Abstract
Recently, four Thiomicrospira strains were isolated from a coastal mud flat of the German Wadden Sea (T. Brinkhoff and G. Muyzer, Appl. Environ. Microbiol. 63:3789–3796, 1997). Here we describe the use of a polyphasic approach to investigate the functional role of these closely related bacteria. Microsensor measurements showed that there was oxygen penetration into the sediment to a depth of about 2.0 mm. The pH decreased from 8.15 in the overlaying water to a minimum value of 7.3 at a depth of 1.2 mm. Further down in the sediment the pH increased to about 7.8 and remained constant. Most-probable-number (MPN) counts of chemolithoautotrophic sulfur-oxidizing bacteria revealed nearly constant numbers along the vertical profile; the cell concentration ranged from 0.93 × 105 to 9.3 × 105 cells per g of sediment. A specific PCR was used to detect the presence of Thiomicrospira cells in the MPN count preparations and to determine their 16S rRNA sequences. The concentration of Thiomicrospira cells did not decrease with depth. It was found that Thiomicrospira strains were not dominant sulfur-oxidizing bacteria in this habitat. Denaturing gradient gel electrophoresis (DGGE) of PCR-amplified 16S ribosomal DNA fragments followed by hybridization analysis with a genus-specific oligonucleotide probe revealed the diversity of Thiomicrospira strains in the MPN cultures. Sequence analysis of the highest MPN dilutions in which the genus Thiomicrospira was detected revealed that there were four clusters of several closely related sequences. Only one of the 10 Thiomicrospira sequences retrieved was related to sequences of known isolates from the same habitat. Slot blot hybridization of rRNA isolated from different sediment layers showed that, in contrast to the concentration of Thiomicrospira cells, the concentration of Thiomicrospira-specific rRNA decreased rapidly in the region below the oxic layer of the sediment. This study revealed the enormous sequence diversity of closely related microorganisms present in one habitat, which so far has been found only by sequencing molecular isolates. In addition, it showed that most of the Thiomicrospira populations in the sediment studied were quiescent.
The rRNA approach (25, 26), which involves using rRNA as a molecular marker to detect and identify particular bacteria in their natural habitats (3) and to explore microbial diversity without cultivation (6, 7), is routinely used in microbial ecological studies. In some studies, the molecular approach has been combined with microbiological methods in an attempt to isolate the relevant microorganisms (13). In other studies molecular biological techniques have been used in combination with geochemical techniques or with microsensors (see reference 4 for an overview) to characterize environmental parameters. However, molecular biological techniques, microbiological methods, and geochemical techniques or microsensors have been used together in only a few studies (28, 37). Nevertheless, combining techniques and concepts from different disciplines is necessary to obtain a better understanding of the interactions between microorganisms and their natural environments, which is the aim of microbial ecology. Here we describe the use of a comprehensive approach to study the functional role of different closely related Thiomicrospira strains in one habitat, an intertidal mud flat.
Thiomicrospira species are chemolithoautotrophic bacteria that use reduced sulfur compounds as energy sources and CO2 as a carbon source; they are obligate aerobes (18). 16S rRNA sequence comparisons have shown that these organisms form a monophyletic group within the gamma subdivision of the class Proteobacteria (8, 22). In a recent study we demonstrated the ubiquity of the genus Thiomicrospira in environments in which reduced sulfur compounds are present (8). In addition, we were able to isolate Thiomicrospira species from most of these habitats and demonstrated that the species diversity within this genus is high. Four isolates, strains JB-A1, JB-A1F, JB-A2, and JB-B2, were obtained from a sample taken from an intertidal coastal mud flat of the Jadebusen Bay, which is part of the German Wadden Sea. Comparative sequence analysis of their 16S rRNA genes demonstrated that these isolates were phylogenetically affiliated with different members of the genus Thiomicrospira. Two of the isolates from the Jadebusen sediment, strains JB-A1 and JB-A2, were biochemically and physiologically characterized and were recently described by Brinkhoff and coworkers (9) as members of two new species of the genus Thiomicrospira (17), Thiomicrospira kuenenii (JB-A1) and Thiomicrospira frisia (JB-A2). Another isolate, strain JB-A1F, had a 16S rRNA sequence that was identical to the 16S rRNA sequence of Thiomicrospira pelophila (17). The fourth isolate, JB-B2, was phylogenetically related to Thiomicrospira crunogena. In addition to our isolates, members of three Thiomicrospira species, T. pelophila (17), Thiomicrospira denitrificans (39), and Thiomicrospira thyasirae (46, 47), were isolated from this habitat. The presence in one habitat of several Thiomicrospira isolates that exhibited only minor differences in their genotypic and phenotypic features prompted us to study the abundance and vertical distribution of the organisms in the sediment in order to determine niche differentiation. To do this, we used tools and techniques from different disciplines. Microsensor measurements were performed with sediment cores to determine environmental parameters, such as oxygen and sulfide contents and pH (19, 31). In parallel, two additional cores were sliced, and the slices were used for molecular biological and microbiological analyses. The most-probable-number (MPN) technique was used to determine the relative abundance of chemolithoautotrophic sulfur-oxidizing bacteria. A genus-specific PCR (8) allowed us to detect Thiomicrospira species in the cultures. Denaturing gradient gel electrophoresis (DGGE) analysis of 16S ribosomal DNA (rDNA) fragments (see reference 24 for an overview) obtained after enzymatic amplification with primers specific for the domain Bacteria, followed by hybridization analysis with a Thiomicrospira-specific oligonucleotide probe (8), was used to identify different Thiomicrospira strains in the MPN tubes. In addition, primers specific for Thiomicrospira 16S rDNA (8) were used to obtain DNA fragments for a sequence analysis. rRNA slot blot hybridization (29, 36) was performed to determine the abundance of 16S rRNA in different sediment layers and to infer the physiological status of the Thiomicrospira populations present.
The results of this polyphasic approach showed that many different Thiomicrospira populations were present down to a depth of 40 mm. As a significant amount of Thiomicrospira-specific rRNA was present only in the oxic layer of the sediment, we concluded that only the Thiomicrospira populations living in the oxic part of the sediment were metabolically active.
MATERIALS AND METHODS
Sampling.
Three cores (diameter, 5 cm; length, 20 cm) were taken from an intertidal coastal sediment of the Jadebusen Bay, which is part of the German Wadden Sea, on 25 September 1996. The sediment cores were taken during low tide. During high tide the water was ca. 35 cm deep at the sampling side. The air, water, and sediment temperature was 8°C when the samples were taken. The salinity was 35‰. The cores were taken directly to the laboratory for further analysis. In addition, seawater was obtained from the same location.
Microsensor measurements.
Microsensor measurements for each sediment core were obtained at 15°C in a flow cell with aeration and circulation of filtered Jadebusen water. The oxygen concentration in the flow cell was kept at the air saturation value. Measurements were obtained at a light intensity of 830 microeinsteins/m2/s. Microsensors mounted on micromanipulators were positioned at the sediment surface by using a dissecting microscope. Profiles were recorded by penetrating the sediment in 100-μm steps with the micromanipulator.
Microsensors were used to measure the oxygen content (32), the hydrogen sulfide content (16, 20), and the pH (31). The oxygen and pH electrodes were calibrated as described previously (32). The H2S microsensor was calibrated for total dissolved sulfide by measuring the signal in a dilution series prepared with a standard solution (sulfide dissolved in Jadebusen water flushed with nitrogen to avoid oxidation of sulfide). The concentration of total dissolved sulfide (H2S, HS−, S2−) in the dilution series was determined by chemical analysis (11).
Slicing of sediment cores.
The cores used for MPN counts and RNA extraction were sliced by pushing the sediment up in the Plexiglas cylinder which was used for sampling. Every 2 mm a layer was scraped off with a sharp spatula. The sediment in each layer was stirred before it was used for MPN counting in order to obtain even distribution of the cells. The sediment samples used for RNA isolation were frozen directly in liquid nitrogen and stored at −80°C until they were used.
MPN counts of sulfur-oxidizing bacteria.
MPN counts were determined for the first four layers of the sediment (i.e., depths of 0 to 2, 2 to 4, 4 to 6, and 6 to 8 mm) and then for every second layer (i.e., depths of 10 to 12, 14 to 16, 18 to 20 mm, etc.). Altogether, 12 layers, spanning a total sediment depth of about 4 cm, were investigated. Tubes containing 9.9 ml of T. pelophila medium (17) were inoculated with 0.1 g of sediment, and the contents were vigorously mixed. Using these tubes, we prepared dilution series with 1:10 steps. The cultures were incubated at 20°C in the dark to avoid growth of phototrophic bacteria. The sulfur-oxidizing bacteria in the different layers were counted by using a threefold MPN dilution series. Cultures were incubated for 4 weeks. Growth was determined by monitoring changes in the color of the pH indicator. The presence of Thiomicrospira cells in the MPN cultures was determined by using a Thiomicrospira-specific primer set (8) in a PCR performed with whole cells taken directly from the cultures as sources of template DNA. The numbers of sulfur-oxidizing bacteria and Thiomicrospira cells were determined by using the MPN index of the American Public Health Association (5) in three parallel experiments.
Oligonucleotides used in this study.
Details concerning the different oligonucleotides used in this study are shown in Table 1. Oligonucleotides TMS128F and TMS849R are specific for the 16S rDNA of bacteria belonging to the genus Thiomicrospira (8) and have been used for PCR detection of Thiomicrospira species in MPN cultures and for sequencing of the PCR products obtained. The primer pair GM5F-907R amplifies the 16S rDNA of members of the domain Bacteria and was used to obtain 550-bp fragments for DGGE analysis (22). The probes used in the rRNA slot blot hybridization analysis were S-*-Univ-1392-a-A-15 (27), which targets the 16S and 18S rRNA of all known forms of life, and S-D-Bact-0338-a-A-18 (2), which is specific for members of the domain Bacteria. Probes TMS849A and TMS849G target the 16S rRNA of members of the genus Thiomicrospira; they are modifications of oligonucleotide TMS849R (Table 1).
TABLE 1.
Oligonucleotides used in this study
| Oligonucleotidea | Sequence (5′ to 3′) | Target site (positions)b | Td/Ta (°C)c | Assay(s)d |
|---|---|---|---|---|
| GM5Fe | CCT ACG GGA GGC AGC AG | 341–357 | —/55 | 1 |
| 907R | CCG TCA ATT CCT TTR AGT TTf | 907–926 | —/55 | 1 |
| TMS128F | GAA TCT RCC CTT TAG TTGf | 128–145 | —/44 | 1,2 |
| TMS849R | CTT TTT AAT AAG RCC AAC AGf | 830–849 | 52/44 | 1,2,3 |
| TMS849A | CTT TTT AAT AAG ACC AAC A | 831–849 | 41/— | 4 |
| TMS849G | CTT TTT AAT AAG GCC AAC A | 831–849 | 41/— | 4 |
| S-*-Univ-1392-a-A-15g | ACG GGC GGT GTG TRCf | 1392–1406 | 44/— | 4 |
| S-D-Bact-0338-a-A-18g | GCT GCC TCC CGT AGG AGT | 338–355 | 54/— | 4 |
The suffixes F and R (forward and reverse, respectively) indicate the primer orientation in relation to the rRNA gene.
E. coli numbering of 16S rRNA nucleotides (10).
Td, dissociation temperature in the hybridization assays; Ta, annealing temperature in the PCR; —, not applicable.
1, PCR; 2, sequencing; 3, DNA hybridization; 4, rRNA slot blot hybridization.
This primer has a GC-clamp attached to its 5′ end (23).
R, adenine-guanine degeneracy.
Designation according to the Oligonucleotide Probe Database (http://www.cme.msu.edu./OPD/) (1).
Td of Thiomicrospira-specific probes.
The temperature of dissociation (Td) of each probe, defined as the temperature at which one-half of the bound probe was released from the hybrid, was determined by using a graded-temperature wash series as previously described (29). RNA isolated from T. frisia (9) and T. crunogena (15) were used in the experiments performed to determine the Td of Thiomicrospira-specific oligonucleotides TMS849A and TMS849G. Probe TMS849A has no mismatch with the 16S rRNA of T. frisia and only one mismatch with the 16S rRNA of T. crunogena. The opposite is true for probe TMS849G; it has no mismatch with the 16S rRNA of T. crunogena and one mismatch with the 16S rRNA of T. frisia. Both probes had a Td of 41°C and were pooled for general detection of Thiomicrospira rRNA in slot blot hybridization experiments. The combination of the two probes matches the 16S rRNA of all known Thiomicrospira species.
rRNA extraction and slot blot hybridization.
Nucleic acids were isolated by bead beating, phenol extraction, and isopropanol precipitation procedures based on the method described by Stahl et al. (36) and MacGregor et al. (21), with slight modifications. Sediment samples were resuspended in 120 mM NaPO4 buffer (pH 8) supplemented with 1% (wt/vol) polyvinylpolypyrrolidone (acid washed as described by Holben et al. [14]) and then were mixed with phenol-chloroform-isoamyl alcohol equilibrated with TE buffer (100 mM Tris [pH 8], 10 mM EDTA) to extract the nucleic acids. After precipitation, the nucleic acids were resuspended in DNase buffer (6 mM MgCl2, 40 mM Tris; pH 7.5), treated with DNase for 30 min, extracted again with phenol-chloroform-isoamyl alcohol, and precipitated with ethanol. The RNA was blotted onto nylon membranes (Magna Charge; Micron Separations, Westborough, Mass.) in triplicate and probed with radioactively labeled oligonucleotides as described previously (36). For the general probes, the membranes were prehybridized, hybridized at 40°C, and washed at 44°C (probe S-*-Univ-1392-a-A-15) or at 54°C (probe S-D-Bact-0338-a-A-18). For the Thiomicrospira-specific probes the membranes were prehybridized and hybridized at 30°C and were washed at 41°C. The intensity of the hybridization signal was measured with a Phosphor Imager (Molecular Dynamics, Sunnyvale, Calif.) and was quantified by using an Escherichia coli rRNA standard (Boehringer, Mannheim, Germany).
PCR amplification of 16S rDNA fragments.
PCR amplification was performed as described by Muyzer et al. (22). A touchdown PCR was performed with primers GM5F and 907R. No touchdown PCR was used for primers TMS128F and TMS849R (annealing temperature, 44°C).
DGGE analysis of PCR products.
DGGE was performed by using the D-Code system (Bio-Rad Laboratories, Inc.). The following conditions were used: 1-mm-thick, 6% (wt/vol) polyacrylamide gels, 1× TAE electrophoresis buffer (pH 8.3), 20 to 70% denaturant, and electrophoresis for 20 h at a constant voltage of 100 V (see reference 23 for details). After electrophoresis, the gels were stained with ethidium bromide and photographed as described previously (23).
Hybridization analysis of blotted DGGE patterns.
Denaturing gradient gel patterns were transferred to nylon membrane filters (Hybond-N+) by electroblotting and were hybridized with digoxigenin-labeled Thiomicrospira-specific probe TMS849R, as described previously (8).
Sequencing of PCR products.
PCR products were purified by using a Qiaquick PCR purification kit (Qiagen Inc., Chatsworth, Calif.). A Taq Dideoxy terminator cycle sequencing kit (Applied Biosystems, Foster City, Calif.) was used to sequence the 16S rDNA fragments. The sequencing primers used were Thiomicrospira-specific primers TMS128F and TMS849R (8). The sequence reaction mixtures were electrophoresed with an Applied Biosystems model 373S DNA sequencer.
Comparative analysis of 16S rRNA sequences.
The 16S rRNA sequences were aligned by using previously described methods (8). A phylogenetic tree was created by using the neighbor-joining algorithm with maximum likelihood as a model of sequence evolution, as implemented in the test version of the program PAUP 4 developed by David Swofford. A bootstrap analysis (1,000 replicates) was used to validate the reproducibility of the branching pattern of the tree.
Nucleotide sequence accession numbers.
The sequences obtained in this study have been deposited in the EMBL Nucleotide Sequence Database under accession no. AJ011066 to AJ011075.
RESULTS
Microsensor measurements.
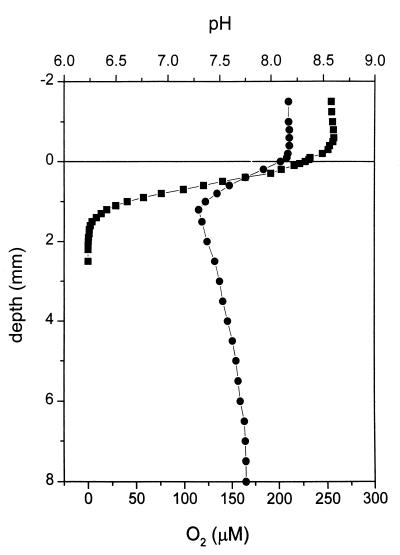
The oxygen measurements obtained for the sediment core showed that the concentration of O2 in the water overlaying the sediment was 250 μM and that the O2 concentration decreased from 225 μM at the sediment surface to zero at a depth of 2.0 mm (Fig. 1). The pH measurements showed that the pH of the overlaying water was about 8.15. From the sediment surface down to a depth of 1.2 mm the pH decreased to about 7.3. From a depth of 1.2 mm to a depth of 6.5 mm the pH increased to about pH 7.8, and the pH remained constant at lower depths (Fig. 1). H2S was not detected in the first 4 cm of the sediment.
FIG. 1.
Oxygen (■) and pH (•) microprofiles for a sediment core from Jadebusen Bay. Note that oxygen was found only in the top 2.0 mm of the sediment. The sulfide content was also examined, but sulfide was not detected. The sediment surface was defined as a depth of 0 mm. Measurements for negative depth values are measurements for the water phase overlaying the sediment; positive depth values indicate the depth in the sediment.
MPN counts of sulfur-oxidizing bacteria.
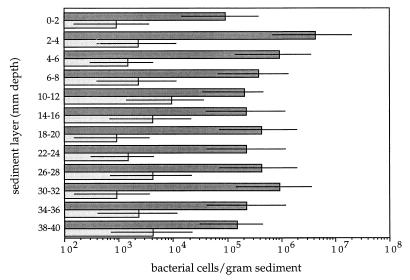
The cultivable chemolithoautotrophic sulfur-oxidizing bacteria in different layers of the Jadebusen sediment were counted by the MPN serial dilution method. The results of growth in the MPN count media are shown in Fig. 2. Between 0.93 × 105 and 9.3 × 105 cells of sulfur-oxidizing bacteria per g of sediment were found in almost all layers. The only exception was the second layer (depth, 2 to 4 mm) which contained slightly more cells (4.3 × 106 cells per g of sediment). However, there was no significant decrease in the number of sulfur-oxidizing bacteria with depth.
FIG. 2.
MPN counts for chemolithoautotrophic sulfur-oxidizing bacteria (dark bars) in sediment samples from Jadebusen Bay, an intertidal coastal mud flat area of the German Wadden Sea. The numbers of Thiomicrospira cells (light bars) were determined by enzymatic amplification of the 16S rDNA with genus-specific primers. The bars indicate 95% confidence intervals.
Screening the MPN cultures for the presence of Thiomicrospira cells.
To determine the numbers of Thiomicrospira cells present in the sediment layers, the MPN cultures were examined by using a Thiomicrospira-specific PCR. The results are shown in Fig. 2 and indicate that Thiomicrospira cells were present in all of the sediment layers which were investigated, down to a depth of 4 cm. Furthermore, similar numbers of Thiomicrospira cells were found in every layer; the concentrations of these bacteria ranged from 0.93 × 103 to 9.3 × 103 cells per g of sediment. A comparison of the concentrations of sulfur-oxidizing bacteria and Thiomicrospira cells showed that the latter accounted for only a minor part (ca. 1%) of the sulfur-oxidizing bacterial population.
DGGE and hybridization analysis of MPN cultures.
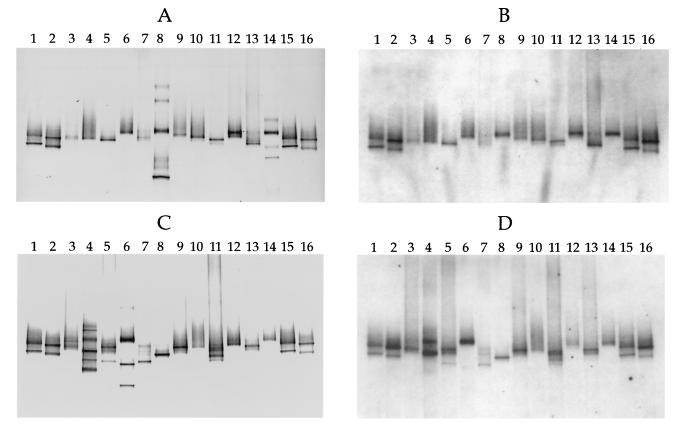
Figures 3A and C show the DGGE results for 16S rDNA fragments obtained from the lowest and highest MPN dilutions at which Thiomicrospira cells were detected. PCR products from known Thiomicrospira strains isolated from the Wadden Sea sediment (8, 9, 18) were applied to both sides of the gels as standards (Fig. 3A and C, lanes 1, 2, 15, and 16). The bacterial diversity in the MPN cultures, as indicated by the number of bands, ranged from one band (Fig. 3A, lanes 3, 4, 6, and 9, and Fig. 3C, lanes 8, 10, 12, and 14) to six bands (Fig. 3A, lane 8). Bands that occurred at the same position in the gels were rare, indicating that the diversity of chemolithoautotrophic sulfur-oxidizing bacteria in the cultures was high.
FIG. 3.
Hybridization analysis of DGGE profiles of 16S rDNA fragments obtained with primers specific for the domain Bacteria and template DNA from MPN cultures and Thiomicrospira isolates from coastal sediments of the Wadden Sea. (A) DGGE patterns. Lanes 1 and 15, T. pelophila (lower band) and Thiomicrospira sp. strain JB-B2 (upper band); lanes 2 and 16, T. kuenenii JB-A1 (lower band) and T. frisia JB-A2 (upper band); lane 3, lowest-dilution MPN culture of the sediment layer at a depth of 0 to 2 mm; lane 4, highest-dilution MPN culture of the sediment layer at a depth of 0 to 2 mm in which Thiomicrospira spp. could still be detected; lanes 5 and 6, equivalent MPN cultures from a depth of 2 to 4 mm; lanes 7 and 8, equivalent MPN cultures from a depth of 4 to 6 mm; lanes 9 and 10, equivalent MPN cultures from a depth of 6 to 8 mm; lanes 11 and 12, equivalent MPN cultures from a depth of 10 to 12 mm; lanes 13 and 14, equivalent MPN cultures from a depth of 14 to 16 mm. (B) Hybridization analysis of the DGGE pattern in panel A performed with the Thiomicrospira-specific, digoxigenin-labeled oligonucleotide, whose target sequence is located within the rDNA amplified. (C) DGGE patterns. Lanes 1, 2, 15, and 16, Thiomicrospira standards (see the explanation above for panel A); lanes 3 and 4, equivalent MPN cultures from a depth of 18 to 20 mm; lanes 5 and 6, equivalent MPN cultures from a depth of 22 to 24 mm; lanes 7 and 8, equivalent MPN cultures from a depth of 26 to 28 mm; lanes 9 and 10, equivalent MPN cultures from a depth of 30 to 32 mm; lanes 11 and 12, equivalent MPN cultures from a depth of 34 to 36 mm; lanes 13 and 14, equivalent MPN cultures from a depth of 38 to 40 mm. (D) Hybridization analysis of the DGGE pattern in panel C. Comparison of panels A and B and panels C and D shows that chemolithoautotrophic bacteria other than Thiomicrospira spp. were present in some of the MPN cultures.
The hybridization analysis of the DGGE patterns with Thiomicrospira-specific probe TMS849R gave positive signals with bands produced by all of the Thiomicrospira species used as standards (Fig. 3B and D, lanes 1, 2, 15, and 16), as well as with one or more bands produced by every MPN culture tested (compare lanes 3 to 14 in Fig. 3A with the same lanes in Fig. 3B and lanes 3 to 14 in Fig. 3C with the same lanes in Fig. 3D). Some of the bands in the DGGE patterns did not hybridize with the Thiomicrospira-specific probe (compare lanes 8 and 14 in Fig. 3A with the same lanes in Fig. 3B and lanes 4 and 6 in Fig. 3C with the same lanes in Fig. 3D), indicating that other bacteria grew under the culture conditions used.
Two or more positive hybridization signals in one lane indicated that two or more Thiomicrospira strains were present in the culture. This was observed mainly with MPN cultures at the lowest dilution (compare lane 11 in Fig. 3A with the same lane in Fig. 3B and lanes 5, 7, 11, and 13 in Fig. 3C with the same lanes in Fig. 3D). However, hybridization signals from two bands were also obtained for one of the highest dilutions (compare lane 4 in Fig. 3C with lane 4 in Fig. 3D).
Phylogenetic analysis of Thiomicrospira sequences.
Although the Thiomicrospira strains from the highest dilutions in which Thiomicrospira cells were present were not isolated in pure culture, partial sequences of the 16S rDNA from nearly all of these MPN cultures were obtained by using the Thiomicrospira-specific primer pair for amplification and sequencing. Both DNA strands of fragments that were approximately 700 bp long were sequenced. Mixed sequences were obtained only with the MPN cultures from two sediment layers (depths, 18 to 20 and 34 to 36 mm). These sequences had ambiguities at positions where differences in Thiomicrospira 16S rDNA occur, indicating that more than one Thiomicrospira strain was present in the cultures. This finding is consistent with the hybridization results for the DGGE pattern of the culture from a depth of 18 to 20 mm, in which two positive signals were obtained (Fig. 3D, lane 4); however, only one signal was obtained for the culture from a depth of 34 to 36 mm (Fig. 3D, lane 12).
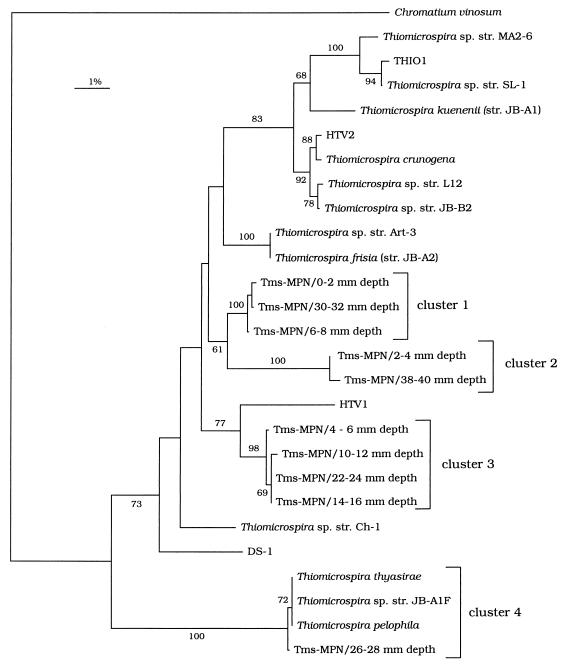
Phylogenetic analysis of the sequences obtained showed that all of the MPN isolates grouped with the known Thiomicrospira isolates (Fig. 4). The sequences obtained in this study occurred in four clusters on the phylogenetic tree. No correlation was found between this phylogenetic grouping and sediment depth; cluster 1 contained sequences from depths of 0 to 2, 6 to 8, and 30 to 32 mm, cluster 2 contained sequences from depths of 2 to 4 and 38 to 40 mm, cluster 3 contained sequences from depths of 4 to 6, 10 to 12, 14 to 16, and 22 to 24 mm, and the sequence from a depth of 26 to 28 mm grouped with the sequences of T. pelophila, T. thyasirae, and Thiomicrospira sp. strain JB-A1F in cluster 4. Less than 1.8% sequence difference was found within each cluster, while more than 3% sequence difference was observed between clusters or with other Thiomicrospira sequences.
FIG. 4.
Unrooted tree showing the phylogenetic relationships of Thiomicrospira strains present in the highest MPN dilutions containing Thiomicrospira cells. The tree is based on partial 16S rRNA sequences and was produced by using the neighbor-joining algorithm with maximum-likelihood correction. The sequences determined in this study are indicated as follows: Tms-MPN/x-y mm depth, where x-y mm depth indicates the depth of the sediment from which a sequence was obtained. The sequence of Chromatium vinosum was used as an outgroup. The numbers on the branches are bootstrap values (1,000 replicates); only values greater than 50% are shown.
rRNA slot blot hybridization.
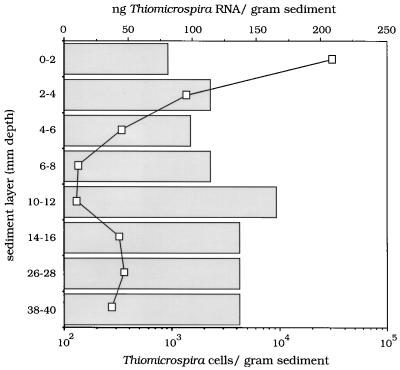
The unexpected presence of relatively high numbers of Thiomicrospira cells in the anoxic part of the sediment prompted us to determine the metabolic state of these bacteria. rRNA was extracted from sliced sediment samples and used in rRNA slot blot hybridization experiments. The samples were hybridized with a probe specific for all known forms of life (universal probe), with probes specific for members of the domain Bacteria, and with a probe specific for Thiomicrospira (Table 1). The following sediment layers were investigated: 0 to 2, 2 to 4, 4 to 6, 6 to 8, 10 to 12, 14 to 16, 26 to 28, and 38 to 40 mm. The results are summarized in Fig. 5. While the concentration of bacterial rRNA was relatively constant, varying with depth from 0.95 to 1.27 μg per g of sediment (data not shown), the vertical concentration profile for Thiomicrospira-specific rRNA revealed a steep decrease in the first 6 mm of the sediment; in the layers measured between depths of 7 and 40 mm the concentration of Thiomicrospira rRNA was never more than 25% of the specific rRNA concentration in the top 2 mm of the sediment.
FIG. 5.
Amounts of Thiomicrospira-specific rRNA (line) in sediment samples from different depths and MPN counts of Thiomicrospira cells (bars) for these samples. The results indicate that a small but active Thiomicrospira population was present in the oxic surface layer of the sediment.
DISCUSSION
Microsensor oxygen measurements indicated that oxygen penetrated into the sediment to a depth of 2.0 mm. However, it must be stated that the Wadden Sea sediment is a very dynamic habitat with diel and seasonal fluctuations in environmental conditions, which have a great influence on the oxygen, sulfide, and pH gradients in the sediments. For instance, Rasmussen and Jørgensen (30) found that oxygen could penetrate to a depth of 1.3 mm in the summer and to a depth of 5.1 mm during the winter in a coastal sediment in Aarhus Bay, Denmark. Also, Visscher and coworkers (40–42) observed diel and seasonal fluctuations in the oxygen and sulfide profiles of microbial ecosystems in coastal mud flats.
Sulfide was not detected in our sediment core; this may have been a result of the low in situ water temperature (8°C) at the time that the core was obtained. It was demonstrated by Vosjan (43) that the sulfate reduction rate in Wadden Sea sediment is temperature dependent and that this rate was 10 times lower at 4°C than at 18°C in an experiment in which lactate was added to the sediment to stimulate sulfate reduction. Thamdrup et al. (38) reported that H2S could not be detected in coastal marine sediment from Aarhus Bay, Denmark, although sulfate reduction proceeded at significant levels. The rapid removal of H2S was attributed to reactions with Fe and Mn oxides. This process might also occur in the Wadden Sea sediment.
The pH profile is consistent with the oxygen profile. There was no net photosynthesis in the sediment, and the initial decrease in the pH was caused by oxidation of organic material; the minimum value in the pH profile corresponded to depletion of oxygen. The increase in pH was probably caused by denitrification or by reduction of iron or manganese.
The comparison of the chemolithoautotrophic sulfur-oxidizing bacterial concentrations with the Thiomicrospira cell concentrations (Fig. 2) indicated that Thiomicrospira strains are not dominant sulfur oxidizers in the Wadden Sea sediments. The Thiomicrospira populations accounted for about 1% of the total sulfur-oxidizing bacteria detected in this study. This finding was confirmed by the absence of hybridization signals in the DGGE profiles of the 16S rDNA fragments obtained after PCR amplification of total community DNA from sediment samples (data not shown). However, as mentioned above, the results described here are the results for just one moment in time; the abundance of Thiomicrospira cells might differ at different seasons. Visscher and van Gemerden (41) obtained different MPN values for different sulfur bacteria, including the colorless sulfur bacteria with which Thiomicrospira cells are associated in the spring and fall, and observed a decrease in the values with depth. Furthermore, Thiomicrospira cells appear to be more abundant in other habitats, such as hydrothermal vent systems (22).
The DGGE-hybridization analysis of the MPN cultures suggested that some Thiomicrospira strains are opportunistic microorganisms that grow rapidly under the culture conditions used (Fig. 3). Growth of sulfur-oxidizing bacteria was determined by a change in the color of the pH indicator present in the culture medium. The presence of Thiomicrospira cells in cultures corresponded with the cultures in which the fastest growth was observed (data not shown). Growth in other tubes was generally observed at least 1 week later. This indicates that Thiomicrospira spp. are the fastest growing sulfur-oxidizing bacteria under the culture conditions used.
A comparison of the hybridized DGGE patterns showed that different Thiomicrospira spp. were present in the MPN cultures from the lowest and highest dilutions from most sediment layers. The Thiomicrospira spp. in both cultures may have been identical only in the cultures from depths of 0 to 2 mm (Fig. 3A and B, lanes 3 and 4) and 30 to 32 mm (Fig. 3C and D, lanes 9 and 10), because the hybridized bands obtained with the lowest and highest dilutions occurred at the same positions on the gel. It has been shown previously that smaller populations may overgrow larger populations that are less well-adapted to the enrichment conditions used (34, 44). This causes problems if only enrichment cultures are used to study bacterial populations, because then only the fast-growing bacteria are obtained. To avoid this problem, dilution of the inoculum to extinction may favor recovery of larger populations that are less adaptive (44). The use of the MPN technique allows not only determination of the number of bacteria which are able to grow with a particular substrate but also makes detection of the most abundant of these bacteria possible. However, it should be mentioned that there might have been Thiomicrospira populations present which were not able to grow under the cultivation conditions used.
A comparison of the sequences described in this paper with the sequences from the same habitat described previously (8, 22) showed that, with one exception, the previously isolated strains were not present in the highest MPN dilutions used for Thiomicrospira spp. This indicates that most of these isolates, all of which were obtained by using enrichment techniques (8, 17, 46, 47), do not belong to the most abundant Thiomicrospira spp. Only one sequence, the Thiomicrospira MPN sequence from a depth of 26 to 28 mm, exhibited high levels of similarity (more than 99.8%) to the sequences of T. pelophila and related species. The other new sequences formed three monophyletic clusters with no close known relatives. The differences between sequences within each cluster were always less than 1.8%; thus, on the basis of the definition of Stackebrandt and Goebel (35), these sequences might be considered sequences which originated from different strains of the same species. However, the differences between sequences in different clusters and other Thiomicrospira strains, which were more than 3%, may that additional new species are present in the habitat studied.
Slot blot hybridization of rRNA extracted from the different sediment layers was used to infer the metabolic activity of the Thiomicrospira populations. The total amount of rRNA depends on the number of cells and the amount of rRNA per cell. Cells which are growing well have more ribosomes and thus more rRNA than cells which are not growing (27). While the numbers of Thiomicrospira cells were relatively constant along the entire vertical profile of the sediment, the concentration of Thiomicrospira-specific rRNA decreased sharply from the oxic zone to suboxic zone of the sediment (Fig. 5). On the basis of the MPN and rRNA slot blot hybridization results, we concluded that Thiomicrospira spp. were metabolically active members of the bacterial community in the first 4 mm of the sediment, from the oxic region to the suboxic region, while the relative activity of these organisms was low in the anoxic zone of the sediment. This finding is consistent with physiological characteristics of Thiomicrospira species, which, with the exception of T. denitrificans, are all obligate aerobes (9, 15, 17). T. denitrificans is able to grow anaerobically with nitrate as an electron acceptor (39). However, 16S rRNA sequence analysis showed that T. denitrificans is phylogenetically affiliated with the genus Thiovulum in the epsilon subdivision of the class Proteobacteria (22). The oligonucleotides designed for detection of the genus Thiomicrospira do not hybridize with T. denitrificans (8).
In recent years many investigations of the microbial diversity of natural samples have revealed that the diversity of closely related sequences in one environment is high (6, 12). However, in these studies no information was obtained about the metabolic state of the microorganisms. Here we also observed a high level of diversity of closely related sequences, all belonging to different Thiomicrospira populations. However, as determined by the polyphasic approach used in this study, we found that these populations have different growth rates depending on their locations in the sediment. This leads to the conclusion that Thiomicrospira cells are metabolically active only in the oxic part of the sediment studied and to the conclusion that most of the Thiomicrospira populations found in the anoxic part of the sediment are probably dormant. How these populations survive under anaerobic conditions is unknown and might be the aim of future studies.
Similar results have been described by Wawer and coworkers (45). Using comparative DGGE analysis of [NiFe] hydrogenase gene fragments obtained by enzymatic amplification of bacterial genomic DNA and total RNA from microbial communities, these authors demonstrated that only one of the five Desulfovibrio populations which were present expressed the [NiFe] hydrogenase gene after hydrogen was added as a substrate, indicating that niche differentiation occurred.
It is still not known why the number of Thiomicrospira cells in the anoxic layers of the sediment is relatively constant. This observation may be partially explained by mixing of the sediment by physical turbation, such as the mixing caused by waves, which can have an impact on the upper few centimeters of the sediment. Furthermore, bioturbation by benthic animals is known to influence particle transport (33, 48) and to have an effect on the distribution of microorganisms in sediment.
A comparison of the physiological features of three Thiomicrospira species, T. pelophila, T. kuenenii, and T. frisia, which were isolated from the Wadden Sea sediment, revealed differences in pH range and salt tolerance (9). Changes in pH values and salt concentrations might occur in an intertidal coastal mud flat. For this reason it is likely that different Thiomicrospira populations are adapted to different environmental conditions and that populations which are inactive at one moment might be active at another, when specific environmental conditions favor their growth.
In summary, we demonstrated that there are many different Thiomicrospira populations in an intertidal mud flat habitat. However, these populations do not represent a dominant proportion of the sulfur-oxidizing bacterial community (they account for ca. 1% of this community), and most of them are quiescent. Only the populations that are present in the oxic sediment layers seem to be metabolically active. On the basis of these results we concluded that the functional role of Thiomicrospira spp. in intertidal mud flats is minor and is limited to the oxic part of the sediment and that other sulfur-oxidizing bacteria might be primarily responsible for oxidation of reduced sulfur compounds produced by sulfate reduction.
ACKNOWLEDGMENTS
We thank Stefan Forster, Eric Epping, and Werner Liesack for helpful discussions. We thank Anja Dittel for helping with sediment sampling and Hendrik Schäfer for critically reading the manuscript. We are grateful to Birgit Rattunde for technical assistance.
This research was financially supported by the Max Planck Society, Munich, Germany, and by the Körber Foundation, Hamburg, Germany.
REFERENCES
- 1.Alm E W, Oerther D B, Larsen N, Stahl D A, Raskin L. The Oligonucleotide Probe Database. Appl Environ Microbiol. 1996;62:3557–3559. doi: 10.1128/aem.62.10.3557-3559.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann R I, Binder B J, Olson R J, Chisholm S W, Devereux R, Stahl D A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amann R I, Kühl M. In situ methods for assessment of microorganisms and their activities. Curr Opin Microbiol. 1998;1:352–358. doi: 10.1016/s1369-5274(98)80041-6. [DOI] [PubMed] [Google Scholar]
- 5.American Public Health Association. Standard methods for the examination of water and wastewater, including bottom sediments and sludge. Washington, D.C: American Public Health Association; 1969. pp. 604–609. [Google Scholar]
- 6.Barns S M, Fundyga R E, Jeffries M W, Pace N R. Remarkable archaeal diversity detected in a Yellowstone National Park hot spring environment. Proc Natl Acad Sci USA. 1994;91:1609–1613. doi: 10.1073/pnas.91.5.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borneman J, Skroch P W, O’Sullivan K M, Palus J A, Rumjanek N G, Jansen J L, Nienhuis J, Triplett E W. Molecular microbial diversity of an agricultural soil in Wisconsin. Appl Environ Microbiol. 1996;62:1935–1943. doi: 10.1128/aem.62.6.1935-1943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brinkhoff T, Muyzer G. Increased species diversity and extended habitat range of sulfur-oxidizing Thiomicrospira spp. Appl Environ Microbiol. 1997;63:3789–3796. doi: 10.1128/aem.63.10.3789-3796.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brinkhoff, T., G. Muyzer, C. O. Wirsen, and J. Kuever. Characterization of Thiomicrospira kuenenii sp. nov. and Thiomicrospira frisia sp. nov., two mesophilic obligately chemolithoautotrophic sulfur-oxidizing bacteria isolated from an intertidal mud flat. Int. J. Syst. Bacteriol., in press. [DOI] [PubMed]
- 10.Brosius J, Dull T J, Sleeter D D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 11.Cline J D. Spectrophotometric determination of hydrogen sulfide in natural waters. Limnol Oceanogr. 1969;14:454–458. [Google Scholar]
- 12.Giovannoni S J, Britschgi T B, Moyer C L, Field K G. Genetic diversity in Sargasso Sea bacterioplankton. Nature (London) 1990;345:60–63. doi: 10.1038/345060a0. [DOI] [PubMed] [Google Scholar]
- 13.Großkopf R, Janssen P H, Liesack W. Diversity and structure of the methanogenic community in anoxic rice paddy soil microcosms as examined by cultivation and direct 16S rRNA gene sequence retrieval. Appl Environ Microbiol. 1998;64:960–969. doi: 10.1128/aem.64.3.960-969.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holben W E, Jansson J K, Chelm B K, Tiedje J M. DNA probe method for the detection of specific microorganisms in the soil bacterial community. Appl Environ Microbiol. 1988;54:703–711. doi: 10.1128/aem.54.3.703-711.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jannasch H W, Wirsen C O, Nelson D C, Robertson L A. Thiomicrospira crunogena sp. nov., a colorless sulfur-oxidizing bacterium from a deep-sea hydrothermal vent. Int J Syst Bacteriol. 1985;35:422–424. [Google Scholar]
- 16.Jeroschewski P, Steuchart C, Kühl M. An amperometric microsensor for the determination of H2S in aquatic environments. Anal Chem. 1996;68:4351–4357. [Google Scholar]
- 17.Kuenen J G, Veldkamp H. Thiomicrospira pelophila, gen. n., sp. n., a new obligately chemolithotrophic colourless sulfur bacterium. Antonie Leeuwenhoek. 1972;38:241–256. doi: 10.1007/BF02328096. [DOI] [PubMed] [Google Scholar]
- 18.Kuenen J G, Robertson L A, Tuovinen O H. The genera Thiobacillus, Thiomicrospira, and Thiosphaera. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K H, editors. The prokaryotes. 2nd ed. Berlin, Germany: Springer; 1992. pp. 2638–2657. [Google Scholar]
- 19.Kühl, M., and N. P. Revsbech. Microsensors for the study of interfacial biogeochemical processes. In B. P. Boudreau and B. B. Jørgensen (ed.), The benthic boundary layer, in press, Oxford University Press, Oxford, United Kingdom.
- 20.Kühl M, Steuckart C, Eickert G, Jeroschewski P. A H2S microsensor for profiling biofilms and sediments: application in an acidic lake sediment. Aquat Microb Ecol. 1998;15:201–209. [Google Scholar]
- 21.MacGregor B J, Moser D P, Alm E W, Nealson K H, Stahl D A. Crenarchaeota in Lake Michigan sediment. Appl Environ Microbiol. 1997;63:1178–1181. doi: 10.1128/aem.63.3.1178-1181.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muyzer G, Teske A, Wirsen C O, Jannasch H W. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch Microbiol. 1995;164:165–172. doi: 10.1007/BF02529967. [DOI] [PubMed] [Google Scholar]
- 23.Muyzer G, Brinkhoff T, Nübel U, Santegoeds C, Schäfer H, Wawer C. Denaturing gradient gel electrophoresis (DGGE) in microbial ecology. In: Akkermans A D L, van Elsas J D, de Bruijn F J, editors. Molecular microbial ecology manual. 3rd ed. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 3.4.4:1–27. [Google Scholar]
- 24.Muyzer G, Smalla K. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Leeuwenhoek. 1998;73:127–141. doi: 10.1023/a:1000669317571. [DOI] [PubMed] [Google Scholar]
- 25.Olsen G J, Lane D J, Giovannoni S J, Pace N R. Microbial ecology and evolution: a ribosomal RNA approach. Annu Rev Microbiol. 1986;40:337–365. doi: 10.1146/annurev.mi.40.100186.002005. [DOI] [PubMed] [Google Scholar]
- 26.Pace N R, Stahl D A, Lane D J, Olsen G J. The analysis of natural microbial populations by ribosomal RNA sequences. Adv Microb Ecol. 1986;9:1–55. [Google Scholar]
- 27.Poulsen L K, Ballard G, Stahl D A. Use of rRNA fluorescence in situ hybridization for measuring the activity of single cells in young and established biofilms. Appl Environ Microbiol. 1993;59:1354–1360. doi: 10.1128/aem.59.5.1354-1360.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramsing N B, Fossing H, Ferdelman T G, Andersen F, Thamdrup B. Distribution of bacterial populations in a stratified fjord (Mariager Fjord, Denmark) quantified by in situ hybridization and related to chemical gradients in the water column. Appl Environ Microbiol. 1996;62:1391–1404. doi: 10.1128/aem.62.4.1391-1404.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raskin L, Stromley J M, Rittmann B E, Stahl D A. Group-specific 16S rRNA hybridization probes to describe natural communities of methanogens. Appl Environ Microbiol. 1994;60:1232–1240. doi: 10.1128/aem.60.4.1232-1240.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rasmussen H, Jørgensen B B. Microelectrode studies of seasonal oxygen uptake in a coastal sediment: role of molecular diffusion. Mar Ecol Prog Ser. 1992;81:289–303. [Google Scholar]
- 31.Revsbech N P, Jørgensen B B. Microelectrodes: their use in microbial ecology. Adv Microb Ecol. 1986;9:293–352. [Google Scholar]
- 32.Revsbech N P. An oxygen microelectrode with a guard cathode. Limnol Oceanogr. 1989;55:1907–1910. [Google Scholar]
- 33.Rhoads D C. Organism sediment relations on the muddy sea floor. Oceanogr Mar Biol Annu Rev. 1974;12:263–300. [Google Scholar]
- 34.Santegoeds C M, Nold S C, Ward D M. Denaturing gradient gel electrophoresis used to monitor the enrichment culture of aerobic chemoorganotrophic bacteria from a hot spring cyanobacterial mat. Appl Environ Microbiol. 1996;62:3922–3928. doi: 10.1128/aem.62.11.3922-3928.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stackebrandt E, Goebel B M. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- 36.Stahl D A, Flesher B, Mansfield H R, Montgomery L. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl Environ Microbiol. 1988;54:1079–1084. doi: 10.1128/aem.54.5.1079-1084.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teske A, Wawer C, Muyzer G, Ramsing N B. Distribution of sulfate-reducing bacteria in a stratified fjord (Mariager Fjord, Denmark) as evaluated by most-probable-number counts and denaturing gradient gel electrophoresis of PCR-amplified ribosomal DNA fragments. Appl Environ Microbiol. 1996;62:1405–1415. doi: 10.1128/aem.62.4.1405-1415.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thamdrup B, Fossing H, Jørgensen B B. Manganese, iron, and sulfur cycling in a coastal marine sediment, Aarhus Bay, Denmark. Geochim Cosmochim Acta. 1994;58:5115–5129. [Google Scholar]
- 39.Timmer-ten Hoor A. A new type of thiosulfate oxidizing, nitrate-reducing microorganism: Thiomicrospira denitrificans sp. nov. Neth J Sea Res. 1975;9:343–351. [Google Scholar]
- 40.Visscher P T, Beukema J, van Gemerden H. In situ characterization of sediments: measurements of oxygen and sulfide profiles with a novel combined needle electrode. Limnol Oceanogr. 1991;36:1476–1480. [Google Scholar]
- 41.Visscher P T, van Gemerden H. Sulfur cycling in laminated marine ecosystems. In: Oremland R S, editor. Biogeochemistry of global change: radioactively active trace gases. New York, N.Y: Chapman and Hall; 1993. pp. 672–690. [Google Scholar]
- 42.Visscher P T, van den Ende F P. Diel and spatial fluctuations of sulfur transformations. NATO Adv Study Inst Ser G. 1994;35:353–359. [Google Scholar]
- 43.Vosjan J H. Ecologische en fysiologische aspecten van bacteriële sulfaatreductie in het waddengebied. Ph.D. thesis. GroningenGroningen, The Netherlands: Rijksuniversiteit; 1975. [Google Scholar]
- 44.Ward D M, Santegoeds C M, Nold S C, Ramsing N B, Ferris M J, Bateson M M. Biodiversity within hot spring microbial mat communities: molecular monitoring of enrichment cultures. Antonie Leeuwenhoek. 1997;71:143–150. doi: 10.1023/a:1000131426164. [DOI] [PubMed] [Google Scholar]
- 45.Wawer C, Jetten M S M, Muyzer G. Genetic diversity and expression of the [NiFe] hydrogenase large-subunit gene of Desulfovibrio spp. in environmental samples. Appl Environ Microbiol. 1997;63:4360–4369. doi: 10.1128/aem.63.11.4360-4369.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wood A P, Kelly D P. Isolation and physiological characterization of Thiobacillus thyasiris sp. nov., a novel marine facultative autotroph and the putative symbiont of Thyasira flexuosa. Arch Microbiol. 1989;152:160–166. [Google Scholar]
- 47.Wood A P, Kelly D P. Reclassification of Thiobacillus thyasiris as Thiomicrospira thyasirae comb. nov., an organism exhibiting pleomorphism in response to environmental conditions. Arch Microbiol. 1993;159:45–47. [Google Scholar]
- 48.Yingst J Y, Rhoads D C. The role of bioturbation in the enhancement of bacterial growth rates in marine sediments. In: Tenore K L, Coull B C, editors. Marine benthic dynamics. Columbia: University of South Carolina Press; 1980. pp. 407–421. [Google Scholar]