SUMMARY
Reconstructing the tempo at which biodiversity arose is a fundamental goal of evolutionary biologists, yet the relative merits of evolutionary-rate estimates are debated based on whether they are derived from the fossil record or time-calibrated phylogenies (timetrees) of living species. Extinct lineages unsampled in timetrees are known to ‘pull’ speciation rates downward, but the temporal scale at which this bias matters is unclear. To investigate this problem, we compare mammalian diversification-rate signatures in a credible set of molecular timetrees (N=5,911 species, ~70% from DNA) to those in fossil genus durations (N=5,320). We use fossil extinction rates to correct or ‘push’ the timetree-based (pulled) speciation-rate estimates, finding a surge of speciation during the Paleocene (ca. 66-56 million years ago, Ma) between the Cretaceous-Paleogene (K-Pg) boundary and the Paleocene-Eocene Thermal Maximum (PETM). However, about two-thirds of the K-Pg-to-PETM originating taxa did not leave modern descendants, indicating that this rate signature is undetectable from extant lineages alone. For groups without substantial fossil records, thankfully all is not lost. Pushed and pulled speciation rates converge starting ca. 10 Ma, and are equal at the present day when recent evolutionary processes can be estimated without bias using species-specific ‘tip’ rates of speciation. Clade-wide moments of tip rates also enable enriched inference, as the skewness of tip rates is shown to approximate a clade’s extent of past diversification-rate shifts. Molecular timetrees need fossil-correction to address deep-time questions, but they are sufficient for shallower time questions where extinctions are fewer.
Keywords: macroevolution, lineage diversification, phylogenetics, mass extinction, tip rates
INTRODUCTION
The last ca. 180-million-years of mammalian evolution has resulted in ~6,400 living species1,2 and many thousands of extinct taxa.3,4 As one of the few large clades with ample paleo- and neontological evidence, mammals are a useful vehicle for investigating whether general patterns of evolutionary-rate variation are detectable through time.5-7 However, even with abundant fossil8 and genomic9 resources, meaningfully integrating these data and their associated biases is challenging.10-12 Deep-time questions, in which rate information among major ‘backbone-level’ lineages is leveraged to test the biological impact of ancient earth-history events (e.g.,13,14), are at the limits of our capacity for inference from neontological data alone. That is because lineages are ‘erased’ by extinctions with greater frequency as one moves back in time.15 This temporal phenomenon causes rate underestimates from time-calibrated molecular phylogenies of living species alone (extant timetrees16-18). Further challenging inferences, fossils at older time intervals tend to be less abundant3 and more spatially biased than more recent paleontological horizons.19 Thus, reconciling the respective biases of fossils and molecules at different timescales may help clarify when their respective insights are expected to be complementary versus confounding.
The limits of inferring diversification rates from extant timetrees were recently formalized by Louca and Pennell,20 who found that, in the absence of additional fossil or demographic information, any given timetree may be equally explained by an infinite number of diversification scenarios. That is, speciation (λ) and extinction (μ) rates are non-identifiable from timetrees alone; however, one class of rate parameters, referred to as ‘pulled’ rates, does emerge as being identifiable: ‘pulled speciation rate’ (λp), ‘pulled diversification rate’ (rp), and ‘sampling fraction × speciation rate at the instantaneous present’ (ρλ020,21). The ‘pulling’ in this context represents how unsampled extinctions, especially at older time intervals, and incomplete modern sampling cause speciation-rate underestimation.20 Indeed, several aspects of diversification tempo should still be estimable from extant timetrees, and made more reliable by reference to parallel evidence sources such as fossils.12,15,22-25
Building on those insights, we suggest that the goal of empirical investigations into questions of deep-time evolutionary history, at least for mammals and other fossil-rich clades, should be framed as a two-part endeavor: (i) leverage as much fossil information as possible for the time period(s) in question, and use that to exclude diversification scenarios that might be indistinguishable by molecular data alone (see also15,26); and (ii) compare parametric estimates of species’ birth and death (e.g.,27,28) with metrics for pulled (λp) and tip (λ0) speciation rates, since the latter should be identifiable when taxon sampling is known and thus instructive about the biases of the former. Making these comparisons in mammals should demonstrate the timescale at which molecular timetree-based inferences can be trusted. Unbiased estimation of ‘tip rates’ of species-specific speciation, λ0, was previously demonstrated to require all extant taxa to be sampled or otherwise modeled (e.g., using the tip DR statistic29,30). However, in mammals, it has only recently become possible to estimate tip rates robustly, thanks to new species-level timetrees that model uncertainty in topology and node ages.2 Speciation and extinction rates through time have not yet been characterized across these ‘backbone-and-patch’ mammal trees,2 nor have they been used to evaluate deep-time questions relative to previous supertree-based inferences (e.g.,31,32) or fossil mammal occurrences.8
Herein, we apply this two-part framework to investigate a key deep-time question in the radiation of mammals: Did early mammals exhibit a burst of lineage diversification coincident with, well before, or well after the Cretaceous-Paleogene (K-Pg) boundary ca. 66-million years ago (Ma)? These hypotheses are known as the Suppression,33,34 Early Rise,13 and Delayed Rise31 models, respectively (reviewed in Grossnickle et al.35 as relates to ecological diversification, but with implications for lineage diversification). The latter model of Delayed Rise emphasizes that recovery from the Paleocene-Eocene Thermal Maximum (PETM) ca. 56 Ma spurred more divergences within crown mammals than did the K-Pg mass extinction or earlier events.31,36,37 Alternatively, the Early Rise model emphasizes that co-diversification with angiosperms in the late Cretaceous (ca. 85–75 Ma) impacted mammal radiations more than the K-Pg event.13,35,38 These Mammalia-wide hypotheses overlap to some degree with Placentalia-specific models of Short/Long Fuse and Hard/Soft Explosive,35,39,55 sparking confusion regarding the extent to which fossil-based conclusions (e.g.,34,38,40) should be used to generate timetree-testable predictions for extant lineages. The K-Pg and PETM events both involved global climatic changes, the former by an extraterrestrial bolide impact and associated volcanism,41 and the latter by a catastrophic release of carbon dioxide and subsequent 5-8 °C spike in temperatures.37 Thus, we here use these climatic perturbations for testing (i) the extent to which inferences based on fossils, timetrees, or both combined illuminate the same or different aspects of mammalian evolutionary response, and (ii) how the reliability of paleo- and neontological information sources varies from deep to shallow time.
Our specific objectives are three-fold. First, we test for branch-specific rate shifts in the mammal timetrees relative to the K-Pg and PETM events to assess whether any residual impact signature has been retained in the branching times of extant lineages. Second, we directly compare diversification rates derived from fossil genus durations versus timetrees, and reconcile their contrasting signals via the formation of a combined rate metric that ‘pushes’ the pulled rates of speciation. Finally, we evaluate how the unbiased estimators of tip speciation rate and pulled speciation rate can be applied to assess clade-wise rate variation using timetrees alone. Overall, we show that while timetree-based rate estimates are highly uncertain at deep timescales, they are increasingly reliable closer to the present. Joining the timetree- and fossil-based inferences helps us narrow the range of possible diversification scenarios for mammals and, more broadly, to demonstrate that, even for clades without fossils, tip rates carry reliable signatures of shallow-time evolutionary processes.
RESULTS
Broad tempo of diversification in the Mammalia timetree.
The timetree of extant mammals shows that while the earliest divergences of crown Marsupialia and crown Placentalia occurred prior to the K-Pg boundary, and are thus consistent with the Early Rise model, the vast majority of intraordinal diversity arose after the PETM ca. 56 Ma (Figures 1, 2A). The first 4 placental divergences unambiguously preceded the K-Pg event, as evidenced by divergence-time 95% highest posterior density (HPD) intervals that do not overlap 66 Ma (Figure 2A; filled circles). However, the next 29 placental divergences have 95% HPDs that overlap the K-Pg event, including the divergences of 12 superordinal lineages (Figure 2A; open circles), 9 crown orders (Figure 3A), and 8 intraordinal splits. In Marsupialia, the first 5 splits after the crown divergence overlap the K-Pg boundary, all at the superordinal level. Of those 34 divergences that overlap the K-Pg boundary, 18 also overlapped the PETM, making their timings statistically indistinguishable from causality by either event. The next 14 mammal divergences overlap the PETM exclusively, including crown ages of Paenungulata (Hyracoidea + Sirenia + Proboscidea), 4 orders (Lagomorpha, Artiodactyla, Monotremata, and Diprotodontia), and 9 intraordinal splits within bats and rodents (Table S1). Thus, the K-Pg and PETM events compare by having the possible coincidence of 34 vs. 32 mammal divergences, respectively, of which 16 vs. 14 are respectively exclusive to those events, providing timetree-based evidence for the Suppression and Delayed Rise models. By contrast, only 7 pre-K-Pg divergences of crown Theria support the Early Rise model using the extant timetree.
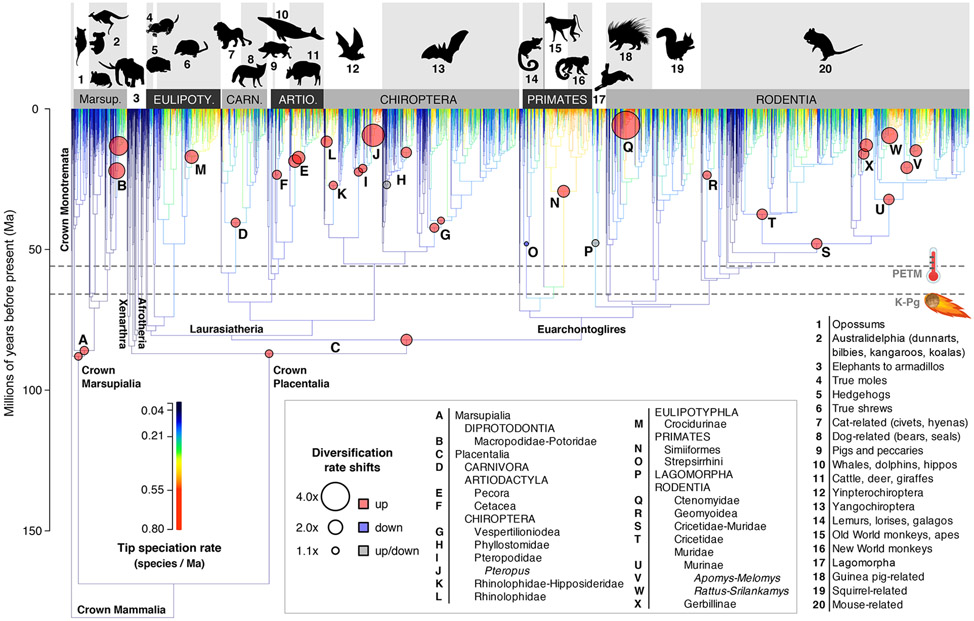
Figure 1. Extant time-calibrated molecular phylogeny for 5,911 species of mammals globally.
The maximum clade credibility topology of 10,000 node-dated timetrees is shown with branches coloured according to tip-level speciation rates (tip DR metric) and marked with 24 shifts in branch-specific net diversification rates inferred using BAMM (nodes A-X; shifts with multiple circles were inferred on either branch, not both, over a sampling of 10 trees from the credible set). Highlighted at 66-million years ago (Ma) is the extraterrestrial bolide impact that coincided with the Cretaceous-Paleogene (K-Pg) boundary, and at 56 Ma is the Paleocene-Eocene Thermal Maximum (PETM). Tip speciation rates are reconstructed to interior branches using Brownian motion for visualization purposes only. Numbered labels correspond to monophyletic groups listed in the plot periphery: Marsup., Marsupialia; Eulipoty., Eulipotyphla; Carn., Carnivora; Artio., Artiodactyla. Artwork is public domain from phylopic.org, open-source fonts, and creazilla.com (see Acknowledgments). See also Figure S1, Table S2.
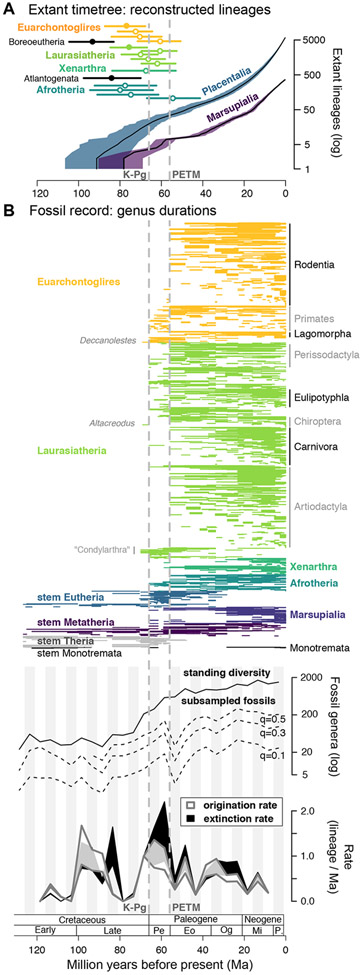
Figure 2. Diversification of mammal lineages relative to the Cretaceous-Paleogene (K-Pg) boundary and Paleocene-Eocene Thermal Maximum (PETM; dashed grey lines).
Temporal dynamics are compared as reconstructed in the fossil-calibrated extant timetree (A) versus observed in the genus-level fossil record (B). (A) Lineage-through-time plot of placentals and marsupials over 100 trees (black line is consensus tree) relative to the timing of the first 16 superordinal divergences of placentals (mean crown ages; filled circle if 95% error interval does not overlap the K-Pg boundary). (B, upper) Paleontological durations from first to last occurrence for all genera in crown Mammalia as recorded in the Paleobiology Database (N = 5,320 unique genera from 72,579 observations). Putative crown placental taxa recovered prior to the K-Pg boundary are highlighted in light gray. (B, middle) The richness of fossil generic diversity is shown through time as binned in 5-million-year, Ma, intervals from 131–1 Ma (taxa spanning boundaries go in both bins) and subsampled using shareholder quorum sizes (q) of 0.5, 0.3, and 0.1 to maximize the uniformity of coverage. (B, bottom) Rates of fossil genus origination and extinction estimated using six different rate metrics and presented as confidence intervals from the low to high estimate in each 5-Ma bin (shaded polygons). Binned richness and rate data are plotted at the midpoint of each 5-Ma bin. Note that fossil coverage was insufficient for estimating rates in the most recent bin (6-1 Ma), and only the ‘second-for-third’ metric could calculate rates for the 71-66 Ma bin. Paleocene, Pe; Eocene, Eo; Oligocene, Og; Miocene, Mi; Pliocene, P. See also Figure S2; Tables S1, S3, S4, and S5.
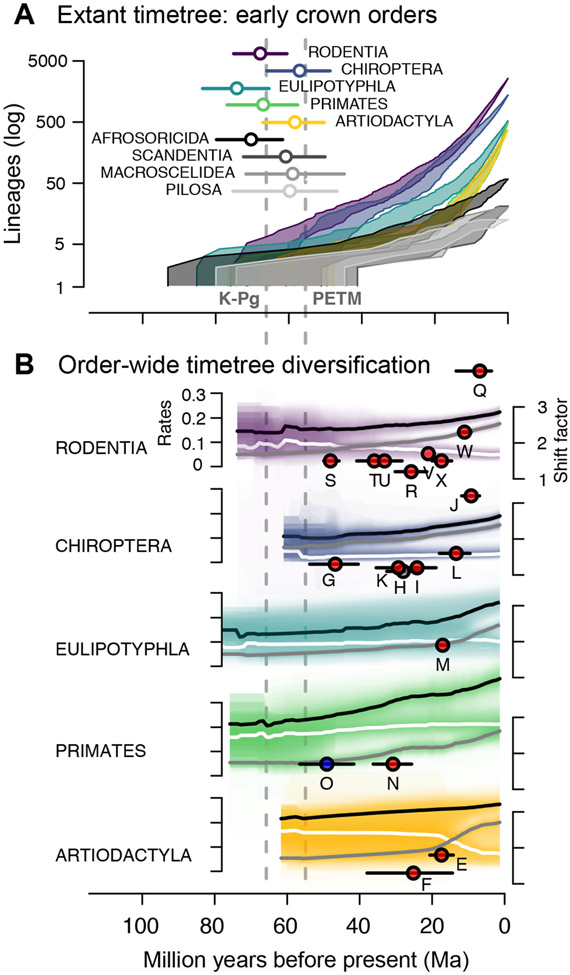
Fig. 3. Diversification of early crown orders of mammals using the extant timetree.
(A) Lineage-through-time plots and divergence times for all placental orders with crown radiations starting near the Cretaceous-Paleogene boundary (K-Pg) and Paleocene-Eocene Thermal Maximum (PETM; dashed grey lines). (B) Rates of speciation (black), extinction (white), and net diversification (grey) through time in the five most speciose early crown orders, as calculated in BAMM, as well as corresponding branch-specific shifts in net diversification (median rates from 10 trees, 95% error intervals in colours). The last 2 Ma are removed to focus on pre-recent rate dynamics. See also Figure S1, Tables S1, S2, and S5.
Branch-specific rate shifts in the extant timetree.
Across 10 timetrees, BAMM estimates a tree-wide mean speciation rate of 0.206 (95% CI: 0.188–0.223) and mean extinction rate of 0.068 (0.053–0.088, units of species/lineage/Ma). The mean number of branch-specific shifts is 36.7 (95% CI for 10 runs: 27.9–43.4; Figure S1). A total of 253 shifts are detected in one or more trees, of which 208 are up-shifts (increases in net diversification rate) and 45 are down-shifts (Figures 3B, 4A). We identify 24 rate shifts that are consistently present in at least five of the 10 trees (Figures 1, 3B, 4A; Table S2), including 9 shifts paired (occurring on adjacent branches in different trees) and 18 non-nested shifts (Figure 1; see Supplemental Data for further summaries of the BAMM runs). Of those consistent shifts, we found substantial variation in the number of shifts per ‘patch clade’ that was used to construct the backbone-and-patch mammal trees, indicating that the location of rate shifts was unrelated to patch clade delimitation (Figure S4).
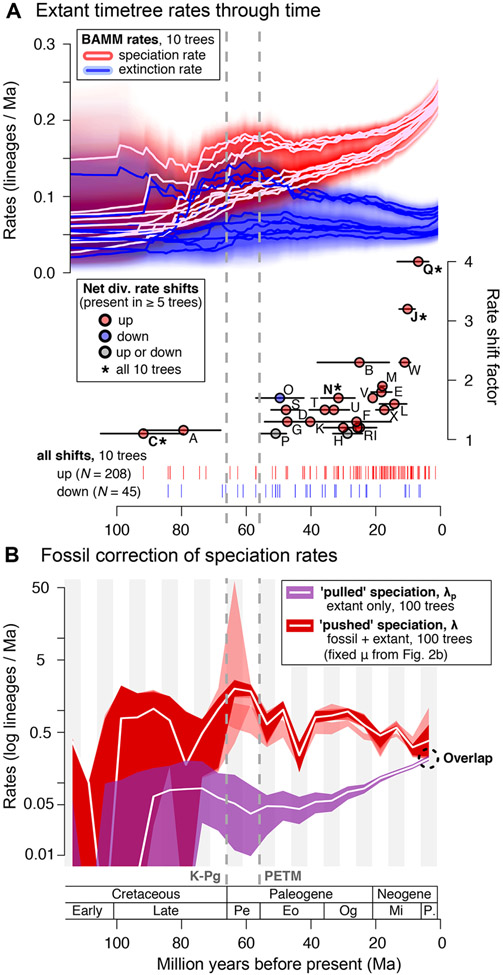
Figure 4. Rates of mammal diversification estimated considering the fossil record or not relative to the Cretaceous-Paleogene (K-Pg) boundary and Paleocene-Eocene Thermal Maximum (PETM; dashed grey lines).
(A, upper) Inferred variation in rates of speciation, extinction, and net diversification upon 10 mammal timetrees using BAMM (shown are median rates and 95% confidence intervals (CIs) from each tree drawn from the credible set; the last 2-million years, Ma, are removed to focus on pre-recent rate dynamics). (A, lower) Of the 253 possible shifts in branch-specific net diversification rates recovered, 24 were present in at least five of the ten trees (A-X; symbols match Figure 1) and 4 shifts were present in all trees (asterisks). (B) Pulled speciation rates, λp, estimated over 100 timetrees at 5-Ma bins (purple; 95% CI and median), and then corrected or ‘pushed’ as an estimate of true speciation rates, λ (shades of red; 95% CI for each of 6 methods used to push the rates, with the grouped median shown). Note the log scale of λp and λ.. The homogenous birth-death modelling of λ across each tree was performed by fixing the extinction rates, μ, according to the 6 metrics of fossil extinction rate displayed in Figure 2B and repeating each estimate of λ across 100 timetrees. See also Figures S1, S2, S3; Tables S2, S4, and S5.
In the case of the Placentalia and 8 other rate shifts, shift location is contingent on the rooting of the tree (see2) and how the concomitant background rate varies (Table S2), highlighting the relevance of considering a sample of phylogenies. Four of the 24 shifts are ever recorded as decreases, and only 1 shift is consistently a decrease across trees: the strepsirrhine primates (lemurs, lorises, and galagos, O), for which we show a 1.7-fold reduction in net diversification relative to the background rate (Figure 1, Table S2). Overall, the 20 consistent up-shifts have larger magnitudes nearer the present, with a 2.2-fold mean shift factor in the Miocene versus 1.3-fold in each the Oligocene and Eocene (Figure 4A; three-way ANOVA: df=2, F=7.772, P=0.003). No branch-specific rate shifts are consistently associated with the K-Pg or PETM events, but the Cretaceous timing of crown Marsupialia and Placentalia shifts is consistent with the Early Rise model (Figure 4A; shifts A, C).
Tempo of the genus-level fossil record.
Following a 100–80 Ma burst of origination and extinction involving lineages of extinct clades (e.g., multituberculates, cimolestans), the ca. 66-Ma K-Pg and ca. 56-Ma PETM events appear to have influenced the fossil record of crown Mammalia (Figure 2B). We find that at least 203 fossil genera originated during the Cretaceous prior to the K-Pg boundary, most of which are assigned to stem lineages of Monotremata, Theria, Marsupialia, and Placentalia. However, 3 Cretaceous lineages are highlighted for being possible members of crown Placentalia (Figure 2B): (i) Deccanolestes (Adapisoriculidae; range: 69.9–66 Ma) as either stem Afrotheria or stem Euarchontoglires;42-44 (ii) Altacreodus (possible “Creodonta”; 69.9–66 Ma) as stem Laurasiatheria;45 and (iii) some members assigned to “Condylarthra” such as Paleoungulatum (69.9–66 Ma), Protungulatum (69.9–65.1 Ma), and Baioconodon (69.9–63.1 Ma) as stem or crown Laurasiatheria.44,46-48 Nevertheless, considerable debate underpins the phylogenetic placement of these fossils (e.g.,49).
Cenozoic fossil diversification rates roughly follow the genus richness curves, with the highest increase of λ occurring in the 66–61 Ma time bin (range of 6 rate metrics: 0.74–1.36 lineages/Ma) and μ in the 61–56 Ma bin (range: 0.89–2.20 lineages/Ma), with both rates stabilizing to ~0.5 (range: 0.16–0.88) lineages/Ma from 20 Ma to the present (Figure 2B, Figure S2, Table S4). The Paleocene (66–56 Ma) surge of λ and μ apparent in the fossil record involves the origination of 495 genera between the K-Pg and PETM events, of which ~82% went extinct before the start of the Eocene ca. 56 Ma (408 genera). Of those extinctions, over 38% were within 1 Ma of the ca. 56 Ma PETM event when per genus durations are examined directly (157 genera). Only 26.9% of the Paleocene-originating taxa (113 genera) are allocated to the stem or crown of extant taxonomic orders (Eulipotyphla, Macroscelidea, Primates, Perissodactyla, and Rodentia each with 10 or more representatives), indicating that nearly three-quarters of the total K-Pg-to-PETM originations did not leave closely related modern descendants. Alternatively, if we conservatively assign all Paleocene “Condylarthra” to stem Artiodactyla or Perissodactyla (e.g., phenacodontids50), then the fraction that are not allocated to extant taxonomic orders deceases to about two-thirds.
Subsampled genus richness reaches an early peak in the 61–56 Ma bin, just prior to the PETM, and then falls by ~80% in the next bin before recovering with a 3-fold increase in the 51–46 Ma bin (Figure 2B; Table S3). Raw richness importantly misses this major drop in taxonomic diversity, indicating non-uniform sampling of these fossil strata. Overall peak subsampled richness is consistently found in the 26–21 Ma time bin (late Oligocene–early Miocene; Figure 2B) as associated with the highest evenness of any interval (Shannon’s H=6.515; Table S3). Subsampled richness then declines by ~15% in the next bin and remains roughly flat until the most recent bin (Figure 2B, Table S3). Raw genus richness remarkably parallels the stability of subsampled richness since the Oligocene-Miocene transition ~23 Ma, underscoring the uniformity Miocene-Recent fossil sampling. Fully 27% of now-extant genera of mammals have fossil records older than 1 Ma (N=351 of 1,283 genera in the timetree taxonomy), demonstrating continuity between fossil- and timetree-based investigations.
Pushing the pulled speciation rates.
The calculation of pulled speciation rates, λp, using extant timetrees recovers per-interval 95% CIs that include zero from 111–76 Ma, indicating that we are only confident of non-zero λp beginning at ca. 71 Ma when a rate of 0.083 lineages/Ma (95% CI: 0.027–0.198) is recovered (Figure 4B, Table S5). After an apparent subsequent decline in the median from 66–56 Ma, pulled speciation rates rise steadily to a modern peak of 0.216 lineages/Ma (0.201–0.237) at 6–1 Ma. By comparison, the tree-wide estimate of λ0 at the present day is 0.217 lineages/Ma (0.197–0.239), and the tree-wide median of species’ tip DR is 0.206 species/Ma (95% CI: 0.055–0.476), with higher variability due to the per-species estimation of values.
Because older values of λp are ‘pulled’ downward by unsampled lineages, they are biased relative to their true values, λ.20 Efforts to ‘push’ the estimated speciation rates back to λ yield differing results depending on whether we fix fossil extinction rates or the total extant lineages through time (Figure S3). We favor fixed extinction rates as they allow estimated uncertainty across 6 different metrics to be integrated into the pushed speciation rate analyses. Using the fixed extinction rates, we importantly find that the K-Pg-to-PETM interval includes both the lowest value of pulled rates (0.037 lineages/Ma; 95% CI: 0.009–0.092) and the highest value of pushed rates (median of 2.0 lineages/Ma; 0.608–24.487; Figure 4B). Moreover, the molecular timetree (pulled) and fossil-corrected (pushed) rates of speciation begin converging ca. 10 Ma (11–6 Ma bin) with estimates that overlap in the 6-1 Ma bin, indicating that they are statistically indistinguishable near the present (Figure 4B). Overall, the pushed rates of speciation are on average 14-fold higher than the pulled rates, ranging from a peak 50-fold difference in medians near the PETM to a 1.7-fold difference toward the present (Table S5), which is consistent with expectations that fewer unsampled extinctions result in less bias nearer to timetree tips.
Comparing pulled rates, tip-level rates, and rate shifts.
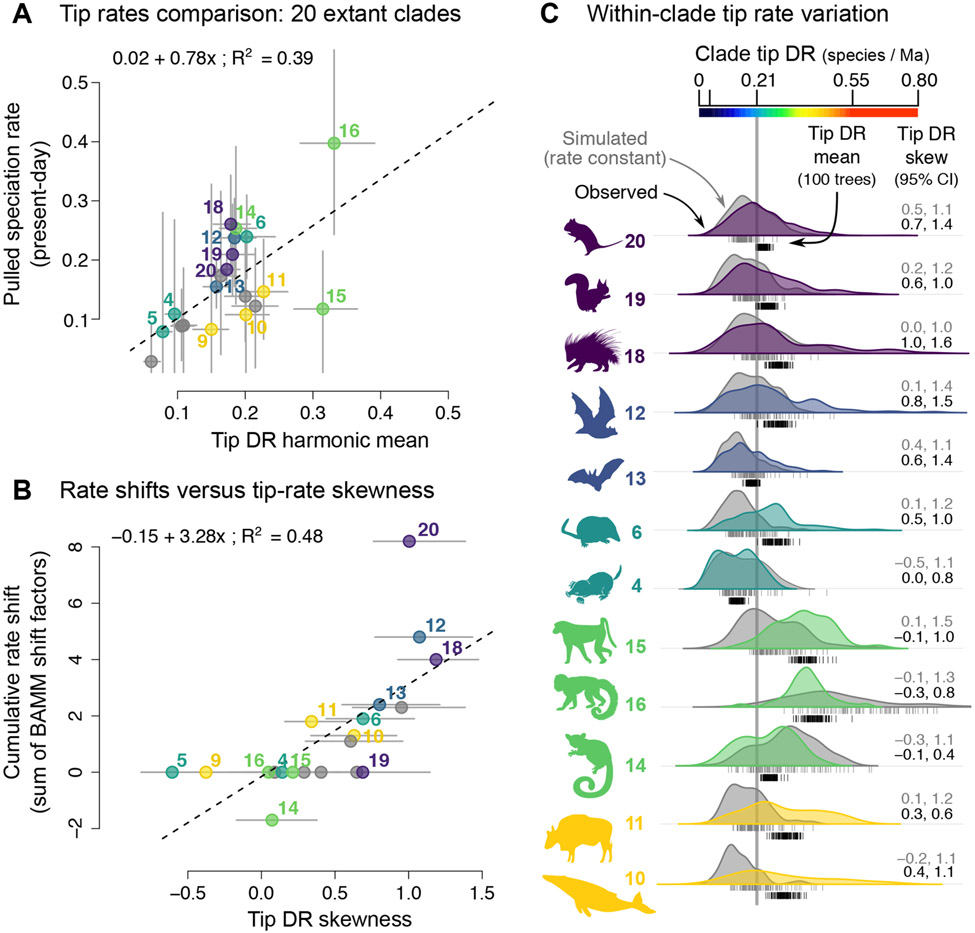
Considering rate summaries across the 20 extant clades delimited in Figure 1, we find that pulled speciation rates at the instantaneous present, λ0, show a strong positive relationship with the clade harmonic mean of species’ tip DR values (R2=0.39, P=0.003; Figure 5A). Hence, tip DR calculated under the modern sampling fraction of 1.0 approximates ρλ0, a quantity which was shown to be identifiable.20,21 Comparing clade-level skewness of tip DR to the cumulative rate shift of each clade (sum of BAMM rate-shift factors for independent shifts; Tables S2, S6) also reveals a strong positive relationship (R2=0.48, P<0.001; Figure 5B). Tip DR skewness thus offers a simple, approximate means of assessing the likelihood that a shift in net diversification rates occurred in that clade’s recent history.
Figure 5. Present-day (tip) rates of speciation across and within mammal clades.
Comparison of 20 mammal clades following the delimitation in Figure 1, showing clades numbered from the five most speciose orders. (A) Pulled speciation rates at the instantaneous present (λ0) for each clade as compared to the harmonic mean of species’ tip DR values in those clades (95% confidence intervals, CIs, in grey; linear model is given). (B) The cumulative total of BAMM rate shift factors within each clade as compared to that clade’s skewness of species’ tip DR values (95% CIs for the clade tip DR skew across 10,000 mammal trees; linear model is given). (C) Rate variation within clades of the five most speciose orders, showing species’ tip DR distributions for the empirically reconstructed phylogenies of mammals (colours) versus simulated trees of the same species richness using a rate-constant birth-death model (grey). Density plots of tip DR from one empirical and one simulated tree are shown as examples relative to the 100-tree calculations of clade tip DR mean (black and grey ticks) and skew (95% CIs). See also Table S6.
The intuition of tip DR skewness is clear upon plotting observed distributions of clade tip DR relative to expected values under rate-constant birth-death processes (Figure 5C; Table S6). The clades with the largest magnitude rate-shifts such as guinea pig-related rodents (clade 18, shift Q) and Yinpterochiroptera (clade 12, shift J) also show the highest tip DR skewness (Figures 5B, 5C). Beyond statistical moments, the tip DR distribution offers an intriguing summary of the clade-wide, among-species variation in speciation rates, ranging from broad in Old and New World monkeys (clades 15, 16) to multi-peaked in true moles and lemur-related primates (clades 4, 14).
DISCUSSION
We find diversification-rate signatures in the mammal fossil record and extant timetrees that are distinct yet complementary in the stories they tell. Fossil rates provide greater insights for deep-time questions while molecular timetree rates are increasingly consistent with the fossil information toward the present. Timetrees offer richer, species-level coverage and associated insights of modern ecological diversity that are lacking in the fossil record. When combined for mammals, the two speciation-rate sources begin to converge starting ca. 10 Ma, but overlap in the most recent time-bin (Figure 4B), consistent with species’ tip rates being unbiased estimators of recent speciation processes (Figure 1, 5A). These results are in accord with theoretical work20,22 regarding the accumulation of unrecorded extinctions in extant timetrees, which causes increasing bias for deeper-time rate dynamics as lineages are progressively erased.
The K-Pg mass extinction ca. 66 Ma is found to precede a surge of fossil genus turnover (origination + extinction; (Figure 2B)) and frame a bevy of timetree divergences that overlap in age with either the K-Pg event or ensuing PETM ca. 56 Ma (Figures 1, 2A). However, none of those timetree divergences are recovered as branch-specific shifts in net diversification rate (Figure 3B, 4A), counter to expectations for novel macroevolutionary regimes (sensu Rabosky28) to be timetree-detectable in response to major geobiotic events. Combining fossil and extant rate information, our analyses support both the Suppression and Delayed Rise models of diversification:31,33,35 diversity recoveries following the K-Pg and PETM events were milestones in the radiation of mammals (Figure 4B). Elevated fossil diversification ca. 100–80 Ma (Figure 2B) is also consistent with the early side of the Early Rise model, but more in the context of lineage turnover and regional effects of now-extinct or -depauperate clades51 rather than the rise of extant mammal radiations. Fossil-corrected rates from this Cretaceous period are highly uncertain (Figure 4B), presumably because only a few ‘early rising’ lineages survived to be represented in the timetree. Comparing fossil- and timetree-based perspectives thus establishes that these models of deep-time diversification are not exclusive from the Mammalia-wide perspective; yet if only extant timetree lineages are considered, the concentration of Eocene and Oligocene rate shifts tells a Delayed Rise-focused story.
As predicted,20,22 rates of speciation estimated across the molecular timetree are ‘pulled’ downward in magnitude for the same intervals in which extinction events have erased lineages from the timetree. The largest extinction-rate increase is recorded in fossil mammals prior to the PETM ca. 56 Ma (Figure 2B), an event apparently triggered by the anomalous spike in global temperatures.37 Concurrent with those extinctions is a substantial dip in the tree-wide pulled speciation rates, which results in rate estimates of 50-fold greater magnitude when those pulled rates are ‘pushed’ by fossil lineage-level extinction rates (Figure 4B). Hence, on their own, the backbone-and-patch timetrees of mammals do not record the ‘Paleocene pulse’ (K-Pg to PETM) in turnover that fossil durations show, a finding that helps to explain the absence of any ‘explosive’ K-Pg signature in most mammal phylogenetic studies (e.g.,13,31,52-55; but see O’Leary et al.46 and Phillips & Fruciano56).
Neontologists have long searched for the ‘smoking gun’ of K-Pg-driven radiations in molecular timetrees, but perhaps they have been looking in the wrong place. Finding such a rate signature is only expected if lineages that originated near the K-Pg event survived until the present. For mammals, the fossil record shows the selective extinction of ecological specialists at the end-Cretaceous, both in North America40,57-60 and globally,34 followed by a Paleocene rebound in taxonomic richness dominated by the now-extinct stem lineages of crown orders (e.g., archaic ungulates, leptictids, plesiadapiforms, creodonts, and mesonychids34,35,40,44,60). The stemward placement of most Paleocene placental fossils44 supports the hypothesis that K-Pg-associated diversification signatures were lost from the branching pattern of mammal molecular timetrees, as does the PETM extinction of nearly one-third of taxa that originated in the Paleocene pulse (Figure 2B). Thus, many aspects of mammalian diversification history are not knowable from extant timetrees alone. Extant timetree-based inference of the rate dynamics near ancient events like the K-Pg and PETM may rightly be viewed as impossible to ascertain,15,26 making the fossil record indispensable for understanding deep-time evolutionary questions.
Tip rates and their clade-wide moments as an identifiable path forward
Thankfully, modern timetrees are not devoid of rate information, despite some important biases15,20 and some headlines to the contrary.61 We show that molecular timetree- and fossil-corrected rate estimates (pulled vs. pushed) are congruent near the present-day (Figure 4B), a finding that reinforces the use of tip rates as an identifiable means of investigating recent evolutionary processes, at least when extant taxon sampling is known. Louca and Pennell20 established a formal proof and clear explication of how unobserved lineages (both extinct and extant) can render timetree rate inferences non-identifiable, extending previous theoretical and empirical work.15,22,26,62,63 By using completely sampled extant phylogenies, tip rates of speciation can be estimated using non-parametric approaches (e.g., the tip DR method we use here, or the coarser metric of node density29,64) that are computationally scalable across samples of timetrees and thus able to account for phylogenetic uncertainty. In contrast, parametric methods of calculating tip rates do not scale well across tree samples but can be more accurate under some diversification scenarios (see30).
Tip-rate insights are importantly not limited to present-day environments: we show that the skewness of tip DR distributions across 20 mammal clades is predictive of their historical extent of branch-specific rate shifts identified using BAMM (Figures 5B,C). That is, the clade-level distributions of species’ tip speciation rates approximate rate dynamics estimated using a formal birth-death process model. This suggests that tip-rate skewness alone can identify timetree rate shifts, analogous to how non-parametric tests of trait-dependent speciation are conducted (e.g., FiSSE, ES-sim65,66). Establishing a null distribution of the test statistic (e.g., the clade-level skewness of tip rates given trees simulated under birth-death) will allow comparisons between observed and expected tip-rate skewness. Our initial trials of this test find that empirical tip DR distributions tend to be more right skewed and with higher mean values than expected by chance (Figure 5C; Table S6). A fuller exploration of simulation parameters is needed to formalize this approach, but these initial results are promising. The most common approaches to measuring clade-level diversification, including BAMM and others, provide summarized rate information only for clades or rate regimes. Capturing skewness and other moments from clade-wise tip DR distributions offers more nuanced avenues for quantitative as well as visual inference that might unmask patterns not readily conveyed using parametric approaches.
Reconciling timetree rate shifts: when are they believable?
The above discussion raises the question: when do branch-specific rate shifts in timetrees reflect historical changes in macroevolutionary regime versus artifacts of unsampled extinction? Our finding that rate shifts are concentrated in the mammal timetrees since ca. 50 Ma, including eight shifts detected from 50–30 Ma (Figures 1, 4A), is consistent with the post-PETM, mid-to-late Eocene radiations of crown Carnivora and suborders of primates, rodents, bats, and marsupials. Collectively, these crown radiations expanded the taxonomic and eco-morphological diversity of modern mammals.35 However, as mentioned above, their stem lineages are known to have originated and gone extinct earlier in the Cenozoic (e.g., creodonts potentially allied with Carnivora, plesiadapiforms allied with Primates). If those stem lineages were sampled in the timetree, presumably each ‘shift’ to higher net diversification rates would appear less abrupt; perhaps showing no detectable change in macroevolutionary regime; or perhaps showing that less frequent extinction rather than faster speciation underpin some rate shifts (e.g.,25). Conversely, groups like horses67 that declined in diversity since the Miocene show diversification-rate stasis from an extant timetree perspective.68,69 Thus, without aid from the fossil record, we find an artifactual view of deep-time diversification dynamics.
By contrast, we expect branch-specific rate shifts to carry a greater signal of real biological processes toward the present. In mammals we see such convergence begin ca. 10 Ma, which is not only when pulled speciation rates start converging with fossil-corrected estimates (Figure 4B) but also when rate shifts show the greatest magnitude and consistency in signal across sampled timetrees (Figure 4A). Strikingly, the two largest rate increases (4.0x and 3.2x) occurred in the last ca. 10 Ma in clades with very disparate life modes: the fossorial tuco-tucos of South America (Ctenomys, Q), and the flying foxes of Indo-Pacific islands (Pteropus, J; Figures 1, 3B, 4A). Small burrowers and large flyers both show similar signatures of recent and rapid net diversification under conditions of insularity, although in subterranean and oceanic realms, respectively, suggesting that their similar propensities for geographic isolation may be driving these dynamics. The role of allopatry in mammal speciation has long been noted as the predominant pattern among closely related species (e.g.,70), and vagility differences appear to be inversely related to speciation rates in some taxa (e.g.,71). However, how traits of low and high vagility (burrowing and flying, respectively) might interact with different scales of insular habitat matrix to produce similar outcomes is less well explored (but see72). We hypothesize that apparently large rate shifts like Q and J may in reality be more common than is detectable in extant timetrees, especially if speciation and extinction rates are temporally coupled.73 Yet with the fragmentary information at hand, including many unsampled extinctions even when fossils are added, all efforts to detect macroevolutionary rate disjunctions will first need to reckon with the likelihood that enough divergence events have been sampled to recover robust rate signals.
We see two main paths of recourse regarding when to trust rate-shift analyses in extant timetrees: (i) evaluate results relative to parallel evidence (e.g., fossils, demography, eco-geographic context), especially for older rate shifts: if multiple lines of evidence support a shift, it is more likely to be real; and (ii) focus on understanding which common causes may underlie shift dynamics in younger clades for which presumably fewer unsampled extinctions are affecting rate estimates. When working with younger clades, the possibility that young clades might actually have inherently faster rates than older clades should also be tested. Although this time-dependent rates hypothesis currently lacks a mechanism, it appears robust to fossil and molecular data types,74 which show gradual, tree-wide increases in both speciation and extinction rates. Alternatively, detecting larger rate shifts nearer to the present, as we did in mammals, may suggest a different picture whereby uncharacterized eco-evolutionary accelerators (e.g., species traits, environmental factors) are causing exceptional clade-specific radiations, either in addition to or instead of time-dependence. In the latter case, apparent time-dependence may be an artifact of rare but exceptional clade-specific radiations relative to a broader taxon. Such exceptional radiations need not have adaptive drivers, though that is a possibility.75
Reconciling previous mammal studies: what do we actually know?
In light of our joint insights from the backbone-and-patch timetrees and fossil record, there is a critical need to re-evaluate what aspects of previous studies of extant mammal diversification can be deemed reliable. Two influential mammal phylogenies have been used to address similar questions of branch-specific rate shifts:32,76 the largely species-level supertree of Bininda-Emonds et al.31 and the family-level supermatrix timetree of Meredith et al.13. These trees differ from backbone-and-patch timetrees investigated here by collapsing topological and age uncertainty into a consensus phylogeny and, in the case of the supertree approach, losing branch lengths whenever information from merged subclades disagreed.2
The 24 branch-specific shifts in net diversification we recover in the backbone-and-patch timetrees (Figures 1, 3B, 4A) compare to 27 shifts detected in the supertree (15 up-shifts, 12 down-shifts32) and 9 up-shifts in the supermatrix timetree.76 To their credit, Purvis et al.32 analyzed only 1,335 bifurcating nodes in the supertree, which avoided some rate artifacts of polytomies. However, both studies returned overconfident estimates by treating the consensus phylogeny as known without error.77 If we only compare up-shifts, given the likely erasure of extant lineages as net diversification slows down,17,78,79 we find three lineages shared by our study, Purvis et al.,32 and Yu et al.76: (i) Placentalia (or one branch forward at Boreoeutheria); (ii) Simiiformes (Primates, New and Old World monkeys); and (iii) Macropodidae (Diprotodontia, kangaroos and wallabies). The commonality of those shifts argues that their evolutionary-rate signatures are robust to different models of phylogenetic reconstruction (supertree vs. supermatrix vs. Bayesian backbone-and-patch) and rate inference (SymmeTree vs. MEDUSA vs. BAMM). Of these, the Macropodidae shift is the youngest, recovered in our study at ca. 15 Ma (12.2–18.0), and thus the least likely to be biased by unsampled extinctions. Previous fossil and molecular analyses suggest that kangaroo genus-level divergences may be driving this shift.80,81
At shallower phylogenetic levels, other studies have found rate shifts similar to those we infer. For example, the Cetacea shift (F in Figure 1) was previously recovered two branches forward on the branch leading to oceanic dolphins.25,28,82 Similarly, the Simiiformes shift (N) was recovered two branches forward (Cercopithecidae83) and the Ctenomyidae shift (Q) two branches back (Octodontoidea excluding Abrocomidae84). In contrast, the six rate shifts we found in bats (G-L, Figure 1) compare to two shifts previously recovered (shifts H, J85). We suggest that high topological uncertainty in bats86 contributes to equivocal modeling of branch-specific processes. Therein lies a paradox: pinpointing rate-shift signatures is difficult in clades that are difficult to resolve, and resolving clades may be hardest in cases of recent, rapid radiation, in which signals of incomplete lineage sorting and hybridization are expected to be strongest.87 Thus, rather than relying on phylogenomic data to yield greater resolution, comparative methods also need to develop more meaningful ways of handling phylogenetic uncertainty.
Caveats to the pushing of pulled speciation rates
Approaches to using the fossil record to correct or ‘push’ the pulled rates of speciation estimated from extant timetrees are nascent, but offer a promising means of parsing plausible diversification scenarios.20 We here applied two approaches to this problem: fixing E(t), the total number of extinct and extant lineages through time, and fixing μ(t), the fossil-estimated extinction rate through time using different rate metrics. While the latter approach returned more realistic speciation-rate estimates (Figure S3), we highlight that both approaches could be substantially improved. For example, among-bin heterogeneity in fossil sampling probabilities could be incorporated into rate models,12,88 which should make the estimation of E(t) and μ(t) more robust. Similarly, using a fossil phylogenetic approach would add expected ghost lineages among genera, even if coarse taxonomic assignments are used as a proxy for cladistic data (e.g.,25,89,90).
There are known issues with the fidelity of stratigraphic and taxonomic assignments in compiled fossil data, including in the Paleobiology Database (e.g.,91), which have added noise to our analyses. Our efforts to improve public data by integrating curated snapshots by geological time interval40,44,92 and fossil taxon45,93,94 are critical, but importantly also highlight the need to incentivize ongoing public curation of paleontological resources. Despite these concerns, our view is that paleontological biases are far less systematic than those that emerge from conducting deep-time rate inferences without considering unsampled extinctions. Hence, any addition of fossil data to extant phylogenetic analyses is likely to provide greater macroevolutionary realism.
Conclusion
Overall, our results indicate that extant timetrees contain sufficient evolutionary-rate information for approximately unbiased investigation at levels of species tips to shallow clades (e.g., ca. 10-Ma stem age or younger). Extant taxon sampling must be complete or at least completely modeled while accounting for non-random (e.g., geographically biased) sampling. When interpreting results, the probability of bias from clade-, region-, or ecotype-specific extinctions must also be considered. We emphasize that fossil and living organisms record signatures of the same evolutionary processes, just from very different temporal viewpoints. Debating ‘rocks versus clocks’ as the ultimate arbiters of evolutionary history misses the point of their interdependence. Timetrees and fossils are like the bow and stern of an evolutionary ship sailing through the sea of time; as the bow cuts through the recent past and probable future of biodiversity processes, it leaves behind fossils in its wake. Traces of the past may or may not help navigate the future, but they nevertheless illuminate our evolutionary trajectory. Harnessing these complementary data sources should allow us to realize the strengths of timetrees (recent processes) alongside those of fossils (ancient processes) toward establishing a fuller understanding of evolutionary history. Future work to query the causal impact of deep-time events like the K-Pg or PETM upon diversification rates should merge fossil and living diversity into phylogenetic analyses, or else be viewed with caution. In turn, fossil-free timetrees should be prioritized for application to shallow-time questions for which species-level lineages can be fully sampled and meaningfully analyzed relative to hypothesized covariates.
STAR METHODS
Lead contact
Further information and requests for resources and data should be directed to and will be fulfilled by the lead contact, Nathan Upham (nathan.upham@asu.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
All data have been deposited on Github (https://github.com/n8upham/MamDiv-fossil-vs-timetree) and Zenodo (https://doi.org/10.5281/zenodo.5059100), and are publicly available as of the date of publication. All original code has been deposited at the same Github and is publicly available as of the date of publication. The DOI is also listed in the key resources table. Source data for mammal phylogenies analyzed in the paper are available at http://vertlife.org/data/mammals/. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-Snail | Cell Signaling Technology | Cat#3879S; RRID: AB_2255011 |
| Mouse monoclonal anti-Tubulin (clone DM1A) | Sigma-Aldrich | Cat#T9026; RRID: AB_477593 |
| Rabbit polyclonal anti-BMAL1 | This paper | N/A |
| Bacterial and virus strains | ||
| pAAV-hSyn-DIO-hM3D(Gq)-mCherry | Krashes et al., 2011 | Addgene AAV5; 44361-AAV5 |
| AAV5-EF1a-DIO-hChR2(H134R)-EYFP | Hope Center Viral Vectors Core | N/A |
| Cowpox virus Brighton Red | BEI Resources | NR-88 |
| Zika-SMGC-1, GENBANK: KX266255 | Isolated from patient (Wang et al., 2016) | N/A |
| Staphylococcus aureus | ATCC | ATCC 29213 |
| Streptococcus pyogenes: M1 serotype strain: strain SF370; M1 GAS | ATCC | ATCC 700294 |
| Biological samples | ||
| Healthy adult BA9 brain tissue | University of Maryland Brain & Tissue Bank; http://medschool.umaryland.edu/btbank/ | Cat#UMB1455 |
| Human hippocampal brain blocks | New York Brain Bank | http://nybb.hs.columbia.edu/ |
| Patient-derived xenografts (PDX) | Children's Oncology Group Cell Culture and Xenograft Repository | http://cogcell.org/ |
| Chemicals, peptides, and recombinant proteins | ||
| MK-2206 AKT inhibitor | Selleck Chemicals | S1078; CAS: 1032350-13-2 |
| SB-505124 | Sigma-Aldrich | S4696; CAS: 694433-59-5 (free base) |
| Picrotoxin | Sigma-Aldrich | P1675; CAS: 124-87-8 |
| Human TGF-β | R&D | 240-B; GenPept: P01137 |
| Activated S6K1 | Millipore | Cat#14-486 |
| GST-BMAL1 | Novus | Cat#H00000406-P01 |
| Critical commercial assays | ||
| EasyTag EXPRESS 35S Protein Labeling Kit | PerkinElmer | NEG772014MC |
| CaspaseGlo 3/7 | Promega | G8090 |
| TruSeq ChIP Sample Prep Kit | Illumina | IP-202-1012 |
| Deposited data | ||
| Raw and analyzed data | This paper | GEO: GSE63473 |
| B-RAF RBD (apo) structure | This paper | PDB: 5J17 |
| Human reference genome NCBI build 37, GRCh37 | Genome Reference Consortium | http://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/human/ |
| Nanog STILT inference | This paper; Mendeley Data | http://dx.doi.org/10.17632/wx6s4mj7s8.2 |
| Affinity-based mass spectrometry performed with 57 genes | This paper; Mendeley Data | Table S8; http://dx.doi.org/10.17632/5hvpvspw82.1 |
| Experimental models: Cell lines | ||
| Hamster: CHO cells | ATCC | CRL-11268 |
| D. melanogaster: Cell line S2: S2-DRSC | Laboratory of Norbert Perrimon | FlyBase: FBtc0000181 |
| Human: Passage 40 H9 ES cells | MSKCC stem cell core facility | N/A |
| Human: HUES 8 hESC line (NIH approval number NIHhESC-09-0021) | HSCI iPS Core | hES Cell Line: HUES-8 |
| Experimental models: Organisms/strains | ||
| C. elegans: Strain BC4011: srl-1(s2500) II; dpy-18(e364) III; unc-46(e177)rol-3(s1040) V. | Caenorhabditis Genetics Center | WB Strain: BC4011; WormBase: WBVar00241916 |
| D. melanogaster: RNAi of Sxl: y[1] sc[*] v[1]; P{TRiP.HMS00609}attP2 | Bloomington Drosophila Stock Center | BDSC:34393; FlyBase: FBtp0064874 |
| S. cerevisiae: Strain background: W303 | ATCC | ATTC: 208353 |
| Mouse: R6/2: B6CBA-Tg(HDexon1)62Gpb/3J | The Jackson Laboratory | JAX: 006494 |
| Mouse: OXTRfl/fl: B6.129(SJL)-Oxtrtm1.1Wsy/J | The Jackson Laboratory | RRID: IMSR_JAX:008471 |
| Zebrafish: Tg(Shha:GFP)t10: t10Tg | Neumann and Nuesslein-Volhard, 2000 | ZFIN: ZDB-GENO-060207-1 |
| Arabidopsis: 35S::PIF4-YFP, BZR1-CFP | Wang et al., 2012 | N/A |
| Arabidopsis: JYB1021.2: pS24(AT5G58010)::cS24:GFP(-G):NOS #1 | NASC | NASC ID: N70450 |
| Oligonucleotides | ||
| siRNA targeting sequence: PIP5K I alpha #1: ACACAGUACUCAGUUGAUA | This paper | N/A |
| Primers for XX, see Table SX | This paper | N/A |
| Primer: GFP/YFP/CFP Forward: GCACGACTTCTTCAAGTCCGCCATGCC | This paper | N/A |
| Morpholino: MO-pax2a GGTCTGCTTTGCAGTGAATATCCAT | Gene Tools | ZFIN: ZDB-MRPHLNO-061106-5 |
| ACTB (hs01060665_g1) | Life Technologies | Cat#4331182 |
| RNA sequence: hnRNPA1_ligand: UAGGGACUUAGGGUUCUCUCUAGGGACUUAGGGUUCUCUCUAGGGA | This paper | N/A |
| Recombinant DNA | ||
| pLVX-Tight-Puro (TetOn) | Clonetech | Cat#632162 |
| Plasmid: GFP-Nito | This paper | N/A |
| cDNA GH111110 | Drosophila Genomics Resource Center | DGRC:5666; FlyBase:FBcl0130415 |
| AAV2/1-hsyn-GCaMP6- WPRE | Chen et al., 2013 | N/A |
| Mouse raptor: pLKO mouse shRNA 1 raptor | Thoreen et al., 2009 | Addgene Plasmid #21339 |
| Software and algorithms | ||
| ImageJ | Schneider et al., 2012 | https://imagej.nih.gov/ij/ |
| Bowtie2 | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| Samtools | Li et al., 2009 | http://samtools.sourceforge.net/ |
| Weighted Maximal Information Component Analysis v0.9 | Rau et al., 2013 | https://github.com/ChristophRau/wMICA |
| ICS algorithm | This paper; Mendeley Data | http://dx.doi.org/10.17632/5hvpvspw82.1 |
| Other | ||
| Sequence data, analyses, and resources related to the ultra-deep sequencing of the AML31 tumor, relapse, and matched normal | This paper | http://aml31.genome.wustl.edu |
| Resource website for the AML31 publication | This paper | https://github.com/chrisamiller/aml31SuppSite |
Method details
Mammalian phylogeny.
We conducted all analyses using the species-level mammal trees of Upham et al.2. Briefly, these phylogenies include 5,911 extant or recently extinct species in credible sets of 10,000 trees. They were built using a ‘backbone-and-patch’ framework consisting of two stages of Bayesian inference, with information from age and topological uncertainty incorporated as well as the probabilistic addition of 1,813 species that lacked DNA characters using taxonomic constraints. We analyzed the credible set of trees that was node-dated using 17 fossil calibrations.
Fossil genus durations.
To assess the congruence of our extant timetree-based rate estimates with rates estimated directly from the fossil record, we analyzed genus-level fossil occurrence data from a variety of sources. Starting from the Paleobiology Database (http://paleobiodb.org) downloaded on 16 August 2018 globally for taxon Mammalia, we grouped by genus after specifying the exclusion of ichnotaxa and uncertain genera, then manually cleaned the taxonomy for consistency relative to expert resources.34,44,92-95 To avoid artifacts from inflated stratigraphic intervals, a known issue in public databases,91 we merged the expert-curated dataset of Pires et al.40 from late Cretaceous–Paleocene fossil assemblages of western North America. That interval, spanning 69.9–55 Ma, covers both the K-Pg and PETM events of interest and is thereby critical for our study. We merged 2,670 occurrences of 289 genera from Pires et al., replacing data for 193 genera from the Paleobiology Database for which genus names matched, and adding data for 96 genera that were unmatched. In total, we recovered 72,579 occurrences of 5,320 fossil genera that are allocated to crown Mammalia and younger than 131 Ma, which was our temporal cutoff point to focus analyses upon the Cretaceous-Recent (earlier Cretaceous mammal fossils are too sparsely known for the planned diversity analyses).
Quantification and statistical analyses
Branch-specific rate shifts.
We performed searches for macroevolutionary shifts using BAMM v2.5,28 a reversible-jump algorithm for sampling birth-death rate regimes. Although extinction biases have been the focus of critiques to BAMM and related rate-shift models,17,18,96 we note that these issues are not unique to BAMM (e.g., state-dependent models suffer similarly16,97), nor do they preclude the model’s utility for detecting rate disjunctions in extant timetrees, regardless of their underlying cause (unsampled extinctions or a regime shift). See also Figure S4 for validation of the BAMM algorithm relative to the location of mammal tree patch clades.
We evaluated the number and location of rate shifts on 10 trees drawn randomly from the credible set, specifying globalSamplingFraction = 1.0, reflecting that the trees are taxonomically complete. Although sampling >50 trees is generally preferred for comparative methods, computation times limited us to 10 trees for BAMM analyses, which is likely >90% accurate (see figure 3 in98), while still accounting for age and topological uncertainty. On each tree, we ran the model targeting 100 million generations, while sampling every 10,000 generations. We ran the models with settings determined using the “setBAMMpriors” function in the R package BAMMtools:99 expectedNumberOfShifts=1.0; lambdaInitPrior=6.446; lambdaShiftPrior=0.00447; and muInitPrior=6.446. We set the model to estimate speciation rates as exponentially varying through time and extinction rates as constant (i.e., similar to an independent ‘SPVAR’ model within each rate regime100). Two of the 10 analyses finished all generations on 1 node before expiration of 168-hours of runtime (the analysis is not parallelizable) of the High Performance Computing Center at Yale University; the other 8 runs completed a mean of 46.1 million generations (range: 29.2–83.1 million) in the same time. The resultant events after a 33% burn-in (mean: 3727.4; range: 1949–6667) were then subsampled to yield 1,000 evenly spaced samples for each of 10 runs with the function “getEventData” in BAMMtools. After burn-in, all BAMM runs returned stable estimates of the log likelihood (ESS mean and range across 10 trees: 585.4, 268.9 – 1200.0; log likelihood: −15743.43, −16266.4 – −15314.5) and the total number of shift events (ESS mean and range: 827.0, 328.0–1498.0). The many nearly-equiprobable shift configurations in each tree’s 95% credible set of shifts prompted us to focus on the maximum shift credibility (MSC) shift sets on a per tree basis. For the rate shifts in each MSC set, we summarized the node and clade contents implicated in the shift, and the mean net diversification rate of all branches inside the shifted clade (clade rate) versus that outside it (background rate). The ratio of clade to background rates provided the rate shift magnitude and direction, whether an increase (up shift), decrease (down shift), or a mix of both among MSC sets (labeled ‘up or down’).
Comparisons with fossil genus diversification.
To calculate fossil diversity curves and diversification rates, we first binned fossil occurrences to 5-Ma intervals from 131–1 Ma, placing any genus that spans a given boundary in both bins. We chose a 5-Ma interval as a well-suited balance between interval length and regularity (e.g., preferred over geological stages), and ended the binning at 1 Ma rather than zero to avoid inflation from late Pleistocene fossils. This strategy also allowed us to examine bin-level diversity dynamics directly surrounding both the K-Pg event (71–66 Ma, 66–61 Ma) and PETM event (61–56 Ma, 56–61 Ma). Binning resulted in 90,548 bin-level occurrences, to which we applied shareholder quorum subsampling (SQS 101) to ensure that uniform coverage was met for different levels of subsampling across time bins. We used quorum sizes (q) of 0.5, 0.3, and 0.1, which correspond to those proportions of frequency distribution coverage measured by Good’s u on the subsampled data 101, and performed 1,000 trials including singletons (‘sqs’ function available at: https://bio.mq.edu.au/~jalroy/SQS-3-3.R). We estimated corresponding origination (λ) and extinction (μ) rates per time bin in the R package divDyn,102 applying six widely used metrics: per-capita,103,104 three-timer and corrected three-timer,105 gap-filler,101 and second-for-third and transformed second-for-third.106 By comparing all six metrics, we obtained rough confidence limits for the estimation of fossil λ and μ, which we then propagated to downstream analyses. Fossil rates were not estimable for the most recent time bin (6–1 Ma), since all metrics incorporate forward-boundary crossing.
Speciation rates.
We calculated two types of speciation-rate metrics using the molecular timetree: (i) pulled speciation rates across all mammals and separately for each of 20 major clades; and (ii) tip speciation rates for each species at the present as summarized for the same 20 clades. The selected clades are monophyletic across all trees selected from the credible set of timetrees, cover nearly the full species diversity of mammals, and divide that diversity more equitably than do orders (e.g., rodents are 3 clades, bats are 2 clades). Pulled speciation rates, λp, were estimated in the R package castor107 using the function ‘fit_hbd_psr_on_grid’ specifying the same 5-Ma bins interval and 10 bootstrap replicates for each of 100 mammal trees sampled randomly from the credible set. We separately calculated λp at the instantaneous present, λ0, for each of the 20 clades using the same function in castor, also on 100 trees. Species-specific ‘tip’ speciation rates were estimated using the tip DR metric,29 which is equivalent to the inverse of the equal splits measure.29,108 This metric has been called ‘DR’ and ‘tip-level diversification rate’ (tip DR) because it approximates the expected pure-birth diversification rates for the instantaneous present,2,109 at least when trees contain 10 or more species.29,110 However, because tip DR is a biased estimator of birth-death net diversification when relative extinction is high (>0.830), it is best viewed as a tip-level speciation-rate metric despite the name ‘tip DR’, which we retain to reflect its common usage. We calculated tip DR across all 10,000 mammal trees using a large-tree optimized R function (https://github.com/n8upham/MamDiv-fossil-vs-timetree/blob/main/source_functions/tipDR_functions_correct.R). To compare these speciation-rate metrics on empirical timetrees, we plotted clade-level summaries of λ0 (median) and tip DR (harmonic mean, skewness) with 95% confidence intervals (CI) using R.
We also simulated tip DR distributions expected under a homogenous birth-death process for comparison to those observed empirically. To do so, we first estimated clade-level λ and μ rates across 100 empirical subtrees for each of the 20 clades (‘birthdeath’ function in ape111), then used each set of empirical rates to simulate 1 tree of the same empirical species richness (‘pbtree’ function in the phytools112). This resulted in 20 sets of 100 simulated trees on which we then calculated tip DR values to generate expected tip DR distributions.
Fossil-correction of pulled speciation rates.
Following the logic of Louca and Pennell,20 we used the mammal fossil record for each of the 5-Ma time bins to correct or ‘push’ the pulled speciation rate, λp, to the true speciation rate, λ. Even though fossil genera not species were examined, using lineage-level rate information derived from fossil genus durations is an established means of inferring origination and extinction rates (e.g.,113,114). We used two approaches: (i) fixing the extinction rate, μ, and then modeling the homogenous birth-death (HBD) process on the timetree to estimate λ; and (ii) fixing the missing fraction of extant lineages through time, E(τ), to estimate λ. In the first approach, we used the castor function ‘fit_hbd_model_on_grid’ to successively set the extinction rate per time bin equal to each of the six metrics of fossil μ. Only the second-for-third metric could calculate extinction for the 71-66 Ma bin, so we used that value (1.0 lineages/Ma) as fixed in all trials. We truncated the most recent 1 Ma from each of 100 mammal trees to align with the 6-1 Ma time bin, and then used each tree to estimate the HBD model. We fixed no other parameters, but set initial values for (i) the present-day sampling fraction, ρ0, as 0.8 to correspond to taxonomically complete extant trees truncated at 1 Ma with an assumed speciation rate of ~0.2 species/lineage/Ma; and (ii) speciation rates per bin as the estimated fossil origination rates. Those initial values were required for the HBD model to successfully converge for all 100 trees using 10 trials.
In the second approach, we took the supplemental equation 8 from Louca and Pennell,20
| (1) |
where E(τ) is the fraction of lineages extant at age τ (time before the present) that are missing from the timetree, either due to extinction or not having been sampled, and then re-arranged it to solve for speciation rate:
| (2) |
We determined E(τ) by adding the total extant lineages, as derived from the lineages through time plot for each of 100 trees (‘fitted LTT’ element in the output from ‘fit_hbd_psr_on_grid’), to the total missing lineages, as derived from the q=0.3 subsampling of fossil genus durations for each of the time bins (chosen as indicative of the general pattern of subsampled fossil richness). Those values were used to solve for pushed λ at each time bin, and then compared to the pushed λ values obtained using the fixed fossil extinction rates.
Supplementary Material
Table S1. Summary of early diverging nodes in the backbone-and-patch timetree of mammals relative to the Cretaceous-Paleogene (K-Pg) boundary and the Paleocene-Eocene Thermal Maximum (PETM), Related to Figures 2A, 3A. The 16 superordinal placental divergences that occurred before or overlap with the K-Pg event are displayed in Figure 2A with their 95% highest posterior density (HPD) age interval. Also given is the node number in the maximum clade credibility (MCC) tree of the credible set of 10,000 node-dated completed timetrees, and the posterior probability for that node in the MCC tree.
Table S2. Summary of diversification rate shift recovered using BAMM on 10 mammal trees, Related to Figures 1, 3B, 4A. For all maximum shift credibility (MSC) shifts present in at least 5 of the 10 trees, the average net diversification rates are summarized across all branches in the rate-shift (clade rate) and all branches outside that clade in Mammalia (background rate). Their ratio gave the rate shift factor. The 24 shifts are given letters A-X, while related shifts have the same letter (even when in < 5 trees). The mean and 95% HPD divergences are given for each rate-shift location. Rate-shifts in tipward clades that are independent from each other (i.e., non-nested) are coded with "1" and used to sum up the per-clade rate-shift factors in Figure 5B.
Table S3. Summary output from the "sqs()" function, Related to Figure 2B. Shown are results for three different levels of quorum subsampling of the fossil record within 5-million-year time bins: quorum (q) of 0.1, 0.3, and 0.5.
Table S4. The output from the 'divDyn' R function for calculating rates of origination and extinction upon the binned fossil genus durations, Related to Figure 2B, 4B. All column values are defined in the glossary below the table.
Table S5. The accumulation of lineages through time (LTT) and rates through time (RTT), Related to Figures 4B, S2, S3. We used 5-million-year time bins to summarize values across 100 timetrees, including the median values and 95% confidence intervals for the extant timetree LTT and pulled speciation rates (lambda). We then performed two types of fossil correction on the pulled rates to 'push' them to estimated true values of lambda, one that fixes extant + extinct lineages through time (Fixed E(t)), and one that fixes extinction rates through time (Fixed Mu). From the Fixed Mu method, we used 6 different metrics of extinction rates to fix the homogeneous birth-death model when estimating lambda-- these were 'extPC', 'ext3t', 'extC3t', 'extGF', 'E2f3', and 'ext2f3' (defined in the Table S4 glossary)
Table S6. Comparison of 20 mammal clades with summaries across 100 observed timetrees, Related to Figure 5. Shown are values for the clade harmonic mean of tip-level speciation rates (tip DR), pulled speciation rates at the present day (λ0), the cumulative magnitude of rate shift factors estimated with BAMM, and the skewness of tip DR values within each clade. Rate-constant birth-death simulations of trees the same age and species richness (100 trees) were performed to provide null expectations for the tip DR skewness values, which were compared using two-sided Mann-Whitney U tests. The observed and simulated clades were only compared across the 16 groups that were monophyletic in all analyses and contained >40 species.
Acknowledgments:
We thank I. Quintero, M. Landis, D. Schluter, A. Mooers, A. Pyron, G. Thomas, D. Greenberg, S. Upham, and E. Florsheim for conceptual discussions that improved this study; B. Patterson, K. Rowe, J. Brown, T. Colston, T. Peterson, D. Field, T. Stewart, and J. Davies for comments on earlier drafts; D. Grossnickle and two anonymous reviewers for insightful critiques; and M. Koo, A. Ranipeta, and J. Hart for database help. Artwork from phylopic.org as CC-BY4 or open source fonts as follows: Didelphis and Sminthopsis, Sarah Werning; Phascolarctos, Gavin Prideaux; Macropus and Balaenoptera, Web Dog [AussieIcons, Freeware]; Stegodon, Zimices; Erinaceus, Claus Rebler; Solenodon, T Michael Keesey after Monika Betley; Sorex, Becky Barnes; Condylura, WindWalker64 [WWFurryFriends, Shareware]; Leo and Sus, Iconian Fonts [Zoologic, Freeware]; Vulpes and Rattus, Rebecca Groom; Bos and Pteropus, Cristopher Silva; Vespertilio, Yan Wong; Papio, Owen Jones; Lagothrix, (uncredited); Galago, Joseph Wolf 1863 vectorization by Dinah Challen; Lepus, Jan A Venter Herbert H T Prins David A Balfour Rob Slotow vectorized by T Michael Keesey; Erethizon, Fonts of Afrika [Afrika Wildlife B Mammals2, Freeware]; and Spermophilus, Alan Carr [Animals, Freeware].
Funding:
The NSF VertLife Terrestrial grant to W.J. and J.E. (DEB 1441737 and 1441634) and NSF grants DBI-1262600 to W.J. and DEB-1754393 to J.E. supported this work. Arizona State University President’s Special Initiative Fund provided additional support to N.S.U.
Footnotes
Declaration of Interests: The authors declare no competing interests.
REFERENCES
- 1.Burgin CJ, Colella JP, Kahn PL, and Upham NS (2018). How many species of mammals are there? J. Mammal 99, 1–14. [Google Scholar]
- 2.Upham NS, Esselstyn JA, and Jetz W (2019). Inferring the mammal tree: species-level sets of phylogenies for questions in ecology, evolution, and conservation. PLOS Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davies TW, Bell MA, Goswami A, and Halliday TJD (2017). Completeness of the eutherian mammal fossil record and implications for reconstructing mammal evolution through the Cretaceous/Paleogene mass extinction. Paleobiology 43, 521–536. [Google Scholar]
- 4.Bennett CV, Upchurch P, Goin FJ, and Goswami A (2018). Deep time diversity of metatherian mammals: implications for evolutionary history and fossil-record quality. Paleobiology 44, 171–198. [Google Scholar]
- 5.Osborn HF (1902). The Law of Adaptive Radiation. Am. Nat 36, 353–363. [Google Scholar]
- 6.Simpson GG (1944). Tempo and mode in evolution (Columbia Univ. Press). [Google Scholar]
- 7.Vrba ES (1992). Mammals as a Key to Evolutionary Theory. J. Mammal 73, 1–28. [Google Scholar]
- 8.Alroy J, Uhen MD, Mannion PD, Jaramillo C, Carrano MT, and van den Hoek Ostende LW (2018). Taxonomic occurrences of Mammalia recorded in Fossilworks, the Evolution of Terrestrial Ecosystems database, and the Paleobiology Database. Fossilworks. http://fossilworks.org. [Google Scholar]
- 9.Scornavacca C, Belkhir K, Lopez J, Dernat R, Delsuc F, Douzery EJP, and Ranwez V (2019). OrthoMaM v10: Scaling-Up Orthologous Coding Sequence and Exon Alignments with More than One Hundred Mammalian Genomes. Mol. Biol. Evol 36, 861–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hunt G, and Slater G (2016). Integrating Paleontological and Phylogenetic Approaches to Macroevolution. Annu. Rev. Ecol. Evol. Syst 47, 189–213. [Google Scholar]
- 11.Hopkins MJ, Bapst DW, Simpson C, and Warnock RCM (2018). The inseparability of sampling and time and its influence on attempts to unify the molecular and fossil records. Paleobiology 44, 561–574. [Google Scholar]
- 12.Silvestro D, Warnock RCM, Gavryushkina A, and Stadler T (2018). Closing the gap between palaeontological and neontological speciation and extinction rate estimates. Nat. Commun 9, 5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meredith RW, Janečka JE, Gatesy J, Ryder OA, Fisher CA, Teeling EC, Goodbla A, Eizirik E, Simão TLL, Stadler T, et al. (2011). Impacts of the Cretaceous Terrestrial Revolution and KPg Extinction on Mammal Diversification. Science 334, 521–524. [DOI] [PubMed] [Google Scholar]
- 14.Oliveros CH, Field DJ, Ksepka DT, Barker FK, Aleixo A, Andersen MJ, Alström P, Benz BW, Braun EL, Braun MJ, et al. (2019). Earth history and the passerine superradiation. Proc. Natl. Acad. Sci, 201813206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marshall CR (2017). Five palaeobiological laws needed to understand the evolution of the living biota. Nat. Ecol. Evol 1, 1–6. [DOI] [PubMed] [Google Scholar]
- 16.Maddison WP, Midford PE, Otto SP, and Oakley T (2007). Estimating a Binary Character’s Effect on Speciation and Extinction. Syst. Biol 56, 701–710. [DOI] [PubMed] [Google Scholar]
- 17.Moore BR, Höhna S, May MR, Rannala B, and Huelsenbeck JP (2016). Critically evaluating the theory and performance of Bayesian analysis of macroevolutionary mixtures. Proc. Natl. Acad. Sci 113, 9569–9574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rabosky DL, Mitchell JS, and Chang J (2017). Is BAMM flawed? Theoretical and practical concerns in the analysis of multi-rate diversification models. Syst. Biol [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benson RBJ, and Mannion PD (2012). Multi-variate models are essential for understanding vertebrate diversification in deep time. Biol. Lett 8, 127–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Louca S, and Pennell MW (2020). Extant timetrees are consistent with a myriad of diversification histories. Nature, 1–4. [DOI] [PubMed] [Google Scholar]
- 21.Louca S, Shih PM, Pennell MW, Fischer WW, Parfrey LW, and Doebeli M (2018). Bacterial diversification through geological time. Nat. Ecol. Evol, 1. [DOI] [PubMed] [Google Scholar]
- 22.Lambert A, and Stadler T (2013). Birth–death models and coalescent point processes: The shape and probability of reconstructed phylogenies. Theor. Popul. Biol 90, 113–128. [DOI] [PubMed] [Google Scholar]
- 23.Heath TA, Huelsenbeck JP, and Stadler T (2014). The fossilized birth–death process for coherent calibration of divergence-time estimates. Proc. Natl. Acad. Sci 111, E2957–E2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stadler T, Gavryushkina A, Warnock RCM, Drummond AJ, and Heath TA (2018). The fossilized birth-death model for the analysis of stratigraphic range data under different speciation modes. J. Theor. Biol 447, 41–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lloyd GT, and Slater GJ (2021). A Total-Group Phylogenetic Metatree for Cetacea and the Importance of Fossil Data in Diversification Analyses. Syst. Biol [DOI] [PubMed] [Google Scholar]
- 26.Quental TB, and Marshall CR (2010). Diversity dynamics: molecular phylogenies need the fossil record. Trends Ecol. Evol 25, 434–441. [DOI] [PubMed] [Google Scholar]
- 27.Morlon H, Parsons TL, and Plotkin JB (2011). Reconciling molecular phylogenies with the fossil record. Proc. Natl. Acad. Sci 108, 16327–16332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rabosky DL (2014). Automatic detection of key innovations, rate shifts, and diversity-dependence on phylogenetic trees. PLoS ONE 9, e89543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jetz W, Thomas GH, Joy JB, Hartmann K, and Mooers AO (2012). The global diversity of birds in space and time. Nature 491, 444–448. [DOI] [PubMed] [Google Scholar]
- 30.Title PO, and Rabosky DL (2019). Tip rates, phylogenies and diversification: What are we estimating, and how good are the estimates? Methods Ecol. Evol 10, 821–834. [Google Scholar]
- 31.Bininda-Emonds ORP, Cardillo M, Jones KE, MacPhee RDE, Beck RMD, Grenyer R, Price SA, Vos RA, Gittleman JL, and Purvis A (2007). The delayed rise of present-day mammals. Nature 446, 507–512. [DOI] [PubMed] [Google Scholar]
- 32.Purvis A, Fritz SA, Rodríguez J, Harvey PH, and Grenyer R (2011). The shape of mammalian phylogeny: patterns, processes and scales. Philos. Trans. R. Soc. Lond. B Biol. Sci 366, 2462–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Osborn HF (1910). The Age of Mammals in Europe, Asia and North America (Macmillan). [Google Scholar]
- 34.Grossnickle DM, and Newham E (2016). Therian mammals experience an ecomorphological radiation during the Late Cretaceous and selective extinction at the K–Pg boundary. Proc R Soc B 283, 20160256. [Google Scholar]
- 35.Grossnickle DM, Smith SM, and Wilson GP (2019). Untangling the Multiple Ecological Radiations of Early Mammals. Trends Ecol. Evol 34, 936–949. [DOI] [PubMed] [Google Scholar]
- 36.Gingerich PD (2006). Environment and evolution through the Paleocene–Eocene thermal maximum. Trends Ecol. Evol 21, 246–253. [DOI] [PubMed] [Google Scholar]
- 37.Bowen GJ, Maibauer BJ, Kraus MJ, Röhl U, Westerhold T, Steimke A, Gingerich PD, Wing SL, and Clyde WC (2015). Two massive, rapid releases of carbon during the onset of the Palaeocene–Eocene thermal maximum. Nat. Geosci 8, 44–47. [Google Scholar]
- 38.Wilson GP, Evans AR, Corfe IJ, Smits PD, Fortelius M, and Jernvall J (2012). Adaptive radiation of multituberculate mammals before the extinction of dinosaurs. Nature 483, 457–460. [DOI] [PubMed] [Google Scholar]
- 39.Archibald JD, and Deutschman DH (2001). Quantitative Analysis of the Timing of the Origin and Diversification of Extant Placental Orders. J. Mamm. Evol 8, 107–124. [Google Scholar]
- 40.Pires MM, Rankin BD, Silvestro D, and Quental TB (2018). Diversification dynamics of mammalian clades during the K–Pg mass extinction. Biol. Lett 14, 20180458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hull PM, Bornemann A, Penman DE, Henehan MJ, Norris RD, Wilson PA, Blum P, Alegret L, Batenburg SJ, Bown PR, et al. (2020). On impact and volcanism across the Cretaceous-Paleogene boundary. Science 367, 266–272. [DOI] [PubMed] [Google Scholar]
- 42.Seiffert ER (2010). The Oldest and Youngest Records of Afrosoricid Placentals from the Fayum Depression of Northern Egypt. Acta Palaeontol. Pol 55, 599–616. [Google Scholar]
- 43.Goswami A, Prasad GVR, Upchurch P, Boyer DM, Seiffert ER, Verma O, Gheerbrant E, and Flynn JJ (2011). A radiation of arboreal basal eutherian mammals beginning in the Late Cretaceous of India. Proc. Natl. Acad. Sci 108, 16333–16338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Halliday TJD, Upchurch P, and Goswami A (2017). Resolving the relationships of Paleocene placental mammals. Biol. Rev. 92, 521–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fox RC (2015). A revision of the Late Cretaceous–Paleocene eutherian mammal Cimolestes Marsh, 1889. Can. J. Earth Sci 52, 1137–1149. [Google Scholar]
- 46.O’Leary MA, Bloch JI, Flynn JJ, Gaudin TJ, Giallombardo A, Giannini NP, Goldberg SL, Kraatz BP, Luo Z-X, Meng J, et al. (2013). The Placental Mammal Ancestor and the Post–K-Pg Radiation of Placentals. Science 339, 662–667. [DOI] [PubMed] [Google Scholar]
- 47.Kelly TS (2014). Preliminary report on the mammals from Lane’s Little Jaw Site quarry: A latest Cretaceous (earliest Puercan?) local fauna, Hell Creek Formation, southeastern Montana. Paludicola 10, 42. [Google Scholar]
- 48.Archibald JD, Zhang Y, Harper T, and Cifelli RL (2011). Protungulatum, Confirmed Cretaceous Occurrence of an Otherwise Paleocene Eutherian (Placental?) Mammal. J. Mamm. Evol 18, 153–161. [Google Scholar]
- 49.Manz CL, Chester SGB, Bloch JI, Silcox MT, and Sargis EJ (2015). New partial skeletons of Palaeocene Nyctitheriidae and evaluation of proposed euarchontan affinities. Biol. Lett 11, 20140911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cooper LN, Seiffert ER, Clementz M, Madar SI, Bajpai S, Hussain ST, and Thewissen JGM (2014). Anthracobunids from the Middle Eocene of India and Pakistan Are Stem Perissodactyls. PLOS ONE 9, e109232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Benson RBJ, Mannion PD, Butler RJ, Upchurch P, Goswami A, and Evans SE (2013). Cretaceous tetrapod fossil record sampling and faunal turnover: Implications for biogeography and the rise of modern clades. Palaeogeogr. Palaeoclimatol. Palaeoecol 372, 88–107. [Google Scholar]
- 52.Stadler T (2011). Mammalian phylogeny reveals recent diversification rate shifts. Proc. Natl. Acad. Sci 108, 6187–6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.dos Reis M, Inoue J, Hasegawa M, Asher RJ, Donoghue PCJ, and Yang Z (2012). Phylogenomic datasets provide both precision and accuracy in estimating the timescale of placental mammal phylogeny. Proc. R. Soc. Lond. B Biol. Sci 279, 3491–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu L, Zhang J, Rheindt FE, Lei F, Qu Y, Wang Y, Zhang Y, Sullivan C, Nie W, Wang J, et al. (2017). Genomic evidence reveals a radiation of placental mammals uninterrupted by the KPg boundary. Proc. Natl. Acad. Sci 114, E7282–E7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Springer MS, Foley NM, Brady PL, Gatesy J, and Murphy WJ (2019). Evolutionary Models for the Diversification of Placental Mammals Across the KPg Boundary. Front. Genet 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Phillips MJ, and Fruciano C (2018). The soft explosive model of placental mammal evolution. BMC Evol. Biol 18, 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wilson GP (2013). Mammals across the K/Pg boundary in northeastern Montana, U.S.A.: dental morphology and body-size patterns reveal extinction selectivity and immigrant-fueled ecospace filling. Paleobiology 39, 429–469. [Google Scholar]
- 58.Longrich NR, Scriberas J, and Wills MA (2016). Severe extinction and rapid recovery of mammals across the Cretaceous–Palaeogene boundary, and the effects of rarity on patterns of extinction and recovery. J. Evol. Biol 29, 1495–1512. [DOI] [PubMed] [Google Scholar]
- 59.Lyson TR, Miller IM, Bercovici AD, Weissenburger K, Fuentes AJ, Clyde WC, Hagadorn JW, Butrim MJ, Johnson KR, Fleming RF, et al. (2019). Exceptional continental record of biotic recovery after the Cretaceous–Paleogene mass extinction. Science 366, 977–983. [DOI] [PubMed] [Google Scholar]
- 60.Alroy J (1999). The Fossil Record of North American Mammals: Evidence for a Paleocene Evolutionary Radiation. Syst. Biol 48, 107–118. [DOI] [PubMed] [Google Scholar]
- 61.Pagel M (2020). Evolutionary trees can’t reveal speciation and extinction rates. Nature 580, 461–462. [DOI] [PubMed] [Google Scholar]
- 62.Nee S, Holmes EC, May RM, and Harvey PH (1994). Extinction rates can be estimated from molecular phylogenies. Philos. Trans. R. Soc. Lond. B-Biol. Sci 344, 77–82. [DOI] [PubMed] [Google Scholar]
- 63.Quental TB, and Marshall CR (2009). Extinction During Evolutionary Radiations: Reconciling the Fossil Record with Molecular Phylogenies. Evolution 63, 3158–3167. [DOI] [PubMed] [Google Scholar]
- 64.Freckleton RP, Phillimore AB, and Pagel M (2008). Relating Traits to Diversification: A Simple Test. Am. Nat 172, 102–115. [DOI] [PubMed] [Google Scholar]
- 65.Rabosky DL, and Goldberg EE (2017). FiSSE: A simple nonparametric test for the effects of a binary character on lineage diversification rates. Evolution 71, 1432–1442. [DOI] [PubMed] [Google Scholar]
- 66.Harvey MG, and Rabosky DL (2018). Continuous traits and speciation rates: Alternatives to state-dependent diversification models. Methods Ecol. Evol 9, 984–993. [Google Scholar]
- 67.Cantalapiedra JL, Prado JL, Fernández MH, and Alberdi MT (2017). Decoupled ecomorphological evolution and diversification in Neogene-Quaternary horses. Science 355, 627–630. [DOI] [PubMed] [Google Scholar]
- 68.Burin G, Alencar LRV, Chang J, Alfaro ME, and Quental TB (2019). How Well Can We Estimate Diversity Dynamics for Clades in Diversity Decline? Syst. Biol 68, 47–62. [DOI] [PubMed] [Google Scholar]
- 69.Kodandaramaiah U, and Murali G (2018). What affects power to estimate speciation rate shifts? PeerJ 6, e5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Baker RJ, and Bradley RD (2006). Speciation in mammals and the genetic species concept. J. Mammal 87, 643–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Claramunt S, Derryberry EP, Remsen JV, and Brumfield RT (2012). High dispersal ability inhibits speciation in a continental radiation of passerine birds. Proc. R. Soc. Lond. B Biol. Sci 279, 1567–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kisel Y, and Barraclough TG (2010). Speciation Has a Spatial Scale That Depends on Levels of Gene Flow. Am. Nat 175, 316–334. [DOI] [PubMed] [Google Scholar]
- 73.Stanley MS (1998). Macroevolution: pattern and process (Johns Hopkins University Press). [Google Scholar]
- 74.Diaz LFH, Harmon LJ, Sugawara MTC, Miller ET, and Pennell MW (2019). Macroevolutionary diversification rates show time dependency. Proc. Natl. Acad. Sci, 201818058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Simões M, Breitkreuz L, Alvarado M, Baca S, Cooper JC, Heins L, Herzog K, and Lieberman BS (2016). The Evolving Theory of Evolutionary Radiations. Trends Ecol. Evol 31, 27–34. [DOI] [PubMed] [Google Scholar]
- 76.Yu W, Xu J, Wu Y, and Yang G (2012). A Comparative Study of Mammalian Diversification Pattern. Int. J. Biol. Sci 8, 486–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huelsenbeck JP, Rannala B, and Masly JP (2000). Accommodating Phylogenetic Uncertainty in Evolutionary Studies. Science 288, 2349–2350. [DOI] [PubMed] [Google Scholar]
- 78.Moore BR, Chan KMA, and Donoghue MJ (2004). Detecting Diversification Rate Variation in Supertrees. In Phylogenetic Supertrees Computational Biology., Bininda-Emonds ORP, ed. (Springer Netherlands; ), pp. 487–533. [Google Scholar]
- 79.Mitchell JS, Etienne RS, and Rabosky DL (2019). Inferring Diversification Rate Variation From Phylogenies With Fossils. Syst. Biol 68, 1–18. [DOI] [PubMed] [Google Scholar]
- 80.Couzens AMC, and Prideaux GJ (2018). Rapid Pliocene adaptive radiation of modern kangaroos. Science 362, 72–75. [DOI] [PubMed] [Google Scholar]
- 81.Brennan IG (2019). Incorporating uncertainty is essential to macroecological inferences: Grass, grit, and the evolution of kangaroos. bioRxiv, 772558. [Google Scholar]
- 82.Steeman ME, Hebsgaard MB, Fordyce RE, Ho SYW, Rabosky DL, Nielsen R, Rahbek C, Glenner H, Sørensen MV, and Willerslev E (2009). Radiation of Extant Cetaceans Driven by Restructuring of the Oceans. Syst. Biol 58, 573–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Arbour JH, and Santana SE (2017). A major shift in diversification rate helps explain macroevolutionary patterns in primate species diversity. Evolution 71, 1600–1613. [DOI] [PubMed] [Google Scholar]
- 84.Upham NS (2014). Ecological diversification and biogeography in the Neogene: Evolution of a major lineage of American and Caribbean rodents (Caviomorpha, Octodontoidea). Unpubl. Ph.D. thesis. [Google Scholar]
- 85.Shi JJ, and Rabosky DL (2015). Speciation dynamics during the global radiation of extant bats. Evolution 69, 1528–1545. [DOI] [PubMed] [Google Scholar]
- 86.Amador LI, Arévalo RLM, Almeida FC, Catalano SA, and Giannini NP (2018). Bat Systematics in the Light of Unconstrained Analyses of a Comprehensive Molecular Supermatrix. J. Mamm. Evol 25, 37–70. [Google Scholar]
- 87.Scornavacca C, and Galtier N (2017). Incomplete Lineage Sorting in Mammalian Phylogenomics. Syst. Biol 66, 112–120. [DOI] [PubMed] [Google Scholar]
- 88.Wagner PJ (2019). On the probabilities of branch durations and stratigraphic gaps in phylogenies of fossil taxa when rates of diversification and sampling vary over time. Paleobiology 45, 30–55. [Google Scholar]
- 89.Smits PD (2015). Expected time-invariant effects of biological traits on mammal species duration. Proc. Natl. Acad. Sci 112, 13015–13020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Soul LC, and Friedman M (2015). Taxonomy and Phylogeny Can Yield Comparable Results in Comparative Paleontological Analyses. Syst. Biol 64, 608–620. [DOI] [PubMed] [Google Scholar]
- 91.Prothero D (2015). Garbage in, garbage out: the effects of immature taxonomy on database compilations of North American fossil mammals. N. M. Mus. Nat. Hist. Sci. Bull 68, 257–264. [Google Scholar]
- 92.Rankin B (2015). Paleobiogeography of Latest Cretaceous and Early Paleocene Mammals from North America. [Google Scholar]
- 93.Eldridge MDB, Beck RMD, Croft DA, Travouillon KJ, and Fox BJ (2019). An emerging consensus in the evolution, phylogeny, and systematics of marsupials and their fossil relatives (Metatheria). J. Mammal 100, 802–837. [Google Scholar]
- 94.Huttenlocker AK, Grossnickle DM, Kirkland JI, Schultz JA, and Luo Z-X (2018). Late-surviving stem mammal links the lowermost Cretaceous of North America and Gondwana. Nature 558, 108. [DOI] [PubMed] [Google Scholar]
- 95.Kielan-Jaworowska Z, Cifelli RL, and Luo Z-X (2005). Mammals from the Age of Dinosaurs: Origins, Evolution, and Structure (Columbia University Press). [Google Scholar]
- 96.May MR, and Moore BR (2016). How Well Can We Detect Lineage-Specific Diversification-Rate Shifts? A Simulation Study of Sequential AIC Methods. Syst. Biol 65, 1076–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Beaulieu JM, and O’Meara BC (2016). Detecting Hidden Diversification Shifts in Models of Trait-Dependent Speciation and Extinction. Syst. Biol 65, 583–601. [DOI] [PubMed] [Google Scholar]
- 98.Nakagawa S, and De Villemereuil P (2019). A General Method for Simultaneously Accounting for Phylogenetic and Species Sampling Uncertainty via Rubin’s Rules in Comparative Analysis. Syst. Biol 68, 632–641. [DOI] [PubMed] [Google Scholar]
- 99.Rabosky DL, Grundler M, Anderson C, Title P, Shi JJ, Brown JW, Huang H, and Larson JG (2014). BAMMtools: an R package for the analysis of evolutionary dynamics on phylogenetic trees. Methods Ecol. Evol 5, 701–707. [Google Scholar]
- 100.Rabosky DL, and Lovette IJ (2008). Explosive evolutionary radiations: decreasing speciation or increasing extinction through time? Evolution 62, 1866–1875. [DOI] [PubMed] [Google Scholar]
- 101.Alroy J (2014). Accurate and precise estimates of origination and extinction rates. Paleobiology 40, 374–397. [Google Scholar]
- 102.Kocsis ÁT, Reddin CJ, Alroy J, and Kiessling W (2019). The r package divDyn for quantifying diversity dynamics using fossil sampling data. Methods Ecol. Evol 10, 735–743. [Google Scholar]
- 103.Foote M (2000). Origination and extinction components of taxonomic diversity: general problems. Paleobiology 26, 74–102. [Google Scholar]
- 104.Alroy J (1996). Constant extinction, constrained diversification, and uncoordinated stasis in North American mammals. Palaeogeogr. Palaeoclimatol. Palaeoecol 127, 285–311. [Google Scholar]
- 105.Alroy J (2008). Dynamics of origination and extinction in the marine fossil record. Proc. Natl. Acad. Sci 105, 11536–11542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Alroy J (2015). A more precise speciation and extinction rate estimator. Paleobiology 41, 633–639. [Google Scholar]
- 107.Louca S, and Doebeli M (2018). Efficient comparative phylogenetics on large trees. Bioinformatics 34, 1053–1055. [DOI] [PubMed] [Google Scholar]
- 108.Redding DW, and Mooers AØ (2006). Incorporating Evolutionary Measures into Conservation Prioritization. Conserv. Biol 20, 1670–1678. [DOI] [PubMed] [Google Scholar]
- 109.Quintero I, and Jetz W (2018). Global elevational diversity and diversification of birds. Nature. [DOI] [PubMed] [Google Scholar]
- 110.Steel M, and Mooers A (2010). The expected length of pendant and interior edges of a Yule tree. Appl. Math. Lett 23, 1315–1319. [Google Scholar]
- 111.Paradis E, Claude J, and Strimmer K (2004). APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics 20, 289–290. [DOI] [PubMed] [Google Scholar]
- 112.Revell LJ (2012). phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol 3, 217–223. [Google Scholar]
- 113.Sepkoski J (1998). Rates of speciation in the fossil record. Philos. Trans. R. Soc. Lond. B. Biol. Sci 353, 315–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jablonski D, and Finarelli JA (2009). Congruence of morphologically-defined genera with molecular phylogenies. Proc. Natl. Acad. Sci 106, 8262–8266. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Summary of early diverging nodes in the backbone-and-patch timetree of mammals relative to the Cretaceous-Paleogene (K-Pg) boundary and the Paleocene-Eocene Thermal Maximum (PETM), Related to Figures 2A, 3A. The 16 superordinal placental divergences that occurred before or overlap with the K-Pg event are displayed in Figure 2A with their 95% highest posterior density (HPD) age interval. Also given is the node number in the maximum clade credibility (MCC) tree of the credible set of 10,000 node-dated completed timetrees, and the posterior probability for that node in the MCC tree.
Table S2. Summary of diversification rate shift recovered using BAMM on 10 mammal trees, Related to Figures 1, 3B, 4A. For all maximum shift credibility (MSC) shifts present in at least 5 of the 10 trees, the average net diversification rates are summarized across all branches in the rate-shift (clade rate) and all branches outside that clade in Mammalia (background rate). Their ratio gave the rate shift factor. The 24 shifts are given letters A-X, while related shifts have the same letter (even when in < 5 trees). The mean and 95% HPD divergences are given for each rate-shift location. Rate-shifts in tipward clades that are independent from each other (i.e., non-nested) are coded with "1" and used to sum up the per-clade rate-shift factors in Figure 5B.
Table S3. Summary output from the "sqs()" function, Related to Figure 2B. Shown are results for three different levels of quorum subsampling of the fossil record within 5-million-year time bins: quorum (q) of 0.1, 0.3, and 0.5.
Table S4. The output from the 'divDyn' R function for calculating rates of origination and extinction upon the binned fossil genus durations, Related to Figure 2B, 4B. All column values are defined in the glossary below the table.
Table S5. The accumulation of lineages through time (LTT) and rates through time (RTT), Related to Figures 4B, S2, S3. We used 5-million-year time bins to summarize values across 100 timetrees, including the median values and 95% confidence intervals for the extant timetree LTT and pulled speciation rates (lambda). We then performed two types of fossil correction on the pulled rates to 'push' them to estimated true values of lambda, one that fixes extant + extinct lineages through time (Fixed E(t)), and one that fixes extinction rates through time (Fixed Mu). From the Fixed Mu method, we used 6 different metrics of extinction rates to fix the homogeneous birth-death model when estimating lambda-- these were 'extPC', 'ext3t', 'extC3t', 'extGF', 'E2f3', and 'ext2f3' (defined in the Table S4 glossary)
Table S6. Comparison of 20 mammal clades with summaries across 100 observed timetrees, Related to Figure 5. Shown are values for the clade harmonic mean of tip-level speciation rates (tip DR), pulled speciation rates at the present day (λ0), the cumulative magnitude of rate shift factors estimated with BAMM, and the skewness of tip DR values within each clade. Rate-constant birth-death simulations of trees the same age and species richness (100 trees) were performed to provide null expectations for the tip DR skewness values, which were compared using two-sided Mann-Whitney U tests. The observed and simulated clades were only compared across the 16 groups that were monophyletic in all analyses and contained >40 species.
Data Availability Statement
All data have been deposited on Github (https://github.com/n8upham/MamDiv-fossil-vs-timetree) and Zenodo (https://doi.org/10.5281/zenodo.5059100), and are publicly available as of the date of publication. All original code has been deposited at the same Github and is publicly available as of the date of publication. The DOI is also listed in the key resources table. Source data for mammal phylogenies analyzed in the paper are available at http://vertlife.org/data/mammals/. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-Snail | Cell Signaling Technology | Cat#3879S; RRID: AB_2255011 |
| Mouse monoclonal anti-Tubulin (clone DM1A) | Sigma-Aldrich | Cat#T9026; RRID: AB_477593 |
| Rabbit polyclonal anti-BMAL1 | This paper | N/A |
| Bacterial and virus strains | ||
| pAAV-hSyn-DIO-hM3D(Gq)-mCherry | Krashes et al., 2011 | Addgene AAV5; 44361-AAV5 |
| AAV5-EF1a-DIO-hChR2(H134R)-EYFP | Hope Center Viral Vectors Core | N/A |
| Cowpox virus Brighton Red | BEI Resources | NR-88 |
| Zika-SMGC-1, GENBANK: KX266255 | Isolated from patient (Wang et al., 2016) | N/A |
| Staphylococcus aureus | ATCC | ATCC 29213 |
| Streptococcus pyogenes: M1 serotype strain: strain SF370; M1 GAS | ATCC | ATCC 700294 |
| Biological samples | ||
| Healthy adult BA9 brain tissue | University of Maryland Brain & Tissue Bank; http://medschool.umaryland.edu/btbank/ | Cat#UMB1455 |
| Human hippocampal brain blocks | New York Brain Bank | http://nybb.hs.columbia.edu/ |
| Patient-derived xenografts (PDX) | Children's Oncology Group Cell Culture and Xenograft Repository | http://cogcell.org/ |
| Chemicals, peptides, and recombinant proteins | ||
| MK-2206 AKT inhibitor | Selleck Chemicals | S1078; CAS: 1032350-13-2 |
| SB-505124 | Sigma-Aldrich | S4696; CAS: 694433-59-5 (free base) |
| Picrotoxin | Sigma-Aldrich | P1675; CAS: 124-87-8 |
| Human TGF-β | R&D | 240-B; GenPept: P01137 |
| Activated S6K1 | Millipore | Cat#14-486 |
| GST-BMAL1 | Novus | Cat#H00000406-P01 |
| Critical commercial assays | ||
| EasyTag EXPRESS 35S Protein Labeling Kit | PerkinElmer | NEG772014MC |
| CaspaseGlo 3/7 | Promega | G8090 |
| TruSeq ChIP Sample Prep Kit | Illumina | IP-202-1012 |
| Deposited data | ||
| Raw and analyzed data | This paper | GEO: GSE63473 |
| B-RAF RBD (apo) structure | This paper | PDB: 5J17 |
| Human reference genome NCBI build 37, GRCh37 | Genome Reference Consortium | http://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/human/ |
| Nanog STILT inference | This paper; Mendeley Data | http://dx.doi.org/10.17632/wx6s4mj7s8.2 |
| Affinity-based mass spectrometry performed with 57 genes | This paper; Mendeley Data | Table S8; http://dx.doi.org/10.17632/5hvpvspw82.1 |
| Experimental models: Cell lines | ||
| Hamster: CHO cells | ATCC | CRL-11268 |
| D. melanogaster: Cell line S2: S2-DRSC | Laboratory of Norbert Perrimon | FlyBase: FBtc0000181 |
| Human: Passage 40 H9 ES cells | MSKCC stem cell core facility | N/A |
| Human: HUES 8 hESC line (NIH approval number NIHhESC-09-0021) | HSCI iPS Core | hES Cell Line: HUES-8 |
| Experimental models: Organisms/strains | ||
| C. elegans: Strain BC4011: srl-1(s2500) II; dpy-18(e364) III; unc-46(e177)rol-3(s1040) V. | Caenorhabditis Genetics Center | WB Strain: BC4011; WormBase: WBVar00241916 |
| D. melanogaster: RNAi of Sxl: y[1] sc[*] v[1]; P{TRiP.HMS00609}attP2 | Bloomington Drosophila Stock Center | BDSC:34393; FlyBase: FBtp0064874 |
| S. cerevisiae: Strain background: W303 | ATCC | ATTC: 208353 |
| Mouse: R6/2: B6CBA-Tg(HDexon1)62Gpb/3J | The Jackson Laboratory | JAX: 006494 |
| Mouse: OXTRfl/fl: B6.129(SJL)-Oxtrtm1.1Wsy/J | The Jackson Laboratory | RRID: IMSR_JAX:008471 |
| Zebrafish: Tg(Shha:GFP)t10: t10Tg | Neumann and Nuesslein-Volhard, 2000 | ZFIN: ZDB-GENO-060207-1 |
| Arabidopsis: 35S::PIF4-YFP, BZR1-CFP | Wang et al., 2012 | N/A |
| Arabidopsis: JYB1021.2: pS24(AT5G58010)::cS24:GFP(-G):NOS #1 | NASC | NASC ID: N70450 |
| Oligonucleotides | ||
| siRNA targeting sequence: PIP5K I alpha #1: ACACAGUACUCAGUUGAUA | This paper | N/A |
| Primers for XX, see Table SX | This paper | N/A |
| Primer: GFP/YFP/CFP Forward: GCACGACTTCTTCAAGTCCGCCATGCC | This paper | N/A |
| Morpholino: MO-pax2a GGTCTGCTTTGCAGTGAATATCCAT | Gene Tools | ZFIN: ZDB-MRPHLNO-061106-5 |
| ACTB (hs01060665_g1) | Life Technologies | Cat#4331182 |
| RNA sequence: hnRNPA1_ligand: UAGGGACUUAGGGUUCUCUCUAGGGACUUAGGGUUCUCUCUAGGGA | This paper | N/A |
| Recombinant DNA | ||
| pLVX-Tight-Puro (TetOn) | Clonetech | Cat#632162 |
| Plasmid: GFP-Nito | This paper | N/A |
| cDNA GH111110 | Drosophila Genomics Resource Center | DGRC:5666; FlyBase:FBcl0130415 |
| AAV2/1-hsyn-GCaMP6- WPRE | Chen et al., 2013 | N/A |
| Mouse raptor: pLKO mouse shRNA 1 raptor | Thoreen et al., 2009 | Addgene Plasmid #21339 |
| Software and algorithms | ||
| ImageJ | Schneider et al., 2012 | https://imagej.nih.gov/ij/ |
| Bowtie2 | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| Samtools | Li et al., 2009 | http://samtools.sourceforge.net/ |
| Weighted Maximal Information Component Analysis v0.9 | Rau et al., 2013 | https://github.com/ChristophRau/wMICA |
| ICS algorithm | This paper; Mendeley Data | http://dx.doi.org/10.17632/5hvpvspw82.1 |
| Other | ||
| Sequence data, analyses, and resources related to the ultra-deep sequencing of the AML31 tumor, relapse, and matched normal | This paper | http://aml31.genome.wustl.edu |
| Resource website for the AML31 publication | This paper | https://github.com/chrisamiller/aml31SuppSite |