A and C) and Kruskal-Wallis test (B and C).
Antifibrotic properties of BET inhibition in dermal fibroblasts.
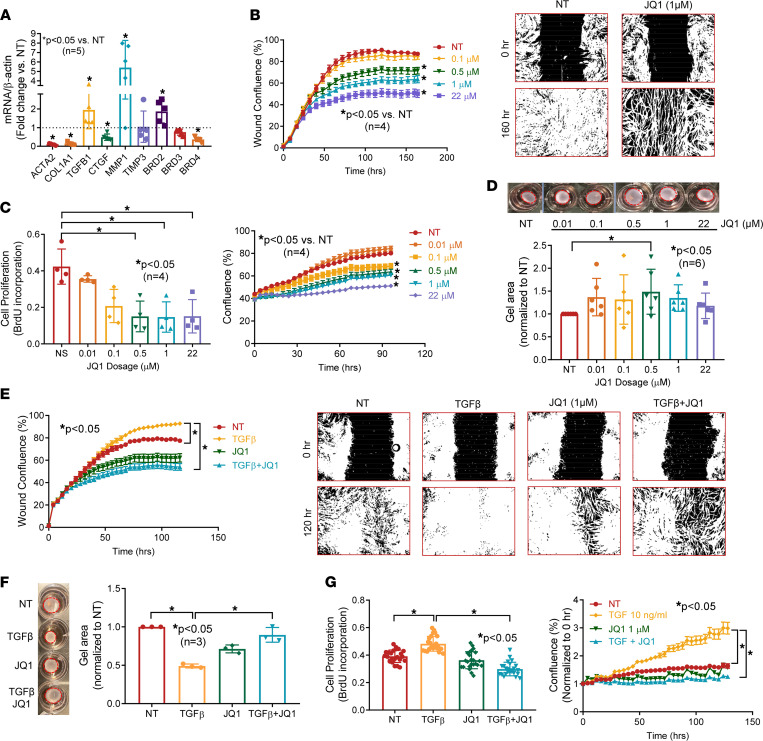
To further evaluate the effect of BET proteins in SSc fibrosis, we treated dcSSc fibroblasts with JQ1 at various doses (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.150871DS1). Given that the highest dose (22 μM) had minimal effect on CTGF but upregulated TGFB1, suggesting a potential off-target effect, we found that 1 μM appears to be the optimal dose to use. We evaluated the expression of profibrotic genes COL1A1, ACTA2, CTGF, and TGFB1, as well as the collagen-degrading enzyme MMP1 and its inhibitor TIMP3. Inhibition of BET by JQ1 led to a dose-dependent decrease in fibrotic markers, including COL1A1, ACTA2, and CTGF (Supplemental Figure 1 and Figure 2A). In addition, JQ1 also increased MMP1 expression, which is critical for collagen turnover, while it had minimal effect on TIMP3. Interestingly, JQ1 significantly increased TGFB1 expression. This might be an off-target effect of the inhibitor at high doses.
Figure 2. Inhibition of BETs shows potent antifibrotic properties in dcSSc fibroblasts.
(A) At 1 μM, JQ1 significantly downregulated ACTA2, COL1A1, CTGF, and BRD4 expression in dcSSc fibroblasts and upregulated MMP1, TGFB1, and BRD2. JQ1 did not affect TIMP1 and BRD3 expression. n = 5 patients. (B) Migration of dcSSc fibroblasts was significantly inhibited by JQ1. Wound confluence indicates the area occupied by cells that migrated into the wound gap. Representative pictures of JQ1 at 1 μM are shown. n = 4 patients. (C) Inhibition of BETs by JQ1 significantly reduced cell proliferation of dcSSc fibroblasts. Cell growth was analyzed by BrdU uptake in cells or monitored by Incucyte live-cell imaging system. n = 4 patients. (D) Gel contraction by dcSSc fibroblasts was inhibited by JQ1 at 0.5 μM. n = 6 patients. (E–G) Cell migration (n = 4), proliferation (n = 4), and gel contraction (n = 3) significantly increased after TGF-β treatment in normal dermal fibroblasts and can be inhibited by coincubation of 1 μM JQ1. Results are expressed as mean ± SD or mean ± SEM (time courses in B, C, E, and F); P < 0.05 was considered significant. Significance was determined by unpaired 2-tailed t test and Mann-Whitney U test (A), 2-way ANOVA (B, C, E, and G), 1-way ANOVA (F), and Kruskal-Wallis test (D and G). NT, no treatment.
To further characterize the antifibrotic effect of BET inhibition in dcSSc fibroblasts, we performed several functional assays. Treatment with JQ1 dose-dependently reduced migration and proliferation of dcSSc fibroblasts (Figure 2, B and C). The mechanism of reduction of cell proliferation by JQ1 might be due in part to its effect on inducing apoptosis, as JQ1 dose dependently increased apoptotic cells, as indicated by green fluorescence released by activated caspase-3/7, measured using Incucyte live-cell imaging system (Supplemental Figure 2A). However, this effect appears to be delayed, because apoptotic cells appeared after 40 hours, whereas differences in cell proliferation became evident after 30 hours of JQ1 treatment (Figure 2C, right panel, and Supplemental Figure 2A). This finding suggests that other mechanisms are involved in the effect of JQ1 on cell growth. To further analyze the effect of JQ1 on cell growth, we used Vybrant DyeCycle Violet stain to analyze cell cycle distribution in dcSSc fibroblasts. JQ1 induced a reduction in cells in the S phase and accumulation of cells in the G1/G0 phase (Supplemental Figure 2B). Treatment of dcSSc fibroblasts with JQ1 also resulted in a decrease in gel contraction (Figure 2D), with 0.5 μM being the most effective concentration.
Because TGF-β is critical in promoting fibrosis in SSc, we treated normal dermal fibroblasts with TGF-β to induce a myofibroblast phenotype. The induction was confirmed by an increase in cell migration and proliferation, as well as gel contraction in these TGF-β–treated normal fibroblasts compared with nontreated controls (Figure 2, E–G). Cotreatment with JQ1 significantly reduced TGF-β–induced migration, proliferation, and gel contraction in normal dermal fibroblasts.
Transcriptome analysis.
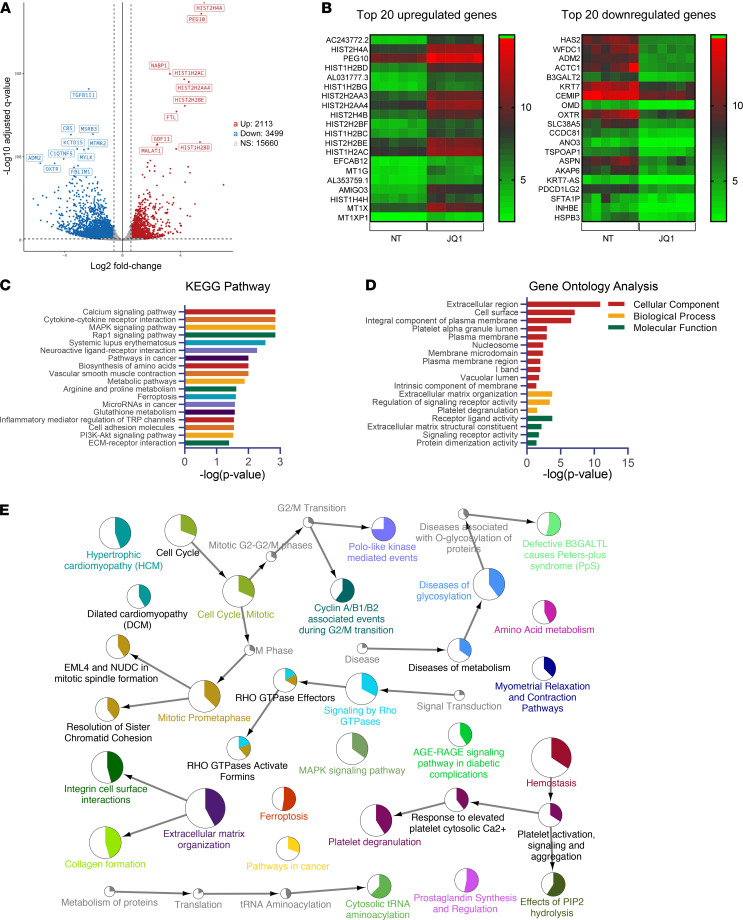
To further explore the mechanisms involved in BET inhibition in SSc fibrosis, mRNA-Seq was performed on dcSSc fibroblasts in the absence or presence of 1 μM JQ1. By this analysis, we identified 2113 upregulated and 3499 downregulated genes affected by JQ1 (Figure 3A and Supplemental Figure 3). The excess number of downregulated genes with BET inhibition indicates that BET proteins primarily act as transcription activators in SSc fibroblasts, echoing the findings in other tissues indicating that histone readers are, indeed, transcription activators. The top 20 upregulated and downregulated genes are shown in Figure 3B. In addition, RNA-Seq results confirmed expression changes observed using quantitative PCR in fibrosis-related genes (Figure 2A): ACTA2 and COL1A1 were significantly downregulated while TGFB1 and BRD2 were significantly upregulated at 1 μM JQ1 treatment (Supplemental Figure 4).
Figure 3. JQ1 treatment followed by RNA-Seq identified targets important for ECM remodeling, cell cycle regulation, and signaling in dcSSc fibroblasts.
(A) A total of 6 patient pairs were used for RNA-Seq analysis. In total, 5612 genes were significantly differentially expressed by JQ1 treatment in dcSSc fibroblasts (3499 downregulated and 2113 upregulated). (B) Top 20 upregulated and downregulated genes were shown. (C) The 18 most significantly enriched KEGG pathways after treatment with JQ1 in dcSSc fibroblasts are shown. (D) Gene Ontology analysis of the differentially expressed genes after JQ1 treatment in dcSSc fibroblasts. (E) Network of functional categories represented by JQ1-associated changes in dcSSc fibroblasts based on functional enrichment analysis. The node size corresponds with the level of significance (P value range, <0.05 to <0.0005) of the term it represents within the network. Colored portions of the pie charts represent the number of genes identified from the RNA-Seq experiment that are associated with the term. NT, no treatment. TRP, transient receptor potential.
Further pathway enrichment analysis of the differentially expressed genes regulated by JQ1 revealed that these genes are enriched in Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, including the Ca2+ signaling pathway, cytokine–cytokine receptor pathway, MAPK and Rap1 signaling pathways, as well as metabolic pathways, among others (Figure 3C and Supplemental Table 1). In addition, Gene Ontology analysis showed that BET proteins are involved in a wide spectrum of cellular components, biological processes, and molecular functions known to play critical roles in fibroblast activation (Figure 3D). These include ECM, plasma membrane, and signaling-receptor activity.
To better visualize the network of functional categories represented by JQ1-associated changes, we performed additional functional enrichment analysis using ClueGO/CluePedia by incorporating the KEGG, Reactome, and WikiPathways. As shown in Figure 3E, networks related to cell cycle regulation, ECM organization, signaling by Rho GTPases, ferroptosis, homeostasis, and diseases of glycosylation, among others, were significantly enriched.
We also examined the subcellular localization of the enriched pathways using the cerebral layout tool implemented in Cytoscape. The subcellular localization of JQ1-affected pathways was skewed toward ECM and plasma membranes (Supplemental Figure 5). This is not surprising, as functions related to ECM and homeostasis, as well as signaling pathways, were among the most enriched (as demonstrated by the size of the nodes). Indeed, when the differentially expressed genes were overlaid on the ECM–receptor interaction pathway (KEGG pathway 04512), many ECM genes and their binding partners were affected by JQ1, with the majority of these genes being downregulated (Supplemental Figure 6). In the intracellular compartment and the nucleus, cell cycle regulation is the main enriched pathway (Supplemental Figure 5). These results echo our functional data shown in Figure 2 and Supplemental Figure 2, as JQ1 showed strong antiproliferative and antifibrotic effects in these cells. Finally, as expected, the predicted upstream regulator analysis showed JQ1 as the most significant upstream chemical: 1521 genes from our transcriptomic analysis overlapped with reported JQ1-target genes (Supplemental Figure 7). This finding not only validates the JQ1 inhibition condition and sequencing analysis used in this study but also shows that genes affected by BET inhibition are common among different biological systems and cell types.
BET inhibition affects the Ca2+ signaling pathways in dcSSc fibroblasts.
As shown in Figure 3C, the most significant enrichment of the differentially expressed genes in the KEGG pathways was in the Ca2+ signaling pathway. To better visualize the location of the differentially expressed genes, we overlaid them in the pathway. JQ1 appeared to affect genes involved in Ca2+ channels on the plasma membrane, Ca2+ receptors that are located in the ER or sarcoplasmic reticulum, Ca2+-related signaling pathways, and Ca2+-dependent downstream effectors (Figure 4A). All the differentially expressed genes enriched in this pathway are listed in Figure 4B. Of the 59 genes, 15 were upregulated by JQ1, again pointing to BET proteins as transcriptional activators.
Figure 4. BET inhibition affects Ca2+-related pathways and intracellular Ca2+ in dcSSc fibroblasts.
(A) The Ca2+ signaling pathway (KEGG: 04020) diagram is overlaid with the expression changes of each gene. The legend describes the values on the gradient. Downregulation is shown in blue, while upregulation is in red. For legibility, 1 gene may be presented at multiple boxes in the diagram. In addition, 1 box may represent multiple genes in the same gene family. For each gene family, the color corresponding to the gene with the highest absolute fold change is displayed. For example, the ORAI box depicts significant upregulation of ORAI1 and ORAI3 and downregulation of ORAI2. The ORAI box is shown in red because ORAI1 shows the most significant fold change among the 3. (B) All the differentially expressed genes in Ca2+ signaling pathway (KEGG: 04020) are ranked based on their absolute value of log fold change. Upregulated genes are shown in red, and downregulated genes are shown in blue. n = 6 patient pairs. (C) BET inhibition by JQ1 significantly reduced intracellular Ca2+ levels in dcSSc fibroblasts. n = 5 patients. Results are expressed as mean ± SD and P < 0.05 was considered significant. Significance was determined by paired 2-tailed t test (C). NT, no treatment.
Because Ca2+ signaling affects fibroblast function and myofibroblast transformation (10), we examined whether JQ1 affects intracellular Ca2+ levels in dcSSc fibroblasts. As shown in Figure 4C, JQ1 significantly reduced intracellular Ca2+ levels in dcSSc fibroblasts.
JQ1 mediates its antifibrotic effect in part by affecting Ca2+ signaling in dcSSc fibroblasts.
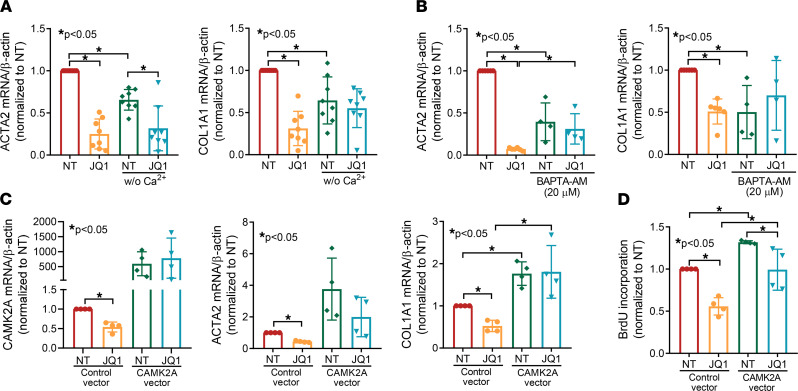
To further examine the involvement of intracellular Ca2+ in the antifibrotic effect of JQ1, we cultured dcSSc fibroblasts in the presence or absence of Ca2+ and treated the cells with or without JQ1. Comparing the nontreated groups in the presence or absence of Ca2+ in culture medium, the expression of ACTA2 and COL1A1 was significantly lower in cells cultured in medium without Ca2+ (Figure 5A). JQ1 significantly downregulated ACTA2 and COL1A1 in Ca2+-containing media, and JQ1’s effect on COL1A1 was diminished in medium without Ca2+. Using BAPTA-AM, an intracellular Ca2+-chelating reagent, we also showed that depleting Ca2+ in cells decreased ACTA2 and COL1A1 expression, and the inhibitory effect of JQ1 on these genes was blocked (Figure 5B). To further demonstrate the involvement of Ca2+ in the antifibrotic effect of JQ1, we focused on CAMK2A, which encodes Ca2+/calmodulin–dependent protein kinase II α (CaMKII-α), in subsequent studies. This gene is central for Ca2+-mediated effects in cells and was significantly downregulated by JQ1 (Figure 4, A and B). We further confirmed the effect of JQ1 on CAMK2A expression by quantitative PCR (Figure 5C). Overexpression of CAMK2A in dcSSc fibroblasts blocked the inhibitory effect of JQ1 on COL1A1 expression (Figure 5C) but not on ACTA2. Because members of the CaMKII family are involved in regulating the cell cycle (11), we examined whether CAMK2A is involved in JQ1-mediated antiproliferative effect in dcSSc fibroblasts. Indeed, we found that CAMK2A appears to be involved, at least in part, in the antiproliferative effect of JQ1, because overexpression of CAMK2A resulted in an approximately 25% reduction in cell growth by JQ1 compared with 50% in the control group (Figure 5D). Notably, CAMK2A overexpression in dcSSc fibroblasts not only resulted in elevated levels of ACTA2 and COL1A1 but also increased cell proliferation, suggesting that this gene plays a critical and nonredundant role in promoting SSc fibrosis. These results support the involvement of Ca2+ and Ca2+-related pathways in the antifibrotic effect of JQ1, specifically on collagen expression and cell proliferation.
Figure 5. The antifibrotic effect of JQ1 is abolished by the sequestration of intracellular Ca2+ and overexpression of CAMK2A in dcSSc fibroblasts.
(A) Cells cultured in Ca2+-free media had lower levels of ACTA2 and COL1A1 compared with cells cultured in Ca2+-containing media. The effect of JQ1 on COL1A1 is blocked in cells cultured without Ca2+. n = 8 patients. (B) BAPTA-AM, an intracellular Ca2+-chelating reagent, significantly decreased ACTA2 and COL1A1 expression in dcSSc fibroblasts. It also blocked the effect of JQ1. n = 4 patients. (C) JQ1 downregulated CAMK2A expression in dcSSc fibroblasts. Overexpression of CAMK2A not only increased ACTA2 and COL1A1 expression, but it also blocked the antifibrotic effects of JQ1. n = 4 patients. (D) CAMK2A is critical for cell proliferation, as overexpression of this gene enhanced cell proliferation. The inhibitory effect of JQ1 on cell proliferation was blocked by CAMK2A overexpression. n = 4 patients. Results are expressed as mean ± SD and P < 0.05 was considered significant. Significance was determined by 1-way ANOVA. NT, no treatment.
BET expression in SSc.
We examined the expression of BET proteins in dermal fibroblasts. Of the 4 BETs, BRD-t is expressed predominantly in the testis and therefore was excluded from our study. At the mRNA level, only BRD4 was significantly upregulated in dcSSc fibroblasts, compared with normal fibroblasts (Supplemental Figure 8A). At the protein level, however, the expression of the BETs was variable in dcSSc fibroblasts, but no difference between normal and dcSSc was seen (Supplemental Figure 8B).
BRD4 is profibrotic.
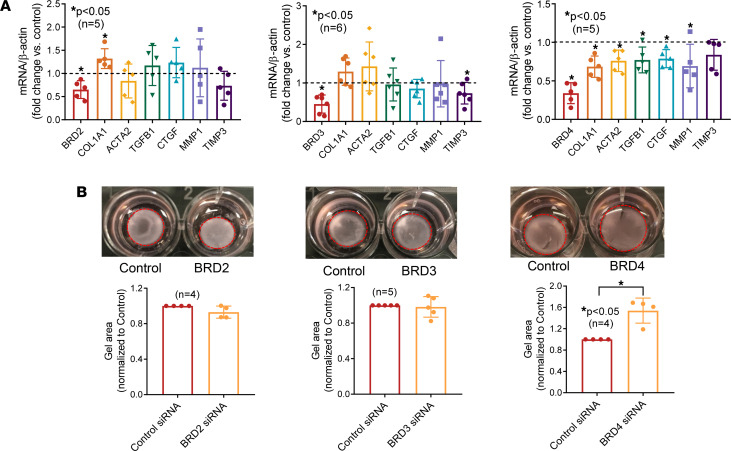
Because JQ1 is a pan-BET inhibitor, we next performed a set of experiments to determine which BET is responsible for the antifibrotic effect of JQ1. JQ1 significantly induced BRD2 and decreased BRD4, whereas it had minimal effect on BRD3 (Supplemental Figure 1 and Figure 2A). We then knocked down BRD2, BRD3, or BRD4 individually and examined the expression of genes involved in fibrosis. As shown in Figure 6A, knockdown of BRD2 resulted in significant elevation of COL1A1 expression, whereas knockdown of BRD4 led to significant reduction of COL1A1, ACTA2, TGFB1, CTGF, and MMP1 expression. BRD3 knockdown decreased TIMP3 expression and had no effect on the other genes examined. In addition, BRD4 knockdown led to significant reduction in gel contraction (Figure 6B), suggesting that BRD4 is profibrotic and that JQ1, by significantly reducing BRD4 expression in dcSSc fibroblasts, inhibits fibrosis in SSc.
Figure 6. BRD4 mediates the profibrotic effect of BETs in dcSSc fibroblasts.
(A) BRD2 knockdown in dcSSc fibroblasts led to upregulation of COL1A1, and BRD3 knockdown resulted in downregulation of TIMP3. Knockdown of BRD4 resulted in downregulation of COL1A1, ACTA2, TGFB, CTGF, and MMP1. n = 5–6 patients. (B) BRD4 knockdown significantly inhibited gel contraction in dcSSc fibroblasts, whereas BRD2 or BRD3 knockdown had minimal effect on gel contraction. n = 4–5 patients. Results are expressed as mean ± SD and P < 0.05 was considered significant. (A and B) Significance was determined by unpaired 2-tailed t test or Mann-Whitney U test.
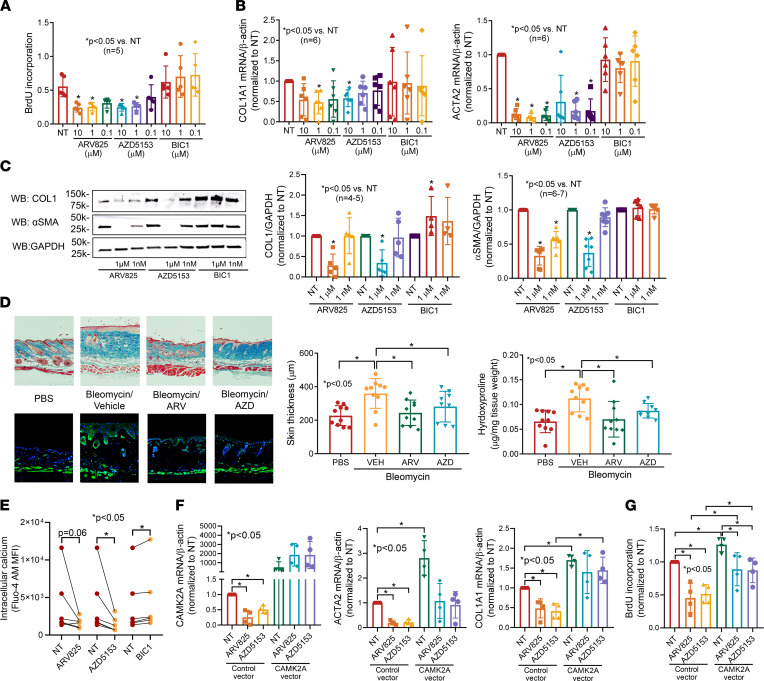
To confirm the involvement of BRD2 and BRD4 in myofibroblast function, we treated dcSSc fibroblasts with a specific BRD2 inhibitor (BIC1) or BRD4 inhibitors (AZD5153 or ARV825). As shown in Figure 7A, inhibition of BRD2 by BIC1 did not affect dcSSc fibroblast proliferation; BRD4 inhibition by either AZD5153 or ARV825 significantly inhibited proliferation at both 1 and 10 μM. In addition, dcSSc fibroblasts treated with BRD4 inhibitors ARV825 or AZD5153 significantly reduced ACTA2 and COL1A1 expression, whereas BRD2 inhibitor BIC1 did not have any effect (Figure 7B). Similar results were observed at the protein level (Figure 7C).
Figure 7. BRD4 inhibitors show prominent antifibrotic effects in vitro and in vivo.
(A) Treating dcSSc fibroblasts with the specific BRD2 inhibitor BIC1 had minimal effect on cell proliferation, whereas inhibition of BRD4 using BRD4 inhibitors AZD5153 or ARV825 significantly reduced cell proliferation at concentrations of 1 and 10 μM. n = 5 patients. (B) Inhibition of BRD4 by ARV825 or AZD5153 in dcSSc fibroblasts significantly reduced both ACTA2 and COL1A1 expression, whereas blockade of BRD2 by BIC1 had no effect. n = 6 patients. (C) BRD4 inhibitors significantly decreased α-SMA (n = 7) and COL1 (n = 5) in dcSSc fibroblasts, whereas BRD2 inhibition by BIC1 had minimal effect on α-SMA (n = 6) but increased COL1 at 1 μM (n = 4 patients). (D) Bleomycin-treated mice had increased dermal thickness and hydroxyproline content in skin, and ARV825 or AZD5153 efficiently prevented skin fibrosis in these mice. Immunofluorescence staining of α-SMA–positive cells (green) is shown. Nuclei were stained with DAPI (blue). Original magnification, ×40–100. n = 9–10 mice. (E) BRD4 inhibitor AZD5153 significantly decreased intracellular Ca2+ and BRD2 inhibitor BIC1 significantly increased it. n = 6 patients. (F) Overexpression of CAMK2A resulted in significant increase in ACTA2 and COL1A1 expression and blocked the effect of AZD5153 on COL1A1 expression. n = 4 patients. (G) CAMK2A overexpression significantly increased cell proliferation while it abolished the effect of BRD4 inhibition on cell proliferation. n = 4 patients. Results are expressed as mean ± SD and P < 0.05 was considered significant. Significance was determined by 1-way ANOVA (A–D, F, and G), Kruskal-Wallis test (A–C), and Wilcoxon’s test (E). NT, no treatment.
To further determine the antifibrotic effect of BRD4 inhibition in vivo, we induced skin fibrosis by bleomycin in mice and dosed the animals with either ARV825 or AZD5153. We found that both BRD4 inhibitors prevented bleomycin-induced skin fibrosis in mice, as indicated by dermal thickness, α-SMA–positive cells, and hydroxyproline content (Figure 7D). These results further confirm the involvement of BRD4 in myofibroblast transformation in dcSSc fibroblasts and in the bleomycin mouse model.
BRD4 mediates its fibrotic properties through Ca2+ signaling in dcSSc fibroblasts.
To further investigate whether BRD4 inhibitors affect intracellular Ca2+ in dcSSc fibroblasts similar to what was observed with JQ1, we measured intracellular Ca2+ levels in the presence or absence of the BRD4 inhibitors. We found that both inhibitors decreased intracellular Ca2+ levels after 48 hours of treatment, with AZD5153 reaching statistical significance (Figure 7E). Interestingly, the BRD2 inhibitor BIC1 significantly increased intracellular Ca2+ levels, suggesting that this BET isoform could act in an opposite way than BRD4 in dcSSc fibroblasts. We further showed that both BRD4 inhibitors downregulated CAMK2A in dcSSc fibroblasts (Figure 7F). In addition, the antifibrotic effects of both AZD5153 and ARV825 in dcSSc fibroblasts were partially dependent on CAMK2A. Overexpression of CAMK2A in these cells appeared to block the inhibitory effect of AZD5153 on COL1A1 (Figure 7F), whereas it had minimal effect on ACTA2 expression. BRD4 inhibitors also blocked cell proliferation by 50% in control cells but only by approximately 25% in the CAMK2A-overexpressing cells, suggesting that CAMK2A is partially involved in BRD4-mediated effects on cell growth (Figure 7G).
Discussion
The increased understanding of the effect of epigenetic aberrations on gene transcription has led the field to a better appreciation of the role of transcriptional dysregulation in initiating and perhaps maintaining SSc fibrosis. In this study, we performed an in-depth examination of BET inhibition in SSc fibrosis. Our in vivo studies showed that administration of JQ1 prevented skin fibrosis induced by bleomycin in mice. We further demonstrated that BET inhibition by JQ1 suppressed expression of many profibrotic genes in dcSSc fibroblasts and reversed the established progression of myofibroblast differentiation in vitro. Transcriptomic analysis of JQ1-treated cells not only showed that differentially expressed genes are enriched in ECM- and cell cycle–related pathways but also revealed the involvement of the Ca2+ signaling pathway as a novel antifibrotic mechanism. Moreover, results from siRNA knockdown experiments confirmed the role of BRD4 in SSc fibrosis. This was further confirmed using BRD4-specific inhibitors. Functional studies showed that Ca2+ and the Ca2+-related gene CAMK2A play critical roles in the antifibrotic effects of JQ1, AZD5153, and ARV825. Together, these results suggest that BRD4 is critically involved in promoting fibrosis in SSc, and that inhibition of BRD4, and perhaps targeting intracellular Ca2+, would be effective treatments for this disease.
JQ1, a first-in-class BET inhibitor, competitively binds to the acetyl-lysine recognition areas of these proteins and displaces them from acetylated chromatin, thereby repressing transcription of target genes. Early studies suggested that JQ1 exerted potent antiproliferative properties in multiple myeloma via cell growth arrest and senescence in a c-MYC–dependent manner (12). Since then, more studies have shown the benefits of JQ1 in other proliferative disorders (5–8). In addition, this pan-BET inhibitor has demonstrated great efficacy in blocking fibrotic progression in a range of fibrosis models (5–8). In a lung fibrosis model, JQ1 significantly reduced collagen deposition in bleomycin-treated mice compared with control mice (8). This drug also ameliorated the phenotypic changes of lung fibroblasts from patients with idiopathic pulmonary fibrosis. We showed in this study that JQ1 inhibits proliferation of dcSSc dermal fibroblasts by inducing cell cycle arrest and apoptosis. It also exhibits potent antifibrotic effects in patient-derived cells and in the bleomycin skin-fibrosis model. The functional changes in cell proliferation were also confirmed from the RNA-Seq results, as genes enriched in cell cycle, mitosis, and ferroptosis pathways were highlighted in the pathway analysis (Figure 3E). Our results are consistent with the findings of Shin et al. (13), who showed that JQ1 repressed collagen expression in SSc skin organ culture, likely due to the increase in MMP1.
BET proteins are located in the nucleus. Because of their involvement in various cellular processes, BETs have been shown to participate in tumor development, autoimmunity, infections, and inflammation (14–17). Specifically, BRD4 has been studied extensively for its role in gene transcription, including regulation (through acetylated histones), initiation (via engaging RNA polymerase II), and elongation (by interaction with P-TEFb) (18). Indeed, BRD4 is reported to control various fibrosis-related genes, including ACTA2 and COL1A1, shown from BRD4 knockdown studies or ChIP analyses (5, 7, 8, 19). This is not surprising, because both genes are reported to possess acetylated histones in their promoter region in fibroblasts (Supplemental Figure 9). In dcSSc fibroblasts, inhibition of BETs by JQ1 downregulated both ACTA2 and COL1A1, and this downregulation appears to be mediated through blockade of BRD4, because BRD4 knockdown or inhibition led to decreased ACTA2 and COL1A1 levels, while knockdown of BRD2 or BRD3 did not. Interestingly, TGFB1, which was significantly upregulated by JQ1, was downregulated in BRD4–knocked-down cells. Similar results were seen in MMP1 expression. This discrepancy suggests that JQ1, as a pan-BET inhibitor, has off-target effects compared with BRD4 knockdown, especially at the higher concentrations. This could potentially explain the U-shaped dose-response curve of JQ1 in the gel contraction assay. Of note, our results showing that BRD2 knockdown leads to COL1A1 upregulation are consistent with those in previous reports (19). Because JQ1 dose-dependently increased BRD2 expression in dcSSc fibroblasts, it is possible that the antifibrotic effect of JQ1 is partly mediated by enhancing the antifibrotic potential of BRD2. However, BRD2 inhibition, although trending to induction of fibroblast migration and profibrotic gene expression (Figure 7, A–C), had no significant effect on enhancing myofibroblast functions. Furthermore, BIC1 significantly increased intracellular Ca2+ levels in dcSSc fibroblasts, in contrast to BRD4 inhibition.
To gain mechanistic insight into the antifibrotic effect of BET inhibition in SSc, we performed mRNA-Seq in JQ1-treated dcSSc fibroblasts. In addition to pathways in cell cycle regulation, ECM, and intracellular signaling, our analysis also revealed potentially novel pathways, including Ca2+ signaling, metabolism, ferroptosis, and prostaglandin synthesis and regulation (Figure 3E). These pathways have been implicated in various fibrotic conditions; thus, it is possible that they are involved in the antifibrotic effect of BET inhibition in dcSSc fibroblasts. Specifically, the Ca2+ signaling pathway, which is the most enriched pathway in our analysis (Figure 3C), is critical in various fibrotic conditions (10, 20, 21). Intracellular Ca2+ levels are controlled by Ca2+-permeable channels in the plasma membrane (voltage-operated channels and receptor-operated channels), Ca2+ release from the ER and sarcoplasmic reticulum, as well as Ca2+ extrusion pumps. The increase in intracellular Ca2+ levels can activate various pathways that are involved in many physiological functions (22–25). Studies show that interference of intracellular Ca2+ levels through blockade of Ca2+ channels or receptors has potential antifibrotic effects in various models (26–29). In dcSSc fibroblasts, JQ1 treatment significantly reduced Ca2+ levels in these cells (Figure 4C). This is possibly mediated by significant downregulation of Ca2+ channels and receptors on the plasma membrane, including P2RX1 and P2RX5 (coding for purinergic receptors P2X1 and P2X5), calcium voltage-gated channel subunit α-1D (CACNA1D), and voltage-dependent Ca2+ channel protein TPC1 (TPCN1), as well as ones on the ER and sarcoplasmic reticulum, such as ryanodine receptor 1 (RYR1) (Figure 4B). Interestingly, certain Ca2+ channels, Ca2+ extrusion pumps, and Ca2+ release transporters that increase intracellular Ca2+ were affected by JQ1. It is possible that these events represent a compensatory mechanism that attempts to antagonize the decreased Ca2+ levels in these fibroblasts. The identification of Ca2+ pumps, carriers, and channels involved in Ca2+ entry, as well as their involvement in mediating the fibrotic effect of BET proteins in SSc, requires further investigation.
In this study, we identified CaMKII, specifically the α isoform, as a critical mediator of the fibrotic effect of BETs and BRD4. CaMKII, composed of 4 isoforms, is a downstream messenger of the Ca2+ signaling pathway. CaMKII activation augmented collagen production in cardiac fibroblasts, while inhibition of CaMKII blocked proliferation and collagen and profibrotic cytokine production (22, 30, 31). We showed similar findings in dcSSc fibroblasts in this study in the context of the BET or BRD4 inhibitors. In addition to cardiac fibrosis, CaMKII is also involved in pulmonary fibrosis (32), ureteral scar formation (33), and renal fibrosis (34). CaMKII is associated with FGFR3/FGF9 in SSc fibroblasts, functioning as a downstream mediator of the profibrotic effect of FGFR3 (25). Interestingly, our results showed that CAMK2A appears to play a more critical role in BET or BRD4-mediated COL1A1 expression in dcSSc fibroblasts, but not as important a role in blocking myofibroblast transformation. CAMK2A only partially explains the proliferative effects of BET or BRD4, as shown in Figures 5 and 7. These results point to the possibility of other mediators being involved in the antifibrotic effects of BET and BRD4 inhibitors. Intriguingly, we found that overexpression of CAMK2A alone led to significant increase in fibrotic gene expression and cell proliferation in dcSSc fibroblasts, suggesting that this gene itself could play a critical role in SSc fibrosis.
Although JQ1 possesses desirable qualities of a small molecular inhibitor, including high target potency and well-characterized selectivity, it is known for its inhibitory effect on lymphoid and hematopoietic tissues (35). JQ1 exhibits linear pharmacokinetics in mice, with an oral bioavailability of approximately 50% and a half-life of 1 hour (35, 36). Due to the short half-life and potential off-target effects, this compound is only used in preclinical studies. Currently, there are several BET inhibitors with better pharmacokinetic properties in clinical trials for various types of cancer (9). Recent trials have also focused on targeting specific BETs, including BRD4 inhibitors AZD5153 (ClinicalTrials.gov NCT03205176), which was used in this study, and ABBC-744 (ClinicalTrials.gov NCT03360006). Based on our results, BETs, in particular BRD4, regulate essential processes involved in SSc fibrosis. We not only revealed an antifibrotic mechanism involving Ca2+ signaling by BET inhibition but also highlighted the potential of epigenetic therapeutic strategies targeting BRD4 for patients with SSc. These data, along with future studies focusing on additional molecular mechanisms underlying the role of BRD4 in SSc, should provide the framework that supports the use of more selective BRD4 inhibitors as a therapeutic option for SSc.
Methods
Bleomycin-induced skin fibrosis.
The procedure to induce fibrosis by bleomycin in mice was published previously (2, 37). In C57BL/6 mice (The Jackson Laboratory, 000664), 100 μL of bleomycin (1 mg/mL) or PBS was injected s.c. into a single location on the shaved back once every day for 2 weeks. JQ1 (50 mg/kg; MedChemExpress) or vehicle control (20% DMSO, 50% PBS, 30% PEG from J.T.Baker) was given daily by oral gavage. In a separate experiment, BRD4 inhibitors AZD5153 (5 mg/kg; Cayman Chemical) or ARV825 (5 mg/kg, MedChemExpress) or vehicle control (22% DMSO, 48% PBS, 30% PEG) were given daily i.p. At the end of the experiment, fixed skin sections were stained with Masson’s trichrome. Dermal thickness was measured by analyzing the distance between the epidermal–dermal junction and the dermal–fat junction in 3 fields in 2 or more skin sections from each animal. Immunofluorescence was performed on sections using anti–α-SMA (Abcam ab5694) or anti-F4/80 (Invitrogen MA5-16630; clone CI:A3-1) Abs after antigen-retrieval. Collagen content in the skin was measured using the Hydroxyproline Kit (Abcam). All animal protocols were approved by the IACUC at the University of Michigan.
Patients and control participants.
All patients recruited from the University of Michigan Scleroderma Program in this study met the American College of Rheumatology and European Alliance of Associations for Rheumatology criteria for the classification of SSc (38). Punch-biopsy specimens from the distal forearm of healthy volunteers and patients with dcSSc were obtained for fibroblast isolation. This study was approved by the University of Michigan IRB. The demographics and clinical characteristics of the enrolled individuals are summarized in Supplemental Table 2.
Cell culture.
Punch-biopsy specimens obtained from healthy study participants and patients with SSc were digested as previously described (2, 39). Briefly, biopsy specimens were placed in 2.4 U/mL dispase (Invitrogen) overnight at 4°C. After removing the epidermal layers, the biopsy specimens were transferred to a 0.2% collagenase solution (MilliporeSigma) and incubated at 37°C for 45 minutes. The dissociated cells were then collected and cultured. After separating out the endothelial cells by magnetic bead selection (CD31 MicroBead Kit from Miltenyi Biotec), the resultant fibroblasts were maintained in RPMI-1640 medium (Hyclone) supplemented with 10% FBS and antibiotics. Cells between passage 3 and 6 were used in all experiments.
Cell treatment and transfection.
Dermal fibroblasts from patients with dcSSc were treated with 0.01 to 22 μM JQ1 (Cayman Chemical), a pan-BET inhibitor, for 48 hours. Normal dermal fibroblasts were treated with TGF-β (10 ng/mL; Cell Signaling Technology) and/or JQ1 (1 μM) for 72 hours to induce a myofibroblast phenotype. In a separate experiment, BRD2 inhibitor BIC1 (Cayman Chemical) and BRD4 inhibitors AZD5153 or ARV825 (0.1–10 μM; Cayman Chemical) were used to treat dcSSc fibroblasts for 48 to 72 hours. BRD2, BRD3, or BRD4 knockdown in dcSSc dermal fibroblasts was performed using BRD2 (200 nM), BRD3 (100 nM), or BRD4 (200 nM) siRNA (ON-TARGETplus siRNA; Dharmacon). Scrambled siRNA (Dharmacon) was used as a control. The cells were transfected for 72 hours before downstream experiments were performed. CAMK2A in dcSSc fibroblasts was overexpressed using CAMK2A vectors from OriGene. CAMK2A vector (0.1 μg/mL) was mixed with Lipofectamine 2000 (Invitrogen) and then added to culture medium. Inhibitors were added 24 hours after the transfection. PCMV6-XL6 vector (OriGene) was used as a negative control. To determine the effect of Ca2+ on myofibroblast transformation, dcSSc dermal fibroblasts were cultured in Ca2+-containing or Ca2+-free DMEM. In a separate experiment, dcSSc fibroblasts cultured in Ca2+-containing RPMI medium were treated with BAPTA-AM (20 μM; Thermo Fisher Scientific) in the presence or absence of 1 μM JQ1 for 2 days.
Western blots.
Western blotting was performed following the protocol as described previously (39). Equal amounts of protein were separated by SDS-PAGE, then transferred to a nitrocellulose membrane. After blocking, the blots were probed with Abs against BRD2 (Abcam ab139690; clone EPR7642), BRD3 (Santa Cruz Biotechnology sc-81202; clone 2088C3a), BRD4 (Abcam ab128874; clone EPR5150[2]), collagen I (Abcam ab6308; clone COL-1), or α-SMA (Abcam ab5694). For loading control, the blots were immunoblotted with Abs against β-actin (MilliporeSigma A2066) or GAPDH (Cell Signaling Technology 2118; clone 14C10) as controls. Band quantification was performed using ImageJ (NIH) (40).
mRNA extraction and quantitative real-time PCR.
Total RNA was extracted using Direct-zol RNA MiniPrep Kit (Zymo Research) before being converted to cDNA. Quantitative PCR was performed in a ViiA 7 Real-Time PCR System.
Cell proliferation assays.
Proliferation of dermal fibroblasts was measured using the BrdU Cell Proliferation Assay Kit (BioVision). After 4 hours of BrdU incubation, cells were fixed before adding Abs and substrate. The absorbance at 450 nm was measured. In a separate experiment, the Incucyte live-cell imaging system (Sartorius) was used to monitor cell proliferation. Cells were seeded and allowed to grow overnight. After adding different treatments, cells were monitored by Incucyte up to 5 days. Cell counts were analyzed by the Incucyte S3 Analysis software (Sartorius).
Gel contraction and cell migration assays.
The Cell Contraction Assay Kit (Cell Biolabs) was used for gel contraction (39). After treatment, cells were suspended and mixed with collagen solution. Solidified gels were lifted after 24 hours, and the areas of the gels were quantified using ImageJ (40). To evaluate the effect of JQ1 on cell migration, we performed a scratch-wound assay using the Incucyte live-cell imaging system. Cell migration was monitored by Incucyte up to 7 days.
Detection of apoptosis.
To assess the effect of JQ1 on cell apoptosis, cells were plated in 96-well plates and treated with JQ1 in the presence of Incucyte Caspase 3/7 Apoptosis Reagent. The apoptotic cells were quantified by measuring cells with green fluorescence staining. The data were presented as the number of green fluorescing cells normalized to total cell count.
Cell cycle analysis.
Fibroblasts were treated with 1 μM JQ1 for 48 hours, and the nontreated dcSSc fibroblasts were used as controls. Both the JQ1-treated and nontreated cells were first synchronized using the double thymidine block method before treatment (41). Cells were then harvested and stained with 7-aminoactinomycin D (7-AAD) viability dye and Vybrant DyeCycle Violet (both from Invitrogen) according to the manufacturer’s protocol. The stained cells were analyzed using a BD FACSCanto II flow cytometer. Cell cycle analysis was performed on gated 7-AAD–live cells using FlowJo 7.0.
RNA-Seq and analysis.
RNA-Seq was performed on mRNA extracted from control or JQ1-treated (1 μM) dcSSc fibroblasts. Samples with an RNA integrity number of more than 7 were subjected to library preparation and sequencing to 151 paired-end cycles on the NovaSeq 6000 platform (Illumina), resulting in approximately 35 million reads per sample. Differential gene expression analysis was performed using DESeq2 (42), using a negative binomial generalized linear model (thresholds: linear fold change greater than 1.5 or less than –1.5, Benjamini-Hochberg FDR-adjusted P < 0.05). Plots were generated using variations of DESeq2 plotting functions and other packages with R, version 3.3.3. Functional analysis, including candidate pathways activated or inhibited in comparison(s) and Gene Ontology (GO) term enrichments, was performed using iPathway Guide (Advaita) (43). Additional functional enrichment analyses to generate networks for visualization were performed using ClueGO (version 2.5.7)/CluePedia (version 1.5.7) (44, 45) and Cytoscape (v3.8.0) (46).
Intracellular Ca2+ measurement.
Dermal fibroblasts from patients with dcSSc were treated with or without 1 μM JQ1, AZD5153, ARV825, or BIC1 for 48 hours before being collected for Ca2+ measurement by flow cytometry. Detached cells (1 × 107/mL) were labeled in loading buffer (HBSS with 1% FBS, 1 mM CaCl2, and 1 mM MgCl2) containing 1 μM Fluo-4 AM (Thermo Fisher Scientific) with 0.02% Pluronic F-127 at 37°C for 30 minutes with gentle agitation every 10 minutes. The labeled cells were washed twice with loading buffer containing 0.1 mM sulfinpyrazone (MilliporeSigma) and resuspended in the same loading buffer with sulfinpyrazone at 1 × 106/mL, incubated at 37°C for 30 minutes before basal fluorescence of Fluo-4 was measured on a BD FACSCanto II flow cytometer. Mean fluorescence intensity of unlabeled cells was subtracted from the mean fluorescence intensity of the labeled cells.
Data availability.
All data relevant to the study are included in the article. The RNA-Seq data are deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus (GSE186961).
Statistics.
A normality test was conducted to determine whether the data were normally distributed or skewed. To determine the differences between groups, unpaired 2-tailed t test, Mann-Whitney U test, paired 2-tailed t test, Wilcoxon’s test, 1-way ANOVA with Holm-Šidák test, Kruskal-Wallis test with Dunn’s test, or 2-way ANOVA with Dunnett’s test were performed using GraphPad Prism, version 8 (GraphPad Software). P < 0.05 was considered statistically significant. Results were expressed as mean ± SD.
Study approval.
This study was approved by the University of Michigan IRB. Written informed consent was received from participants prior to inclusion in the study. All animal protocols were approved for use from the University of Michigan IACUC.
Author contributions
All authors participated in the interpretation of study results and in the drafting, critical revision, and approval of the final version of the manuscript. PST, AHS, DAF, YMD, and DK contributed to study conception and/or design. SV, MGR, MNM, WDB, SM, PJP, MA, PLC, MAA, JHR, QW, ENM, DMR, JLH, and PT contributed to the acquisition of study results. SV, PJP, MA, QW, and PT contributed to the analysis of study results.
Supplementary Material
Acknowledgments
This work was supported by the funds from the American Autoimmune Related Disease Foundation and the Edward T. and Ellen K. Dryer Early Career Professorship (to PST); Chugai, PCORI, Novartis, Sanofi-Genzyme, and Genentech (to YMD); NIH/National Institute of Arthritis and Musculoskeletal and Skin Diseases (grant K24AR063120 to DK, grant T32AR007080 to MGR, and grant R01AR070148 to AHS); and NIH/National Institute of Allergy and Infectious Diseases (grants UM1-AI110557-05 and UM1-AI144298-01 to YMD and grant R01AI097134 to AHS).
Version 1. 03/29/2022
In-Press Preview
Version 2. 05/09/2022
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2022, Vichaikul et al. This is an open access article published under the terms of the Creative Commons Attribution 4.0 International License.
Reference information: JCI Insight. 2022;7(9):e150871.https://doi.org/10.1172/jci.insight.150871.
Contributor Information
Sirapa Vichaikul, Email: sirapav@umich.edu.
Mikel Gurrea-Rubio, Email: mikelg@med.umich.edu.
M. Asif Amin, Email: maamin@med.umich.edu.
Phillip L. Campbell, Email: axilla@med.umich.edu.
Qi Wu, Email: qiw@umich.edu.
Megan N. Mattichak, Email: mnmatt@umich.edu.
William D. Brodie, Email: wbrodie@umich.edu.
Pamela J. Palisoc, Email: pamelajane1994@gmail.com.
Mustafa Ali, Email: muali248@gmail.com.
Sei Muraoka, Email: seih@umich.edu.
Jeffrey H. Ruth, Email: jhruth@umich.edu.
Ellen N. Model, Email: modele@umich.edu.
Dallas M. Rohraff, Email: rohraffd@mail.gvsu.edu.
Jonatan L. Hervoso, Email: jhervoso@g.ucla.edu.
Yang Mao-Draayer, Email: Maodraay@med.umich.edu.
David A. Fox, Email: dfox@umich.edu.
Dinesh Khanna, Email: khannad@umich.edu.
Amr H. Sawalha, Email: asawalha@umich.edu.
Pei-Suen Tsou, Email: ptsou@umich.edu.
References
- 1.Tsou PS. Epigenetic control of scleroderma: current knowledge and future perspectives. Curr Rheumatol Rep. 2019;21(12):69. doi: 10.1007/s11926-019-0877-y. [DOI] [PubMed] [Google Scholar]
- 2.Tsou PS, et al. Inhibition of EZH2 prevents fibrosis and restores normal angiogenesis in scleroderma. Proc Natl Acad Sci U S A. 2019;116(9):3695–3702. doi: 10.1073/pnas.1813006116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergmann C, et al. The histone demethylase Jumonji domain-containing protein 3 (JMJD3) regulates fibroblast activation in systemic sclerosis. Ann Rheum Dis. 2018;77(1):150–158. doi: 10.1136/annrheumdis-2017-211501. [DOI] [PubMed] [Google Scholar]
- 4.Kramer M, et al. Inhibition of H3K27 histone trimethylation activates fibroblasts and induces fibrosis. Ann Rheum Dis. 2013;72(4):614–620. doi: 10.1136/annrheumdis-2012-201615. [DOI] [PubMed] [Google Scholar]
- 5.Xiong C, et al. Pharmacological targeting of BET proteins inhibits renal fibroblast activation and alleviates renal fibrosis. Oncotarget. 2016;7(43):69291–69308. doi: 10.18632/oncotarget.12498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou B, et al. Brd4 inhibition attenuates unilateral ureteral obstruction-induced fibrosis by blocking TGF-β-mediated Nox4 expression. Redox Biol. 2017;11:390–402. doi: 10.1016/j.redox.2016.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ding N, et al. BRD4 is a novel therapeutic target for liver fibrosis. Proc Natl Acad Sci U S A. 2015;112(51):15713–15718. doi: 10.1073/pnas.1522163112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang X, et al. BET bromodomain proteins mediate downstream signaling events following growth factor stimulation in human lung fibroblasts and are involved in bleomycin-induced pulmonary fibrosis. Mol Pharmacol. 2013;83(1):283–293. doi: 10.1124/mol.112.081661. [DOI] [PubMed] [Google Scholar]
- 9.Alqahtani A, et al. Bromodomain and extra-terminal motif inhibitors: a review of preclinical and clinical advances in cancer therapy. Future Sci OA. 2019;5(3):FSO372. doi: 10.4155/fsoa-2018-0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng J, et al. Ca 2+ signaling in cardiac fibroblasts and fibrosis-associated heart diseases. J Cardiovasc Dev Dis. 2019;6(4):34. doi: 10.3390/jcdd6040034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skelding KA, et al. Controlling the cell cycle: the role of calcium/calmodulin-stimulated protein kinases I and II. Cell Cycle. 2011;10(4):631–639. doi: 10.4161/cc.10.4.14798. [DOI] [PubMed] [Google Scholar]
- 12.Delmore JE, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146(6):904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shin JY, et al. Epigenetic activation and memory at a TGFB2 enhancer in systemic sclerosis. Sci Transl Med. 2019;11(497):eaaw0790. doi: 10.1126/scitranslmed.aaw0790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klein K. Bromodomain protein inhibition: a novel therapeutic strategy in rheumatic diseases. RMD Open. 2018;4(2):e000744. doi: 10.1136/rmdopen-2018-000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morgado-Pascual JL, et al. Bromodomain and extraterminal proteins as novel epigenetic targets for renal diseases. Front Pharmacol. 2019;10:1315. doi: 10.3389/fphar.2019.01315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mietton F, et al. Selective BET bromodomain inhibition as an antifungal therapeutic strategy. Nat Commun. 2017;8(1):15482. doi: 10.1038/ncomms15482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andrieu GP, et al. BET proteins in abnormal metabolism, inflammation, and the breast cancer microenvironment. J Leukoc Biol. 2018;104(2):265–274. doi: 10.1002/JLB.5RI0917-380RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Donati B, et al. BRD4 and cancer: going beyond transcriptional regulation. Mol Cancer. 2018;17(1):164. doi: 10.1186/s12943-018-0915-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar K, et al. BET inhibitors block pancreatic stellate cell collagen I production and attenuate fibrosis in vivo. JCI Insight. 2017;2(3):e88032. doi: 10.1172/jci.insight.88032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mukherjee S, et al. Disruption of calcium signaling in fibroblasts and attenuation of bleomycin-induced fibrosis by nifedipine. Am J Respir Cell Mol Biol. 2015;53(4):450–458. doi: 10.1165/rcmb.2015-0009OC. [DOI] [PubMed] [Google Scholar]
- 21.Chung CC, et al. Calcium regulation on the atrial regional difference of collagen production activity in atrial fibrogenesis. Biomedicines. 2021;9(6):686. doi: 10.3390/biomedicines9060686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang W, et al. Inhibition of calcium-calmodulin-dependent kinase II suppresses cardiac fibroblast proliferation and extracellular matrix secretion. J Cardiovasc Pharmacol. 2010;55(1):96–105. doi: 10.1097/FJC.0b013e3181c9548b. [DOI] [PubMed] [Google Scholar]
- 23.Wei C, et al. Calcium flickers steer cell migration. Nature. 2009;457(7231):901–905. doi: 10.1038/nature07577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tupper JT, et al. Membrane transport properties differ following return of serum-deprived versus Ca++-deprived human fibroblasts to a proliferative state. J Cell Physiol. 1982;110(1):29–34. doi: 10.1002/jcp.1041100106. [DOI] [PubMed] [Google Scholar]
- 25.Chakraborty D, et al. Fibroblast growth factor receptor 3 activates a network of profibrotic signaling pathways to promote fibrosis in systemic sclerosis. Sci Transl Med. 2020;12(563):eaaz5506. doi: 10.1126/scitranslmed.aaz5506. [DOI] [PubMed] [Google Scholar]
- 26.Tian X, et al. ANO1 regulates cardiac fibrosis via ATI-mediated MAPK pathway. Cell Calcium. 2020;92:102306. doi: 10.1016/j.ceca.2020.102306. [DOI] [PubMed] [Google Scholar]
- 27.Ding Z, et al. Ryanodine receptor type 2 plays a role in the development of cardiac fibrosis under mechanical stretch through TGFβ-1. Int Heart J. 2017;58(6):957–961. doi: 10.1536/ihj.16-572. [DOI] [PubMed] [Google Scholar]
- 28.Lin BL, et al. In vivo selective inhibition of TRPC6 by antagonist BI 749327 ameliorates fibrosis and dysfunction in cardiac and renal disease. Proc Natl Acad Sci U S A. 2019;116(20):10156–10161. doi: 10.1073/pnas.1815354116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adapala RK, et al. TRPV4 channels mediate cardiac fibroblast differentiation by integrating mechanical and soluble signals. J Mol Cell Cardiol. 2013;54:45–52. doi: 10.1016/j.yjmcc.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kreusser MM, et al. Inducible cardiomyocyte-specific deletion of CaM kinase II protects from pressure overload-induced heart failure. Basic Res Cardiol. 2016;111(6):65. doi: 10.1007/s00395-016-0581-2. [DOI] [PubMed] [Google Scholar]
- 31.Zhong P, et al. Role of CaMKII in free fatty acid/hyperlipidemia-induced cardiac remodeling both in vitro and in vivo. J Mol Cell Cardiol. 2017;109:1–16. doi: 10.1016/j.yjmcc.2017.06.010. [DOI] [PubMed] [Google Scholar]
- 32.Winters CJ, et al. CaMKII inhibition in type II pneumocytes protects from bleomycin-induced pulmonary fibrosis by preventing Ca2+-dependent apoptosis. Am J Physiol Lung Cell Mol Physiol. 2016;310(1):L86–L94. doi: 10.1152/ajplung.00132.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeng MQ, et al. Verapamil inhibits ureteral scar formation by regulating CaMK II-mediated Smad pathway. Chem Biol Interact. 2021;346:109570. doi: 10.1016/j.cbi.2021.109570. [DOI] [PubMed] [Google Scholar]
- 34.Zhou J, et al. NMDA receptor-mediated CaMKII/ERK activation contributes to renal fibrosis. BMC Nephrol. 2020;21(1):392. doi: 10.1186/s12882-020-02050-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee DU, et al. Nonselective inhibition of the epigenetic transcriptional regulator BET induces marked lymphoid and hematopoietic toxicity in mice. Toxicol Appl Pharmacol. 2016;300:47–54. doi: 10.1016/j.taap.2016.03.013. [DOI] [PubMed] [Google Scholar]
- 36.Filippakopoulos P, et al. Selective inhibition of BET bromodomains. Nature. 2010;468(7327):1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kahl DJ, et al. 5-Aryl-1,3,4-oxadiazol-2-ylthioalkanoic acids: a highly potent new class of inhibitors of rho/myocardin-related transcription factor (MRTF)/serum response factor (SRF)-mediated gene transcription as potential antifibrotic agents for scleroderma. J Med Chem. 2019;62(9):4350–4369. doi: 10.1021/acs.jmedchem.8b01772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van den Hoogen F, et al. 2013 classification criteria for systemic sclerosis: an American college of rheumatology/European league against rheumatism collaborative initiative. Ann Rheum Dis. 2013;72(11):1747–1755. doi: 10.1136/annrheumdis-2013-204424. [DOI] [PubMed] [Google Scholar]
- 39.Tsou PS, et al. Identification of cysteine-rich angiogenic inducer 61 as a potential antifibrotic and proangiogenic mediator in scleroderma. Arthritis Rheumatol. 2019;71(8):1350–1359. doi: 10.1002/art.40890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schneider CA, et al. NIH image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen G, Deng X. Cell synchronization by double thymidine block. Bio Protoc. 2018;8(17):e2994. doi: 10.21769/BioProtoc.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Love MI, et al. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Draghici S, et al. A systems biology approach for pathway level analysis. Genome Res. 2007;17(10):1537–1545. doi: 10.1101/gr.6202607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bindea G, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25(8):1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bindea G, et al. CluePedia Cytoscape plugin: pathway insights using integrated experimental and in silico data. Bioinformatics. 2013;29(5):661–663. doi: 10.1093/bioinformatics/btt019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data relevant to the study are included in the article. The RNA-Seq data are deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus (GSE186961).