Abstract
MicroRNAs (miRNAs) belong to a class of endogenous small noncoding RNAs that regulate gene expression at the posttranscriptional level, through both translational repression and mRNA destabilization. They are key regulators of kidney morphogenesis, modulating diverse biological processes in different renal cell lineages. Dysregulation of miRNA expression disrupts early kidney development and has been implicated in the pathogenesis of developmental kidney diseases. In this Review, we summarize current knowledge of miRNA biogenesis and function and discuss in detail the role of miRNAs in kidney morphogenesis and developmental kidney diseases, including congenital anomalies of the kidney and urinary tract and Wilms tumor. We conclude by discussing the utility of miRNAs as potentially novel biomarkers and therapeutic agents.
Introduction
MicroRNAs (miRNAs) are endogenous small noncoding RNAs that are usually 19–22 nucleotides in length. The human genome contains 1917 annotated hairpin precursors and 2654 mature miRNAs (1), which regulate over 60% of human protein-coding genes (2). MiRNAs regulate gene expression at the posttranscriptional level, through both translational repression and mRNA destabilization (3–5). Since the discovery of the function of the first identified miRNA, which was shown to regulate cell lineage decisions in the nematode Caenorhabditis elegans, in 1993 (6, 7), miRNAs have been demonstrated to modulate diverse biological processes, including kidney morphogenesis. Dysregulation of miRNA expression disrupts early kidney development and has been implicated in the pathogenesis of developmental kidney diseases. In this Review, we summarize current knowledge on miRNA biogenesis, function, and targeting. We then focus on the role of miRNAs in kidney morphogenesis and developmental kidney diseases, including congenital anomalies of the kidney and urinary tract (CAKUT) and Wilms tumor. Additional interesting areas of research, including the role of miRNAs in a variety of other kidney diseases, such as acute kidney injury (8–10), polycystic kidney disease (11), and kidney transplant (10), have been extensively addressed in other recent reviews. Finally, we conclude by discussing the utility of miRNAs as potentially novel biomarkers and therapeutic agents.
MiRNA biogenesis and function
Biogenesis of miRNAs begins in the nucleus, where RNA polymerase II transcribes miRNA-encoding genes into capped and polyadenylated hairpin transcripts, named primary miRNAs, or pri-miRNAs (Figure 1) (12, 13). Depending on their genomic location, miRNA-encoding genes can be classified as intragenic (located within the introns of host genes; ref. 14) or intergenic (transcribed independently of a host gene and having their own transcriptional regulatory elements; ref. 15). In addition, some miRNAs exist in clusters and are transcribed as polycistronic transcripts (16).
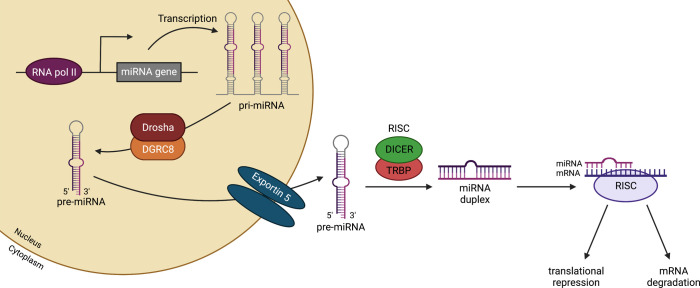
Figure 1. Biogenesis of miRNAs.
MiRNA-encoding genes are transcribed by RNA polymerase II into a primary miRNA (pri-miRNA). Next, a complex formed by the RNA-binding protein DGRC8 and the RNase III enzyme Drosha cleaves the pri-miRNA, generating precursor miRNA (pre-miRNA), which is exported into the cytoplasm through exportin 5. Once in the cytoplasm, the Dicer/TRBP complex cleaves the pre-miRNA, releasing mature miRNA. Finally, the mature miRNA is loaded onto the RISC, driving target mRNA recognition through Watson-Crick base pairing, culminating in gene silencing through translational repression or mRNA degradation. DGRC8, DiGeorge syndrome critical region 8; RISC, RNA-induced silencing complex; TRBP, TAR RNA-binding protein. Created with BioRender.com.
Following transcription, the pri-miRNA is cleaved by the ribonuclease III–like enzyme DROSHA together with the microprocessor complex subunit DGRC8 into a 70-nucleotide hairpin structure, called a pre-miRNA (17–20). The exportin 5/GTP-binding nuclear protein RAN exports the pre-miRNAs to the cytoplasm (21, 22), where the pre-miRNA undergoes cleavage of its terminal loop by the RNase III DICER and TRBP (or TARBP2) to produce a 22-nucleotide miRNA duplex consisting of guide and passenger strands (miRNA:miRNA*, respectively). In the next step, the miRNA duplex is loaded onto an argonaute (AGO) protein to form the RISC (23). Following strand selection and unwinding, the passenger strand is released and degraded (24), while the guide strand remains in the RISC and drives target mRNA recognition through Watson-Crick base pairing.
Most studies show that the domain at the 5′ end of miRNAs (termed the seed sequence, which extends from nucleotide positions 2 to 7) interacts with a specific region at the 3′ untranslated region (3′UTR) of their target mRNAs to induce translational repression and/or mRNA deadenylation and decay (3–5). However, miRNA binding sites have also been identified in other regions, including the 5′UTR (25, 26), coding sequences (27), and gene promoters (28–30). Although miRNAs are primarily associated with gene repression, posttranscriptional upregulation by miRNAs can also occur under certain circumstances (28, 31–33).
There are several unique features associated with miRNA-mediated gene regulation (34, 35). First, a single miRNA can target and repress hundreds of mRNAs, albeit typically to a relatively mild degree for each individual target. Thus, miRNAs are thought to function to fine-tune gene expression. However, as each mRNA can encompass multiple binding sites for the same or different miRNAs, the resultant combined effect is more potent (36–38). Moreover, multiple components within a given signaling pathway can be modulated by individual miRNAs or miRNA clusters (39, 40). Second, miRNA-mediated repression occurs relatively rapidly, as miRNAs block protein synthesis at the ribosome level (41). Third, miRNAs can be concentrated in specific subcellular compartments to regulate site-specific protein translation (42, 43). Finally, a small subset of miRNAs dominates the total miRNA pool in various cell types, suggesting that these may function as master miRNAs (44). In keeping with this idea, a few of the most abundant miRNAs appear to comprise the majority of posttranscriptional regulation mediated by AGO proteins in many cell types (44, 45). For example, in an immortalized human embryonic kidney cell line (HEK293T), miRNAs that were expressed below 100–1000 reads per million did not demonstrate suppressive activity using a high-throughput miRNA sensor library (45).
Biogenesis of miRNAs is under tight spatial and temporal control to ensure appropriate miRNA expression in response to various cellular signals. Regulation of miRNA biogenesis occurs at multiple levels, including transcription factor binding to enhancers and/or promoters of miRNA genes, DROSHA processing of pri-miRNAs, DICER processing of pre-miRNAs, RNA methylation, editing of miRNA precursors, adenylation, uridylation, RNA decay, and many other mechanisms. For an in-depth review, please refer to Ha and Kim (46). Recently, super-enhancers have also emerged as a new class of regulatory elements controlling miRNA biogenesis by enhancing both transcription and DROSHA/DGCR8-mediated pri-miRNA processing. In combination with a broad H3K4me3 signature, super-enhancer activity shapes tissue-specific miRNA expression pattern and function (47).
Kidney development
The mammalian kidney (or metanephros) is a vital organ that plays a critical role in excretion of metabolic wastes, regulation of extracellular fluid volume, and maintenance of electrolyte and acid-base homeostasis. Moreover, the kidney produces important hormones, such as erythropoietin, calcitriol, renin, and prostaglandins (48). The functional capacity of the kidney correlates with the number of functioning nephrons that are formed during kidney development prior to birth, also termed nephron endowment. Each human kidney contains on average 1,000,000 nephrons, although this number varies considerably, with estimates ranging from 200,000 to 2,000,000 nephrons (49, 50). With aging, loss of functional nephron reserve occurs over time (51, 52); therefore, low nephron endowment at birth is associated with an increased risk of developing hypertension and chronic kidney disease (CKD) later in life (53–55). Moreover, CAKUT, which lead to decreased nephron endowment and nephron function, are the leading causes of renal failure in children, resulting in significant morbidity and mortality associated with transplant and dialysis (56, 57). Thus, a better understanding of the cellular and molecular mechanisms underlying the establishment of nephron number and normal nephron formation provides insights into novel avenues to predict, prevent, and treat childhood kidney disease.
Metanephric kidney development starts around embryonic day 10.5 (E10.5) in mice and around the fifth week of gestation in humans (58). In response to inductive signals from the metanephric mesenchyme, the ureteric bud extends from the caudal end of the Wolffian duct and invades into the adjacent mesenchyme (Figure 2). Simultaneously, morphogens emanating from the ureteric bud induce condensation of the metanephric mesenchyme to form the cap mesenchyme (also termed nephron progenitors) around the tips of the ureteric bud. As nephrogenesis progresses, the ureteric bud undergoes successive rounds of branching, elongation, and differentiation to generate the collecting ducts of the kidney. A subpopulation of nephron progenitors undergoes mesenchymal-epithelial transition to form renal vesicles, which after polarization and elongation become comma- and S-shaped body structures. Finally, the distal portion of the S-shaped body fuses with the collecting duct to form a functional nephron (59–61). The S-shaped body undergoes further differentiation to form the mature cell types of the nephron, apart from the collecting duct. Foxd1+ stromal progenitor cells are adjacent to nephron progenitors in the outer cortical or nephrogenic zone of the developing kidney (Figure 2) (62). Signals from the cortical stroma are thought to inhibit nephron progenitor cell expansion and stimulate its differentiation, as ablation of the renal stroma results in impaired nephron progenitor differentiation (63). The Foxd1+ progenitor cells give rise to all stromal cells in the metanephric kidney, including renal cortical and medullary interstitial cells, pericytes, perivascular fibroblasts, mesangial cells, and vascular smooth muscle cells (64, 65). Perturbations in any step of this process can lead to CAKUT, the major cause of childhood CKD (66, 67).
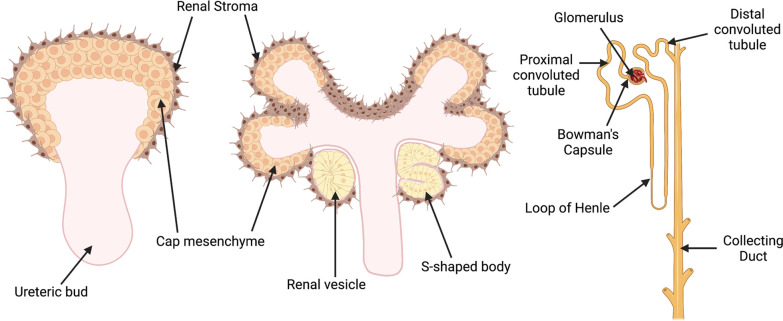
Figure 2. Schematic illustration of the stages of metanephric kidney development.
Signals from the ureteric bud trigger condensation of the metanephric mesenchyme to form a cap of nephron progenitors (cap mesenchyme) around the ureteric bud tips. The cap mesenchyme undergoes a mesenchymal-epithelial transition to form renal vesicles, which develop sequentially into comma- and S-shaped bodies. These structures connect to the ureteric bud stalk, which give rises to the collecting duct. Cells in the proximal domain of the S-shaped body differentiate into specialized epithelial cells of the mature renal corpuscle (i.e., podocytes and Bowman’s capsule cells), while cells in the mid- and distal portions differentiate into the tubular segments of nephron (proximal tubules, loops of Henle, and distal tubules). Created with BioRender.com.
The mature nephron is composed of a glomerulus that acts as the filtration unit and a tubular reabsorption compartment that is subdivided into proximal convoluted tubule, loop of Henle, distal convoluted tubule, and collecting duct (Figure 2). The filtration barrier of the glomerulus (which comprises the fenestrated endothelium, glomerular basement membrane, and foot processes and slit diaphragms of podocytes) allows the filtration of plasma and small solutes, while selectively retaining proteins such as albumin and immunoglobulins in the blood (68, 69). Meanwhile, the tubular reabsorption compartment is responsible for maintenance of water homeostasis, reabsorption of solutes (including sodium, potassium, calcium, phosphorous, magnesium, glucose, and many others), and excretion of acid and other wastes.
MiRNAs in the developing kidney
In studies in conditional transgenic mice, miRNAs have emerged as critical regulators of kidney morphogenesis in multiple cell lineages. The initial studies evaluating a functional role for miRNAs in kidney development utilized conditional deletion of Dicer (70) in different renal lineages. However, Dicer is also known to have miRNA-independent roles (71), which has complicated the interpretation of these models. Conditional deletion of Dicer in early metanephric mesenchyme or nephron progenitors results in augmented apoptosis of nephron progenitors, elevated levels of the proapoptotic protein Bim, and premature cessation of nephrogenesis (72–75) (Table 1). Interestingly, the loss of Bim expression in Dicer-deficient nephron progenitors decreases apoptosis and partially restores nephron formation. Two miRNAs expressed in nephron progenitors, miR-17 and miR-106, were identified as suppressors of BIM expression (76). Together, these findings indicate that miRNAs control the balance between survival and apoptosis in nephron progenitors to ensure the formation of a correct number of nephrons throughout nephrogenesis.
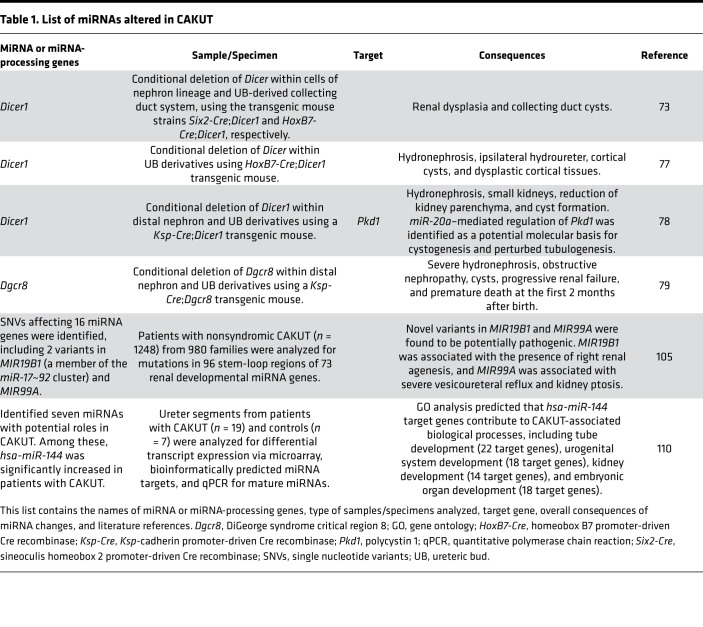
Table 1. List of miRNAs altered in CAKUT.
Conditional deletion of Dicer in the ureteric bud lineage results in a spectrum of abnormalities that strongly resemble CAKUT, including renal dysplasia and the development of collecting duct cysts (73, 77, 78). Premature termination of branching morphogenesis (in response to decreased expression of Wnt11 and c-Ret from the ureteric bud) is likely the major contributing factor for renal dysplasia (73). The onset of cyst formation occurs at around E15.5 and is associated with defects in primary cilia length, increased apoptotic cell death, and excessive cell proliferation (73). As Dicer has important miRNA-independent roles in the cell, conditional deletion of Dgcr8 has been used to confirm that the phenotypes observed in conditional Dicer-knockout models are indeed the result of loss of miRNAs. Animals with Dgcr8 deletion in the distal nephron and derivatives of the collecting duct develop hydronephrosis and collecting duct cysts (79), a CAKUT-like phenotype that resembles loss of Dicer activity in the ureteric bud lineage.
Studies from two independent groups demonstrated that ablation of Dicer from the Foxd1+ renal stroma lineage and its derivatives results in a spectrum of renal anomalies, with consistent findings regarding hypoplastic kidneys, reduced glomerular numbers, abnormal glomerular maturation, and defective vascular patterning (80, 81). Though both groups described largely concordant phenotypes using similar mouse models, two distinct differences were noted. Nakagawa et al. observed a lack of the inner medulla and papilla, as well as a decrease in the nephrogenic zone (80). In contrast, Phua et al. showed an expansion of the nephron progenitor population and preserved renal papilla (81). Nakagawa et al. proposed that these defects are related to disruption of Wnt pathway signaling, resulting in changes in stromal cell migration and proliferation, due to downregulation of the stromal cell miRNAs, miR-214, miR-199a-5p, and miR-199a-3p (80). The study by Phua et al. suggested that changes in apoptotic programs (including augmented expression of Bim and p53 effector genes) contribute to the phenotypic defects (81). It is conceivable that genetic background differences and/or the efficiency of Cre-mediated recombination may be responsible for the differences these studies describe. Nevertheless, the described phenotypes are consistent with the known multifaceted roles of the renal stroma in kidney development, and it is likely that the mechanisms underlying these phenotypes are complex given the nature of a Dicer deletion. Further studies examining specific miRNAs in various stromal subpopulations are needed to better define the regulatory mechanisms at play.
More recent work has addressed the question of the function of specific miRNAs in both the developing kidney and nephron progenitors. Using a human embryonic stem cell model, Bantounas et al. showed that inhibition of the miR-199a~214 cluster results in dysmorphic glomeruli, aberrant proximal tubules, decreased WT1 expression, and increased interstitial capillaries in kidney-like organoids (82). Interestingly, global deletion of hypoxia-responsive miR-210 results in a male-specific nephron deficit (83). For example, conditional deletion of the miR-17~92 cluster in nephron progenitors and their derivatives in mice impairs progenitor cell proliferation and reduces the number of developing nephrons. As a result, mutant mice develop proteinuria, renal fibrosis, and impaired renal function (84). Dysregulated levels of the miR-17~92 target gene, Cftr, are implicated in defective proliferation of progenitor cells and reduced nephron endowment in this mouse model (85).
Small RNA sequencing (smRNA-Seq) has been increasingly used to profile miRNA expression patterns and for the discovery of novel miRNA species. smRNA-Seq of E15.5 nephrogenic mesenchymal cells identified 162 annotated miRNAs that are differentially expressed in this cell population compared with whole kidney and 49 novel miRNA species (86). Interestingly, levels of miR-200 family miRNAs were significantly reduced in nephron progenitors. Given that members of the miR-200 family have been shown to be key regulators of mesenchymal-epithelial transition in the collecting duct (87), we speculate that their expression might be tightly regulated to ensure normal epithelial differentiation of nephron progenitors during kidney development.
MiRNA function in the mature nephron
In addition to their requirement during kidney development, miRNAs regulate numerous biological processes in the major cell lineages that form the mature nephron (69, 88–91). In keeping with this, segment-specific expression of miRNAs along the nephron has been described, including miR-143 and miR-195a in the glomerulus, miR-107 and miR-34a in the proximal tubule, miR-193 and miR-378a in the thick ascending limb, miR-874 and miR-155 in the distal convoluted tubule, and miR-200c in the collecting duct (87). Moreover, functional studies in compartments of the mature nephron support distinct roles for miRNAs.
Mice lacking either Dicer or Drosha in podocytes exhibit marked proteinuria, glomerulosclerosis, and rapid progression to kidney failure, secondary to disruption of the glomerular filtration barrier (90–93). In silico analyses reveal that various upregulated transcripts in mutant glomeruli contain target sequences for miR-30 family members. As all four miR-30 family members (miR-30c-1, miR-30b, miR-30d, and miR-30c-2) are normally highly expressed in podocytes, these miRNAs may be responsible for the podocyte abnormalities and disruption of the glomerular filtration barrier in mutant mice (91).
Somewhat surprisingly, deletion of Dicer from postnatal mammalian proximal tubules does not affect kidney development, histology, or function but does protect against renal ischemia/reperfusion injury. Mutant mice exhibit better kidney function, reduced kidney injury, lower tubular apoptosis, and improved survival compared with their WT littermates (94). This likely reflects the “sum” of the effect of deletion of multiple miRNAs in the proximal tubule, as other work has since demonstrated that the expression of specific miRNAs is protective in renal ischemia/reperfusion injury (e.g., miR-16 and miR-21; refs. 95, 96); whereas others are injurious (e.g., miR-182; ref. 97).
Although miRNAs seem to be dispensable for proximal tubule function, they are essential for distal nephron and collecting duct homeostasis (79, 88, 98). Collecting duct-specific inactivation of Dicer and other critical miRNA biogenesis-associated genes (including Dgcr8, Ago1, 2, 3, and 4) causes renal failure in adult mice because of progressive tubulointerstitial fibrosis and interstitial inflammation (88). This is preceded by a partial epithelial-mesenchymal transition (EMT) of collecting duct cells, and downregulation of miR-200 family members, which inhibit EMT (88). Likewise, ablation of either Dicer or Dgcr8 from distal nephron and ureteric bud derivatives, respectively, results in renal abnormalities and kidney failure (78, 98), which are ultimately associated with downregulation of miR-200 family members (98). Increased expression of miR-200 target gene Pkd1 in these mutant mice disrupts tubulogenesis and produces cyst-like structures (98). These differences in the requirement for functional miRNAs in proximal tubules and distal nephron/collecting duct might be explained by the segmental distribution of miRNAs along the length of the nephron and collecting duct in WT kidneys (88).
Deletion of Dicer in renin-secreting cells in the juxtaglomerular apparatus results in a deficit of juxtaglomerular cells, reduced circulating renin levels with consequent reduction in arterial blood pressure, reduced kidney function, striped pattern of interstitial fibrosis, and vascular abnormalities (89). The reduction in juxtaglomerular cells suggests a requirement for mature miRNAs in the maintenance of their phenotype. Later, miR-330 and miR-125b-5p were identified as potential candidates that either inhibit or promote, respectively, the smooth muscle phenotype of juxtaglomerular cells (99).
Other active areas of research on miRNAs in acute kidney injury (8–10), polycystic kidney disease (11), and kidney transplant (10), have been comprehensively addressed in other recent reviews.
MiRNAs in pediatric kidney diseases
In this section, we provide an overview of the role of miRNAs in developmental kidney diseases, including CAKUT and Wilms tumor. CAKUT are among the most frequent form of malformations at birth, affecting approximately 3–7 out of 1000 live births (100). Disruption of kidney and lower urinary tract development leads to a wide spectrum of clinical manifestations observed in CAKUT, including kidney anomalies (i.e., renal agenesis, renal hypoplasia and dysplasia, and multicystic dysplastic kidneys), ureteropelvic anomalies (i.e., ureteropelvic junction obstruction), duplex collecting system, and anomalies of the bladder and urethra (101–103). This phenotypic heterogeneity is likely due to complex interactions between genetic, epigenetic, and/or prenatal environmental factors that affect kidney and lower urinary tract development, resulting in CAKUT (101). Most of our current knowledge on CAKUT pathogenesis has arisen from mouse models and syndromic forms of CAKUT. These studies have led to the identification of several CAKUT genes, many of which are implicated in early kidney development, including PAX2, SALL1, HNF1B, EYA1, GATA3, RET, WNT4, GDNF, SIX1, SIX2, and others (101, 104, 105). However, single mutations or copy number variants in protein-coding genes do not explain the majority of CAKUT cases (~80%) (101, 106).
As mentioned above, depletion of mature miRNAs from different cell lineages of the developing kidney in mouse models results in renal abnormalities that mimic human CAKUT (73, 74, 78, 79). In addition, germline deletions of MIR17HG, which encodes the miR-17~92 cluster, causes type 2 Feingold syndrome in humans (107). Although a renal phenotype in type 2 Feingold syndrome patients with MIR17HG mutations remains undefined, an 18% incidence of CAKUT has been reported in Feingold syndrome cases associated with MYCN mutations (108, 109). Together, these observations suggest that mutations in miRNAs expressed during kidney development might cause CAKUT in humans, particularly as many miRNAs are highly conserved between mouse and human.
To test this hypothesis, one study investigated 1248 patients with nonsyndromic CAKUT from 980 families and looked for mutations in 96 stem-loop regions of 73 renal developmental miRNA genes (106). Within this cohort, 31 individuals with 17 different single nucleotide variants affecting 16 different miRNA genes were identified. Among these, two novel variants in miRNAs were found to be potentially pathogenic. MIR19B1 (a member of the miR-17~92 cluster) was associated with the presence of right renal agenesis, and MIR99A was associated with severe vesicoureteral reflux and kidney ptosis. This surprisingly low number of candidate pathogenic variants is partly due to limitations of this study, as the analysis only accounted for mutations in miRNA genes that were included in the candidate gene approach and did not detect copy number variations and large DNA rearrangements (106).
In an alternative approach, ureter segments from patients with a variety of CAKUT were analyzed for differential transcript expression via microarray, for the presence of bioinformatically predicted miRNA targets, and for mature miRNAs via qPCR (110). Using this multipronged approach, seven miRNAs were identified with potential roles in CAKUT, and among these, hsa-miR-144 was significantly increased in patients with CAKUT. Gene ontology analysis indicated that predicted hsa-miR-144 target genes contribute to biological processes involved in CAKUT development, including tube development (22 target genes), urogenital system development (18 target genes), kidney development (14 target genes), and embryonic organ development (18 target genes) (110).
Further studies are needed to define the molecular mechanisms underlying the pathogenic roles of miRNAs in CAKUT. Findings from such studies will be critical in improving the care of patients with CAKUT and preventing their progression to CKD, providing appropriate genetic counseling for patients and their families, and developing novel therapeutic strategies.
MiRNAs in Wilms tumor
Wilms tumor, or nephroblastoma, is the most common childhood renal cancer, with an incidence of 1 in 10,000 children in North America (111). It is primarily a sporadic disease, although familial forms occur in approximately 1%–2% of cases (112, 113). Wilms tumors arise from aberrant nephrogenesis, where pluripotent embryonic renal precursors fail to differentiate and persist abnormally into postnatal life (111, 114, 115). These tumors histologically resemble developing kidneys with a disrupted morphology (116), and mutations in genes involved in fetal nephrogenesis, including WT1 (117–119), CTNNB1 (120), SIX1/2 (121, 122), and TP53 (123, 124), are associated with approximately 40% of Wilms tumors (125). Recent studies using whole-genome and whole-exome sequencing of Wilms tumors identified novel mutations in cancer risk genes (REST, CHEK2, PALB2) (126, 127), genes encoding proteins that mediate histone modifications during nephrogenesis (BCOR, MAP3K4) (126), and miRNA-processing genes (121, 122, 128, 129).
Among the miRNA-processing genes, mutations in DROSHA, DGCR8, DICER1, TARBP2, and XPO5 (encodes exportin 5) have been reported in treatment-naive and neoadjuvant chemotherapy-treated Wilms tumors (Figure 3) (121, 122, 128, 129). About 33% of Wilms tumors examined exhibit deleterious mutations in genes of the miRNA-processing pathway (128). A recurrent hotspot mutation (E1147K) in a metal-binding (Mg2+) residue of the RNase IIIb domain of DROSHA, which appears to be unique to Wilms tumor, abolishes the catalytic activity of this domain, resulting in incomplete cleavage of pri-miRNAs and reduced miRNA maturation (128). Somatic hotspot mutations affecting the RNase IIIb domain of DICER1 impair processing of 5p miRNAs (those derived from the 5′-arm of the pre-miRNA hairpin) (129) and are often found as “second hit” mutations that act in tandem with DICER1 germline mutations to induce Wilms tumorigenesis in DICER1 syndrome (a disorder that increases susceptibility to a variety of tumors) (130–132). It remains unclear why impaired DICER1 function in Wilms tumors results in persistent and aberrant nephrogenesis, unlike loss of Dicer1 in mouse nephron progenitors, which causes increased apoptosis and a premature cessation of nephrogenesis (72–75). Some potential possibilities include that the gene dosage activity might be crucial in determining cell survival or that mutations in the RNase IIIb domain might affect the specificity of miRNA binding by DICER1.
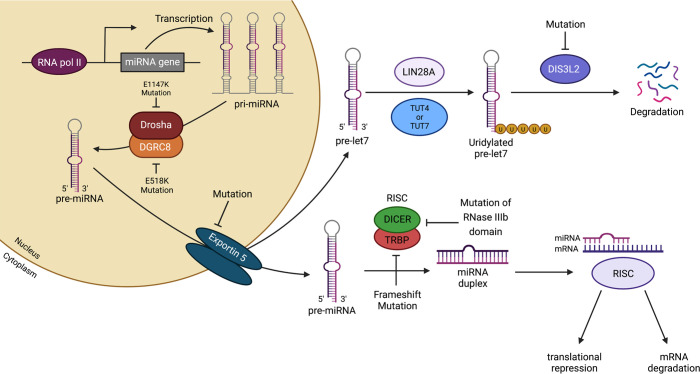
Figure 3. Mutations in miRNA-processing genes result in aberrant miRNA expression and Wilms tumorigenesis.
Recurrent mutations in a metal-binding (Mg2+) residue of the RNase IIIb domain of DROSHA (E1147K) or in the double-stranded RNA-binding domain of DGRC8 (E518K) disrupt the cleavage of pri-miRNAs into pre-miRNAs. Mutations in XPO5 (encodes exportin 5) prevent pre-miRNA export, which culminates in pre-miRNA accumulation in the nucleus. Frameshift mutations in TARBP2 (encodes TRBP) and mutations affecting the RNase IIIb domain of DICER1 can disrupt the processing of pre-miRNAs into mature miRNAs. In stem and progenitor cells, members of the let-7 miRNA family function as tumor suppressors, and their expression is tightly regulated by the RNA-binding protein Lin28. Lin28A binds to the terminal loop of let-7 precursors and recruits the activity of the terminal uridyl transferases TUT4/7 to produce uridylated pre-let-7, which is subsequently degraded by DIS3L2. Overexpression of LIN28 and mutations in DISL3L2 have been associated with aberrant mature let-7 expression and Wilms tumorigenesis. Created with BioRender.com.
Mutations in miRNA-processing genes are associated with downregulation of important miRNAs, including members of the miR-200 (121) and the let-7 families (Figure 3) (121, 129). Let-7 miRNAs and the RNA-binding protein Lin28 function in concert to control the timing of cessation of murine nephrogenesis, possibly via regulation of the growth-promoting gene Igf2 (133). Overexpression of Lin28 during kidney development causes expansion of nephrogenic progenitors, by inhibiting their final wave of differentiation, which culminates in neoplastic transformation that is highly reminiscent of human Wilms tumor (134). Increased DNA copy number of LIN28B and DNA copy loss of let-7a are seen in 25% and 46% of human Wilms tumor samples, respectively (126). In line with these observations, germline mutations in the human DISL3L2 gene, which encodes an exoribonuclease responsible for degrading preprocessed forms of let-7, cause Perlman syndrome and predisposition to Wilms tumor (135, 136). Perlman syndrome is a congenital overgrowth syndrome characterized by macrosomia, polyhydramnios, facial dysmorphology, renal dysplasia, and nephroblastomatosis (a precursor lesion for Wilms tumor) (137). Among infants with Perlman syndrome who survive past the neonatal period, 64% develop Wilms tumor (138). Interestingly, complete or partial DISL3L2 deletions were found in about 30% of sporadic Wilms tumors examined (136).
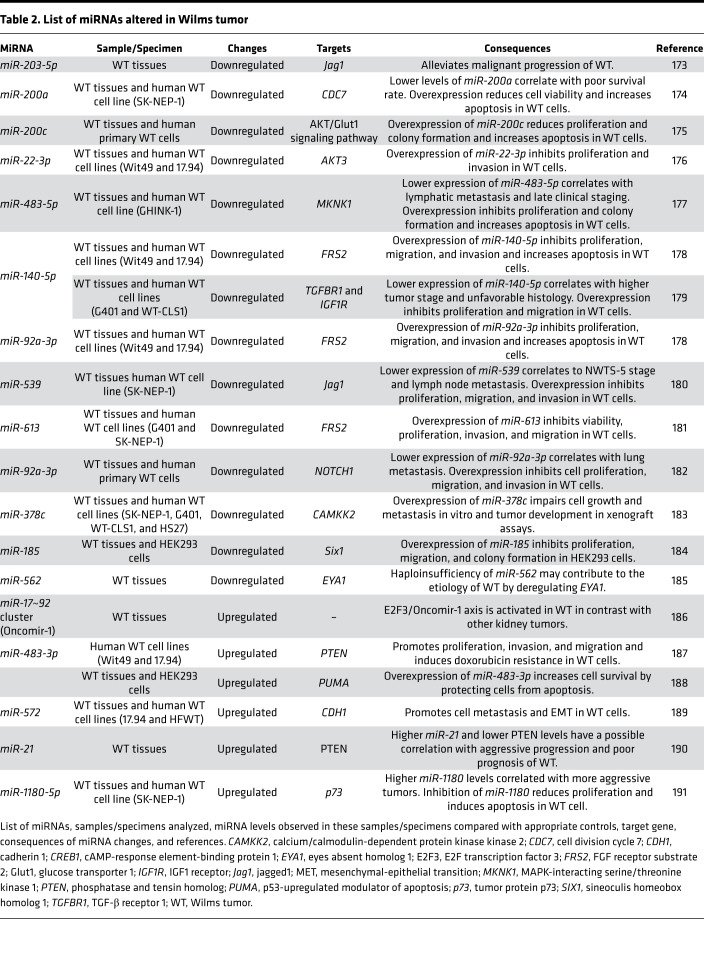
A recent study strengthened the significance of the miRNA regulatory network in the etiology of Wilms tumor. The authors found that pleiomorphic adenoma gene 1 (PLAG1) is one of the most consistently upregulated genes in Wilms tumors with mutations in miRNA-processing genes (125). Ectopic expression of PLAG1 in the developing mouse kidney causes neoplasia, which is accompanied by transactivation of its target gene, the Wilms tumor oncogene IGF2. miR-16 and miR-34, which are downregulated in Wilms tumors, were identified as potential regulators of PLAG1 expression (125). Table 2 summarizes other studies that have reported aberrant expression of specific miRNAs associated with the etiology of Wilms tumor. Interestingly, these miRNAs can function as oncogenes (called oncomiRs) or tumor suppressors in the setting of Wilms tumor development, depending on the nature of their targets.
Table 2. List of miRNAs altered in Wilms tumor.
MiRNAs as potential biomarkers and therapeutic agents
Apart from their intracellular location, miRNAs are also present in significant amounts in biological fluids, including blood, plasma, urine, breast milk, and saliva (139). These circulating miRNAs are found packaged in microparticles (exosomes, microvesicles, and apoptotic bodies) (140, 141), conjugated with AGO (142) or nucleophosmin 1 proteins (143), or loaded into HDL (144), which make them remarkably stable even under unfavorable conditions, such as boiling, extreme variations in pH, extended storage, and multiple freeze-thaw cycles (145, 146). Thus, miRNA signatures in biological fluids can reflect associations with physiological or disease conditions (147). Together, these features make circulating miRNAs attractive for use as noninvasive biomarkers for disease diagnosis and prognosis.
Circulating miRNAs can be extracted directly from unfractionated biological fluids or from extracellular vesicle preparations using commercially available extraction kits (148) or TRIzol (149). Upon isolation, miRNAs can be stored at –70°C and remain stable for up to 1 year (148). There are several platforms available for miRNA profiling, including microarray hybridization, qPCR, and next-generation sequencing (150). Microarray and qPCR are the most frequently used methodologies to investigate the expression of known miRNAs (151). Both methods have the advantages of being simple to use, relatively quick from RNA labeling to data generation, and relatively cost-effective (152). However, they rely on the availability and accurate annotation of miRNA sequences in databases for probe and primer design (150). Although more expensive, next-generation sequencing allows for the simultaneous detection of both known and novel miRNA species and offers high sensitivity (153). Furthermore, the single-nucleotide resolution of next-generation sequencing enables the identification of isomiRs, which are mature miRNA isoforms that differ from canonical ones in length, sequence, or both (154, 155), which change the targeting specificity of the miRNA (156).
Diverse studies have investigated the potential of circulating miRNAs as biomarkers for pediatric kidney diseases (157–160). For instance, one study identified 14 miRNAs that were significantly upregulated in the serum of patients with Wilms tumor. Interestingly, a signature based on miR-100-5p and miR-130-3p expression could differentiate these patients from healthy controls with accuracy, sensitivity, and specificity (159). Although the findings from this study and many other studies have provided compelling motivation to explore the potential of circulating miRNAs as biomarkers, several hurdles in the field need to be overcome before widespread clinical application. First, there is a relative lack of consensus between studies likely due to the absence of standardized methodology for purification (161) and analysis of samples (e.g., differences in miRNA profiling platforms, refs. 162, 163; or differences in smRNA-Seq library preparation methods, ref. 164). Second is the lack of large-sample-size studies and detailed investigations on specific diseases. Another important aspect is that the influence of confounding variables such as age, sex, and external factors (e.g., tobacco, alcohol, etc.) on miRNA profiles has not been fully explored (for an in-depth review, please refer to ref. 165).
On the therapeutic side, several miRNA-based drugs are currently in clinical trials but have not been granted FDA approval yet (166, 167). The main approaches for miRNA therapy involve restoration of miRNA levels using miRNA mimics, or inhibition of specific miRNAs using antagomiRs (168). One of the challenges associated with the development of miRNA-based therapeutics is the identification of miRNA candidates for each disease. Because multiple miRNAs are dysregulated in each disease, a careful analysis of patient samples in combination with in vitro and in vivo assays that address the pathophysiological mechanisms affected by the miRNAs in question should be performed for narrowing down the candidate miRNAs for therapeutic intervention (168, 169). Another challenge involves the development of strategies to improve in vivo stability and site-specific drug delivery with minimal toxicity and off-target effects. RNA molecules are chemically unstable due to the presence of the 2′-hydroxyl group on the pentose ring. To provide higher stability and protection from nucleases present in serum or the endocytic compartment of cells, biotech companies have generated RNA molecules with chemical modifications (2′-O-methyl group, phosphorothioate, or locked nucleic acids) in their backbone (166). As for in vivo delivery, technologies include lipid based (e.g., lipid nanoparticle and neutral liposome) and dendrimers conjugated to a targeting moiety, among many other strategies. Major challenges associated with miRNA delivery systems are immunotoxicity and target-specific affinity toward a disease site (170). Delivery strategies by various methods of administration (intraperitoneal, intravenous, and subcutaneous injections) or by using vectors containing kidney-specific and inducible promoters have been successfully used for selective kidney targeting and to avoid potential adverse effects in other tissues and organs (171, 172).
Summary
There has been an explosion of information regarding miRNA biogenesis, the regulation of miRNA expression, and miRNA function since the initial discovery of miRNAs in 1993 (6, 7). This has been accompanied by an ever-increasing understanding of how miRNAs function both in normal physiology and in the pathophysiology of many diseases. It has become clear that dysregulation of miRNA expression disrupts early kidney development and is implicated in the pathogenesis of developmental kidney diseases, such as CAKUT and Wilms tumor. With recent developments in the use of miRNAs as biomarkers and as novel drug targets, insights into how miRNAs regulate kidney development and disease are critical to understanding how they might be utilized in novel diagnostic and therapeutic approaches to these diseases. To fully realize these efforts, future studies identifying the function of specific miRNAs in kidney development are critical, in addition to technologies to optimize targeting small oligonucleotide therapeutics to the kidney.
Acknowledgments
JH’s laboratory is supported by grants from the NIH National Institute of Diabetes and Digestive and Kidney Diseases (R01DK125015 and R01DK102843). DMC was supported by Nephrotic Syndrome Study Network Career Development Award and Children’s Hospital of Pittsburgh Research Advisory Council Postdoctoral Fellowship. MT was supported by the University of Pittsburgh Summer Research Internship Program (R25DK119180). The figures were created with BioRender.com.
Version 1. 05/09/2022
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2022, Cerqueira et al. This is an open access article published under the terms of the Creative Commons Attribution 4.0 International License.
Reference information: JCI Insight. 2022;7(9):e158277. https://doi.org/10.1172/jci.insight.158277.
Contributor Information
Maliha Tayeb, Email: MAT288@pitt.edu.
Jacqueline Ho, Email: jacqueline.ho2@chp.edu.
References
- 1.Kozomara A, et al. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47(d1):D155–D162. doi: 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friedman RC, et al. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10(2):94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Djuranovic S, et al. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 2012;336(6078):237–240. doi: 10.1126/science.1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12(2):99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 6.Lee RC, et al. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 7.Wightman B, et al. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75(5):855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 8.Brandenburger T, et al. Noncoding RNAs in acute kidney injury. Kidney Int. 2018;94(5):870–881. doi: 10.1016/j.kint.2018.06.033. [DOI] [PubMed] [Google Scholar]
- 9.Liu Z, et al. Non-coding RNAs in kidney injury and repair. Am J Physiol Cell Physiol. 2019;317(2):C177–C188. doi: 10.1152/ajpcell.00048.2019. [DOI] [PubMed] [Google Scholar]
- 10.Ledeganck KJ, et al. MicroRNAs in AKI and kidney transplantation. Clin J Am Soc Nephrol. 2019;14(3):454–468. doi: 10.2215/CJN.08020718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yheskel M, Patel V. Therapeutic microRNAs in polycystic kidney disease. Curr Opin Nephrol Hypertens. 2017;26(4):282–289. doi: 10.1097/MNH.0000000000000333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cai X, et al. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10(12):1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee Y, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23(20):4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11(3):241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monteys AM, et al. Structure and activity of putative intronic miRNA promoters. RNA. 2010;16(3):495–505. doi: 10.1261/rna.1731910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee Y, et al. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21(17):4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee Y, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425(6956):415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 18.Denli AM, et al. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432(7014):231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 19.Gregory RI, et al. The microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432(7014):235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 20.Han J, et al. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18(24):3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lund E, et al. Nuclear export of microRNA precursors. Science. 2004;303(5654):95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 22.Yi R, et al. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17(24):3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hammond SM, et al. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293(5532):1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- 24.Kawamata T, Tomari Y. Making RISC. Trends Biochem Sci. 2010;35(7):368–376. doi: 10.1016/j.tibs.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 25.Lytle JR, et al. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5’ UTR as in the 3’ UTR. Proc Natl Acad Sci U S A. 2007;104(23):9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Brien J, et al. Overview of MicroRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol (Lausanne) 2018;9:402. doi: 10.3389/fendo.2018.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Forman JJ, et al. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc Natl Acad Sci U S A. 2008;105(39):14879–14884. doi: 10.1073/pnas.0803230105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Place RF, et al. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci U S A. 2008;105(5):1608–1613. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim DH, et al. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc Natl Acad Sci U S A. 2008;105(42):16230–16235. doi: 10.1073/pnas.0808830105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Younger ST, Corey DR. Transcriptional gene silencing in mammalian cells by miRNA mimics that target gene promoters. Nucleic Acids Res. 2011;39(13):5682–5691. doi: 10.1093/nar/gkr155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orom UA, et al. MicroRNA-10a binds the 5’UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30(4):460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 32.Valinezhad Orang A, et al. Mechanisms of miRNA-mediated gene regulation from common downregulation to mRNA-specific upregulation. Int J Genomics. 2014;2014:970607. doi: 10.1155/2014/970607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vasudevan S. Posttranscriptional upregulation by microRNAs. Wiley Interdiscip Rev RNA. 2012;3(3):311–330. doi: 10.1002/wrna.121. [DOI] [PubMed] [Google Scholar]
- 34.Hobert O. Gene regulation by transcription factors and microRNAs. Science. 2008;319(5871):1785–1786. doi: 10.1126/science.1151651. [DOI] [PubMed] [Google Scholar]
- 35.Ho J, Kreidberg JA. MicroRNAs in renal development. Pediatr Nephrol. 2013;28(2):219–225. doi: 10.1007/s00467-012-2204-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455(7209):64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Selbach M, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455(7209):58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 38.Shirdel EA, et al. NAViGaTing the micronome--using multiple microRNA prediction databases to identify signalling pathway-associated microRNAs. PLoS One. 2011;6(2):e17429. doi: 10.1371/journal.pone.0017429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uhlmann S, et al. Global microRNA level regulation of EGFR-driven cell-cycle protein network in breast cancer. Mol Syst Biol. 2012;8:570. doi: 10.1038/msb.2011.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mestdagh P, et al. The miR-17-92 microRNA cluster regulates multiple components of the TGF-β pathway in neuroblastoma. Mol Cell. 2010;40(5):762–773. doi: 10.1016/j.molcel.2010.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bhattacharyya SN, et al. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125(6):1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 42.Schratt GM, et al. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439(7074):283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- 43.Ashraf SI, et al. Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell. 2006;124(1):191–205. doi: 10.1016/j.cell.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 44.Landgraf P, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129(7):1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mullokandov G, et al. High-throughput assessment of microRNA activity and function using microRNA sensor and decoy libraries. Nat Methods. 2012;9(8):840–846. doi: 10.1038/nmeth.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15(8):509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 47.Suzuki HI, et al. Super-enhancer-mediated RNA processing revealed by integrative microRNA network analysis. Cell. 2017;168(6):1000–1014. doi: 10.1016/j.cell.2017.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Santoro D, et al. Extraskeletal functions of vitamin D. Biomed Res Int. 2015;2015:294719. doi: 10.1155/2015/294719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bertram JF, et al. Human nephron number: implications for health and disease. Pediatr Nephrol. 2011;26(9):1529–1533. doi: 10.1007/s00467-011-1843-8. [DOI] [PubMed] [Google Scholar]
- 50.Hughson MD, et al. Hypertension, glomerular number, and birth weight in African Americans and white subjects in the southeastern United States. Kidney Int. 2006;69(4):671–678. doi: 10.1038/sj.ki.5000041. [DOI] [PubMed] [Google Scholar]
- 51.Denic A, et al. Structural and functional changes with the aging kidney. Adv Chronic Kidney Dis. 2016;23(1):19–28. doi: 10.1053/j.ackd.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Denic A, et al. The substantial loss of nephrons in healthy human kidneys with aging. J Am Soc Nephrol. 2017;28(1):313–320. doi: 10.1681/ASN.2016020154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brenner BM, et al. Glomeruli and blood pressure. Less of one, more the other? Am J Hypertens. 1988;1(4 pt 1):335–347. doi: 10.1093/ajh/1.4.335. [DOI] [PubMed] [Google Scholar]
- 54.Hoy WE, et al. Nephron number, hypertension, renal disease, and renal failure. J Am Soc Nephrol. 2005;16(9):2557–2564. doi: 10.1681/ASN.2005020172. [DOI] [PubMed] [Google Scholar]
- 55.Baum M. Role of the kidney in the prenatal and early postnatal programming of hypertension. Am J Physiol Renal Physiol. 2010;298(2):F235–F247. doi: 10.1152/ajprenal.00288.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Seikaly MG, et al. Chronic renal insufficiency in children: the 2001 Annual Report of the NAPRTCS. Pediatr Nephrol. 2003;18(8):796–804. doi: 10.1007/s00467-003-1158-5. [DOI] [PubMed] [Google Scholar]
- 57.Smith JM, et al. Contributions of the transplant registry: the 2006 annual report of the North American pediatric renal trials and collaborative studies (NAPRTCS) Pediatr Transplant. 2007;11(4):366–373. doi: 10.1111/j.1399-3046.2007.00704.x. [DOI] [PubMed] [Google Scholar]
- 58.Osathanondh V, Potter EL. Development of human kidney as shown by microdissection. III. Formation and interrelationship of collecting tubules and nephrons. Arch Pathol. 1963;76:290–302. [PubMed] [Google Scholar]
- 59.Lechner MS, Dressler GR. The molecular basis of embryonic kidney development. Mech Dev. 1997;62(2):105–120. doi: 10.1016/S0925-4773(97)00667-9. [DOI] [PubMed] [Google Scholar]
- 60.Saxen L, Sariola H. Early organogenesis of the kidney. Pediatr Nephrol. 1987;1(3):385–392. doi: 10.1007/BF00849241. [DOI] [PubMed] [Google Scholar]
- 61.Dressler GR. Advances in early kidney specification, development and patterning. Development. 2009;136(23):3863–3874. doi: 10.1242/dev.034876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cullen-McEwen LA, et al. The where, what and why of the developing renal stroma. Nephron Exp Nephrol. 2005;99(1):e1–e8. doi: 10.1159/000081792. [DOI] [PubMed] [Google Scholar]
- 63.Das A, et al. Stromal-epithelial crosstalk regulates kidney progenitor cell differentiation. Nat Cell Biol. 2013;15(9):1035–1044. doi: 10.1038/ncb2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kobayashi A, et al. Identification of a multipotent self-renewing stromal progenitor population during mammalian kidney organogenesis. Stem Cell Reports. 2014;3(4):650–662. doi: 10.1016/j.stemcr.2014.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Levinson RS, et al. Foxd1-dependent signals control cellularity in the renal capsule, a structure required for normal renal development. Development. 2005;132(3):529–539. doi: 10.1242/dev.01604. [DOI] [PubMed] [Google Scholar]
- 66.Ardissino G, et al. Epidemiology of chronic renal failure in children: data from the ItalKid project. Pediatrics. 2003;111(4 pt 1):e382–e387. doi: 10.1542/peds.111.4.e382. [DOI] [PubMed] [Google Scholar]
- 67.Sanna-Cherchi S, et al. Renal outcome in patients with congenital anomalies of the kidney and urinary tract. Kidney Int. 2009;76(5):528–533. doi: 10.1038/ki.2009.220. [DOI] [PubMed] [Google Scholar]
- 68.Scott RP, Quaggin SE. Review series: the cell biology of renal filtration. J Cell Biol. 2015;209(2):199–210. doi: 10.1083/jcb.201410017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tryggvason K, Wartiovaara J. How does the kidney filter plasma? Physiology (Bethesda) 2005;20:96–101. doi: 10.1152/physiol.00045.2004. [DOI] [PubMed] [Google Scholar]
- 70.Harfe BD, et al. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci U S A. 2005;102(31):10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Johanson TM, et al. MicroRNA-independent roles of the RNase III enzymes Drosha and Dicer. Open Biol. 2013;3(10):130144. doi: 10.1098/rsob.130144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ho J, et al. The pro-apoptotic protein Bim is a microRNA target in kidney progenitors. J Am Soc Nephrol. 2011;22(6):1053–1063. doi: 10.1681/ASN.2010080841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nagalakshmi VK, et al. Dicer regulates the development of nephrogenic and ureteric compartments in the mammalian kidney. Kidney Int. 2011;79(3):317–330. doi: 10.1038/ki.2010.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chu JY, et al. Dicer function is required in the metanephric mesenchyme for early kidney development. Am J Physiol Renal Physiol. 2014;306(7):F764–F772. doi: 10.1152/ajprenal.00426.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kruber P, et al. Loss or oncogenic mutation of DROSHA impairs kidney development and function, but is not sufficient for Wilms tumor formation. Int J Cancer. 2019;144(6):1391–1400. doi: 10.1002/ijc.31952. [DOI] [PubMed] [Google Scholar]
- 76.Cerqueira DM, et al. Bim gene dosage is critical in modulating nephron progenitor survival in the absence of microRNAs during kidney development. FASEB J. 2017;31(8):3540–3554. doi: 10.1096/fj.201700010R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pastorelli LM, et al. Genetic analyses reveal a requirement for Dicer1 in the mouse urogenital tract. Mamm Genome. 2009;20(3):140–151. doi: 10.1007/s00335-008-9169-y. [DOI] [PubMed] [Google Scholar]
- 78.Bartram MP, et al. Conditional loss of kidney microRNAs results in congenital anomalies of the kidney and urinary tract (CAKUT) J Mol Med (Berl) 2013;91(6):739–748. doi: 10.1007/s00109-013-1000-x. [DOI] [PubMed] [Google Scholar]
- 79.Bartram MP, et al. Loss of Dgcr8-mediated microRNA expression in the kidney results in hydronephrosis and renal malformation. BMC Nephrol. 2015;16:55. doi: 10.1186/s12882-015-0053-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nakagawa N, et al. Dicer1 activity in the stromal compartment regulates nephron differentiation and vascular patterning during mammalian kidney organogenesis. Kidney Int. 2015;87(6):1125–1140. doi: 10.1038/ki.2014.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Phua YL, et al. Renal stromal miRNAs are required for normal nephrogenesis and glomerular mesangial survival. Physiol Rep. 2015;3(10):e12537. doi: 10.14814/phy2.12537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bantounas I, et al. The miR-199a/214 cluster controls nephrogenesis and vascularization in a human embryonic stem cell model. Stem Cell Reports. 2021;16(1):134–148. doi: 10.1016/j.stemcr.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hemker SL, et al. Deletion of hypoxia-responsive microRNA-210 results in a sex-specific decrease in nephron number. FASEB J. 2020;34(4):5782–5799. doi: 10.1096/fj.201902767R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Marrone AK, et al. MicroRNA-17~92 is required for nephrogenesis and renal function. J Am Soc Nephrol. 2014;25(7):1440–1452. doi: 10.1681/ASN.2013040390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Phua YL, et al. Loss of miR-17~92 results in dysregulation of Cftr in nephron progenitors. Am J Physiol Renal Physiol. 2019;316(5):F993–F1005. doi: 10.1152/ajprenal.00450.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Phua YL, et al. Small non-coding RNA expression in mouse nephrogenic mesenchymal progenitors. Sci Data. 2018;5:180218. doi: 10.1038/sdata.2018.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gregory PA, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10(5):593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 88.Hajarnis S, et al. Suppression of microRNA activity in kidney collecting ducts induces partial loss of epithelial phenotype and renal fibrosis. J Am Soc Nephrol. 2018;29(2):518–531. doi: 10.1681/ASN.2017030334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sequeira-Lopez ML, et al. The microRNA-processing enzyme dicer maintains juxtaglomerular cells. J Am Soc Nephrol. 2010;21(3):460–467. doi: 10.1681/ASN.2009090964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Harvey SJ, et al. Podocyte-specific deletion of dicer alters cytoskeletal dynamics and causes glomerular disease. J Am Soc Nephrol. 2008;19(11):2150–2158. doi: 10.1681/ASN.2008020233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shi S, et al. Podocyte-selective deletion of dicer induces proteinuria and glomerulosclerosis. J Am Soc Nephrol. 2008;19(11):2159–2169. doi: 10.1681/ASN.2008030312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ho J, et al. Podocyte-specific loss of functional microRNAs leads to rapid glomerular and tubular injury. J Am Soc Nephrol. 2008;19(11):2069–2075. doi: 10.1681/ASN.2008020162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhdanova O, et al. The inducible deletion of Drosha and microRNAs in mature podocytes results in a collapsing glomerulopathy. Kidney Int. 2011;80(7):719–730. doi: 10.1038/ki.2011.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wei Q, et al. Targeted deletion of Dicer from proximal tubules protects against renal ischemia-reperfusion injury. J Am Soc Nephrol. 2010;21(5):756–761. doi: 10.1681/ASN.2009070718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chen HH, et al. Urinary miR-16 transactivated by C/EBPβ reduces kidney function after ischemia/reperfusion-induced injury. Sci Rep. 2016;6:27945. doi: 10.1038/srep27945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Song N, et al. miR-21 protects against ischemia/reperfusion-induced acute kidney injury by preventing epithelial cell apoptosis and inhibiting dendritic cell maturation. Front Physiol. 2018;9:790. doi: 10.3389/fphys.2018.00790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wilflingseder J, et al. miR-182-5p inhibition ameliorates ischemic acute kidney injury. Am J Pathol. 2017;187(1):70–79. doi: 10.1016/j.ajpath.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 98.Patel V, et al. MicroRNAs regulate renal tubule maturation through modulation of Pkd1. J Am Soc Nephrol. 2012;23(12):1941–1948. doi: 10.1681/ASN.2012030321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Medrano S, et al. Two microRNAs, miR-330 and miR-125b-5p, mark the juxtaglomerular cell and balance its smooth muscle phenotype. Am J Physiol Renal Physiol. 2012;302(1):F29–F37. doi: 10.1152/ajprenal.00460.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.[No authors listed] Birth defects monitoring program (BDMP)/commission on professional and hospital activities (CPHA) surveillance data, 1988-1991. Teratology. 1993;48(6):658–675. doi: 10.1002/tera.1420480608. [DOI] [PubMed] [Google Scholar]
- 101.Nicolaou N, et al. Genetic, environmental, and epigenetic factors involved in CAKUT. Nat Rev Nephrol. 2015;11(12):720–731. doi: 10.1038/nrneph.2015.140. [DOI] [PubMed] [Google Scholar]
- 102.Renkema KY, et al. Novel perspectives for investigating congenital anomalies of the kidney and urinary tract (CAKUT) Nephrol Dial Transplant. 2011;26(12):3843–3851. doi: 10.1093/ndt/gfr655. [DOI] [PubMed] [Google Scholar]
- 103.Nakanishi K, Yoshikawa N. Genetic disorders of human congenital anomalies of the kidney and urinary tract (CAKUT) Pediatr Int. 2003;45(5):610–616. doi: 10.1046/j.1442-200X.2003.01779.x. [DOI] [PubMed] [Google Scholar]
- 104.Weber S, et al. Prevalence of mutations in renal developmental genes in children with renal hypodysplasia: results of the ESCAPE study. J Am Soc Nephrol. 2006;17(10):2864–2870. doi: 10.1681/ASN.2006030277. [DOI] [PubMed] [Google Scholar]
- 105.Hwang DY, et al. Mutations in 12 known dominant disease-causing genes clarify many congenital anomalies of the kidney and urinary tract. Kidney Int. 2014;85(6):1429–1433. doi: 10.1038/ki.2013.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kohl S, et al. Targeted sequencing of 96 renal developmental microRNAs in 1213 individuals from 980 families with congenital anomalies of the kidney and urinary tract. Nephrol Dial Transplant. 2016;31(8):1280–1283. doi: 10.1093/ndt/gfv447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.De Pontual L, et al. Germline deletion of the miR-17 approximately 92 cluster causes skeletal and growth defects in humans. Nat Genet. 2011;43(10):1026–1030. doi: 10.1038/ng.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Celli J, et al. Feingold syndrome: clinical review and genetic mapping. Am J Med Genet A. 2003;122A(4):294–300. doi: 10.1002/ajmg.a.20471. [DOI] [PubMed] [Google Scholar]
- 109.Marcelis CL, et al. Genotype-phenotype correlations in MYCN-related Feingold syndrome. Hum Mutat. 2008;29(9):1125–1132. doi: 10.1002/humu.20750. [DOI] [PubMed] [Google Scholar]
- 110.Jovanovic I, et al. Transcriptome-wide based identification of miRs in congenital anomalies of the kidney and urinary tract (CAKUT) in children: the significant upregulation of tissue miR-144 expression. J Transl Med. 2016;14(1):193. doi: 10.1186/s12967-016-0955-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rivera MN, Haber DA. Wilms’ tumour: connecting tumorigenesis and organ development in the kidney. Nat Rev Cancer. 2005;5(9):699–712. doi: 10.1038/nrc1696. [DOI] [PubMed] [Google Scholar]
- 112.Scott RH, et al. Syndromes and constitutional chromosomal abnormalities associated with Wilms tumour. J Med Genet. 2006;43(9):705–715. doi: 10.1136/jmg.2006.041723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Treger TD, et al. The genetic changes of Wilms tumour. Nat Rev Nephrol. 2019;15(4):240–251. doi: 10.1038/s41581-019-0112-0. [DOI] [PubMed] [Google Scholar]
- 114.Hohenstein P, et al. The yin and yang of kidney development and Wilms’ tumors. Genes Dev. 2015;29(5):467–482. doi: 10.1101/gad.256396.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Beckwith JB, et al. Nephrogenic rests, nephroblastomatosis, and the pathogenesis of Wilms’ tumor. Pediatr Pathol. 1990;10(1–2):1–36. doi: 10.3109/15513819009067094. [DOI] [PubMed] [Google Scholar]
- 116.Breslow NE, et al. Familial Wilms’ tumor: a descriptive study. Med Pediatr Oncol. 1996;27(5):398–403. doi: 10.1002/(SICI)1096-911X(199611)27:5<398::AID-MPO2>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 117.Pelletier J, et al. WT1 mutations contribute to abnormal genital system development and hereditary Wilms’ tumour. Nature. 1991;353(6343):431–434. doi: 10.1038/353431a0. [DOI] [PubMed] [Google Scholar]
- 118.Pritchard-Jones K, et al. The candidate Wilms’ tumour gene is involved in genitourinary development. Nature. 1990;346(6280):194–197. doi: 10.1038/346194a0. [DOI] [PubMed] [Google Scholar]
- 119.Park S, et al. Inactivation of WT1 in nephrogenic rests, genetic precursors to Wilms’ tumour. Nat Genet. 1993;5(4):363–367. doi: 10.1038/ng1293-363. [DOI] [PubMed] [Google Scholar]
- 120.Koesters R, et al. Mutational activation of the beta-catenin proto-oncogene is a common event in the development of Wilms’ tumors. Cancer Res. 1999;59(16):3880–3882. [PubMed] [Google Scholar]
- 121.Walz AL, et al. Recurrent DGCR8, DROSHA, and SIX homeodomain mutations in favorable histology Wilms tumors. Cancer Cell. 2015;27(2):286–297. doi: 10.1016/j.ccell.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wegert J, et al. Mutations in the SIX1/2 pathway and the DROSHA/DGCR8 miRNA microprocessor complex underlie high-risk blastemal type Wilms tumors. Cancer Cell. 2015;27(2):298–311. doi: 10.1016/j.ccell.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 123.Maschietto M, et al. TP53 mutational status is a potential marker for risk stratification in Wilms tumour with diffuse anaplasia. PLoS One. 2014;9(10):e109924. doi: 10.1371/journal.pone.0109924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ooms AH, et al. Significance of TP53 mutation in Wilms tumors with diffuse anaplasia: a report from the children’s oncology group. Clin Cancer Res. 2016;22(22):5582–5591. doi: 10.1158/1078-0432.CCR-16-0985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chen KS, et al. Mutations in microRNA processing genes in Wilms tumors derepress the IGF2 regulator PLAG1. Genes Dev. 2018;32(15–16):996–1007. doi: 10.1101/gad.313783.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gadd S, et al. A children’s oncology group and TARGET initiative exploring the genetic landscape of Wilms tumor. Nat Genet. 2017;49(10):1487–1494. doi: 10.1038/ng.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mahamdallie SS, et al. Mutations in the transcriptional repressor REST predispose to Wilms tumor. Nat Genet. 2015;47(12):1471–1474. doi: 10.1038/ng.3440. [DOI] [PubMed] [Google Scholar]
- 128.Torrezan GT, et al. Recurrent somatic mutation in DROSHA induces microRNA profile changes in Wilms tumour. Nat Commun. 2014;5:4039. doi: 10.1038/ncomms5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rakheja D, et al. Somatic mutations in DROSHA and DICER1 impair microRNA biogenesis through distinct mechanisms in Wilms tumours. Nat Commun. 2014;2:4802. doi: 10.1038/ncomms5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wu MK, et al. Biallelic DICER1 mutations occur in Wilms tumours. J Pathol. 2013;230(2):154–164. doi: 10.1002/path.4196. [DOI] [PubMed] [Google Scholar]
- 131.Robertson JC, et al. DICER1 syndrome: DICER1 mutations in rare cancers. Cancers (Basel) 2018;10(5):143. doi: 10.3390/cancers10050143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Foulkes WD, et al. DICER1: mutations, microRNAs and mechanisms. Nat Rev Cancer. 2014;14(10):662–672. doi: 10.1038/nrc3802. [DOI] [PubMed] [Google Scholar]
- 133.Yermalovich AV, et al. Lin28 and let-7 regulate the timing of cessation of murine nephrogenesis. Nat Commun. 2019;10(1):168. doi: 10.1038/s41467-018-08127-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Urbach A, et al. Lin28 sustains early renal progenitors and induces Wilms tumor. Genes Dev. 2014;28(9):971–982. doi: 10.1101/gad.237149.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chang HM, et al. A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28-let-7 pathway. Nature. 2013;497(7448):244–248. doi: 10.1038/nature12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Astuti D, et al. Germline mutations in DIS3L2 cause the Perlman syndrome of overgrowth and Wilms tumor susceptibility. Nat Genet. 2012;44(3):277–284. doi: 10.1038/ng.1071. [DOI] [PubMed] [Google Scholar]
- 137.Morris MR, et al. Perlman syndrome: overgrowth, Wilms tumor predisposition and DIS3L2. Am J Med Genet C Semin Med Genet. 2013;163C(2):106–113. doi: 10.1002/ajmg.c.31358. [DOI] [PubMed] [Google Scholar]
- 138.Alessandri JL, et al. Perlman syndrome: report, prenatal findings and review. Am J Med Genet A. 2008;146A(19):2532–2537. doi: 10.1002/ajmg.a.32391. [DOI] [PubMed] [Google Scholar]
- 139.Turchinovich A, et al. Extracellular miRNAs: the mystery of their origin and function. Trends Biochem Sci. 2012;37(11):460–465. doi: 10.1016/j.tibs.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 140.Zernecke A, et al. Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Sci Signal. 2009;2(100):ra81. doi: 10.1126/scisignal.2000610. [DOI] [PubMed] [Google Scholar]
- 141.Valadi H, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 142.Arroyo JD, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108(12):5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wang K, et al. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 2010;38(20):7248–7259. doi: 10.1093/nar/gkq601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Vickers KC, et al. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13(4):423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mitchell PS, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105(30):10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Chen X, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18(10):997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 147.Duttagupta R, et al. Impact of cellular miRNAs on circulating miRNA biomarker signatures. PLoS One. 2011;6(6):e20769. doi: 10.1371/journal.pone.0020769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sourvinou IS, et al. Quantification of circulating miRNAs in plasma: effect of preanalytical and analytical parameters on their isolation and stability. J Mol Diagn. 2013;15(6):827–834. doi: 10.1016/j.jmoldx.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 149.Ma W, et al. Effect of long-term storage in TRIzol on microarray-based gene expression profiling. Cancer Epidemiol Biomarkers Prev. 2010;19(10):2445–2452. doi: 10.1158/1055-9965.EPI-10-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Chugh P, Dittmer DP. Potential pitfalls in microRNA profiling. Wiley Interdiscip Rev RNA. 2012;3(5):601–616. doi: 10.1002/wrna.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Tam S, et al. Robust global microRNA expression profiling using next-generation sequencing technologies. Lab Invest. 2014;94(3):350–358. doi: 10.1038/labinvest.2013.157. [DOI] [PubMed] [Google Scholar]
- 152.Pradervand S, et al. Concordance among digital gene expression, microarrays, and qPCR when measuring differential expression of microRNAs. Biotechniques. 2010;48(3):219–222. doi: 10.2144/000113367. [DOI] [PubMed] [Google Scholar]
- 153.Coenen-Stass AML, et al. Evaluation of methodologies for microRNA biomarker detection by next generation sequencing. RNA Biol. 2018;15(8):1133–1145. doi: 10.1080/15476286.2018.1514236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Neilsen CT, et al. IsomiRs--the overlooked repertoire in the dynamic microRNAome. Trends Genet. 2012;28(11):544–549. doi: 10.1016/j.tig.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 155.Lee LW, et al. Complexity of the microRNA repertoire revealed by next-generation sequencing. RNA. 2010;16(11):2170–2180. doi: 10.1261/rna.2225110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Tan GC, et al. 5’ isomiR variation is of functional and evolutionary importance. Nucleic Acids Res. 2014;42(14):9424–9435. doi: 10.1093/nar/gku656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Chen T, et al. Increased urinary exosomal microRNAs in children with idiopathic nephrotic syndrome. EBioMedicine. 2019;39:552–561. doi: 10.1016/j.ebiom.2018.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Murray MJ, et al. Solid tumors of childhood display specific serum microRNA profiles. Cancer Epidemiol Biomarkers Prev. 2015;24(2):350–360. doi: 10.1158/1055-9965.EPI-14-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ludwig N, et al. Circulating serum miRNAs as potential biomarkers for nephroblastoma. Pediatr Blood Cancer. 2015;62(8):1360–1367. doi: 10.1002/pbc.25481. [DOI] [PubMed] [Google Scholar]
- 160.Luo Y, et al. Increased serum and urinary microRNAs in children with idiopathic nephrotic syndrome. Clin Chem. 2013;59(4):658–666. doi: 10.1373/clinchem.2012.195297. [DOI] [PubMed] [Google Scholar]
- 161.Mori MA, et al. Extracellular miRNAs: from biomarkers to mediators of physiology and disease. Cell Metab. 2019;30(4):656–673. doi: 10.1016/j.cmet.2019.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Mestdagh P, et al. Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study. Nat Methods. 2014;11(8):809–815. doi: 10.1038/nmeth.3014. [DOI] [PubMed] [Google Scholar]
- 163.Hong LZ, et al. Systematic evaluation of multiple qPCR platforms, NanoString and miRNA-Seq for microRNA biomarker discovery in human biofluids. Sci Rep. 2021;11(1):4435. doi: 10.1038/s41598-021-83365-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Linsen SE, et al. Limitations and possibilities of small RNA digital gene expression profiling. Nat Methods. 2009;6(7):474–476. doi: 10.1038/nmeth0709-474. [DOI] [PubMed] [Google Scholar]
- 165.Nik Mohamed Kamal NNSB, Shahidan WNS. Non-exosomal and exosomal circulatory microRNAs: which are more valid as biomarkers? Front Pharmacol. 2019;10:1500. doi: 10.3389/fphar.2019.01500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Bonneau E, et al. How close are miRNAs from clinical practice? A perspective on the diagnostic and therapeutic market. EJIFCC. 2019;30(2):114–127. [PMC free article] [PubMed] [Google Scholar]
- 167.Chakraborty C, et al. Therapeutic advances of miRNAs: a preclinical and clinical update. J Adv Res. 2021;28:127–138. doi: 10.1016/j.jare.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16(3):203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 169.Bader AG. miR-34 — a microRNA replacement therapy is headed to the clinic. Front Genet. 2012;3:120. doi: 10.3389/fgene.2012.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Dasgupta I, Chatterjee A. Recent advances in miRNA delivery systems. Methods Protoc. 2021;4(1):10. doi: 10.3390/mps4010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Lv W, et al. Therapeutic potential of microRNAs for the treatment of renal fibrosis and CKD. Physiol Genomics. 2018;50(1):20–34. doi: 10.1152/physiolgenomics.00039.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Trionfini P, et al. MicroRNAs in kidney physiology and disease. Nat Rev Nephrol. 2015;11(1):23–33. doi: 10.1038/nrneph.2014.202. [DOI] [PubMed] [Google Scholar]
- 173.Bao JW, et al. MiRNA-203a-5p alleviates the malignant progression of Wilms’ tumor via targeting JAG1. Eur Rev Med Pharmacol Sci. 2020;24(10):5329–5335. doi: 10.26355/eurrev_202005_21315. [DOI] [PubMed] [Google Scholar]
- 174.Liang XL, et al. MiR-200a with CDC7 as a direct target declines cell viability and promotes cell apoptosis in Wilm’s tumor via Wnt/β-catenin signaling pathway. Mol Cell Biochem. 2021;476(6):2409–2420. doi: 10.1007/s11010-021-04090-9. [DOI] [PubMed] [Google Scholar]
- 175.Zhao GZ, et al. MiR-200c inhibits proliferation and promotes apoptosis of Wilms tumor cells by regulating akt signaling pathway. Eur Rev Med Pharmacol Sci. 2020;24(12):6623–6631. doi: 10.26355/eurrev_202006_21648. [DOI] [PubMed] [Google Scholar]
- 176.Luo B, et al. MiR-22-3p regulates the proliferation and invasion of Wilms’ tumor cells by targeting AKT3. Eur Rev Med Pharmacol Sci. 2020;24(11):5996–6004. doi: 10.26355/eurrev_202006_21493. [DOI] [PubMed] [Google Scholar]
- 177.Liu K, et al. miR-483-5p targets MKNK1 to suppress Wilms’ tumor cell proliferation and apoptosis in vitro and in vivo. Med Sci Monit. 2019;25:1459–1468. doi: 10.12659/MSM.913005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Li JL, Luo P. MiR-140-5p and miR-92a-3p suppress the cell proliferation, migration and invasion and promoted apoptosis in Wilms’ tumor by targeting FRS2. Eur Rev Med Pharmacol Sci. 2020;24(1):97–108. doi: 10.26355/eurrev_202001_19899. [DOI] [PubMed] [Google Scholar]
- 179.Liu Z, et al. miR-140-5p could suppress tumor proliferation and progression by targeting TGFBRI/SMAD2/3 and IGF-1R/AKT signaling pathways in Wilms’ tumor. BMC Cancer. 2019;19(1):405. doi: 10.1186/s12885-019-5609-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Su H, et al. MicroRNA-539 inhibits the progression of Wilms’ tumor through downregulation of JAG1 and Notch1/3. Cancer Biomark. 2019;24(1):125–133. doi: 10.3233/CBM-181972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Wang HF, et al. MicroRNA-613 attenuates the proliferation, migration and invasion of Wilms’ tumor via targeting FRS2. Eur Rev Med Pharmacol Sci. 2017;21(15):3360–3369. [PubMed] [Google Scholar]
- 182.Zhu S, et al. MicroRNA‑92a‑3p inhibits the cell proliferation, migration and invasion of Wilms tumor by targeting NOTCH1. Oncol Rep. 2018;40(2):571–578. doi: 10.3892/or.2018.6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Yu Q, et al. miR-378c suppresses Wilms tumor development via negatively regulating CAMKK2. Acta Biochim Biophys Sin (Shanghai) 2021;53(6):739–747. doi: 10.1093/abbs/gmab047. [DOI] [PubMed] [Google Scholar]
- 184.Imam JS, et al. MicroRNA-185 suppresses tumor growth and progression by targeting the Six1 oncogene in human cancers. Oncogene. 2010;29(35):4971–4979. doi: 10.1038/onc.2010.233. [DOI] [PubMed] [Google Scholar]
- 185.Drake KM, et al. Loss of heterozygosity at 2q37 in sporadic Wilms’ tumor: putative role for miR-562. Clin Cancer Res. 2009;15(19):5985–5992. doi: 10.1158/1078-0432.CCR-09-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Kort EJ, et al. The E2F3-Oncomir-1 axis is activated in Wilms’ tumor. Cancer Res. 2008;68(11):4034–4038. doi: 10.1158/0008-5472.CAN-08-0592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Che G, et al. MicroRNA-483-3p promotes proliferation, migration, and invasion and induces chemoresistance of Wilms’ tumor cells. Pediatr Dev Pathol. 2020;23(2):144–151. doi: 10.1177/1093526619873491. [DOI] [PubMed] [Google Scholar]
- 188.Veronese A, et al. Oncogenic role of miR-483-3p at the IGF2/483 locus. Cancer Res. 2010;70(8):3140–3149. doi: 10.1158/0008-5472.CAN-09-4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Zhang C, et al. MicroRNA-572 targets CDH1 to promote metastasis of Wilms’ tumor. Eur Rev Med Pharmacol Sci. 2019;23(9):3709–3717. doi: 10.26355/eurrev_201905_17794. [DOI] [PubMed] [Google Scholar]
- 190.Cui M, et al. Clinicopathological parameters and prognostic relevance of miR-21 and PTEN expression in Wilms’ tumor. J Pediatr Surg. 2017;52(8):1348–1354. doi: 10.1016/j.jpedsurg.2016.12.005. [DOI] [PubMed] [Google Scholar]
- 191.Jiang X, Li H. MiR-1180-5p regulates apoptosis of Wilms’ tumor by targeting p73. Onco Targets Ther. 2018;11:823–831. doi: 10.2147/OTT.S148684. [DOI] [PMC free article] [PubMed] [Google Scholar]